Study of intraflagellar transport protein IFT20 in male germ cell development shows that it is essential for male fertility and spermiogenesis in mice, playing a role in the building of sperm flagella and the disposal of cytoplasmic components.
Abstract
Intraflagellar transport (IFT) is a conserved mechanism believed to be essential for the assembly and maintenance of cilia and flagella. However, little is known about its role in mammalian sperm flagella formation. To fill this gap, we disrupted the Ift20 gene in male germ cells. Homozygous mutant mice were infertile, with significantly reduced sperm counts and motility. In addition, abnormally shaped, elongating spermatid heads and bulbous, round spermatids were found in the lumen of the seminiferous tubules. Electron microscopy revealed increased cytoplasmic vesicles, fiber-like structures, abnormal accumulation of mitochondria, and a decrease in mature lysosomes. The few developed sperm had disrupted axonemes, and some retained cytoplasmic lobe components on the flagella. ODF2 and SPAG16L, two sperm flagella proteins, failed to be incorporated into sperm tails of the mutant mice, and in the germ cells, both were assembled into complexes with lighter density in the absence of IFT20. Disrupting IFT20 did not significantly change expression levels of IFT88, a component of the IFT-B complex, and IFT140, a component of the IFT-A complex. Even though the expression level of an autophagy core protein that associates with IFT20, ATG16, was reduced in the testis of the Ift20 mutant mice, expression levels of other major autophagy markers, including LC3 and ubiquitin, were not changed. Our studies suggest that IFT20 is essential for male fertility and spermiogenesis in mice, and its major function is to transport cargo proteins for sperm flagella formation. It also appears to be involved in removing excess cytoplasmic components.
INTRODUCTION
Cilia are cell surface organelles that are present on almost all vertebrate cells at some point during development and differentiation (Brown and Witman, 2014). They are formed by a process that requires intraflagellar transport (IFT), a bidirectional transport system directed by IFT protein complexes and motors (Pedersen and Rosenbaum, 2008). IFT involves movement of large protein complexes called IFT particles from the cell body to the ciliary tip (anterograde transport), followed by their return to the cell body (retrograde transport; Blacque et al., 2008). It was first described in Chlamydomonas (Kozminski et al., 1993). IFT protein complexes have been distinguished into complex A and complex B, containing ∼20 IFT proteins in Chlamydomonas (Fort and Bastin, 2014). Complex A is believed to be composed of six subunits, and complex B contains 16 known proteins, including IFT20. IFT proteins are typically found along the ciliary shaft and at the base (Mizuno et al., 2012), and the IFT complexes serve as adaptors that mediate contacts between cargo proteins and motors (Follit et al., 2009). Mutations in both complex A and B IFT genes result in ciliogenesis defects and human diseases (Murcia et al., 2000; Huangfu and Anderson, 2005; Badano et al., 2006; Tran et al., 2008; Cortellino et al., 2009; Stottmann et al., 2009; Friedland-Little et al., 2011; Hildebrandt et al., 2011; Huber and Cormier-Daire, 2012).
IFT20 is the smallest IFT protein. The role of mouse IFT20 has been studied in vitro and in vivo. Of the identified mouse IFT proteins, IFT20 is the only one present in the Golgi body, and the localization is dependent on GMAP210 (Follit et al., 2008). In living cells, fluorescently tagged IFT20 is highly dynamic and moves between the Golgi complex and the cilium, as well as along ciliary microtubules; it functions in the delivery of ciliary membrane proteins from the Golgi complex to the cilium (Follit et al., 2006, 2008).
Disruption of the gene globally by a conventional knockout strategy resulted in embryonic lethality, demonstrating that IFT20 is required for development (Keady et al., 2011). The role of IFT20 in individual cells/tissues has been explored using floxed Ift20 mice crossed to several tissue-specific Cre transgenic mice. A variety of phenotypes relating to cilia dysfunction have been identified (Jonassen et al., 2008; May-Simera et al., 2015). IFT20 was also found to be expressed in nonciliated T-cells and to have functions in immune synapse formation (Finetti et al., 2009, 2014; Yuan et al., 2014). Part of the molecular machinery involved in ciliogenesis also participates in autophagy (Pampliega et al., 2013), the catabolic process that involves degradation of unnecessary cellular components through the action of lysosomes (Kobayashi, 2015). This process is mediated by the autophagy-related genes (ATGs) and their associated enzymes (Kim et al., 2012; Diao et al., 2015; Liu et al., 2015). IFT20 associates with autophagy core protein ATG16L and is believed to play a role in autophagy (Pampliega et al., 2013). IFT proteins usually associate with other proteins to form a complex in order to conduct their functions. Besides ATG16L and GMAP210, IFT20 was reported to associate with several other proteins, including IFT57, a kinesin II subunit, KIF3B, and CCDC41 (Baker et al., 2003; Joo et al., 2013).
Even though IFT20 has been shown to play roles in both ciliated and nonciliated cells, it is not clear whether it also controls sperm tail formation. Of all mammalian cells, sperm have the longest motile cilia. IFT20 is expressed in male germ cells. It is present in the Golgi complex, manchette, and the basal bodies of differentiating germ cells. These are key structures in ciliogenesis (Sironen et al., 2010, 2011, 2012). However, the exact role of IFT20 in sperm development is unknown. To elucidate the function of IFT20 in the testis, particularly in male germ cell development, we generated a male germ cell–specific Ift20 knockout mouse model by crossing floxed Ift20 mice to Stra8-iCre transgenic mice. Our studies demonstrated that IFT20 is essential for spermiogenesis and male fertility. As expected, sperm flagella formation was disrupted in conditional Ift20 mutant mice. The unexpected finding was that some sperm that completed all phases of spermatogenesis retained the cytoplasmic lobe residue. These findings suggest dual functions of IFT20 in male germ cell development: directing sperm flagella formation by IFT mechanisms and helping to remove residual cytoplasmic components.
RESULTS
Mouse IFT20 protein is highly abundant in the testis, and its expression is up-regulated during spermiogenesis
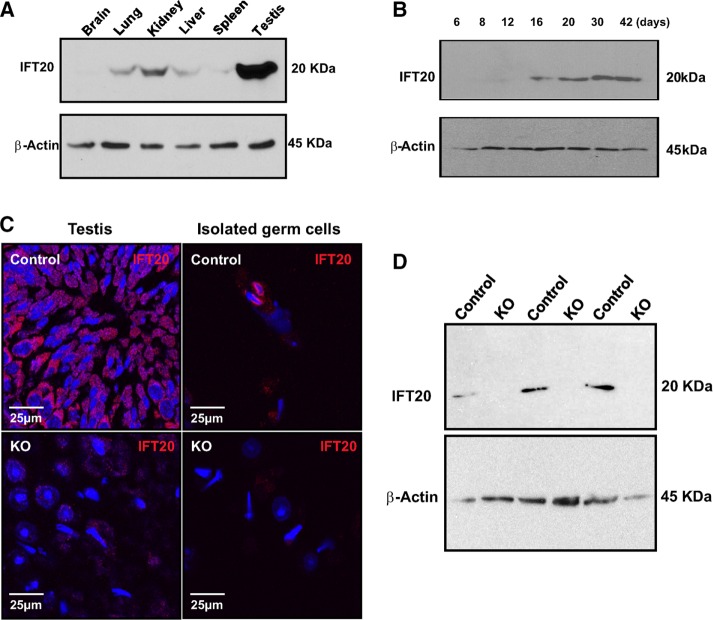
IFT20 protein expression in various tissues was analyzed, including the brain, lung, kidney, liver, spleen, and testis. IFT20 protein was only detected in the testis using the less sensitive Pico ECL system (Supplemental Figure S1). However, when the more sensitive Femto system was used, it was found that IFT20 is expressed also in most somatic tissues, particularly in the kidney (Figure 1A). Next we examined the testicular expression levels during germ cell development. The expression of IFT20 protein was first visible at postnatal day 16 and increased significantly at days 30 and 42, when germ cells enter the condensation/elongation stage in the first wave of spermatogenesis (Figure 1B).
FIGURE 1:
IFT20 protein expression in the wild-type and conditional mutant mice. (A) Analysis of tissue distribution of mouse IFT20 protein by Western blot using a high-sensitivity Femto system. Note that IFT20 is highly expressed in the testis and is also present in most somatic tissues. (B) IFT20 expression levels during the first wave of spermatogenesis. Its expression is significantly increased at day 16 of birth and further increased at day 30 of birth, when germ cells enter spermiogenesis. (C) Examination of IFT20 protein expression by immunofluorescence staining on testicular sections (left) and isolated germ cells (right). Strong IFT20 signals (red) are observed in the control samples (top). However, almost no specific IFT20 signals are observed in the knockout samples (bottom). (D) Analysis of testicular IFT20 expression by Western blot using the Pico system. IFT20 was expressed in the three control mice. However, no specific signal was observed in the three mutant mice analyzed.
Generation of male germ cell–specific Ift20 conditional knockout mice
To study the role of IFT20 in mouse spermatogenesis, particularly spermiogenesis, when germ cells form flagella, we generated male germ cell–specific conditional knockout mice by crossing Stra8-iCre males with Ift20flox/flox females, following the breeding strategy shown in Supplemental Figure S2A (Bao et al., 2013). The resulting mice were genotyped by PCR (Supplemental Figure S2B) using previously described specific primers (Jonassen et al., 2008). To examine IFT20 protein expression in the mutant mice, we conducted immunofluorescence staining and Western blotting. The control mice showed robust IFT20 signals in both testicular sections (Figure 1C, left, top) and isolated germ cells (Figure 1C, right, top). However, specific IFT20 signal was barely detectable in the testicular sections (Figure 1C, left, bottom) and isolated germ cells (Figure 1C, right, bottom) in Ift20 mutant mice. Western blot analysis confirmed that no specific IFT20 signal was detectable in the testes of Ift20 mutant mice when the less sensitive Pico system was used (Figure 1D). However, when a highly sensitive Femto system was used, very low levels of IFT20 were still detected in the mutant mice (Supplemental Figure S3).
Homozygous mutant adult Ift20 males are grossly normal but infertile
All mutant mice survived to adulthood and did not show any gross abnormalities. To test fertility, we bred 2- to 3-mo-old Ift20 mutant males and controls with 3- to 4-mo-old wild-type females. All 12 controls exhibited normal fertility and sired normal-sized litters after 1 mo of breeding. However, none of the 14 Ift20 mutant males tested produced any litters after 3 mo of breeding (Table 1), even though these mice showed normal sexual behavior and vaginal plugs were observed in the females that were bred with the Ift20 mutant males.
TABLE 1:
Fertility, fecundity of control, and conditional Ift20 mutant mice.
| Adult mice | Mice at 6 wk of age | ||||
|---|---|---|---|---|---|
| Genotype | Fertility | Litter size | Genotype | Fertility | Litter size |
| Control | 12/12 | 8.5 ± 0.5 | Control | 10/10 | 8.1 ± 1.5 (n = 10) |
| KO | 0/14 | 0 | KO | 6/16 | 3.1 ± 1.2 (n = 6) |
To test fertility, adult mature and 6-wk-old males were bred to wild-type females for at least 2 mo. Litter size was recorded for each mating.
A fertility test was also performed with a group of control and homozygous conditional Ift20 mutant mice at 6 wk of age to examine the mutant model during the first wave of spermatogenesis. Of interest, even though all adult mutant males were infertile, 37.5% of the 6-wk-old conditional Ift20 mutant males (six of 16) sired pups. However, the litter sizes (two to five pups/litter) were significantly smaller than the controls at the same age (seven–11 pups/litter). The six homozygous mutant males never sired pups again after the first litter (Table 1).
Abnormal sperm morphology, significantly reduced sperm count, and motility
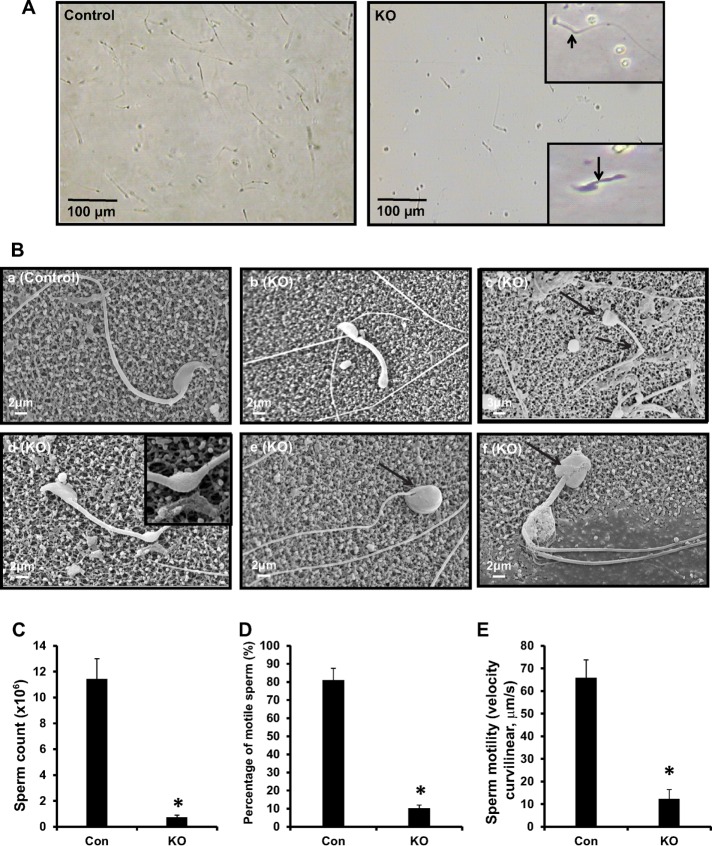
To determine sperm factors in the infertility phenotype, we examined sperm morphology, number, and motility in control and Ift20 mutant mice. Sperm were collected from cauda epididymides of control and conditional Ift20 mutant mice. Under light microscopy, sperm density was remarkably lower in mutant mice compared with that of controls under the same dilution. The tails of many sperm were kinked or shortened (Figure 2A). By scanning electron microscopy (SEM), sperm from control mice had long, smooth tails with normally condensed heads (Figure 2Ba). However, multiple abnormalities were observed in sperm of mutant mice, including short, kinked tails, which were also observed under light microscopy (Figure 2B, b–d). In addition, some sperm showed round, swollen heads (Figure 2B, c, e, and f). Some sperm with kinked tails retained cytoplasm in the kinked area (Figure 2Bd).
FIGURE 2:
Disruption of the Ift20 gene in germ cells leads to abnormal sperm morphology associated with significantly reduced sperm count and motility. (A) Examination of epididymal sperm by light microscopy. Under the same dilution, compared with control (left), very few sperm were seen in conditional Ift20 mutant mice (right). Sperm from mutant mice often had shorter (arrow) or kinked (arrowhead) tails (right, insets). (B) Examination of epididymal sperm by SEM. (a) Representative image of epididymis sperm with normal morphology from a control mouse shows the sperm with a long, smooth tail and condensed head. (b–f) Representative images of epididymal sperm from a mutant mouse. A variety of abnormal sperm morphologies are observed: short (b) and kinked (c, d) tails and round heads (c, e, f). The inset in d shows excess retention of cytoplasm. The arrows point to sperm with round heads; the dashed arrows point to bent tails. (C) The sperm count was significantly reduced in mutant mice. (D) The percentage of motile sperm was significantly lower in the mutant mice. (E) Sperm VCL was significantly reduced in mutant mice compared with controls (*p < 0.05).
Epididymal sperm numbers in mutant mice were significantly lower than in controls (Figure 2C). Movies were taken from swim-out sperm. Most control sperm displayed vigorous flagellar activity and progressive long track-forward movement (Supplemental Movie S1 and Figure 2D). However, only a small percentage of the mutant sperm were motile and showed forward motility, and flagellar movement was limited to a slow, erratic waveform with a very low amplitude (Supplemental Movie S2 and Figure 2D). Even if some sperm were motile, their motility was significantly less than that of controls (Figure 2E).
Very few well-developed epididymal sperm and abnormal spermiogenesis in conditional Ift20 mutant mice
We examined the histology of cauda epididymides of adult control and conditional Ift20 mutant mice. The control mice showed highly concentrated epididymal sperm with aligned sperm tails and normal sperm heads (Figure 3A, left). However, Ift20 mutant mice showed a very low concentration of sperm, with a high incidence of sperm abnormalities, sloughed round germ cells, residual bodies, and sperm with short tails or sperm tails without heads (Figure 3A, right).
FIGURE 3:
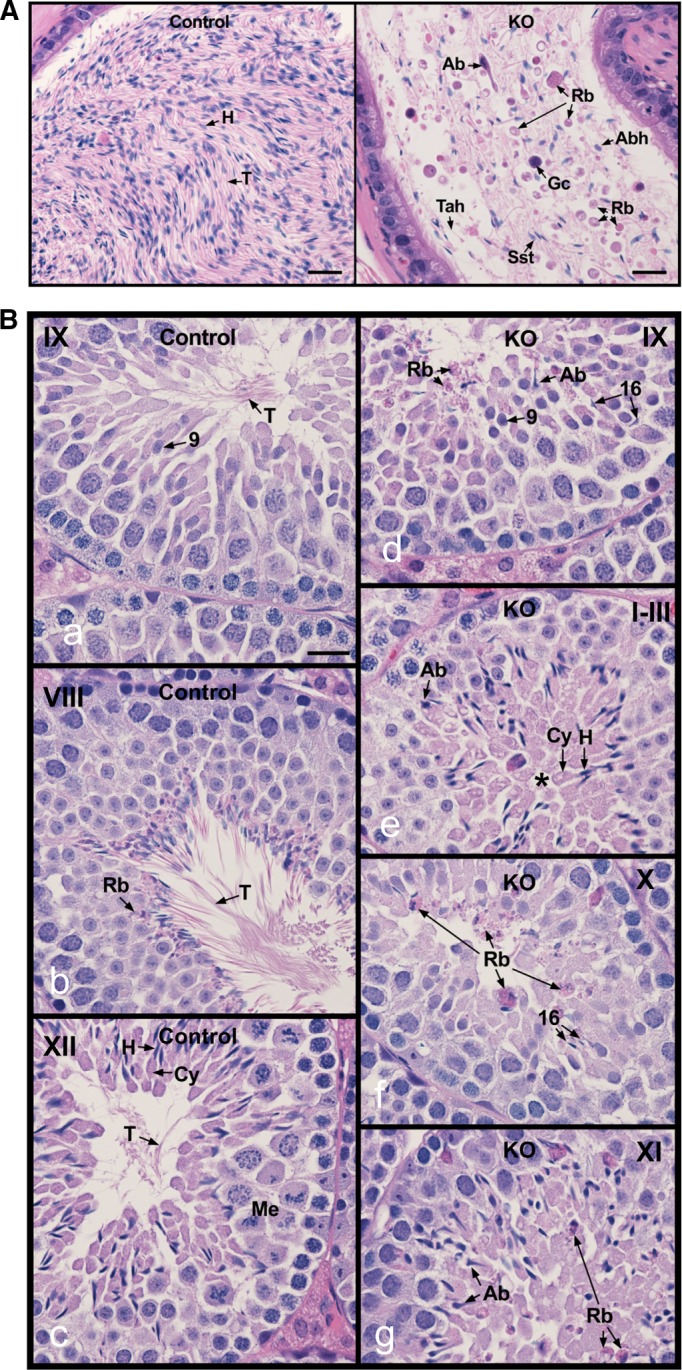
Histology of the cauda epididymis and testis from control and conditional Ift20 mutant mice. (A) Epididymis from control mice showing highly concentrated epididymal sperm with aligned sperm tails (T) and normal sperm heads (H; left). (B) Epididymis from IFT20 knockout mice (right), showing a very low concentration of sperm, with high incidence of sperm abnormalities (Ab), sloughed round germ cells (Gc), residual bodies (Rb), sperm with short tails (Sst), and sperm tails without heads (Tah). Bars, 20 mm. Testes from control (a–c) and IFT20 knockout mice (IFT20 KO; d–g). (a) Stage IX, showing normal step 9 spermatids with long tails (T) extending into the lumen. (b) Stage VIII, showing mature sperm with long tails (T) being released into the lumen. Residual bodies (Rb) of leftover cytoplasm are being formed and phagocytized by Sertoli cells. (c) Stage XII, showing spermatocytes in meiotic division (Me), step 12 elongating spermatids with head (H) of condensed chromatin, and bulges of cytoplasm (Cy). The spermatid tails (T) are long and extend into the lumen. (d) Stage IX, showing abnormal step 16 elongated spermatids (Ab) that were not released into the lumen and residual bodies (Rb) that are being sloughed into the lumen. (e) Stages I–III, showing abnormal elongated spermatids (Ab). The elongated spermatids show some normal heads (H) and cytoplasm (Cy), but their tails do not appear in the lumen (*). (f) Stage X, showing failure of spermiation, with step 16 spermatid heads remaining in the seminiferous epithelium. Residual bodies (Rb) remain at the lumen rather than being phagocytized. (g) Stage XI, showing abnormal elongating spermatids (Ab) and sloughed residual bodies (Rb) remaining from prior stages. Bars, 20 mm.
To understand the testicular factors that contribute to low sperm counts and abnormal sperm morphology, we examined testes of control (Figure 3B, a–c) and Ift20 mutant mice (Figure 3B, d–g). There was no difference between testis weight/body weight between controls and mutant mice (Supplemental Figure S4). We further investigated testis histology (Figure 3B and Supplemental Figure S5). The control mice showed normal completion of spermatogenesis. Stage IX showed normal spermatids with long tails (T) extending into the lumen (Figure 3Ba). Stage VIII showed mature sperm with long tails (T) being released into the lumen. Residual bodies (Rb) of leftover cytoplasm were being formed and phagocytized by Sertoli cells (Figure 3Bb). Stage XII showed spermatocytes in meiotic division (Me), step 12 elongating spermatids with heads (H) of condensed chromatin, and bulges of cytoplasm (Cy). The spermatid tails (T) were long and extended into the lumen (Figure 3Bc). In the mutant testis, stage IX showed abnormal step 16 elongated spermatids (Ab) that were not released into the lumen and residual bodies (Rb) that were being sloughed into the lumen (Figure 3Bd). In mutant stages I–III (Figure 3Be), abnormal elongated spermatids (Ab) were observed along with some normal heads (H) and cytoplasm (Cy), but spermatid tails did not appear in the lumen (*). Mutant stage X (Figure 3Bf) showed failure of spermiation, with step 16 spermatid heads remaining in the seminiferous epithelium and residual bodies (Rb) remaining at the lumen rather than being phagocytized. Mutant stage XI showed abnormal elongating spermatids (Ab) and sloughed residual bodies (Rb) remaining from prior stages (Figure 3Bg).
Ultrastructural changes in the sperm and testis of conditional Ift20 mutant mice
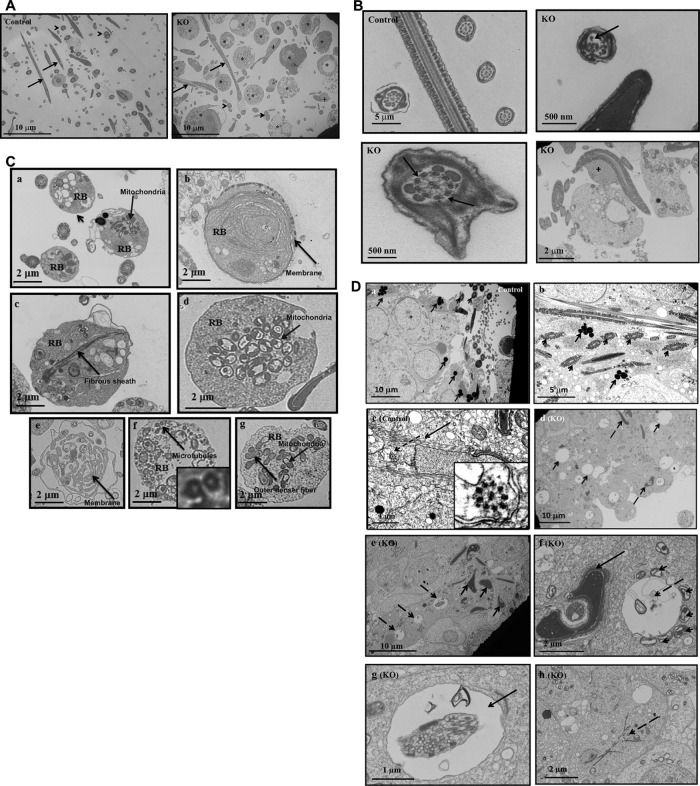
Transmission electron microscopy (TEM) was conducted to examine ultrastructure changes in epididymal sperm (Figure 4, A and B, and Supplemental Figures S6 and S7) and testis (Figure 4C and Supplemental Figure S8) of control and Ift20 mutant mice. Spermatozoa were the predominant cell type in the control mouse seminiferous tubule lumen (Figure 4A, left). However, few sperm were present in mutant mice, and significant numbers of residual bodies were present (Figure 4A, right, and Supplemental Figure S6). Control mice showed normal sperm structure (Figure 4B, left, top). However, abnormal axonemal structures were frequently discovered in mutant mice, including a disrupted “9 + 2” pattern (Figure 4B, right, top), and missing outer dense fibers (ODFs) in the axoneme (Figure 4B, bottom, left). Some sperm retained excess retention of cytoplasm (Figure 4B, bottom, right, and Supplemental Figure S7). High-magnification images of the residual bodies demonstrated that they contained mitochondria, fibrous sheath materials, microtubules, ODFs, and membranes that normally would have been part of the assembled flagella (Figure 4C).
FIGURE 4:
Ultrastructure of sperm from epididymis and testis of control and conditional Ift20 mutant mice. (A) Low- magnification images of cells collected from cauda epididymides of a control mouse and a conditional Ift20 mutant mouse. Notice that sperm cells are the predominant cell type in the control mouse. However, few sperm were present in the mutant mouse, and some of these sperm had excess cytoplasm (+). In addition, significant numbers of residual bodies (*) were present. The arrows point to the sperm axoneme in longitudinal section, and the arrowheads point to the sperm axoneme in cross section. (B) Representative high-magnification images of sperm axonemes from control and conditional Ift20 mutant mice. Note that the sperm axonemes from the control mouse show normal architecture. However, multiple defects were frequently discovered in sperm from the mutant mice. The arrow points to an axoneme with disrupted “9 + 2” structure and the dashed arrows to an axoneme missing three ODFs; the plus sign indicates sperm with excess retention of cytoplasm. (C) Representative high-magnification images of residual bodies obtained from cauda epididymides of conditional Ift20 mutant mice. Residual bodies are normally phagocytosed by Sertoli cells during spermiation and contain components from sperm that failed to assemble correctly; some identifiable structures include mitochondria (a, d, g), fibrous sheath (c), microtubules (f), ODFs (g), and membranes (b, e). (D) Ultrastructure of the testis of control and conditional Ift20 mutant mice. (a) Representative image of the lumen area of a seminiferous tubule from a control mouse. A number of sperm axonemes were present (arrowheads), and mature lysosomes (arrows) were easily seen. (b) Higher-magnification image of a seminiferous tubule from a control mouse. A number of developing axoneme (arrowheads) and mature lysosome (arrows) were observed. (c) Developing spermatid from a control mouse. The arrow points to the manchette; the dashed arrow points to a developing axoneme. The zoomed-in image (inset) shows the “9 + 2” structure of the developing axoneme. (d) Representative image of the lumen area of a seminiferous tubule from a conditional Ift20 mutant mouse. Axoneme structure can hardly be observed. Instead, a large number of large vacuoles (arrows) as well as deformed chromatin (dashed arrows) were discovered. (e) Higher-magnification image of a seminiferous tubule from a conditional Ift20 mutant mouse. Neither axoneme nor mature lysosomes were present. Instead, various cell debris, including degenerating chromatin (arrows) and vacuoles (dashed arrows), appeared throughout the seminiferous tubules. (f) Arrow points to deformed chromatin; dashed arrow points to a large vacuole surrounded by many mitochondria (arrowheads). (g) Arrows point to large vacuoles with various contents inside. (h) Dashed arrow points to some fiber-like structures.
In the control testis, well-developed sperm were present in the lumen of seminiferous tubules, as indicated by the cross section of normal axonemes. Dark lysosomes were seen in the cytoplasm of late-step spermatids (Figure 4D, a and b). The elongating spermatids from the control mice had normally developing manchette microtubules (Figure 4Dc), as well as normally condensed chromatin (Figure 4D). In the mutant testes, flagellar axonemes were rarely seen in seminiferous tubules and lumen (Figure 4Dd). Cell debris, including large vacuoles and abnormally condensed chromatin, were present in the tubule lumen and seminiferous tubules (Figure 4De); some vacuoles were surrounded by mitochondria, some vacuoles had various contents inside, and some cells had fiber-like cytoplasmic structures (Figure 4D, f–h, and Supplemental Figure S8).
Expression levels of an ODF component, ODF2, and a central apparatus component, SPAG16L are normal but not assembled into sperm flagella of Ift20 mutant mice
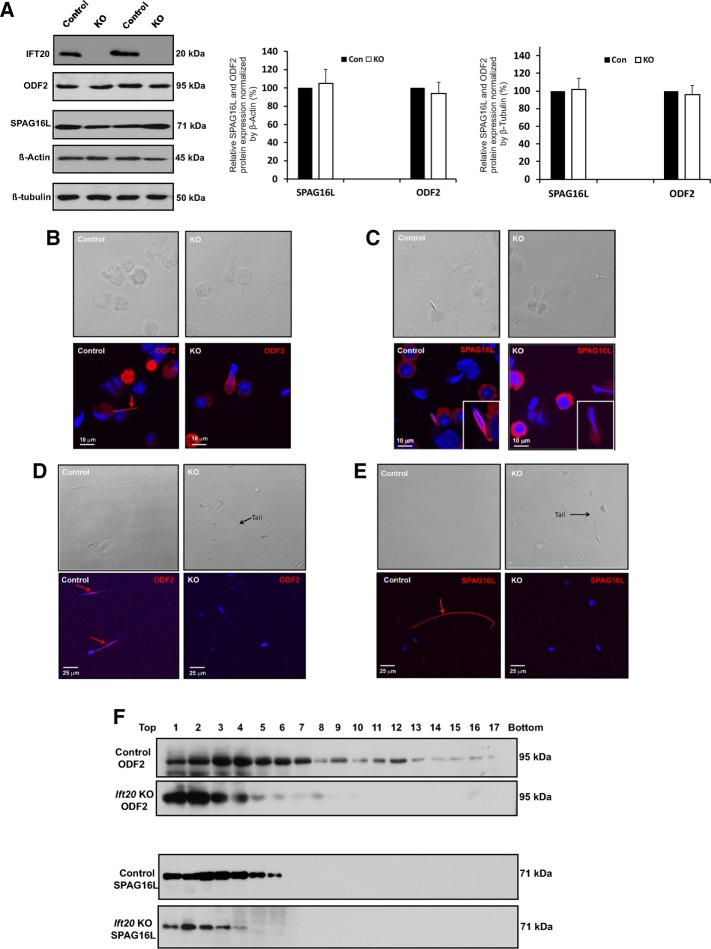
To examine the expression of components of sperm flagella of control and conditional Ift20 mutant mice, we conducted Western blotting and immunofluorescence staining targeting two representative proteins: ODF2, an ODF component, and SPAG16L, a central apparatus component. Western blotting analysis revealed that there was no difference in the testicular expression levels of both proteins between control and Ift20 mutant mice (Figure 5A). Immunofluorescence staining demonstrated that both ODF2 and SPAG16L were present in the developing flagella of elongating spermatids isolated from the control mice (Figure 5, B and C, left). However, even though the two proteins were expressed in the elongating spermatids isolated from the Ift20 mutant mice, the proteins were still in the cytoplasm (Figure 5, B and C, right).
FIGURE 5:
Expression and localization of sperm flagellar proteins in control and Ift20 mutant mice. (A) Testicular expression levels of a major component of sperm tail (ODFs), ODF2, and a central apparatus protein, SPAG16L, are not changed in the Ift20 mutant mice. Left, representative Western blot results using specific antibodies to examine testicular ODF2 and SPAG16L expression levels in control and Ift20 mutant mice. β-Actin and β-tubulin were used as controls. Right, quantitation of relative ODF2 and SPAG16L expression from six control and six Ift20 mutant mice normalized by β-actin and β-tubulin. There is no significant difference in expression levels between control and mutant mice. (B) ODF2 and SPAG16L are not assembled into flagella correctly in Ift20 mutant mice. Representative image of localization of ODF2 in the elongating spermatids of a control mouse (left) and an Ift20 mutant mouse (right). Specific signal (red) was detected in the developing tail of an elongating spermatid of the control mouse. However, ODF2 signal was still in the cytoplasm of an elongating spermatid of Ift20 mutant mouse. (C) Representative image of localization of SPAG16L in the elongating spermatids of a control mouse (left) and an Ift20 mutant mouse (right). Specific signal (red) was detected in the developing tail of an elongating spermatid of the control mouse. However, SPAG16L was still in the cytoplasm of an elongating spermatid of Ift20 mutant mouse. (D) Representative image of localization of ODF2 in the epididymis sperm of a control mouse (left) and an Ift20 mutant mouse (right). Specific signal (red) was observed in the principal piece of the control mouse. However, no specific signal was seen in Ift20 mutant mouse. (E) Representative image of localization of SPAG16L in the epididymis sperm of a control mouse (left) and an Ift20 mutant mouse (right). Specific signal (red) was detected in whole sperm tail of the control mouse. However, SPAG16L signal was not observed in the few developed sperm of the Ift20 mutant mouse. (F) The distribution of testicular ODF2 and SPAG16L in sucrose gradient is altered in Ift20 mutant mice. Note that both ODF2 and SPAG16L had a wider distribution in the gradients. However, distribution of the two proteins appeared to shift to the lighter fractions, indicating that the two proteins were assembled into the complexes with light density in the absence of IFT20.
Localization of the two proteins was also examined in epididymal sperm. In control mice, ODF2 was present in the flagellar midpiece (Figure 5D, left), and SPAG16L was present along the entire flagellum (Figure 5E, left). However, when a tail was formed in the Ift20 mutant sperm, no ODF2 or SPAG16L protein was detected (Figure 5, D and E, right).
Different distributions of ODF2 and SPAG16L in sucrose gradient between control and Ift20 mutant mice
Distribution of testicular ODF2 and SPAG16L in sucrose gradient was compared between the control and Ift20 mutant mice. After centrifugation, the collected gradients were analyzed by SDS–PAGE gels, and the presence of the two proteins in the gradients was examined by specific antibodies. Both ODF2 and SPAG16L proteins were present in most gradients, from lighter density to greater density in the control. However, the two proteins were present only in the gradients with lighter density in mutant testes (Figure 5F), suggesting that these two proteins formed the complexes with lighter density in the mutant mice.
Expression levels of other IFT proteins in the testes of control and Ift20 mutant mice
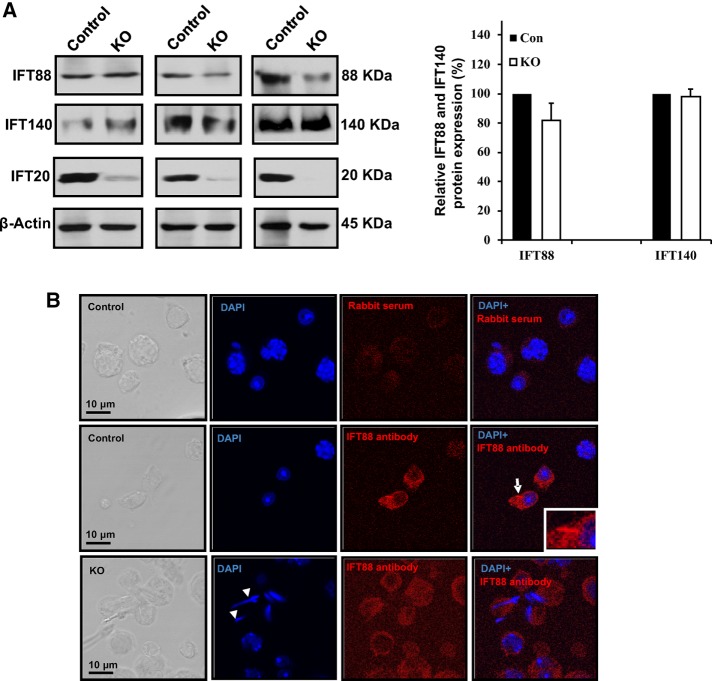
An IFT protein usually forms a complex with other IFT proteins, and these IFT proteins work in a coordinate manner to regulate ciliogenesis. To determine whether disruption of Ift20 affects expression levels of other IFT proteins, we analyzed IFT88, a component of the IFT-B complex, and IFT140, a component of the IFT-A complex,. All of the eight mutant mice studied expressed the same amount of IFT140 in the testis compared with the control mice. However, the Western blot results for IFT88 were variable. Four of the eight mutant mice expressed less IFT88 protein in the testis, and the reduction was appreciable (Figure 6A). Localization of IFT88 was further examined in the control and the mutant mice. In the control mice, the IFT88 signal was localized in the cytoplasm and the developing flagella of germ cells. However, in the mutant mice, the IFT88 signal was observed only in the cytoplasm, and the signal appeared to be weaker than that of the control mice (Figure 6B and Supplemental Figure S9). It is not clear whether IFT140 localization is changed, as the antibody did not work for the immunofluorescence staining.
FIGURE 6:
IFT-B component, IFT88, and IFT-A component, IFT140, in control and Ift20 mutant mice. (A) Left, representative Western blot results showing IFT88 and IFT140 expression in the testis of control and Ift20 mutant mice. Right, quantitation of relative IFT88 and IFT140 expression from eight control and eight Ift20 mutant mice. Note that compared with controls, the expression level of IFT88 was slightly but not significantly reduced in Ift20 mutant mice. There is no significant difference in IFT140 expression level between control and mutant mice. Variable IFT20 residual was also observed in Ift20 mutant mice, which might explain variable IFT80 expression level in the mutant mice. (B) Representative images of localization of IFT88 in isolated germ cells from control and Ift20 mutant mice. Note that IFT88 is present in the cytoplasm and the developing flagella in controls (arrow). In mutant mice, only cytoplasm staining was observed, and the signal appears to be weaker than in control mice. The arrowheads in the KO image point to spermatids with abnormal head.
Expression of autophagy-related proteins in the testes of control and conditional Ift20 mutant mice
IFT20 has been reported to play a role in autophagy, and it recruits autophagy core protein, ATG16L, to the autophagy core complex (Pampliega et al., 2013). Although some germ cells in the mutant testes completed all phases of spermatogenesis, the mutant spermatids were unable to process the redundant cytoplasm components of the cytoplasmic lobes for phagocytosis by the Sertoli cells, a sign of potential autophagy failure. We first examined ATG16L expression by Western blot analysis. Compared to the control mice, the ATG16L expression level was significantly reduced in conditional Ift20 mutant mice (Supplemental Figure S10A). We next analyzed the expression levels of LC3, an autophagy marker (Tanida et al., 2004), and ubiquitin, a conserved polypeptide unit that plays an important role in the ubiquitin-proteasome pathway (Kirkin et al., 2009) and is also considered to be an autophagy marker (Brown and Kaganovich, 2016). There were no differences in LC3 and ubiquitin expression between control and mutant mice (Supplemental Figure S10B).
DISCUSSION
Our laboratory identified a meiosis expressed gene 1 (MEIG1)/parkin coregulated gene (PACRG) complex in male germ cells during the study of cargo transport along the manchette and sperm flagellum formation (Zhang et al., 2009; Teves et al., 2013; Li et al., 2015, 2016). MEIG1 is localized in the whole cell body of spermatocytes, and is recruited to the manchette by PACRG in elongating/condensing spermatids. The manchette is a structure present only in elongating/condensing spermatids, and its major proposed function is to transport cargo proteins to build sperm flagella (Kierszenbaum, 2002; Lehti and Sironen, 2016). It appears that the roles of the MEIG1/PACRG complex and IFT proteins overlap in the transport of cargo during the assembly of cilia and flagella. Therefore, we decided to extend the study of MEIG1 to IFT and investigate the role of IFT in spermatogenesis and mammalian sperm flagella formation.
Even though mammalian male gametes have the longest motile cilia, very few studies have been conducted to investigate the role of IFT in the regulation of mammalian sperm flagellum formation. This is largely due to the fact that IFT is essential for embryonic development, and complete knockout of an IFT component usually causes embryonic lethality (Nonaka et al., 1998; Marszalek et al., 1999).
The IFT20 expression pattern in male germ cells strongly suggests a role in cargo transport for building sperm flagella. Therefore, floxed Ift20 mice were crossed to Stra8-iCre mice for the conditional knockout of Ift20 (Sadate-Ngatchou et al., 2008).
The present study demonstrated that IFT20 is essential for normal spermatogenesis and male fertility. In conditional Ift20 mutant mice, very few germ cells completed spermatogenesis, which resulted in a significantly reduced sperm count in the mutant cauda epididymis. Multiple mechanisms might be involved, including abnormal cells being phagocytized by the Sertoli cell, an increase in apoptosis of abnormal cells, and failure of spermiation. Histologic examination of testis revealed normal mitosis and meiosis in the mutant mice. However, spermiogenesis was disturbed, which is consistent with the predicted role of IFT in carrying cargo for the formation of cilia and flagella. There are at least two explanations for the few spermatids that were able to fully differentiate and complete spermatogenesis. First, given that the deletion was not 100% in this model, the residual IFT20 protein might be sequestered in a few germ cells, through cytoplasmic bridges, at a level sufficient for select spermatid differentiation. Second, other IFT proteins may partially compensate for the loss of IFT20. Even though some mutant germ cells completed spermatogenesis, most had abnormal morphology and did not function well, as most sperm were immotile. Sperm motility in mutant mice was significantly reduced compared with that in control mice, and the sperm showed random movements instead of directional forward movement. The low sperm count and reduced sperm motility fully explain the infertility of adult conditional Ift20 mutant males.
Why were the six mutant males subfertile at 6 wk of age but infertile after the first wave of spermatogenesis? It is likely that the six mutant mice retained more IFT20 protein than the others, which were completely infertile, but the protein level was still not enough for all germ cells to finish normal spermatogenesis, and so fertility was reduced. After the first wave, less IFT20 protein was available, and the mutant mice completely lost fertility. Alternatively, other IFT proteins that could partially compensate for the loss of IFT20 may show differential expression during the first and subsequent waves of spermatogenesis. In addition, it seems that whereas the first wave likely had relatively few normal or functional cells produced, these cells likely were passed along to the epididymis, but in subsequent waves, there would be more cumulative or secondary effects on the germ cells, and Sertoli cells would likely be responding to the abnormal germ cells, which would further reduce the chance of producing functional sperm.
The abnormal sperm function seems to have been caused by disrupted sperm structure. Examination of epididymal sperm by light microscopy and SEM revealed multiple gross abnormalities, including short, kinked tails and round heads. Of interest, some sperm seemed to have redundant cytoplasm attached on the flagella, suggesting a failure to remove the cytoplasmic component of the residual bodies in mutant mice testes. TEM of testis and epididymal sperm further demonstrated abnormal spermiogenesis and sperm structure. Very few axonemes were discovered in seminiferous tubules of the mutant mice. Accumulation of increased amounts of vesicles and fiber-like structures in the mutant cytoplasm suggests a failure in transportation of cargo-containing vesicles to their destinations in these cells and failure of incorporation of flagellar components into sperm tails. We do not know the mechanisms that account for the significant number of sloughed germ cells in the lumen of seminiferous tubules in mutant mice. This might be caused by a defect in Sertoli/germ cell recognition and failure of residual body formation and Sertoli cell phagocytosis of the excess germ cell cytoplasm.
Soon after the beginning of spermiation, specialized structures, tubulobulbar complexes (TBCs), appear between the spermatid and the Sertoli cells; these were suggested to be a modified form of endocytic machinery (Vogl et al., 2014). Numerous proteins have been localized to specific sites within TBCs, including the integral membrane adhesion proteins nectin-2 (Sertoli cell) and nectin-3 (spermatid; Guttman et al., 2004; Young et al., 2009), and associated vesicles that are positive for junction proteins also are positive for endosomal and lysosomal markers (Guttman et al., 2004; Young et al., 2009, 2012). TBCs are believed to be involved in reduction in the volume of the spermatid cytoplasm during spermiation (Russell, 1979, 1993). It remains to be determined whether IFT20 is also involved in TBC formation and function.
In the few epididymal sperm of the mutant mice, many exhibited abnormal “9 + 2” axonemal structures, indicating improper flagella assembly, which is consistent with defective function of an IFT protein. Redundant cytoplasmic components were better illustrated by TEM when cross-sectional images were taken. It is believed that the retained cytoplasmic components mechanically obstructed the straightening of sperm flagella and affected their motility. This phenotype of failure to remove the residual body of leftover cytoplasm from late spermatids was previously reported in Sperm1 mutant mice (Zheng et al., 2007) and several other mutant mice, including Tnp1, Tnp2, Prm1, Prm2, H1t2, Camk4, and Csnk2a2 (Xu et al., 1999; Wu et al., 2000; Cho et al., 2001, 2003; Zhao et al., 2001, 2004; Meistrich et al., 2003; Shirley et al., 2004; Tanaka et al., 2005). It is not clear whether IFT20 and these proteins function along similar pathways.
IFT proteins usually function as adaptors, as they associate with other proteins to form complexes for cargo transport (Lechtreck, 2015). Several IFT20 binding partners were identified (Sironen et al., 2010; Joo et al., 2013), but not ODF2 and SPAG16L, two flagellar proteins examined in this study. Given that localization of these two proteins in the elongating spermatids and epididymal sperm was dependent on IFT20 and their distribution in the sucrose gradient assay was shifted to the gradients with lower density, they appeared to be IFT20 cargo. However, IFT20 may not directly bind the two proteins, as other proteins may mediate the association. It is unlikely that the interaction was mediated through the known IFT20 binding partners. Therefore additional IFT20 binding partners might be present and should be identified in the future.
The mislocalization of ODF2 and SPAG16L could also be the result of other mechanisms. For example, because the ODFs and central pair are not assembled properly in mutant mice, these proteins are fundamentally orphaned. Alternatively, it is possible that defects in flagellar assembly may shut down spermiogenesis, and the intraflagellar transport mechanism is more broadly perturbed.
Even though IFT has been shown to be important for the assembly of cilia in most somatic tissues, it is dispensable for sperm flagellar assembly in both Drosophila and Plasmodium falciparum. It is believed that axonemal assembly occurs in the cytoplasm and the axoneme does not become membrane enclosed until after assembly (Han et al., 2003; Sarpal et al., 2003; Avidor-Reiss et al., 2004). However, the studies of Ift88 and our conditional Ift20 mutant mice revealed that IFT is essential for mouse spermiogenesis (San Agustin et al., 2015). Of interest, ODF2 was affected in both Ift88 and Ift20 mutant mice, suggesting that different IFT proteins may transport the same cargo, or the IFT complex is disrupted by both mutations and the transport defect is secondary. We previously discovered that SPAG16L localization is also dependent on the MEIG1/PACRG complex (Li et al., 2015). It is likely that the IFT20 and MEIG1/PACRG complexes function in the same transport pathway, although another possibility is that transport of one cargo may need multiple transport systems.
Chlamydomonas IFT20 is in the IFT-B complex (Lucker et al., 2005; Taschner and Lorentzen, 2016). The IFT-B complex consists of nine or 10 stably associated core subunits and six “peripheral” subunits that were shown to dissociate from the core structure at moderate salt concentrations (Taschner et al., 2016). Recent studies demonstrated that IFT20 forms a stable tubulin-binding IFT-B2 (core) complex with IFT172, 80, 57, 54, and 38 (Taschner et al., 2016); further, IFT88, together with the N-terminal domain of IFT52, is necessary to bridge the interaction between IFT-B1 and -B2. It is not clear whether this is also true in the mouse. We analyzed expression of two IFT proteins in this study: IFT88, an IFT-B component; and IFT140, an IFT-A component. It appeared that the testicular IFT88 expression level was slightly reduced in some of the Ift20 mutant mice. However, IFT140 expression level was not changed. These observations support the notion that mouse IFT20 might not function together with IFT140. It was reported that IFT20 was not affected in the Ift88 mutant mice (San Agustin et al., 2015). Variation in IFT88 expression levels might be caused by variation in residual IFT20 expression due to incomplete deletion of the Ift20 gene (Figure 6). Reduced IFT88 expression might be the result of reduced germ cell populations enriched in IFT88 in these Ift20 mutant mice. Based on our limited survey of IFT proteins, it appears that IFT20 works independently of the two IFT140 and IFT88, raising the possibility that IFT20 does not function in the same pathway as these other IFT proteins. However, Ift20 and Ift88 mutant mice shared many phenotypes, including reduced sperm count, disrupted flagellar structures—including disrupted axoneme and accessory structures—and ectopically assembled ODFs and microtubules, suggesting that the two IFT proteins might function through a similar mechanism in assembling sperm flagella, even though they might function independently. However, IFT20 mutant mice retained cytoplasmic lobe components that were not observed in IFT88 mutant mice, suggesting that the two proteins have unique functions.
During spermiogenesis, the excess cytoplasmic organelles need to be removed from the cell body. IFT20 not only regulates ciliogenesis through an IFT mechanism, it also associates with autophagy core protein ATG16L and helps to regulate a normal autophagy process required to clear redundant cytoplasmic components (Kobayashi, 2015). Several autophagy core genes, such as Atg7, participate in the control of spermatogenesis (Wang et al., 2014). In medaka fish, Ol-epg5 deficiency correlates with selectively impaired spermatogenesis and low allele transmission rates of the mutant allele caused by failure of germplasm and mitochondrial clearance during the process of germ cell specification and in the adult gonads (Herpin et al., 2015). IFT20 might participate in autophagy in the male germ cell, which involves the completion of the final steps required for the removal of late spermatid cytoplasmic residual body that is subsequently phagocytized by the Sertoli cells (Sprando and Russell, 1987a– c). This is supported by the following facts: 1) the expression of the autophagy core protein ATG16L was significantly reduced in testes of conditional Ift20 mutant mice; 2) sloughed residual bodies were present in the epididymal lumen; 3) epididymal mutant sperm contained excess cytoplasmic components that were not removed during late spermiogenesis; and 4) there was a dramatic reduction in mature lysosomes in the cytoplasm of the mutant mice. However, redundant cytoplasm was not discovered in most epididymal sperm, and the fact that expression levels of two autophagy markers—LC3 and ubiquitin—were not changed in the testis of mutant mice suggested that autophagy is not the major role of IFT20 in spermiogenesis.
In summary, we studied the role of IFT20 in mouse sperm development and found evidence to support the hypothesis that IFT20 has at least two functions. The major one is to transport cargo such as ODF2 and SPAG16L to assemble the sperm flagellum, and it also is involved in removing cytoplasmic components. The molecular pathways involved with these two functions will need to be elucidated by identifying novel binding partners of IFT20.
MATERIALS AND METHODS
Generation of genetically altered mice
All animal work was approved by Virginia Commonwealth University’s Institutional Animal Care and Use Committee (protocol AD10000167) in accordance with federal and local regulations regarding the use of nonprimate vertebrates in scientific research.
Stra8-iCre mice were purchased from Jackson Laboratory (008208; Bar Harbor, ME), and Ift20flox/flox mice were generated previously (Jonassen et al., 2008). Ift20 genotypes were identified as described previously (Jonassen et al., 2008).
Assessment of fertility and fecundity
To test fertility, we paired ≥6-wk-old Stra8-iCre; Ift20flox/Δ (KO) and control males with mature wild-type females for at least 2 mo and recorded the number of mice achieving a pregnancy and the number of offspring from each mating set or pregnancy.
Western blot analysis
Standard procedure was used as described previously. Expression of the indicated proteins was examined using adult mice by the following specific antibodies: anti-IFT20, IFT88, and IFT140 (all in 1:1000 dilution; generated by G.J.P.’s laboratory); anti-SPAG16L antibody (1:1000; generated by Z.Z.’s laboratory); anti-ODF2 (1:800; 12058-1AP; Proteintech, Rosemont, IL); anti–β-actin (1:2000; 4967; Cell Signaling Technology, Danvers, MA); anti-ATG16L (1:1000; AP1817b; ABGENT, San Diego, CA); anti-LC3A/B (1:1000; 4108S; Cell Signaling Technology); anti-ubiquitin (1:1000; 3983; Cell Signaling Technology); and anti–β-tubulin (1:2000; T8328; Sigma-Aldrich, St. Louis, MO). Horseradish peroxidase–labeled anti-rabbit (NA934V; 1:2000) and anti-mouse (NA931V; 1:2000) were obtained from Amersham (Pittsburgh, PA). Super Signal Pico Chemiluminescent (34080) and Femto Maximum Sensitive (34095) systems were purchased from Pierce/Thermo Fisher Scientific (Waltham, MA).
Transmission electron microscopy
Mouse testes and epididymal sperm were fixed in 3% glutaraldehyde/1% paraformaldehyde/0.1 M sodium cacodylate, pH 7.4, at 4°C overnight and processed for electron microscopy. Images were taken with a Hitachi HT7700 (Krefeld, Germany) transmission electron microscope.
Scanning electron microscopy
For SEM analysis, mouse epididymal sperm were collected and fixed in the same fixative solution as for TEM. The samples were processed by standard methods (Jonassen et al., 2008). Images were taken with a Zeiss (Jena, Germany) EVO 50 XVP SEM at Virginia Commonwealth University’s Microscopy Facility.
Spermatozoa counting
Sperm cells were collected from cauda epididymides and fixed in 2% formaldehyde for 10 min at room temperature. Sperm were counted using a hemocytometer chamber under a light microscope, and sperm number was calculated by standard methods (Zhang et al., 2006).
Spermatozoa motility assay
Sperm were collected after swimming out from the cauda epididymides. Sperm motility was observed using an inverted microscope (Nikon, Tokyo, Japan) equipped with 10× objective. Movies were recorded at 30 frames/sec with a SANYO (Osaka, Japan) color charge-coupled device, high-resolution camera (VCC-3972) and Pinnacle Studio HD (version 14.0) software. For each sperm sample, 10 fields were analyzed. Individual spermatozoa were tracked using ImageJ (National Institutes of Health, Bethesda, MD) and the plug-in MTrackJ. Sperm motility was calculated as curvilinear velocity (VCL), which is equivalent to the curvilinear distance (DCL) traveled by each individual spermatozoon in 1 s (VCL = DCL/t).
Histology and immunofluorescence staining on tissue sections
Testes and epididymides of adult mice were fixed in 4% formaldehyde solution in phosphate-buffered saline, paraffin embedded, and sectioned into 5-μm slides. Histology and immunofluorescence staining were carried out using standard procedures. The same anti-IFT20 antibody as used in the Western blot analysis was used, and the dilution was 1:200. To visualize IFT20, Cy3-conjugated anti-rabbit immunoglobulin G secondary antibody (1:5000; Jackson ImmunoResearch Laboratories, West Grove, PA) was used, and the slides were sealed using VectaMount with 4′,6-diamidino-2-phenylindole (H1200; Vector Laboratories, Burlingame, CA). Images were taken by confocal laser-scanning microscopy (Leica TCS-SP2 AOBS; Wetzlar, Germany).
Isolation of spermatogenic cells and immunofluorescence analysis
Spermatogenic cells were isolated as described previously (Li et al., 2015). Immunofluorescence (IF) was conducted and images were captured by confocal laser-scanning microscopy as before. The primary antibodies used were the same as used for Western blot analysis, but the dilutions were 10 times higher. The secondary antibodies were the same as used for IF on tissue sections.
Sucrose gradient analysis of cytoplasmic extracts of testes
Adult control and Ift20 mutant mice were killed by CO2 asphyxiation, and the testes were removed, decapsulated, dissected, and homogenized as rapidly as possible. Cytoplasmic extracts were prepared by lysis of two testes in 0.5 ml HNM buffer (0.1 M NaCl, 3 mM MgCl2, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4) containing 0.5% Triton N-101 with 10 strokes of a motor-driven glass–Teflon homogenizer (Thomas Scientific, Swedesboro, NJ) and centrifugation at 13,000 × g for 2 min. The supernatants were layered on a 5-ml 15–40% (wt/wt) linear sucrose gradient in HNM buffer, centrifuged at 28,000 rpm for 150–175 min in a Beckman (Weinheim, Germany) SW40 rotor, and decelerated with the brake to 8000 rpm. The gradients were collected as 17 0.3-ml fractions (Cataldo et al., 1999).
Supplementary Material
Acknowledgments
We thank Scott C. Henderson, Frances K. White, and Judy C. Williamson for assistance with confocal microscopy and electronic microscopy at the Microscopy Core Facility of Virginia Commonwealth University. This research was supported by National Institutes of Health Grant HD076257, the Virginia Commonwealth University Presidential Research Incentive Program and a Massey Cancer Award (to Z.Z.), National Institutes of Health Grant GM060992 (to G.J.P.), Chenery and Rashkind Grants from Randolph-Macon College (to J.F.), Youth Talents of Health and Family Planning Commission of Hubei Province of China (WJ2015Q026), and the National Natural Science Foundation of China (81571428, 81502792, 81300536, 81172462, and 81671514). Confocal microscopy and SEM were performed in the VCU Microscopy Facility of Virginia Commonwealth University (5P30NS047463). TEM was performed at Randolph-Macon College by Courtney Stevens and Brandon Delpi (NSF1229184).
Abbreviations used:
- ATG
autophagy-related gene
- Cre
Cre recombinase
- GMAP210
Golgi network–associated protein
- IFT
intraflagellar transport
- KIF3B
kinesin family member 3B
- KO
knockout
- LC3
light chain 3
- MEIG1
meiosis expressed gene 1
- ODF
outer dense fiber
- PACRG
parkin coregulated gene
- Rb
residual bodies
- SEM
scanning electron microscopy
- SPAG16L
sperm-associated antigen 16
- Stra8
stimulated by retinoic acid gene 8
- TBCs
tubulobulbar complexes
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0318) on September 28, 2016.
REFERENCES
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Baker SA, Freeman K, Luby-Phelps K, Pazour GJ, Besharse JC. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem. 2003;278:34211–34218. doi: 10.1074/jbc.M300156200. [DOI] [PubMed] [Google Scholar]
- Bao J, Ma HY, Schuster A, Lin YM, Yan W. Incomplete cre-mediated excision leads to phenotypic differences between Stra8-iCre; Mov10l1(lox/lox) and Stra8-iCre; Mov10l1(lox/Delta) mice. Genesis. 2013;51:481–490. doi: 10.1002/dvg.22389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Cevik S, Kaplan OI. Intraflagellar transport: from molecular characterisation to mechanism. Front Biosci. 2008;13:2633–2652. doi: 10.2741/2871. [DOI] [PubMed] [Google Scholar]
- Brown JM, Witman GB. Cilia and diseases. Bioscience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R, Kaganovich D. Look out autophagy, ubiquilin UPS its game. Cell. 2016;166:797–799. doi: 10.1016/j.cell.2016.07.048. [DOI] [PubMed] [Google Scholar]
- Cataldo L, Mastrangelo MA, Kleene KC. A quantitative sucrose gradient analysis of the translational activity of 18 mRNA species in testes from adult mice. Mol Hum Reprod. 1999;5:206–213. doi: 10.1093/molehr/5.3.206. [DOI] [PubMed] [Google Scholar]
- Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, Schultz RM, Hecht NB, Eddy EM. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod. 2003;69:211–217. doi: 10.1095/biolreprod.102.015115. [DOI] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NwB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Wang C, Wang B, Bassi MR, Caretti E, Champeval D, Calmont A, Jarnik M, Burch J, Zaret KS, et al. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev Biol. 2009;325:225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, Rosenbaum JL, Baldari CT. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Patrussi L, Masi G, Onnis A, Galgano D, Lucherini OM, Pazour GJ, Baldari CT. Specific recycling receptors are targeted to the immune synapse by the intraflagellar transport system. J Cell Sci. 2014;127:1924–1937. doi: 10.1242/jcs.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 2008;4:e1000315. doi: 10.1371/journal.pgen.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Xu F, Keady BT, Pazour GJ. Characterization of mouse IFT complex B. Cell Motil Cytoskeleton. 2009;66:457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort C, Bastin P. Elongation of the axoneme and dynamics of intraflagellar transport [in French] Med Sci (Paris) 2014;30:955–961. doi: 10.1051/medsci/20143011008. [DOI] [PubMed] [Google Scholar]
- Friedland-Little JM, Hoffmann AD, Ocbina PJ, Peterson MA, Bosman JD, Chen Y, Cheng SY, Anderson KV, Moskowitz IP. A novel murine allele of Intraflagellar Transport Protein 172 causes a syndrome including VACTERL-like features with hydrocephalus. Hum Mol Genet. 2011;20:3725–3737. doi: 10.1093/hmg/ddr241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman JA, Takai Y, Vogl AW. Evidence that tubulobulbar complexes in the seminiferous epithelium are involved with internalization of adhesion junctions. Biol Reprod. 2004;71:548–559. doi: 10.1095/biolreprod.104.028803. [DOI] [PubMed] [Google Scholar]
- Han YG, Kwok BH, Kernan MJ. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol. 2003;13:1679–1686. doi: 10.1016/j.cub.2003.08.034. [DOI] [PubMed] [Google Scholar]
- Herpin A, Englberger E, Zehner M, Wacker R, Gessler M, Schartl M. Defective autophagy through epg5 mutation results in failure to reduce germ plasm and mitochondria. FASEB J. 2015;29:4145–4161. doi: 10.1096/fj.14-265462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C, Cormier-Daire V. Ciliary disorder of the skeleton. Am J Med Genet C Semin Med Genet. 2012;160C:165–174. doi: 10.1002/ajmg.c.31336. [DOI] [PubMed] [Google Scholar]
- Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol. 2008;183:377–384. doi: 10.1083/jcb.200808137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo K, Kim CG, Lee MS, Moon HY, Lee SH, Kim MJ, Kweon HS, Park WY, Kim CH, Gleeson JG, Kim J. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci USA. 2013;110:5987–5992. doi: 10.1073/pnas.1220927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Le YZ, Pazour GJ. IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol Biol Cell. 2011;22:921–930. doi: 10.1091/mbc.E10-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol Reprod Dev. 2002;63:1–4. doi: 10.1002/mrd.10179. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Zhong Q, Sheng ZH, Yoshimori T, Liang C, Jung JU. Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J Cell Sci. 2012;125:4740–4750. doi: 10.1242/jcs.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;15:259–69. doi: 10.1016/j.molcel.2009.04.026. 34. [DOI] [PubMed] [Google Scholar]
- Kobayashi S. Choose delicately and reuse adequately: the newly revealed process of autophagy. Biol Pharm Bull. 2015;38:1098–1103. doi: 10.1248/bpb.b15-00096. [DOI] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF. IFT-cargo interactions and protein transport in cilia. Trends Biochem Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti MS, Sironen A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction. 2016;151:R43–R54. doi: 10.1530/REP-15-0310. [DOI] [PubMed] [Google Scholar]
- Li W, Tang W, Teves ME, Zhang Z, Zhang L, Li H, Archer KJ, Peterson DL, Williams DC, Jr, Strauss JF., 3rd A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development. 2015;142:921–930. doi: 10.1242/dev.119834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Walavalkar NM, Buchwald WA, Teves ME, Zhang L, Liu H, Bilinovich S, Peterson DL, Strauss JF, 3rd, Williams DC, Jr, Zhang Z. Dissecting the structural basis of MEIG1 interaction with PACRG. Sci Rep. 2016;6:18278. doi: 10.1038/srep18278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Zhi X, Zhong Q. ATG14 controls SNARE-mediated autophagosome fusion with a lysosome. Autophagy. 2015;11:847–849. doi: 10.1080/15548627.2015.1037549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, Cole DG. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Simera HL, Petralia RS, Montcouquiol M, Wang YX, Szarama KB, Liu Y, Lin W, Deans MR, Pazour GJ, Kelley MW. Ciliary proteins Bbs8 and Ift20 promote planar cell polarity in the cochlea. Development. 2015;142:555–566. doi: 10.1242/dev.113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Taschner M, Engel BD, Lorentzen E. Structural studies of ciliary components. J Mol Biol. 2012;422:163–180. doi: 10.1016/j.jmb.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, Woychik RP. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Russell L. Role in spermiation. In: Russell LD, Griswold MD, editors. The Sertoli Cell. Clearwater, FL: Cache River Press; 1993. pp. 269–302. [Google Scholar]
- Russell LD. Spermatid-Sertoli tubulobulbar complexes as devices for elimination of cytoplasm from the head region late spermatids of the rat. Anat Rec. 1979;194:233–246. doi: 10.1002/ar.1091940205. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin JT, Pazour GJ, Witman GB. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell. 2015;26:4358–4372. doi: 10.1091/mbc.E15-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal R, Todi SV, Sivan-Loukianova E, Shirolikar S, Subramanian N, Raff EC, Erickson JW, Ray K, Eberl DF. Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr Biol. 2003;13:1687–1696. doi: 10.1016/j.cub.2003.09.025. [DOI] [PubMed] [Google Scholar]
- Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod. 2004;71:1220–1229. doi: 10.1095/biolreprod.104.029363. [DOI] [PubMed] [Google Scholar]
- Sironen A, Hansen J, Thomsen B, Andersson M, Vilkki J, Toppari J, Kotaja N. Expression of SPEF2 during mouse spermatogenesis and identification of IFT20 as an interacting protein. Biol Reprod. 2010;82:580–590. doi: 10.1095/biolreprod.108.074971. [DOI] [PubMed] [Google Scholar]
- Sironen A, Kotaja N, Mulhern H, Wyatt TA, Sisson JH, Pavlik JA, Miiluniemi M, Fleming MD, Lee L. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol Reprod. 2011;85:690–701. doi: 10.1095/biolreprod.111.091132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironen A, Uimari P, Iso-Touru T, Vilkki J. L1 insertion within SPEF2 gene is associated with increased litter size in the Finnish Yorkshire population. J Anim Breed Genet. 2012;129:92–97. doi: 10.1111/j.1439-0388.2011.00977.x. [DOI] [PubMed] [Google Scholar]
- Sprando RL, Russell LD. Comparative study of cytoplasmic elimination in spermatids of selected mammalian species. Am J Anat. 1987a;178:72–80. doi: 10.1002/aja.1001780109. [DOI] [PubMed] [Google Scholar]
- Sprando RL, Russell LD. A comparative study of Sertoli cell ectoplasmic specializations in selected non-mammalian vertebrates. Tissue Cell. 1987b;19:479–493. doi: 10.1016/0040-8166(87)90042-5. [DOI] [PubMed] [Google Scholar]
- Sprando RL, Russell LD. Germ cell-somatic cell relationships: a comparative study of intercellular junctions during spermatogenesis in selected non-mammalian vertebrates. Scanning Microsc. 1987c;1:1249–1255. [PubMed] [Google Scholar]
- Stottmann RW, Tran PV, Turbe-Doan A, Beier DR. Ttc21b is required to restrict sonic hedgehog activity in the developing mouse forebrain. Dev Biol. 2009;335:166–178. doi: 10.1016/j.ydbio.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, et al. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol. 2005;25:7107–7119. doi: 10.1128/MCB.25.16.7107-7119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. Recombinant reconstitution and purification of the ift-b core complex from Chlamydomonas reinhardtii. Methods Mol Biol. 2016;1454:69–82. doi: 10.1007/978-1-4939-3789-9_5. [DOI] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves ME, Jha KN, Song J, Nagarkatti-Gude DR, Herr JC, Foster JA, Strauss JF, 3rd, Zhang Z. Germ cell-specific disruption of the Meig1 gene causes impaired spermiogenesis in mice. Andrology. 2013;1:37–46. doi: 10.1111/j.2047-2927.2012.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, Herron BJ, Chesebro AL, Qiu H, Scherz PJ, Shah JV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl AW, Du M, Wang XY, Young JS. Novel clathrin/actin-based endocytic machinery associated with junction turnover in the seminiferous epithelium. Semin Cell Dev Biol. 2014;30:55–64. doi: 10.1016/j.semcdb.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, Yang L, Tang H, Zhang X, Duan E, et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:852–869. doi: 10.1038/cr.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet. 2000;25:448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
- Xu X, Toselli PA, Russell LD, Seldin DC. Globozoospermia in mice lacking the casein kinase II alpha’ catalytic subunit. Nat Genet. 1999;23:118–121. doi: 10.1038/12729. [DOI] [PubMed] [Google Scholar]
- Young JS, Guttman JA, Vaid KS, Vogl AW. Tubulobulbar complexes are intercellular podosome-like structures that internalize intact intercellular junctions during epithelial remodeling events in the rat testis. Biol Reprod. 2009;80:162–174. doi: 10.1095/biolreprod.108.070623. [DOI] [PubMed] [Google Scholar]
- Young JS, Takai Y, Kojic KL, Vogl AW. Internalization of adhesion junction proteins and their association with recycling endosome marker proteins in rat seminiferous epithelium. Reproduction. 2012;143:347–357. doi: 10.1530/REP-11-0317. [DOI] [PubMed] [Google Scholar]
- Yuan X, Garrett-Sinha LA, Sarkar D, Yang S. Deletion of IFT20 in early stage T lymphocyte differentiation inhibits the development of collagen-induced arthritis. Bone Res. 2014;2:14038. doi: 10.1038/boneres.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kostetskii I, Tang W, Haig-Ladewig L, Sapiro R, Wei Z, Patel AM, Bennett J, Gerton GL, Moss SB, et al. Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol Reprod. 2006;74:751–759. doi: 10.1095/biolreprod.105.049254. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shen X, Gude DR, Wilkinson BM, Justice MJ, Flickinger CJ, Herr JC, Eddy EM, Strauss JF., 3rd MEIG1 is essential for spermiogenesis in mice. Proc Natl Acad Sci USA. 2009;106:17055–17060. doi: 10.1073/pnas.0906414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Mounsey S, Meistrich ML. Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol Reprod. 2004;71:1016–1025. doi: 10.1095/biolreprod.104.028191. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM, et al. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol. 2001;21:7243–7255. doi: 10.1128/MCB.21.21.7243-7255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Stratton CJ, Morozumi K, Jin J, Yanagimachi R, Yan W. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc Natl Acad Sci USA. 2007;104:6852–6857. doi: 10.1073/pnas.0701669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.