ATP9A is localized to phosphatidylserine-positive early and recycling endosomes, but not late endosomes, in HeLa cells. ATP9A plays a crucial role in recycling of transferrin and glucose transporter 1 from endosomes to the plasma membrane.
Abstract
Type IV P-type ATPases (P4-ATPases) are phospholipid flippases that translocate phospholipids from the exoplasmic (or luminal) to the cytoplasmic leaflet of lipid bilayers. In Saccharomyces cerevisiae, P4-ATPases are localized to specific subcellular compartments and play roles in compartment-mediated membrane trafficking; however, roles of mammalian P4-ATPases in membrane trafficking are poorly understood. We previously reported that ATP9A, one of 14 human P4-ATPases, is localized to endosomal compartments and the Golgi complex. In this study, we found that ATP9A is localized to phosphatidylserine (PS)-positive early and recycling endosomes, but not late endosomes, in HeLa cells. Depletion of ATP9A delayed the recycling of transferrin from endosomes to the plasma membrane, although it did not affect the morphology of endosomal structures. Moreover, depletion of ATP9A caused accumulation of glucose transporter 1 in endosomes, probably by inhibiting their recycling. By contrast, depletion of ATP9A affected neither the early/late endosomal transport and degradation of epidermal growth factor (EGF) nor the transport of Shiga toxin B fragment from early/recycling endosomes to the Golgi complex. Therefore ATP9A plays a crucial role in recycling from endosomes to the plasma membrane.
INTRODUCTION
Membrane traffic is mediated by vesicular and tubular carriers formed at the plasma membrane or organelles by the activities of multiple factors, including small GTPases, coat proteins, SNARE (soluble NSF [N-ethylmaleimide–sensitive factor] attachment protein receptor) proteins, fission proteins such as members of the dynamin family, and other accessory proteins. During formation of the vesicular–tubular carriers and fusion of organelles, dynamic membrane shape changes occur. These shape changes are thought to involve modification in the composition and distribution of lipids and many of the proteins mentioned above (Graham, 2004; Graham and Kozlov, 2010; Shin et al., 2012).
The type IV P-type ATPases (P4-ATPases), a subfamily of P-type ATPases, have been implicated in flipping of aminophospholipids from the exoplasmic (luminal) leaflet to the cytoplasmic leaflet (Tang et al., 1996; Pomorski et al., 2003; Daleke, 2007; Coleman et al., 2009; Zhou and Graham, 2009). Thus P4-ATPases are essential for the regulation and generation of trans-bilayer asymmetry of the plasma membrane: aminophospholipids, phosphatidylserine (PS), and phosphatidylethanolamine (PE) are primarily localized in the cytoplasmic leaflet, whereas phosphatidylcholine (PC) and sphingomyelin (SM) occur primarily in the exoplasmic leaflet (Op den Kamp, 1979; Devaux, 1991; Zachowski, 1993; Murate et al., 2015). We recently showed that ATP8B1, ATP8B2, and ATP10A preferentially flip PC, but not aminophospholipids (Takatsu et al., 2014; Naito et al., 2015), whereas ATP11A and ATP11C flip aminophospholipids (Yabas et al., 2011; Takatsu et al., 2014; Takada et al., 2015). Moreover, the enhanced PC flipping by ATP10A at the plasma membrane is associated with plasma membrane dynamics, including changes in cell shape (Naito et al., 2015; Miyano et al., 2016). Therefore local changes in composition of specific phospholipids by P4-ATPases are actively involved in the membrane-deformation process.
The five yeast P4-ATPases are all involved in protein transport at different stages of the secretory and endocytic pathways (Muthusamy et al., 2009). P4-ATPases also play essential roles in membrane trafficking in other organisms, including Caenorhabditis elegans and Arabidopsis thaliana (Poulsen et al., 2008; Anne-Francoise et al., 2009). PS flipped by ATP8A1 is required for recruitment of EHD1 to endosomes and for endosome-mediated trafficking in COS-1 cells (Lee et al., 2015). In addition, phospholipid flipping itself, as opposed to the flipped phospholipids, is important for vesicle formation in yeast (Takeda et al., 2014). Thus the phospholipid-flipping activities of P4-ATPases are indispensable for membrane trafficking in eukaryotic cells.
The yeast CDC50 proteins Cdc50p, Lem3p, and Crf1p associate with the P4-TPases Drs2p, Dnf1p/Dnf2p, and Dnf3p, respectively (Saito et al., 2004; Furuta et al., 2007). By contrast, Neo1p P4-ATPase does not associate with either Cdc50p or Lem3p (Saito et al., 2004). Most of the 14 mammalian P4-ATPases require association with CDC50 for their exit from the endoplasmic reticulum and subsequent localization (Paulusma et al., 2008; Bryde et al., 2010; van der Velden et al., 2010; Coleman and Molday, 2011; Takatsu et al., 2011). However, ATP9A and ATP9B, the putative mammalian orthologues of Neo1p, do not interact with CDC50 (Takatsu et al., 2011).
Endosomes are multifunctional organelles that regulate membrane transport between the plasma membrane and various intracellular compartments. After endocytosed cargoes such as soluble proteins, membrane proteins, and lipids arrive at early endosomes, some (e.g., recycling receptors and bulk membrane constituents) are returned to the plasma membrane, whereas others (e.g., growth factor receptors and nutrients) are transported to late endosomes and lysosomes for degradation. Some cargoes are transported from early endosomes to the Golgi complex (Grant and Donaldson, 2009; Chia and Gleeson, 2011; Johannes and Wunder, 2011).
In this study, we analyzed the function of ATP9A, which localizes to endosomes and the Golgi complex, in the endocytic pathway. We found that ATP9A plays an important role in recycling of transferrin (Tfn) from endosomes to the plasma membrane, but is dispensable for the lysosomal degradation of epidermal growth factor (EGF) and retrograde transport of Shiga toxin B fragment (StxB) from endosomes to the Golgi complex.
RESULTS
ATP9A localizes to the TfnR-positive recycling endosomes
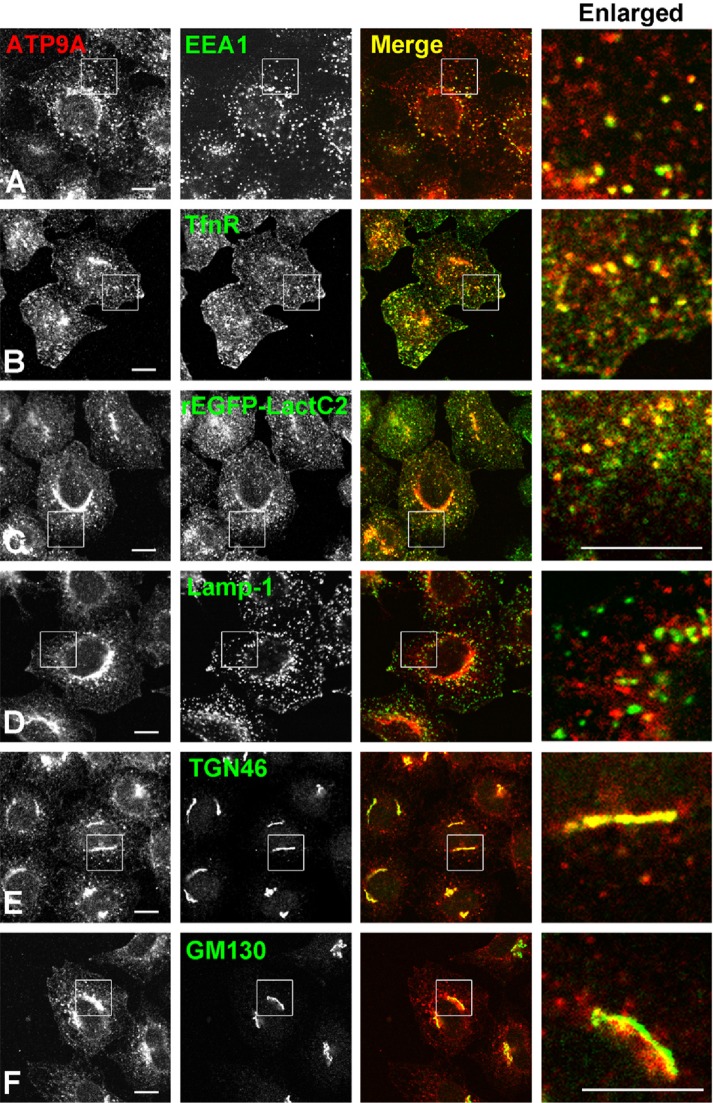
We previously determined the subcellular localization of human P4-ATPases by transient expression in HeLa cells. Among them, ATP9A localizes to early/recycling endosomes and the trans-Golgi network (TGN; Takatsu et al., 2011). To confirm the localization of ATP9A, we established stable cells expressing moderate levels of ATP9A and observed ATP9A colocalization with several organelle markers by confocal microscopy (Figure 1). ATP9A colocalized with EEA1, transferrin receptor (TfnR), and TGN46, which are markers for early endosomes, early/recycling endosomes, and the TGN, respectively (Figure 1, A, B, and E), but not with Lamp-1 and GM130, which are markers for late endosomes and the cis-Golgi, respectively (Figure 1, D and F).
FIGURE 1:
Subcellular localization of ATP9A. HeLa cells stably expressing C-terminally HA-tagged ATP9A were doubly stained for HA and organelle markers EEA1 (early endosome), TfnR (early/recycling endosome), Lamp-1 (late endosome), TGN46 (TGN), and GM130 (cis-Golgi), followed by Cy3-conjugated anti-rat and Alexa Fluor 488–conjugated anti-mouse or anti-rabbit secondary antibodies. The recombinant EGFP-Lactadherin C2 domain (rEGFP-LactC2) was treated with anti-HA antibody after permeabilization of cells with digitonin. Images were obtained by confocal microscopy. Insets are enlarged in the right panels. Scale bars: 10 μm.
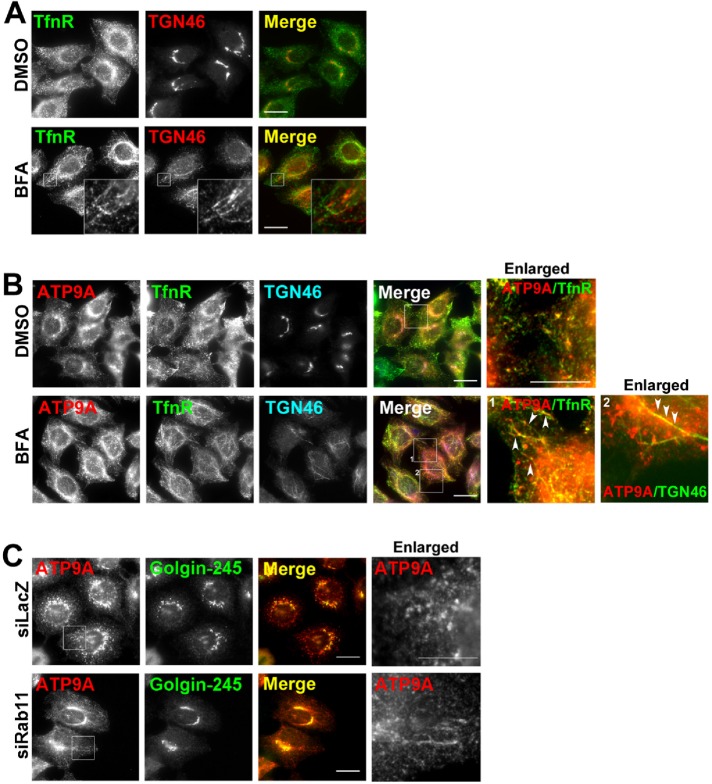
To confirm the recycling endosomal localization of ATP9A, we observed morphological changes of ATP9A-positive compartments following brefeldin A (BFA) treatment of cells stably expressing ATP9A. BFA treatment induces tubulation from recycling endosomes and the TGN but not from early endosomes (Sonnichsen et al., 2000; Shin et al., 2004). As shown in Figure 2A, TfnR-positive endosomes and the TGN were tubulated upon BFA treatment, but the tubules did not overlap. When ATP9A-expressing cells were treated with BFA, ATP9A-positive compartments were tubulated and overlapped with either TfnR (Figure 2B, inset 1, white arrowheads)- or TGN46 (Figure 2B, inset 2, white arrowheads)-positive tubular structures. Knockdown of Rab11a and b also induced tubulation of recycling endosomes (Takahashi et al., 2012). Depletion of Rab11a and b in ATP9A-expressing cells caused tubulation of ATP9A-positive compartments (Figure 2C). These results indicate that ATP9A localizes to the recycling endosomes, as well as to early endosomes and the TGN.
FIGURE 2:
Morphological changes in ATP9A-positive compartments following treatment with BFA or knockdown of Rab11. (A) HeLa cells or (B) HeLa cells stably expressing ATP9A-HA were treated with vehicle (dimethyl sulfoxide) or 5 μg/ml BFA for 15 min. (A) Cells were fixed and doubly stained for TfnR and TGN46, followed by Alexa Fluor 488–conjugated anti-mouse and Alexa Fluor 555–conjugated anti-rabbit secondary antibodies. (B) Cells were triply stained for HA, TfnR, and TGN46, followed by Cy3-conjugated anti-rat, Alexa Fluor 488–conjugated anti-mouse, and DyLight649-conjugated anti-rabbit secondary antibodies. Insets were enlarged. Scale bar: 10 μm. Arrowheads indicate colocalized tubular structures. (C) HeLa cells stably expressing ATP9A-HA were treated with siRNAs against LacZ (control) or Rab11a and Rab11b and doubly stained for HA and Golgin-245, followed by Cy3-conjugated anti-rat and Alexa Fluor 488–conjugated anti-mouse secondary antibodies. Scale bars: 20 μm. Insets were enlarged. Scale bar: 10 μm.
Lactadherin C2 domain (LactC2) is a specific probe for PS (Gagescu et al., 2000; Yeung et al., 2008). Transiently expressed enhanced green fluorescent protein (EGFP)-LactC2 localizes to the endosomal compartments and the plasma membrane (Fairn et al., 2011; Uchida et al., 2011). Therefore we investigated whether ATP9A colocalizes with PS-positive endosomal compartments. Because overexpression of EGFP-LactC2 affects endosomal integrity, we exposed cells mildly permeabilized with digitonin (Figure 1C) to recombinant EGFP-LactC2 protein (rEGFP-LactC2). ATP9A colocalized extensively with rEGFP-LactC2 (Figure 1C), indicating that ATP9A was localized to the PS-enriched endosomes. As shown in Supplemental Figure S1A, rEGFP-LactC2 preincubated with PS-liposomes, but not with PC- or PI-liposomes, was unable to detect subcellular organelles, indicating that the purified rEGFP-LactC2 specifically recognized PS-positive organelles. The rEGFP-LactC2 signals overlapped extensively with TfnR, partially with EEA1 and TGN46, and rarely with Lamp-1 (Supplemental Figure S1B), confirming that PS is enriched in the recycling endosomal compartments and is present at a lower level in other organelles (Fairn et al., 2011) in HeLa cells.
Tfn, but not EGF, localizes to PS-positive endosomes all the way through the transport process
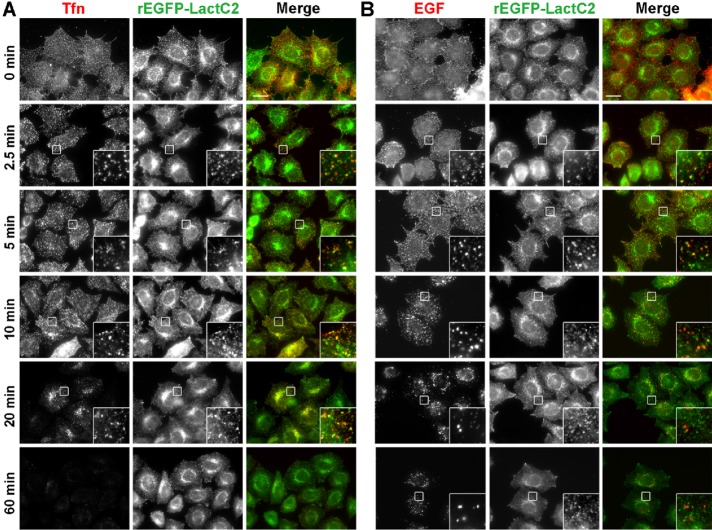
Tfn and EGF were internalized into cells via clathrin-dependent endocytosis and entered early endosomes. At early endosomes, some Tfn-TfnR is recycled to the plasma membrane (fast recycling), whereas some is transported to the recycling endosomes and recycled back to the plasma membrane (slow recycling). By contrast, EGF-EGFR (EGF receptor) is sorted at the early endosomes and then delivered to late endosomes and lysosomes for degradation. We investigated whether Tfn and EGF pass through the PS-positive endosomes during their respective trafficking pathways (Figure 3). Cells were incubated with Alexa Fluor 555–conjugated Tfn or EGF at 4°C for 60 min, washed to remove unbound Tfn or EGF, and then allowed to internalize the surface bound Tfn or EGF at 37°C for various time periods. Cells were then fixed, permeabilized, and incubated with rEGFP-LactC2 to visualize PS-positive endosomes. Internalized Tfn localized to PS-positive endosomes throughout its transport process (Figure 3A). Although internalized EGF localized to the PS-positive endosomes at early time points (Figure 3B, 2.5 and 5 min), it began to be segregated from the PS-positive endosomes at 10 min and was completely segregated at later time points (Figure 3B, 20 and 60 min). Thus EGF was probably sorted and delivered to the PS-poor late endosomes. By contrast, internalized Tfn passed through the PS-positive endosomes throughout the transport process, supporting a previous report that PS is required for Tfn trafficking (Lee et al., 2015).
FIGURE 3:
Tfn, but not EGF, passes through the PS-positive compartments throughout its trafficking process. HeLa cells were serum starved for 3 h and then incubated at 4°C for 50 min with Alexa Fluor 555–conjugated Tfn or EGF. Cells were washed and chased at 37°C for the indicated times, fixed, and mildly permeabilized with digitonin. The cells were then incubated with rEGFP-LactC2, washed, and fixed again before mounting. Scale bars: 20 μm.
Overexpression of ATP9A, but not ATP9B, inhibits the Tfn recycling pathway
In yeast, all five P4-ATPases are involved in the membrane trafficking, and some of them genetically interact with clathrin and the small GTPase Arf, which play major roles in vesicular transport (Chen et al., 1999). Therefore we hypothesized that ATP9A may be involved in vesicular transport at the early/recycling endosomes. To test this idea, we transiently overexpressed ATP9A and ATP9B, which do not localize to endosomes but instead exclusively localize to the Golgi complex (Takatsu et al., 2011), and then monitored the endocytosis and recycling of Tfn (Figure 4). We incubated the cells with Alexa Fluor 555–conjugated Tfn (Tfn-555) at 4°C for 60 min, washed them to remove unbound Tfn, and then allowed Tfn to internalize at 37°C for the indicated time periods in the presence of holo-Tfn. Internalization of Tfn was not affected in ATP9A- or ATP9B-expressing cells (Figure 4, A and B, 5 min). After a 20 min incubation at 37°C, despite the fact that most internalized Tfn was recycled and the intracellular Tfn signal had substantially decreased in nontransfected or ATP9B-overexpressing cells (Figure 4B), the internalized Tfn had accumulated at endosomal compartments in cells overexpressing ATP9A (Figure 4A, 20 min). This result indicates that overexpression of ATP9A, which localizes to early/recycling endosomes, inhibits Tfn trafficking, probably by alteration of endosomal membrane dynamics. Otherwise, it could be an artifact of crowding membrane proteins in endosomes.
FIGURE 4:
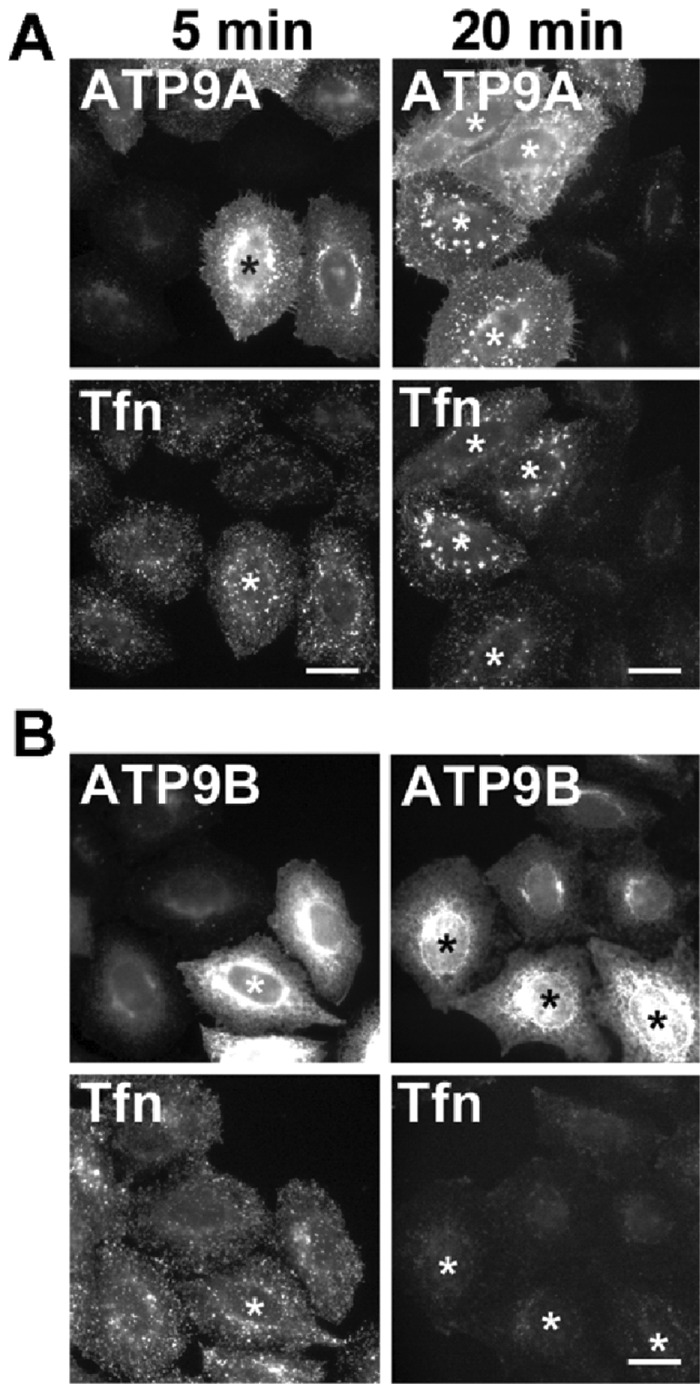
Overexpression of ATP9A affects Tfn trafficking. HeLa cells were transiently transfected with expression vector encoding HA-tagged ATP9A (A) or ATP9B (B), serum starved for 3 h, and then incubated at 4°C for 50 min with Alexa Fluor 488–conjugated Tfn. Cells were washed and chased at 37°C for 5 min and 20 min in the absence of the labeled Tfn. Cells were fixed and incubated with anti-HA antibody followed by Cy3-conjugated anti-rat secondary antibody (asterisks: overexpressing cells). Scale bars: 20 μm.
Depletion of ATP9A does not affect the endosomal structure
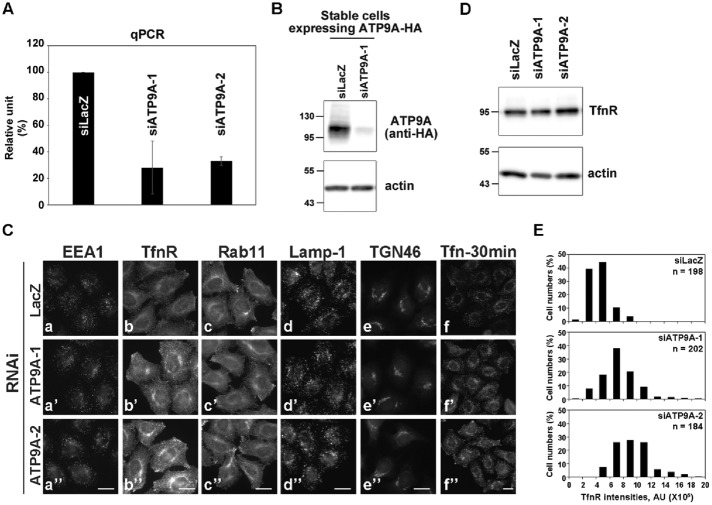
To investigate the role of ATP9A in endosome-mediated membrane trafficking, we next tried to knock down ATP9A using RNA interference (RNAi). To exclude off-target effects, we designed two different small interfering RNAs (siRNAs), one targeting the coding region of ATP9A (ATP9A-1) and the other targeting the noncoding region (ATP9A-2). Efficient depletion of ATP9A mRNA was confirmed by quantitative reverse transcription PCR (RT-PCR; Figure 5A). Although we were unable to raise antibodies capable of detecting endogenous ATP9A, we confirmed knockdown of protein expression by ATP9A-1 siRNA in cells stably expressing hemagglutinin (HA)-tagged ATP9A (Figure 5B).
FIGURE 5:
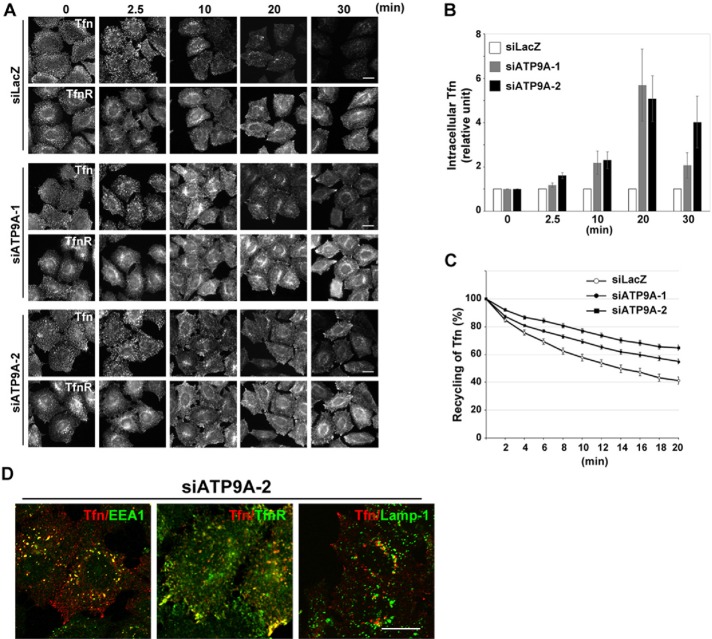
Depletion of ATP9A does not affect the distribution of organelles. (A) HeLa cells were transfected with siRNAs against LacZ (control), ATP9A-1, or ATP9A-2, and total RNAs were isolated from the cells, and RT-qPCR was performed. The results are from three independent experiments (averages ± SD). (B) HeLa cells stably expressing ATP9A-HA were transfected with siRNAs against LacZ or ATP9A-1, and each cell lysate was analyzed by immunoblotting with anti-HA and anti-actin antibodies (as an internal control). (C) siRNA-treated HeLa cells described in A were stained for EEA1 (a), TfnR (b), Rab11 (c), Lamp-1 (d), or TGN46 (e). Cells were treated with Alexa Fluor 555–Tfn for 30 min at 37°C and then fixed (f). Scale bars: 20 μm. (D) siRNA-treated HeLa cells described in A were lysed, and the lysates were analyzed by immunoblotting with anti-TfnR and anti-actin antibodies. (E) Fluorescence intensities of TfnR in (C, f–f″) were quantitated using the MetaMorph software; the frequency distribution of intensities is shown.
We then investigated whether depletion of ATP9A would affect the morphology of endosomal structures. As shown in Figure 5C, knockdown cells did not exhibit any morphological defects in endosomal markers, such as EEA1 for early endosomes (a), TfnR for early/recycling endosomes (b), Rab11 for recycling endosomes (c), Lamp-1 for late endosomes (d), and TGN46 for TGN (e). However, the fluorescent signals of TfnR were higher in ATP9A-knockdown cells than in control cells (Figure 5C, b–b″), although the total TfnR level was comparable to that in control cells (Figure 5D). Quantitation of TfnR signals (Figure 5E) revealed that the proportion of cells with high fluorescence intensities was higher in cells depleted of ATP9A than in control cells. Therefore depletion of ATP9A does not affect the integrity and biogenesis of endosomes or the integrity of the Golgi complex but may influence the trafficking of TfnR.
Depletion of ATP9A inhibits Tfn recycling but not internalization
Next we asked whether depletion of ATP9A affects the endocytic/recycling pathway of Tfn. In these experiments, Tfn-555 was allowed to internalize for 30 min at 37°C. Signals of internalized Tfn-555 were higher in knockdown cells than in control cells (Figure 5C, f–f″), suggesting that knockdown increased the rate of Tfn endocytosis or inhibited Tfn recycling from endosomes to the plasma membrane. To investigate the endocytosis and recycling of Tfn more quantitatively, we incubated control and knockdown cells with Tfn-555 at 4°C for 60 min, washed them to remove unbound Tfn-555, and then allowed Tfn-555 to internalize at 37°C for various time periods in the presence of unlabeled holo-Tfn (Figure 6, A and B). The amount of internalized Tfn-555 at early time points (2.5 min) was not significantly affected by knockdown of ATP9A. In control cells, signals of internalized Tfn-555 markedly decreased after 20 min incubation at 37°C, due to recycling of Tfn/TfnR to the plasma membrane. By contrast, a significant level of Tfn-555 persisted inside cells depleted of ATP9A. After 30 min, Tfn-555 disappeared almost completely from control cells, whereas some Tfn-555 was still retained within the knockdown cells. Thus the endocytosis of Tfn was not affected, but the recycling of Tfn was considerably delayed (but not completely blocked) by the depletion of ATP9A. We calculated the ratios of fluorescence intensity of Tfn-555 between ATP9A-knockdown cells and control cells at each time point (Figure 6B).
FIGURE 6:
Depletion of ATP9A delays the recycling of Tfn. (A) HeLa cells were transfected with siRNAs against LacZ, ATP9A-1, or ATP9A-2; serum starved for 3 h; and then incubated at 4°C for 60 min with Alexa Fluor 555–conjugated Tfn. Cells were washed and incubated at 37°C for the indicated times and then fixed and stained for TfnR. Scale bars: 20 μm. (B) Pixel intensities of Alexa Fluor 555–conjugated Tfn were estimated at the indicated times using MetaMorph software. Data are shown as the ratio of the mean of cellular Tfn intensities between cells depleted of ATP9A and control cells at each time point. The graph is representative of three independent experiments, and 160–200 cells of each sample were analyzed. Graphs show means ± SE. (C) HeLa cells were transfected with siRNAs as described above, incubated at 37°C for 60 min with Alexa Fluor 555–conjugated Tfn, washed, and subjected to time-lapse recording. Images in multiple areas were captured every 2 min for a total of 20 min, and pixel intensities of Alexa Fluor 555–Tfn were quantitated in every image. Graphs show means ± SE from 25 cells for siLacZ, 86 cells for siATP9A-1, and 59 cells for siATP9A-2. Raw data are shown in Supplemental Figure S2. (D) HeLa cells treated with siRNA against ATP9A-2 were serum starved for 3 h, incubated at 4°C for 60 min with Alexa Fluor 555–conjugated Tfn, washed, incubated at 37°C for 20 min, and stained with anti-EEA1, anti-TfnR, and anti-Lamp-1 antibodies, followed by anti-mouse Alexa Fluor 488–conjugated secondary antibody. Images were obtained by confocal microscopy. Scale bars: 20 μm.
To examine the recycling of Tfn more quantitatively in single cells, we incubated cells with Tfn-555 at 37°C for 30 min, washed them to remove unbound Tfn-555, and then incubated them at 37°C in the absence of labeled Tfn. The reduction in the Tfn-555 signal due to recycling was recorded in single cells by time-lapse microscopy (images acquired every 2 min), and the intracellular fluorescence intensity of Tfn-555 was quantitated (Figure 6C). Recycling of Tfn was delayed in cells depleted of ATP9A (Figure 6C; raw data, Supplemental Figure S2). Taken together, these observations indicate that ATP9A is required for efficient recycling of Tfn from endosomes to the plasma membrane but dispensable for the endocytosis of Tfn.
Next we asked whether the internalized Tfn was missorted to late endosomes in ATP9A-knockdown cells. To this end, cells were stained with anti–Lamp-1 antibody at the 20 min points in Figure 6A and observed by confocal microscopy. Signals of internalized Tfn-555 partially overlapped with EEA1 but not with Lamp-1, indicating that the internalized Tfn was not missorted to late endosomes (Figure 6D).
FIGURE 7:
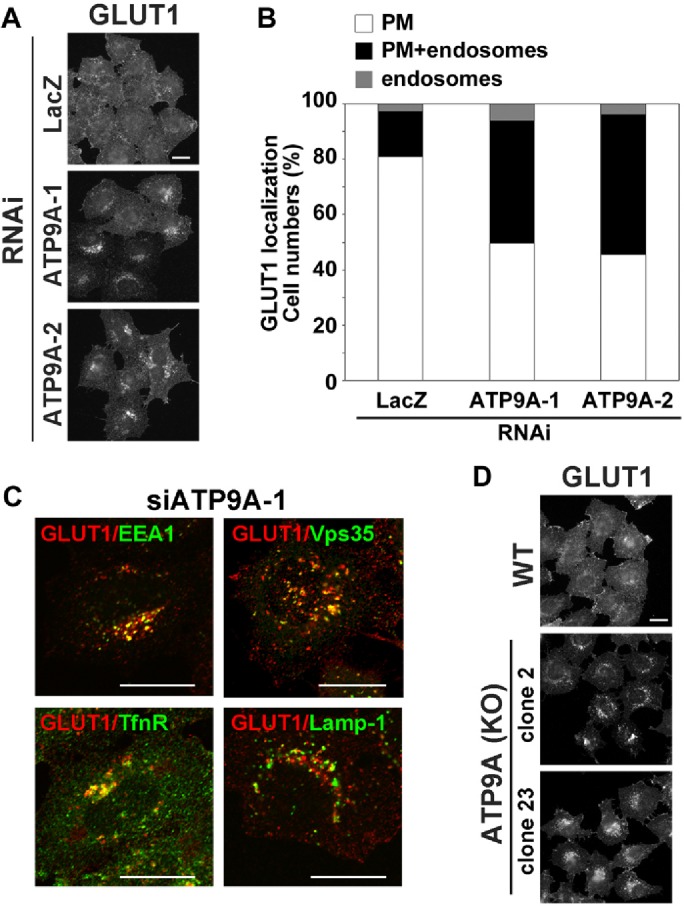
Depletion of ATP9A inhibits recycling of GLUT1. (A) HeLa cells were transfected with siRNAs against LacZ, ATP9A-1, or ATP9A-2, and then stained with anti-GLUT1 antibody. After being fixed, cells were incubated with Alexa Fluor–conjugated secondary antibody. Scale bar: 20 μm. (B) Cells in which GLUT1 localized to the plasma membrane (white), to the plasma membrane and endosomes (black), or to the endosomes (gray) in A were counted; counts were normalized against the total number of counted cells. A total of 214 cells for siLacZ, 199 cells for siATP9A-1, and 180 cells for siATP9A-2 were counted. (C) HeLa cells treated with siRNA against ATP9A-2 were fixed, permeabilized, and incubated with anti-GLUT1 and anti-EEA1, anti-Vps35, anti-TfnR, or anti-Lamp-1 antibodies, followed by Alexa Fluor 555–conjugated anti-rabbit and Alexa Fluor 488–conjugated anti-mouse secondary antibodies. Images were obtained by confocal microscopy. Scale bars: 20 μm. (D) Parental HeLa cells and two clones of ATP9A knockout cells were stained with anti-GLUT1 antibody. After fixation, cells were incubated with Alexa Fluor–conjugated secondary antibody. Scale bar: 20 μm.
We also examined Tfn recycling in ATP9A-knockout cells established using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system (Supplemental Figure S3). Consistent with the result of Figure 6A, recycling of Tfn, but not endocytosis, was delayed in two independent ATP9A-knockout clones (Supplemental Figure S4).
Depletion of ATP9A increases intracellular glucose transporter 1 (GLUT1)
We next investigated whether depletion of ATP9A would affect recycling of other cargo proteins. GLUT1 cycles between endosomes and the plasma membrane (Bridgewater et al., 2012; Steinberg et al., 2013; De Franceschi et al., 2015). In control cells, GLUT1 mainly localized to the plasma membrane and rarely to intracellular compartments (Figure 7A). By contrast, intracellular punctate staining of GLUT1 appeared in cells depleted of ATP9A (Figure 7A). The punctate structures colocalized with EEA1, TfnR, and Vps35 (a component of retromer complex), but not with Lamp-1, indicating that localization of GLUT1 in steady state was shifted to early and recycling endosomes from the plasma membrane in ATP9A-knockdown cells. Counting of cells in which GLUT1 localized to the plasma membrane, plasma membrane and endosomes, or endosomes (Figure 7B) revealed that endosomal localization of GLUT1 was substantially increased in cells depleted of ATP9A. We also confirmed that GLUT1 localization was shifted from the plasma membrane to endosomes in ATP9A-knockout cells (Figure 7D). Thus, in cells depleted of ATP9A, GLUT1 accumulated in endosomes, probably due to the delay of recycling from endosomes to the plasma membrane. Therefore ATP9A is required for the recycling of TfnR and GLUT1 from endosomes to the plasma membrane.
Depletion of ATP9A does not inhibit trafficking of StxB and EGF
We next examined the effects of the ATP9A knockdown on lysosomal transport of endocytosed EGF, which passes through early and late endosomes. After serum starvation, control and knockdown cells were incubated with Alexa Fluor 488–conjugated EGF (EGF-488) at 4°C for 60 min, washed to remove unbound EGF-488, and then allowed to internalize EGF-488 at 37°C for various time periods. The internalized EGF-488 colocalized extensively with the early endosomal marker EEA1 at early time points (Figure 8, 5 and 10 min), and a significant pool of EGF was segregated from early endosomes at later time points. After 120 min, the EGF-488 signal dramatically decreased due to lysosomal degradation. Endocytosis and the degradation of EGF was not significantly affected over the course of incubation in cells depleted of ATP9A (Figure 8A). We calculated the ratios of EGF-488 fluorescence intensity between ATP9A knockdown cells and control cells at each time point (Figure 8B).
FIGURE 8:
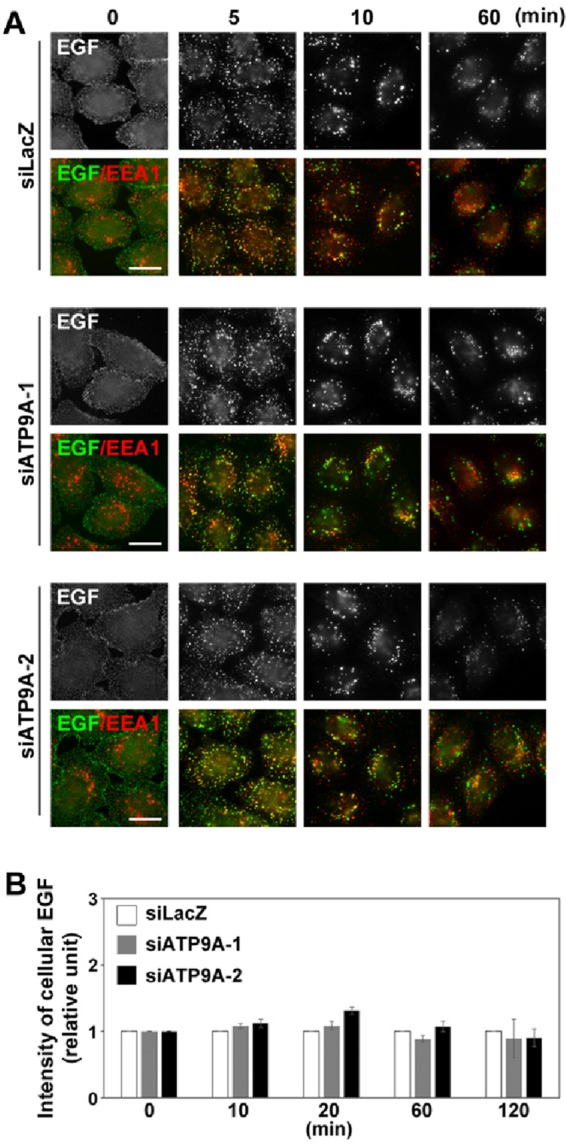
Depletion of ATP9A does not affect EGF trafficking. (A) HeLa cells were transfected with siRNAs against LacZ, ATP9A-1, or ATP9A-2; serum starved for 3 h; incubated at 4°C for 60 min with Alexa Fluor 488–conjugated EGF; washed; and incubated at 37°C for the indicated times. Cells were fixed and stained for EEA1. Scale bars: 20 μm. (B) Pixel intensity of Alexa Fluor 488–conjugated EGF was estimated at the indicated times using MetaMorph software. Data are shown as the ratio of the mean of cellular EGF intensities between cells depleted of ATP9A and control cells at each time point. Graph is representative of three independent experiments, and 100–200 cells of each sample were analyzed. Graphs show means ± SE.
We next examined the retrograde transport of StxB, which traverses through early/recycling endosomes en route to the TGN (Mallard et al., 1998; Shin et al., 2004) and endoplasmic reticulum. Cells treated with siRNAs against either LacZ or ATP9A were incubated for 60 min at 4°C with StxB. After cells were washed with phosphate-buffered saline (PBS), internalization was allowed to proceed at 37°C for the indicated time periods in the absence of StxB. Immunofluorescence and subsequent semiquantitative analysis revealed no significant differences between control and ATP9A-knockdown cells over the course of incubation (Figure 9, A and B). Therefore it is unlikely that ATP9A is required for retrograde transport of StxB from early/recycling endosomes to the Golgi complex. Together these observations suggest that ATP9A is not required for the late endosomal/lysosomal transport of EGF from early endosomes or the retrograde transport of StxB from early/recycling endosomes.
FIGURE 9:
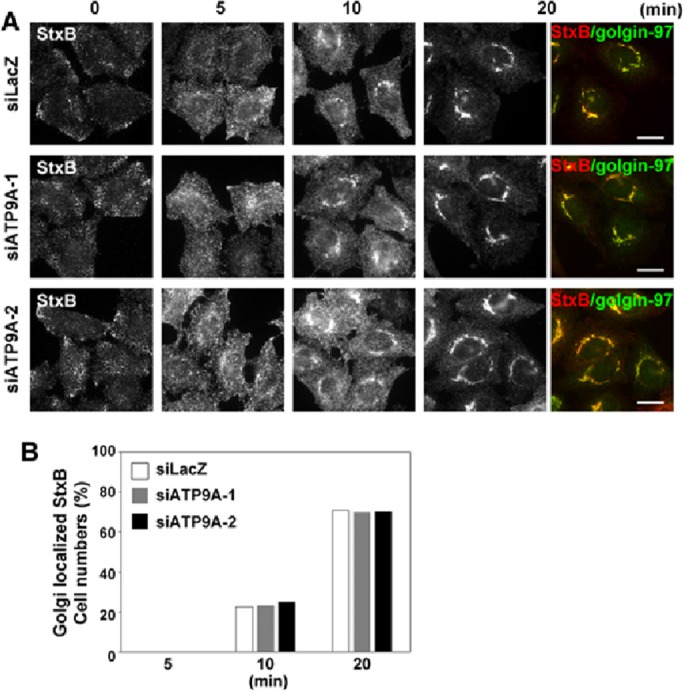
Depletion of ATP9A does not affect StxB trafficking. (A) HeLa cells were transfected with siRNAs against LacZ, ATP9A-1, or ATP9A-2; incubated at 4°C for 60 min with anti-StxB antibody; washed; and incubated at 37°C for the indicated times. The cells were then fixed and stained for golgin-97, followed by Alexa Fluor 555–conjugated anti-mouse and Alexa Fluor 488–conjugated anti-rabbit secondary antibodies. Scale bars: 20 μm. (B) Golgi arrival of internalized StxB was estimated by counting cells in which StxB colocalized with golgin-97. A total of 211 cells for siLacZ, 250 cells for siATP9A-1, and 241 cells for siATP9A-2 were counted.
DISCUSSION
Here we demonstrated that ATP9A, a member of the P4-ATPase family, is localized to the early/recycling endosomes and the TGN and is required for efficient recycling of TfnR and GLUT1 from endosomes to the plasma membrane. Although depletion of ATP9A did not affect the integrity or biogenesis of endosomal compartments, it inhibited efficient Tfn recycling from endosomal compartments to the plasma membrane but not StxB transport from endosomes to the Golgi complex. Thus ATP9A might be localized to a specialized membrane domain of early/recycling endosomes, where it is involved in recycling to the plasma membrane but not retrograde transport to the Golgi complex. In addition, internalized Tfn and accumulated intracellular GLUT1 were not missorted to late endosomes in ATP9A-depleted cells, suggesting that ATP9A may not be involved in cargo sorting at endosomes. We previously showed that, although a pair of the small GTPases Arf1/Arf4 and Arf1/Arf3 are redundantly required for the integrity of recycling endosomes, the former pair is required for StxB transport from endosomes to the TGN, whereas the latter is required for Tfn recycling from endosomes to the plasma membrane, indicating the presence of a specialized membrane domain in endosomes (Kondo et al., 2012; Nakai et al., 2013). ATP9A seems to be localized to the membrane domain in which recycling cargoes accumulate, and thus may play a role in the formation of plasma membrane–destined vesicles or tubules from endosomes. Depletion of ATP9A did not abrogate the Tfn recycling but delayed the recycling rate. Thus we cannot exclude a possibility of the functional redundancy in the P4-ATPase family. Despite the fact that Tfn recycling was inhibited by the overexpression of ATP9A, the recycling was not affected by the overexpression of ATP9B, a close homologue of ATP9A (Figure 4), suggesting that ATP9B may not be involved in the recycling pathway.
In yeast, PS flipping by Drs2p enhances membrane curvature, and the resultant negative charge is required for the function of Gcs1p, which is crucial for protein transport between the TGN and endosomes (Xu et al., 2013; Hankins et al., 2015). In COS-1 cells, PS in the cytoplasmic leaflet of recycling endosomes is required for recruitment of evectin-2, which plays a role in cholera toxin transport from the endosomes to the Golgi (Uchida et al., 2011). Therefore flipping of PS and the presence of PS in the cytoplasmic leaflet is crucial for endosome-mediated protein transport. Although we do not know whether ATP9A flips PS, or whether its flippase activity is required for the recycling pathway, it is tempting to speculate that ATP9A regulates PS translocation in endosomes because 1) ATP9A localizes to LactC2-positive endosomes; 2) internalized Tfn localizes to LactC2-positive endosomes during the course of endocytosis and recycling, and ATP9A is required for Tfn recycling; and 3) internalized EGF is localized to the LactC2-positive endosomes only at early time points but segregates from these compartments at later time points. On one hand, Takeda et al. (2014) proposed that flippase-dependent vesicle formation is mediated by phospholipid flipping, rather than by the flipped phospholipids. Accordingly, it is not clear whether flippase activity itself (causing lipid composition changes between lipid bilayers), flipped PS (causing recruitment of effector proteins, such as evectin-2), or both are required for the membrane-trafficking pathway. In any case, we believe that the flippase activity of ATP9A is involved in the formation of transport vesicles or tubules from a specialized membrane domain in endosomes, although we have not yet identified its lipid substrate.
ATP8A1 is localized to the PS-positive recycling endosomes and is involved in the membrane traffic through endosomes in COS-1 cells (Lee et al., 2015). In HeLa cells, however, ATP8A1 localizes to late endosomes, but not early/recycling endosomes (Takatsu et al., 2011), even in cells stably expressing moderate levels of the protein (Supplemental Figure S5A). Moreover, the ATP8A1 mRNA level in HeLa cells is much lower than in other cell types, whereas the ATP9A mRNA level is comparable (Supplemental Figure S5B). Therefore ATP8A1 may not play a crucial role in the recycling pathway in HeLa cells.
We observed no significant change in the distribution of cytoplasmic PS, which is detected with EGFP-LactC2, in cells depleted of ATP9A (Supplemental Figure S6), although the perinuclear signals of EGFP-LactC2 slightly increased as did those of TfnR in ATP9A-knockdown cells (Figure 5C). This could be explained in several ways: 1) ATP9A may have flippase activity toward other phospholipids but not PS; 2) even if ATP9A could flip PS and the local flippase activity of ATP9A is required for Tfn recycling, local changes of PS in the cytoplasmic leaflet of endosomes are undetectable; or 3) expression of EGFP-LactC2, even at moderate levels, may change PS dynamics by trapping cytoplasmic PS. Consistent with this, overexpression of EGFP-LactC2 alters endosomal integrity.
ATP9A is localized to the TGN as well as to endosomes. It is therefore tempting to speculate that ATP9A plays a role in the formation of transport carriers from the TGN in the secretory pathway. The substrate specificity of ATP9A and the roles of ATP9A at the TGN remain to be elucidated.
MATERIALS AND METHODS
Plasmids
The entire coding sequence of human ATP9A was cloned into vector pENTR3C (Invitrogen, Carlsbad, CA) as described previously (Takatsu et al., 2011). The region encompassing the attR1 site, the ccdB gene, the chloramphenicol resistance gene, and the attR2 site (DEST) was amplified from pcDNA6.2/V5-DEST (Invitrogen). BglII and XhoI restriction sites were introduced at the 5′ and 3′ ends of the DEST region and cloned into the pMXs-neo expression vector (Kitamura et al., 2003) with a C-terminal HA tag (pMXs-neo-DEST-HA). The pMXs-neo vector and pEF-gag-pol plasmid were kind gifts from Toshio Kitamura (University of Tokyo). Transfer of genes to pMXs-neo-DEST-HA was performed using the Gateway system (Invitrogen). The pCMV-VSVG-Rsv-Rev plasmid was a kind gift from Hiroyuki Miyoshi (RIKEN BRC). The pCAG-based vector for expression of ATP9A with a C-terminal HA tag was described previously (Takatsu et al., 2011). The StxB expression vector was a kind gift from Ludger Johannes (Institut Curie; Johannes et al., 1997). A cDNA fragment of lactadherin C2 domain (LactC2) was provided by Hiroyuki Arai (University of Tokyo) and subcloned into the pEGFP vector (Clontech; Takatsu et al., 2013). The lentiviral vector pRRLsinPPT and packaging plasmids (pRSV-REV, pMD2.g, and pMDLg/pRRE; Thomas et al., 2009) were kindly provided by Peter McPherson (McGill University). A fragment of EGFP-LactC2 was subcloned into pRRLsinPPT.
Antibodies and reagents
Antibodies were obtained from the following sources: monoclonal mouse anti-TfnR (H68.4) from Zymed (Carlsbad, CA); monoclonal mouse anti-EEA1 (clone 14), anti-Rab11 (clone 47), anti-Lamp-1 (H4A3), anti-p230 (Golgin-245), and anti-GM130 from BD Biosciences (San Diego, CA); monoclonal rat anti-HA (3F10) from Roche Applied Science (Indianapolis, IN); polyclonal rabbit anti-GLUT1 from Abcam (Cambridge, UK); monoclonal mouse anti-StxB was purified from culture supernatant of a hybridoma clone (13C4) obtained from the American Type Culture Collection (Manassas, VA); Alexa Fluor–conjugated secondary antibodies from Molecular Probes (Eugene, OR); Cy3-, DyLight649-, and horseradish peroxidase–conjugated secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA). Polyclonal rabbit anti-TGN46 (Kain et al., 1998) was a kind gift from Minoru Fukuda (Sanford-Burnham Medical Research Institute). Alexa Fluor–conjugated Tfn and EGF were purchased from Molecular Probes. BFA was purchased from Sigma-Aldrich (St. Louis, MO).
RT-PCR and quantitative RT-PCR (RT-qPCR)
Total RNA was extracted from cells using the RNeasy kit (Qiagen) and subjected to RT-PCR analysis using the SuperScript III One-Step RT-PCR system (Invitrogen). For RT-qPCR, total RNA was subjected to reverse transcription using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). The resultant cDNA was used as a template for PCR using LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche Applied Science). ATP8A1 and ATP9A fragments were amplified using the following primers: ATP8A1 forward, 5′-gaaacgctaatgaaagcagacatc-3′; ATP8A1 reverse, 5′-gtgttgaagtccagggcattc-3′; ATP9A forward, 5′-gcctcaccaagatcctctttgg-3′; ATP9A reverse, 5′-ggttcacgcgcaaactaatggg-3′. ATP9A mRNA expression was normalized against the level of β-actin mRNA in the same sample.
Cell culture and RNAi
HeLa cells were cultured in minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum. pRRLsinPPT-EGFP-LactC2 was transfected into HEK293T cells using Polyethylenimine Max (Polysciences, Warminster, PA) along with the packaging plasmids (pRSV-REV, pMD2.g, and pMDLg/pRRE). The resultant lentiviruses were concentrated as described previously (Takatsu et al., 2013) and then used to infect HeLa cells to establish stable cells. The infected cells were selected in medium containing G418 (1 mg/ml). The HA-tagged ATP9A-expressing stable cell line was obtained by transfection with pcDNA3-ATP9A-HA and selection with G418 (1 mg/ml).
Knockdown of ATP9A was performed as described previously (Yamamoto et al., 2010; Kondo et al., 2012). Briefly, pools of siRNAs targeting the coding region of human ATP9A (PCR amplified using primers 5′-gaacttcaccatcctacagatc-3′ and 5′-ccacgccgcagtcagattcc-3′) or the 3′-untranslated region of human ATP9A (PCR amplified using primers 5′- ttggtggatctttaagagccaag-3′ and 5′-aagtcaacttgtgcacctcaatc-3′) were prepared using the BLOCK-iT RNAi TOPO transcription kit and BLOCK-iT Dicer RNAi kit (Invitrogen) or the Replicator RNAi kit and PowerCut Dicer (Fynnzymes, Vantaa, Finland). Cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) and incubated for 24 h. The transfected cells were then split and seeded onto culture dishes. After 24 h, the cells were transfected again with the siRNAs and incubated for 24 h. The transfected cells were then transferred to new culture dishes containing coverslips, incubated for another 48 h, and processed for immunofluorescence and immunoblot analyses as described previously (Shin et al., 2004; Ishizaki et al., 2008; Nakai et al., 2013).
Gene editing by CRISPR/Cas9
We inactivated the ATP9A gene in HeLa cells using the CRISPR/Cas (CRISPR-associated) 9 system (Cong et al., 2013; Ran et al., 2013). Target sequences for ATP9A gene were designed using the CRISPR Design Tool from the Zhang lab (http://crispr.mit.edu). Complementary oligonucleotides (5′-caccgttctggcgcaccggctgcag-3′ and 5′-aaacctgcagccggtgcgccagaac-3′; target sequences are underlined) were synthesized and introduced into BbsI-digested vector PX459 (Addgene #48139). To edit the ATP9A gene, we used a previously described method (Kimura et al., 2014) with some modifications (unpublished data): cotransfection of the plasmid containing ATP9A target sequences and the Cas9 gene, and a donor plasmid (pDonor-tBFP-NLS-Neo; deposited in Addgene, ID 80766) containing a insertion cassette for the single-guide RNA (sgRNA)-guided cleavage site and reporter genes (TagBFP-3×NLS [nuclear localization signal] and neomycin resistance gene; referred to as Donor in this article; Supplemental Figure S3). Both plasmids were introduced into HeLa cells by transfection using the X-tremeGENE9 DNA Transfection Reagent (Roche). Transfected cells were selected in medium containing G418 (1 mg/ml), and clones were isolated on the basis of expression of the reporter gene Tag-BFP. For confirmation of editing of ATP9A, genomic DNA was extracted from individual clones, and subjected to PCR with three primer sets to amplify ATP9A lacking the Donor integration (GS1, 5′-tagggctgggatgtgattgtg-3′, and GAS1, 5′-ggccggcatcaggctaggtg-3′), ATP9A with Donor integrated in the forward orientation (D-primer, 5′-gttgtccacggtgccctccatgtac-3′ and GS1), and ATP9A with Donor integrated in the reverse orientation (D-primer and GAS1). Among clones with Donor-integrated clones in either orientation, the knockout (KO) was confirmed by direct sequencing of the amplified PCR product, ATP9A without Donor integration, using a specific sequencing primer (GS2, 5′-gagctcgggcccatggtgc-3′) (Supplemental Figure S3). The ATP9A gene was observed in three of eight clones carrying biallelic changes that resulted in Donor integration in forward and frame-shifting indels. Two of these clones (2 and 23) were used in this study.
Uptake of Tfn, EGF, and StxB; immunofluorescence; and time-lapse imaging analysis
Tfn internalization and recycling assays and EGF internalization assay were carried out as described previously (Shin et al., 2004; Kondo et al., 2012). HeLa cells were transfected with a pool of siRNAs targeting LacZ (as a control) or ATP9A. At 120 h posttransfection, cells were serum starved for 3 h in MEM medium containing 0.2% bovine serum albumin and then incubated with Alexa Fluor 488– or Alexa Fluor 555–conjugated Tfn or Alexa Fluor 488–conjugated EGF (Molecular Probes) at 4°C for 60 min. After cells were washed with ice-cold PBS, they were incubated in medium containing unlabeled holo-Tfn or normal medium (for Tfn or EGF uptake, respectively) at 37°C for the indicated times and processed for immunofluorescence analysis (Shin et al., 2004). In some cases, serum-starved cells were incubated with Alexa Fluor 488– or Alexa Fluor 555–conjugated Tfn at 37°C for 30 min, washed, and then incubated in medium containing unlabeled holo-Tfn at 37°C for the indicated times. For StxB uptake assay, cells were incubated with StxB (final concentration of 3.5 μg/ml) at 4°C for 60 min. After being washed, the cells were incubated in normal medium without StxB at 37°C for the indicated times and then processed for immunofluorescence analysis with anti-StxB antibody (Man et al., 2011).
For detection of PS-positive compartments using recombinant EGFP-LactC2, cells were fixed with 3% paraformaldehyde (PFA) and permeabilized with 20 μM digitonin for 5 min at room temperature. After incubation with recombinant EGFP-LactC2 and indicated antibodies, cells were fixed again with 3% PFA before mounting. Immunofluorescence analysis was performed on an Axiovert 200 MAT microscope (Carl Zeiss, Jena, Germany) for epifluorescence imaging or an A1R-MP (Nikon, Tokyo, Japan) or FV1000 (Olympus, Melville, NY) microscope for confocal imaging. The fluorescence intensity of Tfn was quantitated using MetaMorph imaging software (Molecular Devices, Sunnyvale, CA).
For time-lapse recording, siRNA-treated cells were allowed to internalize Alexa Fluor 555–conjugated Tfn for 30 min at 37°C, washed, placed on a microscope stage prewarmed to 37°C, and then observed on an A1R-MP confocal microscope. Images in multiple areas were acquired every 2 min for a total of 20 min; each image was acquired for 2 s. The fluorescence intensity of Tfn was quantitated using the NIS Elements software (Nikon).
Preparation of recombinant EGFP-LactC2
EGFP-LactC2 C-terminally fused to glutathione S-transferase (GST) was expressed in Escherichia coli BL21-Codon Plus (DE3) cells (Stratagene, La Jolla, CA) and purified as described previously (Takatsu et al., 2013). Expression of fluorescent proteins was induced by 0.1 mM isopropyl β-d-thiogalactopyranoside (Nacalai Tesque, Kyoto, Japan) at 20°C overnight. After purification with glutathione–Sepharose 4B beads (GE Healthcare), GST-fusion proteins were cleaved using PreScission protease to elute fluorescent proteins.
Preparation of liposome and competition assay
PS- or PI-liposomes were prepared with cholesterol (10 mol%), PC (45 mol%), PE (35 mol%), and either PS (10 mol%) or phosphatidylinositol (PI; 10 mol%), respectively. PC-liposomes were prepared using PC instead of PS or PI. Each liposome was preincubated with recombinant EGFP-LactC2 (rEGFP-LactC2) for 1 h and then removed by ultracentrifugation at 100,000 × g for 1 h. The supernatant was added to the fixed and permeabilized cells as described above.
Immunoblot analysis
Cells were lysed in lysis buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA) containing a protease inhibitor mixture (Nacalai Tesque) at 4°C for 30 min. The lysates were centrifuged at maximum speed for 20 min at 4°C in a microcentrifuge to remove cellular debris and insoluble materials. Cell lysates (10 μg/sample) were incubated in SDS sample buffer containing β-mercaptoethanol at 37°C for 2 h or at room temperature overnight, and then the samples were subjected to SDS–PAGE and immunoblot analysis using rat anti-HA, mouse anti-actin, or mouse anti-TfnR antibody. Immunoblots were developed using a Chemi-Lumi One L kit (Nacalai Tesque), and recorded on a LAS-3000 bioimaging system (Fujifilm).
Supplementary Material
Acknowledgments
We thank Feng Zhang (MIT), Toshio Kitamura (University of Tokyo), Hiroyuki Miyoshi (RIKEN BioResource Center), Hiroyuki Arai (University of Tokyo), Ludgar Johannes (Institut Curie), Minoru Fukuda (Sanford-Burnham Medical Research Institute), and Peter McPherson (McGill University) for kindly providing materials. We also thank Toyoshi Fujimoto (Nagoya University) for helpful advice. This work was supported by MEXT KAKENHI grant number JP23113714 (to H.-W.S.) and JSPS KAKENHI Grant Numbers JP26460065, JP15H01320, and JP16H00764 (to H.-W.S.); by the Takeda Science Foundation (to H.-W.S.); and by the Research Foundation for Pharmaceutical Sciences (to H.-W.S.).
Abbreviations used:
- BFA
brefeldin A
- EGF
epidermal growth factor
- EGF-488
Alexa Fluor 488–conjugated EGF
- EGFP
enhanced green fluorescent protein
- GLUT1
glucose transporter 1
- GST
glutathione S-transferase
- HA
hemagglutinin
- LactC2
lactadherin C2 domain
- P4-ATPase
type IV P-type ATPase
- PBS
phosphate-buffered saline
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- RNAi
RNA interference
- siRNA
small interfering RNA
- SM
sphingomyelin
- StxB
Shiga toxin B fragment
- Tfn
transferrin
- TfnR
transferrin receptor
- TGN
trans-Golgi network
- SNARE
soluble NSF (N-ethylmaleimide–sensitive factor) attachment protein receptor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-08-0586) on October 12, 2016.
REFERENCES
- Anne-Francoise R, Lars N, Fabrice R, Morten Krog L, Jean-Louis B, Simon T. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 2009;10:88–100. doi: 10.1111/j.1600-0854.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- Bridgewater RE, Norman JC, Caswell PT. Integrin trafficking at a glance. J Cell Sci. 2012;125:3695–3701. doi: 10.1242/jcs.095810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryde S, Hennrich H, Verhulst PM, Devaux PF, Lenoir G, Holthuis JC. CDC50 proteins are critical components of the human class-1 P4-ATPase transport machinery. J Biol Chem. 2010;285:40562–40572. doi: 10.1074/jbc.M110.139543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PZ, Gleeson PA. The regulation of endosome-to-Golgi retrograde transport by tethers and scaffolds. Traffic. 2011;12:939–947. doi: 10.1111/j.1600-0854.2011.01185.x. [DOI] [PubMed] [Google Scholar]
- Coleman JA, Kwok MCM, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Molday RS. Critical role of the b-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem. 2011;286:17205–17216. doi: 10.1074/jbc.M111.229419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke DL. Phospholipid flippases. J Biol Chem. 2007;282:821–825. doi: 10.1074/jbc.R600035200. [DOI] [PubMed] [Google Scholar]
- De Franceschi N, Hamidi H, Alanko J, Sahgal P, Ivaska J. Integrin traffic—the update. J Cell Sci. 2015;128:839–852. doi: 10.1242/jcs.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux PF. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 1991;30:1163–1173. doi: 10.1021/bi00219a001. [DOI] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagescu R, Demaurex N, Parton RG, Hunziker W, Huber LA, Gruenberg J. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell. 2000;11:2775–2791. doi: 10.1091/mbc.11.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell. 2015;26:4674–4685. doi: 10.1091/mbc.E15-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki R, Shin H-W, Mitsuhashi H, Nakayama K. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol Biol Cell. 2008;19:2650–2660. doi: 10.1091/mbc.E07-10-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Tenza D, Antony C, Goud B. Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J Biol Chem. 1997;272:19554–19561. doi: 10.1074/jbc.272.31.19554. [DOI] [PubMed] [Google Scholar]
- Johannes L, Wunder C. Retrograde transport: two (or more) roads diverged in an endosomal tree. Traffic. 2011;12:956–962. doi: 10.1111/j.1600-0854.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- Kain R, Angata K, Kerjaschki D, Fukuda M. Molecular cloning and expression of a novel human trans-Golgi network glycoprotein, TGN51, that contains multiple tyrosine-containing motifs. J Biol Chem. 1998;273:981–988. doi: 10.1074/jbc.273.2.981. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- Kondo Y, Hanai A, Nakai W, Katoh Y, Nakayama K, Shin H-W. ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct Funct. 2012;37:141–154. doi: 10.1247/csf.12015. [DOI] [PubMed] [Google Scholar]
- Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, et al. Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–688. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F, Antony C, Tenza D, Salamero J, Goud B, Johannes L. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of Shiga toxin B-fragment transport. J Cell Biol. 1998;143:973–990. doi: 10.1083/jcb.143.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man Z, Kondo Y, Koga H, Umino H, Nakayama K, Shin H-W. Arfaptins are localized to the trans-Golgi by interaction with Arl1, but not Arfs. J Biol Chem. 2011;286:11569–11578. doi: 10.1074/jbc.M110.201442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyano R, Matsumoto T, Takatsu H, Nakayama K, Shin HW. Alteration of transbilayer phospholipid compositions is involved in cell adhesion, cell spreading, and focal adhesion formation. FEBS Lett. 2016;590:2138–2145. doi: 10.1002/1873-3468.12247. [DOI] [PubMed] [Google Scholar]
- Murate M, Abe M, Kasahara K, Iwabuchi K, Umeda M, Kobayashi T. Transbilayer distribution of lipids at nano scale. J Cell Sci. 2015;128:1627–1638. doi: 10.1242/jcs.163105. [DOI] [PubMed] [Google Scholar]
- Muthusamy B-P, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta. 2009;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Takatsu H, Miyano R, Takada N, Nakayama K, Shin HW. Phospholipid flippase ATP10A translocates phosphatidylcholine and is involved in plasma membrane dynamics. J Biol Chem. 2015;290:15004–15017. doi: 10.1074/jbc.M115.655191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai W, Kondo Y, Saitoh A, Naito T, Nakayama K, Shin H-W. ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol Biol Cell. 2013;24:2570–2581. doi: 10.1091/mbc.E13-04-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Kamp JA. Lipid asymmetry in membranes. Annu Rev Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- Paulusma CC, Folmer DE, Ho-Mok KS, de Waart DR, Hilarius PM, Verhoeven AJ, Oude Elferink RP. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology. 2008;47:268–278. doi: 10.1002/hep.21950. [DOI] [PubMed] [Google Scholar]
- Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JCM. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LR, Lopez-Marques RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG. The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell. 2008;20:658–676. doi: 10.1105/tpc.107.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-Type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H-W, Morinaga N, Noda M, Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol Biol Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H-W, Takatsu H, Nakayama K. Mechanisms of membrane curvature generation in membrane traffic. Membranes. 2012;2:118–133. doi: 10.3390/membranes2010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavaré JM, Cullen PJ. A global analysis of SNX27–retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol. 2013;15:461–471. doi: 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Takatsu H, Miyano R, Nakayama K, Shin HW. ATP11C mutation is responsible for the defect in phosphatidylserine uptake in UPS-1 cells. J Lipid Res. 2015;56:2151–2157. doi: 10.1194/jlr.M062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci. 2012;125:4049–4057. doi: 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, Nakayama K, Shin H-W. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J Biol Chem. 2011;286:38159–38167. doi: 10.1074/jbc.M111.281006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Katoh Y, Ueda T, Waguri S, Murayama T, Takahashi S, Shin H-W, Nakayama K. Mitosis-coupled, microtubule-dependent clustering of endosomal vesicles around centrosomes. Cell Struct Funct. 2013;38:31–41. doi: 10.1247/csf.12028. [DOI] [PubMed] [Google Scholar]
- Takatsu H, Tanaka G, Segawa K, Suzuki J, Nagata S, Nakayama K, Shin HW. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J Biol Chem. 2014;289:33543–33556. doi: 10.1074/jbc.M114.593012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Yamagami K, Tanaka K. Role of phosphatidylserine in phospholipid flippase-mediated vesicle transport in Saccharomyces cerevisiae. Eukaryotic Cell. 2014;13:363–375. doi: 10.1128/EC.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonniere L, McKinney RA, McPherson PS. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, et al. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc Natl Acad Sci USA. 2011;108:15846–15851. doi: 10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden LM, Wichers CGK, van Breevoort AED, Coleman JA, Molday RS, Berger R, Klomp LWJ, van de Graaf SFJ. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J Biol Chem. 2010;285:40088–40096. doi: 10.1074/jbc.M110.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR. Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol. 2013;202:875–886. doi: 10.1083/jcb.201305094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabas M, Teh CE, Frankenreiter S, Lal D, Roots CM, Whittle B, Andrews DT, Zhang Y, Teoh NC, Sprent J, et al. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat Immunol. 2011;12:441–449. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Koga H, Katoh Y, Takahashi S, Nakayama K, Shin HW. Functional cross-talk between Rab14 and Rab4 through a dual effector, RUFY1/Rabip4. Mol Biol Cell. 2010;21:2746–2755. doi: 10.1091/mbc.E10-01-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA. 2009;106:16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.