A small Nxf1 protein, expressed from an NXF1 mRNA with a retained intron is highly expressed in rodent hippocampal and neocortical neurons, colocalizes with Staufen2 proteins in neuronal RNA granules, is present in polysomes, and replaces Nxt1 as an Nxf1 cofactor in export and expression of mRNA with retained introns.
Abstract
The Nxf1 protein is a major nuclear export receptor for the transport of mRNA, and it also is essential for export of retroviral mRNAs with retained introns. In the latter case, it binds to RNA elements known as constitutive transport elements (CTEs) and functions in conjunction with a cofactor known as Nxt1. The NXF1 gene also regulates expression of its own intron-containing RNA through the use of a functional CTE within intron 10. mRNA containing this intron is exported to the cytoplasm, where it can be translated into the 356–amino acid short Nxf1(sNxf1) protein, despite the fact that it is a prime candidate for nonsense-mediated decay (NMD). Here we demonstrate that sNxf1 is highly expressed in nuclei and dendrites of hippocampal and neocortical neurons in rodent brain. Additionally, we show that sNxf1 localizes in RNA granules in neurites of differentiated N2a mouse neuroblastoma cells, where it shows partial colocalization with Staufen2 isoform SS, a protein known to play a role in dendritic mRNA trafficking. We also show that sNxf1 forms heterodimers in conjunction with the full-length Nxf1 and that sNxf1 can replace Nxt1 to enhance the expression of CTE-containing mRNA and promote its association with polyribosomes.
INTRODUCTION
Alternative splicing of mRNAs is an important posttranscriptional mechanism for gene regulation and generation of diversity (for a recent review, see Lee and Rio, 2015). We now know that more than 90% of human genes are subject to alternative splicing and that splicing patterns can vary greatly in time and space. These findings have made it clear that alternative splicing serves important roles in development, differentiation, and tissue-specific expression.
There are many kinds of alternative splicing, but alternative exon usage, in which an exon is either excluded or included in an mRNA isoform, appears to be the most prominent kind in human and other mammalian cells (Pan et al., 2008; Wang et al., 2008). In contrast, intron retention, in which a complete intron is retained between two exons, was thought until recently to be relatively rare in humans and other mammalian cells (Galante et al., 2004; Braunschweig et al., 2014). However, it has long been recognized as a very common mechanism in alternative splicing in plants, in which it has been proposed to be involved in dynamic processes such as regulation of flowering and responses to environmental signals (Ner-Gaon et al., 2004; Jiao and Meyerowitz, 2010; Remy et al., 2014; Vitulo et al., 2014).
For many years, the presence of retained introns in mRNA preparations from mammalian cells was generally viewed as contamination with unspliced nuclear pre-mRNA and was thus often not reported. Transcriptome sequencing has now led to a more unbiased quantitation of both nuclear and cytoplasmic mRNA on a large scale, and many recent reports have demonstrated a large number of mRNAs with retained introns in different mammalian cell types (Buckley et al., 2011; Yap et al., 2012; Braunschweig et al., 2014; Boutz et al., 2015). It is now recognized that intron retention is much more common in mammalian cells than previously appreciated. Specifically, there are recent reports that mRNA with retained introns may play an important role in granulocyte (Wong et al., 2013) and erythroid differentiation (Edwards et al., 2016; Pimentel et al., 2016). Intron retention has also been demonstrated to be widespread in many cancer cells (Dvinge and Bradley, 2015). In spite of these advances, intron retention remains poorly understood, and the fate of the resulting mRNAs remain controversial. For example, it has been proposed that a majority of mRNAs with one or more retained introns are subject to regulated unproductive splicing and transport, in which the mRNAs are “detained” in the nucleus or are exported but are not translated into stable proteins (Green et al., 2003). Many mRNAs with retained introns also fulfill the criteria for being candidates for nonsense-mediated decay (NMD), and it has thus been proposed that this mechanism leads to mRNA degradation in most cases (Ge and Porse, 2014).
The analysis of retroviruses provided the first evidence that mRNAs with retained introns could be successfully exported to the cytoplasm in a regulated manner and translated into stable proteins (Hammarskjold, 1997). Complex retroviruses such as human immunodeficiency virus (HIV) and human T-cell lymphotropic virus (HTLV) were initially shown to encode regulatory proteins (Rev and Rex) that facilitate this process by binding to cis-acting elements in the viral mRNAs that retain introns (for reviews, see Pollard and Malim, 1998; Shida, 2012; Fernandes et al., 2016). Rev and Rex then recruit chromosome region maintenance 1/exportin 1, a cellular export receptor, enabling nucleocytoplasmic export (Fornerod et al., 1997; Booth et al., 2014). It was later shown that an analogous mechanism also exists in the case of simpler retroviruses, such as Mason-Pfizer monkey virus (MPMV). In this virus, the viral mRNA contains an element that we named the constitutive transport element (CTE; Bray et al., 1994). The CTE directly recruits the host-cell protein nuclear RNA export factor 1 (Nxf1; Gruter et al., 1998), which also serves as an important export receptor for spliced cellular mRNA (Herold et al., 2001).
In previous work, we demonstrated that both the human and mouse NXF1 genes harbor a CTE within an intron that is often retained (Li et al., 2006). The NXF1 CTE shows striking sequence homology to the MPMV CTE. Furthermore, we showed that the Nxf1 protein (together with the cellular cofactor nuclear transport factor 2–like export factor 1 [Nxt1]) interacts with the NXF1 CTE to allow nucleocytoplasmic export of the alternative NXF1 mRNA isoform that retains the CTE-containing intron. Finally, we showed that a novel short Nxf1 (sNxf1) protein can be expressed from this isoform in established human cell lines in vitro. More recently, we also showed that the CTE in NXF1 has been conserved in evolution and is present in multiple mammalian species and also in teleost fish (Wang et al., 2015). The fish-derived CTE can efficiently mediate export and expression of a reporter mRNA with a retained intron in mammalian cells, showing functional conservation of the CTE mechanism.
To date, expression of sNxf1 has been reported only in cancer cell lines, whereas the presence of this protein in normal cells in vivo and its potential function have remained unexplored. Here we show that sNxf1 is expressed in several normal tissues in mammals, including brain, where it is present in rodent hippocampal and cortical neurons. In some neurons in the neocortex, the protein is observed in cytoplasmic granules. Furthermore, we demonstrate that sNxf1 can function as an alternative Nxf1 cofactor in export and translation of CTE-containing mRNA, that it partially colocalizes with Staufen2 (Stau2) proteins in cytoplasmic granules, and that it is also present in polyribosomes.
RESULTS
The sNxf1 protein is highly expressed in mouse neuronal cell lines and rat hippocampal and cortical neurons
In a previous publication, we showed that the sNxf1 protein is expressed in several cell lines, including 293T cells (Li et al., 2006). These cells have some characteristics of human neuronal cells and are believed to be of neuroendocrine origin (Shaw et al., 2002; Campbell et al., 2005). However, the potential expression of sNxf1 beyond transformed cell lines in culture has never been explored.
To determine whether sNxf1 is expressed in normal brain tissue and a bona fide neuronal cell line, we performed a Western blot analysis of lysates prepared from whole adult mouse brain and from mouse neuroblastoma N2a cells, using an antibody that is specific for the peptide encoded within NXF1 intron 10 (Figure 1). We have previously shown that this antibody specifically detects the 356–amino acid sNxf1 protein in human cell lines (Li et al., 2006). As a control, we used cells transfected with a plasmid containing a cDNA engineered to specifically express sNxf1. The results of this experiment showed that sNxf1 was highly expressed in both the N2a cells and adult mouse brain. We also tested lysates from several other mouse tissues (see Supplemental Figure 1) and demonstrated expression of the correct-sized protein in both kidney and thymus. A slightly larger protein was observed in liver (as well as in the N2a cells), and additional larger species were also detected in thymus, possibly indicating expression of other alternatively spliced NXF1 mRNA isoforms.
FIGURE 1:
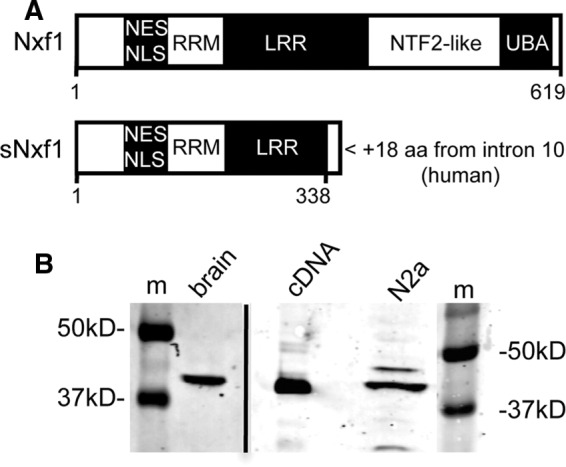
Domain structure of sNxf1 and expression in mouse brain tissue and a neuroblastoma cell line. (A) Functional domains of full-length and sNxf1 proteins. NXF1 mRNA retaining intron 10 encodes a 356-residue protein with a novel 18 amino acids at its carboxy terminus encoded by the intron. NLS, nuclear localization signal; NES, nuclear export signal; RRM, RNA recognition motif; LRR, leucine-rich repeat; and UBA, ubiquitin-associated domain are shown. The NTF-2 like domain binds to Nxt1/p15. (B) Western blot analysis of lysates from mouse brain and mouse neuroblastoma N2a cells using an antibody raised against the unique 18-residue peptide sequence. A control cDNA construct that expresses a protein terminating at the in-frame stop codon in intron 10 of NXF1 was included. m, molecular-weight markers.
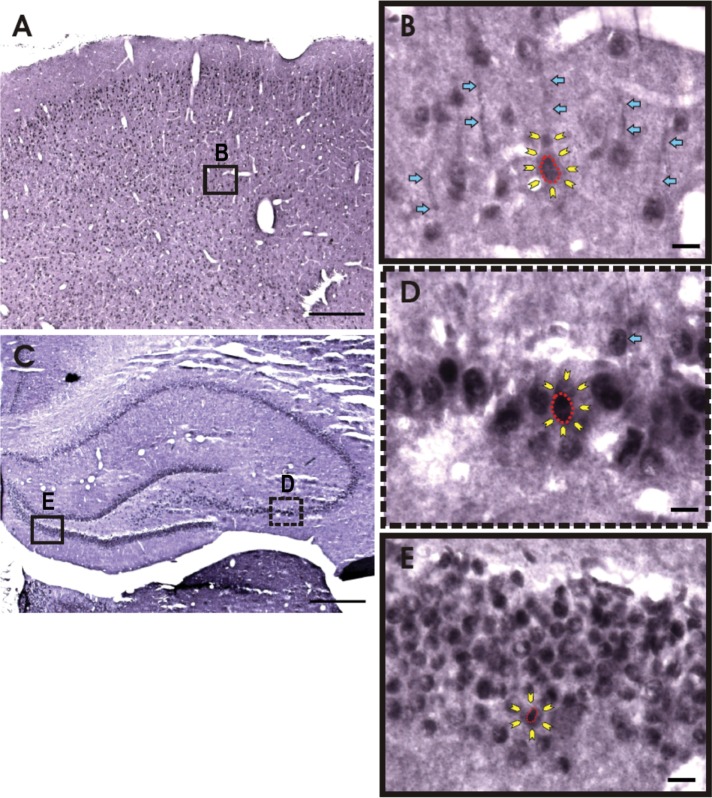
We next analyzed whether sNxf1 could be detected in specific cells in rodent brain. To this end, we performed an immunohistochemistry analysis on brain sections from adult rats, using the sNxf1-specific peptide antibody (Figure 2). This analysis indicated that the sNxf1 protein was highly expressed in neurons in multiple areas of the hippocampus, as well as in the neocortex. Although the staining was primarily nuclear in most cells, we also observed staining of dendritic processes in what appeared to be cytoplasmic granules. This was most easily observed in the neocortex (see Figure 2B). Although Nxf1 proteins have not been previously observed in cytoplasmic RNA granules, proteins expressed from other NXF family genes (hNXF5 and mNXF7), have been proposed to be involved in cytoplasmic neuronal RNA trafficking (Tretyakova et al., 2005; Katahira et al., 2008). Cellular positioning, cell body morphology, and characteristic dendrites indicate that the primary types of sNxf1-positive cells in the neocortex and hippocampus were neurons.
FIGURE 2:
Expression of sNxf1 in adult rodent brain. (A) A low-magnification view of the neocortex shows sNX1-positive cells in multiple layers of the neocortex. Scale bar: 500 μm. (B) Higher magnification view of the boxed area in A. sNxf1 appears to be localized primarily to the nucleus, with some degree of expression in the cytoplasm and apical dendrites of neocortical neurons. The red outline denotes the boundaries of the nucleus of one neuron, while the yellow arrows indicate the cell membrane. Blue arrows highlight staining of apical dendrites. Scale bar: 10 μm. (C) Low-power image of the hippocampus demonstrates sNxf1 expression in the granule cells of the dentate gyrus and in the pyramidal neurons of CA1-CA3. The boxed areas are shown in D and E at higher magnification. Scale bar: 500 μm. (D) Higher-magnification view of the corresponding boxed area in C, showing the expression of sNxf1 in CA3 pyramidal neurons. The red outline denotes the boundaries of the nucleus, while the yellow arrows indicate the cell membrane. The blue arrow indicates a cell with possible nucleolar staining. Scale bar: 10 μm. (E) High magnification of the corresponding boxed area in C, showing expression of sNxf1 in dentate granule cells. The red outline denotes the boundaries of the nucleus, while the yellow arrows indicate the cell membrane. Scale bar: 10 μm.
The sNxf1 protein localizes to cytoplasmic granules in neurites of differentiated N2a cells and shows partial colocalization with the Stau2SS protein
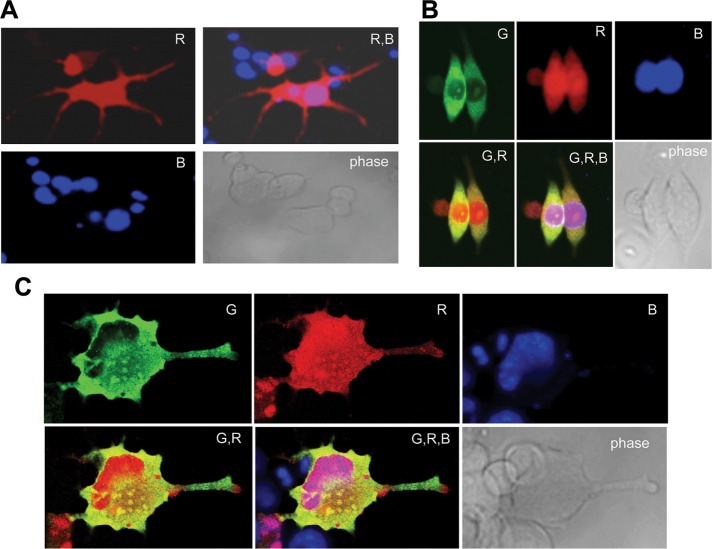
To study the localization of sNxf1 in cultured neuronal cells, we next transfected N2a cells with a plasmid expressing a red fluorescent protein (RFP)-tagged sNxf1 protein and differentiated the transfected cells using N6, 2′-O-dibuturyladenosine 3′,5′-cyclical monophosphate (dibutyryl-cAMP [db-cAMP]). In some experiments, the N2a cells were also cotransfected with a plasmid expressing an enhanced green fluorescent protein (eGFP)-tagged version of the rat Stau2SS protein (a kind gift from Michaela Monshausen and Kenneth Kosik, Harvard Medical School; Monshausen et al., 2004). Stau2 proteins have previously been shown to localize to dendritic neuronal granules in hippocampal and other neurons (Kiebler et al., 1999) and to regulate transport and expression of neuronal mRNAs (Heraud-Farlow et al., 2013). Staufen proteins have also been proposed to play a role in regulation of mRNA stability at the cytoplasmic level (Heraud-Farlow et al., 2013; Park et al., 2013). Furthermore, mammalian Stau2 proteins have been reported to associate with “large” Nxf1 in neuronal cells (Monshausen et al., 2004).
After treatment with db-cAMP, the transfected N2a cells were examined using confocal microscopy (Figure 3). Cells in various states of differentiation were observed. As can be seen in Figure 3A, a significant fraction of the sNxf1 protein was localized to the nuclei of the differentiated transfected cells when RFP-sNxf1 was expressed by itself. However, the protein was also present in cytoplasmic regions outside the nucleus and in the neurite-like processes, consistent with the results from the immune-histochemistry analysis. The image was overexposed to more clearly visualize sNxf1 in the neurites. A similar localization was seen in the cotransfections. Figure 3B shows less differentiated cotransfected cells. In these cells, the protein was again observed in cytoplasmic regions outside the nucleus, where it appeared to localize with Stau2SS. Figure 3C shows a higher-power magnification of a cell that appears to be more differentiated. Again, there was a significant colocalization of sNxf1 and Stau2SS in granular structures outside the nucleus, indicated by the yellow color in the “overlap” panels (marked G,R and G,R,B). In contrast, the proteins appeared to show a largely distinct localization in separate granules in the distal portions of the cell processes.
FIGURE 3:
Localization of sNxf1 in differentiated N2a cells and colocalization with Stau2SS. (A) sNxf1 is present in neurites of differentiated neuroblastoma N2a cells. Cells were transfected with a plasmid expressing an RFP-tagged sNxf1 protein (R, red). Forty-eight hours after transfection, cells were induced to differentiate to form neurites with db-cAMP, fixed with formaldehyde, and RFP visualized using a confocal microscope. Nuclei were stained with DAPI (B, blue). RB, merged pictures of R and B; phase, phase-contrast image of the cells. (B, C) The sNxf1 protein partially colocalizes with the Stau2SS protein in cytoplasmic granules. N2a cells were transiently cotransfected with a plasmid expressing an RFP-tagged sNxf1 protein (R, red) and a plasmid expressing an eGFP-tagged rat Stau2SS protein (G, green). Twenty-four hours after transfection, cells were treated with db-cAMP to differentiate them. They were then fixed, and RFP or GFP was visualized using a confocal microscope. Nuclei were stained with DAPI (B, blue). GR, merged pictures of G and R; GRB, merged pictures of G, R, and B; phase, phase-contrast image of the cells.
The sNxf1 protein can function as alternative cofactor in CTE-mediated mRNA expression
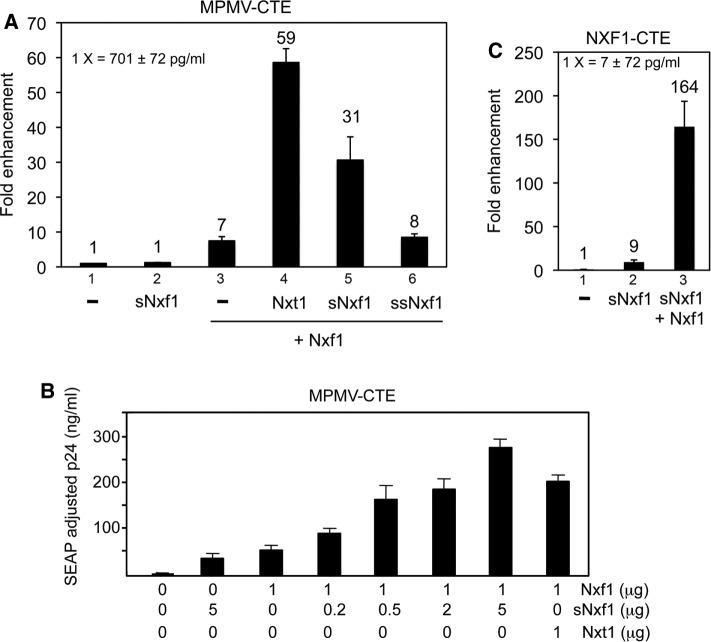
Because the sNxf1 protein retains most of the RNA-binding domains of Nxf1 but lacks the domains shown to interact with the well-established cofactor Nxt1 and nucleoporins (see Figure 1A), this protein would not be expected to serve as an export receptor for mRNA. However, we reasoned that it might work as a regulator of “large” Nxf1 function, possible acting as a “dominant-negative” factor. To investigate this, we cotransfected 293T cells with a plasmid expressing sNxf1 and a plasmid expressing the large Nxf1 protein and tested the effect on expression from an MPMV CTE-containing mRNA reporter (Figure 4). This reporter has been used by us and others in many previous studies of export and expression of mRNA with a retained intron (Bray et al., 1994; Jin et al., 2003). It contains the GagPol region from HIV and expresses the HIV p24 protein in the supernatant of transfected cells. This requires export and expression of a GagPol mRNA that contains a retained intron. We have previously shown that expression of large Nxf1, in conjunction with expression of Nxt1, functions to significantly enhance expression from this mRNA reporter in transfected cells (Jin et al., 2003). This enhancement occurs mainly at the cytoplasmic level and is the result of increased polyribosome association of the CTE-containing mRNA, resulting in significantly increased protein expression from the reporter (Jin et al., 2003; Bor et al., 2006). If Nxf1 is expressed without Nxt1, only low levels of expression from the reporter are observed.
FIGURE 4:
The sNxf1 protein functions as an alternative Nxf1 cofactor to enhance CTE function. (A, B) 293T cells (3 × 106 in a 10 cm culture dish) were cotransfected with 5 μg pCMVGagPol-MPMVCTE (A) or pCMVGagPol-NXF1CTE (B), 0.25 μg pCMVSEAP and 1.0 μg plasmids expressing either Nxf1, sNxf1, ssNxf1, or Nxt1 as indicated. (C) A dose response with the plasmid expressing sNxf1 is shown, using the same amount of pCMVGagPol-MPMVCTE, pCMVSEAP as in A and B and the indicated amounts of the various plasmids shown. Seventy-two hours after transfection, supernatants were harvested and analyzed for p24 levels and SEAP activity. The p24 values were normalized to SEAP activity. In parts A and B the data are plotted as plotted as “fold enhancement” with the raw p24 values from cells transfected with GagPol reporter alone used as the basal level (1×). As previously described, the Nxf1 cellular CTE works less efficiently than the MPMV CTE and gives lower basal levels.
When sNxf1 was expressed alone, it did not have any effect on reporter function (Figure 4A, column 2). However, when the two Nxf1 proteins were expressed together, a significant enhancement was observed (Figure 4A, column 5). Rather than acting as a dominant-negative protein, sNxf1 was able to increase expression from the reporter almost as well as an amount of Nxt1 that was previously shown to be optimal (Jin et al., 2003; compare Figure 4A, columns 4 and 5). Additionally, a dose-dependent increase was observed when a fixed amount of the Nxf1-expressing plasmid was transfected with increasing amounts of the plasmid expressing sNxf1 (Figure 4B). In contrast, expression of a deletion mutant of sNxf1, lacking the first 61 amino acids (ssNxf1), failed to enhance reporter expression (Figure 4A, column 6). Because this protein lacks the domain that has been shown to be important for Nxf1 dimerization (Matzat et al., 2008), this result suggests that function may require heterodimerization of Nxf1 and sNxf1.
We also observed a significant enhancement of p24 protein expression when Nxf1 and sNxf1 were expressed together in conjunction with a reporter containing the CTE from intron 10 in the NXF1 gene (Figure 4C). In this case, a slight enhancement was also observed when sNxf1 protein was expressed by itself. Together these results indicate that the sNxf1 protein can function as an alternative cofactor to enhance large Nxf1 function in conjunction with both viral and cellular CTEs.
Because the Nxt1 protein is expressed endogenously in 293T cells, we next wanted to verify that the enhancement of expression seen with sNxf1 was independent of Nxt1, as endogenous Nxt1 binding to the large Nxf1 protein could potentially have contributed to the observed effects. To address this, we analyzed the effect of sNxf1 in cotransfections with a plasmid expressing a mutant of the large Nxf1 protein, in which the Nxt1-binding region was deleted (Nxf1(∆507–540)). We, and others, have previously shown that Nxf1(∆507–540) lacks the domain necessary for interaction with Nxt1 and that Nxt1 fails to promote Nxf1 function in conjunction with this mutant (Guzik et al., 2001). The results of the experiment using the deletion mutant are shown in Figure 5. As can be seen, Nxt1 is not capable of enhancing Nxf (∆507–540) function (compare columns 3 and 4). However, the short Nxf1 protein was still able to enhance the function of this protein (compare columns 3 and 5), demonstrating that the Nxt1-binding domain in Nxf1 and Nxt1 binding are not necessary for the sNxf1 enhancement. We also retested the plasmid expressing the NH2–terminally deleted version of the sNxf1 protein (ssNxf1) in these experiments. As expected, expression of this protein failed to enhance mutant Nxf1 function (compare columns 3 and 6).
FIGURE 5:
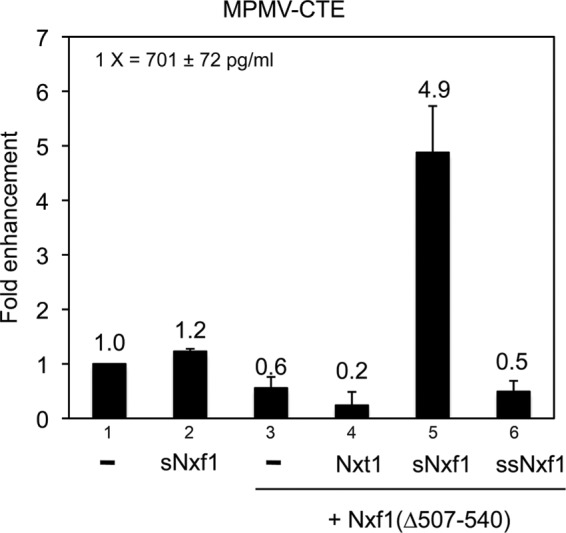
sNxf1 works in conjunction with Nxf1(∆507-540) to enhance CTE function. The 293T cells were transfected with the indicated plasmids. Nxf1(∆507-540) is an internal deletion mutant in Nxf1 that deletes a region essential for Nxt1 binding. Transfection conditions and the analysis were as described in Figure 4.
sNxf1 interacts with Nxf1 through its amino terminal domain
To directly test whether sNxf1 and Nxf1 interact with each other, we next performed immunoprecipitations (IPs), using M2-FLAG beads, on lysates from cells expressing T7-tagged Nxf1(aa1-619) and FLAG-tagged versions of sNxf1(aa1–356) or ssNxf1(aa61–356). Figure 6 shows that a significant amount of Nxf1 was detected by Western blot analysis using T7 tag–specific antibodies in the IP with sNxf1/Nxf1 (lane 1, lower panel). In contrast, no Nxf1 was detected in the ssNxf1/Nxf1 IP (lane 4, lower panel). These results indicate that the sNxf1 protein interacts with Nxf1 through its NH2-terminal domain. This is consistent with the concept that the formation of a heterodimer between these two proteins is essential for sNxf1 cofactor function.
FIGURE 6:
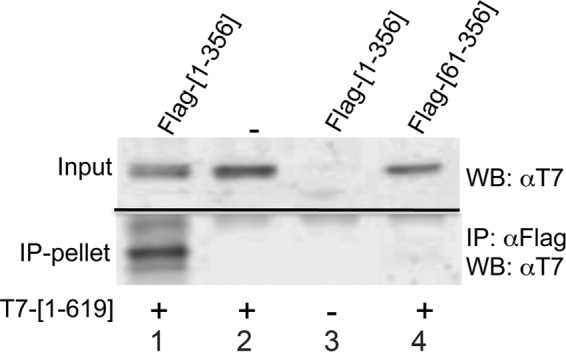
The sNxf1 protein can form heterodimers with Nxf1. The 293T cells were cotransfected with plasmids that express a T7-tagged full-length Nxf1 protein (T7-[1-619]) and plasmids that express either FLAG-tagged sNxf1 (FLAG-[1-356]) or FLAG-tagged ssNxf1 (Flag-[61-356]) Lysates were prepared and immunoprecipitated using M2-FLAG agarose beads. The immunoprecipitated complexes were resolved by SDS–PAGE, and the presence of T7-Nxf1-[1-619] in the IPs was detected using a T7-monoclonal antibody. Input controls show 5% of the total lysate used for IP.
The sNxf1 protein functions to promote mRNA expression at the level of translation
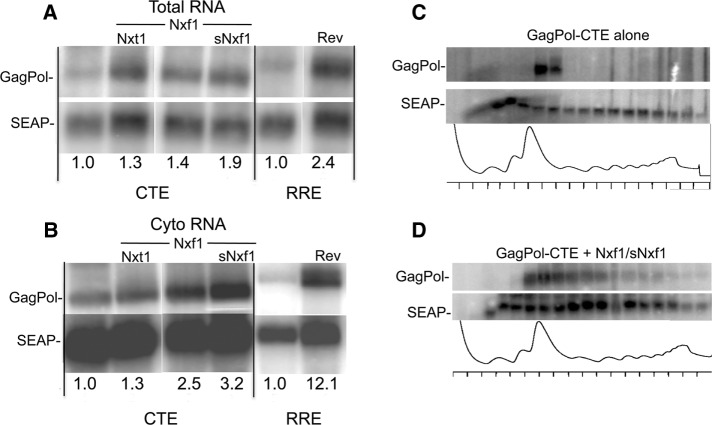
In previously published work, we showed that the enhancement of CTE function seen when Nxf1/Nxt1 is cotransfected with the GagPol-CTE reporter plasmid was primarily an effect on translation of the GagPol mRNA, with only minimal effects on mRNA export (Swartz et al., 2007). To further analyze how sNxf1 enhances Nxf1 function, we performed a Northern blot analysis on total and cytoplasmic mRNA extracted from transfected cells. The result of this analysis showed that the relative levels of total GagPol-CTE reporter mRNA were not significantly increased by expression of Nxf1/sNxf1, compared with the levels seen with Nxf1 alone (Figure 7A). Similar results were obtained using a combination of Nxf1 and Nxt1 (lane 2), confirming previous results. Somewhat bigger differences were observed in the case of cytoplasmic RNA (Figure 7B), indicating that there was a slight enhancement of export, but the differences were much smaller than those observed in the control experiments using Rev and the RRE, in which Rev is known to significantly promote export of the RRE-containing GagPol mRNA (Jin et al., 2003). The small differences in cytoplasmic mRNA levels cannot explain the large increase in p24 expression that was observed with Nxt1 and sNxf1 in conjunction with Nxf1 (compare Figure 7 and Figure 4). Thus the enhancement mainly reflects effects beyond the level of mRNA export.
FIGURE 7:
RNA and polyribosome analysis of cells transfected with Nxf and sNxf1. Northern blot analyses of total (A) and cytoplasmic (B) mRNA from transfected 293T cells. The 293T cells were transfected with pCMVGagPol-CTE and pCMVSEAP in the absence or presence of plasmids that expressed Nxf1 and/or sNxf1 and Nxt1 as indicated. As a control, cells were also transfected with pCMVGagPol-RRE and pCMVSEAP in the absence or presence of a plasmid that expressed Rev. Sixty-five hours after transfection, total and cytoplasmic polyA+ RNA was isolated and analyzed by Northern blot using a 32P-labeled Gag probe. The values shown in the panel marked “RRE,” on the right side of each panel, represent the fold difference in the levels of the GagPol-RRE mRNA bands between transfections without and with Rev. The values under the CTE panel, on the left side of each panel, represent the fold difference in the levels of the GagPol-CTE mRNA in the presence of the indicated proteins compared with the transfection with pCMVGagPol-CTE alone. All values were normalized using the SEAP band. Radioactivity was quantitated using a phosphoimager. (C, D) Polyribosome profile analysis of GagPol-CTE mRNA in transfected cells. The 293T cells were transfected with pCMVGagPol-CTE in the absence of presence of plasmids that expressed Nxf1 and sNxf1. Forty-eight hours posttransfection, cells were harvested and cytoplasmic extracts were subjected to sucrose gradient centrifugation. The gradient was fractionated, and the OD 254 nm was measured using a continuous-flow cell. RNA was isolated from each fraction and analyzed for GagPol-CTE and SEAP mRNA on Northern blots (top panels). The optical density traces of the gradient fractions are shown in the bottom panels.
We previously demonstrated that the mRNA expressed from GagPol-MPMV-CTE reporter in the absence of transfected Nxf1/Nxt1 was largely absent from large polyribosome complexes, whereas association with large polyribosomes was promoted by expression of these proteins (Jin et al., 2003; Bor et al., 2006). To analyze whether this was also true in the case of Nxf1/sNxf1 enhancement, we performed a polyribosome analysis of the reporter RNA in cells transfected with the GagPol-CTE reporter +/− Nxf1/sNxf1, using sucrose gradients. The results of this analysis (shown in Figure 7, C and D) demonstrated that, in the absence of transfected Nxf1/sNxf1, most of the mRNA was in fractions containing monosomes and small polyribosomes (Figure 7C). In contrast, the control secreted alkaline phophatase (SEAP) mRNA that is expressed from a completely spliced mRNA was present throughout the polyribosomal fractions. However, when Nxf1/sNxf1 were coexpressed, the GagPol-CTE mRNA was present in both the small and large polyribosomal fractions (Figure 7D). This supports the hypothesis that Nxf1/sNxf1 expression promotes translation of the reporter mRNA, as previously shown for Nxf1/Nxt1, resulting in the high levels of p24 protein expression shown in Figure 4.
The sNxf1 protein is present in polyribosomal complexes as well as in cytoplasmic complexes that are resistant to EDTA
We previously showed that some of the large Nxf1 protein was present in polyribosomal fractions in 293T cells expressing GagPol-CTE RNA (Jin et al., 2003; Bor et al., 2006). To determine whether sNxf1 could also be detected in these fractions, we performed experiments in which cells were cotransfected with pGAGPol-CTE and plasmids expressing sNxf1/Nxf1 and subjected to sucrose gradient analysis. We then performed Western blot on individual fractions, using an antibody that detects both Nxf1 isoforms. As can be seen in Figure 8A, the large Nxf1 protein was present in all gradient fractions, confirming previous results. sNxf1 was also detected throughout the gradient. However, when the lysates were treated with 15 mM EDTA to disrupt polyribosomes (Figure 8B), the large Nxf1 protein was no longer detected in the “heavier “ fractions, consistent with previous results. In contrast, there was still a significant amount of sNxf1 in the fractions close to the bottom of the gradient after EDTA treatment, whereas the middle fractions were largely devoid of sNxf1. Although 15 mM EDTA causes the disruption of polyribosomes, it has been shown to not disrupt the majority of non-rRNA protein complexes, such as cytoplasmic RNA granules (Calzone et al., 1982; Johannes and Sarnow, 1998). Taken together, these results indicate that some sNxf1 is present in polyribosomes, as previously shown for the large Nxf1 protein (Jin et al., 2003; Bor et al., 2006). The fact that sNxf1 was also present in large nonribosomal cytoplasmic RNA–protein complexes that were not disrupted by 15 mM EDTA is consistent with the results of the immunohistochemistry and GFP/RFP experiments (see Figures 2 and 3), which showed the presences of sNxf1 in cytoplasmic granules in brain sections as well as in neuronal cells in culture.
FIGURE 8:
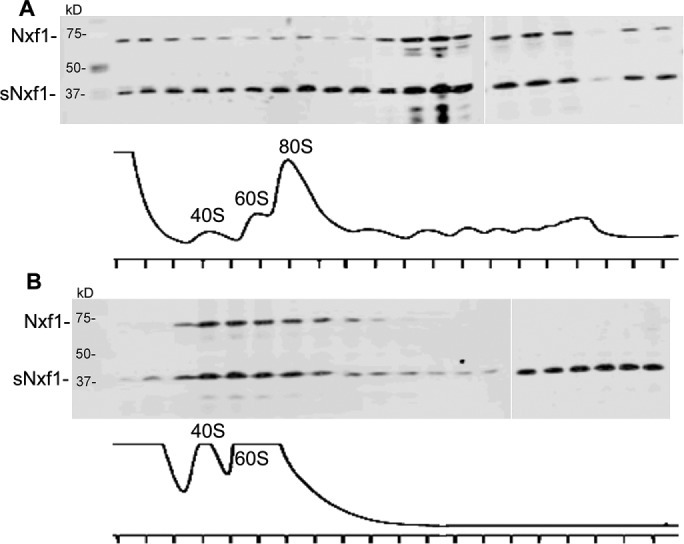
Both sNxf1 and Nxf1 proteins associate with polyribosomes. (A) 293T cells (1 × 107 in a 15 cm culture dish) were transfected with pCMVGagPol-CTE and plasmids expressing Nxf1 and sNxf1 plasmids. Forty-eight hours after transfection, cells were harvested, and cytoplasmic extracts were subjected to sucrose gradient centrifugation. The gradient was fractionated and the OD 254 nm was measured using a continuous-flow cell. Proteins from each fraction were resolved by SDS–PAGE and analyzed by Western blotting using a polyclonal antibody directed against Nxf1. The optical density profiles of the gradient fractions are shown in the bottom panels. (B) The experiment was performed as in A, except that 15 mM EDTA was added to the lysate before loading onto sucrose gradient.
DISCUSSION
Next-generation sequencing (NGS) and other data from many studies published in the past several years have confirmed our original observation of retention of intron 10 in NXF1 mRNA (Braunschweig et al., 2014; Boutz et al., 2015; Cho et al., 2015). It is thus well established that this NXF1 mRNA isoform is expressed in multiple cell lines, as well as in many normal human and mouse tissues. Nevertheless, although our previous work showed that sNxf1, which is translated from this mRNA, could be detected in cancer cell lines, the expression of this protein in normal cells and tissues has remained unexplored, as has its potential function.
Retention of intron 10 in the NXF1 mRNA creates a premature termination codon (PTC), making it a prime candidate for NMD (for a recent review see (Kurosaki and Maquat, 2016). Thus, even though the mRNA that retains intron 10 can be exported to the cytoplasm because of the CTE, NMD mechanisms would be predicted to prevent stable protein expression in the cytoplasm. In spite of this prevailing dogma, the data presented here clearly demonstrate that sNxf1 is expressed in normal mammalian cells and tissues, including several areas of the brain. However, although the mRNA that retains intron 10 appears to be relatively highly expressed in most cells, stable protein expression is only observed in some cells. This suggests the possibility of unknown mechanisms that regulate translation and/or the stability of the expressed protein after export to the cytoplasm.
Our experiments on sections of rat brain, using specific antibodies to visualize sNxf1, demonstrate that the protein is expressed in neocortical neurons and in neurons in multiple areas of the hippocampus. In neocortical regions, the protein is localized to the nucleus and the cytoplasm in structures that appear to be neuronal granules. In addition, the protein colocalizes with Stau2SS in granules in neuronal N2a cells in culture, suggesting a possible role for the protein in cytoplasmic RNA trafficking in the brain after nuclear export.
The sNxf1 protein retains the Nxf1 RNA-binding domain, so it would be expected to be able to bind to CTE and other mRNAs, but it lacks the domains that interact with proteins of the nuclear pore complex. Thus, by itself, it would not be expected to function as a nucleocytoplasmic export receptor. However, we have shown that the small and large Nxf1 proteins can dimerize through their NH2-terminal domains, so we hypothesize that an Nxf1/sNxf1 dimer is the functional unit during export.
When an HIV reporter mRNA with a retained intron is exported to the cytoplasm with the help of a CTE, polyribosome association and protein expression are greatly enhanced by overexpression of Nxf1 and its cofactor Nxt1 (Jin et al., 2003; Bor et al., 2006). This suggests effects of Nxf1/Nxt1 at the cytoplasmic level, beyond the role for this complex in nucleocytoplasmic export. In the experiments presented here, we have shown that coexpression of large and small Nxf1, without Nxt1 expression, can also promote polyribosome association and translation and that sNxf1 is present in polyribosomes. Thus the Nxf1 heterodimer can likely serve to promote both export and cytoplasmic expression of mRNAs containing a retained intron and a CTE (or CTE-like element). Several other genes (e.g., ACTN4 and SIRT7) contain elements that can function as CTEs (Bor et al., 2006). We are currently expanding the vector trap strategy that was used to identify these, in conjunction with NGS, to identify additional functional CTEs and genes that may be regulated in this manner.
There is growing acceptance that mRNAs with retained introns are widespread in mammalian cells (Buckley et al., 2011; Yap et al., 2012; Braunschweig et al., 2014; Boutz et al., 2015). However, because of the well-known restrictions of export of mRNAs with retained introns (Chang and Sharp, 1989; Legrain and Rosbash, 1989; Hammarskjold, 1997), and the fact that many of these mRNAs contain PTCs that make them NMD targets, protein expression has often not been analyzed in studies that report intron retention. Based on the data presented here, it seems reasonable to speculate that, if mRNAs with retained introns contain elements that function as CTEs, they may not only be exported, but would also be likely to be translated into protein, at least in some cells. However, the extent to which this mechanism functions to regulate gene expression remains largely unexplored beyond viral systems.
Although nuclear retention of mRNAs with retained introns has been studied for many years, the molecular mechanism for retention and the links between nuclear export and translation are still not well understood. It has been proposed that excision of introns leaves a “mark” on the RNA that allows recruitment of Nxf1 and that this leads to the formation of an export-competent mRNA (Le Hir et al., 2000; Huang et al., 2004; Muller-McNicoll et al., 2016). However, in the NXF1 mRNA that retains only intron 10, multiple introns both 5′ and 3′ to this intron are removed, leaving multiple “exon junction” marks. Nevertheless, this mRNA is not exported in the absence of a functional CTE that recruits Nxf1. Thus there must be a mechanism that retains the mRNA in the nucleus until all the other introns have been removed. It is possible that all of the introns with the exception of intron 10 are removed cotranscriptionally (Carrillo Oesterreich et al., 2016), while intron 10 is spliced later and more slowly to create the fully spliced NXF1 mRNA. However, this does not explain why, once the mRNA is “released” from the transcription machinery, it is not exported unless the mRNA contains a CTE. We have previously reported that the nuclear basket protein Tpr may be involved in a “proofreading” mechanism to prevent unregulated export of mRNAs with retained introns (Coyle et al., 2011), similar to the role proposed for the budding yeast Myosin-like proteins 1 and 2 (Mlp1/Mlp2; Galy et al., 2004; Bonnet et al., 2015; Saroufim et al., 2015). There have also been reports to suggest that non-POU domain containing octamer-binding protein (Nono/Nrb54) and other paraspeckle proteins may play a role in retention (Chen and Carmichael, 2009). However, there is clearly a need for more studies addressing this issue.
We have recently demonstrated that the NXF1 CTE is highly conserved among mammalian species and that an element with striking primary- and secondary-sequence homology is also found in the NXF1 gene of teleost fish (Wang et al., 2015). The zebrafish CTE functions efficiently to replace Rev/RRE in the context of HIV reporter constructs in human 293T cells, especially in conjunction with Nxf1 and Nxt1 from the same species. This indicates that a CTE in conjunction with Nxf1/Nxt1 is a highly conserved mechanism that has been used throughout vertebrate evolution. We have initiated experiments in mice using CRISPR-Cas9 technology to specifically disrupt NXF1 CTE function by creating small deletions in the CTE to render it nonfunctional. This would be expected to allow Nxf1 “large” protein expression in the absence of sNxf1 and should shed light on the specific functions of sNxf1 in the brain and other organs.
Neuronal cells show very complex and variable alternative splicing patterns (for a recent review, see Vuong et al., 2016), and intron retention is common (Buckley et al., 2011). In addition, it has been suggested that NMD may be down-modulated during neuronal differentiation (Bruno et al., 2011). The NXF1 gene is part of a multigene family of which several members have been shown to function in neuronal cells. Nuclear RNA export factor 5 (NXF5) in humans (called nuclear RNA export factor 7 [NXF7] in rodents) has been linked to hippocampal synaptic plasticity (Callaerts-Vegh et al., 2015) as well as mental retardation (Frints et al., 2003), and proteins expressed from this gene are only present in brain and testes (Jun et al., 2001). To date, these proteins have only been shown to be involved in cytoplasmic mRNA transport. NXF5 is located on the X chromosome, as is nuclear RNA export factor 2 (NXF2) and several other NXF family members (Jun et al., 2001). Nxf2 is highly expressed in hippocampal and other neurons and functions in nucleocytoplasmic export (Herold et al., 2000; Sasaki et al., 2005; Tretyakova et al., 2005; Zhang et al., 2007). This protein has also has been reported to destabilize NXF1 mRNA and only low levels of the “full-length” Nxf1 protein are expressed in hippocampal neurons (Zhang et al., 2007). A recent study identified the NXF1 mRNA isoform that retains intron 10 in polyribosomes isolated from mouse hippocampal neurons using RNASeq analysis (Cho et al., 2015). These results are consistent with the data reported here, showing sNxf1 protein expression in the rat hippocampus. It will be of clear interest to analyze this data set for other mRNAs containing retained introns and to determine whether any of these mRNAs contain functional CTEs or depend on sNxf1 for their export and translation. It may be that the export and expression of proteins from mRNAs with retained introns is an important feature of neuronal gene expression, and Nxf proteins may play important roles in regulating this, ultimately contributing to neuronal plasticity.
MATERIALS AND METHODS
Plasmids and cloning procedures
To facilitate identification, all of the plasmids used in this study were indexed as numbers in the form pHRXXXX. The nomenclature for the plasmids used in this paper has changed, since the TAP gene is now officially named NXF1. The following plasmids have been previously described: pCMVGagPol-MPMVCTE(pHR1361) (Srinivasakumar et al., 1997); pCMVGagPol-NXF1CTE(pHR3405) (Li et al., 2006); pCMVgagpol-RRE(pHR354) (Srinivasakumar et al., 1997); pCMVRev(pHR30) (Smith et al., 1993); pCMVSEAP(pHR1831) (Cullen and Malim, 1992); pcDNANxt1(pHR2283) (Black et al., 1999); and pcDNAFLAG-Nxf1(pHR2352) (Jin et al., 2003). Details of the construction of the following plasmids will be provided upon request: pCMVNxf1(1-619)(pHR3700); pCMVT7-Nxf1(1-619)(pHR3702); pCMVT7-Nxf1(1-619)Δ507-540(pHR4418); pCMVsNxf1(1-356)(pHR3485); pCMVFLAG-sNxf1(1-356)(pHR3487); pCMVRFP-sNxf1(1-356)(pHR4065); pCMVssNxf1(61-356)(pHR3489); and pCMVFLAG-ssNxf1(61-356)(pHR3491). The plasmid expressing rat Staufen 2SS protein, pEGFP-Stau2SS(pHR3052) (Monshausen et al., 2004), was a kind gift from Michaela Monshausen and Kenneth Kosik.
Cell line culture, transient transfections, and differentiation
The 293T/17 cells (Pear et al., 1993) were maintained in Iscove’s modified DMEM supplemented with 10% bovine calf serum. The 293T/17 cells were transfected using a calcium phosphate transfection protocol (Graham and van der Eb, 1973).
Murine neuroblastoma Neuro-2a cells (N2a; American Type Culture Collection No. CCL-131) were grown in DMEM-supplemented 10% fetal bovine serum and antibiotics (50 U/ml penicillin, 50 μg/ml streptomycin). The cells were incubated at 37°C in a humidified 5% CO2 incubator and were split 16 h before transient transfection with pCMVRFP-sNxf1 ± pEGFP-Stau2SS using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
At 24 h posttransfection, N2a cells were differentiated in DMEM with 5 mM db-cAMP (Roche Molecular Biochemicals, Indianapolis, IN) without serum for 48 h at 37°C. Medium containing db-cAMP was exchanged every 16 h. The differentiated N2a cells were fixed with formaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize DNA, and observed under a confocal microscope.
Immunohistochemical staining of rat brain
Adult rats (12 wk) were perfused transcardially with phosphate-buffered saline followed by 4% paraformaldehyde, and their brains were dissected out and cryoprotected in 30% sucrose until sinking. Frozen sections were then cut on a cryostat at 30 μm and processed for immunohistochemistry using a polyclonal antibody against sNxf1 diluted at 1:1000 and developed via the Avidin: Biotinylated Enzyme Complex (ABC method) from Vector Labs (Burlingame, CA) with diaminobenzidine tetrahydrochloride as the chromogen.
p24 enzyme-linked immunosorbent assay (ELISA) and SEAP quantitation
Supernatants from transfected cells were collected at 65–72 h posttransfection and subjected to a short spin in a microcentrifuge to remove residual cells and debris. The p24 expression levels were determined by an ELISA protocol using a p24 monoclonal antibody (183-H12-5C) and pooled human anti-HIV immunoglobulin G (Wehrly and Chesebro, 1997). The p24 antibody was obtained from the AIDS Research and Reference Reagent Program and was contributed by Bruce Chesebro (National Institute of Allergy and Infectious Diseases–Rocky Mountain Laboratories). SEAP activity in the supernatants was measured with the Phospha-Light Chemiluminescent Reporter Kit (Tropix, Bedford, MA).
Western blot analysis
The Western blot analysis was performed essentially as previously described (Hammarskjöld et al., 1986; Guzik et al., 2001). For cell lysates, mouse tissues were dissected, cut into smaller pieces, and homogenized on ice in RIPA buffer with protease inhibitors using a glass homogenizer. After 30 min on ice, the samples were sonicated to shear the DNA and centrifuged to pellet cell debris. Proteins from the lysates were separated on a 13% SDS–PAGE gel, which was transferred to an Immobilon-FL membrane (Millipore, Billerica, MA). For detection of the Nxf1 and sNxf1 proteins, the blots were probed with a rabbit anti-Nxf1 polyclonal antibody (Jin et al., 2003) or a custom-made affinity-purified antibody raised against the sNxf1-specific peptide as previously described (Li et al., 2006). The blots were scanned with an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE).
Co-IP
A plasmid expressing a T7-tagged full-length Nxf1 protein (pCMVT7-Nxf1(1-619)) was cotransfected into 293T cells with FLAG-tagged plasmids expressing FLAG-sNxf1 (pCMVFLAG-sNxf1(1-356)) or FLAG-ssNxf1 (pCMVFLAG-sNxf1(61-356)). Lysates were made and immunoprecipitated with anti-FLAG M2–agarose beads according to the Sigma protocol (Sigma, St. Louis, MO). The immunoprecipitated complexes were resolved by SDS–PAGE, and the presence of T7-Nxf1 (1-619) was detected using a α-T7 monoclonal antibody after Western blotting. Five percent of total lysates were used as input controls.
RNA fractionation and Northern blot analysis
The methods used for nuclear and cytoplasmic RNA extraction, poly(A)+ mRNA selection, and Northern blot analysis were previously described (Hammarskjold et al., 1989, 1994; Swartz et al., 2007).
Polyribosome analysis
Polyribosome profile analyses were performed as previously described (Swartz et al., 2007). Forty-eight hours posttransfection, cells were harvested and cytoplasmic extracts were subjected to sucrose gradient centrifugation. The gradient was fractionated, and the OD 254 nm was measured using a continuous-flow cell. The analysis of Nxf1 and sNxf1 protein expression in polyribosome fractions was performed as previously described (Swartz et al., 2007).
Supplementary Material
Acknowledgments
We thank Susan Prasad for expert technical assistance and Kenneth Kosik and Michaela Monshausen for kindly providing plasmids expressing GFP-tagged Staufen proteins. This work was supported by National Institutes of Health grant R01 GM087651 and funds from the Myles H. Thaler Center research Endowment Fund at the University of Virginia. Salary support for D.R. and M.L.H. was provided by the Myles H. Thaler and Charles H. Ross Jr. Endowments at the University of Virginia.
Abbreviations used:
- CTE
constitutive transport element
- DAPI
4′,6-diamidino-2-phenylindole
- db-cAMP
dibutyryl-cAMP
- eGFP
enhanced green fluorescent protein
- ELISA
enzyme-linked immunosorbent assay
- HIV
human immunodeficiency virus
- HTLV
human T-cell lymphotropic virus
- IP
immunoprecipitation
- MPMV
Mason-Pfizer monkey virus
- NGS
next-generation sequencing
- NMD
nonsense-mediated decay
- PTC
premature termination codon
- RFP
red fluorescent protein
- SEAP
secreted alkaline phophatase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-07-0515) on October 5, 2016.
REFERENCES
- Black BE, Levesque L, Holaska JM, Wood TC, Paschal BM. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol Cell Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A, Bretes H, Palancade B. Nuclear pore components affect distinct stages of intron-containing gene expression. Nucleic Acids Res. 2015;43:4249–4261. doi: 10.1093/nar/gkv280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DS, Cheng Y, Frankel AD. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. Elife. 2014;3:e04121. doi: 10.7554/eLife.04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor Y, Swartz J, Morrison A, Rekosh D, Ladomery M, Hammarskjold ML. The Wilms’ tumor 1 (WT1) gene (+KTS isoform) functions with a CTE to enhance translation from an unspliced RNA with a retained intron. Genes Dev. 2006;20:1597–1608. doi: 10.1101/gad.1402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjold ML. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley PT, Lee MT, Sul JY, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaerts-Vegh Z, Ahmed T, Vermaercke B, Marynen P, Balschun D, Froyen G, D’Hooge R. Nxf7 deficiency impairs social exploration and spatio-cognitive abilities as well as hippocampal synaptic plasticity in mice. Front Behav Neurosci. 2015;9:179. doi: 10.3389/fnbeh.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone FJ, Angerer RC, Gorovsky MA. Regulation of protein synthesis in Tetrahymena: isolation and characterization of polysomes by gel filtration and precipitation at pH 5.3. Nucleic Acids Res. 1982;10:2145–2161. doi: 10.1093/nar/10.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SA, Lin J, Dobrikova EY, Gromeier M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J Virol. 2005;79:6281–6290. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell. 2016;165:372–381. doi: 10.1016/j.cell.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Sharp PA. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Yu NK, Choi JH, Sim SE, Kang SJ, Kwak C, Lee SW, Kim JI, Choi DI, Kim VN, et al. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015;350:82–87. doi: 10.1126/science.aac7368. [DOI] [PubMed] [Google Scholar]
- Coyle JH, Bor YC, Rekosh D, Hammarskjold ML. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA. 2011;17:1344–1356. doi: 10.1261/rna.2616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Malim MH. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 1992;216:362–368. doi: 10.1016/0076-6879(92)16033-g. [DOI] [PubMed] [Google Scholar]
- Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CR, Ritchie W, Wong JJ, Schmitz U, Middleton R, An X, Mohandas N, Rasko JE, Blobel GA. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016;127:e24–e34. doi: 10.1182/blood-2016-01-692764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes JD, Booth DS, Frankel AD. A structurally plastic ribonuceloprotein complex mediates post-transcriptional gene regulation in HIV-1. Wiley Interdiscip Rev RNA. 2016;7:470–486. doi: 10.1002/wrna.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Frints SG, Jun L, Fryns JP, Devriendt K, Teulingkx R, Berghe LV, Vos BD, Borghgraef M, Chelly J, Portes VD, et al. Inv(X)(p21.1;q22.1) in a man with mental retardation, short stature, general muscle wasting, and facial dysmorphism: clinical study and mutation analysis of the NXF5 gene. Am J Med Genet. 2003;119A:367–374. doi: 10.1002/ajmg.a.20195. [DOI] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays. 2014;36:236–243. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, Rio DC, Brenner SE. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19(suppl 1):i118–i121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- Gruter P, Tabernero C, von KC, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Guzik BW, Levesque L, Prasad S, Bor YC, Black BE, Paschal BM, Rekosh D, Hammarskjöld ML. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol Cell Biol. 2001;21:2545–2554. doi: 10.1128/MCB.21.7.2545-2554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold M-L. Regulation of retroviral RNA export. Sem Cell Dev Bio. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- Hammarskjold ML, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold ML, Li H, Rekosh D, Prasad S. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J Virol. 1994;68:951–958. doi: 10.1128/jvi.68.2.951-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjöld ML, Wang S-C, Klein G. High-level expression of the Epstein-Barr virus EBNA1 protein in CV1 cells and human lymphoid cells using a SV40 late replacement vector. Gene. 1986;43:41–50. doi: 10.1016/0378-1119(86)90006-5. [DOI] [PubMed] [Google Scholar]
- Heraud-Farlow JE, Sharangdhar T, Li X, Pfeifer P, Tauber S, Orozco D, Hormann A, Thomas S, Bakosova A, Farlow AR, et al. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013;5:1511–1518. doi: 10.1016/j.celrep.2013.11.039. [DOI] [PubMed] [Google Scholar]
- Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjöld ML. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun L, Frints S, Duhamel H, Herold A, Abad-Rodrigues J, Dotti C, Izaurralde E, Marynen P, Froyen G. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr Biol. 2001;11:1381–1391. doi: 10.1016/s0960-9822(01)00419-5. [DOI] [PubMed] [Google Scholar]
- Katahira J, Miki T, Takano K, Maruhashi M, Uchikawa M, Tachibana T, Yoneda Y. Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shuttling hnRNPs. Nucleic Acids Res. 2008;36:616–628. doi: 10.1093/nar/gkm556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bor YC, Misawa Y, Xue Y, Rekosh D, Hammarskjold ML. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- Matzat LH, Berberoglu S, Levesque L. Formation of a Tap/NXF1 homotypic complex is mediated through the amino-terminal domain of Tap and enhances interaction with nucleoporins. Mol Biol Cell. 2008;19:327–338. doi: 10.1091/mbc.E07-03-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen M, Gehring NH, Kosik KS. The mammalian RNA-binding protein Staufen2 links nuclear and cytoplasmic RNA processing pathways in neurons. Neuromolecular Med. 2004;6:127–144. doi: 10.1385/NMM:6:2-3:127. [DOI] [PubMed] [Google Scholar]
- Muller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, Neugebauer KM. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30:553–566. doi: 10.1101/gad.276477.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci USA. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Parra M, Gee SL, Mohandas N, Pachter L, Conboy JG. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016;44:838–851. doi: 10.1093/nar/gkv1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Remy E, Cabrito TR, Batista RA, Hussein MA, Teixeira MC, Athanasiadis A, Sa-Correia I, Duque P. Intron retention in the 5’UTR of the novel ZIF2 transporter enhances translation to promote zinc tolerance in Arabidopsis. PLoS Genet. 2014;10:e1004375. doi: 10.1371/journal.pgen.1004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saroufim MA, Bensidoun P, Raymond P, Rahman S, Krause MR, Oeffinger M, Zenklusen D. The nuclear basket mediates perinuclear mRNA scanning in budding yeast. J Cell Biol. 2015;211:1131–1140. doi: 10.1083/jcb.201503070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Takeda E, Takano K, Yomogida K, Katahira J, Yoneda Y. Molecular cloning and functional characterization of mouse Nxf family gene products. Genomics. 2005;85:641–653. doi: 10.1016/j.ygeno.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Shida H. Role of nucleocytoplasmic RNA transport during the life cycle of retroviruses. Front Microbiol. 2012;3:179. doi: 10.3389/fmicb.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Srinivasakumar N, Hammarskjöld ML, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjold ML, Rekosh D. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282:19844–19853. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, Felber BK. Nuclear export factor family protein participates in cytoplasmic mRNA trafficking. J Biol Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- Vitulo N, Forcato C, Carpinelli EC, Telatin A, Campagna D, D’Angelo M, Zimbello R, Corso M, Vannozzi A, Bonghi C, et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014;14:99. doi: 10.1186/1471-2229-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Rekosh D, Hammarskjold ML. Evolutionary conservation of a molecular machinery for export and expression of mRNAs with retained introns. RNA. 2015;21:426–437. doi: 10.1261/rna.048520.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Q, Huang Y. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc Natl Acad Sci USA. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.