The yeast casein kinases function upstream of the glucose sensors in the sensor/receptor-repressor signaling pathway. The sensor Rgt2 undergoes Yck-dependent phosphorylation on its C-terminal tail, which is necessary for Mth1 and Std1 binding and downstream signaling.
Abstract
Yeasts have sophisticated signaling pathways for sensing glucose, their preferred carbon source, to regulate its uptake and metabolism. One of these is the sensor/receptor-repressor (SRR) pathway, which detects extracellular glucose and transmits an intracellular signal that induces expression of HXT genes. The yeast casein kinases (Ycks) are key players in this pathway. Our model of the SRR pathway had the Ycks functioning downstream of the glucose sensors, transmitting the signal from the sensors to the Mth1 and Std1 corepressors that are required for repression of HXT gene expression. However, we found that overexpression of Yck1 fails to restore glucose signaling in a glucose sensor mutant. Conversely, overexpression of a glucose sensor suppresses the signaling defect of a yck mutant. These results suggest that the Ycks act upstream or at the level of the glucose sensors. Indeed, we found that the glucose sensor Rgt2 is phosphorylated on Yck consensus sites in its C-terminal tail in a Yck-dependent manner and that this phosphorylation is required for corepressor binding and ultimately HXT expression. This leads to a revised model of the SRR pathway in which the Ycks prime a site on the cytoplasmic tails of the glucose sensors to promote binding of the corepressors.
INTRODUCTION
All organisms must sense nutrient availability and regulate the transport and use of nutrients to optimize growth. Glucose is the primary carbon and energy source for the yeast Saccharomyces cerevisiae, which uses several mechanisms to use it efficiently. One of these is the Snf3/Rgt2-Rgt1 or sensor/receptor-repressor (SRR) glucose-signaling pathway, which detects extracellular glucose and induces expression of glucose transporters that bring glucose into the cell (Özcan and Johnston, 1999). This pathway includes glucose sensors (Rgt2 and Snf3), the Yck1 and Yck2 protein kinases, and the Mth1, Std1, and Rgt1 repressors.
In the absence of glucose, the Rgt1 repressor binds to the promoters of HXT genes and recruits the Mth1 and Std1 corepressors to repress HXT gene expression (Kim et al., 2003; Lakshmanan et al., 2003). Glucose binds to the Rgt2 (low-affinity) and Snf3 (high-affinity) glucose sensors, and this was proposed to activate the yeast casein kinases Yck1 and Yck2 (Ycks), which are functionally redundant for glucose signaling (Moriya and Johnston, 2004). Once activated, the Ycks would be situated to phosphorylate the Mth1 and Std1 corepressors because the Ycks and the corepressors interact with the glucose sensors (Moriya and Johnston, 2004; Özcan et al., 1996, 1998). The phosphorylated corepressors are recognized by the SCFGrr1 ubiquitin ligase and subsequently ubiquitinated, targeting them for proteasomal degradation (Flick et al., 2003). Without its corepressors, Rgt1 is unable to bind the HXT promoters, leading to derepression of HXT gene expression (Polish et al., 2005; Roy et al., 2013).
Our working model of the SRR pathway had the binding of glucose to a glucose sensor activating the Ycks or otherwise enabling them to phosphorylate the Std1 and Mth1 corepressors. This was based on the facts that 1) Yck function is required for signaling, 2) Yck1 is able to phosphorylate the corepressors in vitro, and 3) mutation of Yck consensus phosphorylation sites in the corepressors blocks their degradation (Moriya and Johnston, 2004). However, here we report evidence that suggests that the Ycks act on the glucose sensors in the SRR signaling pathway, which leads us to a revised model of the SRR pathway in which the Ycks first phosphorylate the C-terminal tails of the glucose sensors to establish a functional site for their interaction with the Mth1 and Std1 corepressors, bringing them in the proximity of the glucose sensors to receive and transduce the glucose signal.
RESULTS
The glucose sensors are epistatic to yeast casein kinases
Previous work suggested that the Ycks function downstream of the glucose sensors to transduce the glucose signal from the glucose sensors to the Mth1 and Std1 corepressors via their Yck-dependent phosphorylation. The Ycks are required for glucose signaling and Mth1 and Std1 phosphorylation and destruction, and Yck1 phosphorylates Mth1 and Std1 in vitro on a cluster of conserved serine residues in Yck-consensus phosphorylation sequences (Moriya and Johnston, 2004).
To test the proposed downstream function of the Ycks, we determined the epistatic relationship of YCK2 and RGT2. Because inactivation of the glucose sensors and the Ycks results in the same phenotype (reduced glucose signaling), we tested the effects of overexpression of individual components of the SRR pathway in deletion mutants and assessed the effect on HXT1 expression and Mth1 destruction (typical readouts of SRR pathway function).
If Yck2 functioned downstream of the glucose sensors, then its overexpression might rescue the glucose-sensing defect of the rgt2snf3 mutant. However, expression of HXT1 (measured with an HXT1-lacZ reporter) is not induced by glucose in an rgt2snf3 mutant overexpressing YCK2 (Figure 1A), nor is Mth1 degradation in response to glucose restored (Figure 1B, lane 8). This suggests that the Ycks function upstream of the glucose sensors.
FIGURE 1:
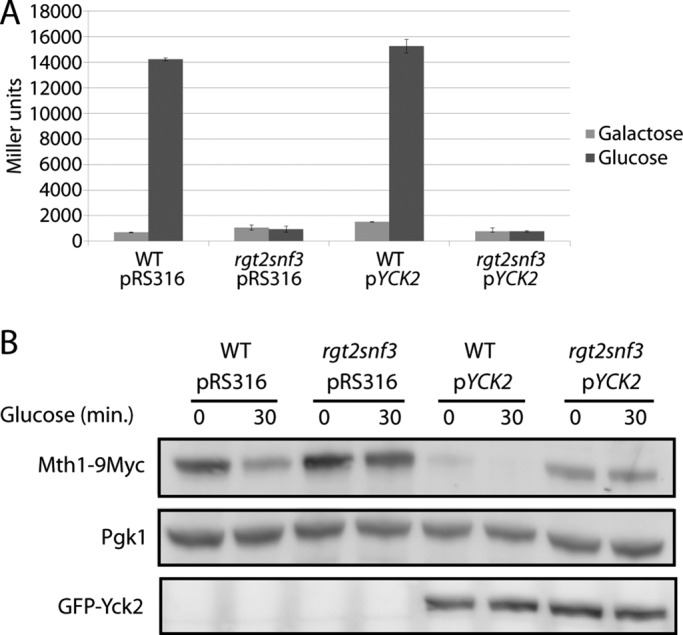
Overexpression of YCK2 does not rescue a sensor knockout. (A) HXT1-lacZ expression (pBM3212) was determined as described in Materials and Methods. BY4742 (WT) or YM6871 (rgt2snf3) with pRS316 or pJB1 (pYCK2) was cultured at 30°C in 2% galactose (Galactose) and spiked with 4% glucose for 90 min (Glucose). (B) Mth1-9myc expressed from its own promoter on a single-copy plasmid (pBM4560) in BY4742 (WT) or YM6871 (rgt2snf3) with pRS316 or pJB1 (pYCK2) was cultured in 2% galactose (time 0) and then spiked with 4% glucose (time 30) at 30°C. The blot was probed with anti-Pgk1 as an internal standard; anti-GFP was used to confirm expression of Yck2.
If the Ycks functioned upstream of the glucose sensors, we would expect RGT2 overexpression to suppress the glucose-signaling defect of a yck1∆yck2ts mutant. Indeed, overexpression of RGT2 results in full glucose induction of HXT1 expression in the yck1∆yck2ts mutant (Figure 2A) and restoration of glucose-induced Mth1 degradation (Figure 2B, lane 6). These results are consistent with the idea that the Ycks act upstream, or at the level of the glucose sensors.
FIGURE 2:
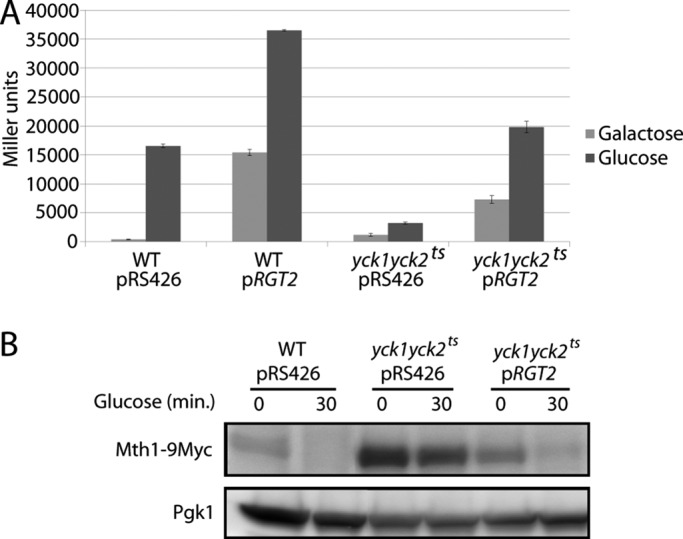
Overexpression of RGT2 suppresses yck1∆yck2ts mutations. (A) HXT1-lacZ expression (pBM3212) was determined as described in Materials and Methods. LRB939 (WT) or LRB1613 (yck1∆yck2ts) with pRS426 or pBM3333 (pRGT2) was precultured overnight in 2% galactose at room temperature, shifted to 30°C for 4 h (Galactose), and then spiked with 4% glucose for 90 min (Glucose) before assay for β-galactosidase activity. (B) Mth1-9myc expressed from its own promoter on a single-copy plasmid (pBM4560) in LRB939 (WT) or LRB1613 (yck1∆yck2ts) with pRS426 or pBM3333 (pRGT2) was precultured overnight in 2% galactose at room temperature, shifted to 30°C for 4 h (time 0), and then spiked with 4% glucose (time 30) at 30°C. The blot was probed with anti-Pgk1 as an internal standard.
The C-terminal tail of Rgt2 undergoes Yck-dependent phosphorylation
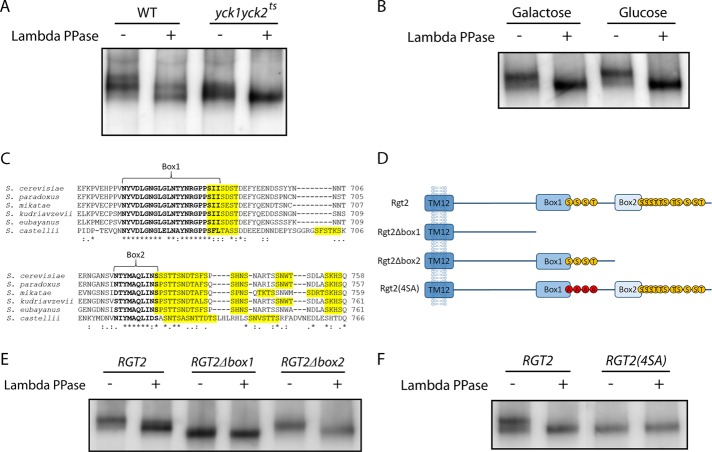
The Ycks are involved in phosphorylation of several nutrient permeases (Estrada et al., 1996; Decottignies et al., 1999; Marchal et al., 2000; Gadura et al., 2006). The glucose sensors Rgt2 and Snf3 share significant sequence similarities with glucose transporters and other nutrient permeases and could be regulated by a similar mechanism. Indeed, phosphatase treatment of Rgt2 increases its mobility in SDS–PAGE (Figure 3A, lanes 1 and 2), suggesting that Rgt2 is phosphorylated. Rgt2 from the yck1∆yck2ts strain migrates similarly to phosphatase-treated Rgt2 from wild-type cells (Figure 3A, lanes 3 and 4), suggesting that Rgt2 phosphorylation depends on Yck function. Phosphorylation of Rgt2 does not appear to be regulated by glucose (Figure 3B).
FIGURE 3:
The C-terminal tail of Rgt2 is phosphorylated in a Yck-dependent manner. (A) Rgt2-13myc expressed from the ADH1 promoter on a multicopy plasmid (pBM6232) in LRB939 (WT) or LRB1613 (yck1∆yck2ts) was cultured in 4% glucose for 4 h and immunoprecipitated and phosphatase treated as described in Materials and Methods. (B) Rgt2-13myc expressed from the ADH1 promoter on a multicopy plasmid (pBM6232) in BY4742 (WT) was cultured in 2% galactose (Galactose) or 4% glucose (Glucose) for 4 h and immunoprecipitated and phosphatase treated as described in Materials and Methods. (C) Multiple sequence alignment (Clustal Omega) of Rgt2 from Saccharomyces species shows sequence conservation in the C-terminal tail. Box1 and box2 are indicated (bold), and Yck consensus phosphorylation sites (S/T-X-X-S/T) are highlighted in yellow. (D) Diagram of three Rgt2 mutants used in this study. Transmembrane 12 of Rgt2 is indicated in the plasma membrane with the C-terminal tail shown extending into the cytoplasm. The Yck consensus phosphorylation sites are indicated with S and T for serine and threonine, respectively. The Rgt2(4SA) shows the point mutations S684A, S687A, S689A, and T690A, represented by A. (E) BY4742 with pBM6232 (RGT2), pBM6233 (RGT2∆box1), or pBM6234 (RGT2∆box2) was cultured in 4% glucose for 4 h and immunoprecipitated and phosphatase treated as described in Materials and Methods. (F) BY4742 with pBM6232 (RGT2) or pBM6235 (RGT2(4SA)) was cultured in 4% glucose for 4 h and immunoprecipitated and phosphatase treated as described in Materials and Methods.
The C-terminal tail of Rgt2 has two clusters of evolutionarily conserved potential Yck phosphorylation sites (Figure 3C). To determine whether these sites are involved in phosphorylation of Rgt2, we generated Rgt2 truncations that remove conserved sequences—box1 and box2—that are required for glucose signaling (unpublished data; Figure 3D). Potential Yck consensus sites cluster immediately downstream of box1 and box2 (Figure 3C). The SDS–PAGE migration shift of these truncated forms of Rgt2 is abolished only in the Rgt2∆box1 mutant (Figure 3E, compare lanes 1 and 2 to lanes 3 and 4); the Rgt2∆box2 mutant shows no change in the relative migration shift upon phosphatase treatment (Figure 3E, lanes 5 and 6). These results suggest that the potential Yck phosphorylation sites immediately downstream of box1 are in vivo targets of the Ycks.
We mutated three serines and one threonine in this cluster to alanine (Figure 3D, bottom). This Rgt2(4SA) shows no shift in migration pattern between phosphatase-treated and -untreated lanes in Western blot analysis (Figure 3F, lanes 3 and 4). Taken together, these results suggest that Rgt2 undergoes Yck-dependent phosphorylation on a cluster of residues immediately downstream of box1 in the C-terminal tail of the sensor.
Yck-dependent phosphorylation of Rgt2 is necessary for glucose signaling
To determine the significance of this phosphorylation of Rgt2 to SRR signaling, we analyzed the effect on glucose signaling of different mutations of Rgt2. HXT1 expression is not induced by glucose in an rgt2snf3 mutant, and this defect is rescued by a single copy of RGT2 (Figure 4A). Rgt2∆box1 does not restore glucose induction of HXT1 expression (Figure 4A) and fails to rescue the Mth1 degradation defect of the rgt2snf3 mutant (see later discussion of Figure 5B). Rgt2(4SA) provides no detectable glucose induction of HXT1 expression (Figure 4A) or Mth1 degradation (Figure 4B). Rgt2∆box2 provides a small but reproducible glucose induction of HXT1 expression (Figure 4A) but fails to restore Mth1 degradation (Figure 4B). Thus phosphorylation of the Yck consensus sites adjacent to box1 is required for glucose signaling.
FIGURE 4:
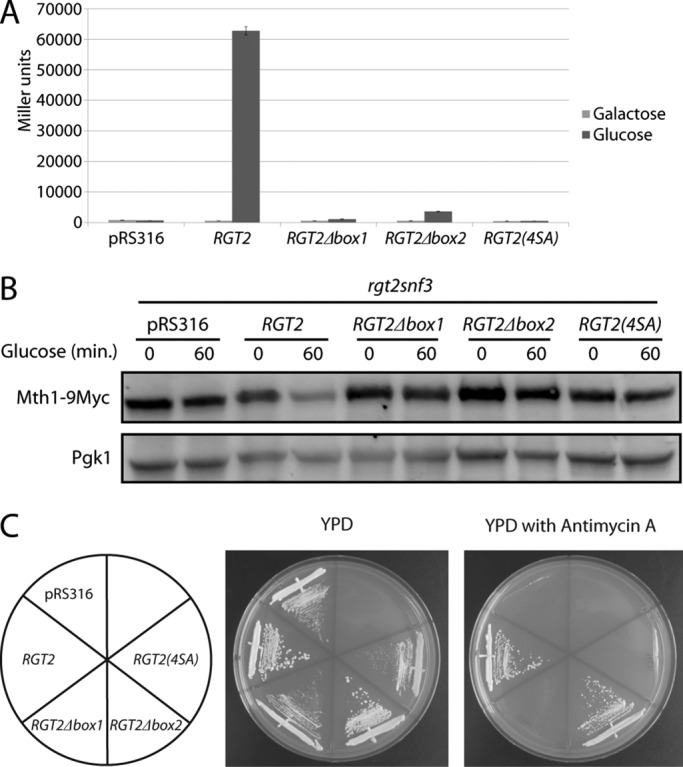
Phosphorylation of the Rgt2 tail is required for SRR signaling. (A) HXT1-lacZ expression (pBM3212) was determined as described in Materials and Methods. YM6871 (rgt2snf3) with pRS316, pBM6236(RGT2), pBM6237 (RGT2∆box1), pBM6238 (RGT2∆box2), or pBM6239 (RGT2(4SA)) was cultured at 30°C in 2% galactose (Galactose) or 4% glucose (Glucose) for 4 h. (B) Mth1-9myc expressed from its own promoter on a single-copy plasmid (pBM4560) in YM6871 (rgt2snf3) with pRS316, pBM6236 (RGT2), pBM6237 (RGT2∆box1), pBM6238 (RGT2∆box2), or pBM6239 (RGT2(4SA)) was cultured at 30°C in 2% galactose (time 0) and then spiked with 4% glucose (time 60). (C) YM6871 (rgt2snf3) containing pRS316, pBM6236 (RGT2), pBM6237 (RGT2∆box1), pBM6238 (RGT2∆box2), or pBM6239 (RGT2(4SA)) was streaked onto YPD or YPD with antimycin A and grown for 3 d at 30°C.
FIGURE 5:
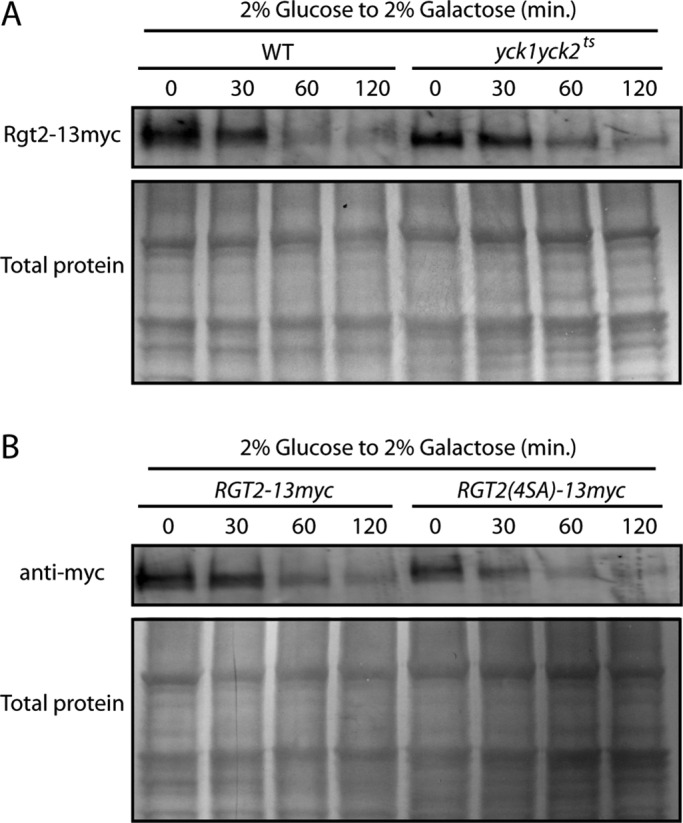
Stability of Rgt2 is not affected by the phosphorylation state of the tail. (A) RGT2-13myc expressed from its own promoter on a single-copy plasmid (pBM6236) in LRB939 (WT) or LRB1613 (yck1∆yck2ts) was cultured in 2% glucose for 4 h (time 0). Cells were washed and resuspended in 2% galactose, and samples were taken at 30, 60, and 120 min. (B) BY4742 with pBM6236 (Rgt2-13myc) or pBM6239 (Rgt2(4SA)-13myc) was cultured in 2% glucose for 4 h (time 0). Cells were washed and resuspended in 2% galactose, and samples were taken at 30, 60, and 120 min. All samples were extracted and blotted as described in Materials and Methods.
Growth on 2% glucose in the presence of antimycin A (an inhibitor of mitochondrial respiration) is another indicator of glucose sensor function. An rgt2snf3 mutant fails to grow on glucose in the presence of antimycin A because it is unable to transport sufficient glucose to support fermentation (Schmidt et al., 1999; Figure 4C). Neither Rgt2∆box1 nor Rgt2(4SA) supports growth of an rgt2snf3 mutant on glucose plus antimycin A, whereas the Rgt2∆box2 mutant is capable of supporting growth (Figure 4C). These results support the conclusion that the Yck-dependent phosphorylation sites in Rgt2 are required for SRR signaling.
Stability of the glucose sensors, like other nutrient transporters that they closely resemble, is regulated. The Rgt2 high-affinity glucose sensor is stabilized by glucose and is degraded in the absence of glucose through its ubiquitination and consequent targeting to the vacuole (Roy and Kim, 2014), and Yck-dependent phosphorylation of the uracil permease Fur4 within a PEST sequence stimulates its ubiquitination and degradation (Marchal et al., 2000). The C-terminal region of Rgt2 near box1, where Yck-dependent phosphorylation occurs, resembles a PEST sequence that we surmised could function to regulate the stability of Rgt2. However, the Ycks have no effect on Rgt2 stability: Rgt2 levels in glucose-grown cells are similar in wild-type and yck1∆yck2ts cells. Rgt2 is degraded similarly after switching them to galactose (Figure 5A), and Rgt2(4SA)-13myc is degraded like the nonmutated form of Rgt2-13myc (Figure 5B). We conclude that Yck-dependent phosphorylation of Rgt2 is required for SRR signaling but does not affect Rgt2 stability.
Phosphorylation of the Rgt2 tail facilitates interaction with Mth1 and Std1
Mth1 and Std1 interact with the tails of the Rgt2 and Snf3 glucose sensors (Schmidt et al., 1999; Lafuente et al., 2000; Moriya and Johnston, 2004). We wondered whether Yck-dependent phosphorylation of the Rgt2 tail affects these interactions. We used the integrated modified yeast two-hybrid (iMYTH) method (Snider et al., 2010) to investigate the interaction of Rgt2 with Mth1 and Std1. We constructed strains with the C-terminal half of ubiquitin (Cub) attached to the C-terminus of either RGT2 or RGT2(4SA). Genes in the SRR pathway fused to the N-terminal half of ubiquitin (Nub) were introduced into these strains. An interaction between the Cub- and Nub-tagged proteins results in expression of a lexA-LacZ reporter gene. This assay reveals that Mth1 and Std1 interact with Rgt2 and that this interaction is significantly reduced (by 86 and 92%, respectively) by the 4SA mutation (Figure 6). Thus the Yck-dependent phosphorylation of the Rgt2 tail potentiates Mth1 and Std1 binding.
FIGURE 6:
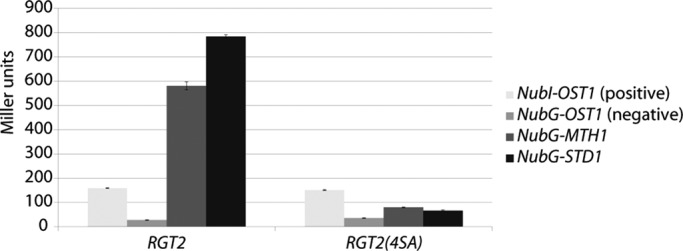
Phosphorylation of the Rgt2 tail is required for its interaction with Mth1 and Std1. THY.AP4 expressing C-terminal Cub fused to the chromosomal copy of RGT2 (YM7807) or RGT2(4SA) (YM7808) and coexpressing pBM6240 (NubI-OST1, positive control), pBM6241 (NubG-OST1, negative control), pBM6242 (NubG-MTH1), or pBM6243 (NubG-STD1) was assayed for β-galactosidase activity as described in Materials and Methods.
DISCUSSION
The Ycks are required for glucose sensing and signaling via the SRR pathway and were believed to function downstream of the glucose sensors to phosphorylate Mth1 and Std1 (Moriya and Johnston, 2004). However, the results of our epistasis tests suggested that the Ycks function upstream of or at the level of Rgt2. Indeed, we discovered that Rgt2 is phosphorylated in a Yck-dependent manner.
Knowing that the Ycks are required for stability of several membrane proteins, including transporters and permeases, and that Rgt2 and Snf3 are turned over in response to low and high glucose levels, respectively (Roy and Kim, 2014), we were surprised to find that Rgt2 is stable in a yck1∆yck2ts mutant and that mutation of the Yck-dependent phosphorylation sites of Rgt2 does not affect its stability (even though the phosphorylation sites on Rgt2 are in a region that resembles a PEST degradation sequence).
Conserved box1 of the Rgt2 tail is required for its interaction with Mth1 and Std1 (Moriya and Johnston, 2004), and we found that the Yck-dependent phosphorylation of a group of residues immediately downstream of box1 stimulates Mth1 and Std1 interaction with the Rgt2 tail. We imagine that this creates a scaffold for bringing the corepressors to the tail of the sensors to facilitate downstream signaling. This also suggests that the protein kinase(s) that phosphorylate the corepressors also localize to the plasma membrane and likely to the sensor tails to bring all components in the same vicinity. It is notable that deletion of conserved box2 of the Rgt2 tail causes a substantial reduction in glucose induction of HXT1 expression without affecting the Yck-dependent phosphorylation of the tail. Thus the Yck phosphorylation sites near box1 are not the only determinants of glucose signaling in the tail.
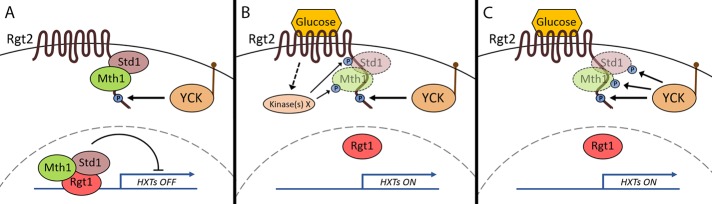
The realization that the Ycks phosphorylate the glucose sensors requires a revision of our model of the SRR pathway. In the absence of glucose (Figure 7A), Rgt1, Mth1, and Std1 form a repressor complex that binds to the promoters of HXT genes to repress their transcription (Kim et al., 2003; Lakshmanan et al., 2003; Polish et al., 2005). Yck-dependent phosphorylation of Rgt2 (and likely also Snf3) facilitates its binding of the Mth1 and Std1 corepressors, effectively priming the system. Binding of glucose to the glucose sensors presumably induces a conformational change in them that activates or recruits a protein kinase(s) to phosphorylate Mth1 and Std1, targeting them for SCF1Grr1-dependent ubiquitination and degradation by the proteasome (Figure 7, B and C). Depletion of the corepressors robs Rgt1 of its ability to repress expression of HXT genes, leading to accumulation of glucose transporters in the plasma membrane.
FIGURE 7:
Revised model of SRR signaling. (A) In the absence of glucose, the repressor complex (Rgt1, Mth1, and Std1) is bound to the promoters of the HXT genes in the nucleus, blocking their transcription. The plasma membrane–bound sensor (Rgt2, and presumably also Snf3) undergoes Yck-dependent phosphorylation on its C-terminal tail. This phosphorylation of the sensor tail generates an interaction site and recruits the corepressors to the plasma membrane. This presumably brings the required signaling components together and primes the system for response to glucose. (B, C) Binding of glucose to Rgt2 activates the downstream signaling cascade. The corepressors are phosphorylated by unknown kinase(s) (B) or possibly by the Ycks (C). The phosphorylated corepressors are then ubiquitinated by the SCFGrr1 ubiquitin–protein ligase and targeted for degradation (shown as faded) by the proteasome. With the depletion of the cellular pool of corepressors, Rgt1 is no longer able to bind to the promoters of the HXT genes, relieving their repression.
This model leaves a potential void in the pathway: the identity of the protein kinase(s) that phosphorylate Mth1 and Std1. Our results suggest that these might not be the Ycks (Figure 7B), because they are not necessary for glucose signaling in a strain overexpressing RGT2 (Figure 2A). However, there may be residual Yck2 protein kinase activity in the yck1∆yck2ts mutant under our experimental conditions that is sufficient for glucose signaling when Rgt2 levels are high, and so we cannot rule out the possibility that the Ycks also act downstream of the sensors to phosphorylate the corepressors (Figure 7C). If this is the case, the residual Yck2 activity must be substantial, because overexpression of RGT2 in the yck1∆yck2ts mutant results in wild-type levels of glucose signaling (Figure 2A). Glucose signaling in the yck1∆yck2ts mutant is negligible (Figure 2A), however, which suggests that Yck2 activity is very low in this mutant under these conditions. In addition, because phosphorylation of Rgt2 by the Ycks does not seem to be regulated by glucose, the glucose-induced event in the SRR pathway that initiates glucose signaling remains to be identified.
MATERIALS AND METHODS
Strains and growth conditions
All cells were cultured in synthetic complete medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate; 2% glucose or 2% galactose; 0.5% ammonium sulfate; and the amino acid supplement mixture [minus histidine, leucine, uracil, and tryptophan]). Antimycin A (A8674; Sigma-Aldrich, St. Louis, MO) was used at a concentration of 1.7 μg/ml on yeast extract/peptone/dextrose (YPD) plates. Cells for analysis of HXT1 expression and Mth1 degradation were inoculated into synthetic medium containing 2% galactose. Cells were grown for 4 h, samples were taken for the zero time point, and glucose was added to the medium to a final concentration of 4%. Cells for immunoprecipitation, Rgt2 turnover analysis, and iMYTH analysis (Paumi et al., 2007) were inoculated from overnight cultures into synthetic medium and grown for 4 h before extraction or analysis. All cultures were grown with shaking at 30°C, with the exception of those containing the yck2 temperature-sensitive mutation, which were cultured overnight at room temperature and switched to 30°C for 4 h before sampling.
Table 1 lists strains used in this study. The Rgt2-Cub strain (YM7807) was constructed by transformation of THY.AP4 (Obrdlik et al., 2004) with a PCR product generated from L2 plasmid (Paumi et al., 2007; primers contained 40 base pairs of homology to RGT2 locus upstream and downstream of the stop codon). The Rgt2(4SA)-Cub strain (YM7808) was constructed by initial disruption of RGT2 in the box1 region of the tail with URA3. The URA3 gene was removed by selection for 5-fluoroorotic acid–resistant cells transformed with an RGT2(4SA)-CUB fragment generated by gap repair in pCMBV4 (Snider et al., 2010). Correct integration was confirmed by the PCR. Table 2 lists plasmids used in this study. All plasmids for this study were constructed by gap repair (Oldenburg et al., 1997) and confirmed by PCR.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Laboratory collection |
| YM6871 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2 rgt2::KanMX::NatMX snf3::KanMX | Laboratory collection |
| LRB939 | MATα his3 leu2 ura3-52 | Lucy Robinson (Department of Biochemistry and Molecular Biology, Louisiana State University Health Sciences Center, Shreveport, LA) |
| LRB1613 | LRB939 yck1::KanMX yck2-2ts | Lucy Robinson |
| THY.AP4 | MATa ura3 leu2 lexA::lacZ::trp1 lexA::HIS3 lexA::ADE2 | Igor Stagljar (Donnelly Centre, University of Toronto, Toronto, ON, Canada) |
| YM7807 | THY.AP4 RGT2-Cub::KanMX | This study |
| YM7808 | THY.AP4 RGT2(4SA)-Cub::KanMX | This study |
TABLE 2:
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pRS316 | Yeast centromere vector with a URA3 marker and an MCS | Sikorski and Hieter (1989) |
| pRS426 | Yeast episomal vector with a URA3 marker and an MCS | Christianson et al. (1992) |
| pJB1 | GAL1 promoter, GFP-YCK2 in YCp50 | Robinson et al. (1999) |
| pBM3212 | HXT1 promoter lacZ fusion, LEU2 | Özcan et al. (1996) |
| pBM3333 | ADH1 promoter, RGT2 in pRS426 | Özcan et al. (1998) |
| pBM4560 | MTH1 promoter, MTH1-9myc, HIS3 | Moriya and Johnston (2004) |
| pBM6232 | pBM3333 with C-terminal 13myc tag | This study |
| pBM6233 | pBM3333 with RGT2deltabox1-13myc (aa 1–664) | This study |
| pBM6234 | pBM3333 with RGT2deltabox2-13myc (aa 1–714) | This study |
| pBM6235 | pBM3333 with RGT2(4SA)-13myc | This study |
| pBM6236 | RGT2 promoter, RGT2-13myc in pRS316 | This study |
| pBM6237 | pBM6236 with RGT2deltabox1-13myc (aa 1–664) | This study |
| pBM6238 | pBM6236 with RGT2deltabox2-13myc (aa 1–714) | This study |
| pBM6239 | pBM6236 with RGT2(4SA)-13myc | This study |
| pBM6240 | pPR3-N (NubI) with OST1 | This study |
| pBM6241 | pPR3-N (NubG) with OST1 | This study |
| pBM6242 | pPR3-N (NubG) with MTH1 | This study |
| pBM6243 | pPR3-N (NubG) with STD1 | This study |
aa, amino acids.
β-Galactosidase assays
The β-galactosidase kit (Pierce, Rockford, IL) was used as per the manufacturer’s specifications, and reactions were stopped with 1 M sodium carbonate after 5 min or 2 h for HXT1 or iMYTH experiments, respectively. All time points were in triplicate, and error bars represent 1 SD.
Protein extraction and Western analysis
For assaying Mth1 degradation, 1 ml of 100% trichloroacetic acid (TCA) was added to 5 ml of cell culture, followed by centrifugation at 3000 × g to pellet cells. The cell pellet was resuspended in 400 μl of 20% TCA and transferred to a microcentrifuge tube. Approximately 0.3 g of glass beads was added, and the samples were vortexed for 5 min. The lysate was cleared by centrifugation, and the beads were washed twice with 200 μl of 5% TCA. Samples were then centrifuged at 3000 × g, and the pellet was resuspended in 50 μl of nonfluorescent sample buffer (Li-Cor, Lincoln, NE) and 25 μl of 1 M Tris. Protein concentrations were determined by Coomassie elution with SDS (Marbach et al., 2001).
Pelleted cells were prepared for immunoprecipitation by first resuspending them in 400 μl of lysis buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM, EDTA, 1 mM phenylmethylsulfonyl fluoride, plus Halt protease inhibitor cocktail [plus or minus phosphatase inhibitors; Thermo Scientific, Waltham, MA]), adding 0.3 g of glass beads, and vortexing for 5 min. Then NP-40 (to 1%) and deoxycholate (to 0.25%) were added, and cells were again vortexed for 5 min. Cell debris was removed by centrifugation, and the lysates were left on ice for immunoprecipitation. Analysis of Rgt2 degradation was performed as described by Roy and Kim (2014).
Protein samples were run on 10% TGS gels (Bio-Rad, Hercules, CA) and transferred to low-fluorescent polyvinylidene fluoride membranes (Bio-Rad). Membranes were probed with mouse anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:1000, rabbit anti–green fluorescent protein (GFP; Sigma-Aldrich) diluted 1:1000, and mouse anti-Pgk1 (Invitrogen, Carlsbad, CA) diluted 1:10,000 in blocking buffer (Rockland, Pottstown, PA). Secondary antibodies used were anti-mouse Dylight 488 (Epitomics, Cambridge, MA) diluted 1:5000 and anti-rabbit Dylight 549 (Epitomics) diluted 1:5000 in blocking buffer. Membranes were imaged on a Bio-Rad imager.
Immunoprecipitation
Cell lysates were incubated with 25 μl of EZview Red anti–c-myc affinity gel (Sigma-Aldrich) suspended in wash buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, pH 8.0) for 90 min at 4°C. Beads were then washed twice with wash buffer, and 50 μl of 1× λ-phosphatase buffer with MnCl2 (New England Biolabs, Ipswich, MA) was added to all samples. Samples without phosphatase inhibitors were treated with 1 μl of λ-protein phosphatase (New England Biolabs) and incubated at 30°C for 20 min. Beads were washed three times with wash buffer and resuspended in 60 μl of nonfluorescent sample buffer (Li-Cor) plus 7.5% β-mercaptoethanol. For Rgt2 turnover analysis, membranes were stained with Coomassie blue after imaging to detect total protein as a loading control.
Acknowledgments
We thank Lucy Robinson and Igor Stagljar for providing strains and plasmids. This work was supported by National Institutes of Health Grant 5R01GM032540.
Abbreviations used:
- SRR
Snf3/Rgt2-Rgt1 or sensor/receptor-repressor
- Yck
yeast casein kinase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0342) on September 14, 2016.
REFERENCES
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Owsianik G, Ghislain M. Casein kinase I-dependent phosphorylation and stability of the yeast multidrug transporter Pdr5p. J Biol Chem. 1999;274:37139–37146. doi: 10.1074/jbc.274.52.37139. [DOI] [PubMed] [Google Scholar]
- Estrada E, Agostinis P, Vandenheede JR, Goris J, Merlevede W, Franc J, Ghislain M. Phosphorylation of yeast plasma membrane H-ATPase by casein kinase I. Biochemistry. 1996;271:32064–32072. doi: 10.1074/jbc.271.50.32064. [DOI] [PubMed] [Google Scholar]
- Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003;14:3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadura N, Robinson LC, Michels C. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics. 2006;172:1427–1439. doi: 10.1534/genetics.105.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente MJ, Gancedo C, Jauniaux JC, Gancedo JM. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol Microbiol. 2000;35:161–172. doi: 10.1046/j.1365-2958.2000.01688.x. [DOI] [PubMed] [Google Scholar]
- Lakshmanan J, Mosley AL, Ozcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003;44 doi: 10.1007/s00294-003-0423-2. 19–25. 2. [DOI] [PubMed] [Google Scholar]
- Marbach I, Licht R, Frohnmeyer H, Engelberg D. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J Biol Chem. 2001;276:16944–16951. doi: 10.1074/jbc.M100383200. [DOI] [PubMed] [Google Scholar]
- Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. Casein kinase 1-dependent phosphorylation within a PEST sequence and ubiquitination nearby lysines signal endocytosis of yeast uracil permease. J Biol Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. K +channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA. 2004;101:12242–12247. doi: 10.1073/pnas.0404467101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Rosenwald AG, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi CM, Menendez J, Arnoldo A, Engels K, Iyer KR, Thaminy S, Georgiev O, Barral Y, Michaelis S, Stagljar I. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol Cell. 2007;26:15–25. doi: 10.1016/j.molcel.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Polish JA, Kim JH, Johnston M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics. 2005;169:583–594. doi: 10.1534/genetics.104.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LC, Bradley C, Bryan JD, Jerome A, Kweon Y, Panek HR. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol Biol Cell. 1999;10:1077–1092. doi: 10.1091/mbc.10.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kim JH. Endocytosis and vacuolar degradation of the yeast cell surface glucose sensors RGT2 and SNF3. J Biol Chem. 2014;289:7247–7256. doi: 10.1074/jbc.M113.539411. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Roy A, Shin YJ, Cho KH, Kim JH. Mth1 regulates the interaction between the Rgt1 repressor and the Ssn6-Tup1 corepressor complex by modulating PKA-dependent phosphorylation of Rgt1. Mol Biol Cell. 2013;24:1493–503. doi: 10.1091/mbc.E13-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wölfl S, Almonte C, Watkins SC. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, Kittanakom S, Damjanovic D, Curak J, Wong V, Stagljar I. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protoc. 2010;5:1281–1293. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]