Abstract
DNA composition dynamics across genomes of diverse taxonomy is a major subject of genome analyses. DNA composition changes are characteristics of both replication and repair machineries. We investigated 3,611,007 single nucleotide polymorphisms (SNPs) generated by comparing two sequenced rice genomes from distant inbred lines (subspecies), including those from 242,811 introns and 45,462 protein-coding sequences (CDSs). Neighboring-nucleotide effects (NNEs) of these SNPs are diverse, depending on structural content-based classifications (genome-wide, intronic, and CDS) and sequence context-based categories (A/C, A/G, A/T, C/G, C/T, and G/T substitutions) of the analyzed SNPs. Strong and evident NNEs and nucleotide proportion biases surrounding the analyzed SNPs were observed in 1−3 bp sequences on both sides of an SNP. Strong biases were observed around neighboring nucleotides of protein-coding SNPs, which exhibit a periodicity of three in nucleotide content, constrained by a combined effect of codon-related rules and DNA repair mechanisms. Unlike a previous finding in the human genome, we found negative correlation between GC contents of chromosomes and the magnitude of corresponding bias of nucleotide C at −1 site and G at +1 site. These results will further our understanding of the mutation mechanism in rice as well as its evolutionary implications.
Key words: CpG dinucleotides, methylation, SNP, transcription-coupled repair
Introduction
DNA compositional dynamics across genomes of diverse taxonomy is a major subject of genome analyses. DNA composition changes are characteristics of both replication and repair machineries that are often lineage- and organelle-specific, such as in cases of chloroplast and nuclear genomes. Nucleotide compositions are also constrained by chromosomal sequence elements that are functional and context-dependent, such as coding sequences (CDSs) and conserved structural elements 1., 2.. One of the approaches to understand DNA compositional dynamics is to analyze signatures of sequence polymorphisms and their rules. There are several factors described to affect mutation spectra of DNA sequences, such as specific mutagens (3), intrinsic enzyme activities (4), and nucleotide contexts 5., 6.. Numerous studies have shown that the degree of transition/transversion (Ts/Tv) is negatively correlated with nucleotide AT account in the immediately adjacent nucleotides in gramineous chloroplast genomes 6., 7.. It was observed recently that AT content favors Tv both within noncoding (6) and silent coding sequences of the chloroplast genome (2). An earlier study has further demonstrated that flanking nucleotide effect extends up to two nucleotides in each direction in the human genes (8). Such nucleotide bias in the human genome was later found extending as far as 200 bp on both sides of an SNP (single nucleotide polymorphism) site (9). However, basic rules and distinctions of such compositional biases across diverse taxonomic groups as well as the underlying molecular mechanisms remain controversial and largely unexplained.
Draft and complete genome sequences of two cultivated rice subspecies (Oryza sativa L. ssp.), indica (93–11) and japonica (Nipponbare), have been published recently 10., 11., 12., 13.. Understanding these sequence quality-related issues, we were able to identify nucleotide substitutions or SNPs between the two subspecies. Despite the fact that these nucleotide substitutions are not strictly speaking the same as those of population-based SNPs, they are, nevertheless, genome-wide nucleotide changes over some evolutionary time scales, and can be traced from the present time all the way back to at least thousands of years. We acquired 3,936,020 SNPs from this exercise and investigated the sequence context effect on nucleotide polymorphisms, shedding lights on mutation mechanisms in molecular details.
Results
Nucleotide compositions in structural contents of the rice genome, genes, and SNPs
Plant genomes are organized differently from those of animals in which most of the genome sequences are transcribed (14). In plant genomes, genes are clustered together and separated by repeat contents that vary from nearly 50% in rice to 90% in barley. Therefore, the discrimination of gene and repeat contents is of essence for any compositional analysis on plant genomes. Based on their locations in the context of genes, we classified the SNPs into three basic categories: genome-wide, intronic, and CDS SNPs, denoted as gSNPs, iSNPs, and cSNPs, respectively. The cSNPs were further divided into P0-SNPs, P1-SNPs, and P2-SNPs. The iSNPs and cSNPs were identified by aligning full-length cDNA sequences with rice genome sequences (see Materials and Methods).
Statistics of the four nucleotides in different regions of the rice genome (indica, 93-11) are summarized in Table 1. There are a couple of noticeable features in the nucleotide content of the genome and genes. Intron sequences (38.17%) have a much lower GC content than that of the exons (54.40%) and the genome average (43.17%). As to the nucleotide context of SNP sites, the average GC content of gSNPs and iSNPs are 46.25% and 44.21%, respectively, which are much lower than that of cSNPs (51.97%). Therefore, GC contents of gSNP and iSNP sites are higher than the genome and intron averages by some substantial margins, 3.08% and 6.04%, respectively. The results may suggest that there is a mutation drive to increase the overall GC content, which is largely contributed by an intronic GC content increase. The opposite was observed in cSNPs, where a slight GC content decrease was found between GC contents of the genome and gSNPs, by a smaller margin of 2.43%. We suspect that this GC content drop is related to selective pressures from codon usage that is often lineage-specific or organism-specific (15). The overall results agree with our earlier observations on a GC gradient that increases from 3′ to 5′ in the rice transcripts 10., 16.. In addition, we are also aware of slight purine content biases in both of the iSNPs and cSNPs, which are absent in the gSNPs. The purine content biases are less pronounced (an order of magnitude smaller) but evident: while the averages of both the genome and gSNPs remain constant, the iSNPs and cSNPs have higher purine contents than pyrimidine contents. The opposite was seen in iSNPs and cSNPs, where the purine contents are actually lower. It is conceivable that selective pressures are at work on both iSNPs and cSNPs, where splicing enhancer elements and codons reside.
Table 1.
The Nucleotide Contents in the Rice Genome, Genes, and SNP Sites (%)
| Nucleotide | Nucleotide content in genome and genes |
Nucleotide content of SNP sites |
||||
|---|---|---|---|---|---|---|
| Genome | Intron | CDS | Genome | Intron | CDS | |
| A | 28.41 | 30.87 | 23.50 | 26.88 | 27.99 | 23.76 |
| C | 21.59 | 19.10 | 26.25 | 23.14 | 22.00 | 25.92 |
| G | 21.58 | 19.07 | 28.15 | 23.11 | 22.21 | 26.05 |
| T | 28.42 | 30.96 | 22.09 | 26.87 | 27.80 | 24.27 |
| GC | 43.17 | 38.17 | 54.40 | 46.25 | 44.21 | 51.97 |
| Purine | 49.99 | 49.94 | 51.65 | 49.99 | 50.20 | 49.81 |
We then calculated SNP frequencies according to a similar content-based classification scheme and further partitioned the statistics of cSNPs into that of three codon phases (Table 2). There are several noticeable sequence signatures. First, transitional SNPs account for 57.23% of gSNPs, 50.65% of iSNPs, and 64.99% of cSNPs; these results suggested that transitional changes are preferred in cSNPs where Tv are constrained both in percentage and in numbers. Second, among transversional SNPs, A/T substitutions are accounted for 13.22% of gSNPs, 16.42% of iSNPs, and only 6.02% of cSNPs. Conversely, C/G substitutions in cSNPs (10.60%) are higher than that of iSNPs (8.71%) and gSNPs (8.08%). The higher C/G substitution rate in CDS regions of the rice genome is in part due to a higher GC content in CDS regions accumulated over evolutional time scales. Third, when cSNPs were separated into three phases, corresponding to codon positions 1, 2, and 3, P2-SNPs (third codon position) showed dominant effects, possessing more SNPs in all categories. In addition, A/C, A/G, and G/T substitutions in P0-SNPs have relatively higher ratios and numbers compared to P1-SNPs, whereas A/T and C/T substitutions of P1-SNPs have higher ratios and numbers. The heterogeneity of nucleotide substitutions reflects codon usage and the nature of codon arrangements. These nucleotide composition variations are of importance for understanding biases in the flanking sequences of SNPs.
Table 2.
The Proportion and Numbers of 6 Types of SNPs
| Type | gSNPs | iSNPs | cSNPs | P0-SNPs | P1-SNPs | P2-SNPs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/C | 10.74% | (387,971) | 12.12% | (29,426) | 8.94% | (4,064) | 10.41% | (1,258) | 10.13% | (1,037) | 7.64% | (1,769) |
| A/G | 28.60% | (1,032,704) | 25.31% | (61,451) | 32.46% | (1,475) | 41.44% | (5,007) | 30.40% | (3,111) | 28.69% | (6,640) |
| A/T | 13.22% | (477,294) | 16.42% | (39,870) | 6.02% | (2,737) | 6.08% | (735) | 7.78% | (796) | 5.21% | (1,206) |
| C/G | 8.08% | (291,724) | 8.71% | (21,145) | 10.60% | (4,821) | 10.23% | (1,236) | 9.82% | (1,005) | 11.15% | (2,580) |
| C/T | 28.63% | (1,033,686) | 25.34% | (61,517) | 32.53% | (14,787) | 20.66% | (2,497) | 34.28% | (3,509) | 37.94% | (8,781) |
| G/T | 10.73% | (387,628) | 12.11% | (29,402) | 9.45% | (4,295) | 11.18% | (1,351) | 7.59% | (777) | 9.36% | (2,167) |
| Total | 100% | (3,611,007) | 100% | (242,631) | 100% | (45,462) | 100% | (12,084) | 100% | (10,235) | 100% | (23,143) |
Neighboring-nucleotide effects of rice SNPs
To study neighboring-nucleotide biases of SNPs, we examined proportions of substitution types (Table 2) and the difference between the observed and the expected proportion (averaged in corresponding region) of flanking nucleotides that surround SNPs (9). If the observed proportion of one nucleotide is higher than expected, we consider it has a positive effect, denoted as “PLUS-bias”, whereas a negative value is believed to have a negative effect and labeled as “MINUS-bias”. The interrogated range is ±12 bp. The “−” and “+” symbols denote 5′ and 3′ directions of the analyzed SNPs, respectively. Numbers following the “−” and “+” symbols indicate the nucleotide positions from an SNP. For example, the value “−10” means the site that is 10 bp upstream the SNP site. We examined the neighboring-nucleotide effects (NNEs) based on six main categories, namely gSNPs, iSNPs, cSNPs, P0-SNPs, P1-SNPs, and P2-SNPs. Each category was subdivided into 3 groups, including whole-genome SNPs, Ts, and Tv, and 6 types, including A/C, A/G, A/T, C/G, C/T, and G/T. Then, we partitioned each type into two parts, for instance, A/G SNPs were divided into gSNPs-A/G-A SNPs and gSNPs-A/G-G SNPs. The last A in gSNPs-A/G-A denotes that the nucleotide at the SNP site is A in the 93-11 genome sequence. In all, the rice SNPs were divided into 90 SNP groups. Taking the position as the x-axis and the neighboring-nucleotides proportion bias as the y-axis, the corresponding figures were plotted (Fig. 1, Fig. 2).
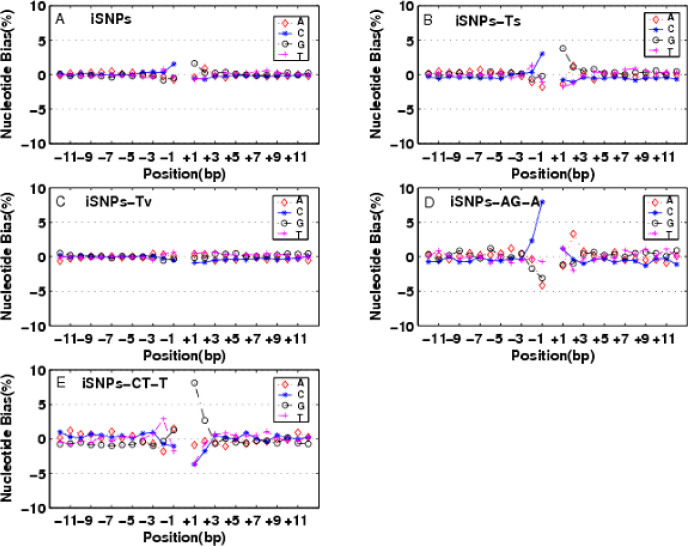
Fig. 1.
A. iSNPs; B. iSNPs-Ts; C. iSNPs-Tv; D. iSNPs-A/G-A; E. iSNPs-C/T-T. PLUS-bias −1 C/+1 G were observed in iSNPs, which was believed to be the effects of the CpG-methylation and deamination process. NNEs extended no more than 3 bp to both sides.
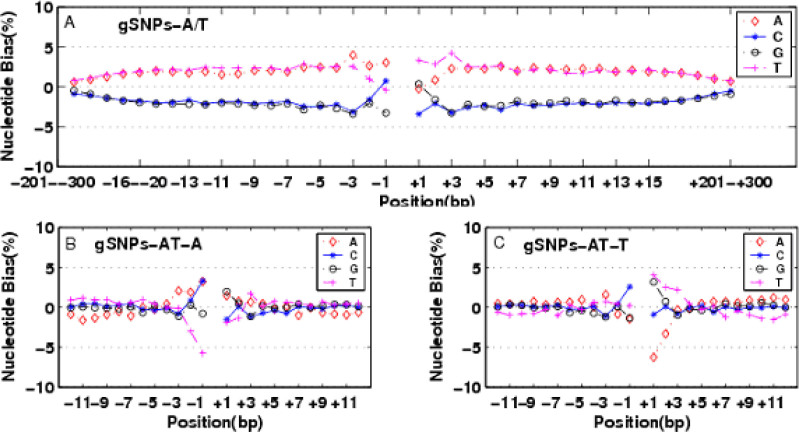
Fig. 2.
The impact of expected nucleotide proportion for NNEs. A. gSNPs-A/T. The expected nucleotide proportion was averaged over the whole rice genome. The PLUS-bias of AT nucleotides extended nearly 300 bp to both sides in gSNPs-A/T; B. gSNPs-A/T-A; C. gSNPs-A/T-T. The expected nucleotide proportion was calculated based on the flanking sequence of the corresponding categories. The long effect range of NNEs disappeared and it was only limited 3 bp to SNP sites in gSNPs-C/T-C and gSNPs-A/T-T.
We observed a similar PLUS-bias of −1 C/+1 G (denoted as −1 C/+1 G) in gSNPs and iSNPs, and this bias extends no more than 3 bp (Figure 1A for iSNPs). The PLUS-bias of −1 C/+1 G was attributed to Ts (Figure 1B) but not Tv (Figure 1C) for iSNPs. Furthermore, we examined this phenomenon on A/G (Figure 1D) and C/T (Figure 1E) types, which were responsible for PLUS-bias −1 C and PLUS-bias +1 G, respectively. It is well agreeable with the CpG-methylation and deamination theory, which shows that the C of CpG dinucleotides is prone to methylation and deamination, and then be transited to T. Unlike the others, for C/T substitutions of P0-SNPs, a PLUS-bias +1 T (P0-C/T-C: 17.17% and P0-C/T-T: 14.80%) and a minor MINUS-bias of +1 G (P0-C/T-C: −0.86% and P0-C/T-T: −0.58%) were observed. For A/G-C/T SNP pairs in intron and genome, there were high MINUS-bias −1 A of A/G SNPs (gSNPs-A/G-A: −6.08% and gSNPs-A/G-G: −6.51%; iSNPs-A/G-A: −4.15% and iSNPs-A/G-G: −5.35%) and high MINUS-bias +1 T of C/T (gSNPs-C/T-C: −6.88% and gSNPs-C/T-T: −5.61%; iSNPs-C/T-C: −5.75% and iSNP-C/T-T: −3.57%). This phenomenon demonstrated that the nucleotide A at −1 site of A/G substitution and the nucleotide T at +1 site of C/T substitution didn’t co-occurred with these mutations. The NNEs extended no more than 3 bp on both sides of rice SNPs.
Immediately adjacent AT content was correlated with transversion
AT-favored Tv in intron and whole genome was confirmed by previous studies in chloroplast (6). To infer the correlation between the ratio of Ts/Tv and immediately adjacent AT content, we divided SNPs into three groups with ±1 (both +1 and −1) AT scores 0, 1, and 2. Then, we calculated the Ts/Tv ratio for each group (Table 3). For genomic and intronic mutations, we observed that the Ts/Tv ratio decreased when AT content increased from 0 to 1 and 2 (±1 AT scores). However, we did not find the same pattern for cSNPs, which endure more selection pressure due to the non-synomouns mutation.
Table 3.
The Ts/Tv and the Numbers of AT Nucleotides in ±1 Regions
| A+T Number* | Genome Ts/Tv | Intron Ts/Tv | CDS Ts/Tv | CDS Phase Ts/Tv |
||
|---|---|---|---|---|---|---|
| P0 | P1 | P2 | ||||
| 0 | 1.626 | 1.301 | 1.818 | 1.490 | 1.921 | 1.969 |
| 1 | 1.404 | 1.093 | 1.965 | 1.795 | 1.867 | 2.107 |
| 2 | 1.113 | 0.862 | 1.651 | 1.548 | 1.520 | 1.767 |
The number of the two immediately neighboring nucleotides (5′ and 3′) that is A or T.
Nucleotide composition asymmetry between coding and noncoding strands in rice intron sequences
In order to avoid the effects of codon usage bias as well as selection pressure, we investigated the nucleotide composition in intron sequences. The proportions of four nucleotides in coding and noncoding strands of intron sequences are listed in Table 4. In coding strands, the proportion of nucleotide T (34.23%) is evidently higher (6.62%) than that of A (27.61%), and the proportion of nucleotide C (18.91%) is slightly less (0.35%) than that of G (19.26%). In addition, the proportion of pyrimidine (53.14%) is apparently higher (6.27%) than that of purine (46.87%) in coding strands.
Table 4.
The Nucleotide Composition Asymmetry Between Coding and Noncoding Strands in Rice Introns
| Strand | A | C | G | T | AG | CT | (T−A)/(A+T) | (C−G)/(C+G) |
|---|---|---|---|---|---|---|---|---|
| Coding | 27.61% | 18.91% | 19.26% | 34.23% | 46.87% | 53.14% | 0.1071 | −0.0092 |
| Noncoding | 34.23% | 19.26% | 18.91% | 27.61% | 53.14% | 46.87% | −0.1071 | 0.0092 |
Nucleotide usage bias in rice cSNPs
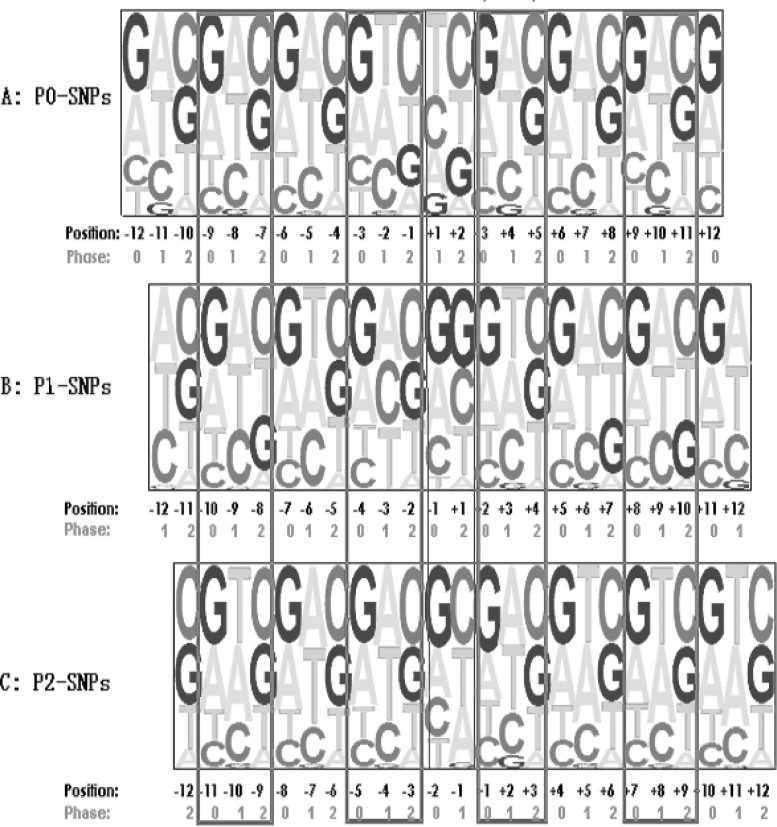
There were 12,084 P0-SNPs, 10,235 P1-SNPs, and 23,143 P2-SNPs in cSNPs. More complex patterns of NNEs in cSNPs were observed. These cSNPs, irrelevant to the phases, displayed high diverse NNE patterns. Most of the flanking nucleotide proportion bias pattern repeats in every “3” nucleotide. The regular bias pattern is correlated with the nucleotide positions in codon (Figure 3). Nucleotides A and G were more often used (PLUS-bias) than T and C (MINUS-bias) in phase 0; in phase 1, we observed PLUS-bias of AT and MINUS-bias of CG; as for the phase 2, PLUS-bias of CG and MINUS-bias of AT were observed.
Fig. 3.
The nucleotide usage bias in rice codon. The magnitude of the letter of nucleotides was used to denote their proportion biases. The nucleotide usage biases were cycled with the nucleotide position in codon. Mostly, it was PLUS-bias of AG and MINUS-bias of TC nucleotides in phase 0; PLUS-bias of AT and MINUS-bias of CG nucleotide in phase 1; PLUS-bias of CG and MINUS-bias of AT nucleotide in phase 2.
The intensity of NNEs among different rice chromosomes
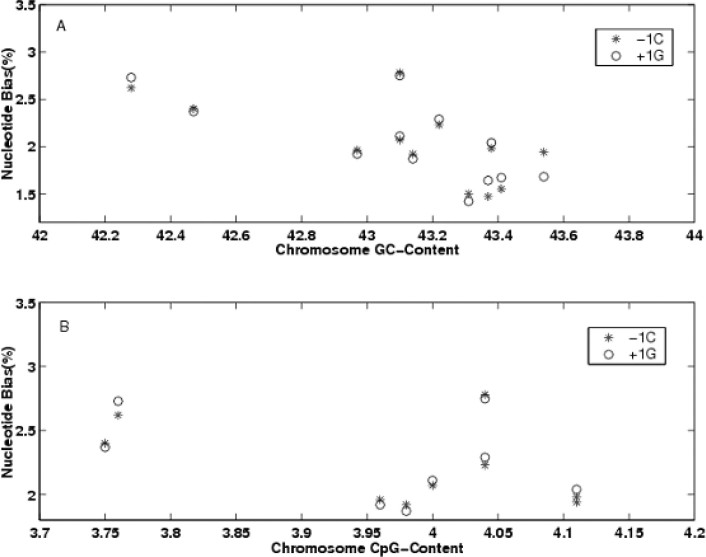
For the 12 chromosomes of the rice genome, the neighboring nucleotide proportion bias (deviation from the chromosome-specific average) was not uniform. For example, the PLUS-bias of −1 C nucleotide at substitution in Chromosome 1 was the least in all the 12 chromosomes (+1.47%), even its GC content (43.37%) ranked as the fourth highest. In contrast, Chromosome 12 (GC content is 42.47%, calculated in 93-11 reference sequences) has the biggest PLUS-bias of −1 C (+2.40%) in all these chromosomes. It is reported that the −1 C/+1 G nucleotide proportion bias is positively correlated with the chromosome’s GC-content in the human genome (9). Whereas in the rice genome, we found that the −1 C/+1 G nucleotide proportion bias is negatively correlated with the GC content (Figure 4A). A statistical significant negative correlation was therefore found; the correlation coefficient was −0.657 (p-value = 0.020) for −1 C and −0.697 (p-value = 0.012) for +1 G. We further studied the correlation between the CG dinucleotide content and the PLUS-bias of −1 C/+1 G nucleotide bias (deviation from the chromosome-specific average) in rice chromosomes (Figure 4B). Similar negative correlation was also found. The correlation coefficient was −0.590 (p-value = 0.0453) for −1 C and −0.596 (p-value = 0.041) for +1 G.
Fig. 4.
The intensity of PLUS-bias of −1 C/+1 G of rice chromosome is negative correlated with the GC content of chromosome. A. −1 C/+1 G nucleotide proportion bias to the GC content in rice chromosome sequences; B. −1 C/+1 G nucleotide proportion bias to CpG dinucleotides in rice chromosome sequences.
Discussion
Minimization of ascertainment biases in SNP analyses
Detailed investigations on nucleotide substitutions and their NNEs are of importance in understanding mutation mechanisms, which are major forces of genome diversity and evolution. The sequences of rice genomes and SNPs discovered therein provide an opportunity for such an analysis. For sequence-based data analyses, especially those focused on variations of nucleotide sequences, it is essential to discuss sequence quality-related issues and ascertainment biases, which are often project-specific (or organism-specific). In this study, we have devised several procedures to minimize ascertainment biases.
First, we employed a community-accepted SNP discovery method (see Materials and Methods) that has been applied in the polymorphism discovery of human (17) and chicken genomes (18). Their validation rates were reported as 95% for human and 94% for chicken. In addition, two independent groups have published SNP finding results on the rice genome utilizing an early published data set. Their SNP validation rates were 98.2% SNPs (19) and 79.8%±7.5% (20), despite the fact that sequence quality information from the raw sequencing traces was ignored in the studies. Furthermore, a small fraction of false positive or negative SNP calls were believed not interfering with our results since both a large number of SNPs and relative ratios were exploited in this study. Second, to reduce possible compositional biases due to extreme AT and GC, SNPs associated with AT content exceeding 70% and GC content above 65% were excluded from our data sets. Third, to avoid side-effects due to gene structures and functional contents, we categorized SNPs and their neighboring sequences according to their locations in a context of genes as well as cSNPs into codon phases (codon positions). We did not use gene-prediction tools for this study; instead, we identified genes and their structures based on alignments to a full-length cDNA collection (21). Finally, the only drawback of this study was that we were unable to obtain the allele frequency for all SNPs and to infer which allele is the wild type, so that the direction of these mutations was not factored in our analyses.
The mechanism of NNEs
In the rice genome, the evident NNEs have been observed, while the exact mechanism of NNEs is still unclear. It can be affected by polymerase fidelity and proofreading (22), mismatch repair efficiency (23), or mismatch stability (24).
For the rice genome, the proportion of Ts and Tv were 57.23% and 42.77%, respectively. The excess of Ts is largely attributed to the CpG-methylation and deamination process. CpG is a signal for methylation by a specific cytosine DNA methyltransferase. It adds a methyl group to the 5′ carbon of the cytosine. The resulted 5′-methylated cytosine is unstable and is prone for deamination, resulting in thymine. Over time, the number of CpG has gradually diminished because the slow but steady conversion of CpG to TpG (and to CpA on the opposite strand). In higher plants, DNA methylation occurs at up to 25% of all cytosine, primarily in the sequences CpG and CpNpG, both of which are more than 80% methylated in wheat and tobacco (25). That is why we witnessed the PLUS-bias of nucleotides C at the −1 site (−1 C) in iSNPs-A/G SNPs, and PLUS-bias of nucleotides G at the +1 site (+1 G) in iSNPs-C/T SNPs (Figure 1, D and E).
MINUS-bias −1 C and PLUS-bias −1 T in iSNPs-C/G-C (−3.45% and 3.28%, respectively) and iSNPs-C/G-G (−6.12% and 4.82%, respectively) were also observed. This bias could be attributed to the CpG-methylation and deamination process. For example, when there is a C at the −1 site of C/G substitution and the two allele frequencies of C/G substitution are similar, the chance that forming a CpG and CpC dinucleotide is nearly 50%. While for CpG dinucleotides, C is prone to be transited to T, thus, the MINUS-bias −1 C and PLUS-bias −1 T is produced.
It has been showed that AT nucleotides in immediately adjacent regions favorite Tv 2., 6., 26.. We also witnessed this phenomenon in rice gSNPs as well as iSNPs. One possible reason is that the mismatch repair efficiency is influenced by the neighboring nucleotide composition. In Escherichia coli, the Tv mismatch repair efficiency increases along with the GC content increasing in the neighboring nucleotide subsequence 23., 27.. Morton (6) pointed that the mis-incorporation and/or proofreading by the DNA polymerase is influenced by the nucleotides upstream the site of incorporation. In Ts of gSNPs, we observed the obvious PLUS-bias of −1 C (4.05%) and PLUS-bias of +1 G (4.06%). Since the PLUS-bias of −1 C/+1 G is due to the CpG-methylation and deamination process, we cannot say that it is G or C that influences the polymorphism activity. Here, we favor the first explanation, which is that the mismatch repair efficiency is influenced by the nearby nucleotide.
The NNEs of codon SNPs
From the genetic code, at each of the three codon positions, there are 64×3=192 possible base substitutions. The numbers of substitutions lead to synonymous changes in terms of the codon position are 8, 2, and 128, respectively (16). That is why the amount of rice SNPs at CDS position ranked as P2 (23,143) > P0 (12,084) > P1 (10,235). Since SNPs at different codon phases endure different selection pressure, we should not expect that one category of SNPs will show similar proportion bias pattern at different CDS positions, such as A/G and C/T.
Under the CpG-methylation and deamination process, there should be a profound PLUS-bias of +1 G content bias of C/T substitutions. However, for P0-C/T SNPs, we observed PLUS-bias of +1 T and MINUS-bias +1 G. Since P0 C/T SNPs is a synonymous mutation that codes for leucine, it will endure less selection pressure compared with these non-synonymous mutations. Because the second nucleotide of leucine is T, so +1 T of P0-C/T SNPs will be observed more frequently than expected. Hereby, we can say that the selective constraint against non-synonymous mutations overtake the CpG-methylation and deamination effects. In total, it is natural selection combined with CpG as well as context nucleotide effect that determine the complex mutation patterns of CDSs.
The NNEs extending range
Krawczak and colleagues studied 7,271 substitutions in the coding regions of 547 genes, and found the NNEs extended no more than 2 bp to the substitution site (8). Zhao and Boerwinkle's study showed that the NNEs extended as far as ±200 bp for SNPs in the human genome (9). Mutations that locate in different regions in the rice genome may endure different mutation mechanisms. Through studying gSNPs, cSNPs (further divided into three phases), and iSNPs separately, we found that the NNEs in gSNPs or iSNPs extended no more than 3 bp to both sides.
The correct determinant of “expected nucleotide proportion” is very important because it can eliminate artificial NNEs. At the beginning, we set the expected nucleotide proportion by averaging the whole rice genome sequences, and the PLUS-bias of nucleotides AT for A/T substitution extended as far as 300 bp (Figure 2A). When the expected nucleotide proportion was calculated only in the flanking subsequences (12 bp to the SNP site), the PLUS-bias of nucleotides AT extended no more than 3 bp of both sides in A/T SNPs (Figure 2, B and C).
The DNA double-strand nucleotide composition asymmetry in intron
In bacteria, the nucleotide composition is asymmetric between coding and noncoding strands 28., 29., 30.. The unequal mutation rate between C→T and G→A mutations may account for this phenomenon (29), where there are two mechanisms. One mechanism is transcription-coupled repair (TCR; ref. 31), which could efficiently repair the C→T mutation in the noncoding strand (template strand), while leave alone the coding strand. Another mechanism is cytosine deamination (32). Cytosine residues involved in a mispairing in DNA are 1–2 orders of magnitude that are more prone to deaminate to uracil than to cytosine in double-strand DNA (33). Because the coding strand will be single when the noncoding strand was transcribed (paired with mRNA), more C→T mutations will occur in the coding strand (28). The difference between these two mechanisms is that TCR reduces the G→A mutation rate in coding strands while cytosine deamination improves the C→T mutation rate in coding strands (34). If TCR is the dominant mechanism, the G→A mutation rate will be reduced, and more nucleotide G will be preserved. The proportion of nucleotide G is higher than that of C in coding strands of rice intron sequences. Even this proportion difference is in a small margin, it still suggests that the TCR may be the fundamental mechanism that caused the DNA double-strand nucleotide composition asymmetry.
In conclusion, for the rice genome, we found apparent evidence to the flanking nucleotide effect on SNPs. The effect is governed by the category and genomic location of SNPs from different chromosomes, which exhibit different flanking nucleotide effect patterns. Generally, the flanking nucleotide effect can extend no more than ±3 bp. The high proportion of Ts and its immediately adjacent proportion bias could be explained largely by the CpG-methylation and deamination theory. The complexity patterns, displayed by different phases of SNPs, are believed to be caused by a combination of selective forces, the CpG-methylation and deamination process, as well as the flanking GC (or AT) content effect. We also found that the Ts/Tv is negatively correlated with the immediately AT nucleotide counts. The proportion discrepancy between nucleotide pairs of A/T and C/G demonstrates that TCR may account for the double-strand nucleotide composition asymmetry in rice intron sequences. Overall, our results provide a full spectrum of rice SNP context effects and their substitution patterns, giving us further knowledge to understand the rice mutation mechanism and evolution history.
Materials and Methods
SNP Data Sets
We used assembled sequences of 93-11 (Oryza sativa L. ssp. indica; ref. 10) and Nipponbare (Oryza sativa L. ssp. japonica; ref. 11) as reference sequences to discover SNPs (BGI-RIS, http://rise.genomics.org.cn/index.jsp). SNPs were identified according to a well-accepted community consensus (17) by using a basic threshold in which the sequence quality must exceed values of Q >23 for an SNP site and Q >15 for its 5-bp flanking sequences (35).
For comparison, an independent analysis (19) reported mean rates of 7.1 SNP/Kb and 2.0 indel/Kb with 98% of these SNPs that were experimentally confirmed. Our SNP rates were two times higher because we aligned more of the intergenic sequence. To eliminate this factor, we restricted our rates to the introns defined by full-length cDNA sequences (21). The rates are 6.1 SNP/Kb and 1.3 indel/Kb, which are actually lower than those from that independent analysis.
To identify iSNPs and gSNPs, we used the following steps. First, we aligned 20,259 full-length cDNA sequences (21) with the two rice genome assemblies (13), using a software tool BLAT (36). A total of 16,412 alignments were selected to determine CDS and intronic sequences from the genome assemblies. SIM4 (37) was used to examine the aligned sequences, and 16,395 alignments were chosen for our analyses. SNPs were then divided into three basic categories according to their positions in the genome regions: (1) gSNPs denote total SNPs regardless their chromosomal locations and positions relative to genes, (2) iSNPs are those located exclusively between two exons, and (3) cSNPs were found from CDSs, excluding 3′ and 5′ untranslated sequences. The cSNPs were further divided into three groups based on their positions in codons, often regarded as phases, as P0-SNPs, P1-SNPs, and P2-SNPs.
To carry out analyses of the neighboring effect of SNPs, we extracted our data sets as follows. The first data set was the flanking sequences of gSNPs, which are 300 bp in length from both up- and down-streams of an SNP. To reduce background compositional biases due to extreme AT or GC contents, we excluded SNPs that have GC or AT contents in the flanking sequences (collectively 600 bp from both sides) exceeding 65% for GC content and 70% for AT content. As a result, 189,904 and 135,040 SNPs corresponding to extreme AT and GC contents were eliminated from the data set, which is about 8% of the total SNPs. From this data set, 45,462 cSNPs and 242,811 iSNPs were subdivided. Since the intron and exon sizes were also vary broadly, with mean sizes of 323 bp (median 161 bp) and 456 bp (median 286 bp), we used a threshold of 15 bp flanking sequences extending toward both directions, so that most of iSNPs and cSNPs became legit, removing 10,836 iSNPs (4.27%) and 3,658 cSNPs (7%) from the unprocessed data. Finally, 3,611,007 gSNPs, 242,811 iSNPs, and 45,462 cSNPs were included in this study.
Data analysis
We took the nucleotide composition at SNP sites of 93-11 reference sequences as the composition of nucleotides at SNP sites.
To calculate the NNEs, we assigned positions and values of the flanking sequences starting from an SNP: negative, or “−”, refers to the 5′ sequence and corresponding values, whereas positive, or “+”, describes the 3′. To infer effects of sequences flanking an SNP, we calculated the expected proportion of each SNP and its surrounding sequences to the observed proportion. The proportion of each nucleotide was calculated by the following formula:
Fij is the proportion of nucleotide j at the ith position of a nucleotide substitution, nij is the number of nucleotide j at the ith position, and Ni is the sum of the four nucleotides at site i. The expected nucleotide proportions were averaged over 24 bp around SNPs (12 bp up- and down-stream the SNP site) of each SNP type.
To demonstrate the NNEs on the rice genome, we calculated the expected proportion of four nucleotides for each SNP group. The nucleotide proportion biases in each group were obtained by subtracting the observed value from the expected proportions (exclusively for this group). A simple script, written in C++, was used to calculate proportions and biases. Then, the proportion biases were plotted as a function of positions around SNPs.
To correlate the Ts/Tv ratio and the neighboring AT content, only two sites immediately flanking an SNP were examined and scored as 0, 1, and 2, corresponding to none, one A/T (only one found from two sites), and two A/T (A or T at both sides), which were tabulated according to different SNP categories.
Acknowledgements
We are grateful to Professor Guo-Ying Li (Academy of Mathematics and Systems Science), Dr. Jun Wang, Jun Li, Pei-Xiang Ni, Yong Zhang, and Heng Li (Beijing Genomics Institute) for their scientific supports. We also thank Dr. Dun-Mei Wang and Mr. Qi Guo for critical reading of the manuscript and suggestive comments. The work was supported partly by the National High-Tech Research and Development Program (863 program; No. 2003AA231050) and the National Natural Science Foundation of China (No.10371126).
Contributor Information
Huan-Ming Yang, Email: yanghm@genomics.org.cn.
Jun Yu, Email: junyu@genomics.org.cn.
References
- 1.Fedorov A. Regularities of context-dependent codon bias in eukaryotic genes. Nucleic Acids Res. 2002;30:1192–1197. doi: 10.1093/nar/30.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morton B.R. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J. Mol. Evol. 2003;56:616–629. doi: 10.1007/s00239-002-2430-1. [DOI] [PubMed] [Google Scholar]
- 3.Seo K.Y. Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 4.Timsit Y. DNA structure and polymerase fidelity. J. Mol. Biol. 1999;293:835–853. doi: 10.1006/jmbi.1999.3199. [DOI] [PubMed] [Google Scholar]
- 5.Zavolan M., Kepler T.B. Statistical inference of sequence-dependent mutation rates. Curr. Opin. Genet. Dev. 2001;11:612–615. doi: 10.1016/s0959-437x(00)00242-2. [DOI] [PubMed] [Google Scholar]
- 6.Morton B.R. Neighboring base composition and transversion/transition bias in a comparison of rice and maize chloroplast noncoding regions. Proc. Natl. Acad. Sci. USA. 1995;92:9717–9721. doi: 10.1073/pnas.92.21.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morton B.R. The influence of specific neighboring bases on substitution bias in noncoding regions of the plant chloroplast genome. J. Mol. Evol. 1997;45:227–231. doi: 10.1007/pl00006224. [DOI] [PubMed] [Google Scholar]
- 8.Krawczak M. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Z., Boerwinkle E. Neighboring-nucleotide effects on single nucleotide polymorphisms: a study of 2.6 million polymorphisms across the human genome. Genome Res. 2002;12:1679–1686. doi: 10.1101/gr.287302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 11.Goff S.A. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 12.Zhao W. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong G.K. Most of the human genome is transcribed. Genome Res. 2001;11:1975–1977. doi: 10.1101/gr.202401. [DOI] [PubMed] [Google Scholar]
- 15.Carlini D.B. Context-dependent codon bias and messenger RNA longevity in the yeast transcriptome. Mol. Biol. Evol. 2005;22:1403–1411. doi: 10.1093/molbev/msi135. [DOI] [PubMed] [Google Scholar]
- 16.Wong G.K. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altshuler D. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 18.Wong G.K. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y.J. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 2004;135:1198–1205. doi: 10.1104/pp.103.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feltus F.A. An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res. 2004;14:1812–1819. doi: 10.1101/gr.2479404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi S. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 22.Petruska J., Goodman M.F. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. J. Biol. Chem. 1985;260:7533–7539. [PubMed] [Google Scholar]
- 23.Jones M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987;115:605–610. doi: 10.1093/genetics/115.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng J.W. Base pairing geometry in GA mismatches depends entirely on the neighboring sequence. J. Mol. Biol. 1992;228:1037–1041. doi: 10.1016/0022-2836(92)90312-8. [DOI] [PubMed] [Google Scholar]
- 25.Gruenbaum Y. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- 26.Morton B.R. The influence of neighboring bases composition on substitutions in plant chloroplast coding sequences. Mol. Biol. Evol. 1997;14:189–194. [Google Scholar]
- 27.Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu. Rev. Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- 28.Beletskii A., Bhagwat A.S. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francino M.P. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science. 1996;272:107–109. doi: 10.1126/science.272.5258.107. [DOI] [PubMed] [Google Scholar]
- 30.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 31.Oller A.R. Transcription-repair coupling determines the strandedness of ultraviolet mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1992;89:11036–11040. doi: 10.1073/pnas.89.22.11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skandalis A. Strand bias in mutation involving 5-methylcytosine deamination in the human hprt gene. Mutat. Res. 1994;314:21–26. doi: 10.1016/0921-8777(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 33.Frederico L.A. Cytosine deamination in mismatched base pairs. Biochemistry. 1993;32:6523–6530. doi: 10.1021/bi00077a005. [DOI] [PubMed] [Google Scholar]
- 34.Francino M.P., Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol. 2001;18:1147–1150. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- 35.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 36.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Florea L. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]