ABSTRACT
DNA guanine (G)-rich 4-stranded helical nucleic acid structures called G-quadruplexes (G4), have been extensively studied during the last decades. However, emerging evidence reveals that 5′- and 3′-untranslated regions (5′- and 3′-UTRs) as well as open reading frames (ORFs) contain putative RNA G-quadruplexes. These stable secondary structures play key roles in telomere homeostasis and RNA metabolism including pre-mRNA splicing, polyadenylation, mRNA targeting and translation. Interestingly, multiple RNA binding proteins such as nucleolin, FMRP, DHX36, and Aven were identified to bind RNA G-quadruplexes. Moreover, accumulating reports suggest that RNA G-quadruplexes regulate translation in cap-dependent and -independent manner. Herein, we discuss potential roles of RNA G-quadruplexes and associated trans-acting factors in the regulation of mRNA translation.
KEYWORDS: Aven, DHX36, FMRP, G-quadruplexes, RNA binding proteins, translation
Structure of G-quadruplexes
In 1910, Bang demonstrated that guanylic acid forms gels at high concentration, providing the first evidence that guanine (G)-rich sequences may form higher-order structures.1 Fifty years later, Gellert et al. (1962) reported that guanylic acid has the ability to form tetrameric structures by self-association.2 These non-canonical structures were identified in conserved DNA sequences of telomeres and shown to form so-called G-quadruplexes (G4) structures in vitro.3,4 Since then, increasing evidence shows that both DNA and RNA containing spaced guanine repeats form G4 structures.5,6 G-quadruplexes are folded in G-quartets that are square planar arrangements formed via Hoogsten pairing of adjacent guanines.7-12 The G-quartets lay on the top of each other to form 4-stranded structures which are stabilized by a cation positioned in the middle of the tetrads with preference for potassium (Fig. 1). The ability of potential G4 sequences to form G-quadruplexes is therefore influenced by the nature of the central cation, the number of stacking G-quartets, the length of the sequences connecting the strands (at least in DNA G-quadruplexes), the direction of the strands, and the presence of an alternative Watson-Crick pair-based stable structure.13,14
Figure 1.
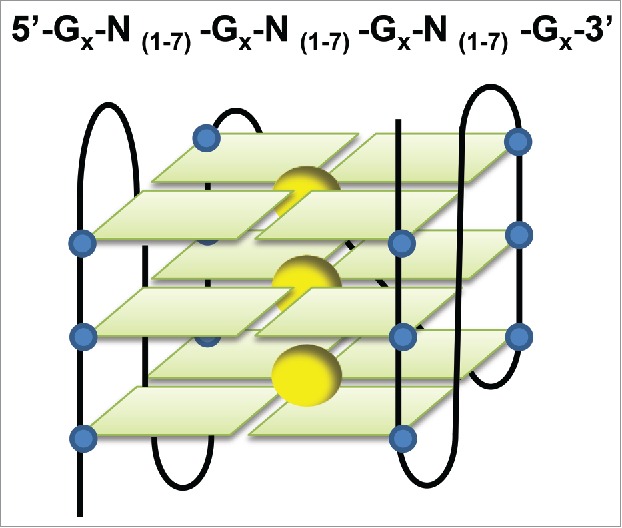
Structure of G-quadruplexes. Guanine-rich sequences fold into G-quadruplex structures (G4 structures), composed of planar G-quartets. This representation is a G-quadruplex parallel structure that could be observed in RNA as well as DNA molecules. The guanines are represented as blue, while cations are yellow.
DNA G-quadruplexes
Bioinformatic search using a script that identifies “regular” G-quadruplexes (i.e. GxN1-7GxN1-7GxN1-7Gx, where G stands for guanine, N can be any nucleotide (A, G, C, U) and x≥3 ) has shown that the human genome harbors ∼376,000 potential G-quadruplex sequences (pG4).15,16 With the development of high resolution sequencing-based methods > 716,000 G4 structures were identified, where ∼451,000 were not predicted by computational methods.17,18 These sequences are highly conserved in mammals, with limited conservation in evolutionarily lower organisms.19,20 Genome-wide searches using G-quadruplex-specific probes and structure-based pull-down strategies have revealed that the pG4 sequences are not randomly distributed, being enriched at telomeres, promoter regions and replication origins.9,21 The highest abundance of pG4 is in human telomeres, where the G4 formation protects the end of chromosomes by inhibiting telomerase activity.16,22,23 Abnormal telomerase overexpression has been observed in > 85% of cancers. Thus, extensive investigations were performed to obtain anti-cancer therapies using small molecules to stabilize telomeric G-quadruplexes as a means to inhibit telomerase.24-27 Moye et al. (2015) characterized the stable human telomeric G-quadruplexes and demonstrated that these G4s are able to extend into parallel, intermolecular conformations, aligning with the intrinsic RNA moiety of the human telomerase RNA (hTR). They also showed that telomerase colocalizes with a subset of telomeric G4 structures in vivo.27 Additionally, structural and computational analysis revealed that pG4 sequences are found in ∼40% of gene promoter regions, mostly acting as transcriptional repressors.23,28,29 It has been shown that pG4 sequences occur with high frequency in the promoter regions of genes encoding oncogenes such as c-Myc, c-Kit, KRAS, PDGF-A (platelet derived growth factor-A), and hTERT (human telomerase reverse transcriptase), while tumor suppressor genes correlate with low pG4 abundance in their promoter regions.12,30,31 Recently, Onel et al. (2016) showed that Bcl-2 forms G-quadruplexes in its promoter by nuclear magnetic resonance (NMR) spectroscopy and dimethylsulfate (DMS) footprinting assays, and these G4s were shown to inhibit transcription by promoter-driven luciferase assay.32 Another study also demonstrated that the human tyrosine hydroxylase (hTH) gene harbors a G4 structure in the 3′ proximal promoter region, acting as a necessary element for transcriptional regulation. Since hTH is linked to several neurological and psychiatric disorders such as Parkinson disease and Schizophrenia,33 these findings suggest that promoter G4 sequences may be linked with these diseases. Furthermore, emerging evidence suggests that ∼90% of DNA replication origins contain pG4 sequences.7,34-36 Recently, it was discovered by performing nascent strand sequencing that G4 sequences may also position nucleosomes at a subset of human replication origins.37 Moreover, it was reported that Potential DNA:RNA Hybrid G-Quadruplex Sequences (PHQS) are present in > 97% of human genes, and these PHQS may modulate transcription.38 Additional studies show that helicases, which are the molecular motors to unwind DNA and RNA, are involved in the active resolution of G4s. The best-characterized DNA G4 helicase are Pif1, RecQ, FANCJ, DDX11, BLM (Bloom syndrome protein) and WRN (Werner syndrome protein).8 Known dual RNA-DNA G4 helicases are DHX9 and RHAU (DHX36).39,40
RNA G-quadruplexes
G-quadruplexes also form in RNA and are more stable than their DNA counterparts.11 RNA G-quadruplexes almost exclusively adopt a parallel conformation in which the 4 strands all have the same directionality. The 2′-hydroxyl group of the ribose locks the RNA in an anti-conformation, which favors the parallel topology. Consequently, RNA G-quadruplexes have less topological diversity than DNA G-quadruplexes.
In eukaryotes, RNA G4 structures are enriched at telomeres and within specific protein encoding transcripts i.e., mRNAs. In telomeres, the G-rich TERRA (telomeric repeat-containing RNAs, or TelRNA) RNA which is transcribed from the human C-rich telomeric DNA, was shown to form G4 structures in vitro41,42 and in cellulo.43 The high resolution of the G4 structure in TERRA revealed that the 2′-hydoxyl group provides intramolecular hydrogen bonding within the parallel-stranded structures, and this is important for ligand targeting and higher-order arrangement.44 The human telomerase RNA (hTR) forms G4 structures at its 5′ end in the presence of potassium, as visualized using gel electrophoresis and UV, CD, and NMR spectroscopy.45,46
RNA G-quadruplexes are frequently present in mRNAs. Kumari et al. (2007) demonstrated that among all the genes in the human transcriptome available at that time, ∼3,000 of the 5′-UTRs were identified to possess at least one pG4 sequence.47 This was revised, when Beaudoin and Perreault (2010) identified 9,979 5′-UTRs to contain at least one pG4 sequence, among the 124,315 transcripts from the human UTRfull dataset based on a “regular” definition of G-quadruplex.48 Subsequently, it was discovered that 1,453 human pG4 sequences possess 2 short distal loops of 1 nucleotide in length and a long central loop of up to 70 nucleotides long in the 5′-UTRs, which significantly expands the number of pG4s in the transcriptome.49 A bioinformatic search for “regular” G-quadruplex in 3′-UTRs of the human transcriptome, detected 8,903 pG4 sequences showing that the enrichment for G-quadruplex is not limited to the 5′-UTRs.50 Additionally, other bioinformatic analyses, in-line probing and luciferase reporter assays revealed the existence of pG4 sequences in both the 5′- and 3′-UTR of transcripts, indicating G4 structures function in the post-transcription regulation in cellulo, and neighboring C-rich sequences that affect G4 folding.14,49,51 Moreover, a bioinformatics search for pG4 sequences in mRNA-coding regions was performed, which revealed that ∼1600 pG4s are present in human ORFs.52 Recently, the existence of G-quadruplex formation in RNA was further confirmed in human cells by using stabilizing ligands that specifically trap RNA G-quadruplexes.53 The authors were able to visualize G4 structures in the cytoplasm of human cells using G-quadruplex-specific antibodies.
RNA G-quadruplexes in TERRA and hTR RNAs regulate telomerase function, whereas G4 formation in mRNAs has been implicated in several gene expression steps, including pre-mRNA processing, mRNA turnover and targeting, and translation.12 While recent reviews have broadly covered the G4 structures in DNA and RNA molecules as well as their functions in telomere maintenance and RNA metabolism in physiology and pathology conditions,9,10,12 we will discuss in detail the emerging areas of RNA G4 structures in mRNA translation and known trans-acting factors that bind these structures.
G-quadruplexes in the control of mRNA translation
mRNA translation is one of the most fundamental processes in RNA metabolism, and its regulation is tightly controlled. Protein synthesis is composed of 4 main steps: translation initiation, elongation, and termination as well as ribosome recycling.54 For most eukaryotic mRNAs, translation initiation involves the association of the 7-methylguanosine cap with the cap-binding complex called eukaryotic initiation factor 4F (eIF4F,Fig. 2A).55,56 eIF4F complex contains the cap-binding protein eIF4E, the scaffold protein eIF4G that bridges interaction between eIF4F and multifactor complex (MFC), thus allowing recruitment of the mRNA to the ribosome, and the ATP-dependent DEAD box RNA helicase eIF4A required to unwind secondary structures in the 5′-UTRs.57,58 The association between the mRNA and eIF4F is the first step of translation, followed by the recruitment of 43S initiation complex, composed of the 40S ribosomal subunit, the eukaryotic initiation factors eIF3, eIF1, eIF1A and eIF5, as well as the ternary complex containing methionine-loaded tRNA, eIF2 and GTP (Fig. 2A).56,59 The 43S complex recognizes the initiation codon, where it is joined by a 60S ribosomal subunit to form the 80S ribosome.57,58 Excessive secondary structures in 5′-UTRs impede mRNA translation in a cap-dependent manner in eukaryotes.60,61 The 3′-UTRs also participate in translational regulation, where the added poly (A) tail is bound by the poly(A)-binding protein (PABP) and eIF4G, resulting in the circularization of mRNAs and enhanced overall initiation rate.57 An alternative mode of translation is driven by IRES (Internal Ribosome Entry Sites), and it occurs in a cap-independent mode.62 Independent of the presence or integrity of several canonical initiation factors (especially eIF4E), IRES directly recruits ribosomes, bypassing the requirement for the 5′cap and eIF4E. Efficient IRES-driven translation is facilitated by the IRES trans-acting factors (ITAFs,Fig. 2B).59,63 Collectively, secondary structure in 5′-UTR is thought to have a major impact on translation efficiency.55
Figure 2.
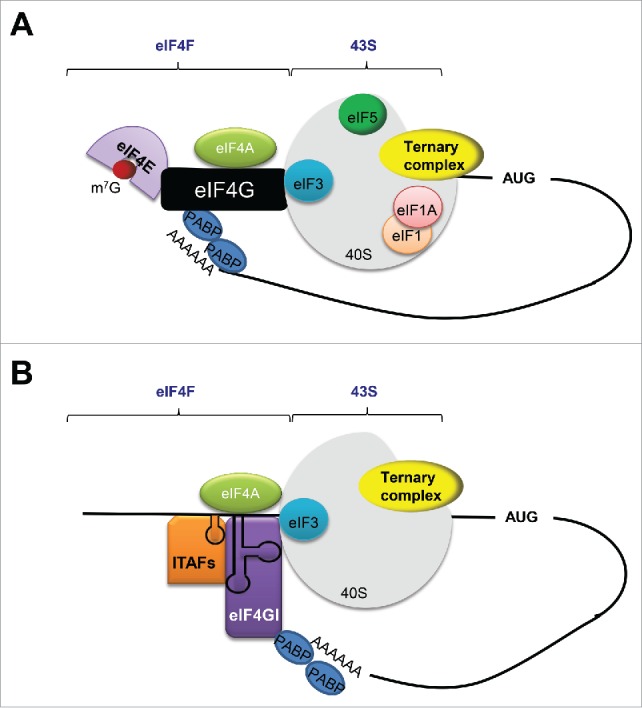
Cap-dependent and Cap–independent translation initiation. (A) In cap-dependent translation, eIF4E binds to the 5′-m7GpppN of the mRNA (m7G). The capped 5′-end is associated with 43S complex by a bridging protein called eIF4G. eIF4G is also bound to eIF4A, the RNA helicase that unwinds 5′ secondary structures. PABP binds the poly (A) tail and brings the 5′-end and 3′-end of the mRNA together through the interaction with eIF4G. eIF3, eIF5, eIF1/eIF1A and ternary complex are shown as represented. (B) ITAFs and eIG4GI (also known as p97/DAP5/NAT1, purple) facilitates IRES cap-independent translation.59
G-quadruplexes in 5′-UTR and translational control
Kumari et al. (2007) reported that pG4 sequences in the human NRAS mRNA are conserved in different organisms.47 They documented the formation of G4 structures in vitro by circular dichroism (CD) spectroscopy and UV-melting experiments, while luciferase reporter assays revealed that the RNA G4 in 5′-UTR of NRAS inhibits translation by ∼80% in rabbit reticulocyte lysates.47 Moreover, it was established that the human ZIC-1 mRNA forms a 27 nucleotide G4 structure within its 5′-UTR and represses protein production by ∼80% in HeLa cells using the dual-luciferase plasmid based assay.64 The presence of G4 structures in 5′-UTR of various human mRNAs and multiple strategies such as bioinformatic analyses, mutagenesis and reporter gene-based expression assays showed that G4s in 5′-UTRs correlate with translational repression of various mRNAs including MT3-MMP,65 ERS1,66 BCL-2,67 TRF2,68 ADAM1069,70 and TGFβ271 (Fig. 3). Moreover, in-depth analysis using CD spectroscopy and in-line probing, identified several 5′-UTRs that harbor pG4 sequences including EBAG9, AASDHPPT, FZD2, BARHL1, NCAM2, and THRA48 (Fig. 3). Most of these genes are involved in transcriptional regulation, protein modification, G-protein-mediated signaling, cation transport and developmental processes. The C-to-A substitution, known to destabilize G4 formation, was able to rescue the repressed translation of all but one gene.48 Recently, G4 structures with longer central loops (> 7 nucleotides) in the HIRA, TOM112 and APC 5′-UTR were also shown to have the ability to repress translation when tested by luciferase reporter assays (Fig. 3).49 Similar conclusions were reached in the study where the “irregular” G4 structures were discovered in the H2AFY and AKIRIN 5′-UTR (Fig. 3).72 It was also shown that antisense oligonucleotides can be used to inhibit or promote the formation of RNA G4 structures.72 Additionally, by using ribosome footprinting on a transcriptome-wide scale, Wolfe et al. (2014) reported that the 12-nucleotide guanine quartet motifs that can form G4 structures in 5′-UTRs rendering mRNAs exceptionally sensitive to eIF4A. As a key factor in cap-dependent translation initiation, eIF4A plays a role in scanning the 5′-UTR of the mRNAs for start codons.73 Notably, eIF4A inhibitors including silvestrol, hippuristanol and pateamine A exhibit potent anticancer activity.74,75 By using silvestrol in murine T-cell acute lymphoblastic leukemia (T-ALL) models and primary human T-ALL samples, Wolf et al. observed that eIF4A promotes the T-ALL development and maintenance by unwinding the G4 structures and stimulating translation of mRNAs encoding oncogenes, superenhancers-associated transcription factors and epigenetic regulators including MYC, NOTCH1, MYB, CDK6, MDM2, CCND3, ETS1, and BCL-2 (Figs. 3, 4B).76 It was, however, suggested that motifs other than 5′-UTR G4 structures may be required to render mRNA translation sensitive to eIF4A.77 To this end, mRNAs with long, but not short 5′-UTRs, appear to exhibit eIF4A-sensitivity, thereby suggesting that the length of 5′-UTR may also determine eIF4A requirement.78,79
Figure 3.
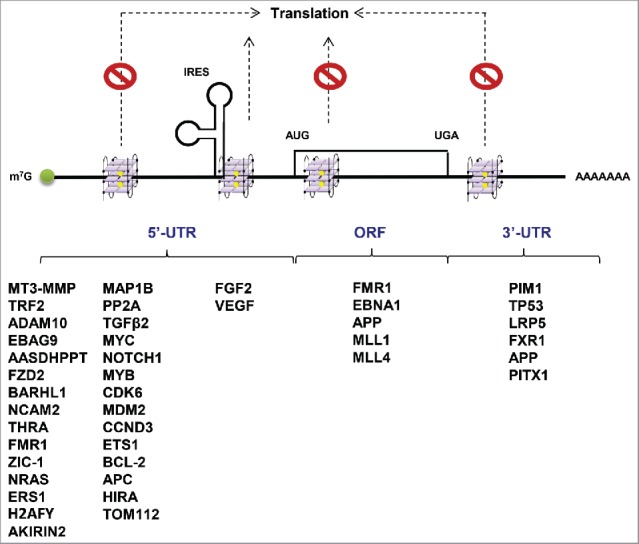
Possible roles of G-quadruplexes in mRNA translation and mRNAs that harbor G4 structures. G-quadruplexes in 5′-UTRs, ORF and 3′-UTRs mainly repress cap-dependent translation, whereas G-quadruplexes in 5′-UTR near IRESs likely enhance the IRES-mediated translation. The genes harboring G4 structures in 5′-UTRs, ORF and 3′-UTRs are listed below.
Figure 4.
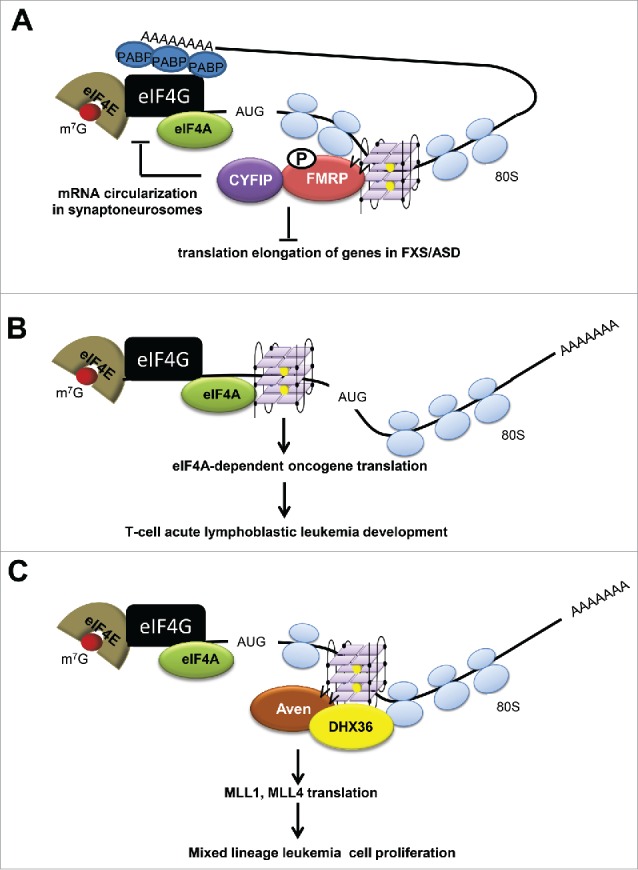
Schematic illustration of the functions of RNA binding proteins that bind RNA G4 structures in mRNAs. (A) Phosphorylated FMRP binds ORFs of mRNAs and inhibits translation. It stalls ribosomes in the elongation stage, resulting in the repressed translation of transcripts related to FXS/ASD. It recruits the co-factor CYFIP in synaptoneurosomes. By interacting with CYFIP, FMRP prevents the complex assembly between eIF4E, eIF4G and PABP, thereby inhibiting translation initiation. RGG/RG motif is denoted as vv. (B) eIF4A unwinds the G4 structures in 5′-UTR of many key transcription factors and oncogenes, thereby contributing to the T cell-acute lymphoblastic leukemia development. (C) Methylated Aven binds G4 structures in the ORFs of MLL1 and MLL4 mRNAs in an RGG/RG motif (denoted as vv) dependent manner. Aven also recruits DHX36 onto the polysomes that may facilitate unwinding of G4 structures. Thus Aven favors the translation of oncogenic proteins to increase leukemic cell proliferation.
G4 structures in 5′-UTRs also influence cap-independent, IRES-driven-translation. The IRES within the 5′-UTR of the FGF2 mRNA forms a G4 structure affecting cap-independent translation.80 Deletion analysis in human liver adenocarcinoma cells showed that the pG4 sequences are sufficient to facilitate IRES activity.81 Another example was shown by Morris et al. (2010) who reported that the hVEGF (human vascular endothelial growth factor) mRNA forms a G4 structure essential for IRES-mediated translation.82 Interestingly, it was also shown that the stabilization of the G4 structure leads to inhibition of IRES-mediated translation of VEGF-A.83 These findings show that G4 structures may influence IRES-mediated cap-independent translation, although the mechanism on how this is achieved is unclear.84
G-quadruplexes in open reading frames and translational control
The role of G4 structures in translational control has been focused mainly on G4 sequences in the 5′-UTRs. However, ORFs also contain G4 sequences and these sequences frequently encode low complexity amino acid sequences, amino acid repeats or short motifs.52 While G-quadruplexes in 5′-UTRs decreases protein expression by inhibiting ribosome scanning processes, G4 structures in ORFs contribute to other translation-related processes, such as elongation,85 ribosomal frameshift,86 no-go mRNA decay87,88 and translational folding of newly synthesized proteins.89-91 Originally, it was demonstrated that the Herpes virus thymidine kinase ORF contains pG4 sequences, leading to the expression of full-length thymidine kinase.92 Subsequently, the ORFs containing G-quadruplexes were found in FMR1 and APP (amyloid precursor protein) mRNAs.93,94 Murat et al. (2014) revealed that the mRNA encoding Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1) forms a G4 in its ORF and using antisense oligonucleotides to G4 sequences to destabilize G4 formation, they observed that the structure impairs translation elongation (Fig. 3).95 Recently, a bioinformatics analysis identified 2 mixed lineage leukemia (MLL) proto-oncogenes KMT2A (lysine methyltransferase 2A or mixed lineage leukemia, MLL1) and KMT2B (MLL4) to harbor pG4 sequences in their ORFs ∼200 nucleotides downstream of the start codon (Fig. 3).52 By performing in-line probing analysis with G-A mutagenesis and the dual luciferase reporter assay, it was confirmed that MLL1 and MLL4 mRNAs form bona-fide G4 structures that block translation by >75%.52 Endoh & Sugimoto (2016) observed that the positioning of the G4 in ORFs, but not in 5′-UTR has a dramatic impact on translational efficiency.96 These results suggest that by acting as ‘roadblocks’, G-quadruplexes in ORFs may play a significant role in protein synthesis by inhibiting ribosomal progression during elongation.
G-quadruplexes in 3′-UTR and translational control
3′ end processing is an essential process in post-transcriptional regulation through which the mRNA is endonucleolytically cleaved and a poly (A) tail is added downstream of a canonical polyadenylation signal (AAUAAA) .97,98 G-quadruplexes are located in the 3′-UTR of some mRNAs and their presence affects translational output. The 3′-UTR of the proto-oncogene PIM1 harbors a conserved pG4 sequence (Fig. 3) and Arora & Suess (2011) showed using reporter assays that this pG4 sequence inhibits translation.99 A pG4 structure in TP53 3′-UTR maintains the 3′-end processing under DNA damage and the G4 formation is critical for p53 protein expression contributing to p53-induced apoptosis.100 More recently, a study combining in silico, in vitro and in cellulo approach, demonstrated that LRP5 and FXR1 mRNAs form G4 structures in their 3′-UTR, affecting the ratio of short/long isoforms produced (Fig. 3).50 Moreover, via a bioinformatic approach and CD spectrophotometry, Crenshaw et al. (2015) identified a candidate G-quadruplex in the 3′-UTR of APP mRNA, whose overexpression leads to Alzheimer disease. The authors also showed that this G-quadruplex inhibits APP protein expression by dual luciferase reporter assay(Fig. 3).101
In addition to translational control, G-quadruplexes in the 3′-UTR also play a role in alternative splicing, polyadenylation and mRNA targeting. For instance, G4 structures in the 3′-UTR of TP53 and hTERT mRNAs regulate alternative splicing.102,103 G-quadruplex in the intron 6 of hTERT mRNA appears to act as a splicing silencer, inasmuch as a potent G-quadruplex-stabilizing agent impaired the splicing of hTERT.102 Similarly, Marcel et al. (2011) reported that the G-quadruplex in intron 3 of TP53 mRNA modulates the splicing of intron 2.103 Additionally, G4 structures within the LRP5 and FXR1 3′-UTRs increase the efficiency of alternative polyadenylation site selection, leading to the expression of shorter transcripts.50 Finally, G4 structures in 3′-UTR of 2 dendritic mRNAs PSD95 and CaMKIIa facilitate their localization in neurites.104
G-quadruplexes and trans-acting proteins
Many cellular RNAs associate with RNA-binding proteins to form ribonucleoprotein (RNP) complexes.105,106 A large number of RNA binding proteins contain RGG/RG motif107 or the related RGG/YGG motif.108 The RGG/RG motifs of nucleolin and fragile X mental retardation protein (FMRP) have been shown to associate with RNA G4 forming sequences.109,110 Some RNA G-quadruplex binding proteins are required for the unwinding of G4 structures during translation progression.111 The trans-acting factors that bind G4 sequences and their links with diseases are discussed below.
Nucleolin
Nucleolin is a 100kDa nucleolar phosphoprotein that contains an RGG/RG motif.109 Nucleolin interacts with both DNA and RNA G-rich sequences and plays a role in translational repression of specific mRNAs.107,112,113 C9orf72, a disease-related gene, harbors the expansion of a (GGGGCC)n(GGCCCC)n repeats within the first intron, leading to amyotrophic lateral sclerosis and frontotemporal dementia.114,115 It has been reported that the DNA/RNA hexanucleotide repeat expansion (HRE), (GGGGCC)n, forms extremely stable G-quadruplex structures and this may play a role in C9orf72 gene activity, protein binding as well as translation into pathologic dipeptides.116,117 It was also elucidated that the DNA of C9orf72 HRE forms antiparallel- and parallel-G-quadruplexes, while the HRE RNA sequence only adopts the parallel-stranded G4s. Nucleolin preferentially binds the HRE G-quadruplexes, which is mislocalized in patient cells carrying the C9orf72 mutation.117 These studies show DNA/RNA G4 sequences bound by nucleolin as a determinant of repeat-associated neurodegenerative diseases.
FMRP
Fragile X syndrome (FXS) is caused by expansion of (CGG)n repeats in the FMR1 gene, and subsequent hypermethylation of the FMR1 gene promoter, leading to the loss of FMRP expression.118,119 FXS is recognized to be the most-frequent heritable syndrome of mental insufficiency.120 This implicates the role of RNA G-quadruplexes in neuronal function.120 In addition to binding to the G4s, the RGG/RG motif of FMRP also modulates its association with polysomes,121 being consistent with its role in translational regulation.120,122 Originally, it was demonstrated that 432 mRNAs were co-immunoprecipitated with the FMRP RNP complex from mouse brain, nearly 70% of which contain a G4 sequence. Dramatic changes in polysome-association of these mRNAs were observed in the absence of FMRP, indicating that translational dysregulation of these mRNAs may underpin FXS.123 Recently, this was redefined by large-scale CLIP (crosslinking immunoprecipitation) studies, where 34,218 consensus FMRP binding sites in 3,703 genes were identified.124 It was reported that a G4 sequence in the FMR1 mRNA provides the docking site for FMRP. Luciferase reporter assays demonstrated that the binding of FMRP to a G4 sequence in 5′-UTR of a reporter gene strongly represses translation initiation in vitro.93 Strikingly, FMR1 G4 structures are bound by several FMRP isoforms, thereby suggesting the presence of a feedback mechanism regulating different FMRP isoform production.125 Additionally, MAP1B, PP2A and Shank1 genes that are essential for neural development have been shown to harbor one or more G-quadruplexes in their 5′-UTRs and/or 3′-UTRs, whereby the absence of FMRP increases production of the corresponding proteins, indicating that FMRP plays pivotal roles in neonatal brain development (Fig. 3).126-128 Furthermore, to explore the functions of FMRP in translational control in neurons, Napoli et al. (2008) found that FMRP inhibits cap-dependent translation by recruiting and/or stabilizing CYFIP1 in synaptoneurosomes. As a novel eIF4E binding protein, CYFIP1 binds eIF4E independently of other factors to impede cap-dependent translation of many mRNAs (Fig. 4A). FMRP is dephosphorylated in response to synaptic activation, resulting in the release from mRNAs encoding synaptic proteins and their derepression.129,130 In contrast, Bechara et al. (2009) reported a novel role of FMRP in translation activation. They observed that FMRP binds SOD1 mRNA through a motif called SoSLIP (SOD1 mRNA Stem Loops Interacting with FMRP), which competes with FMRP binding to G4 structures favoring translation.131 The solution structure of FMRP in complex with the in vitro selected G-rich RNA sequence, sc1 RNA, revealed that arginines within the RGG/RG motif of FMRP are positioned in the major groove of the G4 structure.132 Recently, X-ray crystallography analysis demonstrated that an RGG peptide of human FMRP was able to bind to the selected G-rich RNA in vitro.133 The RGG peptide was shown to stabilize G-quartets and facilitate G4 formation. It was also revealed that the specific binding of these RNAs with FMRP likely also involves the hydrogen binding with RNA duplexes, shown by mutagenesis and footprinting.133 High throughput sequencing of RNAs collected by HITS-CLIP (high-throughput sequencing-crosslinking immunoprecipitation) identified that FMRP also interacts with ORFs and stalls ribosomes on mRNA encoding presynaptic and postsynaptic proteins implicated in autism spectrum disorders (ASD) (Fig. 4A).134 Moreover, FMRP was shown to repress translation by direct binding to the L5 protein on the 80S ribosome,135 and this activity is depended on the integrity of the RGG/RG motif.136
RHAU (DHX36)
RHAU, the RNA helicase associated with AU-rich elements, also known as DHX36, is a member of ATP-dependent DEAH-box RNA helicase family.137,138 DHX36 has been shown to be the predominant G-quadruplex resolving helicase in cellulo, with high activity in unwinding RNA G4 structures.139-141 The solution structure of DHX36 recognizing a G-quadruplex was resolved and identified a 3-anchor-point electrostatic interaction.142 It was also elucidated that DHX36 uses a local, non-processive mechanism to unwind G4 structures, mimicking the DEAD-box RNA helicase eIF4A.39 By applying genome-wide analysis, DXH36 was shown to bind ∼106 RNAs, the majority harboring pG4 sequences including the human telomerase RNA.143,144 Sexton et al. (2011) also discovered the association between DHX36 and TERC (hTR) pG4 sequences in the HEK293 cells.145 Another study confirmed that the interaction between DHX36 and the TERC G-quadruplex disrupts the formation of P1 helix, a structure which defines template boundary for reverse transcription. DHX36 was sufficient to unwind the quadruplexes and promote the formation of a stable P1 helix, and DHX36 depletion led to a reduction in average telomere length.138,141 Besides the function on telomere maintenance, DHX36 regulates translation, although the precise mechanism has not yet been elucidated. By performing a RNA-co-immunoprecipitation screen, Booy et al. (2014) discovered that DHX36 interacts with G-quadruplexes in 3′-UTR of PITX1 mRNA in cellulo, and depletion of DHX36 results in increased PITX1 expression, a transcription factor with roles in development and cancer.146 Finally, it was also demonstrated that the DHX36-mediated regulation of PITX1 involves miRNA regulatory components.146 Moreover, the impact of Aven on mRNA translation regulation is dependent on DHX36 (see below).
Aven
Aven is an anti-apoptotic protein first shown to interact and stabilize the pro-survival factor Bcl-xl, while inhibiting Apaf-1, thereby stimulating survival in particular when the cells are exposed to stress.147 Aven is overexpressed in acute myeloid and acute lymphoblastic leukemia where its expression has been associated with poor prognosis.148,149 Recently, Aven was shown to bind RNAs with G4 structures via its RGG/RG motif.52 The RGG/RG motif of Aven is arginine methylated by PRMT1 and this promotes association with the methylarginine interactors SMN and TDRD3. The arginine methylation and binding to SMN and TDRD3 is required for association of Aven with polysomes. In vitro binding and photocrosslinking immunoprecipitation assays revealed that Aven associates with MLL1 and MLL4 mRNAs. Interestingly, the G4 sequences bound by Aven were in the coding regions regulating their mRNA translation (Fig. 4C). Mechanistically, Aven recruits DHX36 onto the polysomes likely to facilitate unwinding of the G4 structures during translation. Depletion of Aven/DHX36 inhibits KMT2A/KMT2B translation, whereas luciferase assays with G-to-A mutagenesis revealed that both Aven and DHX36 are required to rescue translation in the presence of G4 motifs.52 Thus, Aven represents a G4 interacting protein that recruits DHX36 to unwind G4 structures containing mRNAs during translational elongation. These findings foster our understanding that RNA G4 binding proteins can play a key role in modulating mRNA translation.
Conclusions and perspectives
Increasing evidence shows that G-quadruplexes play essential roles in RNA metabolism. Advances in computational analyses and genome-wide sequencing have facilitated the characterization of RNA quadruplexes and their implication in translational control. Further advancements are required to confirm their occurrence in vivo. Moreover, the trans-acting factors that bind G4 structures are the key to understanding the role of G-quadruplexes in the regulation of mRNA fate. Nucleolin, FMRP, DHX36 and Aven represent the tip of the iceberg of trans-acting factors that recognize G4 RNA structures. Large scale analyses including HITS-CLIP-seq, SHAPE-seq, and HITS-RAP (high-throughput RNA affinity profiling) hold great promise for knowledge advancement as to the role of G4 structures and interacting proteins in vivo.150-154 It is possible that multiple RNA binding proteins recognize one G-quadruplex, and there could be functional interplay. The binding of different RNA binding proteins are likely to: 1) stabilize the G4 structures, 2) unwind the G4s by recruiting helicases, 3) act as chaperones to transport G4 containing mRNAs, and 4) serve as scaffold proteins to recruit other proteins or RNAs.
A growing number of disease-related genes are regulated by G4 structures and their RNA binding proteins. This includes mRNAs encoding the tumor suppressor TP53,100 oncogene NRAS,47,53 oncogenes KMT2A/KMT2B,52 anti-apoptotic Bcl-2,67,76 FMR1,125,155 and the telomerase hTERT.102,156 This indicates that by affecting mRNA translation, splicing, and polyadenylation, G4 structures may be implicated in numerous human diseases, especially cancer. Thus, targeting G4 structures with synthesized small molecules is attractive to modify oncogene expression. Extensive studies have evaluated some “drug-like” molecules including the pyridostatins (PDS) ,157 cationic porphyrins and derivatives158-160 and bisquinolinium compounds.68 Short antisense oligonucleotides affect RNA G4 folding and translation regulation of specific mRNAs72 and may eventually be of therapeutic potential. A new polyaromatic molecule, RGB-1, was recently shown to specifically stabilize RNA G-quadruplexes, but not DNA G-quadruplexes or other RNA structures. RGB-1 inhibits translation in mammalian cells and decreases NRAS expression, providing a new tool to understand G4 structures and therapeutic applications.161 Thus, a better understanding of the various contributions of G4 structures in the human transcriptome may provide important insights into strategies targeting G4s in molecular medicine.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This manuscript was funded by a grant from the Canadian Institute of Health Canada (CIHR) MOP-67070 to S.R.
References
- [1].Bang I. Untersuchungen über die Guanylsäure. Biochemische Zeitschrift. 1910; 26:293-311. [Google Scholar]
- [2].Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A 1962; 48:2013-8; PMID:13947099; http://dx.doi.org/ 10.1073/pnas.48.12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lipps HJ, Gruissem W, Prescott DM. Higher order DNA structure in macronuclear chromatin of the hypotrichous ciliate Oxytricha nova. Proc Natl Acad Sci U S A 1982; 79:2495-9; PMID:6806811; http://dx.doi.org/ 10.1073/pnas.79.8.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989; 342:825-9; PMID:2601741; http://dx.doi.org/ 10.1038/342825a0 [DOI] [PubMed] [Google Scholar]
- [5].Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci U S A 1993; 90:3393-7; PMID:8475087; http://dx.doi.org/ 10.1073/pnas.90.8.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tuesuwan B, Kern JT, Thomas PW, Rodriguez M, Li J, David WM, Kerwin SM. Simian virus 40 large T-antigen G-quadruplex DNA helicase inhibition by G-quadruplex DNA-interactive agents. Biochemistry 2008; 47:1896-909; PMID:18205402; http://dx.doi.org/ 10.1021/bi701747d [DOI] [PubMed] [Google Scholar]
- [7].Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 2012; 13:770-80; PMID:23032257; http://dx.doi.org/ 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mendoza O, Bourdoncle A, Boule JB, Brosh RM Jr., Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res 2016; 44:1989-2006; PMID:26883636; http://dx.doi.org/ 10.1093/nar/gkw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 2015; 43:8627-37; PMID:26350216; http://dx.doi.org/ 10.1093/nar/gkv862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genet Dev 2014; 25:22-9; PMID:24584093; http://dx.doi.org/ 10.1016/j.gde.2013.10.012 [DOI] [PubMed] [Google Scholar]
- [11].Bugaut A, Balasubramanian S. 5'-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res 2012; 40:4727-41; PMID:22351747; http://dx.doi.org/ 10.1093/nar/gks068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA 2012; 3:495-507. [DOI] [PubMed] [Google Scholar]
- [13].Mukundan VT, Phan AT. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J Am Chem Soc 2013; 135:5017-28; PMID:23521617; http://dx.doi.org/ 10.1021/ja310251r [DOI] [PubMed] [Google Scholar]
- [14].Beaudoin JD, Jodoin R, Perreault JP. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res 2014; 42:1209-23; PMID:24121682; http://dx.doi.org/ 10.1093/nar/gkt904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res 2005; 33:2908-16; PMID:15914667; http://dx.doi.org/ 10.1093/nar/gki609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res 2005; 33:2901-7; PMID:15914666; http://dx.doi.org/ 10.1093/nar/gki553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rodriguez R, Miller KM. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nat Rev Genet 2014; 15:783-96; PMID:25311424; http://dx.doi.org/ 10.1038/nrg3796 [DOI] [PubMed] [Google Scholar]
- [18].Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 2015; 33:877-81; PMID:26192317; http://dx.doi.org/ 10.1038/nbt.3295 [DOI] [PubMed] [Google Scholar]
- [19].Frees S, Menendez C, Crum M, Bagga PS. QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs. Hum Genomics 2014; 8:8; PMID:24885782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Konig SL, Evans AC, Huppert JL. Seven essential questions on G-quadruplexes. Biomolecular concepts 2010; 1:197-213; PMID:25961997; http://dx.doi.org/ 10.1515/bmc.2010.011 [DOI] [PubMed] [Google Scholar]
- [21].Lam EY, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun 2013; 4:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Healy KC. Telomere dynamics and telomerase activation in tumor progression: prospects for prognosis and therapy. Oncol Res 1995; 7:121-30; PMID:8555645 [PubMed] [Google Scholar]
- [23].Maizels N, Gray LT. The G4 genome. PLoS genetics 2013; 9:e1003468; PMID:23637633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang Q, Liu JQ, Chen Z, Zheng KW, Chen CY, Hao YH, Tan Z. G-quadruplex formation at the 3' end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res 2011; 39:6229-37; PMID:21441540; http://dx.doi.org/ 10.1093/nar/gkr164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shalaby T, Fiaschetti G, Nagasawa K, Shin-ya K, Baumgartner M, Grotzer M. G-quadruplexes as potential therapeutic targets for embryonal tumors. Molecules 2013; 18:12500-37; PMID:24152672; http://dx.doi.org/ 10.3390/molecules181012500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Crees Z, Girard J, Rios Z, Botting GM, Harrington K, Shearrow C, Wojdyla L, Stone AL, Uppada SB, Devito JT, et al.. Oligonucleotides and G-quadruplex stabilizers: targeting telomeres and telomerase in cancer therapy. Curr Pharm Des 2014; 20:6422-37; PMID:24975605; http://dx.doi.org/ 10.2174/1381612820666140630100702 [DOI] [PubMed] [Google Scholar]
- [27].Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, Lovrecz GO, Beck JL, Bryan TM. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun 2015; 6:7643; PMID:26158869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug discov 2011; 10:261-75; PMID:21455236; http://dx.doi.org/ 10.1038/nrd3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J 2014; 33:732-46; PMID:24521668; http://dx.doi.org/ 10.1002/embj.201387506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J 2010; 277:3459-69; PMID:20670278; http://dx.doi.org/ 10.1111/j.1742-4658.2010.07759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Morgan RK, Batra H, Gaerig VC, Hockings J, Brooks TA. Identification and characterization of a new G-quadruplex forming region within the kRAS promoter as a transcriptional regulator. Biochimica et biophysica acta 2016; 1859:235-45; PMID:26597160; http://dx.doi.org/ 10.1016/j.bbagrm.2015.11.004 [DOI] [PubMed] [Google Scholar]
- [32].Onel B, Carver M, Wu G, Timonina D, Kalarn S, Larriva M, Yang D. A New G-Quadruplex with Hairpin Loop Immediately Upstream of the Human BCL2 P1 Promoter Modulates Transcription. J Am Chem Soc 2016; 138:2563-70; PMID:26841249; http://dx.doi.org/ 10.1021/jacs.5b08596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Farhath MM, Thompson M, Ray S, Sewell A, Balci H, Basu S. G-Quadruplex-Enabling Sequence within the Human Tyrosine Hydroxylase Promoter Differentially Regulates Transcription. Biochemistry 2015; 54:5533-45; PMID:26284527; http://dx.doi.org/ 10.1021/acs.biochem.5b00209 [DOI] [PubMed] [Google Scholar]
- [34].Cayrou C, Coulombe P, Puy A, Rialle S, Kaplan N, Segal E, Mechali M. New insights into replication origin characteristics in metazoans. Cell Cycle 2012; 11:658-67; PMID:22373526; http://dx.doi.org/ 10.4161/cc.11.4.19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al.. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 2011; 21:1438-49; PMID:21750104; http://dx.doi.org/ 10.1101/gr.121830.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 2012; 19:837-44; PMID:22751019; http://dx.doi.org/ 10.1038/nsmb.2339 [DOI] [PubMed] [Google Scholar]
- [37].Foulk MS, Urban JM, Casella C, Gerbi SA. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res 2015; 25:725-35; PMID:25695952; http://dx.doi.org/ 10.1101/gr.183848.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zheng KW, Xiao S, Liu JQ, Zhang JY, Hao YH, Tan Z. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res 2013; 41:5533-41; PMID:23585281; http://dx.doi.org/ 10.1093/nar/gkt264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen MC, Murat P, Abecassis K, Ferre-D'Amare AR, Balasubramanian S. Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase. Nucleic Acids Res 2015; 43:2223-31; PMID:25653156; http://dx.doi.org/ 10.1093/nar/gkv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA repair 2011; 10:654-65; PMID:21561811; http://dx.doi.org/ 10.1016/j.dnarep.2011.04.013 [DOI] [PubMed] [Google Scholar]
- [41].Xu Y, Kaminaga K, Komiyama M. Human telomeric RNA in G-quadruplex structure. Nucleic Acids Symp Ser (Oxf) 2008; 175-6. [DOI] [PubMed] [Google Scholar]
- [42].Takahama K, Takada A, Tada S, Shimizu M, Sayama K, Kurokawa R, Oyoshi T. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol 2013; 20:341-50; PMID:23521792; http://dx.doi.org/ 10.1016/j.chembiol.2013.02.013 [DOI] [PubMed] [Google Scholar]
- [43].Xu Y, Suzuki Y, Ito K, Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc Natl Acad Sci U S A 2010; 107:14579-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Martadinata H, Phan AT. Structure of human telomeric RNA (TERRA): stacking of two G-quadruplex blocks in K(+) solution. Biochemistry 2013; 52:2176-83; PMID:23445442; http://dx.doi.org/ 10.1021/bi301606u [DOI] [PubMed] [Google Scholar]
- [45].Gros J, Guedin A, Mergny JL, Lacroix L. G-Quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembiochem 2008; 9:2075-9; PMID:18683270; http://dx.doi.org/ 10.1002/cbic.200800300 [DOI] [PubMed] [Google Scholar]
- [46].Martadinata H, Phan AT. Formation of a stacked dimeric G-quadruplex containing bulges by the 5'-terminal region of human telomerase RNA (hTERC). Biochemistry 2014; 53:1595-600; PMID:24601523; http://dx.doi.org/ 10.1021/bi4015727 [DOI] [PubMed] [Google Scholar]
- [47].Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol 2007; 3:218-21; PMID:17322877; http://dx.doi.org/ 10.1038/nchembio864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Beaudoin JD, Perreault JP. 5'-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res 2010; 38:7022-36; PMID:20571090; http://dx.doi.org/ 10.1093/nar/gkq557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jodoin R, Bauer L, Garant JM, Mahdi Laaref A, Phaneuf F, Perreault JP. The folding of 5'-UTR human G-quadruplexes possessing a long central loop. RNA 2014; 20:1129-41; PMID:24865610; http://dx.doi.org/ 10.1261/rna.044578.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Beaudoin JD, Perreault JP. Exploring mRNA 3'-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res 2013; 41:5898-911; PMID:23609544; http://dx.doi.org/ 10.1093/nar/gkt265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res 2008; 36:6260-8; PMID:18832370; http://dx.doi.org/ 10.1093/nar/gkn511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Thandapani P, Song J, Gandin V, Cai Y, Rouleau SG, Garant JM, Boisvert FM, Yu Z, Perreault JP, Topisirovic I, et al.. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. eLife 2015; 4; PMID:26267306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem 2014; 6:75-80; PMID:24345950; http://dx.doi.org/ 10.1038/nchem.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol 2012; 4; PMID:23209153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science 2016; 352:1413-6; PMID:27313038; http://dx.doi.org/ 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Ann Rev Biochem 2014; 83:779-812 [DOI] [PubMed] [Google Scholar]
- [57].Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/ 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6:318-27; PMID:15803138; http://dx.doi.org/ 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- [60].Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J 1992; 11:4153-8; PMID:1396596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 1985; 40:515-26; PMID:2982496; http://dx.doi.org/ 10.1016/0092-8674(85)90200-4 [DOI] [PubMed] [Google Scholar]
- [62].Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 2011; 10:229-40; PMID:21220943; http://dx.doi.org/ 10.4161/cc.10.2.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Gen Dev 2001; 15:1593-612; PMID:11445534; http://dx.doi.org/ 10.1101/gad.891101 [DOI] [PubMed] [Google Scholar]
- [64].Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA 2008; 14:1290-6; PMID:18515550; http://dx.doi.org/ 10.1261/rna.1001708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Morris MJ, Basu S. An unusually stable G-quadruplex within the 5'-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry 2009; 48:5313-9; PMID:19397366; http://dx.doi.org/ 10.1021/bi900498z [DOI] [PubMed] [Google Scholar]
- [66].Balkwill GD, Derecka K, Garner TP, Hodgman C, Flint AP, Searle MS. Repression of translation of human estrogen receptor alpha by G-quadruplex formation. Biochemistry 2009; 48:11487-95; PMID:19860473; http://dx.doi.org/ 10.1021/bi901420k [DOI] [PubMed] [Google Scholar]
- [67].Shahid R, Bugaut A, Balasubramanian S. The BCL-2 5' untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry 2010; 49:8300-6; PMID:20726580; http://dx.doi.org/ 10.1021/bi100957h [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gomez D, Guedin A, Mergny JL, Salles B, Riou JF, Teulade-Fichou MP, Calsou P. A G-quadruplex structure within the 5'-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res 2010; 38:7187-98; PMID:20571083; http://dx.doi.org/ 10.1093/nar/gkq563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lammich S, Kamp F, Wagner J, Nuscher B, Zilow S, Ludwig AK, Willem M, Haass C. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5'-untranslated region. J Biol Chem 2011; 286:45063-72; PMID:22065584; http://dx.doi.org/ 10.1074/jbc.M111.296921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dai J, Liu ZQ, Wang XQ, Lin J, Yao PF, Huang SL, Ou TM, Tan JH, Li D, Gu LQ, et al.. Discovery of Small Molecules for Up-Regulating the Translation of Antiamyloidogenic Secretase, a Disintegrin and Metalloproteinase 10 (ADAM10), by Binding to the G-Quadruplex-Forming Sequence in the 5' Untranslated Region (UTR) of Its mRNA. J Med Chem 2015; 58:3875-91; PMID:25822852; http://dx.doi.org/ 10.1021/acs.jmedchem.5b00139 [DOI] [PubMed] [Google Scholar]
- [71].Agarwala P, Pandey S, Mapa K, Maiti S. The G-quadruplex augments translation in the 5' untranslated region of transforming growth factor beta2. Biochemistry 2013; 52:1528-38; PMID:23387555; http://dx.doi.org/ 10.1021/bi301365g [DOI] [PubMed] [Google Scholar]
- [72].Rouleau SG, Beaudoin JD, Bisaillon M, Perreault JP. Small antisense oligonucleotides against G-quadruplexes: specific mRNA translational switches. Nucleic Acids Res 2015; 43:595-606; PMID:25510493; http://dx.doi.org/ 10.1093/nar/gku1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov 2015; 14:261-78; PMID:25743081; http://dx.doi.org/ 10.1038/nrd4505 [DOI] [PubMed] [Google Scholar]
- [74].Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, et al.. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 2011; 208:1799-807; PMID:21859846; http://dx.doi.org/ 10.1084/jem.20110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr., et al.. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Investig 2008; 118:2651-60; PMID:18551192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al.. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014; 513:65-70; PMID:25079319; http://dx.doi.org/ 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Gen Biol 2014; 15:476; PMID:25273840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al.. nanoCAGE reveals 5' UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Gen Res 2016; 26:636-48; PMID:26984228; http://dx.doi.org/ 10.1101/gr.197566.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 2015; 21:479-92; PMID:25738462; http://dx.doi.org/ 10.1016/j.cmet.2015.02.010 [DOI] [PubMed] [Google Scholar]
- [80].Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem 2003; 278:39330-6; PMID:12857733; http://dx.doi.org/ 10.1074/jbc.M305580200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA 2006; 12:1755-85; PMID:16957278; http://dx.doi.org/ 10.1261/rna.157806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc 2010; 132:17831-9; PMID:21105704; http://dx.doi.org/ 10.1021/ja106287x [DOI] [PubMed] [Google Scholar]
- [83].Cammas A, Dubrac A, Morel B, Lamaa A, Touriol C, Teulade-Fichou MP, Prats H, Millevoi S. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biol 2015; 12:320-9; PMID:25826664; http://dx.doi.org/ 10.1080/15476286.2015.1017236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jackson RJ. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 2013; 5:a011569; http://dx.doi.org/ 10.1101/cshperspect.a011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Endoh T, Kawasaki Y, Sugimoto N. Translational halt during elongation caused by G-quadruplex formed by mRNA. Methods 2013; 64:73-8; PMID:23747335; http://dx.doi.org/ 10.1016/j.ymeth.2013.05.026 [DOI] [PubMed] [Google Scholar]
- [86].Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res 2009; 139:193-208; PMID:18621088; http://dx.doi.org/ 10.1016/j.virusres.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006; 440:561-4; PMID:16554824; http://dx.doi.org/ 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Harigaya Y, Parker R. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA 2010; 1:132-41. [DOI] [PubMed] [Google Scholar]
- [89].Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mole Biol 2009; 16:274-80; PMID:19198590; http://dx.doi.org/ 10.1038/nsmb.1554 [DOI] [PubMed] [Google Scholar]
- [90].O'Brien EP, Vendruscolo M, Dobson CM. Prediction of variable translation rate effects on cotranslational protein folding. Nat Commun 2012; 3:868. [DOI] [PubMed] [Google Scholar]
- [91].Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci 2009; 34:16-24. [DOI] [PubMed] [Google Scholar]
- [92].Horsburgh BC, Kollmus H, Hauser H, Coen DM. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell 1996; 86:949-59; PMID:8808630; http://dx.doi.org/ 10.1016/S0092-8674(00)80170-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J 2001; 20:4803-13; PMID:11532944; http://dx.doi.org/ 10.1093/emboj/20.17.4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol 2007; 5:e52; PMID:17298186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Murat P, Zhong J, Lekieffre L, Cowieson NP, Clancy JL, Preiss T, Balasubramanian S, Khanna R, Tellam J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat Chem Biol 2014; 10:358-64; PMID:24633353; http://dx.doi.org/ 10.1038/nchembio.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Endoh T, Sugimoto N. Mechanical insights into ribosomal progression overcoming RNA G-quadruplex from periodical translation suppression in cells. Scientific Rep 2016; 6:22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic Acids Res 2010; 38:2757-74; PMID:20044349; http://dx.doi.org/ 10.1093/nar/gkp1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Gen Dev 1997; 11:2755-66; PMID:9353246; http://dx.doi.org/ 10.1101/gad.11.21.2755 [DOI] [PubMed] [Google Scholar]
- [99].Arora A, Suess B. An RNA G-quadruplex in the 3' UTR of the proto-oncogene PIM1 represses translation. RNA Biol 2011; 8:802-5; PMID:21734463; http://dx.doi.org/ 10.4161/rna.8.5.16038 [DOI] [PubMed] [Google Scholar]
- [100].Decorsiere A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3'-end processing and function during DNA damage. Gen Dev 2011; 25:220-5; PMID:21289067; http://dx.doi.org/ 10.1101/gad.607011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Crenshaw E, Leung BP, Kwok CK, Sharoni M, Olson K, Sebastian NP, Ansaloni S, Schweitzer-Stenner R, Akins MR, Bevilacqua PC, et al.. Amyloid Precursor Protein Translation Is Regulated by a 3'UTR Guanine Quadruplex. PloS one 2015; 10:e0143160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res 2004; 32:371-9; PMID:14729921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Marcel V, Tran PL, Sagne C, Martel-Planche G, Vaslin L, Teulade-Fichou MP, Hall J, Mergny JL, Hainaut P, Van Dyck E. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 2011; 32:271-8; PMID:21112961; http://dx.doi.org/ 10.1093/carcin/bgq253 [DOI] [PubMed] [Google Scholar]
- [104].Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep 2011; 12:697-704; PMID:21566646; http://dx.doi.org/ 10.1038/embor.2011.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Calabretta S, Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem Sci 2015; 40:662-72; PMID:26481498; http://dx.doi.org/ 10.1016/j.tibs.2015.08.012 [DOI] [PubMed] [Google Scholar]
- [106].Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet 2008; 24:416-25; PMID:18597886; http://dx.doi.org/ 10.1016/j.tig.2008.05.004 [DOI] [PubMed] [Google Scholar]
- [107].Thandapani P, O'Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell 2013; 50:613-23; PMID:23746349; http://dx.doi.org/ 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- [108].Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012; 149:1393-406; PMID:22658674; http://dx.doi.org/ 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- [109].Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem 1999; 274:15908-12; PMID:10336496; http://dx.doi.org/ 10.1074/jbc.274.22.15908 [DOI] [PubMed] [Google Scholar]
- [110].Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001;107:489-99; PMID:11719189; http://dx.doi.org/ 10.1016/S0092-8674(01)00566-9 [DOI] [PubMed] [Google Scholar]
- [111].Sissi C, Gatto B, Palumbo M. The evolving world of protein-G-quadruplex recognition: a medicinal chemist's perspective. Biochimie 2011; 93:1219-30; PMID:21549174; http://dx.doi.org/ 10.1016/j.biochi.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM, Selimyan R, Martindale JL, Yang X, Carrier F, et al.. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res 2011; 39:8513-30; PMID:21737422; http://dx.doi.org/ 10.1093/nar/gkr488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Abdelmohsen K, Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol 2012; 9:799-808; PMID:22617883; http://dx.doi.org/ 10.4161/rna.19718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Turner MR, Hardiman O, Benatar M, Brooks BR, Chio A, de Carvalho M, Ince PG, Lin C, Miller RG, Mitsumoto H, et al.. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol 2013; 12:310-22; PMID:23415570; http://dx.doi.org/ 10.1016/S1474-4422(13)70036-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72:245-56; PMID:21944778; http://dx.doi.org/ 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Reddy K, Zamiri B, Stanley SY, Macgregor RB Jr., Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem 2013; 288:9860-6; PMID:23423380; http://dx.doi.org/ 10.1074/jbc.C113.452532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al.. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014; 507:195-200; PMID:24598541; http://dx.doi.org/ 10.1038/nature13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci 2013; 16:1530-6; PMID:23584741; http://dx.doi.org/ 10.1038/nn.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 2014; 343:1002-5; PMID:24578575; http://dx.doi.org/ 10.1126/science.1245831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci 2015; 16:595-605; PMID:26350240; http://dx.doi.org/ 10.1038/nrn4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum Mol Genet 2010; 19:1314-23; PMID:20064924; http://dx.doi.org/ 10.1093/hmg/ddq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet 1997; 6:1465-72; PMID:9285783; http://dx.doi.org/ 10.1093/hmg/6.9.1465 [DOI] [PubMed] [Google Scholar]
- [123].Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al.. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 2001; 107:477-87; PMID:11719188; http://dx.doi.org/ 10.1016/S0092-8674(01)00568-2 [DOI] [PubMed] [Google Scholar]
- [124].Anderson BR, Chopra P, Suhl JA, Warren ST, Bassell GJ. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res 2016; 44(14): 6649-6659; http://dx.doi.org/ 10.1093/nar/gkw593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Blice-Baum AC, Mihailescu MR. Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms. RNA 2014; 20:103-14; PMID:24249225; http://dx.doi.org/ 10.1261/rna.041442.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Lu R, Wang H, Liang Z, Ku L, O'Donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A 2004; 101:15201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet 2005; 14:835-44; PMID:15703194; http://dx.doi.org/ 10.1093/hmg/ddi077 [DOI] [PubMed] [Google Scholar]
- [128].Zhang Y, Gaetano CM, Williams KR, Bassell GJ, Mihailescu MR. FMRP interacts with G-quadruplex structures in the 3'-UTR of its dendritic target Shank1 mRNA. RNA Biol 2014; 11:1364-74; PMID:25692235; http://dx.doi.org/ 10.1080/15476286.2014.996464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al.. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008; 134:1042-54; PMID:18805096; http://dx.doi.org/ 10.1016/j.cell.2008.07.031 [DOI] [PubMed] [Google Scholar]
- [130].Simone R, Fratta P, Neidle S, Parkinson GN, Isaacs AM. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett 2015; 589:1653-68; PMID:25979174; http://dx.doi.org/ 10.1016/j.febslet.2015.05.003 [DOI] [PubMed] [Google Scholar]
- [131].Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, et al.. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol 2009; 7:e16; PMID:19166269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al.. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol 2011; 18:796-804; PMID:21642970; http://dx.doi.org/ 10.1038/nsmb.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Vasilyev N, Polonskaia A, Darnell JC, Darnell RB, Patel DJ, Serganov A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc Natl Acad Sci U S A 2015; 112:E5391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al.. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011; 146:247-61; PMID:21784246; http://dx.doi.org/ 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell 2014; 54:407-17; PMID:24746697; http://dx.doi.org/ 10.1016/j.molcel.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Taha MS, Nouri K, Milroy LG, Moll JM, Herrmann C, Brunsveld L, Piekorz RP, Ahmadian MR. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PloS one 2014; 9:e91465; http://dx.doi.org/ 10.1371/journal.pone.0091465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J Biol Chem 2005; 280:38117-20; PMID:16150737; http://dx.doi.org/ 10.1074/jbc.C500348200 [DOI] [PubMed] [Google Scholar]
- [138].Booy EP, McRae EK, McKenna SA. Biochemical characterization of G4 quadruplex telomerase RNA unwinding by the RNA helicase RHAU. Methods Mol Biol 2015; 1259:125-35; PMID:25579584; http://dx.doi.org/ 10.1007/978-1-4939-2214-7_9 [DOI] [PubMed] [Google Scholar]
- [139].Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J Biol Chem 2008; 283:34626-34; PMID:18842585; http://dx.doi.org/ 10.1074/jbc.M806277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem 2008; 283:35186-98; PMID:18854321; http://dx.doi.org/ 10.1074/jbc.M804857200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Booy EP, Meier M, Okun N, Novakowski SK, Xiong S, Stetefeld J, McKenna SA. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res 2012; 40:4110-24; PMID:22238380; http://dx.doi.org/ 10.1093/nar/gkr1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Heddi B, Cheong VV, Martadinata H, Phan AT. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc Natl Acad Sci U S A 2015; 112:9608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res 2010; 38:6219-33; PMID:20472641; http://dx.doi.org/ 10.1093/nar/gkq372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res 2011; 39:9390-404; PMID:21846770; http://dx.doi.org/ 10.1093/nar/gkr630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Sexton AN, Collins K. The 5' guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol Cell Biol 2011; 31:736-43; PMID:21149580; http://dx.doi.org/ 10.1128/MCB.01033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Booy EP, Howard R, Marushchak O, Ariyo EO, Meier M, Novakowski SK, Deo SR, Dzananovic E, Stetefeld J, McKenna SA. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res 2014; 42:3346-61; PMID:24369427; http://dx.doi.org/ 10.1093/nar/gkt1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Chau BN, Cheng EH, Kerr DA, Hardwick JM. Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol Cell 2000; 6:31-40; PMID:10949025; http://dx.doi.org/ 10.1016/S1097-2765(05)00021-3 [DOI] [PubMed] [Google Scholar]
- [148].Paydas S, Tanriverdi K, Yavuz S, Disel U, Sahin B, Burgut R. Survivin and aven: two distinct antiapoptotic signals in acute leukemias. Ann Oncol 2003; 14:1045-50; PMID:12853345 [DOI] [PubMed] [Google Scholar]
- [149].Choi J, Hwang YK, Sung KW, Kim DH, Yoo KH, Jung HL, Koo HH. Aven overexpression: association with poor prognosis in childhood acute lymphoblastic leukemia. Leukemia Res 2006; 30:1019-25; PMID:16388850; http://dx.doi.org/ 10.1016/j.leukres.2005.11.001 [DOI] [PubMed] [Google Scholar]
- [150].Tome JM, Ozer A, Pagano JM, Gheba D, Schroth GP, Lis JT. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat methods 2014; 11:683-8; PMID:24809628; http://dx.doi.org/ 10.1038/nmeth.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature protocols 2014; 9:263-93; PMID:24407355; http://dx.doi.org/ 10.1038/nprot.2014.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 2009; 106:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nature methods 2014; 11:959-65; PMID:25028896; http://dx.doi.org/ 10.1038/nmeth.3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Xu W, Bolduc F, Hong N, Perreault JP. The use of a combination of computer-assisted structure prediction and SHAPE probing to elucidate the secondary structures of five viroids. Mol Plant Pathol 2012; 13:666-76; PMID:22243942; http://dx.doi.org/ 10.1111/j.1364-3703.2011.00776.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Didiot MC, Tian Z, Schaeffer C, Subramanian M, Mandel JL, Moine H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res 2008; 36:4902-12; PMID:18653529; http://dx.doi.org/ 10.1093/nar/gkn472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bidzinska J, Cimino-Reale G, Zaffaroni N, Folini M. G-quadruplex structures in the human genome as novel therapeutic targets. Molecules 2013; 18:12368-95; PMID:24108400; http://dx.doi.org/ 10.3390/molecules181012368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bugaut A, Rodriguez R, Kumari S, Hsu ST, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org Biomol Chem 2010; 8:2771-6; PMID:20436976; http://dx.doi.org/ 10.1039/c002418j [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Huang XX, Zhu LN, Wu B, Huo YF, Duan NN, Kong DM. Two cationic porphyrin isomers showing different multimeric G-quadruplex recognition specificity against monomeric G-quadruplexes. Nucleic Acids Res 2014; 42:8719-31; PMID:24939896; http://dx.doi.org/ 10.1093/nar/gku526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Morris MJ, Wingate KL, Silwal J, Leeper TC, Basu S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res 2012; 40:4137-45; PMID:22266651; http://dx.doi.org/ 10.1093/nar/gkr1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Faudale M, Cogoi S, Xodo LE. Photoactivated cationic alkyl-substituted porphyrin binding to g4-RNA in the 5'-UTR of KRAS oncogene represses translation. Chem Commun (Camb) 2012; 48:874-6; PMID:22127206 [DOI] [PubMed] [Google Scholar]
- [161].Katsuda Y, Sato S, Asano L, Morimura Y, Furuta T, Sugiyama H, Hagihara M, Uesugi M. A Small Molecule That Represses Translation of G-Quadruplex-Containing mRNA. J Am Chem Soc 2016; 138:9037-40; PMID:27410677; http://dx.doi.org/ 10.1021/jacs.6b04506 [DOI] [PubMed] [Google Scholar]


