Abstract
We have investigated the lipid A of Francisella tularensis subsp. holarctica strain 1547-57, a type B strain, by using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry, nanoelectrospray quadrupole ion-trap mass spectrometry, and chemical methods. In accordance with the previously published structures of the lipid A from F. tularensis live vaccine strain (LVS) (ATCC 29684) (E. Vinogradov et al., Eur. J. Biochem. 269:6112-6118, 2002), all of the major lipid A forms from strain 1547-57 were tetraacylated. As in the LVS strain, the major fatty acids detected in the F. tularensis 1547-57 lipid A sample included 3-hydroxyoctadecanoic acid, 3-hydroxyhexadecanoic acid, hexadecanoic acid, and tetradecanoic acid. However, several of the lipid A components present in strain 1547-57 were of higher molecular weight than the previously published structures. A major component with an Mr of 1,666 was found to contain three C18:0(3-OH) fatty acids, one C16:0 fatty acid, one phosphate group, and one 161-Da moiety. This 161-Da moiety could be removed from the lipid A by treatment with aqueous hydrofluoric acid and was identified as galactosamine following peracetylation and analysis by gas chromatography-mass spectrometry. Detailed investigations of the Mr-1,666 species by ion-trap mass spectrometry with multiple stages of fragmentation suggested that the galactosamine-1-phosphate was linked to the reducing terminus of the lipid A. Similar to the modification of lipid A with arabinosamine, lipopolysaccharide species from F. tularensis containing a phosphate-linked galactosamine could potentially influence its intracellular survival by conferring resistance to antimicrobial peptides.
Francisella tularensis is an encapsulated gram-negative bacterium that causes tularemia, a severe disease of humans and other mammals (12, 24). Severity of disease depends on the host immune response and the route of infection, including intradermal inoculation via zoonotic transmission, ingestion of contaminated meat or water, and inhalation (7). F. tularensis is a facultative intracellular bacterium, and several studies have shown that a cell-mediated immune response may be required for controlling the infection (22, 24). Recently, interest in F. tularensis has increased because of its suitability for use as an agent of biological warfare (6).
F. tularensis possesses a lipopolysaccharide (LPS) that, when compared to Escherichia coli LPS, is not biologically active (1, 8, 23). The only biological activity attributed to F. tularensis LPS in vitro is the ability to activate complement (8). The relatively nonendotoxic nature of the LPS is putatively attributed to the unusual structure of the lipid A molecule. A recent study showed that the lipid A molecule of F. tularensis live vaccine strain (LVS) is not only tetraacylated but also lacks phosphate substituents (27). These structural features may contribute to its low toxicity. It has been shown that mutations affecting the degree of acylation or the acylation pattern of lipid A reduce the endotoxicity of LPS (21).
In this report, we have investigated the structure of the lipid A molecule of F. tularensis subsp. holarctica strain 1547-57, a type B strain. In agreement with the published structure for the LVS strain, we show that the lipid A molecule of this virulent clinical isolate is tetraacylated in vitro. The major lipid A forms described for the LVS strain were seen as components of our F. tularensis strain 1547-57 lipid A preparation. However, in F. tularensis strain 1547-57, we also found several higher-molecular-weight species. These constituents included monophosphorylated lipid A forms and an unusual lipid A species substituted with a galactosamine-1-phosphate moiety. Here we describe the application of various chemical and mass spectrometric methods to the structural characterization of this novel F. tularensis lipid A structure.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
F. tularensis subsp. holarctica strain 1547-57 (type B) was grown on chocolate agar supplemented with IsoVitaleX (Becton Dickinson, Cockeysville, Md.). F. tularensis LVS (ATCC 29684) was grown on cystine heart medium (Difco Laboratories, Detroit, Mich.) supplemented with 9% sheep blood (Colorado Serum Co., Denver, Colo.). Both strains were incubated at 37°C in a 5% CO2 atmosphere for 48 h.
Isolation of LPS.
Bacteria from 10 agar plates were collected by flooding plates with phosphate-buffered saline and scraping colonies from the surface. Samples were centrifuged, and cell pellets were washed twice with 0.15 M NaCl and suspended in 25 ml of deionized water. Cell suspensions and 90% phenol were equilibrated to 65°C, and 25 ml of 90% phenol was added to cell suspensions. Samples were incubated at 65°C for 1 h and on ice for 1 h. To separate the aqueous (containing LPS) and organic phases, samples were centrifuged at 3,300 × g for 10 min at 4°C. The aqueous layer was collected and the organic phase was back-extracted with 25 ml of deionized water. Samples were centrifuged as described above and the aqueous layer was collected and combined with the first extraction. Samples were precipitated with 0.3 M sodium acetate and 3 volumes of cold absolute ethanol, flash cooled in a dry ice-ethanol bath, and incubated for 2 h at −20°C. Samples were centrifuged for 10 min at 12,000 × g at 4°C, pellets were suspended in 6 ml of deionized water, and samples were reprecipitated as described above. Samples were centrifuged and pellets were suspended in 4 ml of 0.06 M Tris base, 10 mM EDTA, and 2.0% sodium dodecyl sulfate (SDS), pH 6.8. Samples were boiled for 5 min and cooled to room temperature. To digest contaminating proteins, samples were incubated at 65°C for 1 h with 10 μg of proteinase K/ml and then incubated overnight at 37°C. To remove SDS, samples were precipitated six times with 0.3 M sodium acetate and 3 volumes of absolute ethanol. After the final precipitation, pellets were suspended in deionized water and centrifuged at 120,000 × g for 1 h 15 min. The glass-like pellet was suspended in high-performance liquid chromatography (HPLC)-grade water and centrifuged at 120,000 × g for 1.5 h. The pellet was suspended in HPLC-grade water and lyophilized overnight in a VirTis BenchTop 2K lyophilizer.
Isolation of lipid A.
LPS from F. tularensis strain 1547-57 (4.6 mg) and F. tularensis LVS (approximately 5 mg) was hydrolyzed in 1 ml of 1% acetic acid at 100°C for 3 h. The samples were centrifuged at 4°C for 30 min, the supernatants were removed, and the lipid A pellets were washed two times with water and then dried under a stream of nitrogen. For further purification, the lipid A pellets were partitioned in CHCl3-CH3OH-H2O (10/5/6), and the bottom organic layers plus interfaces were saved and evaporated to dryness under a stream of nitrogen.
Alternatively, the LPS samples (<0.4 mg) were hydrolyzed under more mild conditions using the method of Caroff et al. (3) with some minor modifications. LPS was treated with 100 μl of 10 mM sodium acetate containing 1% SDS at pH 4.5. The samples were sonicated for approximately 30 s and then heated at 100°C for 1 h. Samples were then frozen and lyophilized in a Speed-Vac concentrator. The dry samples were treated with 100 μl of acidified ethyl alcohol (EtOH) (a 1:200, vol/vol, solution of 4 M HCl in EtOH) and centrifuged at 12,000 × g for 15 min at 4°C. The supernatants were removed, and the pellets were then washed three times with 100 μl of EtOH. The lipid A samples were then extracted with 580 μl of a two-phase Bligh/Dyer mixture consisting of CHCl3-CH3OH-H2O (2/2/1.8 by volume) (28). Bottom organic layers and interfaces were saved and evaporated to dryness under a stream of nitrogen.
GC-MS analysis of fatty acids derived from lipid A.
Approximately 50 μg of the F. tularensis 1547-57 lipid A sample was treated with 0.5 ml of 10% (wt/wt) BF3-methanol (Supelco, Inc., Bellefonte, Pa.) and heated at 100°C for 6 h. After cooling to room temperature, the sample was partitioned between 0.5 ml of saturated NaCl and 0.5 ml of HPLC-grade hexanes (Aldrich, St. Louis, Mo.). The aqueous layer was extracted a second time with 0.5 ml of hexanes, and the combined organic layers were back-extracted with 0.3 ml of H2O. The organic layer was then evaporated to dryness under a stream of nitrogen. The fatty acid methyl esters (FAMEs) were redissolved in hexanes and analyzed by gas chromatography-mass spectrometry (GC-MS) in the electron impact mode using a Hewlett-Packard 5890 gas chromatograph interfaced with a VG70SE mass spectrometer. The gas chromatograph was equipped with an on-column injector (J & W Scientific, Folsom, Calif.). FAMEs were separated on a 30-m by 0.25-mm BPX70 column with a 0.25-μm film thickness (SGE, Inc., Austin, Tex.) with helium as the carrier gas (≈6 lb/in2). The initial oven temperature was 100°C for 5 min, followed by a temperature gradient from 100 to 220°C at 4°C/min. For semiquantitative analysis of the GC-MS data, relative peak areas were measured from the total ion chromatogram and normalized to the C18:0(3-OH) component set at 3.0.
Dephosphorylation of lipid A.
To remove phosphate moieties from the F. tularensis 1547-57 lipid A, approximately 0.5 mg of the sample was treated twice with 100 μl of 48% aqueous hydrofluoric acid (HF) for 20 h. After each treatment, HF was evaporated from the sample under a stream of N2 in a polypropylene desiccator attached to a water aspirator, with an in-line NaOH trap. Samples were then redissolved in 40 μl of deionized H2O and evaporated to dryness in a Speed-Vac concentrator.
Preparation of alditol acetates.
The HF-treated F. tularensis 1547-57 lipid A sample was partitioned in CHCl3-CH3OH-H2O (2:1:2 by volume), and the upper aqueous layer was taken and placed in a clean vessel. The organic layer was then reextracted with a second portion of fresh aqueous layer (prepared separately), and the combined aqueous layers were evaporated to dryness. The dry residue was then reduced by treatment with 150 μl of 10-mg/ml NaBD4 in 1 M NH4OH for 2.5 h at room temperature. The reaction was quenched with 20 μl of glacial acetic acid, and then the sample was evaporated to dryness under a stream of N2 from 0.5 ml of CH3OH containing 1% acetic acid (eight times), followed by pure CH3OH (six times). The reduced sample was dried in a vacuum desiccator over P2O5 for 1 h and then peracetylated by treatment with 300 μl of acetic anhydride (Supelco, Inc.) at 100°C for 3 h, with occasional sonication. After cooling to room temperature, the reaction mixture was evaporated to dryness under a stream of N2, followed by four dryings from 0.5 ml of HPLC-grade toluene (Aldrich, Milwaukee, Wis.). Finally, the product was partitioned between 0.5 ml of CHCl3 and 0.5 ml of H2O, the aqueous layer was extracted two more times with CHCl3, and the combined organic layer was back-extracted with 0.5 ml of H2O and finally evaporated to dryness. As a negative control for this experiment, a sample of Salmonella enterica serovar Typhimurium TV119 Ra mutant lipid A derived from the LPS (Sigma, St. Louis, Mo.) was taken through the same steps as the F. tularensis lipid A.
The derivatized samples were analyzed on the GC-MS system described above, operating in the chemical ionization mode using ammonia gas. The samples were dissolved in CH2Cl2 and injected onto the BPX70 column at 120°C. The initial oven temperature was 120°C for 3 min, followed by a quick ramp to 190°C at 8°C/min, a 1-min hold at 190°C, and then a temperature gradient from 190 to 260°C at 4°C/min.
Reduction of lipid A.
Lipid A samples were subjected to reduction with NaBD4 following a published procedure (15), with some modifications. Portions of the F. tularensis 1547-57 and F. tularensis LVS lipid A samples (approximately 50 μg each) were dissolved in 150 μl of 6.25-mg/ml NaBD4 (Aldrich) in 0.02% triethylamine in deionized water. As controls, equal portions of the lipid A samples were treated with 150 μl of 0.02% triethylamine in deionized water. The samples were allowed to react for 2.5 or 4 h at room temperature and then quenched with 20 μl of glacial acetic acid. Samples were then taken to near dryness by evaporation under a stream of N2, followed by evaporation from 1% acetic acid in methanol (eight times), and finally evaporation from methanol (six times). Dry samples were then partitioned in CHCl3-CH3OH-H2O (10/5/6 by volume) in a total volume of 1,050 μl. The aqueous layer was removed and the organic layer plus interface was evaporated to dryness under a stream of N2. As a control experiment, a commercial sample of monophosphorylated lipid A from Salmonella enterica serovar Minnesota Re 595 (Sigma) was also subjected to the reduction conditions described above.
MALDI-TOF MS.
Lipid A samples were analyzed by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) MS on a Voyager DESTR Plus instrument (Applied Biosystems, Framingham, Mass.) equipped with a 337-nm nitrogen laser. Spectra were obtained in the positive-ion and negative-ion reflectron modes under delayed extraction conditions. In the positive-ion reflectron mode, the delay time was 200 to 275 ns, and the grid voltage was 66.5 to 67.3% of full acceleration voltage (25 kV). In the negative-ion reflectron mode, the delay time was 175 ns, and the grid voltage was 64.5% of full acceleration voltage (20 kV). Samples were dissolved in CHCl3-CH3OH (3:1) at a concentration of approximately 0.1 to 0.5 μg/μl, mixed 1:1 with matrix solution (a saturated solution of 6-chloro-3-mercaptobenzothiazole in CHCl3-CH3OH [3:1 by volume]), spotted on a stainless steel target, and allowed to air dry. Approximately 100 laser shots were recorded for each sample. The spectra were processed with baseline correction and a noise filter. Mass calibration was done externally with a mixture consisting of angiotensin II, renin substrate tetradecapeptide, and insulin chain B (oxidized) or a mixture consisting of angiotensin I, adenocorticotropin (ACTH) 1-17, ACTH 18-39, and ACTH 7-38 (all from Sigma).
Nanoelectrospray tandem MS (MS/MS) and fragmentation MS (MSn).
The F. tularensis 1547-57 lipid A sample, dissolved in CHCl3-CH3OH (1:2 by volume), at a concentration of approximately 0.1 to 0.5 μg/μl, was analyzed by static nanoelectrospray using a LCQDeca quadrupole ion-trap mass spectrometer (ThermoFinnigan, San Jose, Calif.). In the negative-ion nanoelectrospray mode, the needle voltage was typically set to 1.6 to 2.2 kV and the temperature of the heated capillary was at 140 to 180°C. Ions were isolated for collision-induced dissociation with the “isolation width” parameter set to 1 to 3.
RESULTS
The LPS from F. tularensis strain 1547-57 was isolated from plate-grown organisms using phenol-water extraction, followed by proteinase K treatment (13). The lipid A was released from the LPS by mild acid hydrolysis in 1% acetic acid and then further purified by partitioning in CHCl3-CH3OH-H2O (10/5/6).
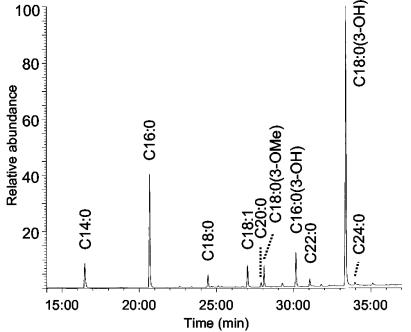
To investigate the fatty acid composition of the lipid A, a small portion of the sample was treated with 10% (wt/wt) BF3-methanol at 100°C for 6 h to prepare FAMEs. The FAMEs were analyzed by GC-MS using a highly polar capillary column optimized for FAME analysis. Figure 1 shows the total ion chromatogram of the FAMEs derived from the F. tularensis lipid A fraction, where the main fatty acids present are hexadecanoic acid (C16:0) and 3-hydroxyoctadecanoic acid [C18:0(3-OH)] in the ratio of 1.2 to 3.0. Minor components seen in the sample include tetradecanoic acid (C14:0), octadecanoic acid (C18:0), octadecenoic acid (C18:1), 3-methoxyoctadecanoic acid [C18:0(3-methoxyl)], and 3-hydroxyhexadecanoic acid [C16:0(3-OH)] in the ratio of 0.3:0.1:0.2:0.2:0.3. Trace amounts of C20:0, C22:0, and C24:0 fatty acids were also detected. Compared to the FAMEs derived from the lipid A of F. tularensis LVS (27), the FAME mixture from strain 1547-57 contains higher proportions of the longer chain fatty acids, i.e., 20 times more C16:0 relative to C14:0 and 4 times more C18:0(3-OH) relative to C16:0(3-OH).
FIG. 1.
GC-MS total ion chromatogram of the FAMEs derived from the lipid A of F. tularensis strain 1547-57.
To evaluate the heterogeneity of the lipid A preparation and measure molecular weights, the lipid A sample was analyzed by MALDI-TOF MS. Both negative-ion (Fig. 2A) and positive-ion (Fig. 2B) spectra were recorded in reflectron mode to allow for the measurement of monoisotopic masses. Table 1 gives the C-12 monoisotopic masses measured from these spectra. As indicated, the major lipid A component with a (M − H)− at m/z 1,665.06 observed in the negative-ion MALDI spectrum (Fig. 2A) is proposed to contain a diglucosamine backbone bearing three C18:0(3-OH) and one C16:0 fatty acids, one phosphate group, and one additional 161-Da moiety (proposed to be a hexosamine, HexN). The species at (M − H)− at m/z 1,503.99 is lacking the additional HexN moiety. In the positive-ion MALDI spectrum (Fig. 2B), the doubly sodiated species (M − H + 2Na)+ at m/z 1,710.92 corresponds to the major component seen in the negative-ion spectrum, and the (M − H + 2Na)+ species at m/z 1,549.88 is the lipid A lacking the additional HexN moiety. Related species lacking both the phosphate and the HexN moieties are also observed in the positive-ion mode [(M + Na)+ ions at m/z 1,447.93 and 1,429.93]. However, the major component observed in the positive-ion spectrum is the (M + Na)+ species at m/z 1,391.88, which fits for a lipid A structure bearing two C18:0(3-OH), one C16:0(3-OH), and one C14:0 fatty acids. This constituent was the major species seen in the lipid A preparation from F. tularensis LVS (27). While this component is only seen in the positive-ion MALDI spectrum of the F. tularensis strain 1547-57 lipid A sample, the novel higher-molecular-weight components present in strain 1547-57 are detected in both the positive-ion and negative-ion modes.
FIG. 2.
Sections of three MALDI-TOF mass spectra of the F. tularensis strain 1547-57 lipid A sample, acquired in reflectron mode. (A) The negative-ion MALDI-TOF MS. (B) The positive-ion MALDI-TOF MS. (C) The positive-ion MALDI-TOF MS of the lipid A sample after HF treatment for 40 h.
TABLE 1.
Molecular weights (Mr) observed for lipid A structures of F. tularensis strain 1547-57 as detected by negative-ion and positive-ion reflectron MALDI-TOF MS
| Mr observed (exact) | Mr calculated (exact) | ΔMd (ppm) | Proposed compositions |
|---|---|---|---|
| 1,666.06,a 1,665.94b | 1,666.17 | 66, 138 | 3× C18:0(3-OH), C16:0, HexN, P |
| 1,638.02,a 1,637.89b | 1,638.14 | 73, 153 | 2× C18:0(3-OH), C16:0(3-OH), C16:0, HexN, P, and/or 3× C18:0(3-OH), C14:0, HexN, P |
| 1,504.99,a 1,504.90b | 1,505.10 | 73, 133 | 3× C18:0(3-OH), C16:0, P |
| 1,476.99,a 1,476.87b | 1,477.07 | 54, 135 | 2× C18:0(3-OH), C16:0(3-OH), C16:0, P, and/or 3× C18:0(3-OH), C14:0, P |
| 1,427.82a | 1,427.94 | 84 | 3× C18:0(3-OH), HexN, P |
| 1,424.94c | 1,425.13 | 133 | 3× C18:0(3-OH), C16:0 |
| 1,406.94c | 1,407.12 | 128 | 3× C18:0(3-OH), C16:0, anhydro |
| 1,396.95c | 1,397.10 | 107 | 2× C18:0(3-OH), C16:0(3-OH), C16:0, and/or 3× C18:0(3-OH), C14:0 |
| 1,368.89c | 1,369.07 | 131 | 2× C18:0(3-OH), C16:0(3-OH), C14:0 |
Species was detected in negative-ion mode as (M − H)−.
Species was detected in positive-ion mode as (M − H + 2Na)+.
Species was detected in positive-ion mode as (M + Na)+.
All masses reported are monoisotopic masses M.
To check for other potentially labile moieties and evaluate the yields of the phosphorylated components, we attempted to hydrolyze the F. tularensis LPS under more mild conditions. The LPS was treated with 10 mM sodium acetate containing 1% SDS at pH 4.5 for 1 h at 100°C (3). The resulting lipid A sample was analyzed by MALDI-TOF MS in both the negative-ion and positive-ion reflectron modes (data not shown). Compared to the lipid A preparation generated by 1% acetic acid hydrolysis, the sample generated under more mild hydrolysis conditions showed slightly higher relative amounts of the phosphorylated lipid A forms by positive-ion MALDI but did not contain any previously unseen components.
As an initial assumption, the 161-Da moiety present on the F. tularensis 1547-57 lipid A was presumed to be a HexN residue linked to the diglucosamine backbone via the phosphate group, in a manner analogous to the way arabinosamine (4-amino-4-deoxy-l-arabinose) is found linked to phosphate in the LPS of other organisms (2, 18, 20, 26). To test for the presence of a hexosamine-1-phosphate linkage, the sample was treated two times with 48% aqueous HF. The MALDI spectrum was recorded after each treatment and was used to monitor the progress of the dephosphorylation reaction. The MALDI spectrum of the sample after two HF treatments (Fig. 2C) shows the complete loss of the HexN moiety (to give the monophosphorylated species at m/z 1,549.89) and the partial removal of the residual phosphate group (to give the species at m/z 1,447.95 which lacks the entire hexosamine-1-phosphate moiety). After only a single HF treatment, the m/z 1,447.95 species was less abundant than after the second HF treatment, suggesting that the hexosamine-phosphate linkage is cleaved more readily than the lipid A-phosphate linkage. Consistent with the published structure for the (M + Na)+ species at m/z 1,391.90, which does not contain any phosphate substituents (27), there was nothing lost from that component upon HF treatment.
Further confirmation of the linkage of the hexosamine-1-phosphate moiety to the lipid A backbone of the novel species was obtained by MS/MS analysis. The lipid A sample, dissolved in CHCl3-CH3OH (1:2) was analyzed by static nanoelectrospray using a quadrupole ion-trap mass spectrometer. This approach has recently proven useful for following multiple stages of MSn of lipid A ions (4, 16). The negative-ion mass spectrum of the fraction gave an abundant (M − H)− ion at m/z 1,665.2 for the component bearing the hexosamine-1-phosphate. The MS/MS spectrum of this species (Fig. 3A) contains ions representing single- and multiple-bond cleavages. The fragment ion arising from the loss of hexosamine from the parent ion (m/z 1,503.9) indicates that the novel hexosamine moiety is in a terminal position. Fragment ions at m/z 1,408.9 and 1,364.9 arise from the losses of C16:0 (−256.2 Da) and C18:0(3-OH) (−300.3 Da) fatty acids from the parent ion, respectively. The major fragment ion at m/z 1,204.1 can be assigned as a two-bond cleavage involving the loss of a C18:0(3-OH) fatty acid (−300.3 Da) and the HexN moiety (−161.1 Da). The ion at m/z 947.9 would appear to arise from three-bond cleavages [losses of C18:0(3-OH), C16:0, and HexN].
FIG. 3.
Negative-ion nanoelectrospray MSn spectra of the lipid A from F. tularensis strain 1547-57. (A) MS/MS of m/z 1,665.2. (B) MS3 of m/z 1,408.9. (C) MS3 of m/z 1,203.9. (D) MS3 of m/z 947.8.
In an effort to fix the locations of the fatty acids and the hexosamine-1-phosphate group on the diglucosamine backbone, various fragment ions in the MS/MS spectrum were selected for further fragmentation (MS3). Both the m/z 1,408.9 and 1,203.9 fragments gave the m/z 947 ion as the base peak in their MS3 spectra (Fig. 3B and C, respectively). In the case of the m/z 1,408.9 fragment (already lacking C16:0), the 947.6 ion arises from two-bond cleavages; the loss of the HexN moiety plus the O-linked C18:0(3-OH) fatty acid (Fig. 3B). In the MS3 spectrum of m/z 1,203.9 [already lacking C18:0(3-OH) and HexN], the 947.7 fragment arises from the loss of the O-linked C16:0 fatty acid (Fig. 3C).
To explore the structure of the m/z 947 fragment, it was selected from the MS/MS spectrum for MS3 analysis as shown in Fig. 3D. Fragmentation of m/z 947.8 led to losses of the N-linked fatty acids as ketenes (fragment ions at m/z 683.5 and 665.5). Also present in the spectrum were fragments at m/z 504.5 and 522.4 that appeared to arise from glycosidic bond cleavages. By using the nomenclature of Costello and Vath (5), these ions could be assigned as B- and C-type fragments if the phosphate group was on the distal glucosamine, and Z- and Y-type fragments in the phosphate group were on the reducing terminal glucosamine. To resolve this ambiguity, corresponding glycosidic bond fragments in the MS3 spectra of m/z 1,408.9 (Fig. 3B) and m/z 1,203.9 (Fig. 3C) were sought. The m/z 983.5 fragment in Fig. 3B can be assigned as a Y-type fragment if the structure of m/z 1,408.9 is as shown in the inset, with the hexosamine-1-phosphate group on the reducing terminal glucosamine. This Y ion also supports the placement of the two C18:0(3-OH) fatty acids on the reducing terminal glucosamine. Correspondingly, the m/z 522.5 fragment in Fig. 3C can be assigned as a Y ion arising from m/z 1,203.9, which is consistent with the assignment of that fragment in Fig. 3D and supports the placement of the one C18:0(3-OH) and one C16:0 fatty acid on the distal glucosamine of the structure. The assignment of the C16:0 fatty acid in an acyloxy linkage to O-3 of the N-linked C18:0(3-OH) fatty acid of the distal glucosamine was suggested by the fragment ion at m/z 683.5 in Fig. 3D (loss of a C18:1 ketene). This fragment ion could only arise from the m/z 947.8 species if the N-linked fatty acid on its distal glucosamine was a C18:1 fatty acid (Fig. 3D, inset), presumably generated by the elimination of C16:0 (as a free acid) from the tetraacylated structure.
Conspicuously lacking from the MS/MS and MS3 spectra are the strong 0,2A2 and 0,4A2 cross-ring fragments generally seen in monophosphorylated lipid A's with the phosphate group on the 4′ position and a free reducing terminus (16, 19). The lack of these A-type fragments may be an indication that the hexosamine-1-phosphate group is located on the reducing terminal glucosamine. An additional curious aspect of these spectra is the ability of many species to lose a 240-Da moiety. This moiety is proposed to be a C16H32O aldehyde eliminated from the N-linked C18:0(3-OH) fatty acid on the reducing terminal glucosamine by a McLafferty rearrangement. This type of fragmentation could potentially result from a charge-driven rearrangement process involving the neighboring reducing terminal phosphate group. When located on the 4′ position of lipid A, phosphate has been proposed to initiate a charge-driven fragmentation process affecting fragmentation at the 3′ position in negative-ion electrospray ionization-MSn (16).
To further confirm the placement of the hexosamine-1-phosphate group on the reducing terminus of the lipid A backbone, the F. tularensis 1547-57 lipid A sample was subjected to reduction by treatment with NaBD4 for 2.5 to 4 h. The reaction mixtures were analyzed by MALDI-TOF MS in both the positive-ion and negative-ion reflectron modes. Figure 4 shows the positive-ion MALDI spectrum of the 4-h reaction. Under the short reaction times used to minimize unwanted cleavage of O-linked fatty acids in the slightly basic solution, the reduction reaction did not go all the way to completion. Thus, species that were susceptible to reduction were seen to split into two sets of molecular ion peaks, with one shifted to higher mass by the addition of 3 Da (Fig. 4). Species that did not undergo reduction were unchanged by the treatment. Control samples that were incubated with solvent lacking NaBD4 did not show any evidence of mass shifts. Referring to Table 1, all of the species containing either hexosamine-1-phosphate (Mr, 1,666 and 1,638) or phosphate alone (Mr, 1,505, 1,477, and 1,427) were not susceptible to reduction, whereas the unphosphorylated species (Mr, 1,425, 1,397, and 1,369) were all able to react. As a control, the monophosphorylated lipid A from S. enterica serovar Minnesota Re 595, which has a phosphate substituent on the 4′ position of the glucosamine disaccharide, was subjected to the same procedures and found to be susceptible to reduction. In all of the reduction experiments, some minor evidence of O-linked fatty acid cleavage was detected, but the resulting molecular ion species had isotope distributions consistent with the corresponding intact structures. Thus, we conclude that the phosphorylated F. tularensis lipid A structures do not have a free reducing terminus available for reaction, supporting the assignment of the hexosamine-1-phosphate on the 1 position of the lipid A.
FIG. 4.
Section of the positive-ion MALDI spectrum of the F. tularensis 1547-57 lipid A sample after reduction by treatment with NaBD4 for 4 h, acquired in reflectron mode. Insets show expansions of the indicated peaks. Asterisks mark the monoisotopic molecular ions of reduced species that have incorporated three deuterium atoms.
To identify the purported hexosamine moiety, the HF-treated lipid A sample that had released its hexosamine (described above) was partitioned in CHCl3-CH3OH-H2O (2:1:2), and the upper aqueous layer was recovered and evaporated to dryness. This material was reduced with NaBD4 and then peracetylated by treatment with acetic anhydride to generate the alditol acetate of the hexosamine. As a control, a sample of HF-treated S. enterica serovar Typhimurium lipid A was taken through the same reaction sequence. The derivatized F. tularensis and S. enterica serovar Typhimurium samples were analyzed by GC-MS using chemical ionization. Samples were compared to a mixture of standards consisting of glucosaminitol acetate, mannosaminitol acetate, and galactosaminitol acetate, which eluted at 37.34, 38.47, and 39.21 min, respectively. The sample derived from the F. tularensis lipid A contained a component with the molecular weight of a hexosaminitol acetate, (M + H)+ at m/z 435.5, that eluted at the retention time of galactosaminitol acetate. No hexosaminitol acetates of any type were detected in the S. enterica serovar Typhimurium sample. Thus, we conclude from these results that the hexosamine-1-phosphate linked to the proximal glucosamine in the novel F. tularensis lipid A species is galactosamine-1-phosphate (Fig. 5).
FIG. 5.
Proposed structure of the 1,666-Mr component of the F. tularensis strain 1547-57 lipid A sample. The diglucosamine backbone bears four fatty acids: three C18:0(3-OH) fatty acids, and one C16:0 fatty acid. The acylation pattern shown is consistent with the arrangement determined for the lipid A of F. tularensis LVS (ATCC 29684) (27). The further modification of the Mr-1,666 component of F. tularensis strain 1547-57 lipid A with a galactosamine-1-phosphate on the reducing terminus represents a novel structural feature not previously observed. The anomeric configuration of the galactosamine-1-phosphate linkage has not been determined.
This modification seen in the Mr-1,666 component of the F. tularensis 1547-57 lipid A mixture was not reported in the lipid A from F. tularensis LVS (27). However, in our laboratory, the same modification was seen, although to a lesser extent, in the lipid A fraction from Francisella philomiragia ATCC 25015 (M. McLendon and B. Schilling, unpublished data). To address this discrepancy, we cultured F. tularensis LVS and prepared the lipid A fraction from its LPS following our protocols as described above. The lipid A from this F. tularensis LVS was analyzed by MALDI-TOF MS in the positive-ion and negative-ion modes (Fig. 6) and compared to the lipid A from the F. tularensis 1547-57 strain (Fig. 2). Under our growth and isolation conditions, the major species that we obtained from the F. tularensis LVS strain was a monophosphorylated lipid A with an Mr of 1,505.1 (Fig. 6). This species was consistent with the tetraacylated, monophosphorylated lipid A seen in the F. tularensis 1547-57 strain [consisting of three C18:0(3-OH) fatty acids, one C16:0 fatty acid, and one phosphate group]. In the positive-ion mass spectrum (Fig. 6B), we observed many of the unphosphorylated species also observed in the F. tularensis 1547-57 strain (m/z 1,391.8, 1,419.9, 1,429.9, and 1,447.9). The species at m/z 1,391.84 and 1,419.87 are consistent with the previously published major structures for the lipid A from F. tularensis LVS (ATCC 29684) (27). The species at m/z 1,565.81 is 16 Da higher than the m/z 1,549.84 species and would appear to differ by the addition of an oxygen atom (possibly a fourth hydroxylated fatty acid), but this has not been determined. Thus, although we did observe monophosphorylated lipid A structures in this preparation of F. tularensis LVS lipid A, we did not see any hint of the Mr-1,666 structure modified with the galactosamine-1-phosphate moiety.
FIG. 6.
Sections of the MALDI-TOF mass spectra of the lipid A from F. tularensis LVS recorded in negative-ion (A) and positive-ion (B) reflectron modes. For comparison to the F. tularensis strain 1547-57 lipid A sample, refer to Fig. 2. For proposed compositions, see the text and Table 1.
To evaluate the relationship of the monophosphorylated lipid A structures in this F. tularensis LVS to the corresponding species in F. tularensis 1547-57, we subjected the F. tularensis LVS sample to reduction with NaBD4. As was seen with the F. tularensis 1547-57 lipid A species, the monophosphorylated structures in this F. tularensis LVS lipid A preparation were not amenable to reduction, whereas the unphosphorylated structures were. Thus, we conclude that under our growth conditions, F. tularensis LVS can add phosphate to the 1 position of the lipid A. However, F. tularensis LVS does not appear to be capable of further modifying the monophosphorylated lipid A structure with the novel galactosamine moiety, as seen in F. tularensis strain 1547-57.
DISCUSSION
The lipid A preparation from F. tularensis strain 1547-57 contained a heterogeneous mixture of lipid A structures. In order to get a complete view of the species present in the mixture, it was necessary to analyze the sample by MALDI-TOF MS in both the positive-ion and negative-ion modes. All of the major lipid A forms detected by MALDI-TOF were tetraacylated, although there was microheterogeneity in the fatty acid constituents. One lipid A form present in F. tularensis strain 1547-57 (Mr, 1,369) had a composition consistent with the major published structure for the lipid A from F. tularensis LVS (ATCC 29684) (27), consisting of two C18:0(3-OH), one C16:0(3-OH), and one C14:0 fatty acids. The other major components found in the F. tularensis 1547-57 strain were of higher molecular weight than this species. Some forms simply contained longer chain fatty acids, whereas others were monophosphorylated as well.
The most striking constituent was a monophosphorylated species with an Mr of 1,666 bearing three C18:0(3-OH) and one C16:0 fatty acids, plus an additional 161-Da moiety that was identified as galactosamine. Through a combination of MSn studies and chemical treatments, the galactosamine was shown to exist as a galactosamine-1-phosphate moiety linked to the reducing terminus of the lipid A. MSn investigations also showed that this tetraacylated structure had two fatty acids on each glucosamine unit of the lipid A, with the nonreducing terminal glucosamine apparently bearing an N-linked acyloxyacyl group (Fig. 5). This acylation pattern is consistent with the architecture of the published structure for F. tularensis LVS (27).
The novel modification of the F. tularensis lipid A with galactosamine-1-phosphate seen in strain 1547-57 was not reported in the previous study of F. tularensis LVS (27). In our hands, a lipid A preparation from F. tularensis LVS was shown to include monophosphorylated species but no species bearing galactosamine-1-phosphate. However, the hexosamine-1-phosphate modification was detected in a lipid A fraction from F. philomiragia (M. McLendon and B. Schilling, unpublished). Thus, it would appear that the lack of this moiety in F. tularensis LVS may result from strain variation. The phosphorylation differences seen in F. tularensis LVS by our group and by Vinogradov et al. (27) may also arise from strain variation or from different lipid A isolation and purification steps. Specifically, our protocols differed in culture conditions (plate versus broth), LPS acetic acid hydrolysis conditions (1 versus 5% acetic acid), and lipid A purification methods (extraction versus extraction plus column chromatography).
To our knowledge, the modification of a lipid A structure with galactosamine-1-phosphate has not been previously observed. However, the addition of arabinosamine to the phosphate group(s) of lipid A has been well characterized (10, 11, 17, 25, 29). In an effort to explore possible connections, we searched the F. tularensis Schu S4 genome for a homolog of the arabinosamine transferase ArnT in S. enterica serovar Typhimurium and YfbI of E. coli (25). A homolog was found on contig 34, nucleotides 16024 to 16950c, and the presumed protein product shares 27% amino acid identity to YfbI and 25% amino acid identity to ArnT. The amino acid sequence has the highest identity (33%) to the arabinosamine transferases of Bordetella parapertussis and Geobacter metallireducens, and 27% identity to the arabinosamine transferases of Pseudomonas fluorescens and Yersinia spp. ArnT and YfbI are members of the protein O-mannosyltransferase family of proteins, involved in posttranslational modification. Thus, it is possible that this homolog may be another member of the protein O-mannosyltransferase protein family.
The modification of lipid A with arabinosamine alters the net negative charge on the lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides (9, 10). The addition of galactosamine-1-phosphate to the lipid A of F. tularensis is likely to have a similar effect on net negative charge. The significance of this modification for F. tularensis is unknown, although it is possible that it could confer some resistance to host antimicrobial peptides in vivo. It is interesting that F. tularensis contains a possible ortholog of a lipid A 1-phosphatase, LpxE, from Rhizobium leguminosarum (14). Thus, dephosphorylation of the lipid A structure in vivo may be a possibility. Preliminary evidence also suggests that F. tularensis may be capable of synthesizing more highly acylated lipid A structures in vivo once infection is established (M. McLendon, unpublished data). Indeed, minor constituents at molecular weights higher than the Mr-1,666 constituent were occasionally detected in the F. tularensis strain 1547-57 lipid A sample by positive-ion MALDI-TOF MS (mainly observed at m/z 1,821.2, 1,842.6, 1,849.2, 1,934.3, and 1,962.3). Thus, the various lipid A structures described in this report may represent only a subset of the lipid A modifications possible in vivo. However, the tetraacylated lipid A structures, with and without the galactosamine-1-phosphate modification, seem likely to contribute to the unusually low endotoxicity of the F. tularensis LPS.
Acknowledgments
We thank Yuequan Sun for assistance with acquisition of GC-MS data, David A. Maltby for helpful input, and the UCSF Mass Spectrometry Facility for access to the GC-MS system (supported by NIH NCRR BRTP RR01614, A. L. Burlingame, Director). We are also grateful to Karen Elkins for supplying F. tularensis LVS and Jeannine Petersen for assistance in typing F. tularensis strain 1547-57. Additionally, we acknowledge William R. Kearney of the University of Iowa for helpful discussions.
Editor: J. N. Weiser
REFERENCES
- 1.Ancuta, P., T. Pedron, R. Girard, G. Sandstrom, and R. Chaby. 1996. Inability of the Francisella tularensis lipopolysaccharide to mimic or to antagonize the induction of cell activation by endotoxins. Infect. Immun. 64:2041-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boll, M., J. Radziejewska-Lebrecht, C. Warth, D. Krajewska-Pietrasik, and H. Mayer. 1994. 4-Amino-4-deoxy-L-arabinose in LPS of enterobacterial R-mutants and its possible role for their polymyxin reactivity. FEMS Immunol. Med. Microbiol. 8:329-341. [DOI] [PubMed] [Google Scholar]
- 3.Caroff, M., A. Tacken, and L. Szabo. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 4.Corsaro, M. M., F. D. Piaz, R. Lanzetta, and M. Parrilli. 2002. Lipid A structure of Pseudoalteromonas haloplanktis TAC 125: use of electrospray ionization tandem mass spectrometry for the determination of fatty acid distribution. J. Mass Spectrom. 37:481-488. [DOI] [PubMed] [Google Scholar]
- 5.Costello, C. E., and J. E. Vath. 1990. Tandem mass spectrometry of glycolipids. Methods Enzymol. 193:738-768. [DOI] [PubMed] [Google Scholar]
- 6.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 7.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulop, M., T. Webber, and R. Manchee. 1993. Activation of the complement system by Francisella tularensis lipopolysaccharide. New Microbiol. 16:141-147. [PubMed] [Google Scholar]
- 9.Gunn, J. S. 2001. Bacterial modification of LPS and resistance to antimicrobial peptides. J. Endotoxin Res. 7:57-62. [PubMed] [Google Scholar]
- 10.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 11.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 12.Hood, A. M. 1977. Virulence factors of Francisella tularensis. J. Hyg. (London) 79:47-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, P. A., N. M. Samuels, N. J. Phillips, R. S. Munson, Jr., J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277:14598-14611. [DOI] [PubMed] [Google Scholar]
- 14.Karbarz, M. J., S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2003. Expression cloning and biochemical characterization of a Rhizobium leguminosarum lipid A 1-phosphatase. J. Biol. Chem. 278:39269-39279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulshin, V. A., U. Zahringer, B. Lindner, C. E. Frasch, C. M. Tsai, B. A. Dmitriev, and E. T. Rietschel. 1992. Structural characterization of the lipid A component of pathogenic Neisseria meningitidis. J. Bacteriol. 174:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kussak, A., and A. Weintraub. 2002. Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from gram-negative bacteria. Anal. Biochem. 307:131-137. [DOI] [PubMed] [Google Scholar]
- 17.Molinaro, A., B. Lindner, C. De Castro, B. Nolting, A. Silipo, R. Lanzetta, M. Parrilli, and O. Holst. 2003. The structure of lipid A of the lipopolysaccharide from Burkholderia caryophylli with a 4-amino-4-deoxy-L-arabinopyranose 1-phosphate residue exclusively in glycosidic linkage. Chemistry 9:1542-1548. [DOI] [PubMed] [Google Scholar]
- 18.Nummila, K., I. Kilpelainen, U. Zahringer, M. Vaara, and I. M. Helander. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271-278. [DOI] [PubMed] [Google Scholar]
- 19.Post, D. M., N. J. Phillips, J. Q. Shao, D. D. Entz, B. W. Gibson, and M. A. Apicella. 2002. Intracellular survival of Neisseria gonorrhoeae in male urethral epithelial cells: importance of a hexaacyl lipid A. Infect. Immun. 70:909-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radziejewska-Lebrecht, J., U. R. Bhat, H. Brade, and H. Mayer. 1988. Structural studies on the core and lipid A region of a 4-amino-L-arabinose-lacking Rc-type mutant of Proteus mirabilis. Eur. J. Biochem. 172:535-541. [DOI] [PubMed] [Google Scholar]
- 21.Rietschel, E. T., T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zahringer, U. Seydel, F. Di Padova, et al. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8:217-225. [DOI] [PubMed] [Google Scholar]
- 22.Sandstrom, G. 1994. The tularaemia vaccine. J. Chem. Technol. Biotechnol. 59:315-320. [DOI] [PubMed] [Google Scholar]
- 23.Sandstrom, G., A. Sjostedt, T. Johansson, K. Kuoppa, and J. C. Williams. 1992. Immunogenicity and toxicity of lipopolysaccharide from Francisella tularensis LVS. FEMS Microbiol. Immunol. 5:201-210. [DOI] [PubMed] [Google Scholar]
- 24.Tarnvik, A. 1989. Nature of protective immunity to Francisella tularensis. Rev. Infect. Dis. 11:440-451. [PubMed] [Google Scholar]
- 25.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 26.Vaara, M., T. Vaara, M. Jensen, I. Helander, M. Nurminen, E. T. Rietschel, and P. H. Makela. 1981. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129:145-149. [DOI] [PubMed] [Google Scholar]
- 27.Vinogradov, E., M. B. Perry, and J. W. Conlan. 2002. Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 269:6112-6118. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, Z., S. Lin, R. J. Cotter, and C. R. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]
- 29.Zhou, Z., A. A. Ribeiro, and C. R. Raetz. 2000. High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-L-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J. Biol. Chem. 275:13542-13551. [DOI] [PubMed] [Google Scholar]