Abstract
Aspergillus fumigatus is an important pathogen of immunocompromised hosts, causing pneumonia and invasive disseminated disease with high mortality. To determine the importance of the cyclic AMP (cAMP) signaling pathway for virulence, the pkaC1 gene encoding a protein kinase A (PKA) catalytic subunit was cloned and characterized. Deletion of pkaC1 led to reduced conidiation and growth. PKA activity was not detectable in ΔpkaC1, ΔgpaB, and ΔacyA mutant strains. gpaB and acyA encode a G protein α subunit involved in cAMP signal transduction and adenylate cyclase, respectively. Addition of cAMP led to PKA activity in crude extracts of both the ΔgpaB and ΔacyA strains but not in crude extracts of the ΔpkaC1 strain. These findings provide evidence that PKAC1 represents the predominant form of PKA under the conditions tested, and GPAB and ACYA are members of the cAMP signaling cascade. Analysis of a pksPp-lacZ gene fusion indicated that the expression of the pathogenicity determinant-encoding pksP gene was reduced in ΔpkaC1 mutant strains compared with the expression of the gene fusion in the parental strain. In a low-dose murine inhalation model, conidia of both the ΔpkaC1 and ΔgpaB mutant strains were almost avirulent. Taken together, these findings indicate that the cAMP-PKA signal transduction pathway is required for A. fumigatus pathogenicity.
Aspergillus fumigatus has become the most important airborne fungal pathogen of humans. Alveolar macrophages form the first line of defense against fungal conidia, the infectious agents, entering the respiratory tract. Neutrophilic granulocytes, the second line of defense, primarily attack hyphae but also attack conidia. An important killing mechanism for both types of immune effector cells consists of the production of reactive oxygen species (ROS) (22, 31, 42). Improvement in transplant medicine and the therapy of hematological malignancies is often complicated by the threat of invasive aspergillosis. A. fumigatus accounts for approximately 90% of invasive aspergillosis cases. Specific diagnostics are still limited, as are the possibilities of therapeutic intervention, which leads to a high mortality rate (30 to 98%) for invasive aspergillosis (reviewed in references 9 and 31).
One of the important questions concerning A. fumigatus is the identification of pathogenicity determinants and their regulation. Recently, the group of J. Kwon-Chung and our group identified a gene that encodes a pathogenicity determinant. This gene was designated pksP (or alternatively alb1) for polyketide synthase involved in pigment biosynthesis (23, 24, 25, 28, 54). Conidia of a pksP mutant strain are white. Based on genetic and biochemical data, the conidial pigment consists of dihydroxynaphthalene (DHN)-melanin (7, 30, 52, 53). pksP mutant strains exhibited reduced virulence in a mouse infection model (23, 54). pksP mutant conidia were 20-fold more sensitive to ROS than wild-type conidia, suggesting that the pigment is able to scavenge ROS, thereby presumably detoxifying ROS (23, 24). Moreover, a key element of antimicrobial activity in macrophages is the formation of functional phagolysosomes that contain a large variety of degrading enzymes in an acidic environment. Recently, we showed that a functional pksP gene is associated with a reduction in phagolysosome fusion in human monocyte-derived macrophages (MDMs). Consistently, the intracellular killing of pksP mutant conidia by MDMs was significantly greater than the intracellular killing of wild-type conidia (25). By using the enhanced green fluorescent protein, it was shown that the pksP gene was expressed during sporulation. Interestingly, this gene was also expressed in outgrowing hyphae isolated from the lungs of infected immunocompromised mice (29). This finding suggested that certain stress conditions have an effect on pksP expression. Therefore, it is essential to understand the communication between host and pathogen, i.e., to elucidate the signaling pathways that enable pathogenic fungi to adapt and survive the drastically altered environmental conditions that they encounter upon infection of the host.
Recently, some elements of the cyclic AMP (cAMP) signaling pathway of A. fumigatus were identified (33, 37). These elements include the adenylate cyclase gene acyA and the G protein α subunit-encoding gene gpaB that was found to be an upstream stimulator [G(s)α] of adenylate cyclase. Deletion mutants with mutations in both genes showed reduced conidiation, and the ΔacyA mutant produced very few conidia. Whereas the growth rate of the ΔacyA mutant was reduced, a reduced growth rate was not observed in case of the ΔgpaB mutant. Addition of 10 mM dibutyryl-cAMP to the agar plates completely restored the wild-type phenotype of both mutant strains. Interestingly, the expression of the pathogenicity determinant-encoding gene pksP measured as a pksPp-lacZ gene fusion was reduced in the ΔgpaB mutant. Moreover, the killing rates of conidia of both deletion strains (the ΔacyA and ΔgpaB strains) by human MDMs were significantly greater than the killing rate of wild-type conidia. Taken together, these findings suggested that cAMP triggers the defense system of A. fumigatus to protect the organism against attack by host immune effector cells.
cAMP signaling was found to control virulence and development in several human-pathogenic fungi, such as Candida albicans and Cryptococcus neoformans, and in plant-pathogenic fungi, such as Ustilago maydis and Magnaporthe grisea (reviewed in references 5, 15, 27, and 32). However, for A. fumigatus final proof was lacking since the mutants generated previously (33) were not tested in a low-dose animal infection model. Therefore, to provide final evidence that the cAMP signaling pathway is required for virulence of A. fumigatus, in this study a gene encoding a putative catalytic subunit of protein kinase A (PKA), designated pkaC1, was cloned and characterized. ΔpkaC1 and ΔgpaB mutants were almost avirulent in a low-dose inhalation mouse infection model.
MATERIALS AND METHODS
Fungal and bacterial strains, media, and growth conditions.
Fungal strains used in this study are listed in Table 1. A. fumigatus ATCC 46645 and CEA17 were used to generate pkaC1 knockout strains by using different selection marker genes. A. fumigatus CEA17 is a uracil-auxotrophic (pyrG1) mutant that was used to generate strains CEA17pksP-lacZ, CEA17ΔpkaC1pksP-lacZ1, and CEA17ΔpkaC1pksP-lacZ2. A. fumigatus strains were cultivated at 37°C in Aspergillus minimal medium (AMM) as previously described (56). Malt extract medium (2% [wt/vol] malt extract, 0.2% [wt/vol] yeast extract, 1% [wt/vol] glucose, 5 mM ammonium chloride, 1 mM dipotassium hydrogen phosphate) and AMM containing 3% (wt/vol) agar were used as solid media. Uridine (5 mM) or uracil (5 mM) was added to the media when required. For transformation of Escherichia coli, strains DH5α (Bethesda Research Laboratories, Gaithersburg, Md.), XL1-Blue (Stratagene, La Jolla, Calif.), INVαF′ (Invitrogen, Groningen, The Netherlands), and TOP10F′ (Invitrogen) were used. E. coli strains were grown at 37°C in Luria-Bertani medium supplemented, when required, with 50 μg of ampicillin per ml or 50 μg of kanamycin per ml.
TABLE 1.
A. fumigatus strains
| Strain | Genotype and/or phenotypea | Source or referenceb |
|---|---|---|
| ATCC 46645 | Wild type | ATCC |
| CEA17 | Derived from CBS144-89; pyrG1 | 13 |
| ΔpkaC1 | pkaC1::hph ΔpkaC1, Hygr | This study |
| ΔgpaB | gpaB::hph ΔgpaB, Hygr | This study |
| ΔacyA | acyA::hph ΔacyA, Hygr | 33 |
| CEA17ΔgpaB | pyrG1 gpaB::(neo-A. niger pyrG-neo) ΔgpaB, PyrG+ | 33 |
| CEA17ΔpkaC1 | pyrG1 pkaC1::(neo-A. niger pyrG-neo) ΔpkaC1, PyrG+ | This study |
| CEA17ΔpkaC1pyrG | pyrG1 pkaC1::(neo) ΔpkaC1 | This study |
| CEA17pksP-lacZ | pyrG1::pyrG2 pksPp-lacZ PyrG+ | 33 |
| CEA17ΔpkaC1pksP-lacZ1 | pyrG1::pyrG2 pksPp-lacZ pkaC1::(neo) ΔpkaC1, PyrG+ | This study |
| CEA17ΔpkaC1pksP-lacZ2 | pyrG1::pyrG2 pksPp-lacZ pkaC1::(neo) ΔpkaC1, PyrG+ | This study |
| CEA17ΔgpaBpksP-lacZ | pyrG1::pyrG2 pksPp-lacZ gpaB::(neo) ΔgpaB, PyrG+ | 33 |
hph, E. coli hygromycin B phosphotransferase gene; neo, Tn5 neomycin phosphotransferase gene.
ATCC, American Type Culture Collection.
Colony radial growth rate determination.
Diameters of A. fumigatus colonies were measured twice a day for at least 10 colonies of each strain for up to 72 h (malt extract medium) 94 h (AMM). The agar plates were inoculated centrally with a 2.5-μl drop of a suspension containing 1 × 106 spores per ml. The colony radial growth rate (51) was calculated from the slope of the line between 40 and 72 h on a plot of colony radius versus time starting from the time of inoculation. Data were processed by least-square regression analysis.
Quantification of sporulation.
Fifty microliters of a spore suspension containing 1 × 105 conidia prepared from a freshly harvested and filtered spore suspension was inoculated onto an AMM agar plate. Because of the high number of conidia the plate was covered by mycelium. The mass of mycelia was about the same for all plates irrespective of which strain was inoculated. Four plates containing each strain were incubated for 3 days, and the conidia produced on each plate were harvested with 10 ml of a saline solution containing 2% (vol/vol) Tween 80 (Merck, Darmstadt, Germany). The spore suspensions were filtered, and the numbers of conidia were determined by using a Thoma chamber.
Microscopic analysis of growth.
For microscopic analysis of mycelial growth, strains were grown for 24 h on slides coated with a thin layer of AMM agar. Microscopic photographs were taken with a confocal laser scanning microscope (Leica, Bensheim, Germany).
Analysis of conidial germination.
Fifty milliliters of AMM was inoculated with 107 conidia. Cultures were incubated at 180 rpm and 37°C. Over a period of 16 h, samples were taken and deposited on microscope slides. To determine germination, at least 100 conidia of each sample were examined.
Standard DNA techniques.
Standard techniques for the manipulation of DNA were carried out as described by Sambrook et al. (41). Chromosomal DNA of A. fumigatus was prepared as previously described for Aspergillus nidulans (3). For Southern blot analysis, chromosomal DNA of A. fumigatus was cut with different restriction enzymes. DNA fragments were separated on an agarose gel and blotted onto Hybond N+ nylon membranes (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Labeling of the DNA probe, hybridization, and detection of DNA-DNA hybrids were performed by using the DIG High Prime labeling and detection system (Amersham Pharmacia Biotech) according to the manufacturer's recommendations. Total RNA for cDNA synthesis was isolated by using an RNeasy plant mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The 5′ end of the pkaC1 transcript was determined by 5′ rapid amplification of cDNA ends (RACE) by using the 5′ RACE system (Invitrogen) according to the manufacturer's recommendations.
Sequence analysis.
Cosmid or plasmid DNA was sequenced on both strands by primer walking by using a Big Dye terminator cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom). The sequencing reaction mixtures were separated with an Applied Biosystems ABI 310 sequencer. DNA sequence data were edited by using the programs Sequence Navigator and Auto Assembler (Applied Biosystems). The analysis of the sequences was carried out by using Gene Works 2.2 (IntelliGenetics Inc.). Amino acid sequence comparisons were performed by using the BLAST 2.0 server (www.ncbi.nlm.gov/BLAST). The programs PepStat and ClustalW (Husar 4.0; DKFZ, Heidelberg, Germany) were used for predicting protein molecular masses and amino acid sequence alignments, respectively. Shading of aligned amino acid sequences was performed with MacBoxShade 2.15 (www.ch.embnet.org/software/BOX_form.html). Features and amino acid motifs of proteins were predicted by using the ISREC server (http://hits.isb-sib.ch/cgi-bin/PFSCAN).
Cloning of the A. fumigatus pkaC1 gene.
A 535-bp DNA fragment encoding part of the pkaC1 gene was obtained by PCR amplification by using degenerate oligonucleotides PPKAC1deg (5′-ACXYTXGGXACXGGXWSXTTYGG-3′) and PPKAC2deg (5′-TARTCXGGXGTXCCRCAXARXGT-3′), which were derived from conserved regions of known fungal homologues. Chromosomal DNA of A. fumigatus wild-type strain ATCC 46645 was used as the template. The DNA fragment obtained by PCR was ligated into plasmid pCR2.1 (Invitrogen). The cloned PCR product was sequenced and used as the probe to screen both a genomic A. fumigatus cosmid library (28) and an A. fumigatus Uni-ZAP cDNA library (Stratagene, Heidelberg, Germany). This resulted in isolation of cosmid cospkaC1-1 and the cDNA-containing plasmid pBKSpkaC1-1.
Generation of pkaC1 knockout plasmids encoding different selection markers.
To construct the pkaC1 deletion plasmids pUCpkaC1hph1 and pUCpkaC1Ura1, a 3.24-kbp PCR product spanning the pkaC1 gene and including a 0.64-kbp upstream sequence and a 0.92-kbp downstream sequence was generated by using oligonucleotides PPKAC12 (5′-TGAAGACGTAGATAGGGTCGA-3′) and PPKAC17 (5′-TGGTGTTTTTCTCGCCCCCT-3′) and DNA of cosmid cospkaC1-1 as the template. The PCR product was ligated into the pCR2.1 vector (TA cloning kit; Invitrogen) to obtain plasmid pTApkaC1-1. The PCR fragment was reisolated by restriction digestion of plasmid pTApkaC1-1 with HindIII/XbaI. This fragment was ligated into pUC18, which was also digested with HindIII/XbaI, yielding plasmid pUCpkaC1-1. So that we could use different selection markers for transformation of A. fumigatus, two different pkaC1 knockout plasmids were generated. One plasmid contained the hygromycin B resistance gene hph from E. coli as a selection marker. The complete hph gene was amplified by PCR by using oligonucleotides PHyg1 (5′-GGTTGAATTTAGAACGTGGC-3′) and PHyg2 (5′-CGCGTGGAGCCAAGAGCGG-3′) and plasmid pUCGH-pksPI (29) as the template. The 2.7-kbp PCR product generated was ligated into plasmid pCR2.1 (TA cloning kit; Invitrogen), yielding plasmid pTAhph1. After verification of the hph-encoding fragment by DNA sequence analysis, it was excised with BamHI/XbaI and cloned into pUC18, resulting in plasmid pUChph1. Plasmid pUChph1 was cut with BamHI/EcoRV, and the resulting 2.7-kbp fragment was ligated into the BclI and MscI restriction sites of pUCpkaC1-1 to obtain plasmid pUCpkaC1hph1. Plasmid pUCpkaC1hph1 contains the E. coli hph gene, which confers hygromycin B resistance, flanked by fragments located upstream and downstream of the part of the pkaC1 gene encoding the protein kinase domain. To obtain a linear fragment for transformation of A. fumigatus, plasmid pUCpkaC1hph1 was cut with HindIII/XbaI, resulting in a 5.0-kbp fragment that was used for transformation of A. fumigatus wild-type strain ATCC 46645.
For construction of a second pkaC1 knockout plasmid, the Ura-blaster was used. The Ura-blaster consists of the Aspergillus niger pyrG gene, which complements the uracil-auxotrophic CEA17 mutant strain. The pyrG gene is flanked by two copies of the neomycin phosphotransferase gene, which allows deletion of the Ura-blaster upon selection with 5-fluoroorotic acid (5-FOA) (13). An 8.6-kbp DNA fragment containing the Ura-blaster was isolated from plasmid pCDA14 (13) by HpaI digestion and ligated into the NruI/MscI-digested pUCpkaC1 plasmid, resulting in plasmid pUCpkaC1Ura1. To obtain a linear fragment for transformation of A. fumigatus strain CEA17, plasmid pUCpkaC1Ura1 was cut at the single NotI site.
Generation of a gpaB deletion strain for use in the animal infection model.
Construction of gpaB deletion mutant CEA17ΔgpaB with A. fumigatus parental strain CEA17 for transformation was described previously (33). In order to compare the virulence of isogenic strains, in this study another gpaB mutant was generated by using strain ATCC 46645 for transformation. Plasmid pUCgpaB1 (33) was cut with XhoI and religated, resulting in deletion of the PstI restriction site from the polylinker. The resulting plasmid, pUCgpaBPst−1, was digested with PstI and ligated with a 3.1-kbp NsiI fragment from plasmid pUCGH-pksP1 (29), which carries the E. coli hph gene conferring hygromycin B resistance to A. fumigatus. The resulting gpaB knockout plasmid was designated pUCgpaBhph1. After transformation of wild-type strain ATCC 46645 with a 5.5-kbp HindIII fragment of plasmid pUCgpaBhph1, a hygromycin B-resistant transformant was identified which showed the same phenotype with respect to growth and sporulation as the gpaB deletion mutant strain CEA17ΔgpaB (33). Southern blot analysis revealed that the transformant strain designated ΔgpaB did not contain the gpaB gene (data not shown).
Transformation of A. fumigatus and deletion of the Ura-blaster.
Transformation of A. fumigatus was carried out by using protoplasts as previously described (56). When selection for hygromycin B resistance was used, 100 μg of hygromycin B per ml was added to agar plates. Isolation of A. fumigatus strains carrying a deletion of the Ura-blaster was achieved by inoculating conidia onto AMM agar plates containing 5 mM uracil, 5 mM uridine, and 1 mg of 5-FOA per ml as previously described (13).
β-Galactosidase activity assays.
A. fumigatus strains were grown in AMM for 24 h at 37°C. β-Galactosidase activities were measured in protein extracts obtained from three A. fumigatus cultures grown in parallel. Specific activities were calculated as previously described for A. nidulans (8).
PKA activity assay and determination of protein concentrations.
A. fumigatus strains were grown in AMM for 24 h at 37°C. After harvesting, mycelia were frozen in liquid nitrogen and ground with a mortar and pestle. Mycelia were suspended in extraction buffer (25 mM Tris-HCl [pH 7.4], 1 mM dithiothreitol, 1 mM EDTA) and incubated on ice for 15 min. Samples were centrifuged at 4°C and 30,000 × g for 10 min. Ten microliters of the protein-containing supernatant adjusted to a protein concentration of 3 mg/ml was used in an assay for PKA activity by using fluorescent dye-coupled kemptide peptide (Promega, Mannheim, Germany) as the phosphoacceptor. Activities of protein extracts obtained from three A. fumigatus cultures grown in parallel were measured. Where indicated, 1 μM cAMP or 0.5 μg of a PKA inhibitor peptide (Promega) was added to the assay mixture. Purified cAMP-dependent PKA catalytic subunit from bovine heart (Promega) was used as the positive control. The incubation period for the phosphorylation reaction was 30 min at room temperature. This was the maximal period during which the control PKA activity increased at a constant rate. Protein concentrations were determined by the method of Bradford (6).
Animal infection model.
The murine low-dose model for invasive aspergillosis (45) as optimized by Liebmann et al. (34) for BALB/c mice was used. Briefly, 18- to 20-g female BALB/c mice were immunosuppressed with 100 mg of cyclophosphamide (Sigma, Taufkirchen, Germany) per ml on days −4, −1, 2, 5, 8, and 11 prior to and after infection on day 0. A single dose of cortisone acetate (200 mg/kg of body weight; Sigma) was injected subcutaneously on day −1. A. fumigatus conidium suspensions were harvested with phosphate-buffered saline containing 0.1% (vol/vol) Tween 80 (Merck) and filtered twice through Miracloth (Calbiochem, Bad Soden, Germany). Mice were anesthetized by intraperitoneal injection of 200 μl of 1% (vol/vol) ketamine (WDTeG, Garbsen, Germany)-0.02% (vol/vol) rompun (Bayer, Leverkusen, Germany) and intranasally infected with a 25-μl drop of a fresh suspension containing 5 × 103 or 104 conidia. Survival was monitored daily, and moribund animals were sacrificed by intraperitoneal injection of 200 μl of 3.2% (vol/vol) narcoren (Rhone Merieux, Laupheim, Germany). The drinking water was supplemented with 0.5 mg of tetracycline (Sigma) per ml to prevent bacterial infections. Lungs were recovered and homogenized in Lysing MatrixD tubes (Q-Biogene, Heidelberg, Germany) containing 1 ml of phosphate-buffered saline (Nunc, Wiesbaden, Germany). To determine the number of CFU, 50-μl portions of homogenates were plated in duplicate on malt agar plates.
Nucleotide sequence accession numbers.
Sequence data for pkaC1 have been deposited in the Swissprot and EMBL databases under accession numbers AJ297841 and Q8J129.
RESULTS
Identification of two genes encoding A. fumigatus PKA catalytic subunits.
To analyze the importance of the cAMP-PKA signaling pathway for A. fumigatus virulence, the pkaC1 gene encoding a homologue of PKA catalytic subunits was identified and isolated (accession no. AJ297841 and Q8J129). This gene was cloned by PCR by using degenerate oligonucleotides whose sequences matched conserved sequences of known fungal homologues (see Materials and Methods). The DNA fragment generated by PCR encoded part of the PKA catalytic subunit, and it was used as a probe to screen both a genomic cosmid library and a cDNA library of A. fumigatus. Starting from the known sequences present on both the isolated cosmid and the cDNA clone, the DNA sequence of the complete gene was determined by primer walking. The pkaC1 gene product is encoded by 1,687 bp, interrupted by three introns that are 65, 53, and 60 bp long. The intron positions were confirmed by sequencing the corresponding regions of the cDNA clone. The isolated cDNA clone contained a truncation at the 5′ end of the cDNA fragment lacking at least 420 bases up to the predicted start codon. Therefore, the 5′ end of the pkaC1 transcript was determined by 5′ RACE, and it was found to be located 136 bp upstream of the putative start codon. The deduced PKAC1 protein consists of 502 amino acid residues which result in a protein with a predicted molecular mass of 57 kDa. The amino acid sequence of PKAC1 revealed significant similarities to other fungal catalytic subunits of PKA (level of amino acid identity to A. nidulans PKAA, 76%; level of amino acid identity to A. niger PKAC, 76%; level of amino acid identity to M. grisea CPKA, 64%). A protein kinase domain characteristic of eukaryotic protein kinases is located between amino acids 191 and 446. Important motifs characteristic of PKA catalytic subunits were found in the amino acid sequence of PKAC1. These motifs include the RDLKPEN sequence of the catalytic loop (amino acids 314 to 320) and the DFGFAKz sequence involved in Mg2+ binding (amino acids 333 to 338). Additional conserved sequences (26) include the nucleotide binding loop GTGSFG (amino acids 198 to 203) and three amino acid residues shown to be required for association of catalytic and regulatory subunits (H235, W344, and L346). An interesting additional feature of PKAC1 is a glutamine-rich stretch (amino acids 122 to 143) consisting of 21 glutamine residues interrupted by a proline residue. Glutamine stretches can be important for protein-protein interactions and are present in some other protein kinases in fungi and slime molds (18, 10, 46). Partial sequences corresponding to both pkaC1 and a gene encoding a putative second PKA catalytic subunit homologue (pkaC2) are present in the A. fumigatus genome sequence database (http://www.tigr.org).
PKAC1 represents the predominant form controlling conidiation and vegetative growth.
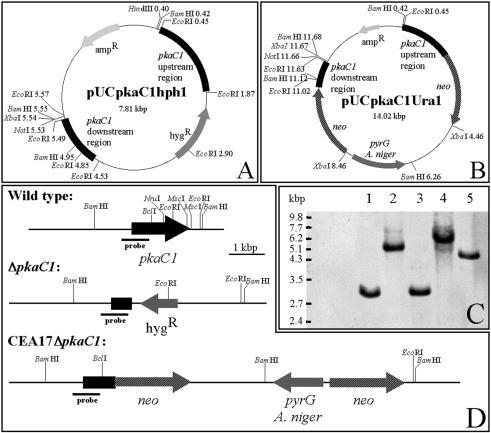
To determine its biological function, the pkaC1 gene was deleted by transformation and homologous recombination. To do this, plasmids pUCpkaC1hph1 and pUCpkaC1Ura1 (Fig. 1A and B) were generated (see Materials and Methods). These plasmids contained different selection markers, the hygromycin B resistance gene hph and the Ura-blaster, respectively. Linearized DNA fragments encoding the selection markers flanked by upstream and downstream sequences of the protein kinase domain of PKAC1 were used for transformation of either A. fumigatus wild-type strain ATCC 46645 or the uracil-auxotrophic pyrG mutant strain CEA17. Fifty hygromycin B-resistant transformants of strain ATCC 46645 were isolated. Ten of these transformants were tested by Southern blot analysis, which resulted in identification of four strains which exhibited the expected gene replacement. Three of these four transformants carried an additional ectopic integration of the knockout construct (data not shown). The remaining transformant exhibited only the hybridization pattern characteristic of a gene replacement event (Fig. 1C and D). This strain was designated ΔpkaC1 and was used for further studies. All of the uracil-prototrophic transformants of strain CEA17 were tested by Southern blot analysis, and five of the strains tested showed a gene replacement event. However, two of these transformants carried an additional ectopic integration of the DNA fragment used for transformation (data not shown). One of the transformants that showed a hybridization pattern characteristic of a gene replacement at the pkaC1 locus was designated CEA17ΔpkaC1 and was used for further studies (Fig. 1C and D).
FIG. 1.
Deletion of A. fumigatus PKA catalytic subunit-encoding gene pkaC1. (A) Schematic map of the pkaC1 knockout plasmid pUCpkaC1hph1. Abbreviations: ampR, ampicillin resistance gene; hygR, hygromycin B phosphotransferase gene hph used as the selection marker gene in A. fumigatus. (B) Schematic map of the pkaC1 knockout plasmid pUCpkaC1Ura1. Abbreviations: ampR, ampicillin resistance gene; pyrG, orotidine 5′-monophosphate decarboxylase gene of A. niger used as the selection marker gene; neo, neomycin phosphotransferase genes. (C) Southern blot analysis of the pkaC1 deletion strains. Chromosomal DNA of the parental strains ATCC 46645 (lane 1) and CEA17pksP-lacZ (lane 3), as well as mutant strains ΔpkaC1 (lane 2), CEA17ΔpkaC1 (lane 4), and CEA17ΔpkaC1pksP-lacZ1 (lane 5), was cut by BamHI. An 830-bp pkaC1-derived PCR fragment was used as the probe. In the ΔpkaC1, CEA17ΔpkaC1, and CEA17ΔpkaC1pksP-lacZ1 mutant strains, the band characteristic of the wild type (lanes 1 and 3) had disappeared. Instead, the bands characteristic of gene replacement at the pkaC1 locus were detected. The difference in the sizes of the bands in lane 2 (ΔpkaC1) and lane 4 (CEA17ΔpkaC1) is due to the different cassettes, hph and Ura-blaster, used to delete the pkaC1 gene. Strain CEA17ΔpkaC1pksP-lacZ1 (lane 5) resulted from a forced recombination in which the Ura-blaster was lost during selection on medium containing 5-FOA. Consequently, a copy of the neo gene remained at the pkaC1 gene locus, which led to a pkaC1 knockout band that migrated more quickly than the band of strain CEA17ΔpkaC1 (lane 4). (D) Schematic representation of the chromosomal pkaC1 locus of the wild type and the ΔpkaC1 deletion mutants. Restriction endonuclease cleavage sites and the position to which the probe hybridizes are indicated. The pkaC1 genes in mutants ΔpkaC1 and CEA17ΔpkaC1 lack the pkaC1 regions encoding amino acids 252 to 502 and 336 to 502, respectively, as well as 58 bp of the 3′ untranslated region.
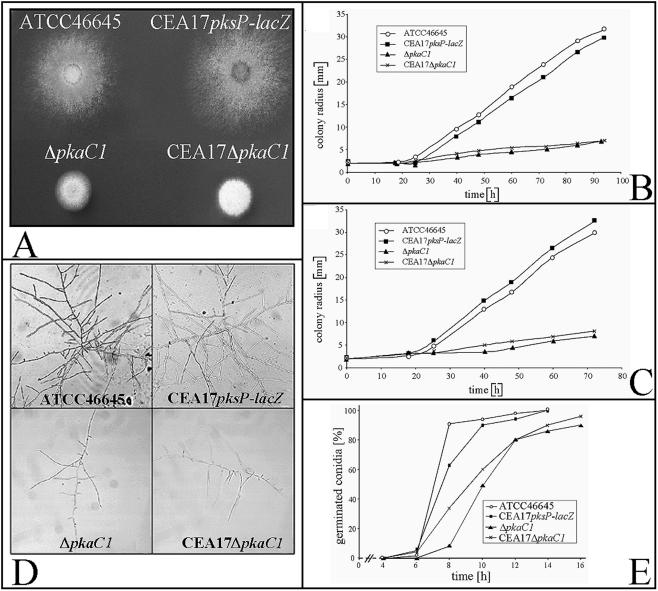
Growth of strains ΔpkaC1 and CEA17ΔpkaC1 on agar plates revealed that the pkaC1 deletion affected growth, sporulation, and germination of A. fumigatus (Fig. 2). pkaC1 deletion strains formed small colonies with few conidiophores (Fig. 2A). The growth rate was quantified on both AMM and malt extract agar plates. AMM was used as a minimal medium to characterize the phenotypes of mutants. Malt extract medium was used as a complete medium in order to analyze whether some phenotypes could be complemented just by the presence of complex nutrients. Quantification of the growth rates on AMM and malt extract agar plates revealed that the specific growth rate of the ΔpkaC1 mutant was strongly reduced (90 and 81%, respectively) compared with the specific growth rate of the wild type (Fig. 2B and C). Accordingly, on AMM and malt extract agar plates, strain CEA17ΔpkaC1 showed reductions in growth of 88 and 80%, respectively, compared with the corresponding parental strains (Fig. 2B and C). It turned out that on both AMM and malt extract agar plates the difference in colony radius after 24 h was only moderate (Fig. 2B and C). Therefore, microscopic inspection was used as a more sensitive method to detect subtle differences in germination and growth. Microscopic inspection revealed that after 24 h of growth on AMM agar, both pkaC1 mutant strains had produced only a few hyphae compared with wild-type strains ATCC 46645 and CEA17pksP-lacZ (Fig. 2D). In order to evaluate the role of pkaC1 during conidial germination, germ tube outgrowth for conidia germinated in AMM was measured. The results (Fig. 2E) demonstrate that the pkaC1 deletion affected the kinetics of germ tube outgrowth. After 10 h of incubation, 94 and 90% of the conidia of parental strains ATCC 46645 and CEA17pksP-lacZ, respectively, had germinated (Fig. 2E). By contrast, only 49 and 60% of the conidia of mutant strains ΔpkaC1 and CEA17ΔpkaC1, respectively, had formed germ tubes after 10 h (Fig. 2E). Prolongation of the incubation time up to a maximum of 16 h led to germination of 90% of the pkaC1 mutant conidia (Fig. 2E). Taken together, these results suggest that pkaC1 deletion resulted in delayed germination kinetics of A. fumigatus. Moreover, compared to the wild-type and parental strains, which produced significant numbers of conidia after incubation on AMM agar plates for 2 to 3 days, macroscopic inspection revealed that the pkaC1 deletion strains produced far fewer conidia (Fig. 2A). Compared with the numbers of conidia produced by wild-type strains, on AMM agar plates the numbers of conidia produced by strains ΔpkaC1 and CEA17ΔpkaC1 were reduced by 89 and 94%, respectively (Table 2). The reduction did not change even when the incubation was extended for 3 days. These data indicate that PKAC1 has a direct effect on formation of conidia. As expected, suppression of the ΔpkaC1 deletion phenotype was not observed upon addition of 10 mM dibutyryl-cAMP to the agar plates (data not shown), suggesting that PKAC1 acts downstream of adenylate cyclase.
FIG. 2.
Phenotypic characterization of mutant strains ΔpkaC1 and CEA17ΔpkaC1 and parental strains ATCC 46645 and CEA17pksP-lacZ. (A) Growth and sporulation on AMM agar plates. Colonies were grown for 72 h at 37°C. (B and C) Growth of A. fumigatus strains on AMM agar plates at 37°C up to 94 h (B) and on malt extract agar plates at 37°C up to 72 h (C). Colony diameters were measured. The data for each strain represent the means for at least 10 independently grown colonies. The standard deviations were in the range from 0.2 to 0.8 mm. (D) Microscopic photographs (magnification, ×200) of A. fumigatus strains grown on AMM agar at 37°C for 24 h. (E) Kinetics of germ tube outgrowth for A. fumigatus conidia incubated in AMM at 37°C. The numbers of conidia showing a germ tube were determined after different times of incubation in at least two microscopic fields. The percentage of germinated conidia (based on the total number of conidia) is shown. The results are representative of the results of two independent experiments.
TABLE 2.
Conidiation of A. fumigatus parental and deletion strains
| Strain | Density of conidia (conidia/ml of suspension) |
|---|---|
| ATCC 46645 | 5.68 × 108 ± 0.41 × 108 |
| ΔpkaC1 | 6.08 × 107 ± 2.47 × 107 |
| CEA17pksP-lacZ | 5.61 × 108 ± 0.94 × 108 |
| CEA17ΔpkaC1 | 3.23 × 107 ± 0.60 × 107 |
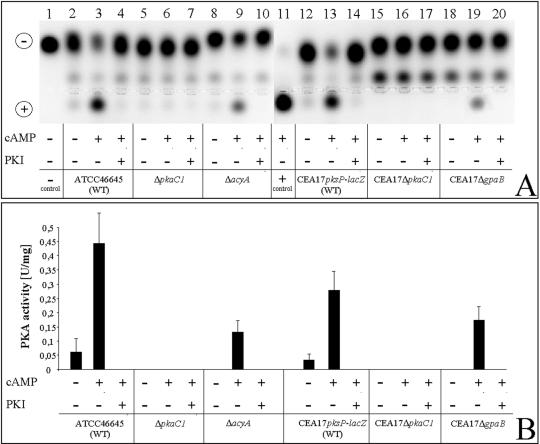
To determine the contribution of PKAC1 to total PKA activity, PKA activity in cell extracts of wild-type strains and both ΔpkaC1 deletion strains was assayed. cAMP-dependent PKA activity was detectable in both wild-type strains (Fig. 3A, lanes 2 and 12, and Fig. 3B). Activation with 5 μM cAMP led to significantly increased PKA activity of protein extracts of the wild-type strains (Fig. 3A, lanes 3 and 13, and Fig. 3B). After addition of the PKA-specific inhibitor, no activity was measured, confirming the validity of the assay method (Fig. 3A, lanes 4 and 14, and Fig. 3B). In contrast to wild-type strains, no PKA activity was detected in the ΔpkaC1 deletion strains ΔpkaC1 and CEA17ΔpkaC1 (Fig. 3A, lanes 5 to 7 and 15 to 17, and Fig. 3B), irrespective of whether cAMP was added to the assay mixture (Fig. 3A, lanes 6 and 16, and Fig. 3B). These findings provided evidence that pkaC1 encodes a PKA catalytic subunit that represents the predominant form of the enzyme under the conditions tested.
FIG. 3.
PKA activities of mutant strains ΔpkaC1, CEA17ΔpkaC1, ΔacyA, and CEA17ΔgpaB and parental strains ATCC 46645 and CEA17pksP-lacZ. (A) Enzyme activity as monitored by gel electrophoresis. A phosphorylated substrate migrated toward the anode. (B) Spots shown in panel A quantified by spectrophotometry. Abbreviations: PKI, PKA inhibitor peptide; WT, wild type.
acyA and gpaB mutants lack PKA activity without activation by cAMP.
PKA activity was not detected in either of the adenylate cyclase and G protein α subunit mutant strains, ΔacyA and CEA17ΔgpaB (Fig. 3A, lanes 8 and 18, and Fig. 3B). However, addition of cAMP to the assay mixture led to significant PKA activity in protein extracts of both strains, although the levels did not reach wild-type levels (Fig. 3A, lanes 9 and 19, and Fig. 3B). This finding indicated that PKA is present but not activated in these mutant strains, most likely due to the lack of significant amounts of cAMP in either strain. This result further confirms the finding that GPAB is an element of the cAMP signaling pathway. Furthermore, it shows that cellular PKAC1 activity depends on cAMP.
cAMP-PKA-dependent expression of a pksPp-lacZ gene fusion.
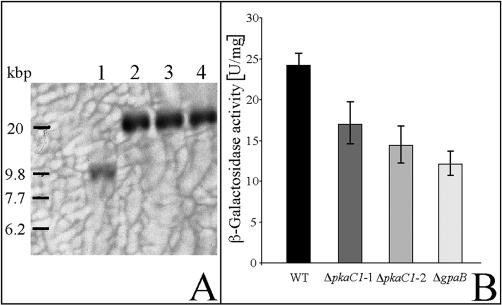
To analyze whether signaling via PKA is involved in the expression of pksP, A. fumigatus pkaC1 deletion strains carrying a pksPp-lacZ gene fusion were generated. Thus, the Ura-blaster of strain CEA17ΔpkaC1, which was initially introduced to delete the pkaC1 gene, was removed by selection on 5-FOA-containing agar plates, resulting in strain CEA17ΔpkaC1pyrG. Strain CEA17ΔpkaC1pyrG was transformed with circular DNA of plasmid pUCpyrG2pksP-lacZ (33), which complemented the uracil auxotrophy of the recipient strain by integration of the plasmid at the chromosomal pyrG locus. The results of a Southern blot analysis of transformants are shown in Fig. 4A. The 8-kbp band characteristic of the wild-type pyrG gene (Fig. 4A, lane 1) had disappeared. Instead, parental strain CEA17pksP-lacZ (Fig. 4A, lane 2) and two ΔpkaC1 transformants (Fig. 4A, lanes 3 and 4) had an 18-kbp band due to the integration of the plasmid at the pyrG gene locus. The newly created transformants were designated CEA17ΔpkaC1pksP-lacZ1 and CEA17ΔpkaC1pksP-lacZ2. In addition, the ΔpkaC1 genotype of transformant CEA17ΔpkaC1pksP-lacZ1 was checked by Southern blot analysis (Fig. 1C). The band characteristic of the wild-type control (Fig. 1C, lane 3) had disappeared in the ΔpkaC1 mutant strain CEA17ΔpkaC1pksP-lacZ1. Instead, a 4.8-kbp DNA fragment caused by deletion of pkaC1 and the subsequent removal of the Ura-blaster by selection on 5-FOA was detected (Fig. 1C, lane 5).
FIG. 4.
Expression of the pksPp-lacZ gene fusion in A. fumigatus ΔpkaC1 and ΔgpaB deletion strains carrying a pksPp-lacZ gene fusion integrated in a single copy at the chromosomal pyrG locus. (A) Southern blot analysis. Chromosomal DNA of the A. fumigatus parental strains ATCC 46645 (lane 1) and CEA17pksP-lacZ (lane 2) and the ΔpkaC1 mutants CEA17ΔpkaC1pksP-lacZ1 (lane 3) and CEA17ΔpkaC1pksP-lacZ2 (lane 4) was digested by BglII. A 450-bp PCR fragment encoding part of the A. fumigatus pyrG gene was used as the probe. Molecular analysis of strain CEA17ΔgpaBpksP-lacZ is shown in Fig. 1. (B) β-Galactosidase specific activities of strains grown in AMM at 37°C for 24 h. The data for each strain and condition are the mean and standard deviation for three independently grown cultures. Abbreviations: WT, parental strain CEA17pksP-lacZ; ΔpkaC1-1, CEA17ΔpkaC1pksP-lacZ1; ΔpkaC1-2, CEA17ΔpkaC1pksP-lacZ2; ΔgpaB, CEA17ΔgpaBpksP-lacZ.
The expression of the pksPp-lacZ gene fusion of the parental strain, the ΔpkaC1 deletion strains, and the ΔgpaB deletion strain was determined. The results are shown in Fig. 4B. There was a moderate but significant difference between the ΔpkaC1 mutants and the wild-type strain. The ΔpkaC1-1 (CEA17ΔpkaC1pksP-lacZ1) and ΔpkaC1-2 (CEA17ΔpkaC1pksP-lacZ2) mutant strains showed 70 and 60%, respectively, of pksPp-lacZ expression measured for the wild-type strain (Fig. 4B). This level of expression is more than that of the ΔgpaB strain, which showed 50% of the wild-type pksP expression (Fig. 4B). These results suggest that the cAMP-PKA signaling network is involved in the regulation of the pksP gene.
Mutants with mutations in elements of the cAMP-PKA signal transduction pathway were almost avirulent.
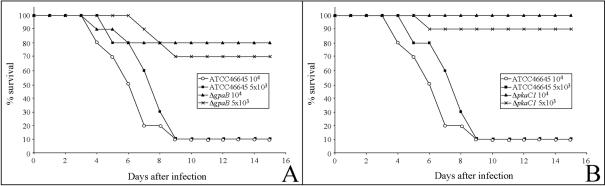
To assess the roles of GPAB and PKAC1 in pathogenesis, the corresponding deletion mutants were tested in an animal infection model of invasive aspergillosis. Groups of 10 immunosuppressed mice were infected by inhalation with 5 × 103 or 104 conidia of the different strains. The results are shown in Fig. 5. In the groups infected with wild-type conidia (strain ATCC 46645), irrespective of the number of conidia given, the mortality was 90% after 9 days (Fig. 5). By contrast, when mice were infected with conidia of the ΔgpaB mutant strain, the mortality was drastically reduced to 30 and 20% in the animal groups infected with 5 × 103 and 104 conidia, respectively (Fig. 5A). When mice were infected with conidia of the ΔpkaC1 mutant, 90 and 100% of the animals survived in the groups infected with 5 × 103 and 104 conidia, respectively (Fig. 5B). The lungs of 15 of 18 animals which died after infection with wild-type conidia were found to be colonized with A. fumigatus at levels of 168 ± 101 CFU/g of tissue. A. fumigatus could not be reisolated from the lungs of either survivor infected with wild-type conidia. With one exception, the lungs of mice infected with ΔpkaC1 mutant conidia were not colonized by A. fumigatus. In the lungs of animals which died after infection with ΔgpaB mutant conidia, we found A. fumigatus at levels of 245 ± 215 CFU/g of tissue. Interestingly, the lungs of five survivors of ΔgpaB infection were also colonized with the fungus at levels of 503 ± 345 CFU/ g of tissue. These results indicate that in some cases, the A. fumigatus gpaB mutant was able to germinate in the lungs of immunosuppressed mice. However, after germination the fungus seemed to persist in the lungs without invasive growth because the corresponding mice did not show symptoms of disease. Taken together, deletion of components of the cAMP signaling pathway of A. fumigatus resulted in significantly reduced virulence. The strains were almost avirulent. However, in contrast to the ΔgpaB mutant, ΔpkaC1 strains grew poorly, which needs to be taken into account when virulence is assessed (see Discussion).
FIG. 5.
Virulence of A. fumigatus wild-type strain ATCC 46645, as well as ΔgpaB (A) and ΔpkaC1 (B) deletion strains, in mice. The members of groups of 10 BALB/c mice were each infected with 5 × 103 or 104 A. fumigatus conidia, as indicated, by nasal inhalation. Survival was monitored for 15 days.
DISCUSSION
Here we analyzed the importance of the cAMP-PKA signal transduction pathway for A. fumigatus growth, development, and virulence. The pkaC1 gene encoding a PKA catalytic subunit was identified. Moreover, the gpaB gene previously shown to encode a putative member of the cAMP signaling network was shown here to be required for virulence. Previously, Oliver et al. (37) reported the isolation of a gene from A. fumigatus designated pkaC. This gene encodes a PKA catalytic subunit with 490 amino acids and a deduced molecular mass of 56 kDa. Until now, the gene and the encoded protein had not been characterized further. Sequence alignments revealed that the previously identified gene is identical to the pkaC1 gene described here. However, PKAC1 is composed of 502 amino acids and has a predicted molecular mass of 57 kDa. Sequence comparisons showed that the PKAC protein described by Oliver et al. (37) lacks 12 glutamine residues present in the N-terminal Q stretch of PKAC1, whose presence was confirmed by repeated sequencing of both pkaC1 genomic DNA and cDNA.
To determine its biological functions, the pkaC1 gene of A. fumigatus was deleted. Deletion was carried out by using two different selection markers. Both mutants with mutations in the PKA catalytic subunit (ΔpkaC1 and CEA17ΔpkaC1) displayed the same phenotype (i.e., delayed germ tube outgrowth and severely impaired sporulation and growth). The ΔpkaC1 phenotypes resembled those of the A. fumigatus adenylate cyclase mutant (ΔacyA), whose growth was also reduced; in contrast, the adenylate cyclase mutant showed almost no conidiation on agar plates (33). With respect to sporulation this phenotype was also observed for the A. fumigatus ΔgpaB mutant, which is defective in a stimulatory Gα subunit, which most likely leads to decreased activation of adenylate cyclase and therefore reduced cAMP levels. This mutant produced only one-half as many conidia as the wild-type strains. Wild-type phenotypes of both ΔacyA and ΔgpaB mutants were restored by addition of dibutyryl-cAMP to the medium (33). As expected, this was not the case for the ΔpkaC1 mutants, because PKA acts downstream of cAMP synthesis. Elevated cAMP levels did not produce an increase in an alternative cellular PKA activity, which could, at least in part, replace the PKAC1 function. This finding was confirmed by data which showed that there was no measurable PKA activity in crude extracts of the ΔpkaC1 deletion strains. Moreover, no PKA activity was detectable in either the ΔgpaB or ΔacyA mutants, most likely due to decreased levels or lack of cAMP in these strains. PKA activity in the ΔacyA and ΔgpaB deletion mutants was partially restored by addition of cAMP to the assay mixtures. These results indicate that PKAC1 activity depends on the cAMP level and, in addition, that PKAC1 acts downstream of GPAB and ACYA in the same pathway. Taken together, our results indicate that pkaC1 encodes an active PKA catalytic subunit which is part of the cAMP-PKA signal transduction pathway regulating growth and sporulation of A. fumigatus.
Interestingly, the observed ΔpkaC1 phenotypes only partially resembled those of, e.g., A. nidulans ΔpkaA and A. niger ΔpkaC mutants, which had mutations in PKA catalytic subunit genes that showed the highest levels of similarity to A. fumigatus pkaC1. The latter two mutants also displayed reduced growth rates on agar plates and delayed germination (17, 43, 47). Interestingly, deletion of pkaA in A. nidulans led to hyperconidiation, while pkaA overexpression resulted in decreased sporulation (43). This is in contrast to the findings reported here for A. fumigatus. ΔpkaC1 strains showed drastically reduced sporulation, indicating that asexual development of conidia is regulated differently at the level of PKA in the two fungi. Disruption of the A. niger PKA regulatory subunit-encoding gene pkaR resulted in very small colonies on plates, the absence of sporulation, and a complete loss of growth polarity during submerged growth (47). These phenotypes were even more drastic than those described for the A. niger ΔpkaC strain, which formed sporulating colonies whose diameters were two- to threefold smaller than those of wild-type colonies (47). Taken together, PKA mutants of fungi closely related to A. fumigatus exhibited similar growth phenotypes but different sporulation phenotypes.
The remaining, weak sporulation of A. fumigatus ΔpkaC1 strains opens up the possibility of a minor role for other catalytic PKA subunits in regulation of sporulation. One of these subunits could be encoded by the pkaC2 gene which was identified in the genome of A. fumigatus. However, in ΔpkaC1 strains this subunit and putative additional existing catalytic subunits did not show any activity under the conditions tested. This result suggests that PKAC1 plays the predominant role in cAMP-PKA signaling. Alternatively, PKAC2 and putative additional existing catalytic subunits were not active under the conditions used or could not be measured with the assay used. It is also conceivable that a cAMP-PKA-independent signal cascade is involved in sporulation, which, under certain conditions, could overlap the PKA-cAMP pathway. For example, a novel transcription factor regulating sporulation of A. nidulans was identified. This transcription factor, designated DevR, was not required for sporulation when the fungus was grown on 0.6 M NaCl but was essential for sporulation when the fungus was grown under standard conditions. There is a homologous gene in A. fumigatus (55). The function of pkaC2, if there is any, will be analyzed in future experiments.
Similar observations were reported for both the human-pathogenic basidiomycete C. neoformans and the rice pathogen M. grisea. Δpka1 and ΔcpkA mutants, respectively, showed almost no detectable PKA activity (1, 14), but a second PKA isoform for each fungus was identified in the corresponding genome databases (14; K. Adachi and J. E. Hamer, EMBL database accession no. AA021201). Until now, however, these PKA isoforms have not been characterized. The existence of multiple isoforms of catalytic PKA subunits has been described for several other fungi, including Saccharomyces cerevisiae, in which PKA is essential for viability (48, 49). The three subunits identified (TPK1 to TPK3) have redundant functions, including regulation of intermediary metabolism, especially carbohydrate metabolism (reviewed in reference 49), response to stress conditions (44), cell cycle control (39), and pseudohyphal morphogenesis (38, 40). In the human pathogen C. albicans two catalytic PKA isoforms, Tpk1 and Tpk2, were found. Both of these isoforms play distinct and redundant roles in morphogenesis, growth, and virulence (4, 46). The corn smut fungus U. maydis also contains two PKA catalytic subunits, Adr1 and Uka1. Only Adr1 is responsible for the main cellular PKA activity and is important for mating and virulence (16).
The key question concerns the importance of cAMP-PKA signaling for virulence. In order to answer this question, two different aspects were considered: (i) a possible influence of PKAC1 on the expression of the pathogenicity determinant-encoding gene pksP; and (ii) the importance of elements of the cAMP signaling cascade for the pathogenicity of the fungus in a low-dose animal model. As shown previously, the pksP gene is expressed mainly during sporulation, but it was also expressed in outgrowing hyphae of A. fumigatus when conidia germinated in the lungs of immunocompromised mice (28, 29, 53, 54). In both ΔpkaC1 mutant strains, reduced pksPp-lacZ expression was measured in hyphae under standard growth conditions. This result agrees well with the previous finding that pksP expression is decreased in an A. fumigatus ΔgpaB mutant (33) (Fig. 4). The ΔgpaB effect is most likely due to the lowered cAMP level and therefore the lack of stimulation of adenylate cyclase activity by the G(s)α subunit GPAB. Lower cAMP levels then lead to decreased activation of PKA. However, the significantly higher pksPp-lacZ expression in the ΔpkaC1 mutants compared with the expression in the ΔgpaB strain suggests that in addition to the GPAB-cAMP-PKA network, there is a GPAB-dependent PKA-independent signaling cascade that influences pksP expression.
Deletion of gpaB or pkaC1 led to drastically attenuated virulence of A. fumigatus. In fact, the mutants were almost avirulent. This is in agreement with the previous finding that conidia of A. fumigatus cAMP signaling mutants (ΔgpaB and ΔacyA) were killed more efficiently by human MDMs than conidia of the wild-type strains were killed (33). However, the decreased virulence of the ΔpkaC1 mutant needs to be seen in the context of its poor growth. We cannot exclude the possibility that the survival of mice infected with the mutant was due to delayed germination or the reduced growth rate of the organism. Similar observations were made for PKA mutants of C. neoformans and U. maydis (14, 16). However, in contrast to the ΔpkaC1 deletion mutants, the ΔgpaB mutant did not show any growth retardation (33). Nevertheless, as shown here, the virulence of the ΔgpaB strain was strongly attenuated, confirming the importance of cAMP signaling for virulence in A. fumigatus. Moreover, the A. fumigatus virulence determinant-encoding gene pksP is also regulated by cAMP-PKA signaling, and decreased expression of pksP, as shown here for both the ΔgpaB and ΔpkaC1 mutants, could contribute to better killing of the mutants in the animal model.
The polyketide synthase PKSP catalyzes the initial step of DHN-melanin biosynthesis in A. fumigatus. In both M. grisea and C. neoformans, melanins play an important role in virulence. This led to the question of whether in melanin-producing fungi the expression of melanin biosynthesis genes is a general mechanism required for fungal pathogenicity (reviewed in reference 30). At least one of the signal transduction pathways triggering the expression of melanin biosynthesis genes in A. fumigatus appears to be similar to the pathways in C. neoformans and M. grisea. In the latter two fungi, both melanin biosynthesis and virulence were affected by deletion of genes encoding Gα protein subunits, adenylate cyclases, and catalytic subunits of PKA (2, 11, 12, 14, 19, 20, 21, 35, 36, 50).
However, the situation in A. fumigatus appears to be rather complex. Analysis of pigmentless pksP mutant conidia which showed reduced virulence in a murine infection model and were more sensitive to ROS indicated that the DHN-melanin-containing pigment is able to scavenge ROS (23, 28, 53, 54). However, the same is true for the green pigment of the nonpathogenic A. nidulans conidia, which does not seem to be synthesized via the DHN-melanin pathway (24). Therefore, both pigments might act as protective agents against oxidant-based host defense mechanisms and thus contribute to the relative resistance of conidia to both alveolar macrophage and neutrophil attack. However, these findings do not explain why A. fumigatus conidia can be pathogenic, whereas this is rarely the case for A. nidulans conidia. An attractive hypothesis is that the pksP gene product of A. fumigatus is involved in the production of a compound which is immunosuppressive. This could be the DHN-melanin pigment, an intermediate or by-product of DHN-melanin, or even another compound in whose biosynthesis PKSP is involved (7, 9). This hypothesis is supported by the notion that the presence of a functional pksP gene in A. fumigatus conidia is associated with inhibition of the fusion of phagosomes and lysosomes in human MDMs (25). As shown here, both ΔpkaC1 and ΔgpaB mutant conidia are grey-green. Therefore, pigment was produced, although the possibility that the amount was lower than that in the wild type cannot be excluded yet. It therefore seems unlikely that ΔpkaC1 and ΔgpaB mutant conidia exhibit a strong increase in ROS sensitivity. This finding suggests that the pigment plays only a minor role in virulence. Consequently, the attenuated virulence of the ΔgpaB mutant in particular supports the alternative model of formation of an immunosuppressive compound by PKSP and, furthermore, strongly suggests that there are genes other than pksP that are involved in pathogenicity and are also regulated by the cAMP-PKA cascade.
In conclusion, the involvement of cAMP signaling in virulence appears to be a common theme in plant- and human-pathogenic fungi. For A. fumigatus it will be interesting to discover which other genes are regulated by the cAMP-PKA network and whether other pathogenicity determinants are found among them.
Acknowledgments
We thank Yvonne Runge for excellent technical assistance and Kim Langfelder for helpful discussions. Christophe d'Enfert kindly provided strain CEA17 and plasmid pCDA14.
This research was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 587, Tp A8, B4).
Editor: T. R. Kozel
REFERENCES
- 1.Adachi, K., and J. E. Hamer. 1998. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell 10:1361-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianopoulos, A., and M. J. Hynes. 1988. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol. Cell. Biol. 8:3532-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bockmühl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243-1257. [DOI] [PubMed] [Google Scholar]
- 5.Bölker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25:143-156. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brakhage, A. A., and B. Jahn. 2002. Molecular mechanisms of pathogenicity of Aspergillus fumigatus, p. 559-582. In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, Dordrecht, The Netherlands.
- 8.Brakhage, A. A., and J. Van den Brulle. 1995. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J. Bacteriol. 177:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brakhage, A. A., and K. Langfelder. 2002. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 56:433-455. [DOI] [PubMed] [Google Scholar]
- 10.Buhr, T. L., S. Oved, G. M. Truesdell, C. Huang, O. Yarden, and M. B. Dickman. 1996. A kinase-encoding gene from Colletotrichum trifolii complements a colonial growth mutant of Neurospora crassa. Mol. Gen. Genet. 251:565-572. [DOI] [PubMed] [Google Scholar]
- 11.Choi, W., and R. A. Dean. 1997. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Jong, J. C., N. McCormack, N. Smirnoff, and N. T. Talbot. 1997. Glycerol generates turgor in rice blast. Nature 389:244-245. [Google Scholar]
- 13.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed] [Google Scholar]
- 16.Dürrenberger, F., K. Wong, and J. W. Kronstad. 1998. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc. Natl. Acad. Sci. USA 95:5684-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fillinger, S., M. K. Chaveroche, K. Shimizu, N. Keller, and C. d'Enfert. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001-1016. [DOI] [PubMed] [Google Scholar]
- 18.Haribabu, B., and R. P. Dottin. 1991. Identification of a protein kinase multigene family of Dictyostelium discoideum: molecular cloning and expression of a cDNA encoding a developmentally regulated protein kinase. Proc. Natl. Acad. Sci. USA 88:1115-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard, R. J. 1994. Cell biology of pathogenesis, p. 3-22. In R. S. Zeigler, S. A. Leong, and P. S. Teng (ed.), Rice blast disease. CAB International, Wallington, United Kingdom.
- 20.Howard, R. J., and M. A. Ferrari. 1989. Role of melanin in appressorium function. Exp. Mycol. 13:403-418. [Google Scholar]
- 21.Howard, R. J., T. M. Bourett, and M. A. Ferrari. 1991. Infection by Magnaporthe grisea: an in vitro analysis, p. 251-264. In K. Mendgen and D. E. Lesemann (ed.), Electron microscopy of plant pathogens. Springer-Verlag, Berlin, Germany.
- 22.Ibrahim-Granet, O., B. Philippe, H. Boleti, E. Boisvieux-Ulrich, D. Grenet, M. Stern, and J.-P. Latgé. 2003. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect. Immun. 71:891-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn, B., A. Koch, A. Schmidt, G. Wanner, H. Gehringer, S. Bhakdi, and A. A. Brakhage. 1997. Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect. Immun. 65:5110-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahn, B., F. Boukhallouk, J. Lotz, K. Langfelder, G. Wanner, and A. A. Brakhage. 2000. Interaction of human phagocytes with pigmentless Aspergillus conidia. Infect. Immun. 68:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahn, B., K. Langfelder, U. Schneider, C. Schindel, and A. A. Brakhage. 2002. PKSP-dependent reduction of phagolysosome fusion and intracellular kill of Aspergillus fumigatus conidia by human monocyte-derived macrophages. Cell. Microbiol. 4:793-803. [DOI] [PubMed] [Google Scholar]
- 26.Knighton, D. R., J. Zheng, L. F. Ten Eyck, V. A. Ashford, N. H. Xuong, S. S. Taylor, and J. M. Sowadski. 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407-420. [DOI] [PubMed] [Google Scholar]
- 27.Kronstad, J., A. D. De Maria, D. Funnell, R. D. Laidlaw, N. Lee, M. M. de Sa, and M. Ramesh. 1998. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch. Microbiol. 170:395-404. [DOI] [PubMed] [Google Scholar]
- 28.Langfelder, K., B. Jahn, H. Gehringer, A. Schmidt, G. Wanner, and A. A. Brakhage. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187:79-89. [DOI] [PubMed] [Google Scholar]
- 29.Langfelder, K., B. Philippe, B. Jahn, J.-P. Latgé, and A. A. Brakhage. 2001. Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 69:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38:143-158. [DOI] [PubMed] [Google Scholar]
- 31.Latgé, J.-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebmann, B., S. Gattung, B. Jahn, and A. A. Brakhage. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the pathogenicity determinant-encoding gene pksP and the defense against killing by macrophages. Mol. Gen. Genomics 269:420-435. [DOI] [PubMed] [Google Scholar]
- 34.Liebmann, B., T. W. Mühleisen, M. Müller, M. Hecht, G. Weidner, A. Braun, M. Brock, and A. A. Brakhage. 2004. Deletion of the Aspergillus fumigatus lysine biosynthesis gene lysF encoding homoaconitase leads to attenuated virulence in a low-dose mouse infection model of invasive aspergillosis. Arch. Microbiol. 181:378-383. [DOI] [PubMed] [Google Scholar]
- 35.Liu, S., and R. A. Dean. 1997. G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10:1075-1086. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell, T. K., and R. A. Dean. 1995. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell 7:1869-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, B. G., J. C. Panepinto, J. R. Fortwendel, D. L. Smith, D. S. Askew, and J. C. Rhodes. 2001. Cloning and expression of pkaC and pkaR, the genes encoding the cAMP-dependent protein kinase of Aspergillus fumigatus. Mycopathologia 154:85-91. [DOI] [PubMed] [Google Scholar]
- 38.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinders, A., N. Bürckert, T. Boller, A. Wiemken, and C. DeVirgilio. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12:2943-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schneemann, M., and A. Schaffner. 1999. Host-defense mechanisms in Aspergillus infections, p. 57-68. In A. A. Brakhage, B. Jahn, and A. Schmidt. (ed.), Contributions to microbiology, vol. 2. Karger, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, A., M. P. Ward, and S. Garret. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, J. M., C. M. Tang, S. Van Noorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restrictocin and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 62:5247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonneborn, A., D. P. Bockmühl, M. Gerads, K. Kurpanek, D. Sanglard, and J. F. Ernst. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35:386-396. [DOI] [PubMed] [Google Scholar]
- 47.Staudohar, M., M. Bencina, P. J. van de Vondervoort, H. Panneman, M. Legisa, J. Visser, and J. G. Ruijter. 2002. Cyclic AMP-dependent protein kinase is involved in morphogenesis of Aspergillus niger. Microbiology 148:2635-2645. [DOI] [PubMed] [Google Scholar]
- 48.Thevelein, J. M., L. Cauwenberg, S. Colombo, J. H. D. de Winde, M. Donation, F. Dumortier, L. Kraakman, K. Lemaire, P. Ma, D. Nauwelaers, F. Rolland, A. Teunissen, P. Van Dijck, M. Versele, S. Wera, and J. Winderickx. 2000. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb. Technol. 26:819-825. [DOI] [PubMed] [Google Scholar]
- 49.Toda, T., S. Cameron, P. Sass, M. Zoller, and M. Wigler. 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277-287. [DOI] [PubMed] [Google Scholar]
- 50.Tolkacheva, T., P. McNamara, E. Piekarz, and W. Courchesne. 1994. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein alpha-subunit homolog. Infect. Immun. 62:2849-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trinci, A. P. J. 1971. Influence of the peripheral growth zone on the radial growth rate of fungal colonies. J. Gen. Microbiol. 67:325-344. [Google Scholar]
- 52.Tsai, H.-F., I. Fujii, A. Watanabe, M. H. Wheeler, Y. C. Chang, Y. Yasuoka, Y. Ebizuka, and K. J. Kwon-Chung. 2001. Pentaketide-melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J. Biol. Chem. 276:29292-29298. [DOI] [PubMed] [Google Scholar]
- 53.Tsai, H.-F., M. H. Wheeler, Y. C. Chang, and K. J. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai, H.-F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tüncher, A., H. Reinke, G. Martic, M. L. Caruso, and A. A. Brakhage. 2004. A basic-region helix-loop-helix protein-encoding gene (devR) involved in the development of Aspergillus nidulans. Mol. Microbiol. 52:227-241. [DOI] [PubMed] [Google Scholar]
- 56.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]