Abstract
Monoallelic expression of the var multigene family enables immune evasion of the malaria parasite Plasmodium falciparum in its human host. At a given time only a single member of the 60-member var gene family is expressed at a discrete perinuclear region called the ‘var expression site’. However, the mechanism of var gene counting remains ill-defined. We hypothesize that activation factors associating specifically with the expression site play a key role in this process. Here, we investigate the role of a GC-rich non-coding RNA (ncRNA) gene family composed of 15 highly homologous members. GC-rich genes are positioned adjacent to var genes in chromosome-central gene clusters but are absent near subtelomeric var genes. Fluorescence in situ hybridization demonstrates that GC-rich ncRNA localizes to the perinuclear expression site of central and subtelomeric var genes in trans. Importantly, overexpression of distinct GC-rich ncRNA members disrupts the gene counting process at the single cell level and results in activation of a specific subset of var genes in distinct clones. We identify the first trans-acting factor targeted to the elusive perinuclear var expression site and open up new avenues to investigate ncRNA function in antigenic variation of malaria and other protozoan pathogens.
INTRODUCTION
Antigenic variation is an effective mechanism for pathogens to evade the immune response of the host organism. It is used by the human malaria parasite Plasmodium falciparum, who remains a major health threat and the cause of death for hundreds of thousands of people per year (1,2). During its 48h-long blood stage cycle the parasite invades and asexually replicates in red blood cells. It goes through three morphologically distinct developmental stages namely ring, trophozoite and schizont. Upon rupture of the infected red blood cell 16–32 newly formed daughter cells are released and start a new infectious cycle.
To promote adhesion to the vascular endothelium and prevent clearance by the spleen, the parasite remodels the infected red blood cells by exporting variant surface proteins (3). The virulence surface adhesion protein P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1) has been highly implicated in pathogenesis and is encoded by the well-studied var variant gene family. The var gene family is subject to monoallelic expression whereby only a single gene of the about 60-member gene family is transcribed while all others remain silenced (4). Switching between var genes allows the parasite to regularly expose surface proteins for which the human immune system is yet naïve. Transcription of the var gene peaks around 10–14 h post invasion (hpi) (5) and is repressed during later developmental stages, but the gene remains in a state ‘poised’ for reactivation in the next cycle (6,7).
Monoallelic expression in P. falciparum involves multiple layers of epigenetic regulation that mediate silencing, activation and poising of a single var gene while still allowing var gene switching rates optimized for efficient parasite infection and transmission (8–11). Post-translational modifications of histones and especially facultative heterochromatin regulated by various histone-modifying enzymes are required to establish a default silencing pathway for var genes (6,12–14). Heterochromatin Protein 1 silences var genes by binding to modified histones (15,16). Further, conserved genetic elements within the var gene play an important role in monoallelic expression. Var genes contain an upstream promoter sequence (ups), an exon 1 coding for the variant extracellular protein domain, a conserved intron and a highly conserved exon 2 containing the intracellular domain (Supplementary Figure S1A). A total of 36 var genes are subtelomeric and driven by subtype-specific promoters termed ups A or B (Supplementary Figure S1A). The 24 central var genes localize in 5 tandem arrays distributed over 4 chromosomes and are driven by ups C (or B/C) promoters (Figure 1A). The promoter contains motifs essential for monoallelic expression (8,17–19). The intron is thought to mediate var gene silencing by strict one-to-one pairing with the promoter and has bidirectional promoter activity (20–23).
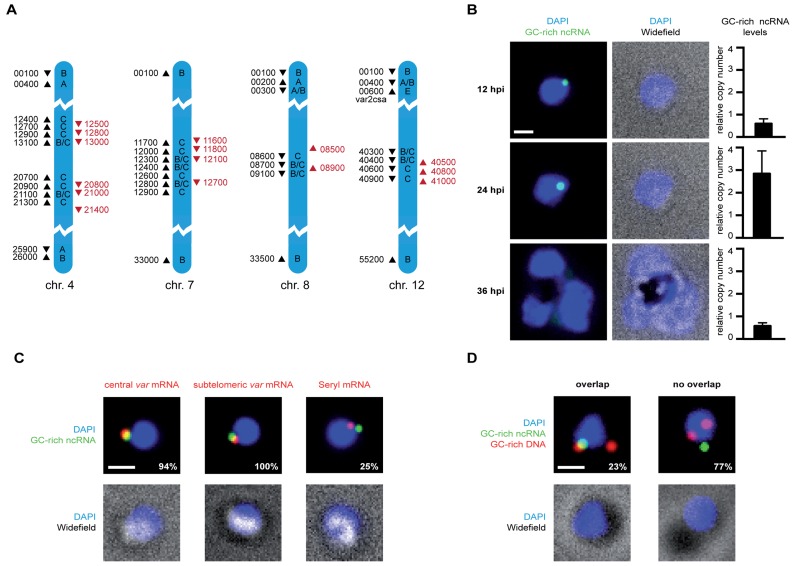
Figure 1.
GC-rich ncRNA localizes to the perinuclear var gene expression site in trans. (A) Genomic organization of all GC-rich ncRNA elements (red arrowheads) and var genes (black arrowheads) on chromosomes 4, 7, 8 and 12. Only five last digits of gene IDs are displayed (PF3D7_chr#xxxxx). Direction of the arrowhead indicates orientation of gene. Drawing is not to scale and only parts chromosomes are shown. Upstream promoter sequence (ups) subtype for each var gene is labeled (A, A/B, B, B/C, C or E). (B) Fluorescent microscopy images of RNA-FISH labeling of parasites at different life cycle stages using a probe targeting all GC-rich ncRNA transcripts (green). All nuclei are stained with DAPI (blue) and overlayed with widefield image. All scale bars, 1 μm. On the right real-time PCR quantification of GC-rich ncRNA levels at the respective stages. (C) Colocalization analysis of GC-rich ncRNA and var gene expression site. Fluorescent microscopy images of ring stage parasites labeled in two colors with RNA-FISH probes targeting GC-rich ncRNAs (green) and mRNA (red) of a central (PF3D7_0412700) var (left), subtelomeric (var2csa) (middle) or Serine-tRNA ligase control gene (right). Percentages indicate frequency of colocalization. (D) Combination of DNA-FISH using a probe against GC-rich element gene loci (red) with RNA-FISH using probe targeting all GC-rich ncRNA transcript (green). Images of one representative cell with overlapping signal (left panels) and with non-overlapping signals (right panels) are shown. Percentages indicate occurrence of overlap or non-overlap within the imaged population.
In addition, spatial organization of var genes is tightly controlled and has been implied in var gene transcriptional control. All silent var genes are targeted to a few repressive clusters at the nuclear periphery except the active var gene, which localizes to a distinct expression site (14,24–27). Perinuclear localization is mediated by telomeric sequences as well as specific intronic motifs (25,28). Lastly, exonuclease activity is involved in controlling cryptic transcription of multiple var genes (29). Despite the tremendous advances in understanding regulation of monoallelic expression key questions remain unanswered. Default silencing and potential poising factors have been identified. The histone methyltransferase, PfSet10, associates with the poised var gene at trophozoite stage when transcription is halted for the rest of the blood stage cycle (7), but a factor specifically regulating the activation of var genes has yet to be identified. Hence, the perinuclear expression site remains undefined and no factors strictly associated with it have been found to date.
Non-coding RNAs (ncRNAs) are emerging as important regulators of eukaryotic gene expression and have been linked to monoallelic expression in other organisms (30). They frequently act as lynchpins for epigenetic regulators targeting non-specific enzymes to specific sites within the genome. Also, in P. falciparum ncRNAs have been associated with regulation of virulence genes (31,32). Various ncRNA transcripts have been detected by different methods (33–37). Telomeres produce long ncRNAs that localize to a distinct nuclear compartment that does not overlap with telomere clusters (38,39). Natural antisense transcripts are very frequent and associate with >24% of all open reading frames (36). Recently, transcription of TR2 ncRNA has been correlated with permeation pathway gene expression (40). Var genes produce ‘sterile’ transcripts from a bidirectional promoter within the intron (20,41). The function of the sense transcript produced from the intron of var genes remains elusive. The natural antisense RNA remains associated with chromatin and its transcription has been correlated with var gene activity (21,31,42). However, it is not yet clear whether this antisense RNA is cause or consequence of transcriptional activation.
The RNA of Unknown Function 6 (RUF-6) gene family encodes 15 ncRNAs with an unusually high GC-content for P. falciparum (>50% versus 20% genome-wide average) (43). Therefore, they have been referred to as GC-rich ncRNA or GC-rich elements (44–46). Their sequences are highly homologous (Supplementary Figure S1B) and interestingly, their gene loci are strictly associated with central var gene clusters where they are interspersed between var genes mostly in a tail-to-tail configuration (Figure 1A). Their transcripts have been detected in various transcriptomic studies, which indicate peak expression around late ring stage (34,36–38,47). Bioinformatical analysis shows conserved promoter elements, namely A- and B-box, within the GC-rich element sequence, suggesting transcription via RNA polymerase III (Supplementary Figure S1B) (48–50). A recent study shows that GC-rich ncRNA expression is clonally variant, although the members are not transcribed in a mutually exclusive fashion (51).
Here, we characterize GC-rich ncRNA using fluorescence in situ hybridization (FISH) and quantitative PCR. We find that even though GC-rich ncRNA genes can form multiple clusters within the nucleus, their transcripts localize to a single perinuclear locus in early ring stages. This locus colocalizes with the var gene expression site of central as well as subtelomeric var genes. Overexpression of distinct GC-rich ncRNAs causes activation of a specific subset of var genes and perturbs monoallelic expression at the single cell level. We uncover the first factor strictly associated with the active var gene expression site and provide functional characterization of a ncRNA gene in P. falciparum. These findings represent an important step toward better understanding of the mechanism of monoallelic expression in the malaria parasite.
MATERIALS AND METHODS
Parasite culture, synchronization and panning
P. falciparum blood stage parasites were cultivated as described previously (14). Parasite culture was synchronized by sorbitol lysis during ring stage, subsequent plasmagel enrichment in schizont stage, followed by another sorbitol treatment at 6 h post invasion. Synchronized parasites were harvested at 4% hematocrit and ∼3–5% parasitemia. Parasite development was monitored by Giemsa staining. To enrich for FCR3 parasites expressing var2csa panning on CSA receptor was carried out as previously described (52). In brief, cell culture flasks were coated with CSA receptor to specifically bind red blood cells infected with parasites expressing the PfEMP1 protein encoded by the var2csa gene.
RNA-FISH, DNA-FISH and immunofluorescence
Infected red blood cells were lysed with 0.015% Saponin in RPMI and the released parasites were fixed in suspension with 4% paraformaldehyde in phophate buffered saline (PBS) over night at 4°C. Parasites were then deposited on #1.5 cover glasses, permeabilized 15 min with 0.1% Triton-X-100 and subjected to DNA or RNA FISH as described previously in a detailed methods chapter (53). For sequential RNA-, DNA-FISH the transcript was labeled first with a GC-rich ncRNA probe. After another fixation step we carried out DNA-FISH using a probe labeling exclusively the non-transcribed 5′ region of all GC-rich genes. FISH probes were PCR amplified from genomic DNA using primers (Supplementary Table S1) and labeled by nick translation using FITC, DIG or Biotin High-Prime (Roche). RNA-FISH combined with immunofluorescence was carried out in an endogenously tagged PfSet10-HA parasite strain using anti-HA antibody (Roche, 3F10) at 1:200 in PBS/3%BSA as described previously (7,53,54).
RNA extraction and quantitative real-time PCR
RNA was extracted after Saponin lysis in 0.06% Saponin in PBS followed by one wash in PBS and resuspension in Trizol. RNA was purified using the miRNeasy kit, which collects RNA fragments down to 20 bp in size (Qiagen). After on-column DNase treatment (Qiagen) reverse transcription of 200 ng of RNA was carried out using Superscript VILO kit (Life technologies) with random hexamer primers, to prevent loss of ncRNAs. cDNA levels were quantified in the CFX384 qPCR machine (Bio-Rad) using gene specific primers (Supplementary Table S2) and 2x Power SYBR Green master mix (Applied biosciences). Data were analyzed using the Bio-Rad CFX manager software after normalization to T-Serine ligase (PF3D7_0717700) transcription levels. GC-rich ncRNA was quantified by using two primer pairs covering small sequence variation in all members of the gene family. To assess total var gene levels expression values for all ups subtype specific primers were added. Quantitative real-time PCR experiments were carried out in triplicates. Limits of detection were calculated as mean of the blank plus three times the standard deviation.
Plasmid construction and transfection
To construct overexpression vectors synthetic gene fragments containing parts of U6 promoter either coding for a GC-rich ncRNA gene PF3D7_1241000, PF3D7_0808500 or a Luciferase control fragment of same size were ordered (Custom gene synthesis, Genscript) and cloned into pL6-egfp vector previously described in (55) using DraII and SwaI restriction site followed by Gibson assembly (In-fusion, HD Clontech). pU6-Luc, pU6-GC12 and pU6-GC08 constructs were transfected as described previously (56) and maintained under WR99210 drug selection pressure.
Image acquisition and analysis
Images were captured using a Nikon Eclipse 80i microscope equipped with 100x NA 1.42 objective and CoolSnap HQ2 camera (Photometrics). Camera pixel size corresponds to 64 nm. NIS Elements 3.0 software (Nikon) was used for acquisition and Fiji (http://fiji.sc) for analysis. Numbers of FISH dots were quantified counting all cells positive for staining. Colocalization was scored in cell nuclei positive for both fluorescent signals when they overlapped while using the same contrast adjustments throughout all images. Scoring was performed by direct optical observation using Fiji (http://fiji.sc). For each analysis images from at least two independent replicas were combined.
RESULTS
GC-rich ncRNA localizes to the perinuclear var gene expression site in trans
GC-rich gene loci are adjacent to central var genes (Figure 1A), which have been reported to cluster at the nuclear periphery (14). Our DNA-FISH analysis confirms that GC-rich ncRNA genes form between 1–4 perinuclear clusters (n = 75, 48, 20 and 9, respectively) in a population of ring stage parasites prior to genome replication (Supplementary Figure S2). To characterize the localization pattern of GC-rich ncRNA transcripts we carried out RNA-FISH using a probe targeting all GC-rich ncRNAs. We analyzed fluorescent images of labeled cells at 12, 24 and 36 hpi (Figure 1B). Notably, at 12 hpi all labeled cells exhibited only a single perinuclear focus (n > 50). At 24 hpi most of the cells still show a single focus of increased fluorescence intensity. However, this signal was no longer particularly associated with the nuclear periphery. All detected signals were exclusively nuclear. In the trophozoite stage, at 36 hpi, we did not detect any cells with a distinct GC-rich ncRNA signal. Fluorescent signal intensities throughout the cell cycle correlate with the GC-rich ncRNA levels quantified by real time PCR carried out on RNA harvested at the same timepoints using primer pairs detecting all GC-rich ncRNA members (Figure 1B). Peak ncRNA levels occur at 24 hpi and drop again at 36 hpi. The lack of distinct GC-rich ncRNA signal at 36 hpi suggests that transcripts do not accumulate at a single site that can be detected by FISH. Next, we investigated the colocalization of GC-rich ncRNA with PfSet10, a histone methyltransferase that associates with the poised var gene during trophozoite stage (7). Due to the different temporal expression we could, however, not test for overlap between the PfSet10 and GC-rich ncRNA signal (Supplementary Figure S3). Importantly, the GC-rich ncRNA localization pattern that does not mimic the clustered distribution of the GC-rich gene loci in ring stage parasites, but suggests targeting to a specific perinuclear site.
Since the localization pattern of GC-rich ncRNA is reminiscent of the var gene expression site, we carried out dual color RNA-FISH of GC-rich ncRNA and var gene mRNA in ring stage parasites to test for colocalization. We used two different parasite lines that express either a central var gene (PF3D7_0412700) or the subtelomeric var2csa gene. As a negative control we used a T-serine ligase RNA (PF3D7_0717700, see Supplementary Figure S1A) probe. We found that GC-rich ncRNA signal overlapped with the central var mRNA in 30 of 32 cells, with var2csa mRNA in 19 out of 19 cells whereas T-Serine ligase mRNA showed colocalization in only 3 out of 16 cases (Figure 1C). While the central var gene could colocalize with GC-rich ncRNA due to spatial proximity of both genes on the chromosome, var2csa mRNA is expressed from a subtelomeric locus distant from any GC-rich element cluster (Supplementary Figure S1A). These data on GC-rich ncRNA localization demonstrate the first specific targeting of a molecular factor to the active var gene expression site.
Colocalization of var mRNA and GC-rich ncRNA could occur by two different mechanisms. The GC-rich ncRNA and active var gene loci could loop together via a long-range chromosomal interaction. Alternatively, GC-rich ncRNA could freely diffuse before being targeted to the var gene expression site. To test these hypotheses we combined DNA-FISH of the GC-rich gene loci with RNA-FISH of the GC-rich ncRNA transcript (Figure 1D). After FISH labeling GC-rich ncRNA in cells primarily expressing the subtelomeric var2csa gene, we labeled the GC-rich gene locus with a DNA-FISH probe exclusively targeting the 5′ region of all gene family members to avoid any recognition of the transcript. We found overlap between signals in only 7 out of 30 cells. These data suggest that GC-rich ncRNA can relocate to the var gene expression site independently of its gene locus.
Overexpression of GC-rich ncRNAs activates distinct subsets of var genes
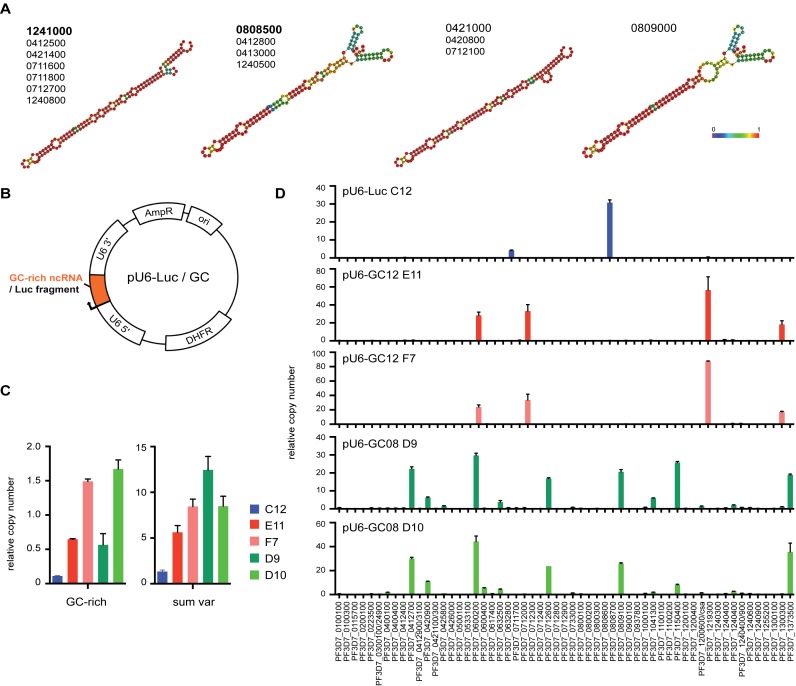
Despite their high sequence similarity (Supplementary Figure S1B), in silico prediction using the RNAfold webserver indicates some structural variability within the gene family resulting in four major ‘classes’ of RNA secondary structures (Figure 2A). While the lower stem loop structure is highly preserved, the ‘head’ region displays much more flexible base pairing configurations. Conservation of specific promoter elements, the A- and B-box, suggests that RNA Polymerase III drives GC-rich ncRNA transcription (Supplementary Figure S1B). Hence, we designed a construct overexpressing GC-rich ncRNA under the highly active Pol III-dependent U6 promoter (Figure 2B). We inserted GC-rich ncRNA sequences of two distinct structural variant types (PF3D7_1241000, PF3D7_0808500) or a Luciferase control fragment of similar length and GC content into the previously described pL6-egfp vector backbone (55), generating pU6-GC12, pU6-GC08 and pU6-Luc, respectively. We transfected these constructs into a 3D7 parasite line and isolated one clone carrying pU6-Luc and two independent clones each carrying pU6-GC12 and pU6-GC08. Quantitative real time PCR analysis of cDNA derived from RNA harvested at 12 hpi demonstrated that pU6-GC12 and pU6-GC08 transfected clones have increased in GC-rich ncRNA levels (Figure 2C). Total var RNA levels, as quantified by a subset of qPCR primers covering the entire gene family, are also increased in GC-rich ncRNA overexpression clones.
Figure 2.
GC-rich ncRNA overexpression causes activation of distinct subsets of var genes. (A) Bioinformatic RNA structure prediction using RNAfold webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Four representative structures covering diversity within entire GC-rich ncRNA gene family are shown. Members with similar structure are listed below. Colored scale represents relative base pairing probabilities. (B) Plasmid map of pU6-GC or pU6-Luc constructs used to overexpress GC-rich ncRNA (PF3D7_1241000 and PF3D7_0808500) or Luciferase control fragment, respectively. (C) Real time qPCR quantification of GC-rich ncRNA and total var mRNA levels in pU6-Luc transfected clone (C12, blue), two pU6-GC12 transfected clones (E11 and F7, red) and two pU6-GC08 clones (D9 and D10, green). (D) Expression profile of all var gene mRNAs as quantified by real time qPCR for pU6-Luc and pU6-GC clones. Relative copy numbers are normalized to T-Serine ligase (PF3D7_0717700) transcription levels. Limit of detection (LOD) for this data set is 0.0114.
Next, we analyzed the RNA levels for all individual var genes in the five clones also using T-Serine RNA ligase levels for normalization (Figure 2D). The pU6-Luc transfected clone expressed only a single var gene, displaying functional monoallelic expression. Notably, both GC12 overexpressing clones primarily transcribed the same four var genes, while both GC08 overexpressing clones transcribed the same set of six var genes. Despite transfections being carried out in the same 3D7 parent strain, the expression profiles for pU6-Luc, pU6-GC12 and pU6-GC08 did not have any var genes in common except for PF3D7_0600200 (Figure 2D and Supplementary Figure S4). RNA levels of up-regulated var genes varied slightly within the same cloned parasite line, but the profile was virtually identical between two clones. Given, that we observe a similar subset of var gene expression in bulk culture before the cloning step, overexpression of distinct GC ncRNA could lead to the activation of that specific var gene subset (Supplementary Figure S4). The temporal regulation of var and GC-rich ncRNA levels was not altered in mutant parasite clones (Supplementary Figure S5A). Lastly, to interfere with GC-rich ncRNA function, we also attempted overexpression of GC-rich antisense RNA in the same vector, which did not cause a significant change in GC-rich ncRNA levels or affected var gene mRNA levels (Supplementary Figure S5B). Taken together, our data suggest that distinct GC-rich ncRNA members activate distinct subsets of var genes establishing a ‘hardwired’ expression profile.
GC-rich ncRNA overexpression perturbs monoallelic var gene expression at single cell level
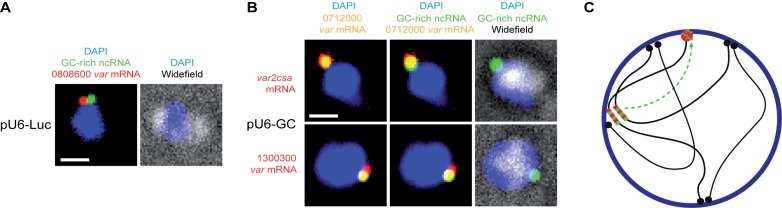
Our quantitative PCR data suggest that GC-rich ncRNA overexpression interferes with monoallelic var gene expression. To test this in individual parasites we carried out multi-color RNA-FISH with gene specific probes targeting three of the highly expressed var mRNAs in the context of GC-rich ncRNA (PF3D7_1241000) overexpression (Figure 2D). Parasites carrying the pU6-Luc control construct show a single var gene (PF3D7_0808600) signal colocalizing with GC-rich ncRNA in 20 out of 21 cells (Figure 3A). When labeling GC-rich ncRNA overexpressing parasites with probes against a central var mRNA (PF3D7_0712000) and var2csa mRNA, we found that 31 out of 59 labeled cells were positive for both signals (Figure 3B). We found similar results when combining the central var mRNA (PF3D7_0712000) probe with another subtelomeric var mRNA (PF3D7_1300300) probe (double staining in 12 out of 19 cells). The lack of double staining in about half of cells could be explained by incomplete labeling of all cells by all probes. Nevertheless, our data show that overexpression of GC-rich ncRNA can disrupt monoallelic expression at single cell level, linking a trans-acting ncRNA to antigenic variation in malaria parasites.
Figure 3.
GC-rich ncRNA overexpression disrupts monoallelic expression. (A) Fluorescent microscopy images of multicolor RNA-FISH labeling. pU6-Luc transfected parasites were labeled with probe against GC-rich ncRNA (green) and var mRNA PFD3D7_0808600 (red). (B) pU6-GC transfected parasites were triple labeled with probe against GC-rich ncRNA (green), var mRNA PFD3D7_0712000 (yellow) and var2csa mRNA (red, middle panels) or var mRNA PFD3D7_1300300 (red, lower panels). Nuclei are stained with DAPI (blue) and overlayed with widefield image. Scale bars, 1 μm. (C) Schematic working model shows relocalization of GC-rich ncRNA (green) from central var gene clusters (red-green stripes) to the distinct perinuclear var expression site (red). Telomeres (black) cluster at the nuclear membrane (blue).
DISCUSSION
In this work, we demonstrate the specific targeting of GC-rich ncRNA to the actively transcribed expression site of var genes. This is the first identification of a marker molecule for a nuclear structure that remains ill-defined despite being a hallmark for monoallelic expression in protozoan pathogens such as P. falciparum and African Trypanosomes (57). It has been postulated that, in the context of monoallelic expression, an as yet unknown limiting factor exists that allows for the activation of only a single var gene locus (10,18). GC-rich ncRNA is a potential candidate for such a limiting factor. Indeed, when we overexpress GC-rich ncRNA, we detect multiple different var transcripts in a single cell. Our study predicts that GC-rich ncRNA knockdown mutants could reduce var gene expression. However, due to the presence of 15 GC-rich ncRNA genes generating mutants by sequential knockout, despite the recent introduction of CRISPR technology to malaria parasites, represents a long-term approach (55,58). A recent study observed only a minor effect of GC-rich ncRNA overexpression on var gene transcription (51). Important technical differences used in their study such as the use of a Pol II instead of a Pol III promoter for the production of the GC-rich ncRNA could explain the observed phenotype. In addition, their study focused only on central var genes, omitting most members of the multigene family represented by the subtelomeric var genes from their transcriptional analysis (51).
Previous studies in P. falciparum described the transcription and localization of various ncRNA species such as subtelomeric RNA and natural antisense RNA, but their mechanisms or biological roles are still unclear (32). In this study, we present experimental data that posits GC-rich ncRNA as a trans-acting factor that contributes to the control of var gene activation at the perinuclear expression site. Because GC-rich ncRNA elements are only in close proximity to central var genes (Supplementary Figure S1), a cis-regulatory function has been suggested for these elements. Furthermore, their positioning in intergenic spaces and recent experimental evidence based on GC-rich element associated silencing of an adjacent GFP gene on episomal constructs has led to speculation that they might act as insulator elements that prevent spreading of epigenetic marks (51). While this hypothesis remains possible, our data strongly support a general trans-acting function linked to monoallelic expression of all var gene members (Figure 3C). The mechanism by which GC-rich ncRNA is targeted to the expression site remains unclear. Bioinformatic prediction, however, suggests a relatively strong secondary RNA structure (Figure 2A), which could mediate specific RNA–RNA and RNA–protein interactions (34,43). Since cryptic var gene transcription has recently been demonstrated (29) GC-rich ncRNA could, hypothetically, bind to nascent var mRNA to target transcriptional activation factors to the expression site. Transcriptional activation would generate more var RNA recruiting more GC-rich ncRNA thereby generating the positive feedback loop that drives singular gene choice. A similar mechanism has recently been described in the Epstein-Barr virus (59). Here, a viral ncRNA, EBER2, recruits a host transcription factor to the viral DNA locus via base-pairing interactions with nascent transcript and thereby modulates transcription. Hence, it is of utmost importance to identify interacting proteins to obtain insight into the underlying molecular mechanism by which this ncRNA regulates monoallelic expression. It is noteworthy that GC-rich ncRNA levels peak only at 24 hpi when var gene transcription is already repressed. GC-rich ncRNA function at this stage remains elusive, but despite its distinct colocalization with var genes, it is possible that it is linked to the expression of other clonally variant genes such as rifins as well (60,61). RNA-seq analysis of parasites that overexpress GC-rich ncRNA may reveal regulation of other target genes or gene families. Interestingly, Plasmodium reichenowi, the only other species carrying a GC-rich ncRNA gene family with almost identical sequences and structures, is also the only other sequenced malaria species encoding the var and rifin gene families (plasmodb.org).
The most striking result of this study is that overexpression of a single member of the GC-rich gene family disrupts monoallelic gene counting. Interestingly, five out of nine of the preferentially activated var genes are subtelomeric, emphasizing that GC-rich ncRNA function is not restricted to central var genes. Further, in distinct overexpression clones, the same subset of var genes is up-regulated, indicating a ‘hardwired’ activation program. A similar observation was made when the histone deacetylase PfSir2a was inactivated, resulting in the de-repression of a specific subset of var genes (62). Considering that GC-rich ncRNAs are clonally variant it is tempting to speculate that individual GC-rich elements could have a preference for specific subsets of var genes and thus, decide on which silent var gene may be activated at the next switch in expression. Given that switching is not understood in malaria parasites at the molecular level, our work points to a promising new research avenue that may lead to a better insight into a process that is key to antigenic variation.
Many years after the discovery of monoallelic var gene expression, we are only now beginning to understand the elusive expression site. These findings expand our knowledge about ncRNA genes as novel players in monoallelic expression and will open avenues to deepen our understanding of antigenic variation in the malaria parasite and other protozoan pathogens.
Supplementary Material
Acknowledgments
The authors thank Allon Weiner, Shruthi Vembar and Jessica Bryant for critical reading of the manuscript, Sebastian Baumgarten for help with data analysis and Jose-Juan Lopez-Rubio for providing reagents.
Footnotes
Present address: Julien Guizetti & Artur Scherf, Unité de Biologie des Interactions Hôte-Parasite, Institut Pasteur, 25 Rue du Dr. Roux, 75724 Paris, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council Advanced [PlasmoEscape 250320, PlasmoSilencing 670301]; Agence Nationale de la Recherche [ANR-11-LABEX-0024-01 ParaFrap, ANR-13-ISV3-0003-01 NSFC MalVir]; Human Frontier Science Program fellowship [LT000620/2012-L to J.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Snow R.W., Guerra C.A., Noor A.M., Myint H.Y., Hay S.I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray C.J., Rosenfeld L.C., Lim S.S., Andrews K.G., Foreman K.J., Haring D., Fullman N., Naghavi M., Lozano R., Lopez A.D. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379:413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 3.Maier A.G., Cooke B.M., Cowman A.F., Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat. Rev. Microbiol. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 4.Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B., Gysin J., Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schieck E., Pfahler J.M., Sanchez C.P., Lanzer M. Nuclear run-on analysis of var gene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 2007;153:207–212. doi: 10.1016/j.molbiopara.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Rubio J.J., Gontijo A.M., Nunes M.C., Issar N., Hernandez Rivas R., Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volz J.C., Bártfai R., Petter M., Langer C., Josling G.A., Tsuboi T., Schwach F., Baum J., Rayner J.C., Stunnenberg H.G., et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe. 2012;11:7–18. doi: 10.1016/j.chom.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S., Wellems T.E., Deitsch K.W. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherf A., Lopez-Rubio J.J., Riviere L. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 10.Guizetti J., Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol. 2013;15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chookajorn T., Ponsuwanna P., Cui L. Mutually exclusive var gene expression in the malaria parasite: multiple layers of regulation. Trends Parasitol. 2008;24:455–461. doi: 10.1016/j.pt.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A.Z., Hartl D.L., Deitsch K.W. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman B.I., Skillman K.M., Jiang R.H.Y., Childs L.M., Altenhofen L.M., Ganter M., Leung Y., Goldowitz l., Kafsack B.F.C., Marti M., et al. A Plasmodium falciparum Histone Deacetylase Regulates Antigenic Variation and Gametocyte Conversion. Cell Host Microbe. 2014;16:177–186. doi: 10.1016/j.chom.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Rubio J.J., Mancio-Silva L., Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Toledo K., Rojas-Meza A.P., Mancio-Silva L., Hernandez-Cuevas N.A., Delgadillo D.M., Vargas M., Martinez-Calvillo S., Scherf A., Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brancucci N.M., Bertschi N.L., Zhu L., Niederwieser I., Chin W.H., Wampfler R., Freymond C., Rottmann M., Felger I., Bozdech Z., et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014;16:165–176. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Brancucci N.M., Witmer K., Schmid C.D., Flueck C., Voss T.S. Identification of a cis-acting DNA-protein interaction implicated in singular var gene choice in Plasmodium falciparum. Cell Microbiol. 2012;14:1836–1848. doi: 10.1111/cmi.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dzikowski R., Frank M., Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss T.S., Healer J., Marty A.J., Duffy M.F., Thompson J.K., Beeson J.G., Reeder J.C., Crabb B.S., Cowman A.F. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 20.Calderwood M.S., Gannoun-Zaki L., Wellems T.E., Deitsch K.W. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 21.Epp C., Li F., Howitt C.A., Chookajorn T., Deitsch K.W. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank M., Dzikowski R., Costantini D., Amulic B., Berdougo E., Deitsch K. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J. Biol. Chem. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gannoun-Zaki L., Jost A., Mu J., Deitsch K.W., Wellems T.E. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot. Cell. 2005;4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duraisingh M.T., Voss T.S., Marty A.J., Duffy M.F., Good R.T., Thompson J.K., Freitas-Junior L.H., Scherf A., Crabb B.S., Cowman A.F. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 25.Figueiredo L.M., Freitas-Junior L.H., Bottius E., Olivo-Marin J.C., Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 27.Ralph S.A., Scheidig-Benatar C., Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q., Huang Y., Zhang Y., Fang X., Claes A., Duchateau M., Namane A., Lopez-Rubio J.J., Pan W., Scherf A. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011;10:451–463. doi: 10.1016/j.chom.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q., Siegel T.N., Martins R.M., Wang F., Cao J., Gao Q., Cheng X., Jiang L., Hon C.C., Scheidig-Benatar C., et al. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature. 2014;513:431–435. doi: 10.1038/nature13468. [DOI] [PubMed] [Google Scholar]
- 30.Yang P.K., Kuroda M.I. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 31.Amit-Avraham I., Pozner G., Eshar S., Fastman Y., Kolevzon N., Yavin E., Dzikowski R. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E982–E991. doi: 10.1073/pnas.1420855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vembar S.S., Scherf A., Siegel T.N. Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr. Opin. Microbiol. 2014;20:153–161. doi: 10.1016/j.mib.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F., Sonbuchner L., Kyes S.A., Epp C., Deitsch K.W. Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J. Biol. Chem. 2008;283:5692–5698. doi: 10.1074/jbc.M707344200. [DOI] [PubMed] [Google Scholar]
- 34.Mourier T., Carret C., Kyes S., Christodoulou Z., Gardner P.P., Jeffares D.C., Pinches R., Barrell B., Berriman M., Griffiths-Jones S., et al. Genome-wide discovery and verification of novel structured RNAs in Plasmodium falciparum. Genome Res. 2008;18:281–292. doi: 10.1101/gr.6836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raabe C.A., Sanchez C.P., Randau G., Robeck T., Skryabin B.V., Chinni S.V., Kube M., Reinhardt R., Ng G.H., Manickam R., et al. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 2010;38:608–617. doi: 10.1093/nar/gkp895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel T.N., Hon C.C., Zhang Q., Lopez-Rubio J.J., Scheidig-Benatar C., Martins R.M., Sismeiro O., Coppee J.Y., Scherf A. Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics. 2014;15:150–165. doi: 10.1186/1471-2164-15-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei C., Xiao T., Zhang P., Wang Z., Chen X., Zhang L., Yao M., Chen R., Wang H. Deep profiling of the novel intermediate-size noncoding RNAs in intraerythrocytic Plasmodium falciparum. PLoS One. 2014;9:e92946. doi: 10.1371/journal.pone.0092946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broadbent K.M., Park D., Wolf A.R., Van Tyne D., Sims J.S., Ribacke U., Volkman S., Duraisingh M., Wirth D., Sabeti P.C., et al. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sierra-Miranda M., Delgadillo D.M., Mancio-Silva L., Vargas M., Villegas-Sepulveda N., Martinez-Calvillo S., Scherf A., Hernandez-Rivas R. Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol. Biochem. Parasitol. 2012;185:36–47. doi: 10.1016/j.molbiopara.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rovira-Graells N., Crowley V.M., Bancells C., Mira-Martinez S., Ribas de Pouplana L., Cortes A. Deciphering the principles that govern mutually exclusive expression of Plasmodium falciparum clag3 genes. Nucleic Acids Res. 2015;43:8343–8357. doi: 10.1093/nar/gkv730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyes S.A., Christodoulou Z., Raza A., Horrocks P., Pinches R., Rowe J.A., Newbold C.I. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K., Yang W., Turner L., Lavstsen T., Theander T.G., et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakrabarti K., Pearson M., Grate L., Sterne-Weiler T., Deans J., Donohue J.P., Ares M.J. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA. 2007;13:1923–1939. doi: 10.1261/rna.751807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall N., Pain A., Berriman M., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., et al. Sequence of Plasmodium falciparum chromosomes 1, 3-9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- 46.Upadhyay R., Bawankar P., Malhotra D., Patankar S. A screen for conserved sequences with biased base composition identifies noncoding RNAs in the A-T rich genome of Plasmodium falciparum. Mol. Biochem. Parasitol. 2005;144:149–158. doi: 10.1016/j.molbiopara.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Otto T.D., Wilinski D., Assefa S., Keane T.M., Sarry L.R., Bohme U., Lemieux J., Barrell B., Pain A., Berriman M., et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dieci G., Conti A., Pagano A., Carnevali D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta. 2013;1829:296–305. doi: 10.1016/j.bbagrm.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Marck C., Kachouri-Lafond R., Lafontaine I., Westhof E., Dujon B., Grosjean H. The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 2006;34:1816–1835. doi: 10.1093/nar/gkl085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 51.Wei G., Zhao Y., Zhang Q., Pan W. Dual regulatory effects of non-coding GC-rich elements on the expression of virulence genes in malaria parasites. Infect. Genet. Evol. 2015;36:490–499. doi: 10.1016/j.meegid.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 52.Pouvelle B., Meyer P., Robert C., Bardel L., Gysin J. Chondroitin-4-sulfate impairs in vitro and in vivo cytoadherence of Plasmodium falciparum infected erythrocytes. Mol. Med. 1997;3:508–518. [PMC free article] [PubMed] [Google Scholar]
- 53.Mancio-Silva L., Scherf A. In situ fluorescence visualization of transcription sites and genomic loci in blood stages of plasmodium falciparum. Methods Mol. Biol. 2013;923:335–351. doi: 10.1007/978-1-62703-026-7_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guizetti J., Martins R.M., Guadagnini S., Claes A., Scherf A. Nuclear pores and perinuclear expression sites of var and ribosomal DNA genes correspond to physically distinct regions in plasmodium falciparum. Eukaryot. Cell. 2013;12:697–702. doi: 10.1128/EC.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghorbal M., Gorman M., Macpherson C.R., Martins R.M., Scherf A., Lopez-Rubio J.J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 56.Hasenkamp S., Merrick C.J., Horrocks P. A quantitative analysis of Plasmodium falciparum transfection using DNA-loaded erythrocytes. Mol. Biochem. Parasitol. 2013;187:117–120. doi: 10.1016/j.molbiopara.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- 58.MacPherson C.R., Scherf A. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat. Biotechnol. 2015;33:805–806. doi: 10.1038/nbt.3291. [DOI] [PubMed] [Google Scholar]
- 59.Lee N., Moss W.N., Yario T.A., Steitz J.A. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160:607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goel S., Palmkvist M., Moll K., Joannin N., Lara P., Akhouri R.R., Moradi N., Öjemalm K., Westman M., Angeletti D., et al. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat. Med. 2015;21:314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 61.Kyes S.A., Rowe J.A., Kriek N., Newbold C.I. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tonkin C.J., Carret C.K., Duraisingh M.T., Voss T.S., Ralph S.A., Hommel M., Duffy M.F., Silva L.M., Scherf A., Ivens A., et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.