Abstract
Mammalian AATF/Che-1 is essential for embryonic development, however, the underlying molecular mechanism is unclear. By immunoprecipitation of human AATF we discovered that AATF forms a salt-stable protein complex together with neuroguidin (NGDN) and NOL10, and demonstrate that the AATF-NGDN-NOL10 (ANN) complex functions in ribosome biogenesis. All three ANN complex members localize to nucleoli and display a mutual dependence with respect to protein stability. Mapping of protein-protein interaction domains revealed the importance of both the evolutionary conserved WD40 repeats in NOL10 and the UTP3/SAS10 domain in NGDN for complex formation. Functional analysis showed that the ANN complex supports nucleolar steps of 40S ribosomal subunit biosynthesis. All complex members were required for 18S rRNA maturation and their individual depletion affected the same nucleolar cleavage steps in the 5′ETS and ITS1 regions of the ribosomal RNA precursor. Collectively, we identified the ANN complex as a novel functional module supporting the nucleolar maturation of 40S ribosomal subunits. Our data help to explain the described role of AATF in cell proliferation during mouse development as well as its requirement for malignant tumor growth.
INTRODUCTION
The apoptosis-antagonizing transcription factor (AATF, Che-1, TRB) has originally been described as an RNA polymerase II-specific transcription factor and was suggested to regulate apoptosis (1–3). Meanwhile, AATF has been linked to many different cellular processes including cell proliferation, DNA damage response and cell cycle control (4). As a transcriptional regulator, AATF plays an important role in stress responses upon DNA damage, including activation of p53 and potentially p21 (5–8). Overexpression of AATF has been observed in several human leukemic cell lines, and, remarkably, depletion of AATF suppresses tumor growth and increases chemotherapy sensitivity of tumors in mouse xenograft models (6,7,9). These findings have led to the proposal that AATF may represent a novel potential target for cancer therapy.
AATF is essential in mice and its mutation affects cell proliferation (10). Interestingly, the mouse protein localizes to nucleoli, where ribosome synthesis takes place, although the functional relevance of this observation has not been elucidated (10,11). Ribosome synthesis and cell proliferation are tightly linked processes, as maintenance of sufficient translational capacity for protein biosynthesis is a prerequisite for cell growth. Rapidly dividing cells, including cancer cells, produce high numbers of ribosomes and, consistently, different oncogenes have been linked to the regulation of ribosome assembly (12,13). At the same time, defects in ribosome maturation have been shown to induce p53 stabilization and p53-dependent stress responses in mammalian cells (14), highlighting that functionality of ribosome biogenesis is crucial for cellular and organismal homeostasis.
The initial steps of eukaryotic ribosome synthesis take place in the nucleolus, where ribosomal RNA (rRNA) precursors are transcribed and premature (pre) ribosomal particles are formed. Nucleolar maturation of pre-ribosomal particles includes rRNA folding, processing and modification as well as incorporation of ribosomal proteins in combination with conformational rearrangements of the pre-ribosomal subunits. Further steps are carried out in the nucleoplasm and cytoplasm to generate functional ribosomes. The complex process of ribosome biogenesis is orchestrated by a great variety of proteins belonging to diverse functional classes and also involves various snoRNAs, which guide site-specific rRNA modifications as part of snoRNPs (15–19). The majority of these trans-acting factors support nucleolar steps of ribosome synthesis, among them a large assembly of trans-acting factors called the small subunit (SSU) processome (20,21). The SSU processome executes essential pre-rRNA cleavage reactions in the 5′ external transcribed spacer (5′ETS) and in the internal transcribed spacer 1 (ITS1), required for 18S rRNA maturation and 40S subunit synthesis.
Recently, we retrieved AATF as a hit in two RNA interference screens performed to identify factors involved in 40S ribosomal subunit production in human cells (20,22). Notably, also the yeast protein Bfr2, which shows sequence homology to AATF, has been demonstrated to function in ribosome synthesis in Saccharomyces cerevisiae as part of the SSU processome (23).
In this study, we define a role for AATF in nucleolar steps of 40S ribosomal subunit synthesis in human cells, and show that AATF forms a novel protein complex with the nucleolar factors NGDN and NOL10. Mapping data illustrate the molecular organization of the complex and verify direct binding between AATF and NGDN. Downregulation of components of the AATF-NGDN-NOL10 (ANN) complex impairs nucleolar 40S subunit maturation, which manifests in the accumulation of a 30S rRNA precursor, reminiscent of defects in SSU processome activity. The function of the ANN complex in 40S subunit synthesis suggests a mechanistic basis for the essential role of AATF in cell proliferation and may help to explain the proposed link of AATF to cancer development.
MATERIALS AND METHODS
Cell lines, antibodies and reagents
The HeLa RPS2-YFP, RPL29-GFP and HEK293 (FlpIn TRex, Invitrogen) HASt-GFP cell lines have been described previously (22,24,25). The HEK293 NOL10-StHA and NGDN-StHA cell lines for tandem affinity purification (TAP) were generated as previously described (25). HeLa K cells were obtained from D. Gerlich (IMBA, Vienna, Austria). HCT116 wt and p53−/− cell lines were kind gifts from B. Vogelstein (Johns Hopkins, Baltimore, USA) and have been described previously (26). All cell lines were grown in DMEM with 10% FCS and 1% penicillin/streptomycin at 37°C, 5% CO2.
α-AATF antibodies were raised against recombinant His-tagged full-length AATF in rabbits and purified using the antigen coupled to SulfoLink beads (Thermo Fisher Scientific). The following antibodies have been described previously: α-ENP1, α-NOB1, α-RPS3 (24); α-CRM1 (27); α-RPL23A (28). α-FBL (sc-166001) and α-NAT10 (sc-271770) were purchased from Santa Cruz Biotechnology; α-NOL10 (ab181161) from Abcam; α-NGDN (STJ24765) from St John's Laboratory; α-actin (A1978) from Sigma and α-HA (ENZ-ABS120) from Enzo Life Sciences. Leptomycin B (L-6100) was purchased from LC Laboratories.
Molecular cloning
The AATF, NOL10 and NGDN coding regions were amplified from HeLa K cDNA and cloned as full-length or truncated versions into the pcDNA5/FRT/TO/cStHA-TAP vector (25) using KpnI, XhoI restriction sites encoding for C-terminally StHA-tagged (Strep-tag II, St; hemagglutinin epitope, HA) protein fusions. GFP was cloned into the pcDNA5/FRT/TO/cStHA-TAP vector using BamHI, NotI restriction sites generating GFP-StHA. A pcDNA5/FRT/TO/cHA vector for C-terminal HA tagging was produced by insertion of an HA-tag into the pcDNA5/FRT/TO vector (Invitrogen) using the restriction sites XhoI, ApaI. Full-length or truncated AATF versions were cloned into KpnI, XhoI sites of the pcDNA5/FRT/TO/cHA vector. RNAi-resistant derivatives of AATF were generated using a synthetic DNA fragment of the AATF gene (GenScript) containing silent mutations in the binding sites for the si-AATF-1 and si-AATF-2 siRNA oligonucleotides.
To purify recombinant His-tagged AATF in Escherichia coli (E. coli) as antigen for antibody production, AATF was cloned into the pQE-30 vector (QIAGEN) using KpnI, HindIII restriction sites. To express recombinant proteins in E. coli for in vitro pull-down experiments (PD), DNA fragments encoding full-length NGDN, NGDN(1–148) and NGDN(149–315) were cloned into NdeI, BamHI restriction sites of the pnEA-pG vector (29) resulting in N-terminal glutathione S-transferase (GST) fusions. AATF full-length and deletion constructs (184–560, 208–552, 422–560, 455–560) were generated using the NdeI, MfeI sites of the pnYC-pM vector, producing fusions harboring maltose binding protein (MBP) at the N terminus.
RNAi and transient transfection
siRNA treatment was performed with INTERFERin transfection reagent (Polyplus-transfection) at a final oligonucleotide concentration of 9 nM. The following siRNA oligonucleotides were used (antisense sequence): si-control (negative control, AllStars, QIAGEN), si-AATF-1 (5′-UAUGAGUUCUCGAAGGAGCUG-3′), si-AATF-2 (5′-UAGUUUGAUCCUUCCUUCCAA-3′), si-NOL10-1 (5′-UUUAAAUCGAUCAUCGGUGAG-3′), si-NOL10-2 (5′-UUGCGAAUGAAAUUCAAUGUA-3′), si-NGDN-1 (5′-UUUCUGUAGGAUAGGCACCAG-3′), si-NGDN-2 (5′-UUAUUAGUAAGGCUCUUUCCA-3′), si-FBL (5′-UUAAUGGAAAUCACAAAGUGU-3′), si-CRM1 (5′-GUAUAAGCAAUUCACCACATT-3′).
Transient transfection of DNA plasmids was carried out using X-tremeGENE 9 transfection reagent (Roche).
Immunofluorescence analysis and confocal microscopy
Immunofluorescence analysis was performed as previously described (24). Imaging was done at a Leica SP2 AOBS microscope using a 63×1.4NA, oil, HCX Plan-Apochromat objective or at a Zeiss LSM 880 upright microscope with a 63×1.4NA, oil, DIC Plan-Apochromat objective.
Immunoprecipitation, tandem affinity purification and pull-down experiments
For preparation of HeLa K and HEK293 cell extracts, cells were detached with PBS containing 0.5 mM EDTA and washed with ice-cold PBS. For immunoprecipitation (IP) of AATF, HeLa K cells were resuspended in IP buffer containing 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 2 mM MgCl2, 0.5% NP-40, protease and phosphatase inhibitors. Lysis was performed using a douncer and the lysate was cleared by centrifugation (16 000 g, 15 min, 4°C). α-AATF antibody was coupled to a 9:1 mixture of protein A/protein G-sepharose (GE Healthcare) and incubated with cell extract for 2 h at 4°C. Beads were washed three times with IP buffer and two times with IP buffer lacking NP-40. Bound proteins were eluted with SDS-PAGE sample buffer without DTT. After addition of DTT, samples were analyzed by SDS-PAGE followed by silver or Coomassie blue staining. For MS analysis, bands were cut out from the Coomassie gel and proteins were digested with trypsin. Nano-LC–ESI-MS/MS analysis was performed using an ACQUITY nano-UPLC system (Waters) coupled to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific).
For tandem affinity purifications (TAPs), expression of HASt- or StHA-tagged bait proteins in stably transfected HEK293 cell lines was induced with tetracycline (0.5 μg/ml) for 48 h. TAP was basically performed as described previously (25). In short, cells were resuspended in lysis buffer containing 20 mM Tris–HCl (pH 7.5), 400 mM NaCl, 2 mM MgCl2, 0.5% NP-40, 1 mM DTT, 2 μM avidin, protease and phosphatase inhibitors. Cells were lysed by douncing and the lysates diluted 1:1 with lysis buffer lacking NaCl. After clearance of the lysates by centrifugation (5000 g, 12 min, 4°C), cell extracts were incubated with StrepTactin sepharose (IBA) for 30 min at 4°C. Beads were washed three times with TAP buffer containing 20 mM Tris–HCl (pH 7.5), 200 mM NaCl, 2 mM MgCl2, protease and phosphatase inhibitors. Bound complexes were eluted with TAP buffer supplemented with 2.5 mM desthio-biotin (Sigma). For the second purification step, eluates were incubated with α-HA agarose (Sigma) for 1 h at 4°C. Beads were washed with TAP buffer three times and bound proteins were eluted with SDS-PAGE sample buffer without DTT. Following the addition of DTT, samples were analyzed by SDS-PAGE.
For purification of StHA-tagged proteins by a single step pull-down using StrepTactin, proteins were expressed for 48 h after transient transfection of HEK293 cells. Generation of cell extracts was done as for TAPs, except that cells were lysed by passing them through a 20 G needle. Extracts were incubated with StrepTactin sepharose for 30 min at 4°C. Beads were washed 4 times with TAP buffer and bound proteins eluted with SDS-PAGE sample buffer.
Protein expression and in vitro binding assays
MBP-AATF proteins were co-expressed with GST-NGDN constructs or GST in E. coli BL21 Star (DE3) cells (Invitrogen) in 50 ml auto-induction medium (30) overnight at 20°C. After harvest, cells were lysed by sonication in 5 ml PD buffer (50 mM Hepes–KOH (pH 7.5), 250 mM NaCl, 2 mM DTT) supplemented with DNase I (5 μg/ml), lysozyme (1 mg/ml) and protease inhibitor cocktail (Roche). Crude lysates were cleared by centrifugation and 1 ml of the supernatant was incubated with 15 μl glutathione sepharose for 30 min at 4°C on a rotating wheel. After three washes with PD buffer, bound proteins were eluted with 30 μl PD buffer containing 25 mM glutathione and analyzed by SDS-PAGE followed by Coomassie blue staining.
Sucrose density gradient analysis
HeLa K cells were incubated with 100 μg/ml cycloheximide in DMEM for 10 min. Cells were detached with PBS containing 0.5 mM EDTA and 100 μg/ml cycloheximide and washed with ice cold PBS including 100 μg/ml cycloheximide. Lysis was performed in 50 mM Hepes-KOH (pH 7.5), 500 mM NaCl, 3 mM MgCl2, 0.5% NP40, 50 μg/ml cycloheximide, 1 mM DTT, protease and phosphatase inhibitors by passing cells through a 27 G needle. The lysate was diluted 1:1.5 in lysis buffer without NaCl and cleared by centrifugation (16 000 g, 5 min, 4°C). Cell extract (1 mg of total protein) was loaded on a linear 10–45% sucrose gradient in 50 mM Hepes–KOH (pH 7.5), 100 mM NaCl, 3 mM MgCl2 and centrifuged (55 000 rpm, 1 h 45 min, 4°C) in a TLS55 rotor (Beckman Coulter). Fractions of 150 μl were collected from the top of the gradient. Proteins were precipitated using TCA, resuspended in SDS-PAGE sample buffer and analyzed by SDS-PAGE followed by immunoblotting.
Northern blot analysis
Total RNA was extracted from HeLa K cells using an RNeasy minikit (QIAGEN). Northern blot analysis was essentially performed as previously described (31). In short: 4 μg RNA were separated on an agarose-formaldehyde gel (75 V, 4.5 h) and stained with GelRed (Biotium). RNA was transferred to a nylon membrane (Hybond-N+, GE Healthcare) by capillary transfer and cross-linked using UV light. rRNA precursors and actin mRNA were analyzed with the following radioactively labeled probes: 5′ETS (5′-CGGAGGCCCAACCTCTCCGACGACAGGTCGCCAGAGGACAGCGTGTCAGC-3′) (32), 5′ITS1 (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′), ITS2 (5′-GCGCGACGGCGGACGACACCGCGGCGTC-3′) (33), actin mRNA (5′-GTGAGGATCTTCATGAGGTAGTCAGTCAGGT-3′).
The membrane was analyzed by phospho-imaging using a Typhoon FLA 9000 (GE Healthcare).
Pulse-labeling experiments
Pulse-labeling of cells with 33P ortho-phosphate to follow pre-rRNA maturation was performed as previously described (20).
RESULTS
AATF is involved in nucleolar steps of ribosome synthesis
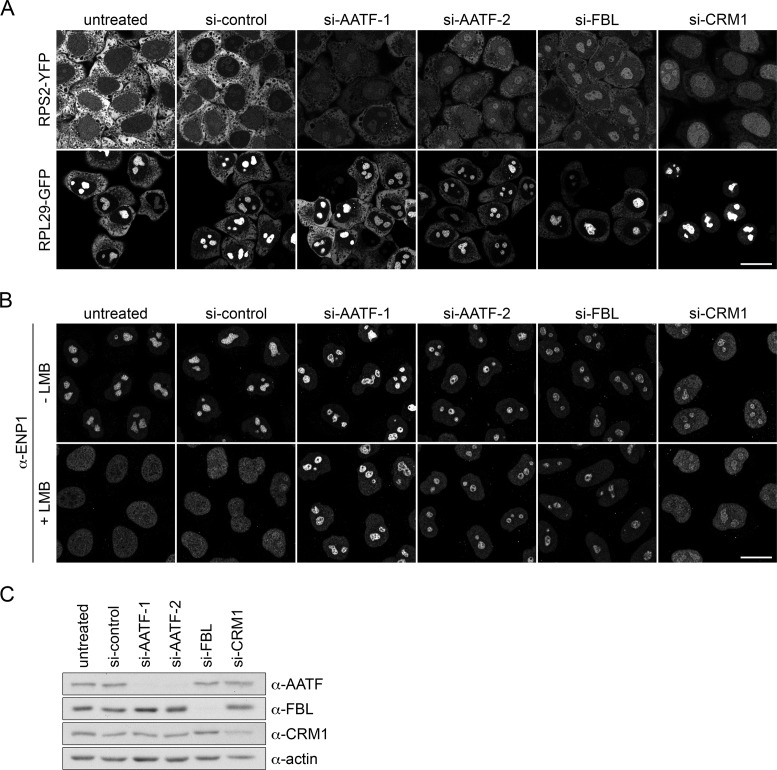
AATF has been identified as a candidate factor involved in 40S ribosomal subunit biogenesis by RNAi-based screening in human cells (20,22). Furthermore, its yeast ortholog Bfr2 has previously been shown to function in ribosome biogenesis (23). To verify the functional link between human AATF and ribosome synthesis, AATF was depleted in a reporter cell line for 40S ribosomal subunit assembly by RNAi. This HeLa cell line expresses RPS2/uS5-YFP in a tetracycline-inducible manner, which allows for monitoring nucleolar and nucleoplasmic steps in the maturation of newly synthesized 40S ribosomal particles (24). Depletion of fibrillarin (FBL), a conserved component of box C/D snoRNPs required for nucleolar steps of 40S subunit maturation (34,35), or of CRM1, a nuclear export receptor for 40S and 60S ribosomal particles (36,37), served as positive controls. As expected, downregulation of FBL and CRM1 caused nucleolar and nucleoplasmic accumulation of the reporter protein, respectively. Consistent with the previous screening results (20,22), RNAi-mediated depletion of AATF led to a reduction of RPS2-YFP in the cytoplasm and an accumulation of the reporter in nucleoli (Figure 1A, upper panel). This nucleolar accumulation of RPS2-YFP indicates a 40S subunit assembly defect, leading to a failure of RPS2-YFP release from nucleoli along with newly synthesized 40S ribosomal subunits. Similarly, the functionality of 60S ribosomal subunit assembly was investigated in a reporter cell line expressing RPL29/eL29-GFP. Downregulation of AATF did not cause a significant relocalization of RPL29-GFP after 2 days of RNAi (Figure 1A, lower panel). Together, these results suggest a specific function of AATF in the synthesis of 40S ribosomal subunits.
Figure 1.
Depletion of AATF impairs nucleolar steps of 40S ribosomal subunit biogenesis. (A) Indicated proteins were depleted by RNAi for 48 h in HeLa cell lines expressing RPS2-YFP or RPL29-GFP in an inducible manner. Expression of RPS2-YFP and RPL29-GFP was induced with tetracycline (125 ng/ml) for 16 h and subsequently chased for 4 h by washing out the tetracycline. Note that the strong nucleolar signal of RPL29-GFP in control cells is due to the slow incorporation of the reporter protein into nucleolar pre-ribosomal particles. Cells were fixed and analyzed by confocal microscopy. Scale bar, 20 μm. (B) siRNA treatment in HeLa cells for 72 h. Localization of ENP1/BYSL was analyzed by immunofluorescence (IF). To visualize early defects in ribosome maturation, CRM1-mediated export of 40S ribosomal subunits was blocked by Leptomycin B treatment (LMB, 20 nM, 2 h). (C) RNAi efficiency was verified by Western blot analysis after 72 h.
As an independent assay, we next used an immunofluorescence (IF) readout to monitor changes in the localization of the endogenous 40S biogenesis factor ENP1/BYSL in HeLa K cells (20,38). ENP1 is localized to nucleoli at steady-state (Figure 1B, upper panel), but accumulates as part of newly made pre-40S subunits in the nucleoplasm upon inhibition of CRM1-dependent pre-40S subunit export (Figure 1B, lower panel). A block of pre-40S export can either be induced by RNAi-mediated depletion of CRM1 or inhibition of CRM1 with Leptomycin B (LMB) (24,36,39). Depletion of AATF did not influence the nucleolar localization of ENP1 in the absence of a nuclear export block (Figure 1B, upper panel). However, when nuclear export of pre-40S subunits was impaired by LMB treatment, downregulation of AATF caused nucleolar retention of ENP1, while in control cells ENP1 relocalized to the nucleoplasm as expected (Figure 1B, lower panel). This demonstrates that ENP1 fails to be released from nucleoli when AATF is missing, consistent with a requirement of AATF for nucleolar steps of 40S subunit synthesis. Similar effects were observed upon depletion of FBL. The efficiency of AATF depletion was verified by immunoblotting using an affinity-purified AATF antibody that we have raised against recombinant full-length AATF (Figure 1C).
AATF has been linked to p53-mediated stress response (8). Consequently, to exclude indirect effects of AATF depletion on ribosome biogenesis via its link to the p53 pathway, we also compared the effects of AATF siRNA treatment in p53 wild-type and p53−/− HCT116 cell lines (26). Depletion of AATF led to nucleolar retention of ENP1 in both wild-type and p53-deficient HCT116 cells after LMB treatment (Supplementary Figure S1). Altogether, these results show that AATF, independently of p53, is required for nucleolar maturation of pre-40S ribosomal particles.
Nucleolar localization of AATF is conferred by its C-terminal subdomain
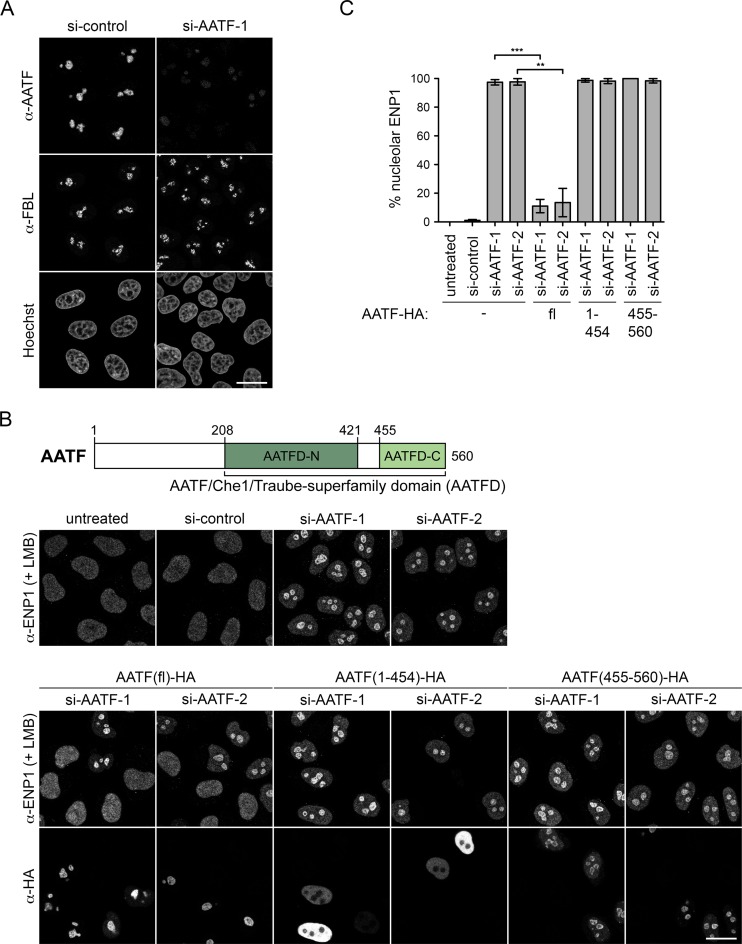
In agreement with a function in ribosome biogenesis, human AATF has been previously identified as a component of the nucleolar proteome (40). However, the protein has also been detected in other subcellular regions in various mouse and human cell types, including the nucleoplasm and cytoplasm (7,10,11,41–43). To define the localization of human AATF in HeLa cells, we performed IF analysis and observed that AATF is strongly enriched in nucleoli, where it co-localizes with FBL (Figure 2A). Downregulation of AATF by RNAi strongly reduced the IF signal, confirming the specificity of the antibody staining.
Figure 2.
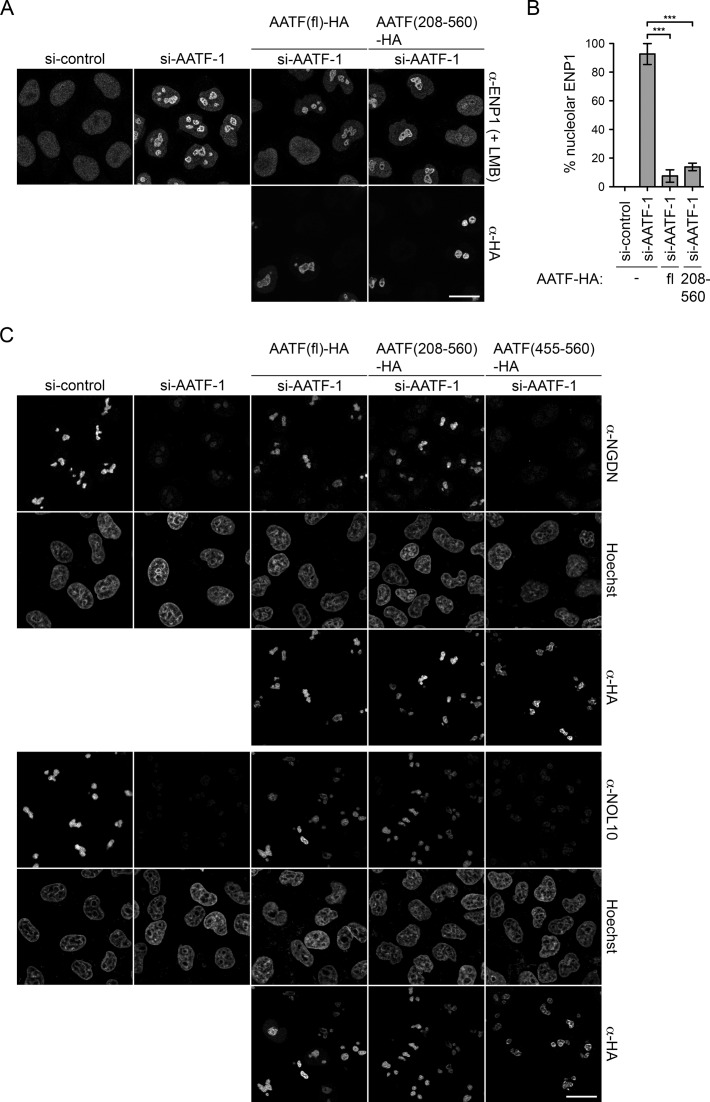
The C-terminal subdomain of human AATF is required for its nucleolar localization and for rescue of the RNAi-induced ribosome biogenesis defect. (A) Localization of endogenous AATF was analyzed by IF in HeLa cells together with the nucleolar marker FBL. Nuclei were visualized by Hoechst staining. Specificity of the anti-AATF antibody was verified by siRNA treatment for 72 h. Scale bar, 20 μm. (B) Illustration of protein domains present in AATF. The AATF protein contains an AATF/Che1/Traube-superfamily domain (AATFD) with an N-terminal (AATFD-N) and C-terminal (AATFD-C) subdomain. An AATF RNAi complementation experiment was performed in HeLa cells using two independent siRNAs transfected for 72 h. The indicated siRNA-resistant versions of HA-tagged AATF were expressed by transient transfection during the last 48 h. Cells were fixed after treatment with 20 nM LMB for 2 h. Localization of ENP1 was visualized by IF and used as readout for nucleolar release of 40S ribosomal subunits. Scale bar, 20 μm. (C) Quantification of RNAi-rescue experiments, performed as in (B), from three independent experiments. Mean intensity of the fluorescent signal derived from ENP1 IF was quantified in nucleoli and the nucleoplasm with ImageJ. To set a stringent threshold for the rescue, all cells with a ratio of mean nucleolar to nucleoplasmic ENP1 signal of ≥2.5 were assigned as ‘nucleolar’. Mean ± SEM (error bars); n ≥ 38; **P ≤ 0.01; ***P ≤ 0.001 (unpaired t-test). Note that AATF-HA full-length (fl) but neither AATF(1-454)-HA nor AATF(455-560)-HA significantly rescued the AATF depletion phenotype (for rescue, only cells transfected with AATF constructs were analyzed).
AATF is a 62 kDa protein that contains an unstructured N-terminal part followed by the AATF/Che1/Traube superfamily-specific domain (AATFD), which consists of two well conserved subdomains (AATFD-N and AATFD-C) of unknown function (Figure 2B, Supplementary Figure S2). Interestingly, a previous study showed that deletion of the C-terminal 72 residues impaired nucleolar localization of mouse AATF, whereas the N-terminal 198 residues were dispensable for nucleolar enrichment (11). To define the part of human AATF required for nucleolar localization, we generated HA-tagged truncation constructs based on sequence similarity to the mouse protein and on secondary structure predictions to avoid disrupting secondary structure elements (44). A small C-terminal fragment of AATF comprising the AATFD-C subdomain (residues 455–560) was sufficient for co-localization with FBL in nucleoli, whereas the remaining N-terminal part of AATF (residues 1–454) displayed nucleoplasmic localization (Supplementary Figure S3A). The integrity of the expressed HA-tagged AATF constructs was verified by Western blot analysis (Supplementary Figure S3B).
To determine whether the portion of AATF required for its nucleolar localization would also suffice for its function in ribosome biogenesis, we tested rescue of the nucleolar ribosome synthesis defect induced by AATF depletion using the dynamic localization of ENP1 as readout. An siRNA-resistant full-length AATF construct efficiently restored nucleoplasmic accumulation of ENP1 in the presence of LMB (Figure 2B and C). However, neither the N- nor the C-terminal truncation construct could rescue the nucleolar defect in 40S subunit synthesis upon downregulation of AATF, apparent by the failure of ENP1 release from nucleoli (Figure 2B and C). In conclusion, the AATFD-C subdomain (aa 455–560) of AATF is necessary and sufficient for its nucleolar localization but not for function in ribosome biogenesis, indicating that the N-terminal region of AATF must also contribute to the functional role of AATF in ribosome biogenesis.
AATF forms a complex with NOL10 and NGDN
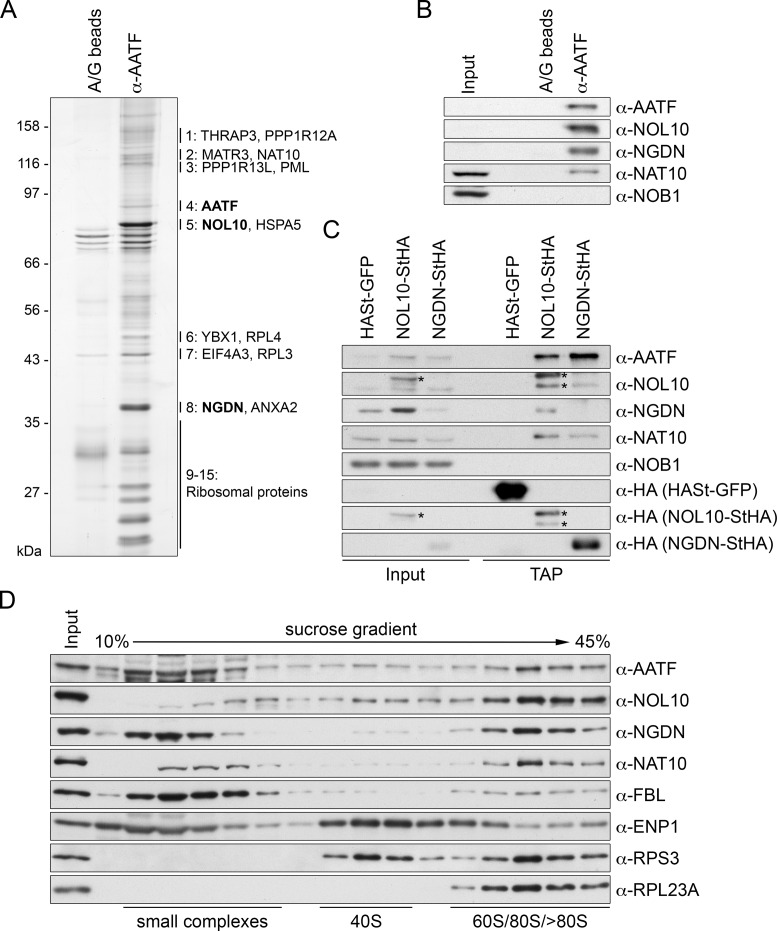
A potential explanation for the requirement of the N-terminal portion of AATF for its functionality could be that it provides a binding platform for other ribosome biogenesis factors. To investigate the protein-protein interaction network of AATF, we performed immunoprecipitation (IP) of endogenous AATF from HeLa cell lysate and determined the identity of the major AATF interacting factors by mass spectrometry (MS). Consistent with the nucleolar function of AATF, the two most prominent binding partners of AATF were identified as the nucleolar factors NOL10 and neuroguidin (NGDN, Ngd, CANu1) (Figure 3A, Supplementary Table S1). Notably, neither inhibition of cellular pre-rRNA synthesis by actinomycin D nor treatment of cell extracts with RNase diminished the association of NOL10 and NGDN with AATF, indicating that their interaction may be independent of RNA (Supplementary Figure S4A, B). NOL10 and NGDN have both been identified as components of the nucleolar proteome (40). NOL10 is a WD40 domain-containing protein enriched in the granular component of nucleoli (45). Also GFP-tagged NGDN has been shown to localize to nucleoli (46). NGDN has been originally implicated in translational control in neuronal cells (47), but belongs to the ‘U three protein’ (UTP) 3 protein family of SSU processome components by sequence similarity. In addition to NOL10 and NGDN, other nucleolar factors were detected in the MS analysis, including the predicted SSU processome component NAT10 (21), an RNA cytidine acetyltransferase already linked to 40S ribosomal subunit biogenesis (31,48,49).
Figure 3.
Identification of NOL10 and NGDN in complex with AATF. (A) AATF was isolated by immunoprecipitation (IP) from HeLa cells using an affinity-purified antibody coupled to protein A/G beads. Beads without antibodies were used as negative control. Co-precipitated proteins were analyzed by SDS-PAGE followed by silver staining. Numbering next to the gel refers to MS analysis of excised bands from a Coomassie Blue-stained gel, which identified AATF, NOL10 and NGDN among the most abundant proteins at the AATF complex (Supplementary Table S1). For band 1 to 8, the two proteins with the highest peptide number detected are listed. Bands 9–15 contained predominantly ribosomal proteins. Note that AATF is inefficiently eluted from the antibody. (B) Western blot analysis of input (0.04%) and eluate (20%) samples from experiment in (A) confirms the strong enrichment of NOL10 and NGDN in the protein complex isolated by AATF IP. (C) Tandem affinity purification (TAP) of NOL10 and NGDN in complex with AATF. Strep-HA (StHA)-tagged NOL10, NGDN or HASt-tagged GFP were induced with tetracycline in HEK293 cell lines and cell extracts (input) subjected to TAP. Inputs (0.008%) and TAP eluates (20%) were analyzed by immunoblotting using the indicated antibodies. Asterisks indicate the StHA-tagged NOL10 bait protein and a degradation product recognized by the anti-NOL10 and anti-HA antibodies. (D) HeLa cell extract was separated by centrifugation on a linear 10–45% sucrose gradient. Proteins present in the input and gradient fractions were analyzed by immunoblotting.
To verify the MS data, the eluate of the AATF IP was analyzed by immunoblotting, confirming the strong enrichment of NOL10 and NGDN in the immunoprecipitate (Figure 3B). The enrichment of NAT10 in the IP was weaker than that of NOL10 and NGDN, suggesting that it is not a core component of the complex. As a control for unspecific protein association, we also probed for NOB1, a component of later nucleoplasmic and cytoplasmic pre-40S particles (25), which was not detected in the MS analysis, and consistently, was not detectable among the AATF-associated proteins by immunoblotting.
To further confirm the existence of a protein complex containing AATF, NOL10 and NGDN, we purified C-terminally Strep-HA (StHA)-tagged NOL10 and NGDN by tandem affinity purification (TAP) from stable, tetracycline-inducible HEK293 cell lines. Immunoblot analysis revealed that AATF indeed efficiently co-purified with both NOL10 and NGDN (Figure 3C). We also observed a reciprocal co-purification of NOL10 and NGDN and verified the association of NAT10 with both proteins. Again, NOB1 could not be detected as part of the protein complex containing AATF, NOL10 and NGDN.
Sucrose gradient analysis was performed to investigate the sedimentation behavior of AATF, NOL10 and NGDN in comparison to ribosomal particles and other factors involved in ribosome assembly. Strikingly, the sedimentation behavior of AATF, NOL10 and NGDN was highly similar (Figure 3D). All three proteins appear in a similar pattern throughout the gradient, with increasing abundance in the bottom fractions containing high molecular weight complexes, with a peak at about 80S. A significant fraction of AATF and NGDN was also found in the upper part of the gradient where free proteins and small protein complexes migrate, likely owed to the use of high salt (500 mM NaCl) needed for the efficient extraction of the proteins from nucleoli, which might cause their dissociation from larger complexes. Interestingly, NOL10 was less abundant in the upper part of the gradient, which could suggest that it is more tightly associated with the 80S-sized protein complex than AATF and NGDN. Nevertheless, when fractions from the upper part of the sucrose gradient, devoid of ribosomal particles, were used as input for IP of AATF, both NGDN and NOL10 efficiently co-precipitated with AATF (Supplementary Figure S4C), suggesting that the three proteins are part of a protein complex independently of ribosomal particles.
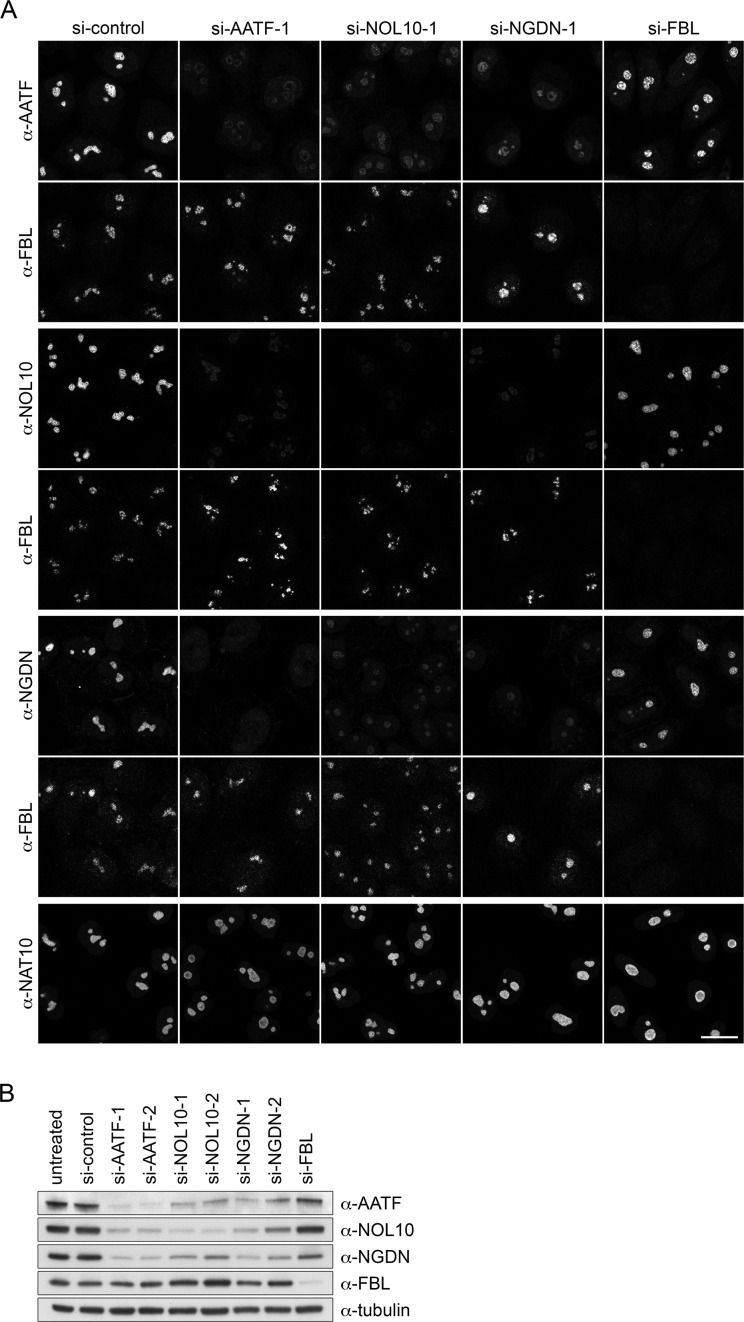
Components of protein complexes often influence each other's stability and/or localization. To examine whether this is the case for the main components of the AATF-containing protein complex, we depleted its core constituents, i.e. AATF, NOL10 and NGDN, by RNAi and analyzed the effects on the respective binding partners by both IF and Western blot analyses. As expected for components of a protein complex, AATF, NOL10 and NGDN displayed a strong interdependency regarding protein abundance (Figure 4A and B). Depletion of each of the three proteins led to a strong reduction in the levels of the other two partners. In contrast, depletion of FBL did not significantly impair stability and localization of the three factors. Notably, NAT10 remained unaffected by depletion of AATF, NOL10 and NGDN (Figure 4A), further supporting the notion that it is not a core constituent of the complex. Overall, these data provide evidence that AATF is part of a nucleolar protein complex containing NOL10 and NGDN, which we will further refer to as the AATF-NGDN-NOL10 or ‘ANN’ complex.
Figure 4.
Co-stabilization of AATF, NOL10 and NGDN. (A) HeLa cells were transfected with the indicated siRNAs. After 72 h, localization of various factors was analyzed by IF. AATF, NOL10 and NGDN were co-stained with FBL as a nucleolar marker. Scale bar, 20 μm. (B) Analysis of RNAi efficiency and protein levels by immunoblotting after 72 h of RNAi. Two independent siRNAs targeting AATF, NOL10 or NGDN and one siRNA for FBL depletion were used.
Characterization of the molecular architecture of the ANN complex reveals direct binding between AATF and NGDN
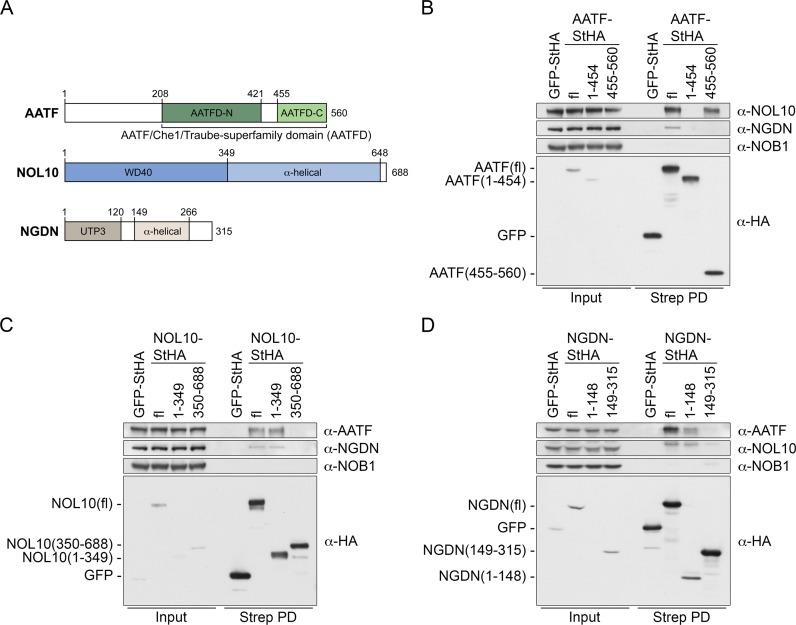
Many nucleolar proteins are known to be recruited to nascent pre-ribosomal subunits sequentially in the form of protein subcomplexes (16). A paradigm of such an assembly line is the assumed co-transcriptional formation of the large SSU processome (19,50–52). A molecular understanding of the assembly and function of individual nucleolar protein modules will require information on the architecture of the distinct building blocks.
The constituents of the ANN complex contain several folded domains, however, their specific functions remain unknown. These domains include the AATF superfamily domain (AATFD), the UTP3/SAS10 domain in NGDN and the WD40 repeats in NOL10 (Figure 5A). To gain insights into which protein domains support the formation of the ANN complex, different StHA-tagged full-length and truncated versions of AATF, NOL10 and NGDN were expressed by transient transfection in HEK293 cells and isolated by a one step pull-down using StrepTactin affinity purification. Expression of the bait proteins and composition of the precipitated complexes were examined by immunoblotting. First, we compared association of binding partners between full-length AATF and the two differentially localized AATF fragments AATF(1-454) and AATF(455-560). Full-length AATF-StHA retrieved NOL10 and also NGDN, albeit less efficiently. Consistent with its nucleoplasmic localization, AATF(1–454) did neither co-precipitate NOL10 nor NGDN. In contrast, AATF(455–560) was able to interact with NOL10 (Figure 5B), whereas NGDN could not be detected. This suggests that the C-terminal part of AATF, which localizes to nucleoli, is sufficient for NOL10 interaction whereas binding of NGDN depends on other parts of AATF.
Figure 5.
Mapping of protein interaction domains between AATF, NOL10 and NGDN. (A) Scheme of protein domains and secondary structure predictions for AATF, NOL10 and NGDN. (B–D) StHA-tagged fl and truncated versions of AATF, NOL10 and NGDN were expressed by transient transfection in HEK293 cells for 48 h. GFP-StHA was used as negative control. StHA-tagged proteins were isolated using StrepTactin sepharose (Strep PD). Protein levels in the inputs (0.45%) and the retrieved protein complexes (30%) were analyzed by immunoblotting using the indicated antibodies.
NOL10 contains a WD40 repeat domain at its N terminus comprising residues 1–349 (Figure 5A) that was sufficient for interaction with both AATF and NGDN (Figure 5C). For NGDN, its N-terminal UTP3/SAS10 homology domain (residues 1–148) was sufficient for the association with both AATF and NOL10, even though immunoblotting indicated lower expression levels of this particular NGDN fragment (Figure 5D). Thus, formation of the ANN complex is mediated by the WD40 domain of NOL10 and the UTP3/SAS10 domain of NGDN.
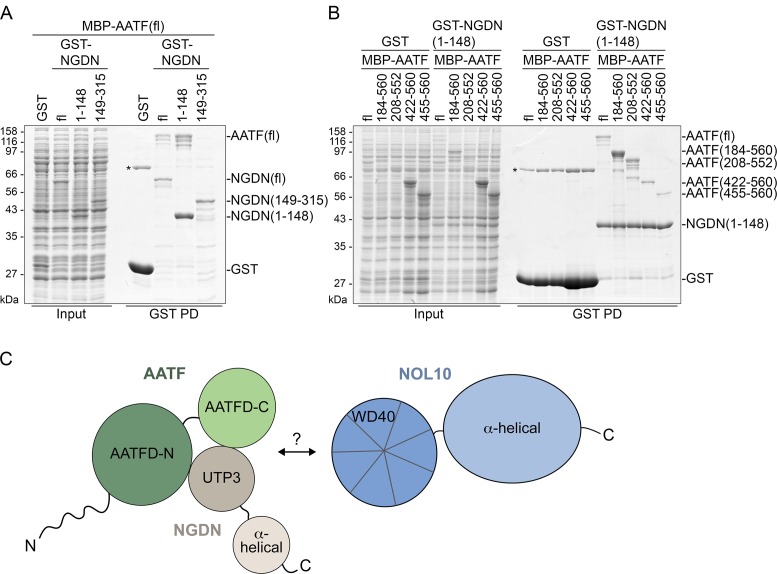
In a next step, direct binding between the mapped interaction domains was tested using recombinant proteins expressed in E. coli. Unfortunately, NOL10 was largely insoluble and could not be included in the analysis. Full-length NGDN or its N- and C-terminal fragments were expressed as GST-fusions together with full-length AATF bearing an N-terminal MBP-tag. GST pull-down experiments from the respective E. coli cell lysates not only revealed direct binding between full-length NGDN and AATF (Figure 6A), but also confirmed that the N-terminal domain of NGDN (residues 1–148) is sufficient for AATF interaction. Consistent with the domain mapping experiments performed in HEK293 cells, an interaction between the C terminus of NGDN and AATF could not be observed.
Figure 6.
AATF binds NGDN directly. (A) NGDN binds AATF directly via its N-terminal UTP3/SAS10 domain. GST or its fusions with full-length NGDN (fl: 1–315) or fragments (1–148 and 149–315) were co-expressed with MBP-tagged AATF in E. coli. GST-tagged proteins were bound to glutathione beads (GST PD). Input samples (0.04%) and bound fractions (30%) were analyzed by SDS-PAGE followed by Coomassie blue staining. The asterisk indicates an unspecific protein co-purifying with GST. (B) AATF residues 208–552 are necessary for efficient binding to NGDN. Full-length and truncated AATF constructs fused to MBP were co-expressed with either GST or GST-tagged NGDN(1–148) in E. coli. Proteins were retrieved from cell extracts by pull-down using glutathione beads. Samples were analyzed as in (A). The asterisk indicates an unspecific protein co-purifying with GST. (C) Schematic depiction of domains involved in protein-protein interactions in the ANN complex.
Reciprocally, to map the NGDN binding region in the AATF protein, MBP-tagged full-length and truncated versions of AATF were co-expressed with GST-tagged NGDN(1–148). Comparison of the expression levels of the different AATF fragments and of GST pull-down efficiency revealed weak direct binding of the C-terminal AATF subdomain (AATFD-C, residues 455–560) to NGDN(1-148) (Figure 6B). Inclusion of the long unstructured loop, which links the C-terminal subdomain of AATF to the N-terminal subdomain (AATFD-N), in the AATF fragment AATF(422–560) did not increase the affinity for NGDN. In contrast, constructs containing both the AATFD-C and AATFD-N subdomains, i.e. AATF(148-560) and AATF(208-552), bound NGDN more strongly. Thus, AATF requires its full AATF superfamily domain for efficient binding to the N terminus of NGDN (Figure 6C).
Our functional analysis had demonstrated that the AATFD-C fragment is inactive (Figure 2), which could be a consequence of inefficient binding to NGDN. To investigate the functional activity of an extended AATF fragment with superior NGDN binding ability, we performed an RNAi-rescue experiment with AATF(208-560) using nucleolar release of ENP1 as readout. Indeed, AATF(208–560) could complement the depletion of AATF and promote nucleolar release of ENP1 (Figure 7A and B). Furthermore, AATF(208–560) efficiently stabilized NGDN and NOL10 protein levels in vivo (Figure 7C), in contrast to AATFD-C. It is conceivable that the functional activity of the AATF(208-560) fragment is linked to its ability to efficiently bind NGDN, but it is also possible that it provides a functionally important binding platform for other factors.
Figure 7.
AATF(208–560) is functionally active and stabilizes NGDN and NOL10 levels in vivo. (A) An RNAi-rescue experiment was performed in HeLa cells using the localization of ENP1 after LMB treatment (20 nM, 2 h) as readout for nucleolar steps in ribosome synthesis. siRNA treatment was done for 72 h. 24 h after siRNA delivery, cells were transfected with RNAi-resistant constructs encoding for either HA-tagged full-length AATF (fl) or AATF(208-560). Localization of ENP1 and AATF constructs was visualized by IF. Scale bar, 20 μm. (B) Three independent experiments were performed as in (A) and quantified as described in Figure 2C. Mean ± SEM (error bars); n ≥ 58; ***P ≤ 0.001 (unpaired t-test). AATF-HA (fl) and AATF(208-560)-HA significantly complement the AATF depletion effect on nucleolar ribosome biogenesis. (C) RNAi and transfection were performed as in (A), including the AATF(455–560) truncated version. Rescue of NGDN and NOL10 levels was analyzed by IF and nuclei were stained with Hoechst.
Collectively, our mapping analysis of protein interaction domains in AATF, NOL10 and NGDN demonstrates that ANN complex formation involves direct binding between the N-terminal part of NGDN comprising its UTP3/SAS10 domain (aa 1–148) and the AATF superfamily domain of AATF (Figure 6C). The interaction between AATF-NGDN and NOL10 is supported by the N-terminal WD40 domain of NOL10 (aa 1–349), although a direct association between NOL10 and AATF-NGDN remains to be verified.
The ANN complex is required for nucleolar steps of 40S subunit synthesis by supporting pre-rRNA processing of the 5′ETS and ITS1
The depletion of individual members of the ANN complex affected the stability of the other complex components (Figure 4), predicting similar functional consequences of their downregulation. To test this hypothesis, we first examined how depletion of NOL10 and NGDN affects 40S subunit synthesis. Similar to depletion of AATF, knockdown of NOL10 and NGDN led to a failure of ENP1 release from nucleoli and a 40S biogenesis defect based on the RPS2-YFP reporter (Figure 8A, Supplementary Figure S5A). Notably, the observed nucleolar signal for ENP1 in presence of LMB was less pronounced for downregulation of NGDN, consistent with its less efficient depletion (Figure 4B).
Figure 8.
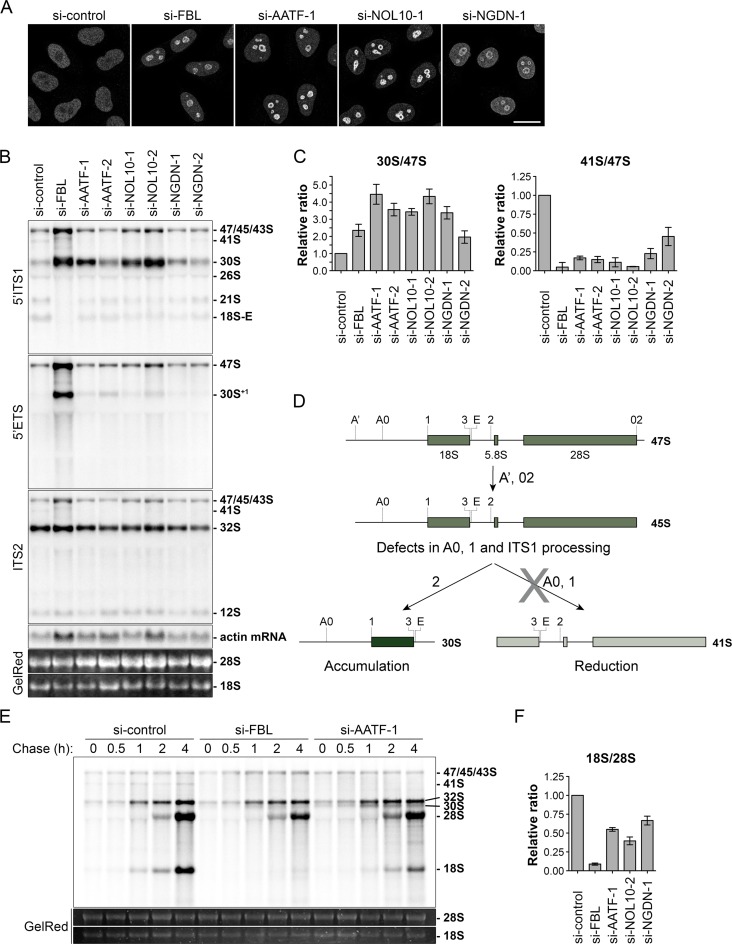
AATF, NOL10 and NGDN depletion causes pre-rRNA processing defects at cleavage sites A0, 1 and in the ITS1 region. (A) Indicated proteins were depleted by RNAi treatment for 72 h in HeLa cells. ENP1 localization was visualized by IF analysis after 2 h of LMB treatment (20 nM). Scale bar, 20 μm. (B) Northern blot analysis of total RNA extracted from HeLa cells after siRNA treatment for 72 h. Radioactively labeled probes binding to the 5′ITS1, 5′ETS, ITS2 or actin mRNA were used to detect the indicated rRNA precursors or mRNA. Mature 18S and 28S rRNA were visualized by GelRed staining on the membrane. (C) Quantification of three independent experiments using the 5′ITS1 probe as shown in (B) using ImageJ. The ‘47S’ (47S, 45S and 43S pre-rRNAs) pre-rRNA signal intensity was used for normalization. Mean ± SD (error bars). (D) Scheme illustrating pre-rRNA processing defects observed in (B). Depletion of AATF, NOL10 and NGDN leads to increased levels of 30S and reduced levels of 41S pre-rRNA, indicating lower efficiency in cleavage of 18S pre-rRNA at the sites A0, 1 and in the ITS1 region. (E) HeLa cells were subjected to RNAi for 48 h. Processing of newly synthesized pre-rRNA was analyzed after 33P pulse-labeling of HeLa cells. At the end of the indicated chase periods, RNA was extracted and separated by gel electrophoresis. Total mature 18S and 28S rRNA were stained with GelRed. (F) Quantification of three independent pulse-labeling experiments as shown in Supplementary Figure S5D. FBL, AATF, NOL10 and NGDN were depleted by siRNA treatment for 48 h and newly synthesized mature 18S and 28S rRNA were quantified after a chase period of 4 h. Mean ± SD (error bars).
For further functional investigation of the ANN complex and its role in 40S subunit maturation, we analyzed pre-rRNA processing by Northern blotting after depletion of AATF, NOL10 or NGDN. Precursors of 18S rRNA were detected using a probe binding to the 5′ internal transcribed spacer 1 (5′ITS1) pre-rRNA region (Supplementary Figure S5B), which is removed as one of the last steps of 40S subunit maturation and thus present in all 18S pre-rRNA processing intermediates. In addition, we used probes complementary to the 5′ external transcribed spacer (5′ETS) region and ITS2, which allow for detection of defects in early steps of pre-rRNA processing and 28S pre-rRNA maturation, respectively. Depletion of AATF, NOL10 and NGDN led to a striking increase of 30S pre-rRNA accompanied by a reduction of the 41S precursor, a low abundant early processing intermediate, relative to 47S pre-rRNA or to actin mRNA (Figure 8B, C, Supplementary Figure S5C). Processing of the precursors to 28S rRNA, detected by the ITS2 probe, was not significantly affected by depletion of any of the studied factors. Intriguingly, the observed defects in pre-rRNA maturation indicate reduced efficiency of endonucleolytic cleavage at the sites A0, 1 and E, an activity attributed to the SSU processome complex (18,53,54), and impaired processing of the ITS1 region (Figure 8D) (38,53,55). Similar to depletion of ANN complex members, downregulation of FBL, which is part of both the SSU processome and other snoRNPs (21,56), induced a strong accumulation of a 30S-sized precursor. However, analysis of cleavage at site A’, a site present in animals and plants (18), revealed strongly increased amounts of both 47S pre-rRNA and of the 5′ extended 30S+1 precursor upon FBL depletion. In comparison, the 30S+1 pre-rRNA was only detected to a very low extent after knockdown of AATF, NOL10 and NGDN, indicating that depletion of ANN complex members does not majorly affect processing of the very 5′ end of the 5′ETS.
Kinetic analysis of the processing efficiency of newly synthesized pre-rRNA by pulse-labeling experiments confirmed the requirement of AATF for efficient pre-18S rRNA maturation (Figure 8E, F). Depletion of AATF reduced the synthesis rate of mature 18S rRNA and, consistent with the Northern blot data, induced an accumulation of 30S pre-rRNA and a reduction of the 41S precursor. Relative to newly made 28S rRNA, the levels of freshly synthesized mature 18S rRNA decreased upon knockdown of AATF, NOL10 or NGDN (Figure 8F, Supplementary Figure S5D). The similarities between the pre-rRNA processing defects induced by downregulation of AATF, NOL10 and NGDN indicate that they are involved in the same steps of pre-18S rRNA maturation, as expected for members of a protein complex. In summary, our analysis shows that the ANN complex supports the synthesis of 40S ribosomal subunits by promoting the endonucleolytic cleavage at sites A0, 1 and ITS1 processing in the nucleolus.
DISCUSSION
A large variety of factors conduct the nucleolar assembly of 40S ribosomal subunits. Many of these factors are known to be organized in protein complexes that collectively assist the formation of premature ribosomal particles in nucleoli (16). In this study, we have identified a novel nucleolar protein complex required for 40S subunit biogenesis in human cells that we have termed the ANN complex, referring to its core components AATF, NGDN and NOL10. The complex was isolated by immunoprecipitation of AATF and the interaction between the identified core components was verified by reciprocal affinity purification of NGDN and NOL10. The association between AATF, NGDN and NOL10 is resistant to RNase and high salt treatment, indicating a tight interaction between these factors. Further, the similar sedimentation behavior and interdependence with respect to protein stability provided supporting evidence for complex formation between AATF, NGDN and NOL10.
Functional analysis revealed that AATF, NOL10 and NGDN are required for efficient nucleolar maturation of 40S ribosomal subunits. Their individual depletion inhibited nucleolar release of pre-40S subunits and induced the accumulation of 30S pre-rRNA, a precursor to 18S rRNA, due to pre-rRNA cleavage defects in the 5′ETS and ITS1 regions. The cleavage defects in the 5′ETS region are further corroborated by the observed reduction in 41S pre-rRNA. Collectively, these data support a function of AATF in 40S ribosomal subunit synthesis and pre-18S rRNA processing, in agreement with recent large-scale screening approaches (20,22,31).
The ANN complex is evolutionary conserved in yeast
The majority of factors involved in eukaryotic ribosome synthesis is conserved between metazoans and fungi. Likewise, AATF (Supplementary Figure S2), NOL10 and NGDN possess yeast orthologs of significant sequence similarity that are encoded by the essential genes BFR2 (34% protein similarity), ENP2 (52%) and LCP5 (29%), respectively, in Saccharomyces cerevisiae. Consistent with our analysis in human cells, budding yeast Bfr2, Enp2 and Lcp5 localize to nucleoli and are required for synthesis of 40S ribosomal subunits (23,57). Like their human counterparts, they are necessary for nucleolar pre-rRNA processing, i.e. for cleavage at sites A0, A1 and A2 of the yeast 35S pre-rRNA. These endonucleolytic cleavage reactions are executed by enzymes associated with the SSU processome. Notably, both Bfr2 and Enp2 have been proposed to be SSU processome components (23). In fact, Bfr2 and Enp2 interact with each other directly and have been isolated in association with an 80S-sized ribonucleoprotein complex containing SSU processome subunits and the U3 snoRNA (23), a major structural and functional component of the SSU processome (21). Similarly, Lcp5 (lethal with conditional pap1 allele), originally isolated in a synthetic lethal screen with a temperature-sensitive allele of poly(A)-polymerase PAP1, has been found in association with the U3 snoRNA. Thus, the conserved yeast orthologs of all ANN complex members function in early pre-rRNA processing, likely as structural components or regulators of the SSU processome. This view is supported by recent analysis of the Chaetomium thermophilum 90S pre-ribosome, which identified Bfr2, Enp2 and Lcp5 as unclassified core members (58). Interestingly, also AATF, NOL10 and NGDN sediment in sucrose gradients in dense fractions of about 80S to 100S, indicating that they might also be part of a large, SSU processome-like particle in human cells that still awaits its purification and molecular characterization.
The ANN complex—a structural module of the SSU processome?
The SSU processome is required for some of the initial key steps in the synthesis of 40S ribosomal subunits. The main functions of the SSU processome are the modification, folding and processing of the pre-rRNA that occur in conjunction with the incorporation of early ribosomal proteins (19,21). Impaired SSU processome activity is associated with defects in nucleolar steps of 40S biogenesis and impaired endonucleolytic cleavage in the 5′ETS and ITS1 regions as observed for AATF, NOL10 and NGDN depletion, supporting the assumption that the ANN complex is required for SSU processome activity.
The SSU processome is thought to dynamically assemble on the nascent pre-rRNA from a number of smaller subcomplexes that altogether comprise more than 70 different proteins (21). Many factors of such a large macromolecular assembly are expected to perform structural roles. Such scaffolding proteins will foster the formation of the intricate network of protein-protein, protein-rRNA and U3 snoRNA-rRNA interactions governing SSU processome architecture. AATF, NOL10 and NGDN all lack domains with identifiable enzymatic activity and may thus fulfill a structural role as a building block of the larger assembly, although it cannot be excluded that AATF, NOL10 and NGDN might bear so far unidentified enzymatic activities.
As potential scaffolding proteins, ANN complex members could facilitate the recruitment, activity or positioning of SSU processome-associated enzymes. One obvious candidate is the DEAD-box RNA helicase DDX10, the human ortholog of S. cerevisiae Dbp4, which is required for SSU processome activity (59,60). Yeast Dbp4 has been shown to interact with Bfr2 and Enp2 in an RNA-dependent manner and all three factors are part of 80S-sized complexes containing U3 snoRNA, indicating that these particles represent SSU processomes. We did not detect peptides for DDX10 in our MS analysis of AATF immunoprecipitates, which might be due to the stringent salt conditions used for the respective experiments. Notably, the association of yeast Dbp4 with the SSU processome is sensitive to high salt (60).
In contrast, we identified other enzymes in complex with AATF, including the RNA helicases DDX3 and eIF4A3 as well as the RNA cytidine acetyltransferase NAT10 (Supplementary Table S1). NAT10 and eIF4A3 have been previously linked to 40S subunit synthesis and rRNA processing in mammalian cells (20,22,31,48,49,61). While the association of NAT10 with AATF, NGDN and NOL10 was verified by TAP, NAT10 is unlikely to be a core constituent of the ANN complex since neither NAT10 protein levels nor its localization changed upon AATF, NOL10 or NGDN depletion. Yet, it is conceivable that NAT10 can dynamically associate with the ANN complex, for instance to promote its function in pre-rRNA modification. It will be interesting to decipher whether the function of any of the candidate enzymes in 40S biogenesis is linked to the integrity of the ANN complex.
Protein–protein interactions required for ANN complex formation
Most of the constituents of the SSU processome lack enzymatic activity but frequently harbor domains implicated in protein-protein interactions such as WD40 repeat domains (21). WD40 domains have long been appreciated for their function as binding platforms in macromolecular assemblies, e.g. in nucleoporins of the nuclear pore complex (62,63). The WD40 domain of NOL10 carries out a similar function within the ANN complex, where it is required for association with AATF and NGDN. In agreement with our interaction data, the yeast homologs of NOL10 and AATF, Enp2 and Bfr2, bind each other directly (23), although their interacting domains have not yet been delineated. The WD40 domains of NOL10 and Enp2 are highly similar (∼45% identity, 59% similarity). This high sequence conservation between the yeast and human proteins indicates that the involved binding interface might be conserved, although a direct interaction between human AATF and NOL10 remains to be experimentally confirmed. Interestingly, WD40 repeats have also been implicated in protein–RNA interactions (64–66). We can thus not exclude that the WD40 domain of NOL10 may contact RNA in addition. However, as the association between core members of the ANN complex is RNase-resistant, RNA may not be involved in mediating interactions between NOL10, AATF and NGDN, although the presence of structurally hidden, RNase-inaccessible RNA within the ANN complex can not be excluded.
Our mapping analysis demonstrated that AATF binds NGDN directly including contacts between the AATF superfamily domain and the N-terminal UTP3/SAS10 domain of NGDN. It has been speculated that the UTP3/SAS10 domain might serve as an RNA interaction platform (67). However, our data classify the UTP3/SAS10 domain of NGDN as a protein-protein interaction module. Similarly, the N-terminal UTP3/SAS10 domain of the yeast exosome component Rrp47 makes protein-protein contacts with both Rrp6 and an N-terminal peptide of RNA helicase Mtr4 (68,69). In analogy to the Rrp6–Rrp47 complex, the UTP3/SAS10 domain of NGDN could in principle be an exosome recruitment domain. However, our data show that NGDN's UTP3/SAS10 domain is involved in direct binding to AATF. It remains to be determined whether the UTP3/SAS10 domain of NGDN employs a structurally similar mode of binding to AATF as the analogous domain in Rrp47 in the exosome complex.
Surprisingly, most components of the SSU processome lack canonical RNA binding domains and so do AATF, NOL10 and NGDN. However, both NGDN and AATF contain conserved stretches of basic residues in their C termini. At this point, one can only speculate that these residues could be involved in RNA binding, similarly to the interaction mode of ribosomal proteins with rRNA by the use of non-canonical RNA binding motifs (70).
Altogether, our analysis has identified conserved domains involved in AATF/NGDN/ NOL10 complex formation, providing first insights into the molecular architecture of the nucleolar ANN complex. Importantly, our results predict that the yeast homolog of NGDN, Lcp5, is an additional component of the yeast Bfr2/Enp2 complex.
AATF is an essential protein for cell proliferation and possibly tumor growth
Functional analyses have linked AATF, NOL10 and NGDN to 40S ribosomal subunit synthesis and pre-rRNA processing. First and foremost, these findings expand the functional repertoire of AATF, which has so far been described as a transcriptional co-factor of RNA polymerase II. The identified nucleolar role of AATF in ribosome production can also provide a mechanistic explanation for its essential role in cell proliferation and embryonic development of mice (10).
AATF and NGDN have been described as factors regulating transcription and neuronal translation, respectively (1–3,47). Our analysis does not exclude that both proteins play additional roles. However, a translational function of NGDN in HeLa cells is unlikely as NGDN is strongly enriched in nucleoli and appears to be absent from the cytoplasm. Also AATF is a nucleolar protein and could in principle support pre-rRNA transcription. However, nucleoli do not disintegrate upon AATF depletion, as expected for impaired pre-rRNA synthesis. Also ENP1 and RPS2-YFP show defects in leaving nucleoli rather than a failure in accumulation. Further, downregulation of AATF does not prevent pre-rRNA synthesis. Thus, we conclude that AATF is not a bona fide pre-rRNA transcription factor.
In its function as RNA polymerase II-specific transcription factor, AATF has been previously suggested to bind the promoter region of the TP53 gene, modulating the transcriptional activity of p53 protein and regulating cellular stress responses (5–8). Importantly, we show that the function of AATF in ribosome synthesis is independent of p53. Still, the described role of AATF as a transcriptional regulator and p53 interactor (71) is highly interesting in light of the fact that ribosome synthesis and cellular stress response pathways are strongly interwoven. Defects in ribosome synthesis, often referred to as ‘nucleolar stress’, are known to elicit p53 stabilization by a post-transcriptional mechanism. This involves an interaction between H/MDM2, an E3 ubiquitin ligase that targets p53 for proteasomal degradation, and the 5S rRNP composed of ribosomal proteins RPL5/uL18, RPL11/uL5 and newly synthesized 5S rRNA (72–74). The identified function of AATF in ribosome synthesis together with the previously described role of AATF in p53 regulation positions AATF at the interface between ribosome production and cellular stress surveillance pathways. It will thus be interesting to explore in future whether the previously described transcriptional activity of AATF is exploited by cells as part of a cross-talk mechanism between ribosome biogenesis and transcriptional responses to defects thereof. Conversely, it remains worth considering whether a transcriptional function of AATF along with RNA polymerase II contributes to the role of AATF in ribosome synthesis. Finally, as depletion of AATF has been shown to inhibit tumor growth (6), our study further sustains the strategy of targeting ribosome synthesis as a promising approach for cancer treatment (75).
Supplementary Material
Acknowledgments
We thank Dr. J. Pfannstiel and B. Wuertz at the University of Hohenheim, Germany, for providing excellent service in mass spectrometry. We also thank Caroline Ashiono for superb technical assistance, Dr. Pierre-Emmanuel Gleizes for insightful advice, Dr. François Dragon for communication of unpublished data, Dr. Ivo Zemp, Christian Montellese and Philipp Leu for critical reading of the manuscript, and the members of the Kutay lab for helpful discussions. Microscopy was performed on instruments of the ETHZ Microscopy Center (ScopeM).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (31003A_166565 to U.K.); FEBS long-term fellowship (to S.J.). Funding for open access charge: Swiss National Science Foundation, ETH Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fanciulli M., Bruno T., Di Padova M., De Angelis R., Iezzi S., Iacobini C., Floridi A., Passananti C. Identification of a novel partner of RNA polymerase II subunit 11, Che-1, which interacts with and affects the growth suppression function of Rb. FASEB J. 2000;14:904–912. doi: 10.1096/fasebj.14.7.904. [DOI] [PubMed] [Google Scholar]
- 2.Lindfors K., Halttunen T., Huotari P., Nupponen N., Vihinen M., Visakorpi T., Mäki M., Kainulainen H. Identification of novel transcription factor-like gene from human intestinal cells. Biochem. Biophysi. Res. Commun. 2000;276:660–666. doi: 10.1006/bbrc.2000.3480. [DOI] [PubMed] [Google Scholar]
- 3.Page G., Lödige I., Kögel D., Scheidtmann K.H. AATF, a novel transcription factor that interacts with Dlk/ZIP kinase and interferes with apoptosis. FEBS Lett. 1999;462:187–191. doi: 10.1016/s0014-5793(99)01529-x. [DOI] [PubMed] [Google Scholar]
- 4.Iezzi S., Fanciulli M. Discovering Che-1/AATF: a new attractive target for cancer therapy. Front. Genet. 2015;6:141. doi: 10.3389/fgene.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruno T., De Nicola F., Iezzi S., Lecis D., D'Angelo C., Di Padova M., Corbi N., Dimiziani L., Zannini L., Jekimovs C., et al. Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell. 2006;10:473–486. doi: 10.1016/j.ccr.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Bruno T., Desantis A., Bossi G., Di Agostino S., Sorino C., De Nicola F., Iezzi S., Franchitto A., Benassi B., Galanti S., et al. Che-1 promotes tumor cell survival by sustaining mutant p53 transcription and inhibiting DNA damage response activation. Cancer Cell. 2010;18:122–134. doi: 10.1016/j.ccr.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 7.Höpker K., Hagmann H., Khurshid S., Chen S., Hasskamp P., Seeger-Nukpezah T., Schilberg K., Heukamp L., Lamkemeyer T., Sos M.L., et al. AATF/Che-1 acts as a phosphorylation-dependent molecular modulator to repress p53-driven apoptosis. EMBO J. 2012;31:3961–3975. doi: 10.1038/emboj.2012.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruno T., Iezzi S., Fanciulli M. Che-1/AATF: a critical cofactor for both wild-type- and mutant-p53 proteins. Front. Oncol. 2016;6:34. doi: 10.3389/fonc.2016.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul D., Mehrotra A. Functional characterization of AATF transcriptome in human leukemic cells. Mol Cell Biochem. 2007;297:215–220. doi: 10.1007/s11010-006-9317-1. [DOI] [PubMed] [Google Scholar]
- 10.Thomas T., Voss A.K., Petrou P., Gruss P. The murine gene, Traube, is essential for the growth of preimplantation embryos. Dev. Biol. 2000;227:324–342. doi: 10.1006/dbio.2000.9915. [DOI] [PubMed] [Google Scholar]
- 11.Ferraris S.E., Isoniemi K., Torvaldson E., Anckar J., Westermarck J., Eriksson J.E. Nucleolar AATF regulates c-Jun-mediated apoptosis. Mol. Biol. Cell. 2012;23:4323–4332. doi: 10.1091/mbc.E12-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggero D., Pandolfi P.P. Does the ribosome translate cancer. Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 13.van Riggelen J., Yetil A., Felsher D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Publishing Group. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 14.Bursac S., Brdovcak M.C., Donati G., Volarevic S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim. Biophys. Acta. 2014;1842:817–830. doi: 10.1016/j.bbadis.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Kressler D., Hurt E., Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Woolford J.L., Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Cruz J., Karbstein K., Woolford J.L. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henras A.K., Plisson-Chastang C., O'Donohue M.-F., Chakraborty A., Gleizes P.-E. An overview of pre-ribosomal RNA processing in eukaryotes. WIREs RNA. 2015;6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turowski T.W., Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. WIREs RNA. 2015;6:129–139. doi: 10.1002/wrna.1263. [DOI] [PubMed] [Google Scholar]
- 20.Badertscher L., Wild T., Montellese C., Alexander L.T., Bammert L., Sarazova M., Stebler M., Csucs G., Mayer T.U., Zamboni N., et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep. 2015;13:2879–2891. doi: 10.1016/j.celrep.2015.11.061. [DOI] [PubMed] [Google Scholar]
- 21.Phipps K.R., Charette J.M., Baserga S.J. The small subunit processome in ribosome biogenesis—progress and prospects. WIREs RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I., Kozak K., Csucs G., Lund E., Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soltanieh S., Lapensée M., Dragon F. Nucleolar proteins Bfr2 and Enp2 interact with DEAD-box RNA helicase Dbp4 in two different complexes. Nucleic Acids Res. 2014;42:3194–3206. doi: 10.1093/nar/gkt1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zemp I., Wild T., O'Donohue M.-F., Wandrey F., Widmann B., Gleizes P.-E., Kutay U. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 2009;185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyler E., Zimmermann M., Widmann B., Gstaiger M., Pfannstiel J., Kutay U., Zemp I. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2011;17:189–200. doi: 10.1261/rna.2325911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 27.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 28.Pool M.R., Stumm J., Fulga T.A., Sinning I., Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 29.Diebold M.-L., Fribourg S., Koch M., Metzger T., Romier C. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011;175:178–188. doi: 10.1016/j.jsb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Tafforeau L., Zorbas C., Langhendries J.-L., Mullineux S.-T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L.J. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Hölzel M., Orban M., Hochstatter J., Rohrmoser M., Harasim T., Malamoussi A., Kremmer E., Längst G., Eick D. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J. Biol. Chem. 2010;285:6364–6370. doi: 10.1074/jbc.M109.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouquette J., Choesmel V., Gleizes P.-E. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baserga S.J., Yang X.D., Steitz J.A. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen R.P., Hurt E.C., Kern H., Lehtonen H., Carmo-Fonseca M., Lapeyre B., Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas F., Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- 37.Trotta C.R., Lund E., Kahan L., Johnson A.W., Dahlberg J.E. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carron C., O'Donohue M.-F., Choesmel V., Faubladier M., Gleizes P.-E. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 2011;39:280–291. doi: 10.1093/nar/gkq734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E.P., Wolff B., Yoshida M., Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad Y., Boisvert F.-M., Gregor P., Cobley A., Lamond A.I. NOPdb: nucleolar proteome database–2008 update. Nucleic Acids Res. 2009;37:D181–D184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbato C., Corbi N., Canu N., Fanciulli M., Serafino A., Ciotti M., Libri V., Bruno T., Amadoro G., De Angelis R., et al. Rb binding protein Che-1 interacts with Tau in cerebellar granule neurons. Modulation during neuronal apoptosis. Mol. Cell. Neurosci. 2003;24:1038–1050. doi: 10.1016/j.mcn.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Guo Q., Xie J. AATF inhibits aberrant production of amyloid beta peptide 1-42 by interacting directly with Par-4. J. Biol. Chem. 2004;279:4596–4603. doi: 10.1074/jbc.M309811200. [DOI] [PubMed] [Google Scholar]
- 43.Di Certo M.G., Corbi N., Bruno T., Iezzi S., De Nicola F., Desantis A., Ciotti M.T., Mattei E., Floridi A., Fanciulli M., et al. NRAGE associates with the anti-apoptotic factor Che-1 and regulates its degradation to induce cell death. J. Cell Sci. 2007;120:1852–1858. doi: 10.1242/jcs.03454. [DOI] [PubMed] [Google Scholar]
- 44.Buchan D.W.A., Minneci F., Nugent T.C.O., Bryson K., Jones D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–W57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wada K., Sato M., Araki N., Kumeta M., Hirai Y., Takeyasu K., Furukawa K., Horigome T. Dynamics of WD-repeat containing proteins in SSU processome components. Biochem. Cell Biol. 2014;92:191–199. doi: 10.1139/bcb-2014-0007. [DOI] [PubMed] [Google Scholar]
- 46.Sihn C.-R., Lee Y.-S., Jeong J.-S., Park K., Kim S.H. CANu1, a novel nucleolar protein, accumulated on centromere in response to DNA damage. Genes Cells. 2008;13:787–796. doi: 10.1111/j.1365-2443.2008.01205.x. [DOI] [PubMed] [Google Scholar]
- 47.Jung M.-Y., Lorenz L., Richter J.D. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol. Cell. Biol. 2006;26:4277–4287. doi: 10.1128/MCB.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito S., Horikawa S., Suzuki T., Kawauchi H., Tanaka Y., Suzuki T., Suzuki T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA) J. Biol. Chem. 2014;289:35724–35730. doi: 10.1074/jbc.C114.602698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma S., Langhendries J.-L., Watzinger P., Kötter P., Entian K.-D., Lafontaine D.L.J. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 51.Dutca L.M., Gallagher J.E.G., Baserga S.J. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011;39:5164–5180. doi: 10.1093/nar/gkr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez-Fernandez J., Martín-Marcos P., Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res. 2011;39:8105–8121. doi: 10.1093/nar/gkr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J.M., Watkins N.J. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013;200:577–588. doi: 10.1083/jcb.201207131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomecki R., Labno A., Drazkowska K., Cysewski D., Dziembowski A. hUTP24 is essential for processing of the human rRNA precursor at site A1, but not at site A0. RNA Biol. 2015;12:1010–1029. doi: 10.1080/15476286.2015.1073437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preti M., O'Donohue M.-F., Montel-Lehry N., Bortolin-Cavaillé M.-L., Choesmel V., Gleizes P.-E. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013;41:4709–4723. doi: 10.1093/nar/gkt160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watkins N.J., Bohnsack M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. WIREs RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 57.Wiederkehr T., Prétôt R.F., Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R., et al. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016;166:380–393. doi: 10.1016/j.cell.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 59.Koš M., Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 60.Soltanieh S., Osheim Y.N., Spasov K., Trahan C., Beyer A.L., Dragon F. DEAD-box RNA helicase Dbp4 is required for small-subunit processome formation and function. Mol. Cell. Biol. 2015;35:816–830. doi: 10.1128/MCB.01348-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexandrov A., Colognori D., Steitz J.A. Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex. Genes Dev. 2011;25:1078–1090. doi: 10.1101/gad.2045411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brohawn S.G., Partridge J.R., Whittle J.R.R., Schwartz T.U. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stirnimann C.U., Petsalaki E., Russell R.B., Müller C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Granneman S., Pruijn G.J.M., Horstman W., van Venrooij W.J., Lührmann R., Watkins N.J. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in Vitro. J. Biol. Chem. 2002;277:48490–48500. doi: 10.1074/jbc.M206631200. [DOI] [PubMed] [Google Scholar]
- 65.Wegrecki M., Marcin W., Neira J.L., Bravo J. The carboxy-terminal domain of Erb1 is a seven-bladed ß-propeller that binds RNA. PLoS ONE. 2015;10:e0123463. doi: 10.1371/journal.pone.0123463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loedige I., Jakob L., Treiber T., Ray D., Stotz M., Treiber N., Hennig J., Cook K.B., Morris Q., Hughes T.R., et al. The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation. Cell Rep. 2015;13:1206–1220. doi: 10.1016/j.celrep.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 67.Mitchell P. Rrp47 and the function of the Sas10/C1D domain. Biochem. Soc. Trans. 2010;38:1088–1092. doi: 10.1042/BST0381088. [DOI] [PubMed] [Google Scholar]
- 68.Schuch B., Feigenbutz M., Makino D.L., Falk S., Basquin C., Mitchell P., Conti E. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 2014;33:2829–2846. doi: 10.15252/embj.201488757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makino D.L., Schuch B., Stegmann E., Baumgärtner M., Basquin C., Conti E. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015;524:54–58. doi: 10.1038/nature14865. [DOI] [PubMed] [Google Scholar]
- 70.Ramakrishnan V., Moore P.B. Atomic structures at last: the ribosome in 2000. Curr. Opin. Struct. Biol. 2001;11:144–154. doi: 10.1016/s0959-440x(00)00184-6. [DOI] [PubMed] [Google Scholar]
- 71.Desantis A., Bruno T., Catena V., De Nicola F., Goeman F., Iezzi S., Sorino C., Gentileschi M.P., Germoni S., Monteleone V., et al. Che-1 modulates the decision between cell cycle arrest and apoptosis by its binding to p53. Cell Death Dis. 2015;6:e1764. doi: 10.1038/cddis.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donati G., Peddigari S., Mercer C.A., Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sloan K.E., Bohnsack M.T., Watkins N.J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishimura K., Kumazawa T., Kuroda T., Katagiri N., Tsuchiya M., Goto N., Furumai R., Murayama A., Yanagisawa J., Kimura K. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 2015;10:1310–1323. doi: 10.1016/j.celrep.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 75.Quin J.E., Devlin J.R., Cameron D., Hannan K.M., Pearson R.B., Hannan R.D. Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta. 2014;1842:802–816. doi: 10.1016/j.bbadis.2013.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.