Abstract
DNA methylation of cytosine in eukaryotic cells is a common epigenetic modification, which plays an important role in gene expression and thus affects various cellular processes like development and carcinogenesis. The occurrence of 5-methyl-2′-deoxycytosine (5mC) as well as the distribution pattern of this epigenetic marker were shown to be crucial for gene regulation and can serve as important biomarkers for diagnostics. DNA polymerases distinguish little, if any, between incorporation opposite C and 5mC, which is not surprising since the site of methylation is not involved in Watson–Crick recognition. Here, we describe the development of a DNA polymerase variant that incorporates the canonical 2′-deoxyguanosine 5′-monophosphate (dGMP) opposite C with higher efficiency compared to 5mC. The variant of Thermococcus kodakaraensis (KOD) exo- DNA polymerase was discovered by screening mutant libraries that were built by rational design. We discovered that an amino acid substitution at a single site that does not directly interact with the templating nucleobase, may alter the ability of the DNA polymerase in processing C in comparison to 5mC. Employing these findings in combination with a nucleotide, which is fluorescently labeled at the terminal phosphate, indicates the potential use of the mutant DNA polymerase in the detection of 5mC.
INTRODUCTION
The most abundant epigenetic mark in vertebrates is the modification of cytosines at position 5 with a methyl group (1). So-called CpG islands are potential marks for 5-methyl-2′-deoxycytosine (5mC) and 75% of such dinucleotides are in fact methylated in promoter regions of mammalian cells (2). Cytosine methylation within CpG islands is generally associated with gene silencing and thus plays a crucial role in gene regulation (3), as well as developmental processes (4,5). A tight regulation of this DNA modification is essential and even slight changes in methylation patterns may have profound consequences for an organism, such as the development of cancer (6,7). While methylation of cytosines outside of CpG islands has been known to be abundant in plants, non-CG methylation in mammals has mostly been viewed as an artifact of sequencing technologies (8). The occurrence of 5mC followed by bases other than G has since been confirmed in mice (9) and more recently in humans (10). Non-CG methylation is predominantly found in stem and germ cells, but their function is still a source of controversy (8). Furthermore, DNA methylation is also found in the genomes of many bacteria and archea (11). Three types of methylated DNA bases - N6-methyladenine and N4-methylcytosine and 5-methylcytosine - are found and known to play a role in biological processes. Firstly, in the restriction–modification system that enables prokaryotes to distinguish between their own and foreign DNA (for extensive review see (12) and (13); also within the mismatch repair pathway that is best understood in the E. coli Dam system (14) and in the control of DNA replication and its coupling to cell cycle progression (15). Similarly to eukaryotes, DNA methylation is also thought to regulate bacterial gene expression. However, this is facilitated exclusively by methylated adenines, which therefore serve as the predominant signal of bacterial epigenetics in contrast to 5mC in the eukaryotic world (16). Nevertheless, it is quite clear that identification and localization of methylated bases are crucial for our understanding of genome organization and function.
The detection of 5mC has emerged as an important factor for molecular diagnostics as well as for choosing effective treatment strategies (17,18). Different schemes are applied to locate this modification throughout the genome. The most common approach for 5mC detection at single nucleotide resolution relies on bisulfite treatment of sample DNA, the unmodified cytosines of which are converted into uracils, whereas methylated cytosines remain unaffected (19). Followed by sequencing, this method shows U where there is a C and C where there is a 5mC (20). Combined with next generation sequencing, whole genomes can be mapped using this method (21). However, this method carries some disadvantages. It is laborious and time consuming since two sequencing runs are required. Conditions used in bisulfite conversion are harsh and prone to destroy about 95% of the sample DNA (22). Furthermore, there are common errors that may occur during the procedure: for example inappropriate conversion of 5mC to thymine or the failure to convert cytosine to uracil (23,24). Single molecular real time sequencing (SMRT) is one sequencing technique, which exploits the potential of DNA polymerases for high catalytic rates as well as high processivity. By using a DNA polymerase mutant in combination with dNTPs that are conjugated to either of four distinguishable fluorescent tags at the terminal phosphate moiety, this method allows continuous observation of DNA synthesis over thousands of bases (25–27). One interesting development is the recent approach of Tet-assisted bisulfite sequencing (28). Tet enzymes convert 5-methylcytosine to 5-formyl- and 5-carboxylcytosine, a naturally occuring oxidation cascade that is thought to be the basis of active DNA demethylation (29–31). Each conversion of 5mC carried out by Tet1 results in a moiety with a unique kinetic behavior that directly enhances both sensitivity and specificity of 5mC detection by SMRT sequencing (32). Due to the complex treatment procedures and the error-prone nature of bisulfite sequencing, there is a need for broadly applicable methods that allow for DNA methylation profiling in diagnostics and prognosis in a direct and accurate manner.
Targeting the DNA polymerases or the nucleotides used for sequencing is a promising approach for the development of alternative methods to detect DNA methylation. DNA polymerases that are widely employed in nucleic acids diagnostics (33,34) fail to discriminate significantly between dGMP incorporation opposite C and 5mC directly (35). This is to be expected since 5-methylation does not affect Watson–Crick base pairing. However, Shen et al. reported the ability of AMV reverse transcriptase to discriminate in dGMP incorporation opposite C and 5mC. But nevertheless, the observed discrimination is accompanied by an increased error rate opposite 5mC that might prevent applications (36). Additionally, archeal DNA polymerases from at least two different sequence families have been found to discriminate U from T in DNA (37–39). Nevertheless, this effect has been shown not to result from the incorporation process per se, but derives from a combination of different events like proofreading and exonuclease activity. We have engineered a variant of Thermococcus kodakaraensis (KOD) exo- DNA polymerase that enables the direct discrimination between C and 5mC at single sites when primer extension is performed from mismatched primer termini while matched primer template complexes failed to show significant discrimination (40). Furthermore, we explored the potential of modified nucleotides to be employed in site specific 5mC detection (35,41). We found that O6-alkylated dGTP-analogues are processed opposite C and 5mC with different efficiencies by KOD exo- DNA polymerase. However, there is no description yet, of a DNA polymerase that can directly discriminate between C and 5mC without the use of modified nucleotides or mismatched primers.
Here, we describe the discovery and characterization of KOD exo- DNA polymerase variants that are able to discriminate processing of unmodified dGTP between C and 5mC in the template. The mutant proteins were identified by a combination of site-directed mutagenesis and screening.
MATERIALS AND METHODS
Construction of KOD exo- DNA polymerase libraries
DNA oligonucleotides were purchased from biomers.net GmbH. Sequences of forward primers harbored a codon mismatch to introduce the desired mutation. Reverse primers were 5′-phosphorylated to allow for ligation of the polymerase chain reaction (PCR) product. Oligonucleotides were dissolved in deionized water to a concentration of 100 μM. Sequences of oligonucleotides used in this study are listed in Supplementary Table S1. To obtain a defined library of all 19 possible mutants at each site investigated, 19 single PCR reactions were performed per site using Pfu Turbo DNA Polymerase (Agilent). Following mutagenesis PCR using a pET21a plasmid (Novagen) containing the KOD exo- DNA polymerase wild-type sequence, template DNA was digested with the methylation sensitive endonuclease DpnI (NEB). PCR products were purified from agarose gels with a NucleoSpin® Gel and PCR Clean-up (Macherey–Nagel) and ligated with T4 DNA ligase (NEB) for either 2 h at room temperature or overnight at 16°C. Chemically competent Escherichia coli BL21 (DE3) cells (Novagen) were transformed with 5 μl of a ligation reaction and positive clones were selected via carbenicillin resistance. Single clones were picked, plasmids prepared using QIAprep® Spin Miniprep Kit (Qiagen) and sequenced (GATC Biotech AG) to ensure correct mutagenesis. Libraries were established essentially as described previously (42). Briefly, cells containing mutant plasmids were grown overnight in 384-well plates, containing 150 μl LB-medium supplemented with 100 μg/ml carbenicillin per well at 37°C on a plate shaker (180 rpm). Single clones were picked per well and each library contained four wild-type clones serving as controls and one clone harboring empty vector as negative control. After overnight incubation, cultures were supplemented with glycerol to a final concentration of 25% and stored at -80°C.
Expression of KOD exo- DNA polymerase mutants and lysate preparation
KOD exo- DNA polymerase and its respective mutants were recombinantly expressed in E. coli BL21 (DE3) (Novagen) in duplicates as described before (43). Briefly, 500 μl cultures were grown in 96 deep well plates, induced with IPTG and cells harvested after expression. Bacterial pellets were lysed in 96 deep well plates in lysis buffer (120 mM TrisHCl pH 8, 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton, 1.5 mM MgCl2, 1 mM PMSF, 1 mg/ml lysozyme), followed by heat denaturation of host proteins at 75°C for 40 min and centrifugation at 4.000 x g for 60 min. Expression levels were checked via SDS-PAGE and cleared lysates were directly used for screening.
Screening of KOD exo- DNA polymerase mutants for activity
Typical reactions consisted of 10 μl and contained 200 μM dNTPs, 100 nM forward (5′-d(CTT GGT GAG ACT GGT AGA CG)-3′) and reverse (5′-d(TTA GAC CCA CCC CTC CTG GCG)-3′) primer respectively, 100 pM template DNA and 1x SYBR Green I (Sigma) in buffer (120 mM TrisHCl pH 8, 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton, 1.5 mM MgCl2). Real time PCR data were collected using a Roche LightCycler 96 system with an initial denaturation at 95°C for 2 min followed by amplification over 50 cycles with denaturation at 95°C for 10 s and annealing and elongation at 68°C for 20 s. Melting curves were measured immediately after PCR amplification. Two independent experiments were conducted, using lysates from independent library expressions.
Screening of KOD exo- DNA polymerase mutants for 5mC detection
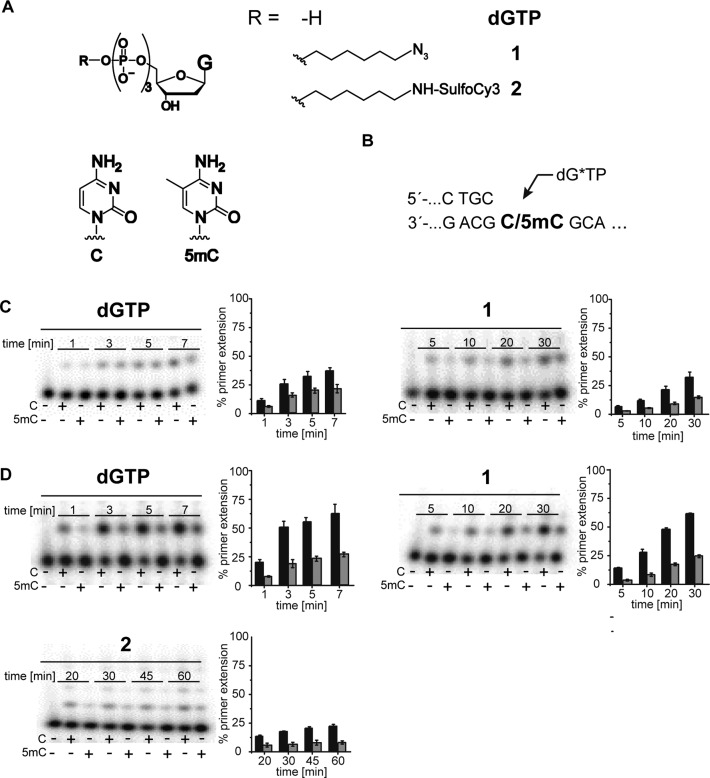
Typical reactions consisted of 15 μl and contained 150 nM [γ-32P]-labeled primer (5′-d(CGA AAT GAT CCC ATC CAG CTG C)-3′), 200 nM of either template (5′-d(CCG CTG CCC ACC AGC CAT CAT GTC GGA CCC CGC GGT CAA CGX GCA GCT GGA TGG GAT CAT TTC GGA CT)-3′), X = C/5mC/T) and 1.5 μl of the respective cleared lysate (diluted 1/50 in ddH2O) or 5 nM purified enzyme for processing of dGTP and 20 nM enzyme for nucleotides 1 and 2 in 1x reaction-buffer (50 mM Tris-HCl pH 8.0, 16 mM (NH4)2SO4, 2.5 mM MgCl2, 0.1% Tween 20). The reaction mixtures were heated to 95°C for 2 min and subsequently cooled to 4°C for annealing. The reaction was started by addition of 100 μM of dGTP/dGT*P at 55°C. Reactions were stopped after the desired incubation time by addition of 2 μl of the respective reaction mixture to 10 μl stop solution (80% (v/v) formamide, 20 mM EDTA, 0.25% (w/v) bromophenol blue, 0.25% (w/v) xylene cyanol) and analyzed by 12% or 15% denaturing polyacrylamide gel electrophoresis (PAGE). Visualization was performed by phosphorimaging.
Expression and purification of KOD exo- DNA polymerase wildtype and mutants
KOD exo- DNA polymerase wildtype and interesting mutants were expressed in 50 ml cultures and purified with complete His-Tag Purification Resin from Roche, essentially following the procedures described elsewhere (42). Briefly, 50 ml of expression culture were harvested and lysed in 20 ml lysis buffer (120 mM TrisHCl pH 8, 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton, 1.5 mM MgCl2, 1 mM PMSF, 1 mg/ml lysozyme) at 37°C for 20 min, followed by heat denaturation of host proteins at 75°C for 40 min and centrifugation at 14.000 x g for 30 min. Supernatants were incubated with 50% (v/v) Ni-beads slurry. Beads were washed and proteins were eluted with elution buffer (100 mM TrisHCl pH 8, 3 mM MgCl2, 200 mM imidazole). Imidazole was removed by centrifuging eluates in Vivaspin columns from Sartorius and purified enzymes were stored in 120 mM TrisHCl pH 8, 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton, 1.5 mM MgCl2, 50% glycerol. Protein concentrations were determined by absorbance measurements and purified enzymes were stored at −20°C.
Enzyme kinetics
Steady-state kinetics of KOD exo- DNA polymerase wild type and G245D were measured under single completed hit conditions in triplicates (44–46). Single-nucleotide incorporation for the respective dNTP in combination with either template (C/5mC) was performed as described, analyzed by 12% denaturing PAGE and visualized by phosphorimaging. Concentrations of the respective DNA polymerase were chosen in a way that less than 20% of the applied primer was extended. The rate of single-nucleotide incorporation was determined at various dNTP concentrations for different incubation times varying from 10 s to 120 s. The amount of extended primer was plotted against incubation time for each examined dNTP concentration. The arithmetic mean and standard deviation of the reaction velocities were calculated from independent triplicates. For kinetic analysis, the arithmetic mean of reaction velocities divided through DNA polymerase concentrations were plotted against the dNTP concentrations used. Using OriginPro8, experimental data were fitted to the Michaelis–Menten equation velocity = vMax[dNTP]/(KM+[dNTP]) to determine KM and vMax. Kcat = vMax/[pol] and kcat/KM were calculated. The corresponding deviation was determined by propagation of uncertainty after Gauß.
RESULTS
Design of substitution sites
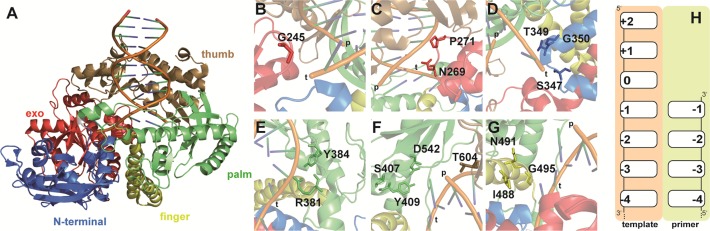
To assess residues of the KOD exo- DNA polymerase that might contribute to enzyme function, we first analyzed a crystal structure of the enzyme bound to a primer–template duplex (Figure 1) (47). Residues that contact both the primer and template strand around the site of nucleotide addition (for nucleotide numbering see Figure 1H), were considered as most promising for amino acid substitutions. Additionally amino acids, potentially influencing the orientation of the nucleobase rotated out from the DNA backbone by 180° (“+2” position indicated in Figure 1H), were targeted. Interesting amino acids contacting the primer strand were N269, P271 (Figure 1C), located in the exonuclease domain, which interact with the phosphate backbone between the −1 and −2 nucleotides. Threonine 604 (Figure 1F) is included in a β-sheet closest to the 3′-end of the primer potentially involved in stabilizing the primer. Contacts of a loop in the palm domain to the phosphate backbone of the template strand at the −1 position are facilitated through R381 and Y384. Thereby, Y384 is extending into the space below the nucleobases and is thus potentially involved in stabilizing the orientation of the newly formed nucleobase pair. The structure alludes to how the palm domain residues S407, Y409 and D542 (Figure 1F) are positioned around the incoming nucleotide (“0”) which is next to be added to the primer strand. Substitution of these amino acids might potentially be interesting for acceptance of modified nucleotides by the enzyme. Residues I488, N491 and G495 (Figure 1G) are part of a finger domain α-helix, which is positioned under the bases extending from the single stranded template, and are thus possibly responsible for stabilizing the template. Changes in the amino acid composition in this area might make room for unnatural base pairings. Located next to it, the structure shows how S347, T349 and G350 (Figure 1D) contact the phosphate backbone at the template where the +2 base is currently flipped out for template reading. Along those lines, residue G245 (Figure 1A) is located in a loop that extends away from the phosphate backbone.
Figure 1.
View of KOD exo- DNA polymerase and investigated residues in detail. (A) Crystal structure of KOD with bound primer–template duplex (PDB ID 4K8Z). Protein domains are labeled: exo (red), N-terminal (blue), palm (green), thumb (brown) and finger (yellow). (B–G) Close up view of amino acid sites used for mutant library generation. Residues are shown as sticks and depicted in a color corresponding to protein domains. DNA strands are labeled “p” or “t” for primer and template strand respectively. (H) Schematic depiction of nucleotide numbering in primer and template strand.
Generation of mutant libraries
In order to achieve the library of all 19 possible mutants at each targeted site, we used single PCR reactions to introduce defined mutations at the respective chosen sites. Each PCR reaction introduced one mutation at a given site, thus providing the identity of the mutant already during the screening process. This greatly reduced laborious library generation, e.g. used for saturation mutagenesis, as only 19 mutants per site had to be expressed and screened as opposed to the handling of hundreds of clones, without knowing whether all amino acid exchanges were covered. Our workflow was further streamlined by amplifying the full plasmid in the PCR reactions, thus allowing us to omit classical cloning of the mutant gene sequence into the expression vector. Instead, PCR products were purified from the reaction and the linear plasmids were directly ligated. With the highly reduced number of clones, high throughput expression, lysate preparation and screening was carried out efficiently. However, three mutants at three different sites, namely S347L, T349E and G350Y, were elusive even after several attempts. As amino acid substitutions at these sites turned out to be less promising (vide infra), no further efforts to obtain these mutants were made.
Screening KOD exo- DNA polymerase libraries for activity
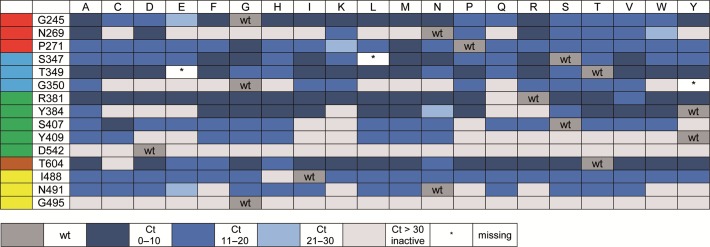
To distinguish between active and inactive mutants, real-time PCR was performed, using the bacterial lysates and SYBR Green I for visualization. Therefore, we employed a fragment spanning 92 nucleotides of the human NANOG gene and corresponding primers to generate an 86 bp long PCR product (40,48). The data output directly showed active versus inactive mutants, and the evaluation of threshold-crossing points (Ct values) of each reaction allowed us to generate a semi-quantitative overview of the mutants’ screening results at the 15 investigated sites of KOD exo- DNA polymerase (Figure 2). Reactions containing wild type polymerase showed a clear amplification curve during the first 10 cycles. Interestingly, we found that mutation at sites G495 and D542 exclusively led to variants with significantly reduced activity, despite expected protein content of the lysates. Glycine at position 495 is located in an α-helix, and steric crowding by any other amino acid at this site might either distort the α-helix or dislocate the neighboring loop. Both effects seem to be involved in stabilizing the template strand. Replacing aspartic acid at position 542 also results in inactive KOD exo- DNA polymerase variants. The interactions of aspartic acid with the incoming nucleotide seem to be essential for the enzyme. Interestingly, even the addition of a single methylene group of the glutamic acid moiety was deleterious for the activity of the enzyme. It should be noted that while expression levels were checked via SDS-PAGE and seen as similar for the libraries (e.g. Supplementary Figure S2), this cannot ensure an equal amount of active protein in the lysates used for activity screening.
Figure 2.
Activity chart of generated KOD exo- DNA polymerase mutants. Mutated sites are listed on the left, sorted according to protein domain (see color code, compare Figure 1A), amino acid exchanges are depicted in the top lane. Color coding indicates activity of mutants in bacterial lysate as detected by Ct values in real time PCR screening assays. Dark blue indicates lowest Ct, light grey indicates inactive mutants. Mutants marked with an asterisk (*) could not be obtained during cloning and are missing in the library.
Screening KOD exo- DNA polymerase libraries for discrimination between C and 5mC
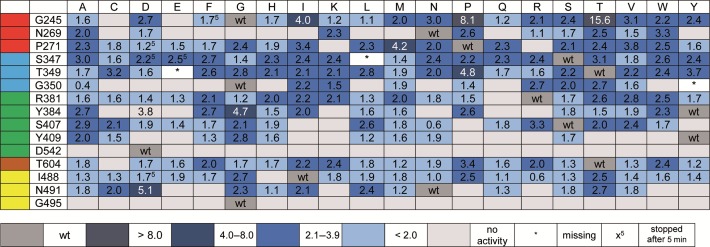
To reveal those variants that are most interesting for the depicted approach, we performed single-nucleotide incorporation primer extension experiments followed by analysis via denaturing PAGE and visualization by autoradiography. Therefore, cleared bacterial lysates of the enzymes identified as active (Figure 2) were employed in further studies. Since previous experiments (35) proved that decreased selectivity between C and T could be problematic, we employed three different templates containing C, 5mC or T (see Supplementary Figure S3) in the template position 0 (see Figure 1H). Reactions were stopped after four different time points, analyzed by PAGE and autoradiography and discrimination was calculated by the quotient of % primer extension opposite C divided by % primer extension opposite 5mC. Screening, done without replicates, was carried out to identify the most promising candidates for further characterisation – results are shown in Figure 3 and Supplementary Figure S3. Characterization of purified enzymes was done in independent triplicates. These results, summarized in Figure 3 and Supplementary Figure S3, show that mutation in some cases led to increased differences in incorporation efficiencies of dGMP opposite C or 5mC, while for some mutants, no increase in discrimination could be observed for the purified enzymes anymore.
Figure 3.
Discrimination for processing of dGTP employing different KOD exo- DNA polymerase mutants; reactions were stopped after 10 min. Discrimination ratios were determined by calculating the quotient of % primer extension opposite C and % primer extension opposite 5mC. Discrimination ratios were color coded as indicated. Results derive from single experiments (note: hits were purified and further evaluated in experiments done in replicates (vide infra)).
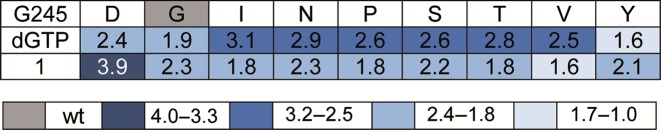
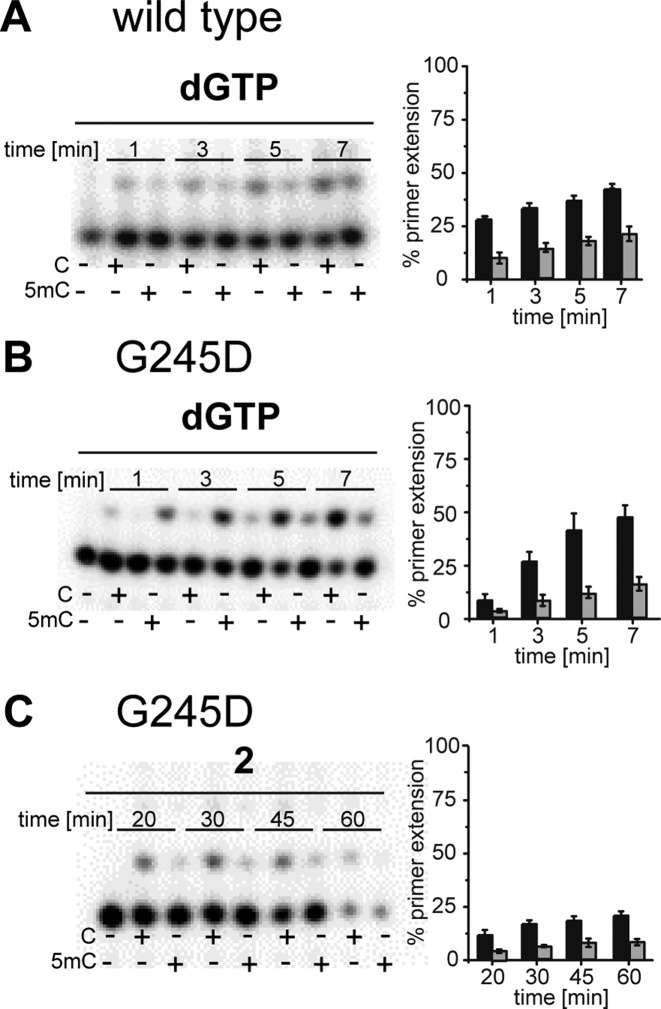
Next, we focused on the most interesting mutants for further investigation. The first semi-quantitative experiments clearly point to site G245 as the most promising position for further investigation. In addition, no remarkable incorporation opposite T could be observed (see Supplementary Figure S3). Along these lines, we focused on G245 mutants with the highest discrimination, namely: G245D, G245I, G245N, G245P, G245S, G245T, G245V and G245Y (see Figure 3). These variants, along with the wild-type enzyme, were expressed and purified for further analysis. The studies using the purified enzymes verified improved discrimination for all variants in comparison to the wild-type enzyme, except for G245Y (see Figure 4). Discrimination ratios between 2 (wt) and 3 (G245I) could be observed after 5 min under the chosen conditions, proving a remarkable improvement of discrimination. Again, selectivity of the variants in combination with dGTP opposite C was ensured, in comparison to T, A and G (see Supplementary Figure S5). These findings show that the substitution of an amino acid of KOD exo- DNA polymerase not directly contacting the templating nucleobase nor the incoming nucleotide, alters the ability of the enzyme to process methylated DNA.
Figure 4.
Discrimination for processing of dGTP and nucleotide 1 employing purified KOD exo- DNA polymerase mutants; reactions were stopped after 5 min. A total of 5 nM DNA polymerase was used for processing of dGTP, 20 nM enzyme was employed for processing of nucleotide 1. Discrimination ratios were determined by calculating the quotient of % primer extension opposite C and % primer extension opposite 5mC. Discrimination ratios were color coded as indicated. Experiments were repeated four times in independent experiments, errors are given in the Supplementary Figure S4.
Employing modified nucleotides in combination with the G245D variant
SMRT is one next generation sequencing technique that exploits the potential of DNA polymerases by using a DNA polymerase in combination with dNTPs, which are conjugated to either of four distinguishable fluorescent tags at the terminal phosphate moiety (25). To elucidate whether the identified KOD exo- DNA polymerase variant has potential for applications along the depicted lines of SMRT sequencing, a dGTP analogue with modifications at the terminal phosphate was synthesized following known procedures (49). First, we synthesized dGTP analogue 1 bearing a 1-azidohexyl-residue attached to the γ-phosphate (see Supplementary Figure S1). Employing nucleotide 1 with the purified enzymes in single-nucleotide incorporation primer extension studies, we observed decreased incorporation efficiencies compared to the processing of unmodified dGTP (see Supplementary Figure S4). In addition, we found decreased C versus 5mC discrimination for most mutants as well (Figures 4 and 5). But when employing nucleotide 1 with G245D, we found remarkably increased discrimination (factor 4) between C and 5mC. Single-nucleotide incorporation primer extension reactions clearly showed that nucleotide 1 could be incorporated more efficiently opposite C than opposite 5mC (Figure 5C). Next, we synthesized the sulfo-Cy3-dye-labeled dGTP analogue 2 and conducted incorporation studies with the KOD exo- DNA polymerase mutant G245D. Those experiments showed slightly decreased discrimination in comparison to nucleotide 1, but still a discrimination of three between C and 5mC (Figures 4 and 5).
Figure 5.
(A) Chemical structure of dGTP and modified nucleotides 1 and 2 (left) and of C and 5mC (right); (B) partial primer/template sequence used (C) PAGE analysis of single-nucleotide incorporation primer extension experiments using dGTP and nucleotide 1 opposite a template containing C in comparison to a template containing 5mC employing KOD exo- wt. (D) PAGE analysis of single-nucleotide incorporation primer extension experiments of dGMP and nucleotides 1 + 2 opposite a template containing C in comparison to a template containing 5mC employing KOD exo- G245D. A total of 5 nM of the respective DNA polymerase were used for processing of dGTP; 20 nM for nucleotides 1 + 2, 100 μM dGTP/ dGT*P. Reactions were stopped after indicated time points. Results are from four independent experiments.
To further verify the observed effect of this nucleotide in combination with the DNA polymerase mutant G245D, we determined steady-state kinetics (44–46) for processing of 2 in comparison to dGTP and opposite C and 5mC (Table 1 and Supplementary Figure S6). Comparison of the catalytic efficiencies (kcat/KM) observed for usage of dGTP opposite C or 5mC in the template strand, verified the decreased incorporation efficiencies for dGMP opposite 5mC in comparison to the unmodified C. For the wild-type enzyme no remarkable discrimination could be observed, as already reported before (35). However, when employing the DNA polymerase mutant G245D, the discrimination between C and 5mC increased to a factor of over 2 when comparing the catalytic efficiencies. Interestingly, this level of discrimination could also be maintained when using the fluorescently modified dGTP analogue 2, where the kcat is similarly decreased for incorporation opposite 5mC compared to the unmodified C.
Table 1. Steady-state kinetic analysis of single nucleotide incorporation primer extension opposite C or 5mC by KOD exo- DNA polymerase wt and G245D.
| dNTP | enzyme | template | KM [μM] | kcat [s−1] | kcat/KM [s−1 μM−1] |
|---|---|---|---|---|---|
| dGTP | wt | C | 4.0 ± 0.3 | 5.9 ± 0.1 | 1.5 ± 0.1 |
| 5mC | 3.3 ± 0.3 | 3.5 ± 0.1 | 1.1 ± 0.6 | ||
| dGTP | G245D | C | 9.2 ± 2.2 | 7 ± 0.9 | 0.76 ± 0.1 |
| 5mC | 9.6 ± 1.9 | 2.7 ± 0.3 | 0.28 ± 0.41 | ||
| 2 | G245D | C | 344.0 ± 49.6 | 2.42 ± 0.22 | 0.0070 ± 0.0006 |
| 5mC | 203.8 ± 32.2 | 0.585 ± 0.043 | 0.0029 ± 0.0002 |
Ratios calculated by the quotient of kcat (C)/kcat(5mC) and kcat/KM (C) and kcat/KM (5mC), respectively.
It has been shown before that exonuclease activity and pyrophosphorolysis affect DNA synthesis of archaeal DNA polymerases when encountering U in DNA (37–39). During these studies an exonuclease deficient variant of the KOD DNA polymerase (KOD exo-) was employed. Thus, exonuclease activity can be ruled out. In order to gain insights into the effect of pyrophosphorolysis we conducted an additional experiment in which we added inorganic pyrophosphatase to the reaction. This enzyme hydrolyses pyrophosphate to phosphate and thereby removes the formed pyrophosphate preventing pyrophosphorolysis (50–52). As it can be seen in Figure 6, addition of a incorganic pyrophosphatase does not change incorporation efficiencies nor observed discriminations in any reaction. Therefore, we assume that the described effects derive from the incorporation event itself and are independent from exonuclease activity and pyrophosphorolysis.
Figure 6.
PAGE analysis of single-nucleotide incorporation primer extension experiments including thermostable inorganic pyrophosphatase. (A) Processing of dGTP by the KOD exo- wild type; (B) employing dGTP and KOD exo- G245D; (C) incorporation of nucleotide 2 by KOD exo- G245D. A total of 5 nM of the respective DNA polymerase were used for processing of dGTP; 20 nM for nucleotide 2, 100 μM dGTP/ dGT*P; 0.5 U/ml thermostable inorganic pyrophosphatase. Reactions were stopped after indicated time points. Results are from four independent experiments.
Employing the DNA polymerase KOD exo- and its G245D variant in running start experiments
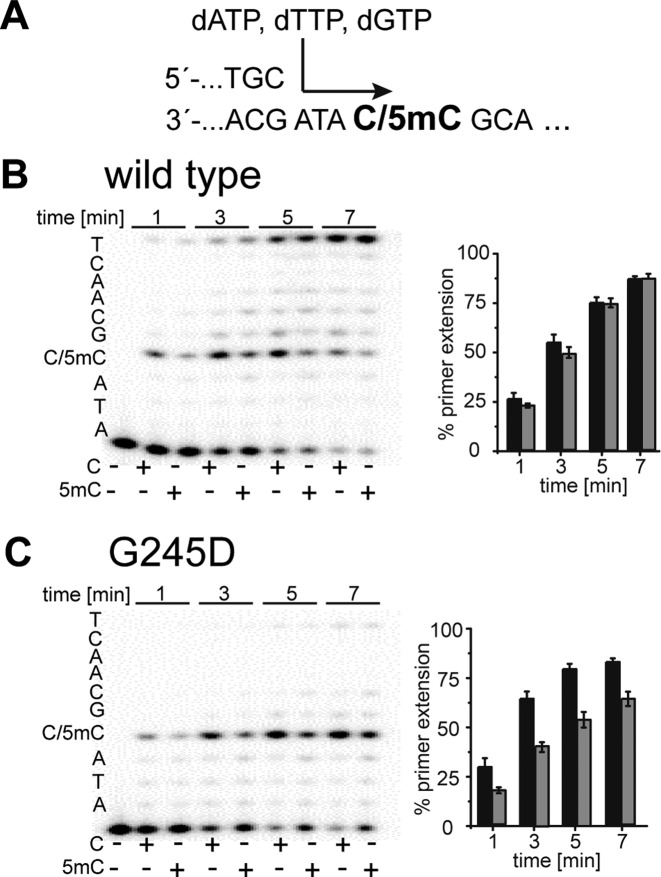
Next, we performed running start experiments to further study the observed discrimination between C and 5mC. To ensure that no sequence bias influences the results and to guarantee comparability between standing start and running start experiments, we employed the same primer but inserted three nucleobases between the 3′-primer end and the investigated C/5mC in the template strand. Running start experiments (see Figure 7) verified the improved ability of the KOD exo- variant G245D for discrimination of 5mC in comparison to the wild-type enzyme.
Figure 7.
PAGE analysis of running start primer extension experiments. (A) partial primer/template sequence used; (B) Processing of dATP, dTTP and dGTP by KOD exo- wild type; (C) Processing of dATP, dTTP and dGTP by KOD exo- G245D. A total of 5 nM of the respective DNA polymerase were used for processing of nucleotides. Reactions were stopped after indicated time points. Results are from four independent experiments.
DISCUSSION
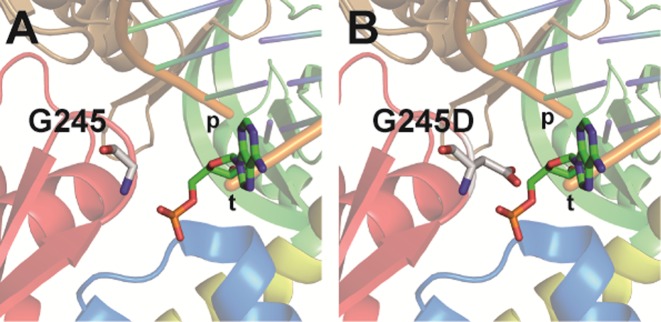
We report on the generation and evaluation of a rationally designed library of KOD exo- DNA polymerase variants, targeting amino acids that were selected because of their contacts with either the primer or template strand in proximity to the active site. This library was screened to select for PCR activity and in a second step for different nucleotide incorporation efficiencies opposite C in comparison to 5mC. Our approach was straightforward and allowed for extensive screening in parallel. We could identify a single site (G245) which, if glycine was substituted, led to significant changes in the ability of KOD exo- DNA polymerase to process C and 5mC containing DNA. The amino acid residue G245 is located in a loop of the exonuclease domain close to the 5′-end of the template strand (Figure 8). This hairpin is also found in other family B DNA polymerases and has been shown to be potentially important for proofreading or more specifically for strand separation associated with pol-to-exo switching of T4 (53,54) and RB69 DNA polymerases (55). The study on RB69 DNA polymerase reasoned that neither the polymerase, nor the exonuclease activities are influenced by deletion of the hairpin. It rather seems that the hairpin facilitates stabilization of the exonuclease complex formed by the enzyme and the separated DNA strands (55). A glycine in this hairpin has been substituted with a serine, which led to a mutant polymerase with an increased tolerance for replication mistakes, because the strand separation cannot be stabilized in a way to ensure sufficient exonuclease activity (54). However, whether this hairpin plays a similar role in other family B DNA polymerases remains speculative, as hairpins among these polymerases show variations in their amino acid sequence (56). Moreover, there is evidence that the hairpin is not generally required for proofreading by family B DNA polymerases, as shown for Saccharomyces cerevisiae polymerases δ (57) and ϵ (58). In our study, a higher tolerance for processing of the mismatched primer–template complex is already ensured by the fact that only the exo- variants were used (exhibited through D141A and E143A substitutions). Higher processivity and especially better tolerance of the 5mC modification cannot be seen as a general effect of tampering with the exonuclease activity associated with the hairpin that contains G245. Thus, the mechanism, by which the identified amino acid substitution contributes to the altered efficiency for incorporation opposite C and 5mC, remains elusive. Obviously, in comparison to the wild-type glycine, altered amino acids at this site have an increased potential to interact with the substrate, e.g. by van der Waals or polar interactions. The aspartic acid in the most promising mutant G254D might be able to form hydrogen bonding with the nucleobase in position +2 of the template strand (Figure 8). This interaction could lead to conformational alterations in the template orientation within the active site and thereby affect differences in the nucleotide incorporation opposite C and 5mC in the template strand.
Figure 8.
(A) Crystal structure of KOD exo- DNA polymerase. Detailed view of amino acid G245 and its proximity to the template strand. (B) Modeled view of G245D mutant to highlight potential interactions with template strand upon steric crowding. Protein domains are colored: exo (red), N-terminal (blue), palm (green), thumb (brown) and finger (yellow). DNA strands are labeled “p” or “t” for primer and template strand, respectively. Amino acid at position 245 as well as the templating base in position +2 are shown as sticks.
It has been known that archaeal DNA polymerases are able to detect uracil in DNA (37–39). Since uracil and the natural DNA base thymine differ by only one methyl group, this system is reminiscent to C and 5mC. The mentioned studies show how family B and D DNA polymerases stall upon encountering uracil in the template strand. If uracil gets close to the active site of the polymerase, the primer-template complex is unwound and the 3′ end of the primer is translocated to the exonuclease domain of the enzyme (39). This unwinding is facilitated by amino acid Y261 of Pfu DNA polymerase and alanine substitution at this site results in a polymerase mutant with drastically lowered fidelity. Despite the similarities, the systems are fundamentally different: while the “missing methyl” in U leads to decreased incorporation efficiency in comparison to T in the T/U system, the “additional methyl” in 5mC results in decreased incorporation efficiency in comparison to C in the C/5mC system. This suggests fundamentally different mechanistical origins for the observed effects.
In order to broaden potential future applications, we also investigated the processing of γ-phosphate modified nucleotides by the identified mutants. The G245D mutant was shown as the most promising variant of KOD exo- DNA polymerase. By analyzing steady state kinetics of this mutant in combination with dGTP and the dye-labeled nucleotide 2, we could demonstrate a more than 2-fold bias in incorporation efficiency (kcat/KM) opposite C compared to opposite 5mC. Further comparison of those kinetic data show a 4-fold discrimination in kcat. In addition, we could ensure selectivity for this incorporation opposite C in comparison to the other nucleobases T, A and G. Taken together, by employing a systematic approach to mutate KOD exo- DNA polymerase and subsequent screening, we identified mutants at one site of KOD exo- DNA polymerase that are capable to discriminate between C and 5mC. Interestingly, the herein developed system keeps the selectivity for incorporation according to the Watson–Crick rule which is in stark contrast to earlier approaches that exploit modified nucleotides (35,41). A multiple sequence alignment of several family-B DNA polymerases (Supplementary Figure S7) shows that the glycine identified in this study is conserved among several family B DNA polymerases. Our results might indicate a potential to improve existing sequencing approaches – like Pacific Biosciences using the Phi29 DNA polymerase (25). Maybe mutating the corresponding glycine in the loop of a beta hairpin of other family-B DNA polymerases could improve their ability to discriminate between methylated and unmethylated cytosines and broaden the sprectrum of tools for SMRT sequencing approaches.
Future attempts to improve this very promising finding will aim at further DNA polymerase engineering and synthesis of γ-phosphate modified nucleotides with longer phosphate chains.
Supplementary Material
Acknowledgments
The authors thank Martina Adam for technical assistance in library preparation and protein purification. We acknowledge support by the European Research Council [Project EvoEPIGEN, Grant 339834] and Konstanz Research School Chemical Biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council [Project EvoEPIGEN, Grant 339834]. Funding for open access charge: ERC [Advanced Grant 339834].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bird A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich M., Wang R.Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 3.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Weber M., Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Ooi S.K., O'Donnell A.H., Bestor T.H. Mammalian cytosine methylation at a glance. J. Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 7.Estecio M.R., Gallegos J., Vallot C., Castoro R.J., Chung W., Maegawa S., Oki Y., Kondo Y., Jelinek J., Shen L., et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010;20:1369–1382. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y., Ecker J.R. Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 2015;16:55–77. doi: 10.1146/annurev-genom-090413-025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray I.A., Clark T.A., Morgan R.D., Boitano M., Anton B.P., Luong K., Fomenkov A., Turner S.W., Korlach J., Roberts R.J. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson G.G., Murray N.E. Restriction and modification systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 13.Roberts R.J., Macelis D. REBASE–restriction enzymes and methylases. Nucleic Acids Res. 1993;21:3125–3137. doi: 10.1093/nar/21.13.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 15.Lu M., Campbell J.L., Boye E., Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 16.Casadesus J., Low D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyn H., Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 18.Sandoval J., Esteller M. Cancer epigenomics: beyond genomics. Curr. Opin. Genet. Dev. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Hayatsu H., Wataya Y., Kai K., Iida S. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry. 1970;9:2858–2865. doi: 10.1021/bi00816a016. [DOI] [PubMed] [Google Scholar]
- 20.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tost J., Gut I.G. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 22.Grunau C., Clark S.J., Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison J., Stirzaker C., Clark S.J. Cytosines adjacent to methylated CpG sites can be partially resistant to conversion in genomic bisulfite sequencing leading to methylation artifacts. Anal. Biochem. 1998;264:129–132. doi: 10.1006/abio.1998.2833. [DOI] [PubMed] [Google Scholar]
- 24.Genereux D.P., Johnson W.C., Burden A.F., Stoger R., Laird C.D. Errors in the bisulfite conversion of DNA: modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res. 2008;36:e150. doi: 10.1093/nar/gkn691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 26.Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark T.A., Murray I.A., Morgan R.D., Kislyuk A.O., Spittle K.E., Boitano M., Fomenkov A., Roberts R.J., Korlach J. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munzel M., Lercher L., Muller M., Carell T. Chemical discrimination between dC and 5MedC via their hydroxylamine adducts. Nucleic Acids Res. 2010;38:e192. doi: 10.1093/nar/gkq724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G.L., Luo C., Jiang H., He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark T.A., Lu X., Luong K., Dai Q., Boitano M., Turner S.W., He C., Korlach J. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kranaster R., Marx A. Engineered DNA polymerases in biotechnology. Chembiochem. 2010;11:2077–2084. doi: 10.1002/cbic.201000215. [DOI] [PubMed] [Google Scholar]
- 34.Erlich H. HLA DNA typing: Past, present, and future. Tissue Antigens. 2012;80:1–11. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- 35.von Watzdorf J., Leitner K., Marx A. Modified nucleotides for discrimination between cytosine and the epigenetic marker 5-methylcytosine. Angew. Chem. Int. Ed. Engl. 2016;55:3229–3232. doi: 10.1002/anie.201511520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen J.C., Creighton S., Jones P.A., Goodman M.F. A comparison of the fidelity of copying 5-methylcytosine and cytosine at a defined DNA template site. Nucleic Acids Res. 1992;20:5119–5125. doi: 10.1093/nar/20.19.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fogg M.J., Pearl L.H., Connolly B.A. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 38.Richardson T.T., Gilroy L., Ishino Y., Connolly B.A., Henneke G. Novel inhibition of archaeal family-D DNA polymerase by uracil. Nucleic Acids Res. 2013;41:4207–4218. doi: 10.1093/nar/gkt083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson T.T., Wu X., Keith B.J., Heslop P., Jones A.C., Connolly B.A. Unwinding of primer-templates by archaeal family-B DNA polymerases in response to template-strand uracil. Nucleic Acids Res. 2013;41:2466–2478. doi: 10.1093/nar/gks1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aschenbrenner J., Drum M., Topal H., Wieland M., Marx A. Direct sensing of 5-methylcytosine by polymerase chain reaction. Angew. Chem. Int. Ed. Engl. 2014;53:8154–8158. doi: 10.1002/anie.201403745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Watzdorf J., Marx A. 6-Substituted 2-Aminopurine-2′-deoxyribonucleoside 5′-Triphosphates that trace cytosine methylation. Chembiochem. 2016;17:1532–1540. doi: 10.1002/cbic.201600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gloeckner C., Sauter K.B., Marx A. Evolving a thermostable DNA polymerase that amplifies from highly damaged templates. Angew. Chem. Int. Ed. Engl. 2007;46:3115–3117. doi: 10.1002/anie.200603987. [DOI] [PubMed] [Google Scholar]
- 43.Sauter K.B., Marx A. Evolving thermostable reverse transcriptase activity in a DNA polymerase scaffold. Angew. Chem. Int. Ed. Engl. 2006;45:7633–7635. doi: 10.1002/anie.200602772. [DOI] [PubMed] [Google Scholar]
- 44.Petruska J., Goodman M.F., Boosalis M.S., Sowers L.C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boosalis M.S., Petruska J., Goodman M.F. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 46.Creighton S., Huang M.M., Cai H., Arnheim N., Goodman M.F. Base mispair extension kinetics. Binding of avian myeloblastosis reverse transcriptase to matched and mismatched base pair termini. J. Biol. Chem. 1992;267:2633–2639. [PubMed] [Google Scholar]
- 47.Bergen K., Betz K., Welte W., Diederichs K., Marx A. Structures of KOD and 9 degrees N DNA polymerases complexed with primer template duplex. Chembiochem. 2013;14:1058–1062. doi: 10.1002/cbic.201300175. [DOI] [PubMed] [Google Scholar]
- 48.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 49.Hacker S.M., Mex M., Marx A. Synthesis and stability of phosphate modified ATP analogues. J. Org. Chem. 2012;77:10450–10454. doi: 10.1021/jo301923p. [DOI] [PubMed] [Google Scholar]
- 50.Meyer P.R., Matsuura S.E., So A.G., Scott W.A. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyer P.L., Gao H.Q., Clark P.K., Sarafianos S.G., Arnold E., Hughes S.H. YADD mutants of human immunodeficiency virus type 1 and Moloney murine leukemia virus reverse transcriptase are resistant to lamivudine triphosphate (3TCTP) in vitro. J. Virol. 2001;75:6321–6328. doi: 10.1128/JVI.75.14.6321-6328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao M., Phong A., Lum K.L., Greene R.A., Buzby P.R., Kwok P.Y. Role of excess inorganic pyrophosphate in primer-extension genotyping assays. Genome Res. 2004;14:1749–1755. doi: 10.1101/gr.2833204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stocki S.A., Nonay R.L., Reha-Krantz L.J. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3′ –>5′ exonuclease activities. J. Mol. Biol. 1995;254:15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- 54.Marquez L.A., Reha-Krantz L.J. Using 2-aminopurine fluorescence and mutational analysis to demonstrate an active role of bacteriophage T4 DNA polymerase in strand separation required for 3′ –>5′-exonuclease activity. J. Biol. Chem. 1996;271:28903–28911. doi: 10.1074/jbc.271.46.28903. [DOI] [PubMed] [Google Scholar]
- 55.Hogg M., Aller P., Konigsberg W., Wallace S.S., Doublie S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 56.Darmawan H., Harrison M., Reha-Krantz L.J. DNA polymerase 3′–>5′ exonuclease activity: Different roles of the beta hairpin structure in family-B DNA polymerases. DNA Repair (Amst) 2015;29:36–46. doi: 10.1016/j.dnarep.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 57.Hadjimarcou M.I., Kokoska R.J., Petes T.D., Reha-Krantz L.J. Identification of a mutant DNA polymerase delta in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics. 2001;158:177–186. doi: 10.1093/genetics/158.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hogg M., Osterman P., Bylund G.O., Ganai R.A., Lundstrom E.B., Sauer-Eriksson A.E., Johansson E. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat. Struct. Mol .Biol. 2014;21:49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.