Abstract
We showed that the deoK operon, which confers the ability to use deoxyribose as a carbon source, is more common among pathogenic than commensal Escherichia coli strains. The expression of the deoK operon increases the competitiveness of clinical isolates, suggesting that this biochemical characteristic plays a role in host infectivity.
Both pathogenic and nonpathogenic strains of Escherichia coli exist. The nonpathogenic strains are found in the normal intestinal flora of humans and animals, and the pathogenic strains are a leading cause of death and morbidity worldwide, particularly in developing countries. Pathogenicity islands (PAIs) carrying virulence genes have been characterized. However, most sequences within PAIs are still of unknown function. PAI IAL862 from the human blood E. coli isolate AL862 was previously described (18). The afa-8 operon is the only region encoding a virulence factor that has been identified in this new PAI (18). Here, we demonstrate the presence of the deoK operon in this PAI. This operon codes for the use of deoxyribose, a sugar that is not fermented by E. coli K-12.
We observed that partially sequenced regions of PAI IAL862 (18) showed similarities with the deoK operon from Salmonella enterica serovar Typhi (2, 30). Using primers deduced from these sequences, we amplified and sequenced a 5,840-bp segment from pILL1272, a cosmid from the AL862 library (Table 1). A 4,375-bp region was 78% identical to the deoK operon from S. enterica, which is composed of four genes (deoQ, deoK, deoP, and deoM) (Fig. 1). Few bacteria are able to catabolize deoxyribose. Deoxyribokinase (product of the deoK gene), which catalyzes the ATP-dependent phosphorylation of 2-d-deoxyribose, has only been identified in Lactobacillus plantarum, Selenomonas ruminantium, and S. enterica (12, 16, 26). We showed that E. coli strains AL862 and MG1655(pILL1272) were able to grow on K5 minimal medium (11) containing 0.1% 2-d-deoxyribose ribose (vol/vol) as the sole carbon source for 24 to 48 h at 37°C, meaning that they express the deoK operon.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source | Deoxy- ribosea |
|---|---|---|---|
| Nonpathogenic E. coli strains | |||
| K-12 MG1655 | Nonpathogenic reference strain (genome sequenced) | 5 | − |
| MG1655 Rif | Spontaneous rifampicin-resistant derivative of MG1655 | This study | − |
| MG1655 Nal | Spontaneous nalidixic acid-resistant derivative of MG1655 | This study | − |
| EC185 | Isolate from feces of a healthy volunteer | 15 | + |
| ExPEC strains | |||
| AL862 | Human blood isolate | 18 | + |
| CFT073 | Isolate from blood of a patient with symptomatic pyelonephritis (genome sequenced) | 31 | + |
| CFT073deoK | CFT073 isogenic mutant with deoK gene deleted | This study | − |
| CFT073 Nal | Spontaneous nalidixic acid-resistant derivative of CFT073 | This study | + |
| InPEC strains | |||
| 55989 | Enteroaggregative strain producing AAF-III fimbriae (tetracycline resistant) | 3 | + |
| 55989deoK | 55989 isogenic mutant with of deoK gene deleted | This study | − |
| AL866 | 55989deoK carrying pILL1314 | This study | + |
| AL868 | 55989deoK carrying pUC18 | This study | − |
| 55989 Nal | Spontaneous nalidixic acid-resistant derivative of 55989 | This study | + |
| AL867 | 55989 Nal carrying pUC18 | This study | + |
| 55989 Rif | Spontaneous rifampicin-resistant derivative of 55989 | This study | + |
| Plasmids | |||
| pILL1272 | 45-kb Sau3A fragment containing the deoK operon of AL862 cloned into pHC79 (carbenicillin resistant) | 18 | + |
| pILL1307 | pILL1272 containing a deoK gene mutated by partial deletion and insertion of the kanamycin resistance cassette (carbenicillin and kanamycin resistant) | This study | − |
| pILL1287 | 45-kb Sau3A fragment containing the deoK operon of 55989 cloned into pHC79 (carbenicillin resistant) | 3 | + |
| pUC18 | E. coli cloning vector (carbenicillin resistant) | 32 | − |
| pILL1314 | BamHI-KpnI PCR product corresponding to the deoK gene from 55989 inserted into pUC18 (carbenicillin resistant) | This study | − |
Growth on minimal medium supplemented with 0.1% deoxyribose and hybridized with probe A.
FIG. 1.
Genetic organization of the deoK region in E. coli. (A) Genetic organization of the deoK operon in S. enterica serovar Typhi strain CT18. The deoK operon spans 4,403 bp. (B) Schematic diagram of the genomic regions carrying the deoK operon from three E. coli strains. The regions from AL862 used as probes A and B are shown. The size of the common region carrying the deoK operon in the three E. coli strains is indicated (5,390 bp). The 4,730-bp region conserved in the E. coli and S. enterica strains is indicated by dashed lines. Boxes indicate the coding sequences, showing their orientations and positions. Noncoding regions are represented by lines. Identical symbols on boxes and lines indicate regions with similarities.
Database searches showed that the deoK operon from AL862 is highly conserved in the uropathogenic E. coli isolate CFT073 (99% identity over 4,378 bp) (31). However, the genomes of the commensal strain MG1655 of E. coli K-12 (5) and the enterohemorrhagic E. coli O157:H7 strains EDL933 and Sakai do not contain this operon (14, 25). We investigated 354 pathogenic and commensal isolates from various collections (Table 2). Colony hybridization assays (performed as described in reference 13) and growth assays on deoxyribose minimal medium with 130 clinical isolates showed that the presence of the deoK operon was always correlated with the use of this sugar. Probes A and B (Fig. 1) were amplified from AL862 DNA with previously described primers (18). Although we found that both pathogenic and commensal strains harbored the deoK operon, our results strongly suggested that this operon is associated with the pathogenicity of the strain (45.4% of pathogenic strains versus 22.9% of commensal strains harbored deoK; P < 0.01). Approximately half of the deoK-positive commensal isolates studied carried sequences encoding at least one virulence factor produced by extraintestinal pathogenic E. coli (ExPEC) (data not shown) and, consequently, could be considered potential ExPEC strains resident in the bowel.
TABLE 2.
Use of deoxyribose by nonpathogenic and pathogenic E. coli strains
| Strain type (no. of isolates) | Reference(s) or source | No. (%) of isolates with deoxyribose as a carbon sourcec |
|---|---|---|
| Nonpathogenic E. colia (61) | 14 (22.9) | |
| ECOR collection (25) | 24 | 6 (24) |
| French collection (36) | 15 | 8 (22.2) |
| Pathogenic E. coli (293) | 133 (45.4)* | |
| ExPEC (202) | 87 (43.1)* | |
| Archetypal (CFT073, J96, 536, RS218)b (4) | 6, 8, 28, 31 | 2 (50) |
| Pyelonephritis isolatesb (88) | 1 | 42 (47.7) |
| Sepsis isolates (110) | 15 | 43 (39) |
| InPEC (91) | 46 (50.5)** | |
| Enterotoxigenic (5) | 29 and this study | 3 (60) |
| Enterohemorrhagic (9) | 7, 25, 27, 33 and C. Martin | 1 (11) |
| Enteropathogenic (11) | 20 and E. Oswald and A. Aidara-Kane | 3 (27) |
| Diffusely adherent enteropathogenic (11) | 17 | 11 (100) |
| Diffusely adherent (20) | 4, 19 | 9 (45) |
| Enteroaggregative (27) | 3, 22, 23 | 15 (55.5) |
| Adherent invasive (8) | 21 | 4 (50) |
Isolates from feces of healthy individuals.
Similar results were obtained by hybridization and growth assays.
The data marked with asterisks were compared by chi-square analysis. The prevalences were significantly different from that in nonpathogenic strains (*, P < 0.01; **, P < 0.001).
We created nonpolar mutations by replacing the deoK gene with a PCR product containing a kanamycin resistance cassette as described previously (9). PCR analysis was performed to confirm the replacement of the gene. The deletion of the deoK gene did not result in any apparent growth defects. A study of parental strains, mutants, and transcomplemented mutants showed that the deoK gene is involved in use of the sugar (Table 1). These data were confirmed by determination of the deoxyribokinase specific activity and by comparison of the proteomes of deoK-positive [AL862, 55989, CFT073, and MG1655(pILL1287)] and deoK-negative (55989deoK and MG1655) strains, as previously described (10, 30). Deoxyribokinase activity (0.005 to 0.014 U/mg) and deoxyribokinase (identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry [data not shown]) could only be detected in soluble extracts from deoK-positive isolates that had been grown in minimal medium in the presence of deoxyribose.
Sequencing and comparison of the deoK operons from three pathogenic (AL862, 55989, and CFT073) and one commensal (EC185) E. coli strain showed that this operon is conserved (98% identity), as is an ∼1-kb DNA region surrounding it (750 bp on the left side and 280 bp on the right side) in all the strains tested (Fig. 1). A few base pairs (242 bp upstream and 115 bp downstream) directly flanking the operon corresponded to partial sequences of the ilvN and uhpA genes, between which the deoK operon is inserted in S. enterica (30). The sequences following these two truncated genes did not share similarity with sequences in the databases. These data suggested that E. coli acquired the deoK operon by horizontal transfer, possibly from S. enterica.
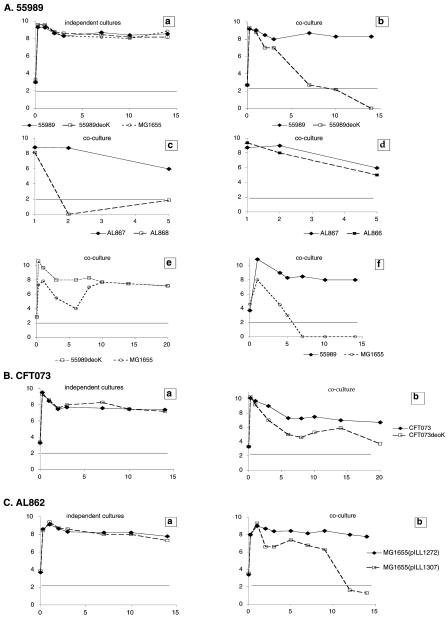
Deoxyribose is a sugar that is exclusively derived from DNA degradation, and the human diet leads to a high concentration of DNA in the intestine. We therefore hypothesized that deoxyribose catabolism plays a role in the colonization of the intestine by both ExPEC (where they are resident) and intestinal pathogenic E. coli (InPEC) strains, by conferring the ability to use a limiting nutrient. In agreement with this hypothesis, we demonstrated that deoK-positive strains were able to ferment deoxyribose. As the intestine is a complex organ that contains various and variable limiting nutrients, we carried out coculture experiments (with equal numbers of wild-type and deoK mutant colonies) in rich and minimal medium (K5 broth containing pyruvate as a constant carbon source) supplemented with or without deoxyribose. Although the growth rates were similar in independent cultures, the deoK mutants were less competitive than the corresponding wild-type strains in the coculture experiments. This effect clearly depends on the presence of both an active deoK gene and deoxyribose in the medium (Fig. 2 and 3). The 55989deoK mutant was totally eliminated after several days in rich medium but only suffered from a loss of fitness in minimal medium. Levels of enzyme activity after the growth of strain 55989 in the different media (data not shown) suggested that competitive differences are probably related to differences in deoxyribokinase activities. Although the MG1655 strain and the 55989deoK mutant showed similar fitness patterns in our coculture experiments, the parental 55989 isolate totally outcompeted the commensal strain after 6 days. Thus, the acquisition of the deoK operon by E. coli strains might confer an evolutionary fitness advantage, especially for pathogenic strains. In conclusion, our results agree with other reports, suggesting that metabolic functions specific for pathogenic strains play a role in host infectivity.
FIG. 2.
Experiments conducted with isolates 55989 (A) and CFT073 (B) and with isolate MG1655 carrying the deoK operon from AL862 (C). Bacteria were grown separately or in coculture for 2 to 3 weeks. Samples were taken periodically, and viable counts were determined by plating out serial dilutions on Luria-Bertani agar supplemented with antibiotic or not, as appropriate. Each assay was performed at least twice. The detection limit of the titration method is <100 CFU/ml. Wild-type strains were cocultured with their respective deoK mutants (Ab, Bb, and Cb). Similar results were obtained in coculture experiments with either the parental 55989 isolate or its Nal derivative. The 55989deoK mutant was transcomplemented with the cloned deoK gene (AL866) and cocultured with AL867 (Ad; compare to Ac as a negative control). Strain 55989 carrying the deoK operon or without it was also cocultured with the commensal strain MG1655 (Af or Ae, respectively). Cocultures were performed with the parental 55989 and MG1655 isolates, the 55989 Nal and MG1655 Rif derivatives, or the 55989 Rif and MG1655 Nal derivatives. Similar results were also obtained when 55989 Nal was cocultured with MG1655 Rif at a 1:100 ratio.
FIG. 3.
Survival of 55989 derivatives in coculture experiments in K5 minimal medium containing deoxyribose or without it. Strains 55989 and 55989deoK were grown separately for two weeks (A and B), in coculture for two weeks (C and D), or in coculture for 24 h (E and F).
Nucleotide sequence accession numbers. The GenBank accession numbers for the 5,840-bp region of pILL1272, the 6,105-bp region of pILL1287, and the 6,921-bp region of strain EC185 reported in this paper are AY299335, AY298765, and AY299336, respectively.
Acknowledgments
We thank Agnès Labigne, Antoine Danchin, and Octavian Bārzu, in whose units this work was carried out, for their continuing interest and helpful discussions. We thank J. Hacker for the gift of strains CFT073 and 536, E. Bingen for the gift of RS218, J. Nataro for the gift of JM221 and 042, S. Moseley for the gift of C1845, T. Baldwin for the gift of 2348-69, L. Riley for the gift of EDL1493, and A. O'Brien for the gift of EDL933. We thank A. Darfeuille-Michaud, C. Martin, E. Oswald, and A. Aidara-Kane for the gift of adherent invasive, enterohemorrhagic, and enteropathogenic E. coli isolates. We thank J. M. Ghigo and S. Da Re for their help with allelic exchange experiments. We also thank Joëlle Ferdinand for excellent technical assistance.
Editor: J. T. Barbieri
REFERENCES
- 1.Archambaud, M., P. Courcoux, and A. Labigne-Roussel. 1988. Detection by molecular hybridization of pap, afa, and sfa adherence systems in Escherichia coli strains associated with urinary and enteral infections. Ann. Inst. Pasteur Microbiol. 139:575-588. [DOI] [PubMed] [Google Scholar]
- 2.Assairi, L., T. Bertrand, J. Ferdinand, N. Slavova-Azmanova, M. Christensen, P. Briozzo, F. Schaeffer, C. T. Craescu, J. Neuhard, O. Bārzu, and A.-M. Gilles. 2004. Deciphering the function of an ORF: Salmonella enterica DeoM protein is a new mutarotase specific for deoxyribose. Protein Sci. 13:1295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier, C., P. Gounon, and C. Le Bouguénec. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70:4302-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonacorsi, S. P., O. Clermont, C. Tinsley, I. Le Gall, J. C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derzelle, S., S. Ngo, E. Turlin, E. Duchaud, A. Namane, F. Kunst, A. Danchin, P. Bertin, and J. F. Charles. 2004. AstR-AstS, a new two-component signal transduction system, mediates swarming, adaptation to stationary phase and phenotypic variation in Photorhabdus luminescens. Microbiology 150:897-910. [DOI] [PubMed] [Google Scholar]
- 11.Epstein, W., and B. S. Kim. 1971. Potassium transport loci in Escherichia coli K-12. J. Bacteriol. 108:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginsberg, A. 1959. A deoxyribokinase from Lactobacillus plantarum. J. Biol. Chem. 235:1292-1298. [PubMed] [Google Scholar]
- 13.Grunstein, M., and D. S. Hogness. 1975. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 72:3961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 15.Hilali, F., R. Ruimy, P. Saulnier, C. Barnabé, C. Lebouguénec, M. Tibayrenc, and A. Andremont. 2000. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect. Immun. 68:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffee, P. A. 1968. 2-Deoxyribose gene-enzyme complex in Salmonella typhimurium. I. Isolation and enzymatic characterization of 2-deoxyribose-negative mutants. J. Bacteriol. 95:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller, R., J. G. Ordonez, R. R. de Oliveira, L. R. Trabulsi, T. J. Baldwin, and S. Knutton. 2002. Afa, a diffuse adherence fibrillar adhesin associated with enteropathogenic Escherichia coli. Infect. Immun. 70:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lalioui, L., and C. Le Bouguénec. 2001. afa-8 Gene cluster is carried by a pathogenicity island inserted into the tRNAPhe of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bouguénec, C., L. Lalioui, L. du Merle, M. Jouve, P. Courcoux, S. Bouzari, R. Selvarangan, B. J. Nowicki, Y. Germani, A. Andremont, P. Gounon, and M. I. Garcia. 2001. Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coli isolates: PCR assays for detection of Afa adhesins that do or do not recognize Dr blood group antigens. J. Clin. Microbiol. 39:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1:1119-1122. [DOI] [PubMed] [Google Scholar]
- 21.Masseret, E., J. Boudeau, J. F. Colombel, C. Neut, P. Desreumaux, B. Joly, A. Cortot, and A. Darfeuille-Michaud. 2001. Genetically related Escherichia coli strains associated with Crohn's disease. Gut 48:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathewson, J. J., R. A. Oberhelman, H. L. Dupont, F. Javier de la Cabada, and E. V. Garibay. 1987. Enteroadherent Escherichia coli as a cause of diarrhea among children in Mexico. J. Clin. Microbiol. 25:1917-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nataro, J. P., M. M. Baldini, J. B. Kaper, R. E. Black, N. Bravo, and M. M. Levine. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152:560-565. [DOI] [PubMed] [Google Scholar]
- 24.Ochman, H., and J. G. Lawrence. 1996. Phylogenetics and the amelioration of bacterial genomes, p. 2627-2637. In F. C. Neidhart, R. Curtiss III, E. C. Ingraham, C. C. Lin, B. K. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 25.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 26.Rasmussen, M. A. 1993. Isolation and characterization of Selenomonas ruminantium strains capable of 2-deoxyribose utilization. Appl. Environ. Microbiol. 59:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 28.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tornieporth, N. G., J. John, K. Salgado, P. de Jesus, E. Latham, M. C. Melo, S. T. Gunzburg, and L. W. Riley. 1995. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J. Clin. Microbiol. 33:1371-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tourneux, L., N. Bucurenci, C. Saveanu, P. A. Kaminski, M. Bouzon, E. Pistotnik, A. Namane, P. Marlière, O. Bārzu, I. L. de la Sierra, J. Neuhard, and A.-M. Gilles. 2000. Genetic and biochemical characterization of Salmonella enterica serovar Typhi deoxyribokinase. J. Bacteriol. 182:869-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 33.Yokoyama, K., K. Makino, Y. Kubota, M. Watanabe, S. Kimura, C. H. Yutsudo, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, M. Yoh, T. Iida, M. Ohnishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 2000. Complete nucleotide sequence of the prophage VT1-Sakai carrying the Shiga toxin 1 genes of the enterohemorrhagic Escherichia coli O157:H7 strain derived from the Sakai outbreak. Gene 258:127-139. [DOI] [PubMed] [Google Scholar]