Abstract
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by using the pluripotency factors Oct4, Sox2, Klf4 and c-Myc (together referred to as OSKM)1. iPSC reprogramming erases somatic epigenetic signatures—as typified by DNA methylation or histone modification at silent pluripotency loci—and establishes alternative epigenetic marks of embryonic stem cells (ESCs)2. Here we describe an early and essential stage of somatic cell reprogramming, preceding the induction of transcription at endogenous pluripotency loci such as Nanog and Esrrb. By day 4 after transduction with OSKM, two epigenetic modification factors necessary for iPSC generation, namely poly(ADP-ribose) polymerase-1 (Parp1) and ten-eleven translocation-2 (Tet2), are recruited to the Nanog and Esrrb loci. These epigenetic modification factors seem to have complementary roles in the establishment of early epigenetic marks during somatic cell reprogramming: Parp1 functions in the regulation of 5-methylcytosine (5mC) modification, whereas Tet2 is essential for the early generation of 5-hydroxymethylcytosine (5hmC) by the oxidation of 5mC (refs 3,4). Although 5hmC has been proposed to serve primarily as an intermediate in 5mC demethylation to cytosine in certain contexts5–7, our data, and also studies of Tet2-mutant human tumour cells8, argue in favour of a role for 5hmC as an epigenetic mark distinct from 5mC. Consistent with this, Parp1 and Tet2 are each needed for the early establishment of histone modifications that typify an activated chromatin state at pluripotency loci, whereas Parp1 induction further promotes accessibility to the Oct4 reprogramming factor. These findings suggest that Parp1 and Tet2 contribute to an epigenetic program that directs subsequent transcriptional induction at pluripotency loci during somatic cell reprogramming.
We performed a functional screen for epigenetic modification factors that promote OSKM-mediated somatic cell reprogramming. Overexpression of a single pool of 29 candidate epigenetic modification factors, selected on the basis of a proteomic analysis of iPSCs (Fig. 1a–c and Supplementary Tables 1 and 2), promoted iPSC colony production in mouse embryonic fibroblast (MEF) cultures transduced with OSKM (OSKM-MEFs; relative to green fluorescent protein vector control transduced MEF; Fig. 1d). Candidate epigenetic modification factors were retested in successive subpools, and Parp1 was identified as a potent inducer of OSKM-MEF reprogramming (Fig. 1d). iPSCs generated by OSKM-MEFs with Parp1 were confirmed as pluripotent by immunocytochemistry (Supplementary Fig. 2a), gene expression (Supplementary Fig. 2b–f and Supplementary Table 3) and pyrosequencing (Supplementary Fig. 2g–i and Supplementary Table 4) analyses at multiple pluripotency loci. Parp1 overexpression did not alter the proliferation rate of transduced cultures (as determined by bromodeoxyuridine (BrdU) incorporation; Supplementary Fig. 2j).
Figure 1. Parp1 promotes OSKM-mediated iPSC generation.
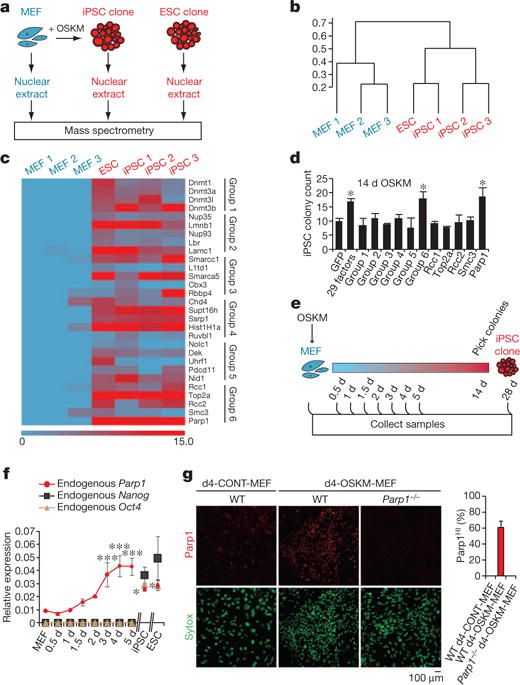
a, Diagram of proteomic strategy to identify candidate epigenetic modification (EM) factors. b, Unsupervised hierarchical clustering analysis (Spearman rank correlation) of mass spectrometry data from nuclear extracts of MEFs (n = 3), iPSCs (n = 3) and ESCs (n = 1). The scale bar represents the correlation height (= 1 − Abs[correlation]). c, Dual-colour heat map for expression levels of 29 proteins highly enriched in both the iPSC and ESC samples (relative to MEFs). The colour scale bar represents the spectral count. Candidate EM factors were divided into six groups for further functional testing. d, Functional screen of candidate EM factors for promotion of somatic cell reprogramming in OSKM-MEFs. EM candidates were transduced together as a single pool of 29 genes, as 6 subpools, or as individual factors from group 6 (as in c). Alkaline phosphatase-positive (AP+) iPSC colonies were counted at day 14 after transduction with OSKM. e, Diagram of time-course analyses of iPSC reprogramming. f, Gene expression time course of endogenous Parp1, Nanog and Oct4. g, Immunocytochemistry analysis of WT or Parp1−/− d4-OSKM-MEFs and d4-CONT-MEFs with an antibody against Parp1 (upper panels; red), and counterstained with Sytox nuclear marker (lower panels; green). Increased Parp1 expression in d4-OSKM-MEFs is quantified on the right (Parp1HI; defined as mean plus 2 s.d. or greater than the expression level in d4-CONT-MEFs); modified nuclear morphology apparent in d4-OSKM-MEFs is as described previously30. Results in d, f and g are shown as means and s.d. for three independent experiments. Asterisk, P < 0.05; three asterisks, P < 0.001.
Parp1 is a broadly expressed nuclear protein involved in the detection and repair of DNA damage, the remodelling of chromatin and the regulation of transcription9. A time course of endogenous Parp1 expression revealed detectable levels even in wild-type (WT) MEFs and a peak at day 4 (d4) after transduction with OSKM (Fig. 1e, f and Supplementary Figs 2k and 3a), thus preceding expression of the endogenous pluripotency loci including Nanog, Oct4 and Esrrb (Fig. 1f and Supplementary Fig. 3b). Immunocytochemistry analysis at d4 after transduction of WT MEFs with OSKM (WT d4-OSKM-MEFs) demonstrated Parp1 accumulation in a majority of nuclei in comparison with d4 control vector-transduced MEFs (WT d4-CONT-MEFs; Fig. 1g). The increased accumulation of Parp1 holoprotein during reprogramming was not paralleled by a corresponding accumulation of cleaved Parp1, a marker of apoptosis (Supplementary Fig. 3a).
In view of the effect of Parp1 overexpression in somatic cell reprogramming, we next tested the impact of Parp1 deficiency. Reprogramming of iPSCs was suppressed in the context of Parp1−/− OSKM-MEFs relative to WT OSKM-MEFs (Fig. 2a and Supplementary Fig. 2k, l). Resupplying WT Parp1 partly rescued iPSC generation in Parp1−/− OSKM-MEFs. In contrast, expression of Parp1 mutants10,11, compromising either the catalytic activity or the DNA-binding activity, failed to rescue iPSC generation, indicating that both activities are required for iPSC reprogramming (Fig. 2a and Supplementary Fig. 2l).
Figure 2. Parp1 activities during iPSC reprogramming.

a, Functional analysis of Parp1 mutants for rescue of iPSC colony formation in Parp1−/− OSKM-MEFs. Cultures were transduced with green fluorescent protein (GFP), WT Parp1, or Parp1 mutants encoding a catalytic domain missense mutation (CAT mutant; E988K), deletion of the catalytic domain (ΔCAT), deletion of the DNA-binding and automodification domains (CAT-only), or triple missense mutation of the DNA-binding domain (DBD mutant; C21G/C125G/L139P). Zn, zinc fingers; BRCT, BRCA1 carboxy terminus. b, Schematic representation of the Nanog locus transcription start site (TSS) region. Indicated are HpaII/MspI sites (green bars) and amplicons for ‘exon 1/intron 1’ and ‘intron 1’ regions(thick greylines). bp,base pairs. c, Parp1 ChIP analyses of the cultures as indicated, presented as the relative enrichment to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). d, Content of 5hmC assessed by GlucMS-qPCR (as a percentage of total cytosine). e, Content of 5mC, quantified by subtraction of 5hmC content (as in d) from the total methylated cytosine (5mC + 5hmC, as determined by HpaII digestion insensitivity; see Supplementary Fig. 3f). Results in a and c–e are shown as means and s.d. for three independent experiments. Asterisk, P < 0.05; two asterisks, P < 0.01; three asterisks, P < 0.001.
The early expression and functional role of endogenous Parp1 in the reprogramming process suggested the possibility of a direct interaction at pluripotency loci. Consistent with this notion, chromatin immunoprecipitation (ChIP) at the Nanog and Esrrb pluripotency loci, in the transcription start site regions (Supplementary Table 5), demonstrated increased Parp1 binding in d4-OSKM-MEFs and iPSCs (compared with d4-CONT-MEFs; Fig. 2b, c and Supplementary Fig. 3g, h, k).
Parp1 has been broadly implicated in the regulation of epigenetic remodelling events7,12, but its role during the reprogramming of pluripotency loci is unclear. We therefore investigated the impact of modified Parp1 expression on two distinct forms of cytosine methylation at pluripotency loci, 5mC and 5hmC, during somatic cell reprogramming. 5hmC is a more recently described DNA modification that has been suggested to participate in the maintenance of pluripotency at regulatory elements5,13–17 in ESCs, but its role during somatic cell reprogramming has not previously been described. We quantified total cytosine methylation (5mC plus 5hmC; either by HpaII digestion sensitivity assays or pyrosequencing of bisulphite-treated DNA) or 5hmC alone (either by MspI sensitivity assay (glucosylation-coupled methylation-sensitive quantitative polymerase chain reaction; GlucMS-qPCR)16,18 or hydroxymethylated DNA immunoprecipitation (hMeDIP); see Supplementary Tables 6 and 7 and Supplementary Methods) at regulatory regions of the Nanog or Esrrb loci. Both d4-OSKM-MEFs and iPSCs showed a significant and consistent increase in 5hmC relative to d4-CONT-MEFs (Fig. 2d and Supplementary Fig. 3c, d, n–q) at the pluripotency loci. In contrast to 5hmC, 5mC was not accumulated at the early time point (in d4-OSKM-MEFs, relative to d4-CONT-MEFs) at either locus (Fig. 2e and Supplementary Fig. 3j, m). The two pluripotency loci differed with respect to the overall pattern of 5mC during reprogramming, as previously described2,19: the Nanog locus showed a canonical pattern of hypermethylation (elevated 5mC) in MEFs and became demethylated (low 5mC) in reprogrammed iPSCs (Fig. 2e and Supplementary Fig. 3e, f), whereas the Esrrb locus showed a relatively low level of methylation even in MEFs, and remained hypomethylated in fully reprogrammed iPSCs (Supplementary Fig. 3i, j, l, m). Thus among these pluripotency loci, 5hmC rather than 5mC seemed to be an early and predictive epigenetic mark for subsequent activation. Immunocytochemistry analysis with an antibody against 5hmC showed increased nuclear staining in most cells in d4-OSKM-MEFs (relative to d4-CONT-MEF cultures; Fig. 3a).
Figure 3. Tet2 is required for 5hmC formation at the Nanog locus.

a, Immunocytochemistry of d4-OSKM-MEFs and d4-CONT-MEFs with an antibody against 5hmC (ref. 5) (upper panels; red) and counterstained with Sytox nuclear marker (lower panels; green). Representative images show increased 5hmC in d4-OSKM-MEFs, as quantified on the right (5hmCHI; defined as mean plus 2 s.d. or greater above the level in d4-CONT-MEFs). b, Time course of Tet1 and Tet2 gene expression assessed by qPCR (relative to ESC level). c, OSKM-mediated iPSC colony formation assay (AP+) in shRNA-mediated Tet2 knockdown (Tet2 KD; blue) and non-silencing control shRNA (mock KD; black)-treated MEFs. d, Diagram of the Nanog locus; regions are the same as in Fig. 2b. e, Tet2 ChIP-qPCR at the exon 1/intron 1 amplicon. f, g, Content of 5hmC in the cultures indicated, assessed by hMeDIP of the exon 1/intron 1 region (f, relative to GAPDH) or GlucMS-qPCR intron 1 amplicon (g, as a percentage of total cytosine). h, Content of 5mC at the intron 1 amplicon, quantified by subtraction of 5hmC (as in g) from the total methylated cytosine levels (as in Supplementary Fig. 4k; determined by HpaII sensitivity assay; as a percentage of total cytosine). Results in a–c and e–h are shown as means and s.d. for three independent experiments. Asterisk, P < 0.05; two asterisks, P < 0.01; three asterisks, P < 0.001.
We next sought to determine the role of Parp1 in the regulation of 5hmC and 5mC epigenetic marks at the pluripotency loci. Parp1 deficiency (which suppressed iPSC reprogramming) led to a consistent, large increase in 5mC accumulation in Parp1−/− d4-OSKM-MEFs, relative to WT d4-OSKM-MEF cultures at both the Nanog and Esrrb loci (Fig. 2e and Supplementary Fig. 3j, m). The increased 5hmC in WT d4-OSKM-MEFs was not suppressed in Parp1−/− d4-OSKM-MEFs; rather, in the context of Parp1 deficiency, 5hmC induction seemed similar to that of WT cells, for example at the Nanog locus, or modestly further increased, for example at the Esrrb locus (Fig. 2d and Supplementary Fig. 3d, n–q). Parp1 overexpression (which promoted iPSC reprogramming) did not consistently modify 5mC or 5hmC in d4-OSKM-MEFs, although a modest increase in 5hmC levels was observed at the Esrrb locus but not at the Nanog locus (Fig. 2d and Supplementary Fig. 3o, q). Taken together, these data implicate Parp1 in the regulation of 5mC; in contrast, we speculate that the variable impact of Parp1 on 5hmC may be indirect.
Given the potential role of 5hmC early in somatic cell reprogramming, we obtained an expression time-course analysis of the TET enzymes, a family of Fe II and 2-oxoglutarate-dependent enzymes that generate 5hmC from 5mC (ref. 4). Expression of Tet2, but not Tet1 or Tet3, was significantly induced in WT d4-OSKM-MEFs and remained elevated in iPSCs (Fig. 3b and Supplementary Fig. 4a). Consistent with a functional role for Tet2, short hairpin RNA (shRNA)-mediated Tet2 knockdown (KD; Supplementary Table 8) abolished iPSC colony formation (Fig. 3c and Supplementary Fig. 4b, c). ChIP analysis with an antibody against Tet2 showed a direct interaction with the Nanog and Esrrb pluripotency loci in WT d4-OSKM-MEFs; this was not altered in Parp1−/− d4-OSKM-MEFs (Fig. 3d, e and Supplementary Fig. 4d, e, l, m). Furthermore, Tet2 KD in d4-OSKM-MEFs suppressed the typical induction of 5hmC at both the Nanog and Esrrb pluripotency loci (Fig. 3f, g and Supplementary Fig. 4f, g). In contrast, the effect of Tet2 KD in d4-OSKM-MEFs on 5mC seemed variable: 5mC seemed to be decreased at the Nanog locus but mildly increased at the Esrrb locus (which is typically hypomethylated even in MEFs19; Fig. 3h and Supplementary Fig. 4h–k). Given the early induction of 5hmC but not 5mC at the pluripotency loci, as well as the consequences of Tet2 deficiency in preventing transcriptional activation and 5hmC induction even in the absence of 5mC reduction, these data support a primary role for 5hmC as a distinct epigenetic mark in the somatic cell reprogramming process, and argue against an alternative model in which 5hmC functions simply as an intermediate in the 5mC demethylation process20. Epigenetic marks with 5hmC may recruit select chromatin modification factors to the pluripotency loci21.
To further probe the roles of Tet2 and Parp1 in early epigenetic events, we evaluated the chromatin state of the Nanog and Esrrb loci in d4-OSKM-MEFs. ChIP analysis revealed an enrichment in the occupancy of these loci by the activation-associated marker histone H3 lysine 4 dimethylation (H3K4me2)22,23 and a parallel decrease in the transcriptional-silencing-associated marker histone H3 lysine 27 trimethylation (relative to d4-CONT-MEFs; H3K27me3 (refs 24–26); Fig. 4a–d and Supplementary Fig. 5a, b). Deficiency of either Parp1 or Tet2 diminished the H3K4me2 chromatin mark at the pluripotency loci of d4-OSKM-MEFs (Fig. 4a, c and Supplementary Fig. 5a). The effects on H3K27me3 were variable: Parp1 deficiency did not significantly alter H3K27me3 at either locus, whereas Tet2 KD led to a decrease at the Nanog locus but not at the Esrrb locus (Fig. 4b, d and Supplementary Fig. 5b). We speculated that altered chromatin states in d4-OSKM-MEFs may affect chromatin accessibility at the pluripotency loci, for example to the transduced Oct4 pluripotency factor. Oct4 occupancy, as quantified by ChIP of d4-OSKM-MEFs, was significantly diminished in the context of Parp1 deficiency at both pluripotency loci, whereas Tet2 KD did not diminish Oct4 occupancy (Fig. 4e, g and Supplementary Fig. 5c). Consistent with this, Parp1 overexpression potentiated Oct4 binding at both pluripotency loci of d4-OSKM-MEFs (Fig. 4e, g and Supplementary Fig. 5c). Furthermore, Parp1 overexpression robustly promoted exogenous Oct4 binding to the pluripotency loci even in the absence of transduction of the other pluripotency factors necessary for somatic cell reprogramming (d4-OMEFs without SKM; Fig. 4f, h and Supplementary Fig. 5d).
Figure 4. Impact of Parp1 and Tet2 on chromatin state and Oct4 accessibility at the Nanog and Esrrb loci.
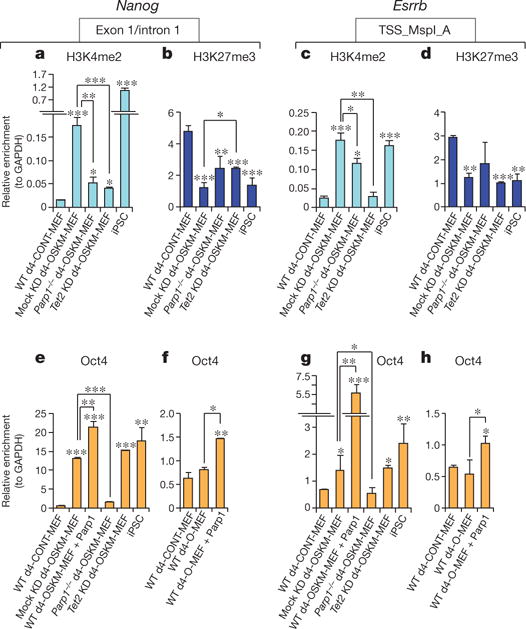
a–d, H3K4me2 (a, c) and H3K27me3 (b, d) ChIP-qPCR at Nanog or Esrrb amplicons in cultures as indicated. e–h, Oct4 ChIP-qPCR. Results are shown as means and s.d. for three independent experiments. Asterisk, P < 0.05; two asterisks, P < 0.01; three asterisks, P < 0.001.
Taken together, these data support necessary but distinct roles for Tet2 and Parp1 in the regulation of epigenetic marks and local chromatin structure at pluripotency loci during an early stage of somatic cell reprogramming that precedes their transcriptional activation (Supplementary Fig. 1a, b). The data further suggest that 5hmC, generated by Tet2, does not simply represent an intermediate in the 5mC demethylation process, but functions as an epigenetic mark, possibly recruiting trans-acting factors that promote chromatin remodelling4,21. The induction of Parp1 and Tet2 gene expression early in the course of reprogramming may reflect their direct activation by OSKM factors (Supplementary Fig. 5e, f)27. Finally, loss of Tet2 function is strongly implicated in human malignancies8, and thus Tet2-mediated chromatin remodelling may affect tumour risk associated with potential iPSC therapies.
METHODS SUMMARY
Cell culture and generation of iPSCs
MEF were prepared from WT or Parp1−/− embryonic day 13.5 embryos (129S-Parp1tm1Zqw/J; Jackson Laboratories)28. Tail-tip fibroblasts were prepared from WT and Tet2−/− mice29. For iPSC generation, MEFs were transduced by incubation with OSKM retroviral supernatants for 24 h as described1. Subsequently, cells were cultured in ESC medium and samples were collected at the time points indicated. Epigenetic modification factors were cloned into lentiviral vectors (pLenti6.3; Invitrogen) for lentiviral production and subsequent transduction.
Tet2 knockdown
Tet2 knockdown was achieved by using a cocktail of five shRNA lentiviral vectors (Open Biosystems) specific for Tet2, or control non-silencing shRNA.
Mass spectrometry
Multidimensional protein identification technology (MudPIT) was performed to analyse nuclear fractions of MEFs, iPSC clones and ESC clones as detailed in Supplementary Methods.
Chromatin immunoprecipitation
ChIP was performed with the Magnify kit (492024; Invitrogen) and the following antibodies: anti-dimethyl K4 of H3 (2 μg; 07–030; Millipore), anti-trimethyl K27 of H3 (2 μg; ab6002; Abcam), anti-Oct4 (2 μg; sc-8628 X; Santa Cruz), anti-Parp1 (2 μg; sc-74469 X; Santa Cruz), anti-Tet2 (2 μg; sc-136926; Santa Cruz), normal goat IgG (2 μg; 005-000-003; Jackson ImmunoResearch), normal rabbit IgG (1 μg; Magnify; Invitrogen) and normal mouse IgG (1 μg; Magnify; Invitrogen).
Digestion with HpaII and MspI
Detection of 5hmC and 5mC as percentages of total cytosine species was performed with the EpiMark Kit (E3317S; NEB). The technique has been described in detail in ref. 16.
Pyrosequencing
Genomic DNA (1 μg) was bisulphite-converted with the EZ DNA Methylation Kit (D5001; Zymo Research), followed by amplification by polymerase chain reaction with the PyroMark PCR Kit (978703; Qiagen). PCR products were sequenced with the PyroMark Q24 instrument (Qiagen).
Supplementary Material
Acknowledgments
We thank G. Q. Daley, A. P. Feinberg, A. Doi, R. M. Santella and M. A. Kappil for reagents and for technical assistance with pyrosequencing; A. Califano and A. Lachmann for assistance with the bioinformatics analyses; E. O. Mazzoni for assistance with the ChIP analyses; and O. Hobert for critical reading of the manuscript. This work was supported by New York State Stem Cell Science (NYSTEM) grants C024402 and C024403 and National Institutes of Health (NIH) grant RO1 NS064433 to A.A., NYSTEM Institution Development Grant N08G-071 to E.I.C., NIH grant RO1 138424 to R.L.L, and a shared NIH/National Center for Research Resources instrument grant for mass spectrometry, 1 S10RR023680-1.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions C.A.D. and A.A. designed the experiments and analysed data. C.A.D., D.B.R., S.T., R.F. and W.B.V. conducted molecular and cellular experiments. T.Y., G.B. and K.I. performed and analysed murine in vivo studies. R.L.L. and A.S. supplied essential reagents. P.G. performed bioinformatics analyses. S.N. and E.I.C. conducted proteomics. C.A.D. and A.A. wrote the manuscript.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo JU, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2011;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wacker DA, et al. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langelier MF, et al. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajkova P, et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis T, Vaisvila R. High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue. J Vis Exp. 2011;(48):e2661. doi: 10.3791/2661. http://dx.doi.org/10.3791/2661. [DOI] [PMC free article] [PubMed]
- 19.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZQ, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 29.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nature Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


