Abstract
Transcarboxylase is a 1.2 million Dalton (Da) multienzyme complex from Propionibacterium shermanii that couples two carboxylation reactions, transferring CO2− from methylmalonyl-CoA to pyruvate to yield propionyl-CoA and oxaloacetate. Crystal structures of the 5S metalloenzyme subunit, which catalyzes the second carboxylation reaction, have been solved in free form and bound to its substrate pyruvate, product oxaloacetate, or inhibitor 2-ketobutyrate. The structure reveals a dimer of β8α8 barrels with an active site cobalt ion coordinated by a carbamylated lysine, except in the oxaloacetate complex in which the product's carboxylate group serves as a ligand instead. 5S and human pyruvate carboxylase (PC), an enzyme crucial to gluconeogenesis, catalyze similar reactions. A 5S-based homology model of the PC carboxyltransferase domain indicates a conserved mechanism and explains the molecular basis of mutations in lactic acidemia. PC disease mutations reproduced in 5S result in a similar decrease in carboxyltransferase activity and crystal structures with altered active sites.
Keywords: carboxyltransferase, crystal structure, multienzyme complex, pyruvate carboxylase, transcarboxylase
Introduction
Nature has selected biotin as a cofactor to be a carrier for carbon dioxide (CO2). In the form of carboxybiotin, CO2 is less reactive and at the site of delivery the transferred carboxyl group can be released as CO2 or directly to an acceptor nucleophile (Knowles, 1989). Biotin-dependent carboxylases function through two partial reactions and fall into three classes (Wood and Barden, 1977). Class I enzymes, which include all eukaryotic biotin enzymes, first use ATP, Mg(II), and bicarbonate to form carboxybiotin, and then transfer the carboxyl group to an acceptor molecule such as pyruvate or acetyl-CoA. Class II enzymes couple decarboxylation of β-keto acids and their thioesters with sodium transport in anaerobic prokaryotes. Transcarboxylase (TC) from Propionibacterium shermanii is the only member of Class III, and couples two carboxylation reactions.
To date, only four mammalian biotin-dependent carboxylases have been identified: acetyl-CoA carboxylase (ACC), methylcrotonoyl-CoA carboxylase (MCC), propionyl-CoA carboxylase (PCC), and pyruvate carboxylase (PC) (Moss and Lane, 1971; Samols et al, 1988). These Class I carboxylases play central roles in such metabolic pathways as oxidation of odd-chain fatty acids, catabolism of branched amino acids, fatty acid synthesis, and gluconeogenesis. As the first enzyme in the gluconeogenic pathway, PC catalyzes the ATP-driven conversion of pyruvate to oxaloacetate using HCO3− and Mg2+ (Wexler et al, 1994). Deficiencies of PC result in lactic acidemia and may present as developmental delay, severe mental retardation or death by 3 months of age (Robinson 2001).
TC has historically served as a model system for biotin-dependent carboxylases because its subunits share significant sequence homology with the important related human enzymes, are easily isolated, and form stable substrate complexes in the absence of the other subunits. TC is a 1.2 million Da multienzyme complex containing 30 polypeptide chains: a catalytic 336 kDa 12S hexameric core, six catalytic 116 kDa 5S dimers, and twelve 12 kDa 1.3S biotinylated linkers (Wood and Zwolinski, 1976) (Figure 1A). The overall TC transcarboxylation reaction consists of two half reactions (Wood and Kumar, 1985). In the first half reaction, 12S transfers CO2− from MMCoA to biotin on 1.3S. 5S transfers the CO2− from the 1.3S biotin to pyruvate in the second half reaction (Figure 1B). TC's organization, in which intermediates are shuttled between different catalytic subunits by a flexible carrier, places it in the general group of multienzyme complexes such as pyruvate dehydrogenase (Coppel et al, 1988; Ho and Patel, 1990; Koike et al, 1990) and glycine decarboxylase (Kume et al, 1991). High-resolution structures of their individual subunits may provide some insight into the mechanism and holo enzyme organization as ambitious efforts continue to crystallize complete multienzyme complexes.
Figure 1.
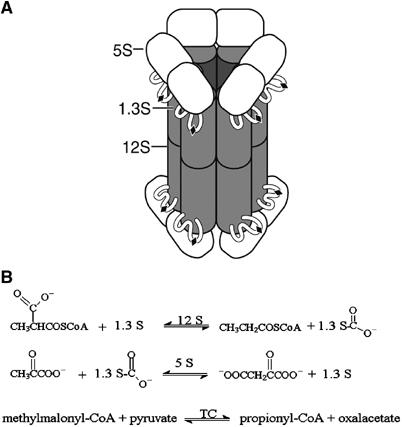
The TC multienzyme complex. (A) EM-based model (adapted from Wrigley et al, 1977). (B) The two half reactions catalyzed by the 12S and 5S subunits, and the overall full reaction.
Crystal structures of TC subunits are important for a mechanistic insight into mammalian Class I enzymes, which have been difficult to express in large quantities and to crystallize. We have previously described the 12S crystal structure and its relevance to PCC (Hall et al, 2003). The other large TC subunit, 5S, is functionally and sequentially homologous (27% identity) to the C-terminal carboxyltransferase region of human pyruvate carboxylase (EC 6.4.1.1) (Thorton et al, 1993; Wexler et al, 1994). No high-resolution three-dimensional structure is currently available for a mammalian PC; thus, elucidation of the 5S crystal structure will provide a mechanistic insight into both TC and related carboxyltransferases such as PC.
We have solved the crystal structure of TC wild-type 5S as free protein and as complexes bound to its substrate pyruvate, product oxaloacetate or competitive inhibitor 2-ketobutyrate. These structures reveal the protein fold, dimer organization, and active site features important for substrate binding and catalysis. The 5S structure serves as a scaffold for the homology modeling of the human PC carboxyltransferase domain, which supports the expectation of conserved fold, active site, and mechanism. We have designed two 5S mutants which mimic PC disease mutations. They have the same level of reduced enzymatic activity as their PC counterparts, explained by their crystal structures, thus reinforcing the value of TC as a model for mammalian biotin-dependent carboxylases.
Results and discussion
We have determined the crystal structures of TC 5S in free form and bound to pyruvate (5S-pyr), oxaloacetate (5S-oxal), and 2-ketobutyrate (5S-2keto), as well as of two single-site mutants (5S-A59T, 5S-M186I) (Table I). The free 5S structure was solved first, by selenomethionine MAD-phasing methods, and refined to 1.9 Å resolution. Its protein coordinates were then used to solve the other 5S structures at either 2.0 Å (5S-pyr, 5S-2keto), 2.5 Å (5S-oxal, 5S-A59T), or 2.8 Å (5S-M186I) resolution. All six 5S structures are very similar: superpositions of the complex and mutant structures on free 5S result in an r.m.s. fit of 0.2–0.3 Å for 471 equivalent Cα atoms. Unless otherwise noted, the highest resolution structure, of free 5S, is described in the following sections.
Table 1.
Data collection, phasing, and refinement statistics
| MAD phasing | SeMet-5S | SeMet-5S | SeMet-5S | |||
| Data processing | ||||||
| Source | X25 | X25 | X25 | |||
| Wavelength (Å) | 1.0734 | 0.9803 | 0.9808 | |||
| Unit cell (Å3) | 96.3 | 96.3 | 96.3 | |||
| 147.1 | 147.2 | 147.2 | ||||
| 79.1 | 79.1 | 79.1 | ||||
| Resolution (Å) | 50–2.0 (2.07–2.0) | 50–2.0 (2.07–2.0) | 50–2.0 (2.07–2.0) | |||
| 〈I〉/〈σ(I)〉 | 19.5 (5.3) | 17.1 (3.7) | 16.4 (3.4) | |||
| Rmerge (%) | 5.3 (14.9) | 5.6 (18.9) | 4.5 (17.7) | |||
| Completeness (%) | 93.3 (83.2) | 90.2 (77.4) | 66.8 (54.3) | |||
| Phasing | ||||||
| Resolution (Å) | 20–2.5 | |||||
| Number of sites | 23 | |||||
| Mean f.o.m. | 0.66 | |||||
| Structures | 5S | 5S-pyr | 5S-oxal | 5S-2keto | 5S-M186I | 5S-A59T |
| Data collection | ||||||
| Source | X25 | Rigaku | Rigaku | Rigaku | Bruker | Rigaku |
| Wavelength (Å) | 0.9803 | 1.5413 | 1.5413 | 1.5413 | 1.5413 | 1.5413 |
| Unit cell (Å3) | 96.5 | 96.0 | 95.9 | 96.2 | 95.7 | 96.3 |
| 145.9 | 145.7 | 145.5 | 146.1 | 147.0 | 146.3 | |
| 79.0 | 78.7 | 78.2 | 78.7 | 78.8 | 78.7 | |
| Resolution (Å)a | 50–1.9 | 30–2.0 | 30–2.5 | 30–2.0 | 30–2.8 | 30–2.5 |
| (1.97–1.9) | (2.07–2.0) | (2.59–2.5) | (2.07–2.0) | (2.93–2.8) | (2.59–2.5) | |
| 〈I〉/〈σ(I)〉 | 19.5 (6.5) | 16.3 (4.3) | 22.8 (7.2) | 22.0 (4.5) | 5.6 (2.7) | 22.0 (6.9) |
| Rmerge (%) | 7.0 (16.4) | 8.7 (21.8) | 6.6 (15.3) | 8.4 (23.4) | 10.0 (22.4) | 6.9 (17.7) |
| Completeness (%) | 95.7 (87.0) | 92.8 (81.0) | 93.5 (86.3) | 85.3 (71.8) | 99.3 (99.5) | 94.8 (87.5) |
| Refinement | ||||||
| Rwork, Rfree | 16.24, 19.07 | 16.42, 19.63 | 15.25, 22.59 | 17.31, 20.45 | 20.78,26.92 | 20.52, 25.00 |
| r.m.s.d. bond lengths (Å) | 0.0072 | 0.0072 | 0.0073 | 0.0071 | 0.0078 | 0.0075 |
| Bond angles (deg) | 1.28 | 1.22 | 1.35 | 1.27 | 1.33 | 1.31 |
| Number of atoms: protein | 3664 | 3677 | 3661 | 3672 | 3657 | 3666 |
| Ions, substrate | 1, 0 | 1, 6 | 1, 9 | 1, 7 | 1, 0 | 1, 0 |
| Solvent | 372 | 415 | 188 | 260 | 31 | 1 |
| B-factor, mean (Å)b | ||||||
| Protein | 25.2 | 31.9 | 31.2 | 34.9 | 34.3 | 19.6 |
| Bound ligand | 20.7 | 33.0 | 26.2 | |||
| Protein within 8 Å of ligand | 21.6 | 18.9 | 24.1 | |||
| Co+2 ion | 12.0 | 28.6 | 21.1 | 24.2 | 35.2 | 29.9 |
| Co+2 ligands | 11.5 | 23.6 | 20.8 | 16.1 | 28.6 | 16.2 |
| Solvent | 32.4 | 37.3 | 29.3 | 35.4 | 20.3 | 13.2 |
| r.m.s.d., bonded | 2.4 | 1.9 | 2.2 | 2.1 | 2.2 | 0.7 |
| r.m.s.d., angled | 3.1 | 2.6 | 3.3 | 2.9 | 3.4 | 1.3 |
| Ramachandran (%) most fav., disallow. |
92.3, 0.0 |
92.0, 0.0 |
88.4, 0.0 |
92.0, 0.0 |
86.4, 0.2b |
89.1, 0.0 |
| Values in parentheses are for the highest resolution shell. | ||||||
| aSame resolution limits were used for data processing and refinement. | ||||||
| bLeu97 refines into a disallowed region in one structure; the corresponding density and structural environment do not explain this observation and this residue is in the allowed region in all other structures. |
Overall structure
Native 5S monomer has 505 residues; the recombinant 5S which was crystallized contains extended termini due to cloning artifacts and the C-terminal His6 tag. The asymmetric unit contains a monomer with overall dimensions ∼50 Å × 60 Å × 56 Å. No density is observed for N- and C-terminal residues, which are presumed to be flexible. The final refined structure includes protein residues 3–474, one cobalt ion, and 372 water molecules (Table I). 5S contains the canonical TIM-barrel fold (Banner et al, 1975), which consists of a core β8α8 motif with the eight parallel β strands forming an enclosed barrel surrounded by eight α helices (Figure 2). In addition, 5S has an N-terminal β strand (β1), two short α helices between β1 and α1 and between β2 and α2 (termed α1a and α2a, respectively), and a large C-terminal extension starting at residue Gly265 that contains two β strands and nine α helices. The C-terminal extension forms a funnel leading to the active site at the C-termini of the parallel β strands.
Figure 2.
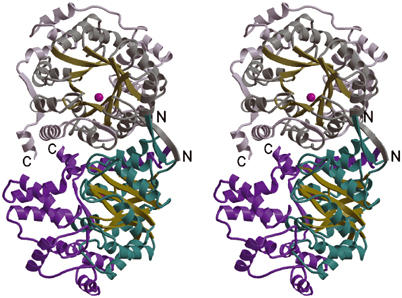
Stereoview ribbon diagram of the 5S dimer. The bottom monomer is colored with its core eight-stranded β-barrel in yellow, the surrounding α helices in cyan, the C-terminal extension in purple, and the active site cobalt ion shown as a pink sphere. The second (top) monomer is generated from the first by a 90° rotation about the vertical axis and a 180° rotation about an axis perpendicular to the page, and is shown in paler colors. The N- and C-termini for the two monomers are labeled.
Structural similarity to other TIM-barrel proteins
Not surprisingly, a DALI search (Holm and Sander, 1998) matched 5S with a large number of proteins with similar folds (Table II). The TIM-barrel domain is estimated to be present in ∼10% of known enzyme structures (Farber and Petsko, 1990) and is utilized by at least 15 different enzyme families, with the active site always located at the C-termini of the β strands (Wierenga, 2001; Nagano et al, 2002). The top DALI match is 4-hydroxy-2-oxovalerate aldolase, which has, in addition to the TIM barrel, a C-terminal extension (Gly250–His340) corresponding to the first half of the 5S C-terminal extension (Gly265–Gly366). The remaining DALI matches share only the TIM barrel with 5S. Of these, seven structures are of particular interest since they are (de)carboxylases, although none are biotin-dependent. The active sites of these seven TIM-barrel (de)carboxylase structures show little conservation with 5S, with the exception of ribulose-1,5 bisphosphate carboxylase/oxygenase (Rubisco), whose active site structure will be discussed in more detail later.
Table 2.
TIM-barrel proteins structurally similar to 5S
| Protein | DALI Z-score | Sequence identity (%) | r.m.s.d. (Å), # equivalent Cα | Reference |
|---|---|---|---|---|
| 4-hydroxy-2-oxovalerate aldolase | 28.6 | 15 | 3.4, 308 | Manjasetty et al, 2003 |
| Orotidine monophosphate decarboxylase | 14.9 | 10 | 3.1, 203 | Wu et al, 2000 |
| 3-keto-L-gulonate 6-phosphate decarboxylase | 13.8 | 11 | 3.0, 194 | Wise et al, 2002 |
| Ribulose-1,5bisphosphate carboxylase | 9.2 | 7 | 3.9, 193 | Lundqvist and Schneider, 1991 |
| Uroporphyrinogen decarboxylase | 9.2 | 7 | 3.9, 188 | Whitby et al, 1998 |
| Ornithine decarboxylase | 7.3 | 4 | 6.2, 188 | Kern et al, 1999 |
| Diaminopimelate decarboxylase | 7.2 | 6 | 4.7, 196 | Gokulan et al, 2003 |
| Phosphoenolpyruvate carboxylase | 7.0 | 7 | 6.1, 246 | Kai et al, 1999 |
| Selected from a total of 247 hits with Z-scores >2.0. |
Dimer organization
The 5S subunit is a homodimer of 116 kDa that is generated by applying crystallographic symmetry to the monomer in the asymmetric unit. Residues from both the TIM-barrel domain and C-terminal extension form the extensive dimerization interface, which buries an average of 3921 Å2 total accessible surface area and is 56% hydrophobic (Figure 2). The dimerization interface also has a high shape complementarity index of 0.63 (Lawrence and Colman, 1993). Eight hydrogen bonds align the two N-termini to form an antiparallel intermolecular β-sheet. To confirm that the crystallized dimer is also observed in solution, we prepared the single-site mutants Lys229Glu and Glu232Lys to remove the key Lys229–Glu232 salt bridge at the dimer interface, and found that the mutants were a mixture of monomer and dimer in solution, while wild-type 5S and two control mutants altering other crystal contacts are essentially completely dimeric (data not shown).
Cobalt-binding site
5S is a cobalt-dependent metalloenzyme (Northrop and Wood, 1969). The presence of a bound metal ion was shown by anomalous difference density, and its identification as cobalt was confirmed by ICP-MS. In the 5S active site, there is a single cobalt ion octahedrally coordinated by the side chains of His215, His217, and Asp23, a water molecule (WatA), and the CO2− from the carbamylated Lys184 (denoted LysC184; Figure 3A). All cobalt ligands are conserved in PC, as expected if the two proteins have a conserved mechanism (Figure 4A). The tight coordination sphere of the cobalt ion, with an average ligand distance of 2.15 Å, is consistent with reports that the 5S metal can be removed only under protein denaturing conditions (Ahmad et al, 1972). The cobalt ion is found at the base of a deep active site cavity; its His215, His217, and Asp23 ligands are in mostly well-packed environments with little freedom for substantial movement, while in contrast LysC184 and WatA are relatively solvent-accessible and have space to move in and out of the cobalt coordination sphere.
Figure 3.
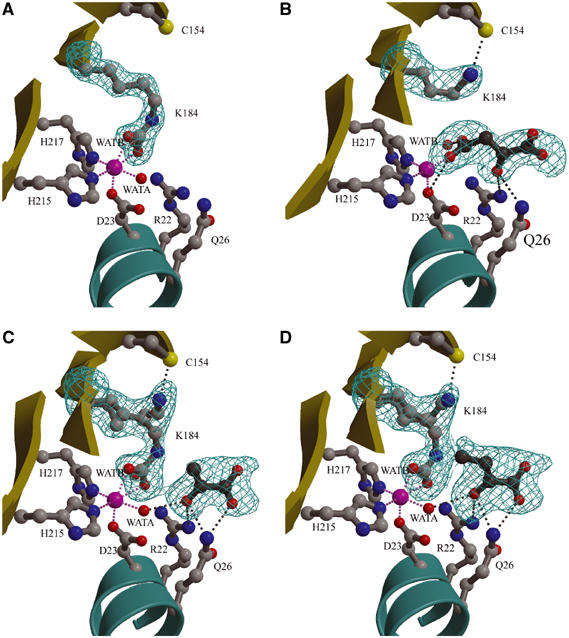
5S active site. The cobalt ion (pink sphere), its water ligands, and side chains of residues which interact with either the cobalt ion or substrate (ball-and-stick) are shown. Potential interactions involving the metal ion and active site ligands are represented by pink and gray dashed lines, respectively. (A) Free 5S: 50–1.9 Å resolution simulated annealing ∣Fo∣–∣Fc∣ omit density contoured at 7σ shows a carbamylated LysC184 that coordinates the cobalt ion. (B) 5S-oxal: 30–2.5 Å resolution simulated annealing ∣Fo∣–∣Fc∣ omit density contoured at 4σ shows bound oxaloacetate interacting with the cobalt ion and a noncarbamylated Lys184 interacting with Cys154. (C) 5S-pyr: 30–2.0 Å resolution simulated annealing ∣Fo∣–∣Fc∣ omit density contoured at 4σ shows bound pyruvate (refined at half occupancy) and two conformations for Lys184/LysC184 (both refined at half occupancy). (D) 5S-2keto: 30–2.0 Å resolution simulated annealing ∣Fo∣–∣Fc∣ omit density contoured at 3σ shows bound 2-ketobutyrate (refined at half occupancy) and two conformations for Lys184/LysC184 (both refined at half occupancy).
Figure 4.
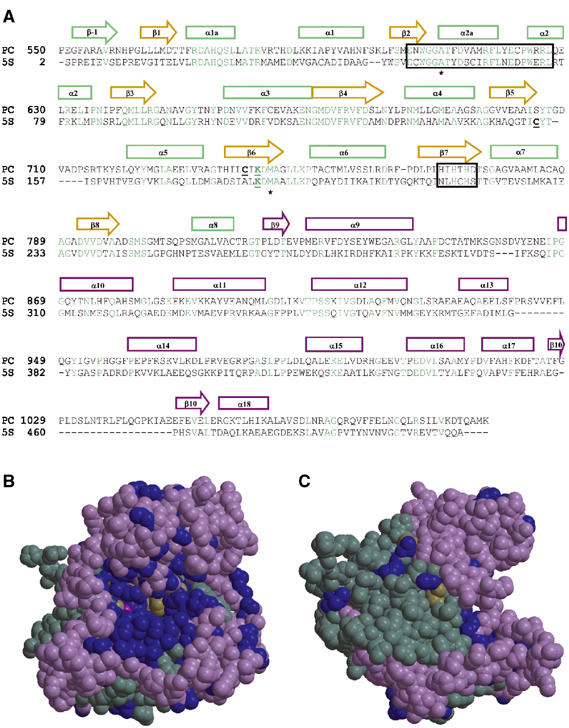
5S homology with PC. (A) Structure-based sequence alignment of 5S with the carboxyltransferase region of human PC (SwissProt accession number P11498), with 5S secondary structure elements colored similarly to their representation in Figure 2. Conserved residues (cyan), sites of PC missense disease mutations (*), the carbamylated Lys184 and the known/potential Cys partner residues (underlined), and the aligned putative pyruvate binding and metal-binding motifs (boxed) are highlighted. (B) Space-filling representation of 5S monomer with view into the active site. Residues conserved in PC are in blue, cobalt ion in pink, and the remaining residues colored by secondary structure and domain location as in Figure 2. (C) As in (B), but rotated 90° about the vertical axis to show the dimerization surface with its few conserved residues.
Free 5S active site
The most intriguing feature of the free 5S active site is the unexpected carbamylation of residue LysC184 (Figure 3A). In addition to coordinating the cobalt ion with distances of 2.2 and 2.3 Å, the CO2− modification of the LysC184 side chain also forms hydrogen bonds with His215, Arg22, Asp23, and two waters, WatA and WatB. Lys184, its amino-acid hydrogen-bonding partners, and nearly all the solvent-exposed active site residues are conserved in PC (Figure 4B). Some of these are of particular interest and will be discussed in more detail later: Gln26, which is located in helix α1a along with Arg22 and Asp23; Ala59 on α2a, Cys154 on strand β5, and Met186 on β6.
5S-oxal complex
In the 5S-oxal structure, clear density is observed for the product oxaloacetate bound in the 5S active site (Figure 3B). Oxaloacetate was refined at three-quarter occupancy to minimize residual difference electron density; as a result, the oxaloacetate average atomic temperature factor of 33 Å3 is higher than that of 19 Å3 for the surrounding protein atoms (Table I). Attempts were made to fit oxaloacetate in two opposing orientations; one clearly fit the electron density better. In the correct orientation, the ‘transferred' CO2− of oxaloacetate coordinates the active site cobalt, with the carboxylate oxygen atoms 1.6 and 3.0 Å from the metal ion. Oxaloacetate thus effectively replaces LysC184 as a cobalt ligand. With the two crystal structures superimposed, the ‘transferred' oxaloacetate CO2− carbon in 5S-oxal is nearly coincident with (only 1.0 Å from) the corresponding carbon atom of LysC184 in free 5S, and is 2.7 Å from the cobalt ion. In the middle of oxaloacetate, the carbonyl oxygen forms hydrogen bonds with Arg22 and Gln26; the adjacent carboxylate group is partially solvent-exposed and not involved in any electrostatic interactions. There are two additional pronounced differences between the 5S-oxal and free 5S active sites. First, in 5S-oxal, Lys184 is not carbamylated as in free 5S; it is pointed away from the cobalt and its amine forms a 2.8 Å hydrogen bond with the sulfhydryl of Cys154. Second, there is no WatA equivalent in 5S-oxal and, as a result, the cobalt ion in the oxaloacetate complex has an incomplete coordination sphere.
5S-pyr complex
The isolated 5S subunit does not have measurable catalytic activity; it is only active in the presence of the other TC subunits (Xie et al, 1993). This made it possible to crystallize the complex of 5S bound to its pyruvate substrate. In the 5S-pyr structure, pyruvate is not directly bound to the cobalt—the closest approach is its C3 methyl carbon, the target for carboxylation during the 5S reaction, which is 5.2 Å from the metal ion (Figure 3C). This distance, along with cobalt to pyruvate carbonyl carbon and carboxylate carbon distances of 5.3 and 6.8 Å, are near those reported earlier (6.3, 5.0, and 6.3 Å, respectively) using EPR and NMR (Fung et al, 1974). In effect, pyruvate is bound in the same position and orientation as oxaloacetate in 5S-oxal, such that there is space between the pyruvate and cobalt for the insertion of the CO2− group to take place during the carboxyltransferase reaction; the pyruvate C3 atom is 1.4 Å from the equivalent C2 of oxaloacetate when the 5S-pyr and 5S-oxal structures are superimposed. As also seen for the oxaloacetate carbonyl, the pyruvate carbonyl group forms hydrogen bonds with Arg22 and Gln26. New ligand interactions include hydrogen bonds between the pyruvate carbonyl oxygen and WatA, a solvent molecule observed in free 5S but not in 5S-oxal, and between the pyruvate carboxylate and Gln26. The electron density indicates two conformational and chemical states for Lys184, corresponding to half occupancies of noncarbamylated Lys184 as observed in 5S-oxal and carbamylated LysC184 as observed in free 5S. In 5S-pyr, the half-occupied LysC184 has its CO2− carbon 3.7 Å from the C3 atom of pyruvate. The half-occupied noncarbamylated Lys184 has shifted 3.7 Å to form a 2.6 Å long hydrogen bond with Cys154. The short pyruvate C3 to LysC184 distance can be explained by partial pyruvate occupancy; pyruvate was refined at half occupancy to minimize the residual difference electron density. As a result, the average atomic temperature factors for the ligand and surrounding protein atoms are both around 21 Å3 (Table I).
5S-2keto complex
The competitive inhibitor 2-ketobutyrate blocks transcarboxylation due to its ethyl group in place of the methyl moiety in pyruvate. In the 5S-2keto structure, 2-ketobutyrate is positioned in the 5S active site similarly to pyruvate and oxaloacetate in 5S-pyr and 5S-oxal, respectively. The 2-ketobutyrate inhibitory carbon C4 is 4.6 Å from the cobalt, and the blocked transcarboxylation ‘target' carbon C3 is 5.7 Å from the metal ion (Figure 3D). Thus, it is clear how 2-ketobutyrate is chemically and sterically incapable of functioning as substrate. 2-ketobutyrate is bound in 5S-2keto in essentially the same manner as pyruvate is bound in 5S-pyr (Figure 5). 5S-2keto also has half occupancies of noncarbamylated Lys184 forming a 2.8 Å hydrogen bond to Cys154 and of carbamylated LysC184 coordinated to the cobalt ion. As speculated for 5S-pyr, in 5S-2keto the short 3.2 Å distance between the 2-ketobutyrate C3 atom and the CO2− carbon of LysC184 can be explained by partial 2-ketobutyrate occupancy. This interpretation is consistent with the refinement of 2-ketobutyrate at half occupancy to minimize residual difference electron density, with the result that the average atomic Bs for the ligand and surrounding protein atoms are both around 25 Å3 (Table I).
Figure 5.
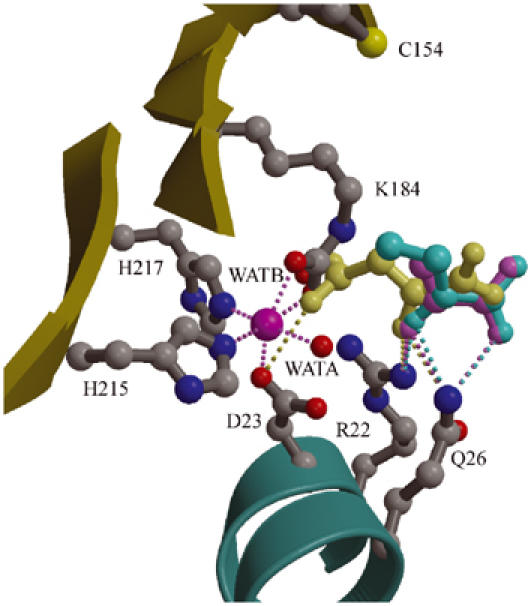
Active sites of 5S complexes. The free 5S active site is shown with its LysC184 (ball-and-stick with gray carbon atoms and bonds), cobalt ion (pink sphere), water ligands (red spheres), and side chains of residues which interact with either the cobalt ion or bound ligand. Superimposed are oxaloacetate (yellow), pyruvate (pale pink), and 2-ketobutyrate (cyan) ligands from their respective complexes.
Homology modeling of pyruvate carboxylase
The gluconeogenic enzyme PC is a homotetramer in which each 125 kDa subunit has a covalently bound biotin and binding sites for acetyl CoA, ATP, HCO3−, and pyruvate (Bardin et al, 1975; Wallace and Easterbrook-Smith, 1985). PC and TC 5S share functional homology and the requirement for a bivalent metal ion (Jitrapakdee and Wallace, 1999). The 5S sequence is 27% identical to that of the PC carboxyltransferase region, with the two most conserved motifs predicted to be the pyruvate-binding site (Samols et al, 1988) and metal-binding site (HXHXH) (Jitrapakdee and Wallace, 1999) (Figure 4A). In 5S, the His215 and His217 cobalt ligands correspond to the second and third histidine residues in the putative PC metal-binding site. In contrast, 5S residues Glu54–Leu76 correspond to the putative PC pyruvate-binding site, but do not interact with any of the ligands in our 5S crystal structures. We have constructed a 5S-based homology model of the carboxyltransferase domain of human PC. Most of the conserved residues are in the TIM barrel, with many of these solvent-inaccessible and likely to be structurally important. Of the solvent-exposed conserved residues, most are in the active site cavity, including Lys184 that is carbamylated in 5S (Lys741 in PC). Nearly all these solvent-accessible active site cavity residues are conserved, including all the amino acids involved in binding cobalt, oxaloacetate, pyruvate, or 2-ketobutyrate (Figure 4B). This observation is consistent with 5S and the PC carboxyltransferase domain having similar substrate-binding sites and catalytic mechanisms. In contrast, very few of the residues at the 5S dimer interface are conserved in PC (Figure 4C), which is not surprising since the TC multienzyme and homotetrameric PC have very different oligomeric organizations.
Structural insight into PC disease mutations
PC deficiency is an autosomal recessive disorder (Atkin et al, 1979) that presents in a number of forms ranging from mild to severe lactic acidemia (Robinson 2001). Of the four single-site missense mutations identified in PC deficiency, two occur in the carboxyltransferase region of PC. These PC mutations, A610T and M743I, alter residues corresponding to the conserved 5S amino acids A59 and M186 (Figure 4A). To confirm that 5S is a useful model to study PC disease mutants, we prepared the 5S mutants A59T and M186I that are homologous to the two PC disease mutations. Activity assays of 5S-A59T and 5S-M186I showed low relative specific activities ranging up to 3.6% of wild type (Table III). These values are similar to published activity measurements of the two PC disease mutants ranging from 0 to 17% that of control values (de Vivo et al, 1977; Atkin et al, 1979; Murphy et al, 1981; Robinson et al, 1984; Robinson, 2001).
Table 3.
Enzymatic activity of 5S mutants
| Relative specific activity (s.d.)a | %Wild-type activity | |
|---|---|---|
| Wild type | 1.93 (0.38) | 100 |
| Ala59Thr | 0.07 (0.04) | 3.6 |
| Met186Ile | 0.04 (0.03) | 2.2 |
| Lys184Ala | −0.02 (0.13) | 0 |
| Lys184Glu | −0.13 (0.21) | 0 |
| Cys154Ala |
0.96 (0.51) |
49.6 |
| Statistics are based on experiments performed in triplicate on two separate protein preparations, and are representative of experiments performed on multiple 5S preparations. |
To obtain a structural view of the molecular consequence of the two PC disease mutants, we have determined the crystal structures of 5S-A59T and 5S-M186I (Table I). Both mutations alter exposed active site residues and do not change the local protein fold (Figure 6). In addition, as observed in wild-type free 5S, LysC184 is observed only in its carbamylated, cobalt-coordinating conformation in the two unliganded 5S mutant structures. In the wild-type 5S crystal structures, the Met186 side chain is found in two conformations: it packs against LysC184 in free 5S, and against the bound ligands in 5S-oxal, 5S-pyr, and 5S-2keto. Thus, Met186 appears to play a role in both binding substrate and stabilizing LysC184 in its cobalt-coordinating conformation. The 5S-M186I crystal structure shows no alteration of the LysC184 interactions or conformation, but instead it may affect ligand binding. By superimposing the 5S-M186I on the 5S-oxal and 5S-pyr structures, the Ile186 side chain comes within 3.5 Å of the ligands. Unlike Met186, Ile186 has a branched side chain and it is not possible to relieve short contacts with bound ligand by simply rotating the side chain. In the 5S-A59T structure, instead of an unfavorable short contact, the mutation introduces a new hydrogen bond, between the Thr59 hydroxyl and the Gln26 side chain. Thr59 appears to compete for electrostatic interactions with Gln26, whose side chain forms hydrogen bonds with oxaloacetate, pyruvate, and 2-ketobutyrate in the complex crystal structures. Thus, an explanation for the lower specific activity of the M186I and A59T mutants is that both compromise substrate/product binding, albeit for different reasons.
Figure 6.
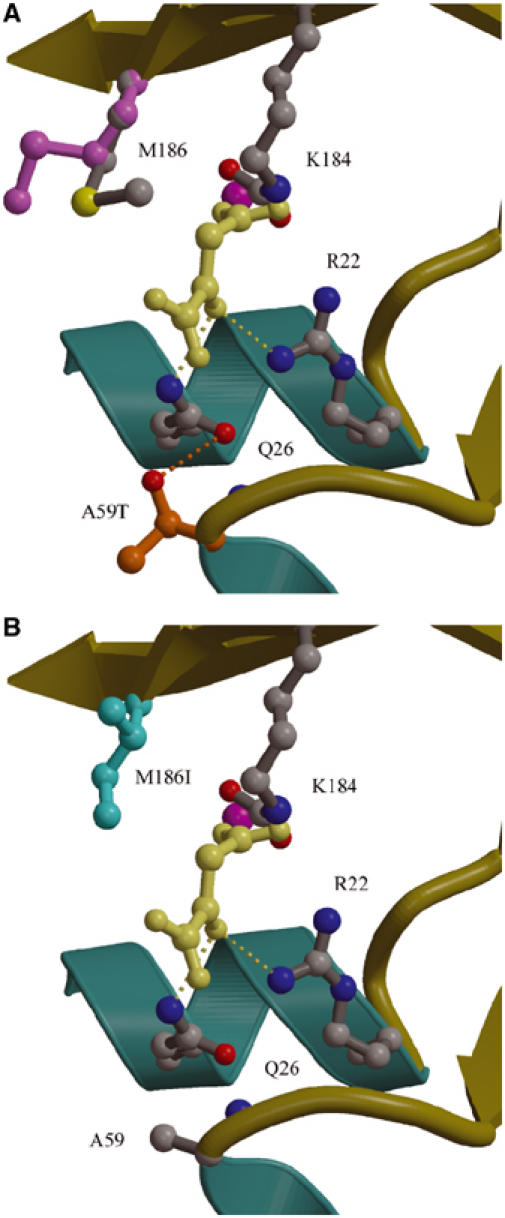
Active sites of 5S mutants. (A) Composite 5S active site showing the 5S-A59T crystal structure. The Thr59 side chain (orange and red ball-and-stick) interacts with Gln26, which in the complex structures forms hydrogen bonds with the bound ligands. For reference, oxaloacetate (yellow, as bound in 5S-oxal) and residues with which it interacts (Arg22 and Gln26), Met186 (gray, conformation in free 5S; pale pink, conformation in 5S-oxal), cobalt ion (pink sphere), and carbamylated LysC184 are also shown. The orientation of this figure is related to that of Figure 5 by ∼90° rotation about the vertical axis combined with ∼90° rotation about the horizontal axis. (B) Composite 5S active site showing the 5S-M186I crystal structure. The I186 side chain (cyan ball-and-stick structure) would be expected to make close contacts with substrate or product; oxaloacetate (yellow, as bound in 5S-oxal) and Ala59 are shown for reference.
Mechanism of carboxyl transferase
The two most prominent features of the 5S active site are the cobalt ion and the carbamylated LysC184. Oxaloacetate binds 5S such that the product's carboxylate group, added as a result of the carboxylation reaction, coordinates the cobalt. Pyruvate binds 5S such that there is space between the substrate carbon atom modified during the carboxylation reaction and the cobalt. The 5S-oxal and 5S-pyr structures thus show that cobalt's role in catalysis is not to participate in substrate binding, but is likely to be to bind the CO2− and transfer it to the substrate. This indication is supported by the 5S-2keto structure, which shows that the competitive inhibitor is bound in the same position and orientation as the substrate, and thus does not directly interact with the cobalt ion.
The carbamylation of LysC184 is the most unexpected and interesting feature of the 5S active site. This lysine modification has been observed in a number of protein structures, but in the active sites of only four other enzymes. The best studied of these is Rubisco (Chapman et al, 1988; Knight et al, 1989), whose Lys191 is slowly carbamylated by CO2 (Lorimer and Miziorko, 1980; Lorimer, 1981) in an activation step required for active site Mg2+ binding (Lorimer, 1979; Lundqvist and Schneider, 1991). This Rubisco activator CO2 is different from its substrate CO2 (Lorimer, 1981). In Klebsiella aerogenes urease (Jabri et al, 1995) and Pseudomonas diminuta phosphotriesterase (Benning et al, 1995), the carbamylated lysine residues bridge a binuclear metal center. Since neither of these two enzymes is a (de)carboxylase, the lysine carbamylation is likely important only for stabilization of the metal ion binding and does not play a direct role in catalysis. In the fourth example, Pseudomonas aeruginosa class D OXA-10 β-lactamase, carbamylated LysC70 acts as a catalytic base, and is the only known example of an active site carbamylated lysine which does not coordinate a metal ion (Maveyraud et al, 2000; Golemi et al, 2001). As revealed by our crystal structures, TC 5S is the fifth example of a carbamylated lysine in an enzyme's active site. However, 5S is particularly notable since its carbamylated LysC184 coordinates the active site cobalt ion in the same way as the transferred carboxylate of the oxaloacetate product. This raises the possibility that the carbamylation CO2− is the same molecule transferred during catalysis.
In the 5S-oxal, 5S-pyr, and 5S-2keto structures, Lys184 forms a hydrogen bond with Cys154 when it is not carbamylated. This is an interesting interaction given the possible importance of a Lys–Cys pair in the homologous enzyme PC. Chicken liver PC is inactivated by o-phthalaldehyde, which crosslinks Lys–Cys pairs in a pH-dependent manner (Werneburg and Ash, 1993). The PC Lys–Cys pair has been proposed to stabilize the enol form of biotin, and proton transfer between pyruvate and biotin is speculated to be facilitated by the cysteine's thiol group (Attwood and Cleland, 1986; Attwood, 1995). The 5S Lys184 corresponds to Lys741 in both human and chicken PC (Figure 4A). 5S Cys154 is not conserved in the PC enzymes, which have a serine residue instead, but in our PC homology model the nearby Cys739 presents itself as the likely partner for Lys741 in the o-phthalaldehyde crosslinking.
To investigate the importance of Lys184 and Cys154 in 5S catalytic function, we have prepared Lys184Glu, Lys184Ala, and Cys154Ala mutants. Both mutants with Lys184 replaced had no detectable enzymatic activity, while the Cys154Ala mutant had modestly reduced specific activity (up to 50% of wild type; Table III). It is clear that Cys154 probably does not play a direct and crucial role in catalysis, and that its likely function is to stabilize Lys184 when noncarbamylated in a conformation such that it does not interfere with substrate or product binding. In contrast, Lys184 is unambiguously indispensable for 5S catalytic activity. This may well be due to its ability to be carbamylated during the catalytic cycle and this intriguing hypothesis will be tested in future mechanistic studies.
The 5S substrate and cobalt-binding sites, lysine carbamylation, and thus catalytic mechanism are expected to be similar in PC. Lys184, the cobalt ligands, and residues involved in substrate or product binding in 5S are all conserved in PC. The similar functional consequences of PC disease mutations reproduced in 5S, and the correlation of the decreased catalytic activity of these mutants with their corresponding crystal structures, further emphasize the high conservation of 5S and PC active sites and the value of 5S as a model system for PC. By yielding comprehensive details of active site structure and binding, these 5S crystal structures provide significant new insight into catalytic mechanism, for not only itself but also eukaryotic biotin-dependent enzymes such as human PC.
Materials and methods
Protein expression and purification
The sequence of the TC 5S subunit from the P. shermanii strain used here differs from the sequence originally reported (Thorton et al, 1993) and has been deposited in the EMBL database (ID AJ606310). Due to cloning artifacts in the pETBlue-2 plasmid and a C-terminal His6 tag, the 505-residue 5S was expressed as a 539-residue polypeptide. The additional residues, 11 inserted after the N-terminal Met (AISRQLVDPNS) and 23 at the C-terminus (TRASQPELAPEDPEDLEHHHHHH), did not affect enzyme activity. 5S was expressed and purified as described previously (Xie et al, 1993; Hall et al, 2004). Briefly, Escherichia coli Tuner(DE3)pLacI cells transformed with the 5S construct were grown in LB medium at 310K until an OD600 of 0.6. Following induction with 1 mM IPTG and addition of 0.1 mM CoCl2 and 0.1 mM ZnCl2, incubation continued for 10–12 h. Cells were harvested by centrifugation and disrupted by passage through a French press. The partially purified lysate was subjected to nickel affinity and size exclusion columns to obtain pure dimeric 5S that was then concentrated to 10 mg/ml in 10 mM Hepes, pH 7.0, 0.1 mM PMSF, 0.1 mM EDTA, and 1 mM DTT for crystallization. SeMet-5S was prepared with the same expression system by inhibition of methionine biosynthesis (van Duyne et al, 1993; M Simmons, personal communication).
Site-directed mutagenesis
The 5S gene in pETBlue-2 plasmid was mutated using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. 5S Met186Ile and Ala59Thr mutants based on the PC Met743Ile and Ala610Thr disease mutations were prepared using the primers 5′-ATCGCCCTGAAGGACATCGCCGCCCTGCTCAAG-3′ and 5′-GAGTGTTGGGGTGGTACCACGTATGACTCGTG-3′, respectively. Active site mutants were prepared with the following primers:
5′-GCGCAGGGCACCATTGCCTACACGATCAGCCCG-3′ for Cys154Ala,
5′-GATTCCATCGCCCTGGCGGACATGGCCGCCCTG-3′ for Lys184Ala, and
5′-GATTCCATCGCCCTGGAGGACATGGCCGCCCTG-3′ for Lys184Glu.
Activity assay
TC activity was assayed in the forward direction as described previously (Wood et al, 1969; Xie et al, 1993). Reconstitution of the recombinant 12S, 5S, and 1.3S subunits to form active TC was achieved by incubating purified proteins in a subunit ratio of 1:6:12 on ice in 0.5 mM potassium phosphate buffer, pH 6.5. A coupled assay using malate dehydrogenase detected the formation of oxaloacetate by spectrophotometrically monitoring the decrease in NADH absorbance. The assay mixture contained the following in a total volume of 300 μl: 2.14 μmol pyruvate, 4.5 U malate dehydrogenase, 0.09 μmol NADH, 90 nmol MMCoA, 75 μmol potassium phosphate buffer, pH 6.5, and the assembled enzyme. Relative specific activities are expressed as oxaloacetate formed in the forward reaction in μmol/min/mg of 5S.
Crystallization, data collection, and processing
Wild-type and mutant (5S-A59T and 5S-M186I) 5S crystals of space group C2221 were grown by vapor diffusion at 293K using equal volumes of the protein sample and well solution containing 18–32% PEG 4K and 0.1 M Tris, pH 7.0 or 7.3. Crystals were cooled by dunking in liquid nitrogen after stabilizing in cryoprotectant containing 34–35% w/v PEG 4K. A potential platinum derivative of the SeMet-5S crystal form was prepared by soaking in 32% PEG 4K, 0.1 M Tris, pH 7.0, containing 10 mM K2PtCl4 for 10 min. Complex crystals were prepared by soaking wild-type 5S crystals in 32–34% PEG 4K, 0.1 M Tris, pH 7.0, containing 10 mM ligand (pyruvate, oxaloacetate, or 2-ketobutyrate at neutral pH) for 4–12 h. Both derivative and complex crystals were subjected to a 30 s back-soak in cryoprotectant before cooling.
Native diffraction data for a SeMet-5S crystal and MAD data from a SeMet-5S-K2PtCl4 crystal were measured on NSLS beamline X25. Data for mutant and complex crystals were measured using either an in-house Rigaku R-AXIS IV imaging plate detector or a Bruker Proteum/R CCD detector, mounted on a rotating copper anode source. All data were processed with HKL (Otwinowski and Minor, 1997), except for data measured on the Bruker system, which were processed with vendor software (Table I).
Structure determination and refinement
The first structure solved was of SeMet-5S (5S) by MAD phasing methods with SOLVE (Terwilliger and Berendzen, 1999), locating 23 out of a possible 24 Se atoms (the N-terminal methionine is disordered). Density modification and automated model building were performed with RESOLVE (Terwilliger 2000, 2002), and the model was refined against a higher-resolution data set collected at the Se anomalous peak wavelength. The isomorphous complex structures 5S-pyr, 5S-oxal, and 5S-2keto, and the two mutant structures 5S-A59T and 5S-M186I, were solved using the free 5S protein coordinates (Table I). For each structure, iterative cycles of model building with O (Jones et al, 1991) and refinement calculations with CNS (Brünger et al, 1998) were carried out until convergence. Five percent of the reflections were used for Rfree calculations, and DDQ (van den Akker and Hol, 1999) was employed to identify local model errors and solvent molecules. Ramachandran analysis using PROCHECK (Laskowski et al, 1993) showed only one residue, in the lowest resolution structure (5S-M186I), in a disallowed region. The final refined crystal structures each include 471–472 residues for one 5S monomer, one cobalt atom, one ligand molecule (for complex structures), and a range of water molecules (Table I). The second half of the 5S dimer is related by crystallographic symmetry. The cobalt ion corresponded to the highest peak of anomalous difference density calculated with diffraction data measured using an in-house source, for example, a 15.5σ anomalous difference peak is found at the cobalt position in the 5S-oxal crystal. The presence of cobalt in 5S crystals was confirmed by ICP-MS analysis; in contrast, neither manganese nor zinc could be detected in the 5S crystals (University of Missouri-Columbia Research Reactor Center).
Homology modeling and structural analysis
A homology model for the carboxyltransferase region of human pyruvate carboxylase was constructed using an alignment of its sequence with 5S and the InsightII software package (MSI). Surface area, charge, and hydrophobicity calculations were carried out with MSCON (Connolly 1983) and GRASP (Nicholls et al, 1991). Molecular figures were generated using MOLSCRIPT (Kraulis, 1991), BOBSCRIPT (Esnouf, 1999), and Raster3D (Merritt and Bacon, 1997).
Coordinates and structure factors
Coordinates and structure factors for the 5S structures have been deposited with the RCSB PDB under the following accession codes: free 5S 1RQB, 5S-pyruvate 1RQH, 5S-oxaloacetate 1RQE, 5S-2-ketobutyrate 1RR2, 5S-M186I 1U5J, and 5S-A59T 1S3H.
Acknowledgments
Diffraction data were measured on beamline X25 of the National Synchrotron Light Source, Brookhaven National Laboratory, supported by the US Department of Energy and National Institutes of Health. We thank F van den Akker and R Hartmann for many helpful discussions, A Miketa for technical assistance, and Michael Becker for beamline support. PRH was partially supported by NIH training grant GM08803. Research in the VCY and PRC laboratories is funded by NSF MCB0401654 and NIH DK053053, respectively.
References
- Ahmad F, Lygre DG, Jacobson BE, Wood HG (1972) Transcarboxylase: XII. Identification of the metal-containing subunits of transcarboxylase and stability of the binding. J Biol Chem 247: 6299–6305 [PubMed] [Google Scholar]
- van den Akker F, Hol WGJ (1999) Difference density quality (DDQ): a method to assess the global and local correctness of macromolecular crystal structures. Acta Crystallogr D 55: 206–218 [DOI] [PubMed] [Google Scholar]
- Atkin BM, Utter MF, Weinberg MB (1979) Pyruvate carboxylase and phosphoenolpyruvate carboxykinase activity in leucocytes and fibroblasts from a patient with pyruvate carboxylase deficiency. Pediatr Res 13: 38. [DOI] [PubMed] [Google Scholar]
- Attwood PV (1995) The structure and mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol 27: 231–249 [DOI] [PubMed] [Google Scholar]
- Attwood PV, Cleland WW (1986) Decarboxylation of oxalacetate by pyruvate carboxylase. Biochemistry 25: 8191–8196 [DOI] [PubMed] [Google Scholar]
- Banner DW, Bloomer AC, Petsko GA, Phillips DC, Pogson CI, Wilson IA, Corran PH, Furth AJ, Milman JD, Offord RE, Priddle JD, Waley SG (1975) Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 Å resolution using amino acid sequence data. Nature 255: 609–614 [DOI] [PubMed] [Google Scholar]
- Bardin RE, Taylor BL, Osohashi I (1975) Structural properties of pyruvate carboxylase from chicken liver and other sources. Proc Natl Acad Sci USA 72: 4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning MM, Kuo JM, Raushel FM, Holden HM (1995) Three-dimensional structure of the binuclear metal center of phosphotriesterase. Biochemistry 34: 7973–7978 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chapman MS, Suh SW, Curmi PM, Cascio D, Smith WW, Eisenberg DS (1988) Tertiary structure of plant RuBisCO: domains and their contacts. Science 241: 71–74 [DOI] [PubMed] [Google Scholar]
- Connolly ML (1983) Analytical molecular surface calculation. J Appl Crystallogr 16: 548–558 [Google Scholar]
- Coppel RL, McNeilage LJ, Surh CD, van de Water J, Spithill TW, Whittingham S, Gershwin ME (1988) Primary structure of the human M2 mitochondrial autoantigen of primary biliary cirrhosis: dihydrolipoamide acetyltransferase. Proc Natl Acad Sci USA 85: 7317–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duyne GD, Standaert RF, Karplus PA, Schreiber L, Clardy J (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol 229: 105–124 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D 55: 938–940 [DOI] [PubMed] [Google Scholar]
- Farber GK, Petsko GA (1990) The evolution of α/β barrel enzymes. Trends Biochem Sci 15: 228–234 [DOI] [PubMed] [Google Scholar]
- Fung C-H, Mildvan AS, Leigh JS Jr (1974) Electron and nuclear magnetic resonance studies of the interaction of pyruvate with transcarboxylase. Biochemistry 13: 1160–1169 [DOI] [PubMed] [Google Scholar]
- Gokulan K, Rupp B, Pavelka M, Jacobs W, Sacchettini J (2003) Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis. J Biol Chem 278: 18588–18596 [DOI] [PubMed] [Google Scholar]
- Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S (2001) Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc Natl Acad Sci USA 98: 14280–14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall PR, Wang YF, Rivera-Hainaj RE, Zheng X, Pustai-Carey M, Carey PR, Yee VC (2003) Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core. EMBO J 22: 2334–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall PR, Zheng R, Pusztai-Carey M, van den Akker F, Carey P, Yee V (2004) Expression and crystallization of several forms of the Propionibacterium shermanii transcarboxylase 5S subunit. Acta Crystallogr D 60: 521–523 [DOI] [PubMed] [Google Scholar]
- Ho L, Patel MS (1990) Cloning and cDNA sequence of the β-subunit component of human pyruvate dehydrogenase complex. Gene 86: 297–302 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res 26: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri E, Carr MB, Hausinger RP, Karplus PA (1995) The crystal structure of urease from Klebsiella aerogenes. Science 268: 998–1004 [PubMed] [Google Scholar]
- Jitrapakdee S, Wallace JC (1999) Structure, function and regulation of pyruvate carboxylase. Biochem J 340: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kai Y, Matsumura H, Inoue T, Terada K, Nagara Y, Yoshinaga T, Kihara A, Tsumura K, Izui K (1999) Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition. Proc Natl Acad Sci USA 96: 823–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, Oliveira MA, Coffino P, Hackert ML (1999) Structure of mammalian ornithine decarboxylase at 1.6 Å resolution: stereochemical implications of Plp-dependent amino acid decarboxylases. Struct Fold Des 7: 567–581 [DOI] [PubMed] [Google Scholar]
- Knight S, Andersson I, Branden C-I (1989) Reexamination of the three-dimensional structure of the small subunit of RuBisCo from higher plants. Science 244: 702–705 [DOI] [PubMed] [Google Scholar]
- Knowles JR (1989) The mechanism of biotin-dependent enzymes. Ann Rev Biochem 58: 195–221 [DOI] [PubMed] [Google Scholar]
- Koike K, Urata Y, Matsuo S, Koike M (1990) Characterization and nucleotide sequence of the gene encoding the human pyruvate dehydrogenase α subunit. Gene 93: 307–311 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Kume A, Koyata H, Sakakibara T, Ishiguro Y, Kure S, Hiraga K (1991) The glycine cleavage system. Molecular cloning of the chicken and human glycine decarboxylase cDNAs and some characteristics involved in the deduced protein structures. J Biol Chem 266: 3323–3329 [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Lorimer GH (1979) Evidence for the existence of discrete activator and substrate sites for CO2 on ribulose-1,5-bisphosphate carboxylase. J Biol Chem 254: 5599–5601 [PubMed] [Google Scholar]
- Lorimer GH (1981) Ribulosebisphosphate carboxylase: amino acid sequence of a peptide bearing the activator carbon dioxide. Biochemistry 20: 1236–1240 [DOI] [PubMed] [Google Scholar]
- Lorimer GH, Miziorko HM (1980) Carbamate formation on the epsilon-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by CO2 and Mg2+. Biochemistry 19: 5321–5328 [DOI] [PubMed] [Google Scholar]
- Lundqvist T, Schneider G (1991) Crystal structure of the ternary complex of ribulose-1,5-bisphosphate carboxylase, Mg(II), and activator CO2 at 2.3 Å resolution. Biochemistry 30: 904–908 [DOI] [PubMed] [Google Scholar]
- Manjasetty AB, Powlowski J, Vrielink A (2003) Crystal structure of a bifunctional aldolase-dehydrogenase: sequestering a reactive and volatile intermediate. Proc Natl Acad Sci USA 100: 6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maveyraud L, Golemi D, Kotra LP, Tranier S, Vakulenko S, Mobashery S, Samama JP (2000) Insights into class D β-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Struct Fold Des 8: 1289–1298 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. In Methods in Enzymology, Carter Jr CW, Sweet RM (eds), Vol. 277, Part A, pp 505–524. California: Academic Press [DOI] [PubMed] [Google Scholar]
- Moss J, Lane MD (1971) The biotin-dependent enzymes. Adv Enzymol Relat Areas Mol Biol 35: 321–442 [DOI] [PubMed] [Google Scholar]
- Murphy JV, Isohashi F, Weinberg MB, Utter MT (1981) Pyruvate carboxylase deficiency—an alleged biochemical cause of Leigh's disease. Pediatrics 88: 401–404 [PubMed] [Google Scholar]
- Nagano N, Orengo CA, Thorton JM (2002) One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol 321: 741–765 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp K, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Northrop DB, Wood HG (1969) Transcarboxylase. V. The presence of bound zinc and cobalt. J Biol Chem 244: 5801–5807 [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Carter Jr CW, Sweet RM (eds), Vol. 276, Part A, pp 306–326. California: Academic Press [DOI] [PubMed] [Google Scholar]
- Robinson BH (2001) Lactic acidemia: disorders of pyruvate carboxylase and pyruvate dehydrogenase. In Metabolic and Molecular Basis of Inherited Disease, Scriver CR, Sly WS, Childs B, Beaudet AL, Valle D, Kinzler KW, Vogelstein B (eds), 8th edn., pp 2275–2295. New York: MacGraw-Hill [Google Scholar]
- Robinson BH, Oei J, Sherwood WG, Applegarth D, Wong L, Haworth J, Goodyer P, Casey R, Zaleski LA (1984) The molecular basis for the two different clinical presentations of classical pyruvate carboxylase deficiency. Am J Hum Genet 36: 283–294 [PMC free article] [PubMed] [Google Scholar]
- Samols D, Thornton CG, Murtif VL, Kumar GK, Hase FC, Wood HG (1988) Evolutionary conservation among biotin enzymes. J Biol Chem 263: 6461–6464 [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum likelihood density modification. Acta Crystallogr D 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2002) Automated main-chain model-building by template-matching and iterative fragment extension. Acta Crystallogr D 59: 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorton CG, Kumar GK, Shenoy BC, Haase FC, Phillips NF, Park VM, Magner WJ, Hejlik DP, Wood HG, Samols D (1993) Primary structure of the 5S subunit of transcarboxylase as deduced from the genomic DNA sequence. FEBS Lett 330: 191–196 [DOI] [PubMed] [Google Scholar]
- de Vivo D, Haymond MW, Leckie MP, Bussmann YL, McDougal DB, Pagliara AS (1977) Clinical and biochemical implications of pyruvate carboxylase deficiency. J Clin Endocrinol Metab 45: 1281–1296 [DOI] [PubMed] [Google Scholar]
- Wallace JC, Easterbrook-Smith SB (1985) Substrate binding to pyruvate carboxylase subunits. In Pyruvate Carboxylase, Keech DB, Wallace JC (eds), p 65 FL, Boca Raton: CRC [Google Scholar]
- Werneburg BG, Ash DE (1993) Chemical modifications of chicken liver pyruvate carboxylase: evidence for essential cysteine–lysine pairs and a reactive sulfhydryl group. Arch Biochem Biophys 303: 214–221 [DOI] [PubMed] [Google Scholar]
- Wexler ID, Du Y, Lisgaris MV, Mandal SK, Freytag SO, Yang B-S, Liu T-C, Kwon M, Patel MS, Kerr DS (1994) Primary amino acid sequence and structure of human pyruvate carboxylases. Biochim Biophys Acta 1227: 46–52 [DOI] [PubMed] [Google Scholar]
- Whitby FG, Phillips JD, Kushner JP, Hill CP (1998) Crystal structure of human uroporphyrinogen decarboxylase. EMBO J 17: 2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga RK (2001) The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett 492: 193–198 [DOI] [PubMed] [Google Scholar]
- Wise E, Yew WS, Babbitt PC, Gerlt JA, Rayment I (2002) Homologous (β/α)8-barrel enzymes that catalyze unrelated reactions: orotidine 5′-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase. Biochemistry 41: 3861–3869 [DOI] [PubMed] [Google Scholar]
- Wood HG, Barden RE (1977) Biotin enzymes. Ann Rev Biochem 46: 385–413 [DOI] [PubMed] [Google Scholar]
- Wood HG, Jacobson B, Gerwin BI, Northrop DB (1969) Oxalacetate transcarboxylase from Propionibacterium. Meth Enzymol 8: 215–230 [Google Scholar]
- Wood HG, Kumar GK (1985) Transcarboxylase: its quaternary structure and the role of the biotinyl subunit in the assembly of the enzyme and in catalysis. Ann NY Acad Sci 447: 1–21 [DOI] [PubMed] [Google Scholar]
- Wood HG, Zwolinski GK (1976) Transcarboxylase: role of biotin, metals, and subunits in the reaction and its quaternary structure. CRC Crit Rev Biochem 4: 47–122 [DOI] [PubMed] [Google Scholar]
- Wrigley NG, Chiao J-P, Wood HG (1977) Electron microscopy of the large form of transcarboxylase with six attached subunits. J Biol Chem 252: 1500–1504 [PubMed] [Google Scholar]
- Wu N, Mo Y, Gao J, Pai EF (2000) Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc Natl Acad Sci USA 97: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Shenoy BC, Magner WJ, Hejlik DP, Samols D (1993) Purification and characterization of the recombinant 5 S subunit of transcarboxylase from Escherichia coli. Protein Expr Purif 4: 456–464 [DOI] [PubMed] [Google Scholar]


