Abstract
Gas chromatography-mass spectrometry (GC/MS) has long been considered one of the premiere analytical tools for small molecule analysis. Recently, a number of GC/MS systems equipped with high-resolution mass analyzers have been introduced. These systems provide analysts with a new dimension of information – accurate mass measurement to the third or fourth decimal place; however, existing data processing tools do not capitalize on this information. Beyond that, GC/MS spectral reference libraries, which have been curated over the last several decades, contain almost exclusively unit resolution MS spectra making integration of accurate mass data dubious. Here we present an informatic approach, called High-Resolution Filtering (HRF), which bridges this gap. During HRF, high-resolution mass spectra are assigned putative identifications through traditional spectral matching at unit resolution. Once candidate identities have been assigned, all unique combinations of atoms from these candidate precursors are generated and matched to m/z peaks using narrow mass tolerances. The total amount of measured signal that is annotated is used as a metric of plausibility for the presumed identification. Here we demonstrate that the HRF approach is both feasible and highly specific towards correct identifications.
INTRODUCTION
Gas chromatography-mass spectrometry (GC/MS) is a premiere analytical tool for small molecule analysis1–3. Highly reproducible chromatographic separations combined with conserved molecular fragmentation lend this technique to both targeted and discovery assays, and has become particularly useful in the area of metabolite profiling4,5. Since the metabolome is closest to phenotype, metabolic profiling has great potential to propel biomedical research and is quickly emerging as a field of interest for both systems biologists and clinical researchers6,7. The ability to rapidly and comprehensively monitor metabolites will doubtless facilitate basic research into disease pathogenesis and also provide new opportunities for disease diagnosis. Moreover, metabolite screens are highly desirable in the clinical setting as they often rank among the least invasive biological assays. As an emergent field there is critical need for the development of advanced tools and technologies to enable deeper small molecule profiling in shorter time spans.
In traditional discovery experiments, volatile analytes are separated by GC and ionized using electron ionization (EI) prior to mass analysis. EI is a “hard” ionization technique and causes molecules to fragment in characteristic patterns. Spectra containing fragments from individual analytes, which may or may not include an intact molecular ion, are extracted and then compared to databases of unit-resolution reference spectra8. Matches with sufficiently high spectral similarity are often presumed to be correct identifications. Identifying all of the observed spectral features resulting from a GC/MS experiment is a formidable challenge,9,10 so often the majority of features often remain unidentified. For those compounds where putative identifications have been assigned, subsequent validation often necessitates analysis of a pure reference standard. This process is laborious, especially when considering that for many spectral features there exist a large number of putative identifications. As such, any auxiliary information which can be used to discriminate between candidate precursors is highly valuable11.
Unit resolution GC/MS instruments have been, and continue to be, the most widespread and commonly used mass spectrometers in the world. Given that, the largest publically available reference libraries are comprised of spectra exclusively acquired on these systems12,13. In the last few years, however, several GC/MS systems possessing mass analyzers capable of high-resolution and accurate mass measurement have become commercially available – i.e., time-of-flight and, most recently, Orbitrap. Despite these exciting technological advances and their potential impact on metabolomic research, data analysis tools have remained largely unchanged14–17. We conclude that, if coupled with novel informatic capability, this new generation of GC/MS systems offers considerable opportunity to drive small molecule discovery. This nascent promise is reminiscent of the revolution that occurred in LC/MS-based proteomics following the introduction of high-resolution/accurate mass measurement. In this case, existing peptide-spectral matching algorithms were easily adapted to achieve a concomitant reduction in search space while affording increased precursor/product ion matching specificity; unfortunately, leveraging the specificity enabled by accurate mass GC/MS data with existing small molecule algorithms is not straightforward. The major EI reference databases comprise unit resolution spectra, precluding the ability to directly compare measured exact masses against their reference counterparts. An alternative route is to generate theoretical EI spectra in silico, though this has proven to be exceptionally challenging18–20. Of course, another approach is to generate new accurate mass libraries which would ostensibly allow for increased discrimination against spurious matches as fragments which are nominally the same but not equivalent within a narrow mass tolerance would no longer be matched. This increased specificity in spectral matching would hopefully make it easier to identify correct matches. Generating new reference databases is an admirable goal but one that will take years, if not decades, to achieve given that current spectral libraries have been compiled over the past fifty years from hundreds of thousands of individual analyses.
Here we describe a new approach to harness the existing unit resolution EI mass spectral databases while simultaneously exploiting the accurate mass measurement capabilities of high-resolution GC/MS systems. In this method accurate mass GC/MS data is searched via spectral matching to existing unit resolution EI spectral libraries as normal. Next, EI-MS top scoring putative identifications are tested for plausibility based on comparison of the experimentally measured accurate mass fragments to combinatorially generated theoretical fragments constrained by the atomic composition of the assigned precursor. This method avoids the pitfalls of theoretical EI spectral prediction by simply generating and testing all possible combinations of atoms, as theoretical fragments, in a precursor. We demonstrate that although this method makes minimal approximations it remains highly specific toward correct precursor identifications. By enabling discrimination between candidate molecular precursors on the basis of both measured fragmentation profiles and accurate mass, this method effectively bridges the current technology gap between high-resolution spectral acquisition and unit resolution mass spectral libraries.
EXPERIMENTAL SECTION
Materials and Reagents
Unless otherwise specified all standard reference materials were purchased from Sigma-Aldrich (St. Louis, MO) with the exception of the 37 pesticide reference standards analyzed which were contained in the Organonitrogen Pesticide Mix #1 – EPA Method 525.2 and purchased from Restek (Bellefonte, PA). Methanol, ethyl acetate, acetone, hexane, dichloromethane, and isopropyl alcohol reagents were also purchased from Sigma-Aldrich. The N-methyl-N-trimethylsilytrifluoroacetamide with 1% trimethylchlorosilane derivatization reagent (MSTFA + 1% TMCS) was purchased from Pierce Biotechnology (Rockford, IL). Compressed gases (methane, helium, and nitrogen) were ultrahigh purity grade and purchased from Airgas (Madison, WI). 200 mg Clean Screen® Extraction Columns were purchased from United Chemical Technologies (Bristol, PA).
Sample Preparation and GC/MS Acquisition
Stock solutions of the reported standards were prepared individually at a concentration of 1 mg/mL in appropriate solvents. Standards were processed in batches containing ~5-10 individual analytes. The EPA 525.2 pesticide mixture was diluted from 500 μg/mL to a working concentration of 3 ng/μL in acetone prior to mass spectral analysis. For the drug-spiked urine experiments, stock solutions of all drugs were first prepared at 1 mg/mL in methanol. These stock solutions were combined and diluted (again in methanol) to appropriate concentrations. For each gradient data point, 100 μL of the drug mixture was added to raw urine prior to extraction using the 200 mg Clean Screen extraction columns. Acidic and basic drug/metabolite fractions were extracted according to manufacturer protocols.21 Yeast metabolites were extracted by first washing cultured cells with buffered saline and submerging into a precooled 1.5 mL plastic tube containing 2:2:1 acetonitrile/methanol/H2O mixture. For all materials (not including the pesticide mixture) 25 μL aliquots were resuspended in 25 μL of pyridine and vortexed. 25 μL of N-methyl-N-[trimethylsilyl]trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) was added and samples were incubated at 60° C for 30 minutes. All samples were analyzed using a GC/MS instrument comprising a Trace 1310 GC coupled to a Q Exactive Orbitrap mass spectrometer. For the yeast metabolite extracts a linear temperature gradient ranging from 50 °C to 320 °C was employed spanning a total runtime of 30 minutes. Analytes were injected using a 1:10 split at a temperature of 275 °C and ionized using electron ionization (EI). The mass spectrometer was operated in full scan mode using a resolution of 30,000 (m/Δm) relative to 200 m/z. Instrumental parameters and specifications for all other experiments are provided in the Supporting Information. All MS experiments utilized Automatic Gain Control (AGC)22–24 and all data was acquired in profile mode.
GC/MS Data Processing
All GC/MS data processing was done using in-house algorithms designed to facilitate spectral deconvolution, spectral matching against a unit resolution reference database, and high-resolution filtering. The details of each algorithmic component are described at length in the Supporting Information. Briefly, following mass spectral acquisition deconvolved spectra were extracted from raw data files. A pseudo-unit resolution copy of each spectrum was made by combining the intensities of peaks falling in the same nominal mass range, setting the measured m/z to the nearest integer value, and normalizing peak intensities relative to the base peak (set to 999). All 212,961 unit resolution reference spectra in the NIST 12 MS/EI Library were exported to a .JDX file through the NIST MS Search 2.0 program and converted to a format suitable for matching against acquired Q Exactive CG spectra. Extracted spectra were submitted for database searching and spectral similarity was measured using the following dot product equation:
Following candidate identification retrieval the high-resolution filtering algorithm was employed by first generating all unique atomic combinations from a given precursor using the most abundant isotope of each considered atom. Starting with the smallest measured m/z peak, peaks were matched to theoretical fragments using a narrow ±10 ppm tolerance centered around the m/z value. To account for isotopic clusters a variant of each matched theoretical fragment was created containing substituted heavy isotopes was placed back on the list of all candidate theoretical fragments. This process was repeated until every measured peak in a given spectrum had been considered. The total amount of measured signal that could be annotated as calculated by:
was returned in the form of an HRF score.
RESULTS AND DISCUSSION
The HRF method is founded on one central tenet – all m/z peaks in a pure EI spectrum are derived from a single molecular precursor and, therefore, contain a subset of the atoms from the molecular precursor. For example, the EI mass spectrum of 3-methyl-3-hexanol (C7H16O) exhibits prominent features at m/z values 73, 87, and 10125. Expert annotation of this spectrum revealed the chemical identity of these fragments as C4H9O, C5H11O, and C6H13O, respectively25. Note each of these formulas is a subset of the molecular precursor, supporting our guiding supposition. Without regard for chemical structure feasibility, there are 271 unique atomic combinations of the parent molecule C7H16O. First consider the m/z peak at 73, only three of these combinations have this nominal mass – C6H, C5H13, and C4H9O; however, only one (C4H9O) has an exact mass within ±10 ppm of the correct annotation. Such is also the case for the other expertly annotated fragments. Given that we can now routinely measure all m/z peaks in an EI mass spectrum with low ppm mass accuracy, we implement this annotation strategy on a large scale.
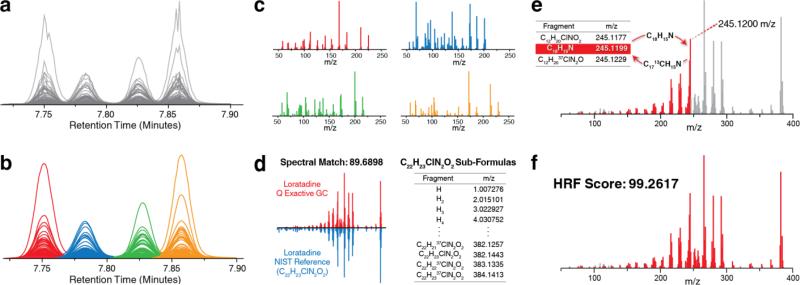
Figure 1 presents an outline of the HRF workflow. The process consists of three main steps: deconvolution, spectral matching, and high-resolution filtering. Spectral deconvolution is a standard part of processing GC/MS data; however, accurate mass is highly advantageous as it reduces, or eliminates, interference between nominally isobaric fragments. Extracted spectral features are subsequently grouped based on corresponding elution apex and a spectrum containing only fragments arising from a singular precursor is derived from each group (Figure 1a-c). Next, by rounding accurate mass m/z peaks to the nearest integer value, a pseudo-unit resolution copy of each spectrum is created and then submitted for spectral matching against a unit resolution reference database. The intent is to retrieve candidate identifications based on spectral similarity. These steps represent a traditional workflow for spectral assignment in a discovery-based GC/MS experiment. In the HRF method, this workflow is further augmented to leverage accurate mass for discrimination between putative identifications.
Figure 1.
High-resolution filtering workflow with spectral matching. (a) Peaks observed across consecutive scans are condensed into data features. (b) Features are smoothed and grouped based on elution apex. All features within a group are assumed to arise from a singular precursor. (c) Individual spectra are derived from feature groups (using average m/z and apex intensity) and can then be submitted for spectral matching. (d) A strong spectral match of an experimentally-derived spectrum of loratadine against the corresponding NIST reference spectrum. All sub-formulas from C22H23ClN2O2 are generated and sorted by exact formula mass less an electron. (e) Sub-formulas are matched to peaks in ascending order based on m/z. For each matched theoretical fragment a variant containing appropriate heavy isotopes is created and placed into the list of sub-formulas in sorted-order. (f) For the high-resolution spectrum of loratadine 99.2617% of the measured ion current can be annotated with a sub-formula of C22H23ClN2O2.
The HRF method attempts to annotate every measured m/z peak in an EI mass spectrum using some combination of atoms from a putative precursor's chemical formula. The amount of total ion current that can be successfully annotated can be used as metric of confidence in that putative identification. Figure 1 d-f illustrates the HRF strategy using an EI mass spectrum of loratadine, a popular over-the-counter antihistamine, collected using a Q Exactive GC mass spectrometer. A unit resolution database search, returns a reference spectrum of loratadine as a strong candidate match. To evaluate the quality of this putative identification we next employ the HRF strategy. With the chemical formula of loratadine (C22H23ClN2O2) all non-repeating combinations of atoms (i.e., sub-formulas) are generated and ordered by ascending exact mass less an electron (Figure 1d). Note that the theoretical fragment search space is restricted by the atomic composition of loratadine. Starting with the smallest measured m/z peak, sub-formulas are matched based on exact mass. To accommodate isotopic clusters present in spectra, a variant containing an appropriate number of heavy isotopes is created for each matched theoretical fragment and placed back onto the list of sub-formulas. For example, once the highlighted m/z peak at 245.1200 is matched to C18H15N (theoretical m/z: 245.1199) a formula containing a substituted 13C isotope (C1713CH15N) is added to the list of candidate sub-formulas (Figure 1e). This strategy of onthe-fly theoretical isotopic fragment generation enables annotation of non-monoisotopic fragments without unduly increasing sub-formula search space. Once every m/z peak in the spectrum has been considered the total percentage of measured ion current that has been annotated is returned in the form of a HRF score. In the example case of loratadine we find that 99.2617% of all measured ion current can be annotated using a sub-formula of its true parent precursor (Figure 1f). Here we demonstrate that the HRF method is viable, enables discrimination between putative identifications, is highly robust even in times of diminished signal-to-noise, and is uniquely enabled with high-resolution GC/MS. Finally, we establish that this approach stands to greatly improve how unknowns are identified in discovery-based analyses.
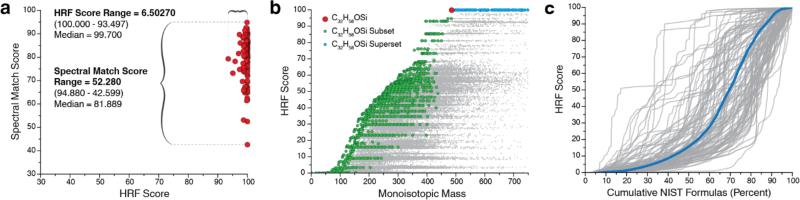
Reference Standard Analysis. To ensure broad utility we benchmarked performance of the HRF algorithm as applied to spectra collected from a diverse array of small molecules. For this work, a data set of high-resolution Q Exactive GC spectra collected from 105 pure reference standards covering many classes of small molecules including metabolites, pesticides, pharmaceuticals, drugs of abuse, among others, was constructed. Following GC/MS analysis of all reference standards, individual spectra were extracted from raw data files using the described deconvolution algorithm. Each extracted spectrum was compared against its corresponding NIST reference spectrum and a weighted dot product score was calculated to measure spectral similarity. For these 105 spectra, a median spectral match score of 81.889, minimum of 42.599, and standard deviation of 9.587 was achieved. Following spectral matching, each spectrum was then subjected to our HRF approach using the chemical formula of the true parent molecule. Considering all spectra in the data set, we report a median HRF score of 99.700, minimum of 93.497, and standard deviation of 1.022 (Fig. 2a. and Supplementary Table 1). From these data we conclude that performance of the HRF method is well-conserved across many different classes of small molecules. Next we wondered whether similar results could be obtained from other chemical formulas in the reference library. To test specificity, 60,560 HRF scores – all from unique formulas residing in the NIST database – were calculated for each of the 105 spectra. Figure 2b presents the results of this experiment for the spectrum of trimethylsilyl-derivatized beta-sitosterol (C32H58OSi). Note the true parent chemical composition is the smallest formula that can produce the maximal HRF score. We were curious as to the scores generated by subset formulas (some but not all of the atoms contained within the precursor formula) as well as superset formulas (all of the atoms contained by the precursor and then some) which are also highlighted. The annotated subsets lack the proper combination of atoms to achieve the same score. Not surprisingly, all supersets of C32H58OSi produce similarly high scores. This is expected as all subformulas from the true parent will also be included in the subformula sets generated by these superset precursors. We note that in some cases very large formulas which are not true supersets but share a large percentage of atoms with the correct parent can also produce high scores (Supplementary Table 2).
Figure 2.
High-resolution filtering results. (a) Spectral match and HRF scores are shown for the 105 spectra in the dataset. (b) HRF scores for a spectrum of beta-sitosterol (TMS) using 60,560 different formulas are shown. The true parent (C32H58OSi) is shown in red. Sub- and supersets of C32H58OSi are shown in green and blue respectively. (c) Cumulative distributions of the 60,560 HRF scores calculated for all 105 spectra are shown in gray. A representative distribution generated by combining all results is shown in blue.
For a global view of the method's specificity we plot the cumulative distributions of HRF scores to all 105 spectra along with the average distribution of all cumulative distributions (Fig. 2c. and Supplementary Figure 1). Note that this analysis provides a worst-case scenario given all 60,560 formulas considered have an equal chance of being selected as a putative parent for an acquired spectrum. In most cases this is not the case, as either spectral matching or a priori information held by the analyst allows discrimination against the majority of these candidates. Still, these data reveal that on average ~86.9% of considered formulas will return a HRF score ≤ 90 and that only 3.560% of candidate formulas will produce a score greater than or equal to the median calculated HRF score (99.700). We also note that specificity is dependent on the complexity of the analyte in question, for example, increases in elemental complexity and atom count will often result in spectra which a smaller number of precursors can successfully annotate.
Urine Drug Testing. Most analytical applications demand the identification of low level analytes, often present within complex matrices. In these situations spectral quality is eroded – manifested by the loss of key diagnostic fragments with diminished signal and increased chemical noise – limiting the ability to correctly assign identifications through traditional spectral matching. To test the benefits of HRF in such situations we next analyzed a panel of drugs at varying concentration in a biological matrix. GC/MS is the ideal platform to test for drugs of abuse, pharmaceuticals, sports dopants, and their metabolites in human urine. These assays are highly desirable in the clinical setting as they are minimally invasive.
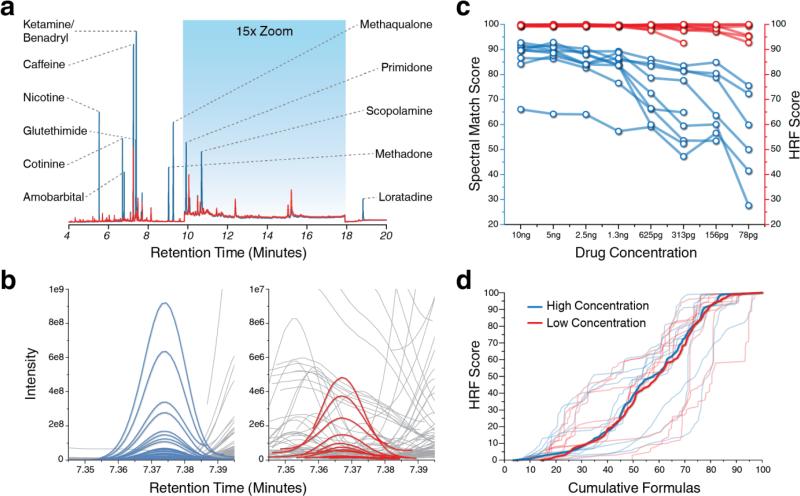
As a proof-of-concept, twelve drugs (amobarbital, Benadryl, caffeine, cotinine, glutethimide, ketamine, loratadine, methadone, methaqualone, nicotine, primidone, and scopoloamine) were spiked into human urine at eight concentations (10 ng/μL to ~78 pg/μL) and extracted prior to GC/MS analysis (Fig. 3a.). Chromatographic resolution was insufficient to separate Benadryl and ketamine, and high native levels of caffeine diminished the ability to analyze the compound through a range of concentrations. As such, further analysis was not carried out and here we report results for nine of the twelve drugs.
Figure 3.
Analysis of drugs spiked into human urine at variable concentration. (a) GC-MS TIC chromatograms from the most concentrated (blue) and least concentrated (red) spiked samples are shown. (b) Deconvolved feature groups for the drug Glutethimide at high (blue) and low (red) concentrations. Background features are shown in gray. (c) Spectral match and HRF scores for each drug analyzed at all concentrations where analyte abundance was sufficient to produce a spectrum. (d) Two spectra were isolated for each drug (one at the most concentrated point, the other at the least) and 55,290 HRF scores were calculated using unique formulas (0-500 Da) in the NIST database. Cumulative HRF results are shown for each drug using a spectrum acquired at high and low concentration (blue and red, respectively). A combined distribution is also shown for each population of drug spectra.
The analysis of compounds in a complex background matrix, such as urine, presents two considerable challenges – extracting high-quality spectra and assigning confident identifications – with the latter being highly dependent on the former. Ideally, extracted spectra should retain all fragment m/z peaks stemming from the eluting precursor while eliminating all other chemical background, which can be of higher abundance. Deconvolution is the core technique for spectral extraction and this process, as we report here, is considerably improved by use of the FT-MS systems. Accurate mass measurement largely eliminates interferences between nominally isobaric fragments and allows extraction of chromatographic profiles using narrow mass tolerances (~ ± 10 ppm). Furthermore, the rapid scan rate (> 18 Hz) provides sufficient temporal resolution enabling more precise detection of chromatographic apex. Note that spectral deconvolution assumes that all peaks are derived from a singular precursor and if two compounds completely overlap the resulting EI-MS spectrum will be chimeric which may impede spectral identification. Figure 3b highlights spectral deconvolution of glutethimide at high and low concentrations. Note the numerous co-eluting interferants in the low concentration chromatogram that are easily distinguished. We conclude that spectral deconvolution is a key parameter for successful downstream identification and is improved by collection of spectra with high-resolution and accurate mass.
Extraction of high-quality spectra from raw data files is only the first step in assigning confident identifications. Mapping these spectra to structure is then commonly done by spectral matching against a library, which is most effective when experimental spectra very closely resemble those contained in the library. The specificity of this approach, however, is reduced as analyte abundance decreases and diagnostic fragments fall below the limit of detection. We surmised that the HRF approach could provide an orthogonal metric, allowing greater discrimination between putative identifications.
To test this hypothesis we applied the HRF approach to analyze the standard drug compounds spiked into the urine matrix across a wide range of concentrations. We required that all spectra contain at least 10 m/z peaks, eliminating 5 of 72 data points (Supplementary Figure 2). In these instances the compound in question was at a sufficiently reduced concentration such that the extracted spectrum was either non-existent or of too low quality for any further processing. Extracted spectra were then compared to their corresponding NIST reference spectrum to generate both spectral match and HRF scores for each (Figure 3c, Supplementary Table 3). As expected, the spectral match score decreases with diminishing analyte abundance, primarily due to the loss of low abundance peaks at decreased concentrations. HRF performance, however, is remarkably consistent, independent of analyte concentration, and remains high (> 92) for all observed spectra. From these data we draw two primary conclusions: First, FT-MS mass analyzers provide robust mass accuracy measurements, even for signals occurring at low S/N26; and second, unlike the conventional spectral matching strategy, the HRF scoring metric is conserved across a wide range of analyte concentrations.
While the experiment described above demonstrates strong HRF scoring performance, we wondered whether the method would maintain the ability to discriminate between candidate precursors, when provided with lower quality spectra. To determine if the HRF scoring method had diminished specificity for spectra containing a reduced number of diagnostic m/z peaks, i.e., those collected at lower abundance, we calculated HRF scores from 55,290 unique formulas in the NIST spectral library (0-500 Da) using two EI-MS spectra for each drug analyzed (one from the most concentrated data point, the other from the least). These high and low concentration spectra present a striking spectral quality difference as the low abundance spectra contain only about 25% of the m/z peaks found in the higher quality analog (23 v. 96, on average). Figure 3d presents the cumulative distributions of these calculated HRF scores for either high (blue) or low (red) concentration spectra (Supplementary Figure 3). The average distribution for each set of spectra is also displayed and no difference is readily observed. It is apparent that, whether analyzing low or high quality spectra, HRF specificity is maintained. The fundamental driving force for this indifference to spectral quality, as compared to traditional spectral matching, is the discriminatory power of mass accuracy which is retained even within low-quality spectra. Based on these data we surmise that the HRF strategy is less dependent upon input spectral quality – a characteristic that will propel the emergent area of small molecule discovery and profiling applications.
Application to Discovery Metabolomic Analyses. High chromatographic resolution, excellent sensitivity, and conserved fragmentation of molecular precursors render GC/MS a fitting method for discovery-based metabolic profiling. In recent years there has been a marked decrease in the time required to comprehensively sequence genomes, transcriptomes, and proteomes. These increases in throughput have largely come as a result of coincident improvements in instrumentation and informatics enabling faster sequencing than ever before. Discovery metabolomics has lagged behind these other “omics” technologies due in large part to the difficulty in assigning confident identifications to analyzed compounds. We assert that by coupling the recent advances in high-resolution GC/MS instrumentation with new data processing schemes, the depth and speed at which metabolomes can be fully characterized can be greatly increased. One approach to realizing this potential is to utilize the HRF approach as a data reduction strategy for eliminating spurious hits, and retaining only those which are chemically plausible.
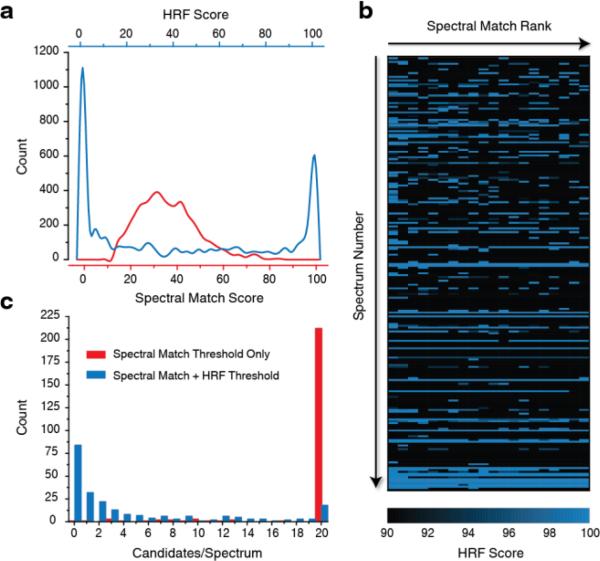
To characterize the utility of the HRF approach for metabolomic applications the algorithm was applied to a discovery analysis of a yeast metabolite extract. Here a TMS-derivatized yeast metabolome and solvent blank were analyzed on a Q Exactive GC system in tandem. Following data acquisition individual spectra were extracted from both raw files using the described in-house deconvolution algorithm. Spectral deconvolution yielded 19,367 spectral features which were placed into 554 feature groups – each group containing fragments which are assumed to stem from a singular precursor. Deconvolution results were manually validated and additional curation was employed where necessary (Supplementary Figure 4). EI-MS spectra that were common to both the yeast extract and solvent blank were eliminated from consideration. In total, 232 EI-MS spectra (all containing no fewer than 10 m/z peaks) were considered for this analysis, post background subtraction. These spectra were then searched against the NIST database (~ 213,000 compounds) at unit resolution. The 20 highest scoring spectral matches were returned and HRF scores were then calculated for each – generating 4,640 HRF scores in total. Figure 4a displays the distributions for both scores. The orthogonality between these two metrics is readily apparent. While the majority of spectral match scores cluster around 30-40 with a skew towards higher scores – again, this distribution represents the 20 best hits to each spectrum, many of which were derived from lowly abundant precursors - the HRF distribution is bimodal with large clusters at both extremes. These clusters (greater than 90 and less than 10) comprise 60.69% of all returned results. Supplementary Figure 5 highlights the uniqueness of matched chemical formulas, assigned HRF scores, and theoretical fragment annotations arising from these top 20 hits to each EI-MS spectrum.
Figure 4.
Discovery yeast metabolomic analysis. (a) Distributions of the top 20 spectral match/corresponding HRF scores to 232 spectra extracted from a yeast metabolomics experiment. (b) HRF scores corresponding to the 20 best spectral matches (left to right) for all 232 spectra (top to bottom) are shown in the blue heat map. The intensity of each pixel reflects the percentage of total ion current that can be annotated with an exact chemical formula. (c) Viable candidates/spectrum when applying spectral match and HRF score thresholds.
In the analysis of reference standards we observed no instances where a correct identification yielded spectral match or HRF scores lower than 20 or 90, respectively. To visualize these data we present a heat map (Figure 4b)displaying each EI-MS spectrum as a row with the calculated HRF score for each of the 20 putative spectral matching assignments as the columns. This plot reveals that top scoring spectral matches are not always consistent with the chemical formula information gleaned by the HRF calculation. We find that 76.00% of returned identifications are eliminated after applying an HRF threshold (90) including 58.62% of all number one spectral match hits. We also note many instances in which lower spectral match scores to a given spectrum yield higher HRF scores suggesting that joint consideration of both metrics is advantageous. To determine the value of the HRF method to eliminate from consideration incorrect putative assignments we plotted the number of candidate identifications per spectrum before and after application of HRF scoring (Fig. 4c). Imposing a spectral match score cutoff of 20, eliminates only 5.28% (245) of hits, leaving the analyst to sort through the remaining 4,240 candidates. Application of the HRF score threshold in addition to the spectral match score threshold, however, allows dismissal of the majority of the putative candidate identifications – 79.78% (3,720). In fact, the HRF method allows the analyst to reduce the number of viable candidate structures with confidence; for example, 65.09% of spectra retain three or fewer valid candidates. While analysts will still find it useful to confirm candidate identifications by sampling pure reference standards, the tremendous reduction in candidate identifications will expedite the process of small molecule identification and provide a means to accelerate the pace of metabolomic discovery.
CONCLUSIONS
Small molecule analysis and discovery remains at the core of many fields – e.g., toxicology, sports doping, environmental analysis, food safety, clinical research, etc. – and is emerging as a key technology in the expanding area of metabolomics. GC/MS is a robust and mature method for profiling small molecules, but has recently undergone a transformation with the introduction of state-of-art mass analyzer capabilities that deliver routine high-resolution and accurate mass measurement. The new type of GC/MS data created by these modern systems has transformative potential – realizing this promise, however, requires new and innovative data processing approaches.
Here we describe a simple and straightforward method, HRF, which leverages accurate mass to both improve spectral deconvolution and increase confidence in small molecule identifications. The HRF approach can be used in conjunction with traditional spectral matching and effectively extends the utility of currently available unit-resolution reference libraries. Moreover, information provided by this approach is orthogonal to traditional spectral matching. In the future we predict this method will be of high value for the analysis of novel compounds, where a suitable reference spectrum is unavailable. In this application users would simply provide suspected chemical formulae and/or structures and utilize the HRF scoring method to test candidate plausibility. No such technology currently exists. We note the HRF approach facilitates rapid annotation of EI-MS spectra, has potential for LC-MS/MS applications, and may prove useful for automated false-discovery rate calculations. In summary, by enabling discrimination between candidate molecular precursors on the basis of both measured fragmentation profiles and accurate mass, the HRF method capitalizes on new high-resolution GC/MS instrumentation and the large, existing unit resolution EI-MS spectral libraries.
Supplementary Material
ACKNOWLEDGMENT
We thank Alan J. Higbee for assistance in curating and acquiring a set of reference standards to fully characterize algorithmic performance. We also thank John Voss, Amelia C. Peterson, Graeme C. McAlister, John E. P. Syka, Jae C. Schwartz, Alexander Makarov, and Jens Griep-Raming for many thoughtful discussions and feedback.
Funding Sources
This work was funded by the National Institute of General Medical Sciences (NIGMS) grant (1R01GM107199) to J.J.C.
Footnotes
ASSOCIATED CONTENT
A compiled executable for HRF analysis along with a reference dataset has been made available on our website (http://coon.chem.wisc.edu/). Supplementary material is available free of charge via the internet at http://pubs.acs.org
NWK, DJB, MSW, and JJC are co-inventors on a patent application (US 14671199) related in part to the material presented here.
REFERENCES
- 1.Westerhoff P, Yoon Y. Environ. Sci. Technol. 2005;39(17):6649–6663. doi: 10.1021/es0484799. [DOI] [PubMed] [Google Scholar]
- 2.Tareke E, Rydberg PJ. Agric. Food Chem. 2002:4998–5006. doi: 10.1021/jf020302f. [DOI] [PubMed] [Google Scholar]
- 3.Kataoka H, Lord HL, Pawliszyn JJ. Chromatogr. A. 2000;880(1-2):35–62. doi: 10.1016/s0021-9673(00)00309-5. [DOI] [PubMed] [Google Scholar]
- 4.Yang C, Park AC, Davis N. a, Russell JD, Kim B, Brand DD, Lawrence MJ, Ge Y, Westphall MS, Coon JJ, Greenspan DS. J. Biol. Chem. 2012;287(48):40598–40610. doi: 10.1074/jbc.M112.406850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiehn O, Kopka J, Dörmann P. Nat. Biotechnol. 2000:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 6.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Trends Biotechnol. 2004;22(5):245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. Nat. Biotechnol. 2003;21(6):692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 8.Stein SJ. Am. Soc. Mass Spectrom. 1999;0305(99) [Google Scholar]
- 9.Fiehn O. Trends Analyt. Chem. 2008;27(3):261–269. doi: 10.1016/j.trac.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiehn O, Kopka J, Trethewey RN, Willmitzer L. Anal. Chem. 2000;72(15):3573–3580. doi: 10.1021/ac991142i. [DOI] [PubMed] [Google Scholar]
- 11.Pluskal T, Uehara T, Yanagida M. Anal. Chem. 2012;84(10):4396–4403. doi: 10.1021/ac3000418. [DOI] [PubMed] [Google Scholar]
- 12.NIST Mass Spectral Library; 2012. [Google Scholar]
- 13.Wiley Registry of Mass Spectral Data. 2010.
- 14.Lewis S, Kenyon CN, Meili J, Burlingame a. L. Anal. Chem. 1979;51(8):1275–1285. doi: 10.1021/ac50044a038. [DOI] [PubMed] [Google Scholar]
- 15.Peterson AC, Balloon AJ, Westphall MS, Coon JJ. Anal. Chem. 2014;86(20):10044–10051. doi: 10.1021/ac5014755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson AC, Hauschild J-P, Quarmby ST, Krumwiede D, Lange O, Lemke R. a S., Grosse-Coosmann F, Horning S, Donohue TJ, Westphall MS, Coon JJ, Griep-Raming J. Anal. Chem. 2014;86(20):10036–10043. doi: 10.1021/ac5014767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson AC, McAlister GC, Quarmby ST, Griep-Raming J, Coon JJ. Anal. Chem. 2010;82(20):8618–8628. doi: 10.1021/ac101757m. [DOI] [PubMed] [Google Scholar]
- 18.Wolf S, Schmidt S, Müller-Hannemann M, Neumann S. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill DW, Kertesz TM, Fontaine D, Friedman R, Grant DF. Anal. Chem. 2008;80(14):5574–5582. doi: 10.1021/ac800548g. [DOI] [PubMed] [Google Scholar]
- 20.Kerber A, Laue R, Meringer M, Varmuza K. Adv. Mass Spectrom. 2001;15:939–940. [Google Scholar]
- 21.Solid Phase Extraction Applications Manual. 2008:42–44. [Google Scholar]
- 22.Michalski A, Damoc E, Hauschild J-P, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mol. Cell. Proteomics. 2011;10(9):M111.011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. Mol. Cell. Proteomics. 2009;8(12):2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Anal. Chem. 2009;81(18):7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vetter W. Biol. Mass Spectrom. 1994;23(6):379. [Google Scholar]
- 26.Wenger CD, McAlister GC, Xia Q, Coon JJ. Mol. Cell. Proteomics. 2010;9(5):754–763. doi: 10.1074/mcp.M900541-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.