Abstract
Objective(s)
To assess the frequency and function of HIV-1-specific HLA-G+ CD8 T cells in HIV-1 controllers and progressors.
Design
We performed an observational cross-sectional cohort analysis in untreated (n=47) and treated (n=17) HIV-1 patients with different rates of disease progression and n=14 healthy individuals.
Methods
We evaluated the frequency, the proportion and the function of total and virus-specific HLA-G+ CD8 T cells by tetramer or intracellular cytokine staining, followed by flow cytometric analysis. Cytokine secretion of sorted CD8 T cell subsets was evaluated by Luminex assays.
Results
The proportion and the absolute frequency of HLA-G+ HIV-1-specific CD8 T cells were directly associated with CD4 T cell counts and inversely correlated with viral loads, while total or HLA-G-negative HIV-1-specific CD8 T cells were not. In functional assays, HLA-G+ CD8 T cells from HIV-1-negative individuals had higher abilities to produce the antiviral CCR5 ligands MIP-1β, MIP-1α and Rantes.
Conclusions
HLA-G+ HIV-1-specific CD8 T cells may represent a previously-unrecognized correlate of HIV-1 immune control.
Keywords: HIV-1, HLA-G, CD8 T cells, controllers, antiviral mechanisms, chemokines
Introduction
A large number of studies suggest that natural HIV-1 disease progression in untreated patients can be modulated by T cell-mediated immune responses [1–3]. In individuals with progressive disease, HIV-1-specific CD8 T cells mostly consist of IFN-γ secreting effector-memory cells, and although these immune responses can exert antiviral immune pressure and influence viral sequence evolution, they are not very effective in suppressing HIV-1 replication [4]. In persons with natural control of HIV-1 infection, HIV-1-specific CD8 T cells have a more polyfunctional profile that includes a higher proportion of IFN-γ/IL-2 co-secreting central-memory CD8 T cells [5]. Cellular immune responses in persons receiving suppressive antiretroviral therapy seem to be enriched for HIV-1-specific CD8 T cells with a stem cell memory phenotype [6].
Surface expression of HLA-G, a non-classical HLA class Ib molecule typically expressed on placental trophoblasts, denotes a subset of CD4 and CD8 T cells with immunoregulatory properties that do not express the Forkhead Box P3 transcription factor [7]. HLA-G-expressing T cells have the ability to suppress T cell proliferation and reduce bystander immune activation, most likely through direct interactions between HLA-G and the inhibitory HLA-G ligand LILRB1 [8, 9]. HLA-G+ CD4 T cells are reduced during untreated progressive HIV-1 infection, and are inversely associated with levels of cellular immune activation, suggesting that these cells may have beneficial effects on HIV-1 disease outcome [9]. In the present study, we analyzed the expression of HLA-G+ HIV-1-specific CD8 T cells in untreated patients with different stages of HIV-1 disease progression. Our results indicate an increase of HLA-G+ HIV-1-specific CD8 T cells in patients with controlled HIV-1 disease, an inverse association between proportions of HLA-G+ HIV-1-specific CD8 and viral loads, and an increased ability of HLA-G+ CD8 T cells from HIV-1-negative individuals to secrete CCR5-binding chemokines, such as Rantes, MIP-1β and MIP-1α. Together, these results suggest that HLA-G-expressing antigen-specific cytotoxic T cells can represent a previously-unrecognized component of antiviral immune defense.
Material and Methods
Patients
Samples from 27 patients with chronic progressive HIV-1 infection, (median viral loads 39,200 HIV-1 RNA copies/ml and CD4 T cell counts 505 cells/ul), 20 controllers (median viral loads 62.5 RNA copies/ml and CD4 T cell counts 829.5 cells/ul) and 17 ART-treated patients (median viral loads 50 RNA copies/ml and CD4 T cell counts 784.5 cells/ul) were used for this study. 14 HIV-1-negative individuals were also recruited. All subjects gave written informed consent and the study was approved by the Institutional Review Board of Massachusetts General Hospital/Partners Healthcare.
Peptide-MHC class I multimer complexes
MHC class I multimers refolded with epitopic HIV-1 (n=6) or CMV/EBV (n=2) peptides were purchased from ProImmune (Oxford, UK). A list of all class I multimers included in this study is included in Table S1.
Flow cytometry
Cryo-preserved blood mononuclear cells were stained with blue viability dye (Life Technologies, 4°C for 20’), followed by incubation with appropriately titrated peptide-MHC class I multimer complexes at room temperature for 20 min in Ca2+-free media as described [10]. Cells were then washed and stained with antibodies against CD3, CD8, CD4, HLA-G at 4°C for 20 min. For intracellular cytokine staining, cells were stimulated overnight with optimal CD8 T cell peptides in presence of brefeldin A. Cells were then stained with blue viability dye (Life Technologies, 4°C for 20’), followed by incubation with appropriately titrated antibodies against CD3, CD4, CD8, HLA-G. After fixation and permeabilization for 20 min at 4°C using a commercial kit (Caltag), cells were stained intracellularly for IFN-γ, TNF-α, MIP-1β and IL-2. Cells were acquired on a Fortessa flow cytometer (Becton Dickinson) and analyzed using FlowJo X (Tree star). Analysis and presentation of cell distributions were performed using Graph Pad prism (version 6) and SPICE version 5.32, downloaded from <http://exon.niaid.nih.gov/spice/>.
Cell sorting
Cryo-preserved blood mononuclear cells were thawed and CD3+ T cell isolation was performed using immunomagnetic enrichment (Miltenyi Biotech). T cells were stained with blue viability dye (Life Technologies) at 4°C for 20min. Afterwards, cells were stained with antibodies directed against CD3, CD8, CD4, HLA-G, CD25, CD127. The following T-cell populations were live-sorted, after exclusion of CD25+ CD127− CD4+ Tregs, using an Aria cell sorting device (Becton Dickinson): CD4+CD25−/lowHLA-G−, CD4+CD25−/lowHLA-G+, CD8+CD25−/lowHLA-G−, CD8+CD25−/lowHLA-G+.
Luminex assays
The sorted cell populations were stimulated in 96-well plates with anti-CD3 (plate-bound) and anti-CD28 (soluble) antibodies or were left unstimulated for 4 days. Afterwards, cell supernatants were analyzed by Luminex assays (Millipore) for the following cytokines: Eotaxin, G-CSF, GM-CSF, IFN-α2, IL-10, IL-12p70, IFN-γ, IL-13, IL-17α, IL-1RA, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7,IL-8, IP10, MCP1, TNF-α, TNF-β, VEGF.
Statistics
Data are plotted as means or medians, as indicated. Statistical significance was tested using Mann-Whitney U tests or paired Wilcoxon tests, as appropriate. Correlations have been tested using Spearman test.
Results
HLA-G is expressed on a small number of bulk CD8 T cells [7], however, the expression of HLA-G on HIV-1-specific CD8 T cells is unknown. To investigate this, we analyzed proportions of HLA-G-positive cells within 36 HIV-1-specific CD8 T cell populations targeting different CTL epitopes (Table S1) and 23 CMV/EBV-specific CD8 T cell populations in cohorts of untreated HIV-1 patients. The frequency of HLA-G+ HIV-1-specific cells within total CD8 T cells was higher in persons with spontaneous control of HIV-1 infection (n=19) than in patients with progressive disease (n=17) (Figure 1A). This was also true when the proportion of HLA-G+ CD8 T cells within a given population of multimer+ HIV-1-specific CD8 T cells was evaluated (Figure S1A). No difference was found between the frequencies or the proportions of CMV/EBV-specific HLA-G-positive CD8 T cells in these two study groups (Figure 1A and figure S1A). Moreover, the frequencies of total or HLA-G-negative HIV-1-specific CD8 T cells were also not significantly different (Figure 1A and S1B). Of interest, the frequencies, the relative proportions and the absolute numbers of HIV-1-specific HLA-G-expressing CD8 T cells were positively associated with total CD4 T cell counts and inversely with viral loads (Figure 1B and S1C). There was no association between total or HLA-G-negative HIV-1-specific CD8 T cells (Figure 1B and S1D) or HLA-G-positive CMV/EBV-specific CD8 T cells and CD4 T cell counts or HIV-1 viral loads (Figure S1E). Interestingly, quantitative differences of HLA-G+ HIV-1-specific CD8 T cells between progressors and controllers were most obvious for immune responses restricted by protective HLA class I alleles (HLA-B57/B27), but not detectable for responses restricted by HLA-A2 or –B8 (Figure 1C and S2A); however, this was not true for total or HLA-G-negative HIV-1-specific CD8 T cells (Figure S2B and 1C).
Figure 1. Association between HIV-1-specific HLA-G+ CD8 T cells and HIV-1 disease progression.
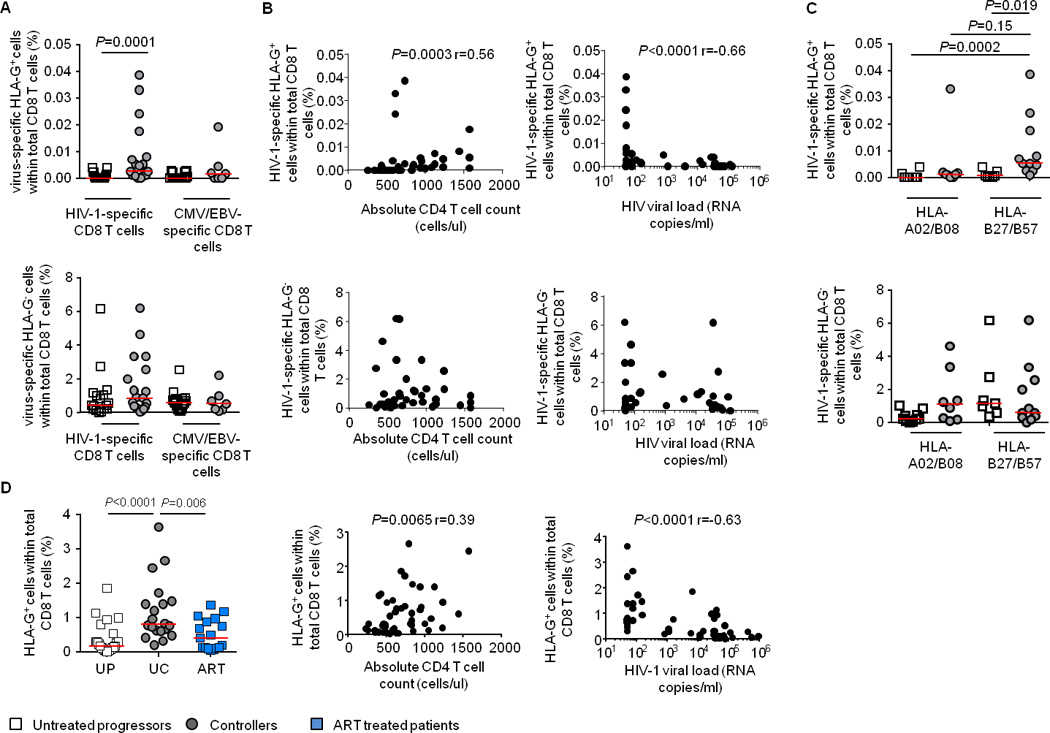
A. Frequency of HIV-1- or CMV/EBV-specific (multimer-positive) HLA-G+ or HLA-G− CD8 T cells within the total CD8 T cell pool in HIV-1-infected patients with untreated progressive disease or in controllers. B. Association between the frequency of HIV-1-specific HLA-G+ or HIV-1-specific HLA-G− CD8 T cells within the total CD8 T cell pool, and CD4 T cell counts or HIV-1 viral load. C. Frequency of HIV-1-specific HLA-G+ or HLA-G− CD8 T cells restricted by non-protective and protective HLA in indicated HIV-1-infected patients. D. (left panel): Frequency of HLA-G+ CD8 T cells in untreated progressors (UP), untreated controllers (UC) and ART-treated HIV-1-infected patients. (middle and right panel): Association between the frequency of HLA-G+ CD8 T cells and CD4 T cell counts or HIV-1 viral load.
For comparative purposes, we analyzed the proportions of total HLA-G-positive CD8 T cells in the two study cohorts. HIV-1-infected controller patients had significantly higher frequencies of HLA-G-expressing CD8 T cells than progressors, while frequencies of total and HLA-G-negative CD8 T cells were highest in progressors (Figure 1D and S3A–C). Notably, the frequencies and the absolute counts of HLA-G-expressing CD8 T cells were positively associated with total CD4 T cell counts and inversely with viral loads (Figure 1D and S3D), as opposed to total or HLA-G-negative CD8 T cells that were unrelated to CD4 T cell counts, and positively associated with HIV-1 viral loads (Figure S3E–F). Initiation of antiretroviral therapy did not restore the frequency and number of HLA-G+ CD8 T cells in chronic progressive patients (Figure 1D and S3A).
Production of chemokines and cytokines by HLA-G+ CD8 T cells was analyzed in 14 HIV-1-negative individuals, using Luminex assays. For these studies, CD8+CD25−/lowHLA-G− (Tc1) and CD8+CD25−/lowHLA-G+ (HLA-G+ CD8 T cells) T cell populations were sorted after exclusion of classical Tregs (CD25high/CD127− CD4 T cells). CD4+CD25−/lowHLA-G− (Th1) and CD4+CD25−/lowHLA-G+ (HLA-G+ CD4 T cells) were sorted as controls. Cells were then stimulated for 4 days with anti-CD3/anti-CD28 antibodies. HLA-G+ CD8 T cells produced higher levels of IL-10 and IL-2 than Tc1 cells (Figure 2A). Moreover, HLA-G-positive CD8 T cells produced higher levels of the CC chemokines MIP-1α, MIP-1β and RANTES (Figure 2A), which can act as competitive entry inhibitors for R5-tropic HIV-1 isolates [11]. No difference was observed between the cytokine profiles of Th1 and HLA-G+ CD4 T cells (data not shown). To extend these investigations, we analyzed secretion of IFN-γ and IL-2 in HLA-G+ and HLA-G− CEF (CMV-, EBV-, Flu-)-specific CD8 T cells from HIV-1-negative donors as well as in HIV-1-specific CD8 T cells from controller patients using intracellular cytokine staining after overnight stimulation with peptides spanning HIV-1 gag (Figure 2 B and C). These data confirmed that virus-specific HLA-G+ CD8 T cells produced more IL-2, consistent with a relative phenotypic enrichment for more immature CCR7+ T cell subsets (Figure S4), while IFN-γ secretion was higher in virus-specific HLA-G− cells compared to HLA-G+ CD8 T cells (Figure 2 B and C).
Figure 2. HLA-G+ CD8 T cells produce cytokines involved in HIV-1-infection control.
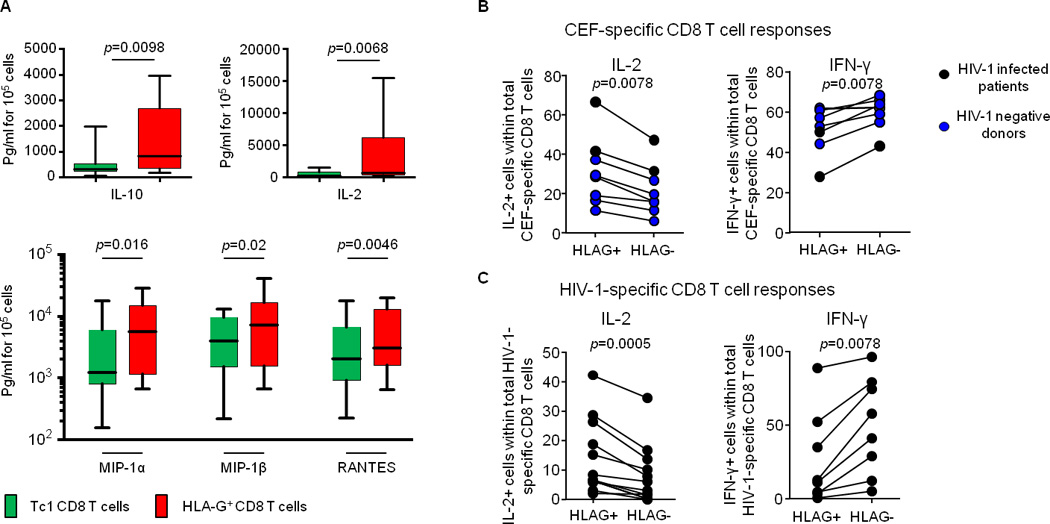
A. Amount (pg/ml for 105 cells) of IL-10, IL-2, MIP-1α, MIP-1β and RANTES produced by Tc1 CD8 T cells and HLA-G+ CD8 T cells quantified by Luminex in 13 HIV-1 negative donors. B. Proportion of IL-2 (left) or IFN-γ (right) producing CEF-specific HLAG+ or HLA-G− CD8 T cells in n=6 HIV-1 negative donors (blue dots) and in n=2 HIV-1 infected patients (black dots). C. Proportion of IL-2 (left), or IFN-γ (right) producing HIV-1-specific HLA-G+ or HLA-G− CD8 T cells in n=8 HIV-1 infected patients.
Discussion
A large number of studies [1] show that T cell-mediated immune responses can influence HIV-1 disease progression. However, a perplexing aspect in HIV-1 immunobiology is that large numbers of HIV-1-specific CD8 T cells are encountered in almost all untreated HIV-1 infected patients, but do not represent a correlate of HIV-1 immune control [12]. This suggests that not all antigen-specific CD8 T cells contribute to antiviral immune defense, and that specific subpopulations of antigen-specific CD8 T cells may have improved antiviral effector functions. In previous studies, “polyfunctional” central-memory HIV-1-specific CD8 T cells with increased antigen-specific secretion of IL-2, stronger proliferative activities and enhanced expression of the Th1 master transcription factor T-bet were found to be associated with HIV-1 immune control [13, 14], but it is likely that alternative, as of yet unidentified subsets of antigen-specific T cells are also involved in HIV-1 immune defense. Here, we described a subset of HIV-1-specific CD8 T cells expressing HLA-G that is associated with immune control in untreated HIV-1 patients, and at least in HIV-1-negative individuals, these cells are able to secrete higher levels of the CC chemokines MIP-1α, MIP-β and Rantes, the physiological ligands for CCR5 [11]. These cells may therefore be able to actively participate in antiviral immune defense by competitively blocking CCR5, the viral co-receptor used by the majority of circulating viruses. Future studies will be necessary to determine if enhanced secretion of these chemokines can also be detected in virus-specific HLA-G+ CD8 T cells from HIV-1-infected patients, and whether these cells indeed have increased functional abilities to restrict HIV-1 replication steps or kill HIV-1-infected cells through enhanced cytotoxic properties. Given the extreme rarity of HIV-1-specific HLA-G+ CD8 T cells, which in many cases include less than 0.01% of all CD8 T cells, such a functional analysis will be technically challenging, and will require large quantities of patient cells. Notably, prior studies suggested that HLA-G positive T cells have elevated expression levels of lymphoid tissue homing markers [15], and for this reason may have higher abilities to penetrate lymph nodes and increase immune surveillance in this anatomical compartment. A closer examination of HLA-G+ CD8 T cells in lymphoid tissues, specifically in germinal centers that may serve as hotspots for viral replication [16], therefore also represents a highly informative future research perspective. Finally, it is noteworthy that HLA-G+ expressing T cells also have immunosuppressive properties [8], consistent with our observation of higher IL-10 secretion in this cell subset. Although IL-10 has been associated with immune dysfunction and accelerated immune exhaustion in prior studies [17], it is also possible that elevated regulatory properties of HLA-G+ CD8 T cells may improve HIV-1 disease progression by inhibition of immune over-activation [9]. An improved understanding of the complexity and heterogeneity of the CD8 T cell-mediated immune responses against HIV-1 may ultimately allow the development of improved immune interventions for HIV-1 treatment and prevention.
Supplementary Material
Acknowledgments
Selena Viganò designed and performed experiments, analyzed data, interpreted the data and wrote the paper.
Jordi J. Negrón, Samantha Tse, Fatema Z. Chowdhury performed experiments.
Mathias Lichterfeld, Xu G. Yu designed the study, interpreted the data and wrote the paper.
Xu G. Yu supervised all aspects of this study.
This work was supported for the US National Institutes of Health (grants R01 AI078799, R56 AI098484 and R01 AI089339 to XGY). PBMC sample collection was supported by the Bill and Melinda Gates Foundation, the Mark and Lisa Swartz Foundation, the Ragon Institute of MGH, MIT and Harvard, and the International HIV Controller Consortium. Ragon Institute Imaging Core is supported by the Harvard University Center of AIDS research (HU CFAR NIH/NIAIS fund 5P30AI060354-10).
Footnotes
Data presented previously at CROI, March 2014, Boston, MA.
Potential conflicts of interest. None of the authors have reported conflicts of interest.
References
- 1.Carrington M, Walker BD. Immunogenetics of spontaneous control of HIV. Annu Rev Med. 2012;63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perreau M, Levy Y, Pantaleo G. Immune response to HIV. Curr Opin HIV AIDS. 2013;8:333–340. doi: 10.1097/COH.0b013e328361faf4. [DOI] [PubMed] [Google Scholar]
- 4.Liu MK, Hawkins N, Ritchie AJ, Ganusov VV, Whale V, Brackenridge S, et al. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest. 2013;123:380–393. doi: 10.1172/JCI65330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vigano S, Negron J, Ouyang Z, Rosenberg ES, Walker BD, Lichterfeld M, et al. Prolonged Antiretroviral Therapy Preserves HIV-1-Specific CD8 T Cells with Stem Cell-Like Properties. J Virol. 2015;89:7829–7840. doi: 10.1128/JVI.00789-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feger U, Tolosa E, Huang YH, Waschbisch A, Biedermann T, Melms A, et al. HLA-G expression defines a novel regulatory T-cell subset present in human peripheral blood and sites of inflammation. Blood. 2007;110:568–577. doi: 10.1182/blood-2006-11-057125. [DOI] [PubMed] [Google Scholar]
- 8.Huang YH, Zozulya AL, Weidenfeller C, Schwab N, Wiendl H. T cell suppression by naturally occurring HLA-G-expressing regulatory CD4+ T cells is IL-10-dependent and reversible. J Leukoc Biol. 2009;86:273–281. doi: 10.1189/jlb.1008649. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Toth I, Schulze Zur Wiesch J, Pereyra F, Rychert J, Rosenberg ES, et al. Functional characterization of HLA-G(+) regulatory T cells in HIV-1 infection. PLoS Pathog. 2013;9:e1003140. doi: 10.1371/journal.ppat.1003140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cellerai C, Harari A, Stauss H, Yerly S, Geretti AM, Carroll A, et al. Early and prolonged antiretroviral therapy is associated with an HIV-1-specific T-cell profile comparable to that of long-term non-progressors. PLoS One. 2011;6:e18164. doi: 10.1371/journal.pone.0018164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 12.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 13.Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011;117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaelsson J, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10:e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YH, Zozulya AL, Weidenfeller C, Metz I, Buck D, Toyka KV, et al. Specific central nervous system recruitment of HLA-G(+) regulatory T cells in multiple sclerosis. Ann Neurol. 2009;66:171–183. doi: 10.1002/ana.21705. [DOI] [PubMed] [Google Scholar]
- 16.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter K, Perriard G, Behrendt R, Schwendener RA, Sexl V, Dunn R, et al. Macrophage and T cell produced IL-10 promotes viral chronicity. PLoS Pathog. 2013;9:e1003735. doi: 10.1371/journal.ppat.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


