Abstract
Background
A defined Microbial Ecosystem Therapeutic (MET-1, or “RePOOPulate”) derived from the feces of a healthy volunteer can cure recurrent C. difficile infection (rCDI) in humans. The mechanisms of action whereby healthy microbiota protect against rCDI remain unclear. Since C. difficile toxins are largely responsible for the disease pathology of CDI, we hypothesized that MET-1 exerts its protective effects by inhibiting the effects of these toxins on the host.
Methods
A combination of in vivo (antibiotic-associated mouse model of C. difficile colitis, mouse ileal loop model) and in vitro models (FITC-phalloidin staining, F actin Western blots and apoptosis assay in Caco2 cells, transepithelial electrical resistance measurements in T84 cells) were employed.
Results
MET-1 decreased both local and systemic inflammation in infection and decreased both the cytotoxicity and the amount of TcdA detected in stool, without an effect on C. difficile viability. MET-1 protected against TcdA-mediated damage in a murine ileal loop model. MET-1 protected the integrity of the cytoskeleton in cells treated with purified TcdA, as indicated by FITC-phalloidin staining, F:G actin assays and preservation of transepithelial electrical resistance. Finally, co-incubation of MET-1 with purified TcdA resulted in decreased detectable TcdA by Western blot analysis.
Conclusions
MET-1 intestinal microbiota confers protection against C. difficile and decreases C. difficile-mediated inflammation through its protective effects against C. difficile toxins, including enhancement of host barrier function and degradation of TcdA. The effect of MET-1 on C. difficile viability seems to offer little, if any, contribution to its protective effects on the host.
Keywords: Microbiota, C. difficile, TcdA
Introduction
There is considerable interest in the therapeutic potential of the gut microbiota to treat recurrent C. difficile infection (rCDI) [1, 2]. Fecal microbiota transplantation (FMT) has proven effective at eradicating rCDI where traditional antibiotic regimens have failed [3, 4]. FMT is thought to protect against CDI primarily through restoration of colonization resistance [5–7]. Although not entirely understood, proposed mechanisms of action for FMT include modulation of host immune function [8], effects on bile salt metabolism that in turn affect C. difficile sporulation [9, 10], and effects on sialic acid metabolism that influence C. difficile viability in the host [11]. However, there are limited studies directly investigating the effects of FMT on C. difficile toxins A (TcdA) and B (TcdB). It is well established that TcdA and TcdB are largely responsible for the pathophysiology of the disease [12–14]. A recent large multicenter study has shown that the presence of toxins in patient stool samples correlates most closely with patient clinical outcomes [15, 16], and C. difficile strains lacking these toxins generally do not cause disease [17].
Although FMT appears promising for the treatment of rCDI, its limitations include the undefined nature of donor stool, the need to intensively screen donors to address safety concerns and the lack of understanding of what components of donor stool are beneficial. The use of defined microbial populations prepared under controlled laboratory conditions is very appealing, and the concept of using defined microbial ecosystems has been recognized as a potential improvement on current methods of FMT [18]. Recently a defined microbial ecosystem (MET-1), derived from the feces of a healthy human volunteer, was successfully used to cure patients with rCDI [19]. However, like FMT, its precise mechanism of action remains to be fully understood. We hypothesized that MET-1 exerts its therapeutic benefits by affecting toxins produced by C. difficile. The purpose of this study was to determine whether MET-1 could provide protection specifically against purified toxin TcdA from ‘hypervirulent’ ribotype 027 and 078 C. difficile strains isolated from hospitalized rCDI patients. Additionally, we sought to identify potential mechanisms whereby MET-1 may confer protection against C. difficile disease in a preventative mouse model of C. difficile infection.
Methods
Ethics statement
This study was carried out in accordance with the Canadian Council of Animal Care guidelines and approved by the Queen’s University Animal Care Committee.
Growth of C. difficile
Each Clostridium difficile ribotype strain (027 or 078) was grown on cycloserine cefoxitin fructose agar (CCFA), a C. difficile-selective medium under anaerobic conditions (90 % N2, 5 % CO2, and 5 % H2) at 37 °C for 48 h. A single colony was then used to inoculate 5 ml of brain heart infusion (BHI) broth (Difco Laboratories, USA). C. difficile cultures were incubated anaerobically at 37 °C for 24 h or for 72 h for large-volume preparation of TcdA toxin (see below). All C. difficile isolates were obtained from stool samples of confirmed clinical cases of C. difficile infection at Kingston General Hospital and were toxinotyped and ribotyped as previously described [20].
C. difficile TcdA preparation
Clostridium difficile TcdA was purified according to Sullivan et al. [21]. In brief, a C. difficile strain 027 or 078 clinical isolate was inoculated with 50 ml BHI for 24 h and then transferred to dialysis bags (12–14 kDa exclusion, Fisher Scientific, USA) and grown for 72 h anaerobically in 1 l BHI. The contents were centrifuged to remove the bacteria, and the supernatant was filtered through a 0.22-μm filter. The culture supernatant was then concentrated using Centricon plus-70 membrane filters (>30 kDa, Millipore, Germany), loaded onto a DEAE-Sepharose CL-6B column (Sigma Aldrich, Canada) and equilibrated (50 mM Tris, pH 7.5), followed by elution with a gradient of 0.05–0.25 M NaCl at 1 ml/min. Fractions were assayed for bioactivity using a fibroblast cytotoxicity assay (described below), and presence of TcdA was verified by Western blot analysis (anti-TcdA antibody, 1:1000 dilution, Santa Cruz Biotechnology, USA). Fractions with TcdA were pooled, concentrated (Amicon ultra-15 filters, Millipore, Germany), aliquotted and stored at −80 °C.
MET-1 preparation
MET-1 was developed by culturing the stool of a healthy 41-year-old female donor, and 33 isolates of commensal bacteria were selected for the final formulation using predetermined ratios as previously described [19]. To prepare the MET-1 mixture, each isolate was cultured on fastidious anaerobe agar (FAA, Lab M Ltd., UK) supplemented with or without 5 % defibrinated sheep blood (Hemostat Laboratories, USA) under strict anaerobic conditions in an Anaerobe chamber (Ruskinn Bugbox, USA) and then formulated in pre-reduced sterile 0.9 % normal saline to an approximate total concentration of 3.5 × 109 colony-forming units (CFU)/ml. A table of MET-1 composition with updated nomenclature has been included in the Supplementary Materials section (Table 1).
Table 1.
Composition of MET-1
| Closest species match (by % identity) | |
|---|---|
| Acidaminococcus intestinia | Eubacterium rectale—2 |
| Bacteroides ovatus | Eubacterium rectale—3 |
| Bifidobacterium adolescentis—1 | Eubacterium rectale—4 |
| Bifidobacterium adolescentis—2 | Eubacterium ventriosum |
| Bifidobacterium longum—1 | Eubacterum rectale—1 |
| Bifidobacterium longum—2 | Fecalibacterium prausnitzii |
| Blautia lutie | Lachnospira pectinoschiza |
| Blautia stercoris | Lactobacillus casei |
| Butyricicoccus pullicaecorumb | Lactobacillus paracasei |
| Clostridium cocleatum | Parabacteroides distasonis |
| Collinsella aerofaciens | Roseburia faecisd |
| Dorea longicatena—1 | Roseburia intestinalis |
| Dorea longicatena—2 | Ruminococcus obeum |
| Enterobacter aerogenesc | Ruminococcus torques—1 |
| Escherichia coli | Ruminococcus torques—2 |
| Eubacterium eligens | Streptococcus mitisf |
| Eubacterium limosum | |
Updated strain identification of the MET-1 mixture, as determined by full-length 16S sequencing. Using BLAST, sequences were matched against the Greengenes database (July 2015)
Formerly identified as Acidaminococcus intestinalis
Formerly identified as Eubacterium desmolans
Formerly identified as Raoultella ornithinolytica
Formerly identified as Roseburia faecalis
Formerly identified as Ruminococcus obeum
Formerly identified as Streptococcus parasanguinis
C. difficile colitis mouse model
Based on a murine model of C. difficile infection [22], 6–8-week-old C57BL/6 mice were first acclimatized for a week and then given an antibiotic mixture of kanamycin (0.4 mg/ml, Sigma Aldrich, USA), gentamicin (0.035 mg/ ml, Amresco, USA), colistin (850 U/ml, Sigma Aldrich, USA), metronidazole (0.215 mg/ml, Sigma Aldrich, USA) and vancomycin (0.045 mg/ml, Sigma Aldrich, USA) in their drinking water for 3 days. Mice were then switched to regular autoclaved water for 2 days. On the 2nd day mice were gavaged with MET-1 (approximately 3.5 × 108 cells) or vehicle control (saline) or BioK+ (Bio-K+ International, Canada), a commercial probiotic, at the same dose of 3.5 × 108 cells (see also supplementary appendix Fig. S1 for details). After 24 h, mice were gavaged with 1 × 105 CFU (vegetative cells) of C. difficile strain 027 or vehicle control (BHI broth). Mice were monitored and weighed daily and euthanized at 52 h, and blood/tissues were harvested and processed as described below.
C. difficile bacterial enumeration
Cecal contents were collected, resuspended in PBS and weighed. The samples were serially diluted and plated on CCFA. Plates were incubated at 37 °C under anaerobic conditions (90 % N2, 5 % CO2, and 5 % H2) for 48 h before enumeration.
Multiplex bead assay to measure serum cytokine levels
Blood was collected by cardiac puncture when the animals were killed. Serum was assessed using a mouse cytokine magnetic bead kit to measure IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17A, eotaxin, G-CSF, GM-CSF, INF-γ, KC, MCP-1, MIP-1α, MIP-1β, and TNF-α, as per the manufacturer’s instructions (Bio-Rad, USA).
Histological staining/scoring and immunohistochemistry
Tissues (colon, cecum, small intestine) were fixed in 10 % formalin followed by 70 % ethanol, 18 h in each, then processed and embedded in paraffin, and 4-μm-thick sections were stained with hematoxylin and eosin (H&E, Thermo Fisher Scientific, USA). Stained sections were examined blinded by a certified gastrointestinal pathologist, using an established scoring method [22]. A graded scoring system was used taking into consideration: neutrophil migration and tissue infiltration, hemorrhagic congestion, and edema of the mucosa and epithelial cell damage. A score between 1 and 3 was assigned for each parameter, and the overall score was the sum [22]. Immunofluorescence staining of myeloperoxidase (MPO) and claudin-1 was carried out on formalin-fixed cecal tissues as previously described [23].
TcdA measurement in mouse stool samples
Quantification of TcdA toxin was performed using an ELISA kit, as per the manufacturer’s instructions (tgcBIOMICS, Germany). In brief, 50 mg of mouse feces was homogenized in 450 μl dilution buffer and centrifuged at 2500×g for 20 min. The supernatants were added to the anti-toxin A antibody-coated ELISA plate (100 μl per well) for 1 h; after washing with the supplied wash buffer, 100 μl anti-TcdA-HRP conjugate was added to each well (30 min). After three more washes, 100 μl of substrate was added and incubated 15 min. All incubation steps were performed at room temperature. Color development was arrested with 50 μl Stop solution per well and absorbance measured at 450 nm using a microplate reader (Bio-Tek μQuant MQX200, USA).
Fibroblast assay with mouse fecal extracts
Fifty mg of mouse fecal material was resuspended in 0.5 ml of PBS, stool samples were centrifuged at 16,000×g for 30 min, the pellet was discarded, and supernatants were frozen at −80 °C until ready for use. Next, NIH 3T3 fibroblasts (ATCC, USA) were grown in DMEM media (GIBCO, Thermo Fischer Scientific, USA) with 10 % fetal calf serum (GIBCO, Thermo Fischer Scientific, USA) in 24-well tissue culture plates with 500 μl media in each well. Assay controls (untreated, PBS, or crude C. diff extract), or freshly thawed stool supernatants (10 μl) from each treatment group (Untreated, MET-1+ C. difficile and C. difficile mice) were then added to each well of fibroblasts and incubated for 1 h at 37 °C with 5 % CO2. Cells were washed twice with PBS, fixed with 10 % phosphate-buffered formalin for 30 min, washed again with PBS, and stained with Giemsa (Sigma Aldrich, USA) followed by two quick rinses with PBS. The stained cells were imaged using the OLYMPUS BX71 microscope at 10× total magnification equipped with a QIMAGING RETIGA-2000RV camera and Image-Pro Plus software (Media Cybernetics, Inc., USA). The numbers of rounded cells and total cells were counted by using Image J 1.51a software (NIH, USA).
Ileal loop model
Using 6–8-week-old C57BL/6 mice, ileal loop surgery was established based on work published by others [24–26]. Mice were orally gavaged with vancomycin (0.1 mg, Sigma Aldrich, USA) and streptomycin (20 mg, SteriMax, Canada). After 24 h, mice were gavaged with 3.5 × 108 cells of MET-1 for 2 consecutive days or vehicle control (0.9 % saline). On the day of surgery, mice were anesthetized using isoflurane (PPC Animal Health, Canada). Using sterile technique, the abdomen was incised, the small intestine was exposed in the abdominal cavity, and 4 cm of distal ileum was sutured using silk thread (Ethicon Inc., Johnson & Johnson, USA) to create ileal loops [25, 26]. Once the loop was formed, 50 μg of purified C. difficile TcdA diluted in 150 μl or vehicle control (matched volume of PBS) was injected in the ileal loop lumen. The small intestine was reinserted into the abdominal cavity; the incision was sutured closed and the animal allowed to regain consciousness. After 4 h, animals were euthanized with isoflurane followed by cervical dislocation. The loops were excised and tissue prepared for histological staining and scoring (see above).
F actin studies
For F and G actin analysis, subconfluent Caco2 human intestinal epithelial cells (ATCC, USA) were grown in 100-mm plates in DMEM with 10 % fetal bovine serum (FBS, Sigma Aldrich, USA), pretreated with 100 μl MET-1 or vehicle control overnight. The media were removed, and cells were washed with fresh media. Cells were then treated with TcdA (1 μg/ml) for 6 h, washed with PBS and harvested by centrifugation. Cell pellets were flash-frozen and stored at −80 °C until Western blot analysis, performed according to the manufacturer’s instructions (Cytoskeleton Inc, USA). For F actin visualization, cells were seeded onto coverslips in 24-well tissue culture plates and subconfluent cells pretreated with MET-1 (1:10 dilution, v/v) or control overnight. Media were removed, and the cells were washed with fresh media and treated with TcdA (1 μg/ml) for 6 h. Cells were washed again, fixed with 3.75 % PBS-buffered formalin, permeabilized with 0.1 % Triton X-100 and stained with Alexa 488-phalloidin, which binds to F-actin (1:50 dilution, Invitrogen, USA) for 60 min at 37 °C. Coverslips were washed and mounted onto slides with VECTaSHIELD mounting medium (Vector Laboratories Inc., USA); nuclei were stained with Hoechst (Sigma Aldrich, USA). Slides were imaged using an Olympus BX71 microscope equipped with a QIMA-GING RETIGA-2000RV camera and Image-Pro Plus software (Media Cybernetics Inc., USA).
Transepithelial electrical resistance (TER)
T84 human intestinal epithelial cells (ATCC, USA) were grown in 50 % DMEM and 50 % Ham’s F12 media (Fisher Scientific, USA) with 10 % FBS on 5-μm pore size, 6.5-mm diameter transwell permeable supports (Corning Inc., USA). Cells were incubated for 4 h in a 1:10 dilution of MET-1, followed by addition of purified ribotype 027 TcdA (6 μg/ml). Treatment was carried out in triplicate and transepithelial resistance was measured hourly for 4 h using a voltohmmeter (World Precision Instruments Inc., USA).
Apoptosis ELISA
Caco2 cells were seeded in 24-well tissue culture plates (4 × 104 cells/well; Corning Inc., USA) and grown for 72 h. Cells were then treated with MET-1 (1:10 dilution, v/v) overnight. Media were removed, and cells were treated with TcdA (6 μg/ml) for 6 h. Treatment was carried out in triplicate wells. Cell lysis and ELISA were performed following the manufacturer’s instructions (Roche Diagnostics, USA). Camptothecin (Sigma Aldrich, USA) was used as the positive control at a final concentration of 2 μg/ ml.
Co-incubation of TcdA with MET-1
Purified TcdA (1 μg) was co-incubated with MET-1 (25 μl) for 2 h at 37 °C. Samples were centrifuged at 10,000×g for 5 min to remove the MET-1 bacteria. Supernatants were then loaded onto 4–15 % gradient SDS-PAGE gels and either silver-stained or transferred to PVDF membranes (EMD Millipore, Germany) for Western blot analysis. Membranes were blocked in 5 % (w/v) nonfat milk in TBS-Tween [Tris-buffered saline, 150 mM NaCl, 5 mM KCl, 10 mM Tris, pH 7.4, with 0.05 % (v/v) Tween 20] prior to incubation with TcdA antibody (1:2500 dilution, Santa Cruz Biotechnology, Inc., USA) overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated secondary antibody was added (1:5000 dilution, Pierce, Thermo Fisher Scientific, USA) and incubated with the membranes for 1 h at room temperature; after three washes, an enhanced chemiluminescence reagent (ECL, Thermo Scientific, USA) was applied before imaging. A Gel Dock MP Imager (BioRad Laboratories Ltd., Canada) was used to visualize the bands. Silver staining for total protein staining was performed using a Pierce silver stain kit, as per the manufacturer’s instructions (Thermo Scientific, USA).
DNA isolation and microbial composition analysis
DNA was extracted from samples using bead beating and modified protocols from a commercially available kits, as previously described [27]. Primer sequences, barcodes used, Perl scripts and full protocols are provided at https://github.com/ggloor/miseq_bin/Illumina_SOP.pdf. In brief, 1 μl of DNA sample (1–5 ng) was PCR amplified using the Earth Microbiome universal primers (named 515F, 806R) using Promega GoTaq hot start colorless master mix reagent (Promega, USA) for 25 cycles with an annealing temperature of 52 °C. Inline 8-mer barcodes similar to what has been described previously [28] were preceded by four randomly synthesized nucleotides. Sequencing was carried out on the Illumina MiSeq platform at the London Regional Genomics Centre, with the 600 cycle v3 chemistry kit (Illumina, USA) with a 2 × 220 cycle profile and 5 % PhiX-174 spike in [29]. Forward and reverse reads were overlapped with PandaSeq [30] disallowing overlapping segments that contained ambiguous bases (N characters). Custom Perl scripts were used to separate and enumerate the reads from each sample as described [28]. Barcodes were chosen to contain a minimum edit distance of 4, and sequences were demultiplexed requiring perfect barcode matching. USEARCH [31, 32] version 7 for MacOSX was used to identify and remove chimeric sequences and to cluster sequences de novo with a 97 % identity threshold into operational taxonomic units (OTUs). Singleton OTUs and OTUs that occurred at an abundance of less than 1 % in any sample were discarded prior to analysis. Sequences were taxonomically assigned with the classify.seqs function of mothur 1.3.4 [33] using the Silva v119 database [34].
OTU frequencies are relative abundances; such data are constrained by a constant sum and have an inherent internal correlation structure, and subsets of the data can give different results than the full set [35–37]. These problems were minimized by transforming the data using the centered log ratio [35], which converts the data into unconstrained data where the distances between OTU abundance are linearly related, less affected by the subset problem and directly comparable between samples [35–37]. Exploratory analysis of the data was performed in R [38] according to the protocols contained in the compositions R package described in [39]. When necessary, zero replacement of the sparse data was performed using the CZM approach from the zCompositions R package [40].
Statistical analysis
Statistical analyses were performed with GraphPad Prism version 4.0 (GraphPad Software, USA), and results are expressed as the mean value with standard error of the mean (SEM). T test with Mann-Whitney correction was used where indicated for individual comparisons. One- or two-way ANOVA with a Tukey correction was used for multiple comparisons. Statistical significance was set at p <0.05.
Results
MET-1 is protective in an antibiotic-associated C. difficile colitis animal model
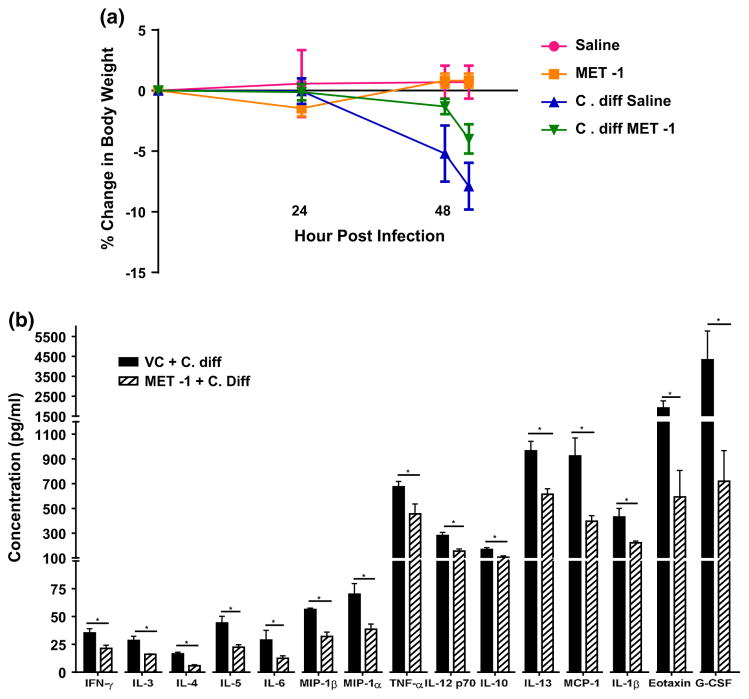
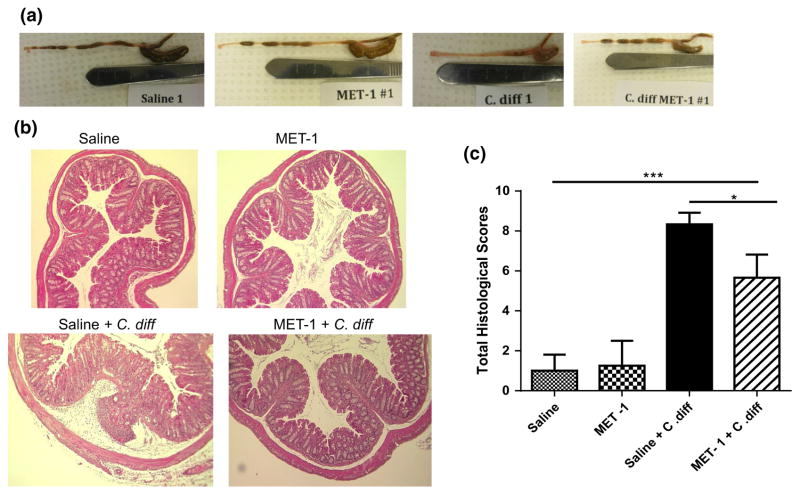
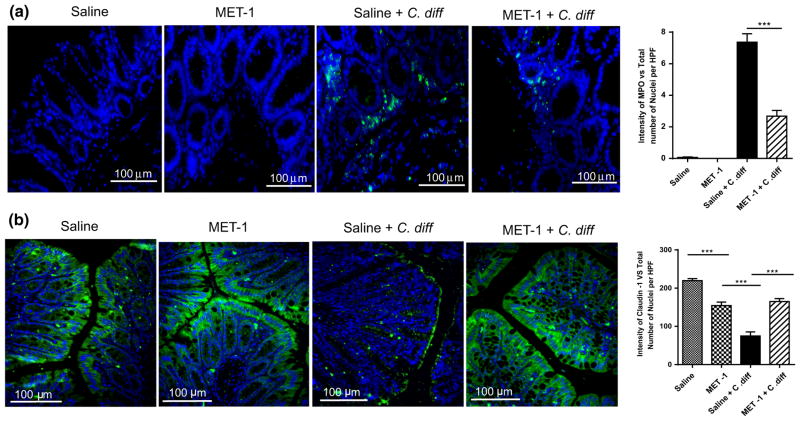
Using a previously described model [22], C57/Bl6 mice were gavaged with an antibiotic cocktail prior to treatment with MET-1 and then gavaged with C. difficile ribotype 027. We chose the more virulent ribotype (027) for this experiment to establish whether MET-1 would provide protection in an animal model of C. difficile. Gavage of BioK+, a commercially available probiotic commonly used for antibiotic-associated diarrhea, was used as a microbe comparator control. MET-1 pretreatment decreased both local and systemic inflammation in a mouse model of C. difficile infection. Animals receiving MET-1 had lower levels of systemic serum cytokines compared to C. difficile-infected vehicle controls (Fig. 1). Cytokines such as eotaxin, known to be elevated in human plasma in severe CDI [41], were markedly reduced in infected animals that received MET-1. The MET-1 animals also showed less colonic inflammation and histologic damage (Fig. 2), including less MPO (marker for neutrophils) by immunohistochemical staining and less loss of claudin-1 staining (tight junction protein), than C. difficile-infected vehicle controls (Fig. 3a, b, respectively). In contrast to MET-1, BioK+ did not confer any survival benefit against C. difficile, prevent weight loss or protect against histologic damage by C. difficile when gavaged in a similar manner (see Fig. S1, additional supplementary materials in appendix).
Fig. 1.
C. difficile-infected mice gavaged with MET-1 have similar weight loss but decreased inflammatory cytokine release compared to mice gavaged with vehicle control in an antibiotic-mediated C. difficile colitis model. C57BL/6 mice were treated with a mixture of broad-spectrum antibiotics and then orally administered MET-1 or saline, followed by 105 C. difficile strain 027 or saline 24 h later, as described in “Methods”. a Mice were weighed every 24 h, and the percent change in body weight is shown. Saline+ C. diff mice and MET-1+ C. diff mice demonstrated similar weight loss by 48 h, and the experiment was terminated at 52 h once saline+ C. diff mice lost 10 % weight. Data were analyzed by two-way ANOVA with Tukey correction, n = 3 for panel (a) and n = 4 for panel (b) for each group (*p <0.05). b Serum cytokine levels were significantly reduced in C. difficile-infected mice pretreated with MET-1 compared to infected mice pretreated with vehicle control (VC + C. difficile). Serum cytokine levels were measured using a Bio-Plex Pro mouse cytokine magnetic bead kit. There were no significant differences in cytokine concentrations between uninfected mice pretreated with MET-1 or vehicle control (data not shown). Data were analyzed by t-Student test with Mann-Whitney correction, n = 3 for panel A and n = 4 for panel B for each group (*p <0.05)
Fig. 2.
C. difficile-infected mice gavaged with MET-1 have less histological damage compared to infected mice gavaged with saline vehicle control. C57BL/6 mice were treated with a mixture of broad-spectrum antibiotics and then orally administered MET-1 or saline, followed by 105 C. difficile strain 027 or saline 24 h later, as described in “Methods”. a Representative images show gross morphology of the cecums and colons of infected mice differed between groups administered MET-1 and saline. Saline+ C. diff mice (“C. diff”, third panel) had smaller cecums and colons that were devoid of stool, while cecums and colons from MET-1 + C. diff mice (far right panel) appeared similar to the uninfected mice (first and second panels). b Representative images showing no histological differences in the colons of saline or MET-1 groups. Saline + C. diff mice showed signs of colonic inflammation including PMN infiltration and epithelial desquamation and submucosal edema. Epithelial cell integrity and crypt architecture were preserved in MET-1 + C. diff mice. c Histological scoring of the colon. The MET-1 + C. diff group had significantly lower histological scores (based on assessment of edema, PMN infiltration and epithelial cell damage) than the saline + C. difficile group. Data were analyzed using one-way ANOVA with Tukey correction, n = 4 for control groups and n = 3 for C. difficile-infected groups (*p <0.05, ***p <0.001)
Fig. 3.
Altered MPO and claudin-1 epithelial staining after C. difficile exposure is prevented by MET-1 in an antibiotic-mediated C. difficile colitis model. Representative immunofluorescence in ceca for a myeloperoxidase (MPO) or b claudin-1 staining. C57BL/6 mice were treated with broad-spectrum antibiotics and then gavaged with either saline vehicle or MET-1. Mice were then gavaged with either 105 C. difficile or saline, and the ceca were harvested and processed, as described in “Methods”. Images were obtained at ×400 total magnification (sizing bar shown). Nuclei were stained with Hoechst. Secondary antibody control slides for each group showed no immunostaining (data/images not shown). Quantification of each respective immunostaining is shown in the graphs to the left by the ratio of the area of green staining to total number of cells (blue nuclei) per high power field (HPF), for MPO (a) and claudin-1 (b). Less MPO staining was observed, and claudin-1 expression was preserved in the MET-1 + C. diff as compared to the saline vehicle + C. diff mice. Data were analyzed using one-way ANOVA with Tukey correction, ***p <0.001, n = 3 mice per group
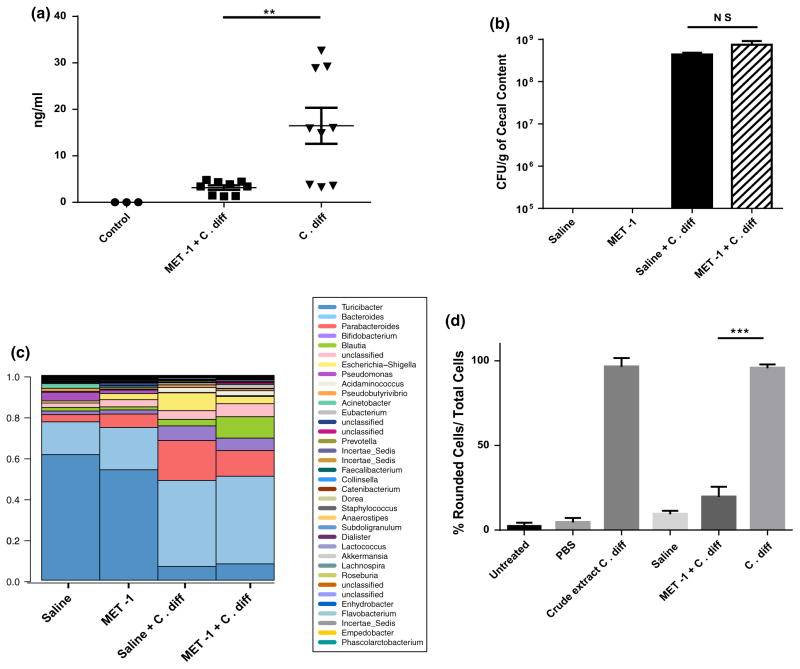
Next, a series of studies on the mouse fecal pellets were performed. When assessed by ELISA, less TcdA was detected from the stool of MET-1 pretreated, C. difficile-infected mice when compared to the same mg weight of stool from C. difficile-infected mice (Fig. 4a). However, when the C. difficile intestinal burden in the animals was assessed, there were no differences in the C. difficile colony-forming units (CFUs) found in the ceca of MET-1 mice as compared to vehicle controls (Fig. 4b), suggesting that the protective effects of MET-1 microbiota were not due to an effect of C. difficile viability or on its ability to propagate in the gut. The effect of the MET-1 and C. difficile treatments on the murine gut microbiota was then investigated using 16S rRNA gene sequencing from fecal samples. A bar plot of the microbiome composition on harvest day for the different treatment groups is shown in Fig. 4c. We did not observe C. difficile in this analysis since the number of CFUs of this organism was 4–5 orders of magnitude lower than the total number of bacteria in the system and was below the limit of detection of this methodology. However, when we analyzed the samples collected over the course of the experiment, as opposed to simply comparing the samples on harvest day, we found that the samples from the mice clustered together into two main groups: one cluster contained all the day 1 and non-MET-1 treated samples, whereas the other cluster contained all MET-1 treated day 2, 3 and harvest samples, regardless of the C. difficile treatment. These data indicated that treatment of the mice with MET-1 altered the murine gut microbiota and that this alteration was independent of the C. difficile treatment status (Fig. S2, see additional supplementary materials in appendix). Finally, a fibroblast cell rounding assay was used to test the cytotoxicity of the stool extracts from the mouse fecal pellets. The use of cell rounding assays, which were instrumental in the identification of C. difficile as the causative agent of this disease in humans [42, 43], is a well-established functional assay for assessing the cytopathic effects of C. difficile toxins [44]. When fibroblasts were treated with stool samples from mice gavaged with MET-1, markedly less cell rounding was observed compared to stool extracts from C. difficile-infected controls (Fig. 4d, see also Fig. S3 supplementary figure). Since the cytopathic effect in the fibroblast assay is toxin-mediated [44], these data further support that MET-1 is likely acting on the C. difficile toxins rather than affecting the viability of C. difficile bacteria in the gut.
Fig. 4.
MET-1 gavage does not decrease the colonic burden of C. difficile but decreases detectable toxin and cytotoxic effects in an antibiotic-mediated C. difficile colitis model and fibroblast rounding assay, respectively. C57BL/6 mice were treated with antibiotics and then gavaged with either MET-1 or saline, followed by 105 C. difficile strain 027 or saline 24 h later, as described in “Methods”. a Stool from untreated, MET-1 + C. diff and C. diff mice was collected at 24 h post-infection, and TcdA quantification was perfomed by ELISA. TcdA levels were decreased in the feces from MET-1 + C. diff mice compared to the C. diff mice at 24 h (**p <0.01). Data were analyzed using one-way ANOVA with Tukey correction (n = 3 untreated, n = 6 MET-1 and MET-1 + C. diff groups). b Cecal contents were cultured at 48 h, as described in “Methods”. Mice pretreated with MET had similar cecal counts of C. difficile compared to the vehicle control-infected mice. Data were analyzed using a one-way ANOVA with Tukey correction, n = 3 mice per group. c Compositional bar plot based on percentage of OTUs (operational taxonomic units) from mouse stool samples. Fecal pellets were collected, and DNA purified from the fecal pellets from the different treatment groups at harvest was compared. The bar plot shows the proportional abundance of each genus in the four samples indicated at the time of harvest [65]. C. difficile did not separate out in this analysis since the numbers of CFUs of this organism were 4–5 orders of magnitude lower than total number of bacteria in the system and were below the limit of detection of this methodology. d Reponse to toxins from fecal extracts of mice infected with C. difficile ribotype 027 using a fibroblast cytotoxicity assay. NIH 3T3 fibroblasts were incubated 1 h with mouse stool extracts (untreated, MET-1 + C. diff and C. diff mice). Cells were fixed with formalin and stained with Giemsa. Treatments were done in duplicate. Rounded and total cells were counted as decribed in “Methods”. Less cytotoxic effect was observed with stool extracts from MET-1 + C. diff mice compared to stool samples from C. diff control mice (***p <0.001). A crude extract culture C. difficile control group is also shown. Data were analyzed using one-way ANOVA with Tukey correction (2 repetitions per group). (Note: representative images can be found in supplementary figure S3)
MET-1 is protective against TcdA in an ileal loop model
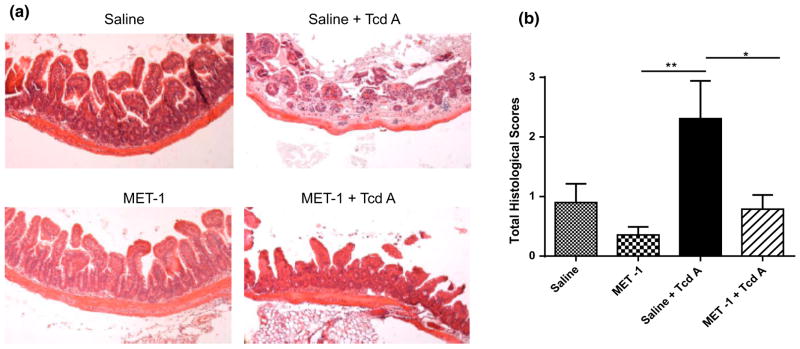
To determine whether MET-1 protected specifically against C. difficile toxin TcdA, we used an ileal loop model that had been employed by others to study the effects of TcdA on the gut [24–26, 41]. Since no live C. difficile bacteria are used in this model, these data would indicate whether the defined microbiota in MET-1 exerts protective effects against TcdA toxin. Mice were gavaged with either MET-1 microbiota or vehicle control, and then, using the ileal loop model, 50 μg of purified TcdA from ribotype strain 027 was injected into the ileal loops. Intestinal loops of vehicle control-gavaged animals injected with TcdA showed signs of epithelial destruction and inflammatory cell infiltration. In contrast, MET-1-gavaged animals showed significantly less histologic damage as compared to animals receiving vehicle control, indicating that MET-1 exerted a protective effect against TcdA in the absence of live C. difficile (Fig. 5a, b). No histological differences between PBS-injected control mice gavaged with MET-1 or vehicle control were noted.
Fig. 5.
Mice that received MET-1 treatment were protected when intestinal loops were exposed to TcdA from C. difficile. Mice were pretreated with antibiotics and gavaged with either MET-1 microbiota or saline vehicle control, and, using an ileal loop model, purified C. difficile toxin A (TcdA) was injected into the ileal loops, as described in “Methods”. a Representative H&E staining shows destruction of the villous tip architecture after 60-min incubation with TcdA in the intestine of the saline-gavaged mice (top right panel). Mice that were gavaged with MET-1 showed no villous loss, and surface epithelial cell integrity was preserved (bottom right panel), n = 3. b Histological scoring from the intestine demonstrates the MET-1 + TcdA treatment group has significantly less damage than the saline + TcdA group. Histological scores were obtained by measuring edema, PMN infiltration and epithelial cell damage in the loops. Scoring data were analyzed by one-way ANOVA with Tukey correction, n = 3 for the Saline, MET-1 and Saline+ TcdA groups and n = 4 for the MET-1 + TcdA group (*p <0.05, **p <0.01)
MET-1 protects against TcdA-mediated disruption of cytoskeletal structure and barrier function in human intestinal epithelial cell lines
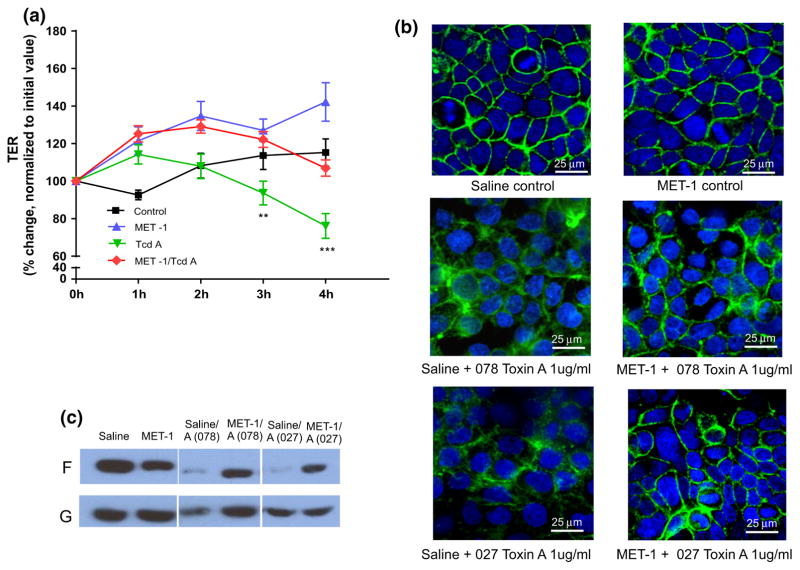
Two different human intestinal epithelial cell lines were then employed to test for effects of MET-1 on TcdA-mediated disruption of barrier integrity. First, T84 cells were grown on transwells, and then TER (transepithelial electrical resistance), a measure of epithelial barrier function integrity that is disrupted by exposure to TcdA [45], was measured. MET-1 preserved barrier function in cells treated with TcdA compared to vehicle control (Fig. 6a). To determine whether MET-1 conferred protective effects on the cytoskeletal structure, another human intestinal epithelial cell line (Caco2) was treated with MET-1 and TcdA and then stained with FITC-phalloidin, which binds to filamentous (F)-actin. This assay has been used by others to show TcdA-mediated disruption of the actin cytoskeleton [46] and provides visualization of the effects of TcdA on the integrity of the actin cytoskeleton. It was again found that MET-1 protected against disruption of F-actin by TcdA purified from two different hypervirulent strains (ribotype 078 and ribotype 027) of C. difficile (Fig. 6b). TcdA is known to mediate disassembly of filamentous actin [45, 47], and when the amount of F-actin decreases, mostly G-actin monomers or globular (G)-actin remain [48]. Western blot analysis of the filamentous to globulin actin ratios confirmed that MET-1 preserved F-actin in Caco2 cells treated with TcdA (Fig. 6c).
Fig. 6.
MET-1 protects Caco-2 intestinal epithelial cells against damage from C. difficile TcdA. a T84 cells were grown to confluence on transwells and then treated with MET-1 for 4 h, followed by addition of ribotype 027 TcdA. Transepithelial electrical resistance (TER), a measure of barrier function, was monitored over 4 h. MET-1 protected against a TcdA-mediated drop in TER, as compared to TcdA without MET-1 pretreatment (**p <0.01 at 3 h and ***p <0.001 at 4 h). Data were analyzed using two-way ANOVA and Tukey correction (n = 3). b Representative images of confocal FITC-phalloidin [which binds to filamentous (F) actin] of Caco-2 cells pretreated overnight with saline or MET-1 prior to TcdA 027 or TcdA 078 exposure for 3 h. Note the more preserved “chicken wire” appearance in the MET-1 + toxin groups compared to saline + toxin controls where the perijunctional actin ring is disrupted (n = 3). c Representative Western blot of filamentous (F) and globular (G) actin, another measure of actindynamics, showing the effect of MET-1 pretreatment on the distribution of actin in F versus G pools in cells treated with C. difficile TcdA (indicated by the ratio of F-to-G actin). MET-1 pretreatment protected against actin depolymerization (conversion of F to G actin) caused by exposure to C. difficile toxin A (n = 3)
MET-1 decreases TcdA-mediated apoptosis and degrades TcdA
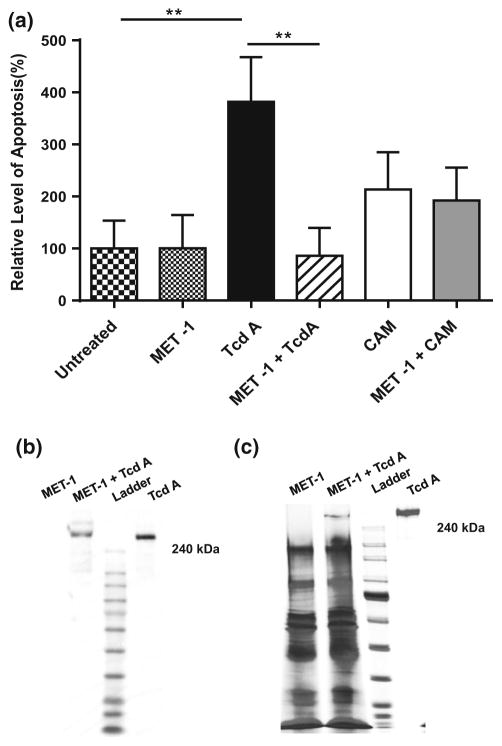
Since TcdA is known to induce apoptosis [49], and apoptosis is a recognized contributor to loss of barrier function [50–52], the effects of MET-1 on TcdA-mediated apoptosis were investigated. Caco2 cells that were pretreated with MET-1 or saline overnight were washed with fresh media and then incubated with TcdA for 6 h (camptothecin was used as an apoptosis-inducing positive control). Using a histone deacetylase assay, it was found that MET-1 protected against TcdA-mediated apoptosis (Fig. 7a). Finally, to determine whether MET-1 had a direct effect on TcdA, MET-1 was directly co-incubated with purified TcdA, and then samples were resolved by Western blot. Co-incubation of MET-1 with purified TcdA resulted in the disappearance of the higher molecular weight band and appearance of lower molecular weight bands by Western blot analysis, indicating that MET-1 is capable of degrading TcdA (Fig. 7b). A silver stain gel of total protein is also shown (Fig. 7c).
Fig. 7.
MET-1 inhibits C. difficile TcdA-mediated apoptosis and degrades TcdA. a Caco2 cells grown to confluence were pretreated with MET-1 overnight and then incubated with TcdA purified from C. difficile ribotype strain 027 for 6 h. Cells were harvested and apoptosis evaluated by histone deacetylase ELISA. The relative level of apoptosis was obtained by normalizing the absorbance at 405 nm to the untreated group. MET-1 inhibited apoptosis caused by TcdA (**p <0.01). In contrast, MET-1 did not protect against CAM [CAM = camptothecin (2 μg/ml), apoptosis-inducing positive control]. Data were analyzed using one-way ANOVA with Tukey correction (n = 3). b Representative blot of three independent experiments shown of purified TcdA (1 μg) co-incubated with MET-1 (25 μl), showing degradation of TcdA. c Representative 4–15 % SDS-PAGE silver staining of three independent experiments shown of purified TcdA (1 μg) co-incubated with MET-1 (25 μl) for 2 h at 37 °C, showing a decrease in the large MW band corresponding to the same size as TcdA, in the MET-1 + TcdA lane. Samples were pelleted to remove MET-1 bacteria
Discussion
It has been reported that the gut microbiota protect against C. difficile, and a variety of mechanisms have been proposed including effects on host inflammatory pathways and bile salt and sialic acid metabolism [9–12]. Most of these studies demonstrate a mechanism of action that involves killing C. difficile, competing with C. difficile or inhibiting its growth. Previously, using a defined microbial ecosystem derived from the stool of a healthy human volunteer (MET-1), we had shown therapeutic efficacy in humans against C. difficile infection [19]. This study shows that mice pre-treated with MET-1 were also protected against C. difficile colitis, showing efficacy of MET-1 as a prevention model. MET-1 decreased inflammation both locally and systemically and decreased the overall amount of detectable TcdA present in mouse stool, but did not decrease the intestinal C. difficile bacterial burden. These data indicate that gut microbiota can protect against C. difficile colitis even without actually killing the pathogen. Our MET-1/TcdA co-incubation data further demonstrate the novel finding that the protective effects of the gut microbiota in C. difficile infection may also be due to effects on the toxins themselves.
There is some precedent for this in the literature with S. boulardii, a yeast probiotic [53, 54]. S. boulardii produces a protease that cleaves TcdA and may also have an effect on the receptor of TcdA [53, 54]. However, although S. boulardii has been used successfully as an adjunct therapy with vancomycin to treat C. difficile [55], particularly in the current clinical setting of more hypervirulent C. difficile strains, it has not shown the same dramatic effects as FMT in treating CDI. MET-1, a more complete ecosystem that more closely resembles the native intestinal gut microbiota, may have the advantage of contributing mechanisms of action that are more complex and likely multifactorial in nature.
To our knowledge, the current study is the first of its kind demonstrating that human-derived gut microbes degrade C. difficile toxins, which is likely a major contributor to their protective mechanism of action. A large multicenter study of CDI demonstrated that positive cytotoxin assays detecting free toxins in stool samples were a better predictor of clinical outcome compared to either stool culture for toxigenic C. difficile or DNA amplification detection methods [15]. These data suggest that the intestinal toxin load is likely the important clinical driver in this disease, and a more recent prospective observational cohort study of over a thousand hospitalized patients supported these findings [16].
Our study focused on the effects of TcdA, isolated from different C. difficile ribotype strains. We chose to study TcdA rather than TcdB, since the effects of TcdA had previously been well characterized by others with the ileal loop model that we used in the current study [24–26, 41]. In addition, TcdA has been identified as an important contributor to the pathogenesis of CDI in humans [56], and its toxicity has been well described in a number of different animal models [49, 57]. However, TcdB is also of major clinical importance in this disease [14, 58]. Clinically relevant TcdA(−) strains of C. difficile exist (notably the 017 ribotype), particularly well described in Asian countries [59–61], and evidence suggests they are becoming increasingly common [62]. It is possible that MET-1 may not protect against these TcdA(−) strains of C. difficile. However, given the similarities in toxin structure and mechanism of action, it is tempting to speculate that MET-1 may also display activity against TcdB and that a broader and more diverse consortium of microbiota, such as is found in FMT, may confer bioactivity against both TcdA and TcdB. The effects of MET-1 and other microbial communities on TcdB activity and on binary toxin (CDT) of C. difficile are currently under investigation.
Finally, our data point toward enhancement of host barrier function as another protective mechanism of MET-1. TcdA has long been known to disrupt the intestinal epithelial barrier [45], and MET-1 prevented a drop in transepithelial electrical resistance (TER), a measure of barrier function integrity, in TcdA-exposed intestinal epithelial cells. MET-1 also attenuated TcdA-mediated apoptosis, another factor known to contribute to loss of gut barrier integrity [45], and enhanced tight junction protein expression. MET-1 has been shown to protect the expression of tight junction proteins important in the maintenance of host barrier function in an antibiotic-associated colitis animal model using Salmonella typhimurium [23], a pathogen that does not depend on toxin production for its pathogenicity. Of interest, MET-1 also preserves tight junction protein expression and attenuates systemic and local inflammation in a DSS mouse model of colitis [63]. Dextran sulfate sodium (DSS), a sulfated dextran molecule that is administered to mice in their water supply, is one of the most commonly used animal models of IBD [64]. DSS chemically compromises barrier function but does not rely on any specific pathogen to induce colitis, and MET-1-mediated enhancement of gut barrier function may be important irrespective of its ability to degrade TcdA. In conclusion, our data show for the first time that degradation of C. difficile toxins by intestinal bacteria and protection of the gut barrier integrity likely play a major role in their protective mechanism of action against C. difficile disease. In our pretreatment model the effect of MET-1 on C. difficile viability seems to offer little, if any, contribution to its protective effects on the host.
Supplementary Material
Acknowledgments
This study was supported by the Canada Foundation for Innovation and the Southeastern Ontario Academic Medical Organization (E.O.P) by NIH Grant No R21AI121575 (E.O.P, E.A.V., G.B.G) and by a Queen’s University CTAQ award (E.O.P and D.J.H). M.G.R. was supported by postdoctoral fellowship CONACYT-263618.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00535-016-1232-y) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest E.O.P and E.A-V. are co-founders of Nubiyota and have filed a patent for MET-1 through Parteq Innovations (Queen’s University). The other authors have no conflict of interests to declare.
References
- 1.Rohlke F, Stollman N. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Ther Adv Gastroenterol. 2012;5(6):403–20. doi: 10.1177/1756283X12453637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 4.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 5.Britton RA, Young VB. Interaction between the intestinal microbiota and host in Clostridium difficilecolonization resistance. Trends Microbiol. 2012;20(7):313–9. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seekatz AM, Theriot CM, Molloy CT, et al. Fecal microbiota transplantation eliminates Clostridium difficile in a murine model of relapsing disease. Infect Immun. 2015;83(10):3838–46. doi: 10.1128/IAI.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson KH. The microecology of Clostridium difficile. Clin Infect Dis. 1993;16(Suppl 4):S214–8. doi: 10.1093/clinids/16.supplement_4.s214. [DOI] [PubMed] [Google Scholar]
- 8.Li M, Liang P, Li Z, et al. Fecal microbiota transplantation and bacterial consortium transplantation have comparable effects on the re-establishment of mucosal barrier function in mice with intestinal dysbiosis. Front Microbiol. 2015;6:692. doi: 10.3389/fmicb.2015.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–8. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190(7):2505–12. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–9. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuehne SA, Cartman ST, Heap JT, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467(7316):711–3. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 13.Kuehne SA, Collery MM, Kelly ML, et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209(1):83–6. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyras D, O’Connor JR, Howarth PM, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458(7242):1176–9. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect Dis. 2013;13(11):936–45. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polage CR, Gyorke CE, Kennedy MA, et al. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med. 2015;175(11):1792–801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerding DN, Meyer T, Lee C, et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA. 2015;313(17):1719–27. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 18.Lawley TD, Clare S, Walker AW, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8(10):e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrof EO, Gloor GB, Vanner SJ, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1(1):3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidet P, Barbut F, Lalande V, et al. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175(2):261–6. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan NM, Pellett S, Wilkins TD. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35(3):1032–40. doi: 10.1128/iai.35.3.1032-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135(6):1984–92. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Martz SL, McDonald JA, Sun J, et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep. 2015;5:16094. doi: 10.1038/srep16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castagliuolo I, Riegler M, Pasha A, et al. Neurokinin-1 (NK-1) receptor is required in Clostridium difficile-induced enteritis. J Clin Invest. 1998;101(8):1547–50. doi: 10.1172/JCI2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Kokkotou EG, Mustafa N, et al. Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. J Biol Chem. 2006;281(34):24449–54. doi: 10.1074/jbc.M605200200. [DOI] [PubMed] [Google Scholar]
- 26.Ishida Y, Maegawa T, Kondo T, et al. Essential involvement of IFN-gamma in Clostridium difficile toxin A-induced enteritis. J Immunol. 2004;172(5):3018–25. doi: 10.4049/jimmunol.172.5.3018. [DOI] [PubMed] [Google Scholar]
- 27.McDonald JA, Fuentes S, Schroeter K, et al. Simulating distal gut mucosal and luminal communities using packed-column biofilm reactors and an in vitro chemostat model. J Microbiol Methods. 2015;108:36–44. doi: 10.1016/j.mimet.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Gloor GB, Hummelen R, Macklaim JM, et al. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One. 2010;5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masella AP, Bartram AK, Truszkowski JM, et al. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 32.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yilmaz P, Parfrey LW, Yarza P, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucl Acid Res. 2014;42:D643–8. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aitchison J. The statistical analysis of compositional data. New Jersey: Chapman and Hall; 1986. [Google Scholar]
- 36.Lovell D, Pawlowsky-Glahn V, Egozcue JJ, et al. Proportionality: a valid alternative to correlation for relative data. PLoS Comput Biol. 2015;11(e):1004075. doi: 10.1371/journal.pcbi.1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandes AD, Reid JN, Macklaim JM, et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:15. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Boogaart K, Tolosana-Delgado R. Analyzing compositional data with R. New York: Springer; 2013. [Google Scholar]
- 39.R Core Team. R: a language and environment for statistical computing. Vienna, Austria: 2015. Available from: http://www.R-project.org/ [Google Scholar]
- 40.Palarea-Albaladejo J, Martín-Fernández JA. zCompositions: R package for multivariate imputation of left-censored data under a compositional approach. Chemometr Intell Lab. 2015;143:85–96. [Google Scholar]
- 41.Rao K, Erb-Downward JR, Walk ST, et al. The systemic inflammatory response to Clostridium difficile infection. PLoS One. 2014;9(3):e92578. doi: 10.1371/journal.pone.0092578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.George RH, Symonds JM, Dimock F, et al. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br Med J. 1978;1(6114):695. doi: 10.1136/bmj.1.6114.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartlett JG, Chang TW, Gurwith M, et al. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298(10):531–4. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 44.Chang TW, Lauermann M, Bartlett JG. Cytotoxicity assay in antibiotic-associated colitis. J Infect Dis. 1979;140(5):765–70. doi: 10.1093/infdis/140.5.765. [DOI] [PubMed] [Google Scholar]
- 45.Hecht G, Pothoulakis C, LaMont JT, et al. Clostridium difficile toxin A perturbs cytoskeletal structure and tight junction permeability of cultured human intestinal epithelial monolayers. J Clin Invest. 1988;82(5):1516–24. doi: 10.1172/JCI113760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu TS, Musch MW, Sugi K, et al. Protective role of HSP72 against Clostridium difficile toxin A-induced intestinal epithelial cell dysfunction. Am J Physiol Cell Physiol. 2003;284(4):C1073–82. doi: 10.1152/ajpcell.00134.2002. [DOI] [PubMed] [Google Scholar]
- 47.Moore R, Pothoulakis C, LaMont JT, et al. C. difficile toxin A increases intestinal permeability and induces Cl− secretion. Am J Physiol. 1990;259(2 Pt 1):G165–72. doi: 10.1152/ajpgi.1990.259.2.G165. [DOI] [PubMed] [Google Scholar]
- 48.Musch MW, Petrof EO, Kojima K, et al. Bacterial superantigen-treated intestinal epithelial cells upregulate heat shock proteins 25 and 72 and are resistant to oxidant cytotoxicity. Infect Immun. 2004;72(6):3187–94. doi: 10.1128/IAI.72.6.3187-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirota SA, Iablokov V, Tulk SE, et al. Intrarectal instillation of Clostridium difficile toxin A triggers colonic inflammation and tissue damage: development of a novel and efficient mouse model of Clostridium difficile toxin exposure. Infect Immun. 2012;80(12):4474–84. doi: 10.1128/IAI.00933-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders DS. Mucosal integrity and barrier function in the pathogenesis of early lesions in Crohn’s disease. J Clin Pathol. 2005;58(6):568–72. doi: 10.1136/jcp.2004.021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Z, Wang X, Wallen R, et al. The influence of apoptosis on intestinal barrier integrity in rats. Scand J Gastroenterol. 1998;33(4):415–22. doi: 10.1080/00365529850171053. [DOI] [PubMed] [Google Scholar]
- 52.Zeissig S, Bojarski C, Buergel N, et al. Downregulation of epithelial apoptosis and barrier repair in active Crohn’s disease by tumour necrosis factor alpha antibody treatment. Gut. 2004;53(9):1295–302. doi: 10.1136/gut.2003.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castagliuolo I, LaMont JT, Nikulasson ST, et al. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun. 1996;64(12):5225–32. doi: 10.1128/iai.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castagliuolo I, Riegler MF, Valenick L, et al. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun. 1999;67(1):302–7. doi: 10.1128/iai.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surawicz CM, McFarland LV, Greenberg RN, et al. The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin Infect Dis. 2000;31(4):1012–7. doi: 10.1086/318130. [DOI] [PubMed] [Google Scholar]
- 56.Islam J, Taylor AL, Rao K, et al. The role of the humoral immune response to Clostridium difficile toxins A and B in susceptibility to C. difficile infection: a case-control study. Anaerobe. 2014;27:82–6. doi: 10.1016/j.anaerobe.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyerly DM, Saum KE, MacDonald DK, et al. Effects of clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47(2):349–52. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Savidge TC, Pan WH, Newman P, et al. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology. 2003;125(2):413–20. doi: 10.1016/s0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 59.Kim H, Riley TV, Kim M, et al. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J Clin Microbiol. 2008;46(3):1116–7. doi: 10.1128/JCM.01188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rupnik M, Kato N, Grabnar M, et al. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J Clin Microbiol. 2003;41(3):1118–25. doi: 10.1128/JCM.41.3.1118-1125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin BM, Kuak EY, Yoo HM, et al. Multicentre study of the prevalence of toxigenic Clostridium difficile in Korea: results of a retrospective study 2000–2005. J Med Microbiol. 2008;57(Pt 6):697–701. doi: 10.1099/jmm.0.47771-0. [DOI] [PubMed] [Google Scholar]
- 62.Cairns MD, Preston MD, Lawley TD, et al. Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative clostridium difficile sublineage PCR Ribotype 017 strain in London, England. J Clin Microbiol. 2015;53(10):3141–7. doi: 10.1128/JCM.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munoz S, Guzman-Rodriguez M, Sun J, et al. Rebooting the Microbiome. Gut Microb. 2016;13:1–11. doi: 10.1080/19490976.2016.1188248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wirtz S, Neufert C, Weigmann B, et al. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2(3):541–6. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 65.Aitchison J, Greenacre M. Biplots of compositional data. J Ry Stat Soc: Ser C (Appl Stat) 2002;51:375–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.