Abstract
Objective:
We tested whether brain gray matter (GM) imaging measures can differentiate between multiple sclerosis (MS) and neuromyelitis optica (NMO) using random-forest classification.
Methods:
Ninety participants (25 patients with MS, 30 patients with NMO, and 35 healthy controls [HCs]) were studied in Tehran, Iran, and 54 (24 patients with MS, 20 patients with NMO, and 10 HCs) in Padua, Italy. Participants underwent brain T1 and T2/fluid-attenuated inversion recovery MRI. Volume, thickness, and surface of 50 cortical GM regions and volumes of the deep GM nuclei were calculated and used to construct 3 random-forest models to classify patients as either NMO or MS, and separate each patient group from HCs. Clinical diagnosis was the gold standard against which the accuracy was calculated.
Results:
The classifier distinguished patients with MS, who showed greater atrophy especially in deep GM, from those with NMO with an average accuracy of 74% (sensitivity/specificity: 77/72; p < 0.01). When we used thalamic volume (the most discriminating GM measure) together with the white matter lesion volume, the accuracy of the classification of MS vs NMO was 80%. The classifications of MS vs HCs and NMO vs HCs achieved higher accuracies (92% and 88%).
Conclusions:
GM imaging biomarkers, automatically obtained from clinical scans, can be used to distinguish NMO from MS, even in a 2-center setting, and may facilitate the differential diagnosis in clinical practice.
Classification of evidence:
This study provides Class II evidence that GM imaging biomarkers can distinguish patients with NMO from those with MS.
Neuromyelitis optica (NMO) shares similar clinical and imaging characteristics with multiple sclerosis (MS), but a correct and timely diagnosis is essential for managing patients as treatment options differ considerably. Differential imaging characteristics include more pronounced brain atrophy, especially in the cortical gray matter (GM), in patients with MS than in patients with NMO 1; more severe thalamic changes in MS than NMO2,3; and a lack of cortical lesions in NMO.1,4,5
Machine-learning algorithms have shown promise in classifying MRI scans of patients with neurologic disorders.6 Their advantage over human observers is that they can handle a large number of variables from each patient and lack inconsistencies. Hence, they offer potential in the clinical setting to support the diagnostic process, and have been used successfully in Alzheimer disease,7 traumatic brain injury,8 and clinically isolated syndromes suggestive of MS.9
Random-forest classification is a powerful machine-learning approach for classification problems, such as distinguishing patients with different diseases, or separating neurologic patients from healthy controls (HCs), on the basis of their MRI scans, especially in multicenter studies.7 A random forest is a set of decision trees trained to answer a decision problem, for example: To which of the 2 groups does a patient belong? It starts with a training phase to construct a set of trees, each using a random subset of the training data, to characterize differences between groups. The testing (or validation) phase then predicts to which of the groups new cases belong by combining votes from the set of decision trees.10
We tested whether GM measures, obtained from MRI scans acquired as part of clinical protocols, can distinguish MS from NMO. In particular, we calculated the thickness, volume, and surface area of the cortex, and the volume of the basal ganglia, which reflect the underlying GM pathology.11 To test whether this can be used in a multicenter setting, we used data from 2 different centers. Finally, we investigated the GM measures that contributed most to the discrimination of MS from NMO, in order to obtain insights into the underlying pathology of these diseases.
METHODS
Our primary question was whether imaging biomarkers extracted from routine MRI measures discriminate between MS and NMO. The case-control design of this work provides Class II evidence.
Participants.
In this retrospective study, we collected all data from consecutive patients with relapsing-remitting MS and NMO. They were recruited in 2 tertiary centers, one in Iran (January 2009–December 2012)12 and one in Italy (June 2013–December 2013). The diagnosis of MS was made according to the McDonald criteria, which were revised in 2005,13 and that of NMO according the Wingerchuk criteria, which were revised in 2006.14 This study started and was completed before the publication of the new NMO criteria, but a retrospective evaluation showed that all patients with NMO fulfilled the 2015 criteria for NMO spectrum disorder (NMOSD).15 Other inclusion criteria were the absence of (1) concomitant neurologic or psychiatric disorders and (2) clinical relapses or IV methylprednisolone administration at least 6 weeks prior to the study entry.
Standard protocol approvals, registrations, and patient consents.
All participants gave written informed consent16 and for each cohort the local ethics committee approved the project.
Clinical assessment and MRI protocol.
All patients were clinically examined and the Expanded Disability Status Scale (EDSS) was scored on the day of MRI scan.17 All participants underwent MRI scans that included high-resolution 3D T1 and T2 fluid-attenuated inversion recovery (FLAIR) imaging, at 1.5T in Padua and 3T in Tehran. The MRI protocol for each cohort is shown in table e-1 at Neurology.org.
Image analysis.
We analyzed the scans of both centers in the same way. The aim was to calculate cortical volume, thickness, and surface area in 25 cortical regions for each hemisphere defined by LONI Probabilistic Brain Atlas (LPBA),18 and the volumes of deep GM nuclei (the accumbens, amygdala, caudate, pallidum, putamen, and thalamus). We performed image analysis in ANTs software (v1.9) and FSL (v5.0) (fsl.fmrib.ox.ac.uk/fsl/).
We manually constructed binary lesion maps on FLAIR/T2 scans, coregistered them with the T1 scan, and used this to fill hypointense lesions of T1 scans to avoid segmentation errors in patients.19 Next, we constructed a study-specific template as a common space for registration.20 We calculated transformation matrices from the atlas space to the common template, and then to each participant's native space, and applied it to LPBA labels to transfer them into native space to extract the imaging measures.
To calculate the cortical thickness, we used a new registration-based approach that has recently been validated.21,22 We calculated the surface area, at each cortical region, by dividing cortical volume by the thickness. We used FSL FIRST to calculate volumes of the deep GM structures.23
Classification analysis.
We performed all statistical analysis in R version 3.1.0 and Scientific Python 2.7 libraries (Scikit-Learn package).24,25 We calculated 157 variables, which were the cortical thickness, surface area, and volume of each cortical region, and the volume of each deep GM nucleus. We divided regional volumes by the total intracranial volume to adjust for different head sizes.26 We adjusted each GM measure separately for the effect of age. We fitted a linear regression line where age was the independent variable and each GM measure was the dependent variable. We calculated the amount of the GM measure that remained unexplained by the regression model (residual of the fit) and used it for all the subsequent analyses.
To investigate the effect of different centers and MRI scanners on the classification, we added the variable “center” to the analysis. Next, we constructed 3 models to differentiate between each pair of groups: (1) MS vs NMO; (2) MS vs HCs; and (3) NMO vs HCs. For each model, we randomly assigned participants from both centers to either the training or test set, so that each set contained half of the participants. Next, we performed the training step, and then the cross-validation on the left-out half (with 5,000 repetitions). The mean and SD of the accuracy of 5,000 trained and cross-validated models were calculated. Clinical diagnosis of MS or NMO was the gold standard against which the classification accuracy was calculated.
We used default values of random forest parameters as provided by the Scikit learn package, and calculated the importance of variables according to the original random-forest algorithm.10 Moreover, when distinguishing between MS and NMO, we built models using cortical region volume, thickness, surface area, and deep GM nuclei volumes, first on their own, and then in combination, to assess the effect of each measure on the accuracy, using a cross-validation approach as explained above. Since a random subset of variables is chosen for each decision tree in a forest of decision trees, random-forest classifiers are not affected by collinearity (correlation among surface, thickness, and volume).10 In a post hoc analysis, we simplified the model to assess the accuracy in distinguishing NMO from MS by including the most discriminating GM variable and white matter lesion load. Finally, we trained the NMO vs MS classifier with the scans of patients with NMO who were AQP4-positive and tested it to distinguish the scans of patients with NMO who were AQP4-negative from those with MS.
To gain a better understanding of GM changes, boxplots of the median and 75th percentile of cortical thickness, surface area, and volumes of the subcortical regions were calculated.
RESULTS
Participants.
We included 144 participants; 90 participants (25 patients with MS, 30 patients with NMO, and 35 HCs) were recruited in Tehran, Iran,12 and 54 (24 patients with MS, 20 patients with NMO, and 10 HCs) in Padua, Italy. The clinical and demographic characteristics are shown in table 1 (for treatment information, see table e-2).
Table 1.
Demographic characteristics of participants from each country
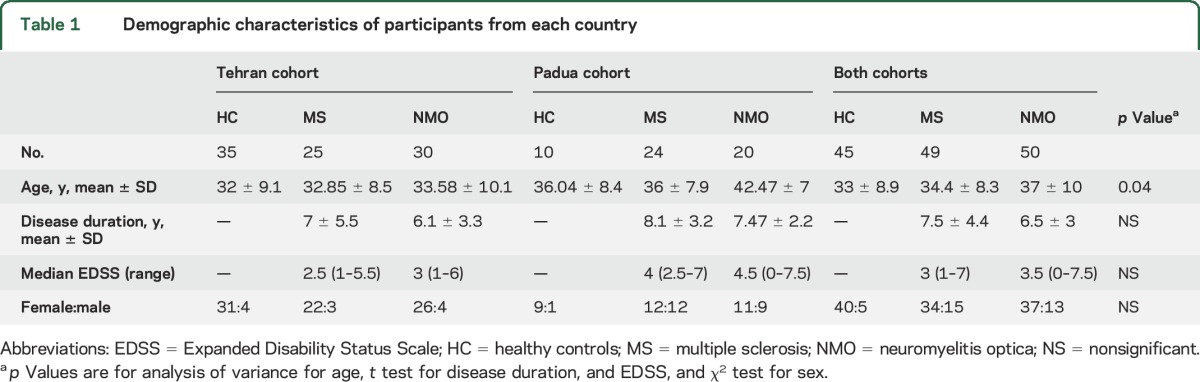
Seventeen out of 30 patients diagnosed with NMO from the Tehran cohort tested positive for aquaporin-4 autoantibody (AQP4-Ab) (immunofluorescence method, bioscientia.de/en/), while 18 out of 20 patients from the Padua cohort were AQP4-Ab positive (cell-based assay, euroimmun.com). Patients in the Italian cohort had significantly higher disability than those studied in Iran (MS: median EDSS 4.0 vs 2.5, p < 0.05; NMO: 4.5 vs 3, p < 0.05, respectively). Disease duration was not significantly different between the 2 cohorts. There was a significant difference in age between groups (table 1).
Classification results.
The average accuracy of the model for NMO vs MS trained with all GM imaging features was 74%, with a sensitivity of 77% (i.e., 77% of patients with MS were classified as MS) and a specificity of 72% (i.e., 72% of patients without MS, but with NMO, were classified correctly) (p < 0.01 calculated against a classifier with randomly changed, or permuted, labels and corrected for multiple comparisons with Bonferroni method) (table 2 and figure e-1). When cortical volume, cortical thickness, and surface area were used on their own to classify the 2 groups of patients, the average accuracy was lower (59%, 62%, and 66%, respectively) (figure 1). The results of the analysis remained the same when not adjusting for age.
Table 2.
Classification results
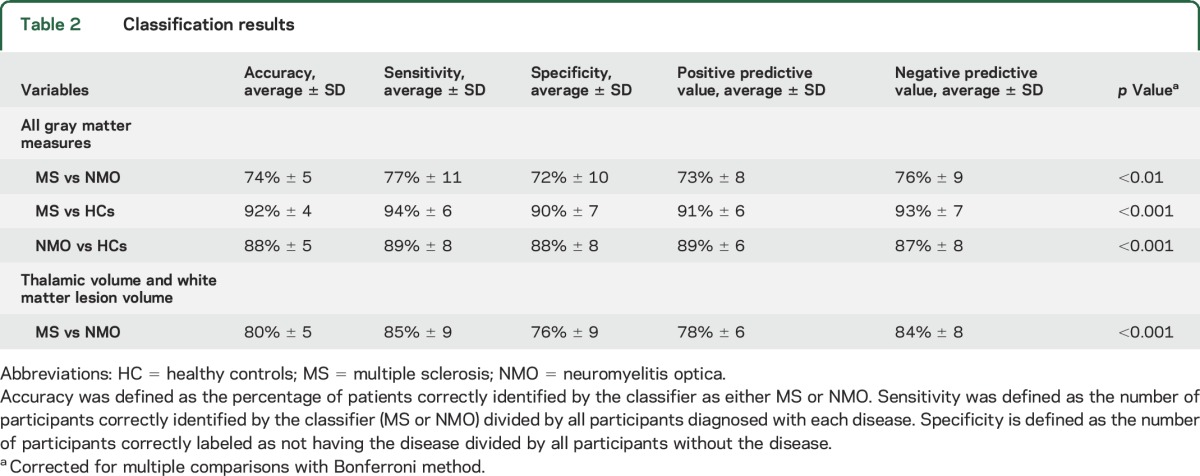
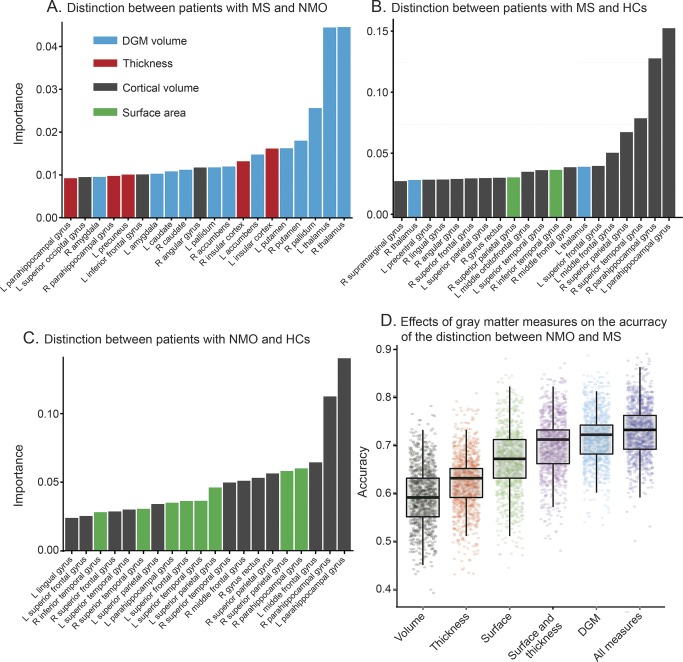
Figure 1. Importance of variables in the classifiers differentiating multiple sclerosis (MS), neuromyelitis optica (NMO), and healthy controls (HCs).
(A–C) The importance of each variable to classification inside random-forest algorithm. Importance is a relative measure, and is normalized to sum to 1 for each model, and should be used to compare the importance of variables inside each model (not among models). (D) The accuracy of models with different gray matter measures, including cortical volumes, cortical surface area, cortical thickness, thickness and surface area in combination, and subcortical volumes. The combination of surface area and thickness obtains a higher accuracy than volume. Each model has been trained and tested 1,000 times, after shuffling participants from 2 centers. DGM = deep gray matter.
A high accuracy (92%) was obtained when distinguishing MS from HCs (sensitivity/specificity 94/90; p < 0.001); the accuracy of classifying NMO vs HCs was also high (88%; sensitivity/specificity 89/88; p < 0.001).
Variable importance.
The volumes of the deep GM structures were the most important GM measures to distinguish MS from NMO, while the volumes of cortical regions were more important in distinguishing patients from HCs. In general, the GM volumes were more useful to the classification than the cortical thickness and surface area.
MS vs NMO.
The volumes of the deep GM (the thalami, right pallidum, and putamina) followed by the insular thickness, which showed reduction in MS compared to NMO (figures 1 and 2), were the most important GM measures to distinguish MS from NMO.
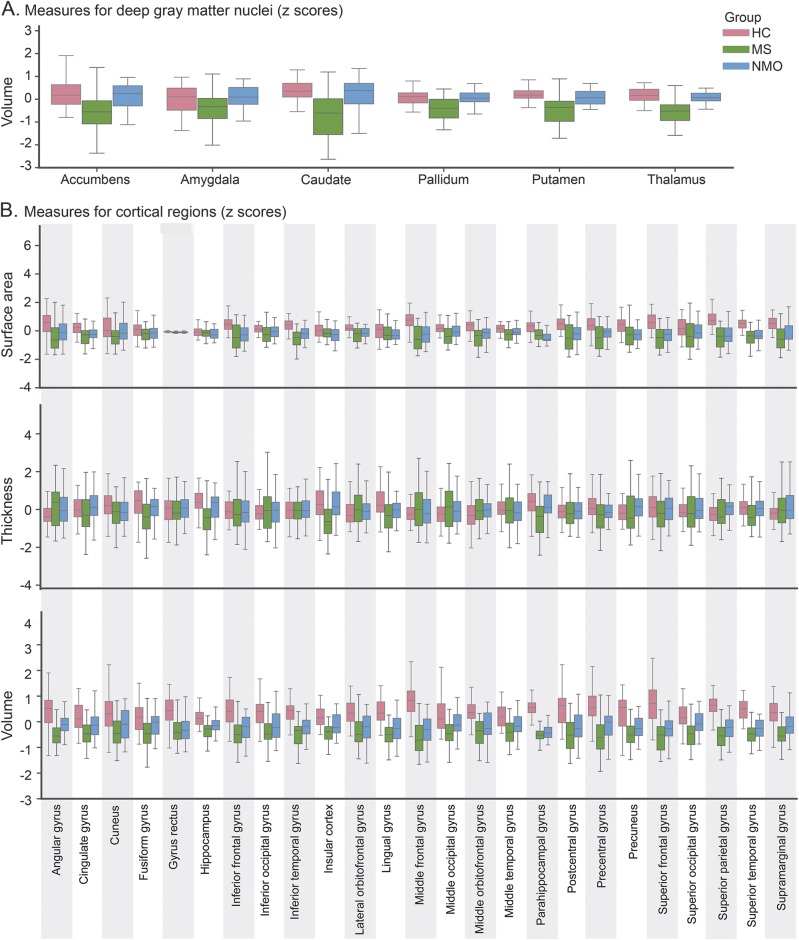
Figure 2. Descriptive statistics of the thickness, surface, and volume of deep gray matter (DGM) nuclei and cortical regions.
(A) DGM nuclei. (B) Cortical regions. Left and right hemisphere values are averaged for better visualization. All measures have been normalized, and the Z scores were calculated. HC = healthy control; MS = multiple sclerosis; NMO = neuromyelitis optica.
When looking at individual effects of GM measures (cortical and subcortical volumes, surface area, and thickness) to distinguish between NMO and MS, the combination of all measures gave the best accuracy, which was similar to the accuracy of the classification obtained using only the deep GM volumes (figure 1D), but higher than that obtained using each measure on its own.
When we used the thalamic volume (the most discriminating GM measure) together with the white matter lesion volume (see figure e-2), the average accuracy of the classification of MS vs NMO (over 5,000 permutations) was 80% (sensitivity/specificity 85%/76%, p < 0.001).
When we trained the NMO vs MS classifier using the data from people with NMO who were AQP4-positive and tested on people with NMO who were AQP4-negative, the performance of the model was similar to that of the model that trained and tested randomly selected scans from all people with NMO regardless of serostatus (average ± SD accuracy 77% ± 12, sensitivity 74% ± 19, specificity 80% ± 15).
NMO vs HC.
The volumes of the parahippocampal gyri and the left middle frontal gyrus were the most important variables for this classification; these regions showed smaller volumes in patients with NMO as compared to HCs (figure 2).
MS vs HC.
The volumes of the bilateral parahippocampal gyri and the right superior temporal gyrus were the most important variables for this classification (these regions showed reduced volumes in MS when compared to HCs). The variable “center” (i.e., Tehran and Padua) was consistently found to be the least important feature for all the classifications.
DISCUSSION
Previous imaging studies have highlighted differences in MRI characteristics between NMO and MS1,12,27,28, but an automatic distinction is still challenging. Here, we automatically classified patients with MS and NMO on the basis of their brain MRI scans routinely acquired with clinical protocols, using a random-forest classifier. Our findings showed that GM imaging measures, such as cortical thickness, cortical surface area, and subcortical GM volumes, led to an accuracy of 74% when classifying the 2 patient groups, which is higher than that obtained with each GM measure on its own. When we used both the most discriminating GM measure (the thalamic volume) and the white matter lesion volume, the accuracy of the model in classifying NMO vs MS was higher (80%) than the original model. Random-forest classification accuracy was higher for distinguishing MS from HCs (92%) and NMO from HCs (88%), because the classification task is less challenging than discriminating the 2 diseases. Therefore, this automatic approach may support the diagnostic process in clinical practice, for example in patients with NMOSD without AQP4. The variable “center” did not significantly affect the classification. Moreover, the contribution of imaging features to the classification was more relevant than the differences in clinical characteristics, which may exist between patient cohorts.
We demonstrated that the volumes of the deep GM, which were lower in people with MS than NMO (figure 2A), were more important features for discriminating NMO and MS than cortical thickness and volume. Our results are in line with a recent study that failed to show significant thalamic atrophy in NMO when compared to HCs,2,3 while another study detected a mild reduction in thalamic volume in NMO when compared to HCs.1 The thalamic volume loss in MS may be related to secondary neurodegeneration occurring in cortical areas or may represent primary neurodegeneration,1,2,11 whereas in NMO a milder neurodegeneration secondary to axonal degeneration in spinal cord or visual pathways is present.29
Cortical thickness represents the number of neurons in each cortical column.30 The thickness of the insular cortices had the second highest discriminating value (after the deep GM volumes) in distinguishing MS from NMO. This region shows a predilection for atrophy, especially as patients with MS progress.31 The insula is deep in the temporal lobe and may be more exposed to inflammatory mediators than other cortical areas.31
While atrophy in regions such as the thalamus and insula was only seen in MS, both demyelinating diseases seemed to affect the parahippocampal gyrus in a similar fashion (figure 2). The parahippocampus is part of the limbic system and has extensive connections to temporal, frontal, and deep GM regions. Transection of these connections, which occurs as a result of white matter lesions, could lead to atrophy of this region via retrograde neurodegeneration in both diseases.32,33 In addition, in MS, GM damage might be the result of either noninflammatory or inflammatory processes that have occurred within the GM itself.32,34 In NMO, the GM loss is more selective and depends on AQP4 expression, and the ratio of M1 to M23 proteins in astrocytes.33 The parahippocampus may show a different pattern of AQP4 expression or may be connected to areas with high AQP4 expression, although the dynamic pattern of AQP4 expression in the brain remains unclear.35,36
Patients with NMO may have severe relapses affecting the motor or visual system.37 However, GM measures of the occipital or motor cortices did not distinguish between NMO and MS or HCs. These findings are consistent with previous voxel-based morphometry studies in NMO that failed to show detectable changes in the GM volume of these regions when compared to MS or HCs.2,38,39 Another study found mild thinning of the calcarine, precentral, and postcentral cortices in NMO when compared to HCs, but no significant differences between NMO and MS.1
One strength of our study is that we included participants from 2 centers. We previously demonstrated, in a 1-center setting using scans from the same Iranian patients used in this study, that a combination of advanced MRI modalities (functional MRI and diffusion tensor imaging) distinguishes between MS and NMO with high accuracy, using a technique called data fusion with multikernel learning.12 The limitations of this technique are that it is computationally intensive and its application in multicenter studies is limited. The present work, using a random-forest classification, is more robust in a multicenter setting, and is less likely to overfit.10 We included center as a variable in our analysis and found that it had the lowest importance in all 3 classifications: NMO vs MS, MS vs HC, and NMO vs HC. We repeated the cross-validation 5,000 times, which allowed the inclusion of all patients in both training and test sets. This could be generalized to other centers, provided that sufficient participants from new centers are included in the training sample. Moreover, for an easier application we simplified the model including only thalamic volume and white matter lesion volume; this achieved 80% accuracy in distinguishing between MS and NMO. In the future, greater accuracy may be achieved by including additional features, such as lumbar puncture results, antibody status and quantitative spinal cord measures.40,41
In this study, patients with NMO were significantly older than patients with MS, and therefore we adjusted GM measures for age. NMO has a later disease onset than MS,37 so after we matched 2 groups according to disease duration, age was higher in NMO than MS. However, despite older age, which has the effect of reducing GM volume,42 patients with NMO had higher GM volume than those with MS, which suggests that age is unlikely to be a confounder in our analysis. Moreover, we have built our model using scans of patients with a secure diagnosis of MS or NMO, with a mean disease duration of 7.5 years. This is because the clinical diagnosis of MS or NMO was the gold standard against which the classification accuracy was calculated. An important question to be addressed in future studies is whether this algorithm can help to classify difficult radiologic cases or to predict the outcome after the first attack. For example, in a previous study we found that machine learning correctly predicted the presence (or absence) of clinically definite MS in 71.4% of patients within 1 year of onset of a clinically isolated syndrome.9
This study is not without limitations. First, the NMO group, which was recruited prior to the revised criteria for the diagnosis of NMOSD,15 included both AQP4-negative and AQP4-positive patients (15 negative and 50 positive). Recent work has highlighted the possible different clinical and radiologic characteristics between these 2 groups that may indicate distinct subtypes.43 There were no notable differences in age, sex, or clinical disability between NMO without AQP4 (2 from Italy, 13 from Iran) and NMO with AQP4. However, the group of seronegative patients was small. In the future, this technique could help to identify patients with NMOSD without AQP4, since in this cohort they show similarities with the NMOSD with AQP4 cases; in particular, the algorithm correctly classified them in 77% of NMOSD without AQP4 cases after having learned from only the NMOSD with AQP4 cases. Second, the role of GM lesions in distinguishing between the 2 diseases was not assessed,1 because the sequences that allow GM lesion detection are not routinely acquired in the clinical setting. Further studies will clarify these issues.
We showed that the random-forest classification robustly and automatically discriminates between MS and NMO on the basis of MRI scans in a 2-center setting with up to 80% accuracy. Furthermore, deep GM volumes and cortical thickness of specific key regions may give increased power to detect subtle GM features, which may facilitate the differential diagnosis between MS and NMO.
Supplementary Material
GLOSSARY
- AQP4-Ab
aquaporin-4 autoantibody
- EDSS
Expanded Disability Status Scale
- FLAIR
fluid-attenuated inversion recovery
- GM
gray matter
- HC
healthy control
- LPBA
LONI Probabilistic Brain Atlas
- MS
multiple sclerosis
- NMO
neuromyelitis optica
- NMOSD
neuromyelitis optica spectrum disorder
Footnotes
Supplemental data at Neurology.org
AUTHOR CONTRIBUTIONS
A.E. designed the project, acquired data from the Tehran cohort, analyzed the data, interpreted the analysis, and wrote the manuscript. V.W. contributed to research design, data analysis, and revised the manuscript. M.C. acquired data for the Padua cohort, supervised the project, and revised the manuscript. M.A.S. contributed to patient recruitment in the Tehran cohort, revised the manuscript, and supervised the project. A.J.T. supervised the project and revised the manuscript. D.C.A. and O.C. designed the project, supervised the project, and revised the manuscript.
STUDY FUNDING
A.E. has received the McDonald Fellowship from Multiple Sclerosis International Federation (msif.org). This study was funded by the Sina MS Research Centre and National Institute for Health Research (NIHR), UCL Hospitals (UCLH) Biomedical Research Centre (BRC).
DISCLOSURE
A. Eshaghi holds a McDonald Fellowship from Multiple Sclerosis International Federation (MSIF) and a MAGNIMS Fellowship from the European Committee for Treatment and Research in Multiple Sclerosis (ECTRIMS). V. Wottschel and R. Cortese report no disclosures relevant to the manuscript. M. Calabrese serves on the advisory boards of Merck-Serono, Bayer-Schering, Genzyme, Roche, and Biogen and as a consultant for Biogen, Merck-Serono, Bayer-Schering, and Roche. M. Sahraian received travel support and lecture fees from Biogen Idec, Merck Serono, Bayer-Schering, Novartis, and Cinnagen. A. Thompson has received honoraria, support for travel, or consultancy from Biogen Idec, Eisai, Excemed, MedDay, and Novartis; has received support for travel from the MS International Federation as Chair of their Medical and Scientific Advisory Board, the International Progressive MS Alliance as chair of their Scientific Steering Committee, and the National MS Society (USA) as member of their Research Programs Advisory Committee; and receives an honorarium from SAGE Publishers as Editor-in-Chief of Multiple Sclerosis Journal. D. Alexander reports no disclosures relevant to the manuscript. O. Ciccarelli receives research grant support from the EPSRC, Multiple Sclerosis Society of Great Britain and Northern Ireland, and the Department of Health NIHR Comprehensive Biomedical Centre; serves as consultant for GE, Novartis, and Biogen (all payments are made to the Institute); and is an Associate Editor of Neurology®. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Calabrese M, Oh MS, Favaretto A, et al. No MRI evidence of cortical lesions in neuromyelitis optica. Neurology 2012;79:1671–1676. [DOI] [PubMed] [Google Scholar]
- 2.Matthews L, Kolind S, Brazier A, et al. Imaging surrogates of disease activity in neuromyelitis optica allow distinction from multiple sclerosis. PLoS One 2015;10:e0137715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finke C, Heine J, Pache F, et al. Normal volumes and microstructural integrity of deep gray matter structures in AQP4+ NMOSD. Neurol Neuroimmunol Neuroinflamm 2016;3:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinnecker T, Dorr J, Pfueller CF, et al. Distinct lesion morphology at 7-T MRI differentiates neuromyelitis optica from multiple sclerosis. Neurology 2012;79:708–714. [DOI] [PubMed] [Google Scholar]
- 5.Kister I, Herbert J, Zhou Y, Ge Y. Ultrahigh-field MR (7 T) imaging of brain lesions in neuromyelitis optica. Mult Scler Int 2013;2013:398259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klöppel S, Stonnington CM, Chu C, et al. Automatic classification of MR scans in Alzheimer's disease. Brain 2008;131:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebedev AV, Westman E, Van Westen GJ, et al. Random forest ensembles for detection and prediction of Alzheimer's disease with a good between-cohort robustness. NeuroImage Clin 2014;6:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lui YW, Xue Y, Kenul D, Ge Y, Grossman RI, Wang Y. Classification algorithms using multiple MRI features in mild traumatic brain injury. Neurology 2014;83:1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wottschel V, Alexander DC, Kwok PP, et al. Predicting outcome in clinically isolated syndrome using machine learning. NeuroImage Clin 2015;7:281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Liaw A, Breiman L. Using Random Forest to Learn Imbalanced Data. Berkeley: University of California; 2004. [Google Scholar]
- 11.Minagar A, Barnett MH, Benedict RH, et al. The thalamus and multiple sclerosis: modern views on pathologic, imaging, and clinical aspects. Neurology 2013;80:210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eshaghi A, Riyahi-Alam S, Saeedi R, et al. Classification algorithms with multi-modal data fusion could accurately distinguish neuromyelitis optica from multiple sclerosis. NeuroImage Clin 2015;7:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polman CH, Reingold SC, Edan G, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria.” Ann Neurol 2005;58:840–846. [DOI] [PubMed] [Google Scholar]
- 14.Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Revised diagnostic criteria for neuromyelitis optica. Neurology 2006;66:1485–1489. [DOI] [PubMed] [Google Scholar]
- 15.Wingerchuk DM, Banwell B, Bennett JL, et al. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology 2015;85:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Medical Association. Declaration of Helsinki, Ethical Principles for Medical Research Involving Human Subjects. Edinburgh: 52nd WMA General Assembly; 2000. [PubMed] [Google Scholar]
- 17.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 1983;33:1444–1452. [DOI] [PubMed] [Google Scholar]
- 18.Shattuck DW, Mirza M, Adisetiyo V, et al. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 2008;39:1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sdika M, Pelletier D. Nonrigid registration of multiple sclerosis brain images using lesion inpainting for morphometry or lesion mapping. Hum Brain Mapp 2009;30:1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avants BB, Yushkevich P, Pluta J, et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 2010;49:2457–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das SR, Avants BB, Grossman M, Gee JC. Registration based cortical thickness measurement. Neuroimage 2009;45:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tustison NJ, Cook PA, Klein A, et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage 2014;99:166–179. [DOI] [PubMed] [Google Scholar]
- 23.Patenaude B, Smith SM, Kennedy DN, Jenkinson MA. Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 2011;56:907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliphant TE. Python for scientific computing. Comput Sci Eng 2007;9:10–20. [Google Scholar]
- 25.Abraham A, Pedregosa F, Eickenberg M, et al. Machine learning for neuroimaging with Scikit-learn. Front Neuroinform 2014;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peelle JE, Cusack R, Henson RN. Adjusting for global effects in voxel-based morphometry: gray matter decline in normal aging. Neuroimage 2012;60:1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HJ, Paul F, Lana-Peixoto MA, et al. MRI characteristics of neuromyelitis optica spectrum disorder: an international update. Neurology 2015;84:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan Y, Liu Y, Liang P, et al. Comparison of grey matter atrophy between patients with neuromyelitis optica and multiple sclerosis: a voxel-based morphometry study. Eur J Radiol 2012;81:e110–114. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Fu Y, Schoonheim MM, et al. Structural MRI substrates of cognitive impairment in neuromyelitis optica. Neurology 2015;85:1491–1499. [DOI] [PubMed] [Google Scholar]
- 30.Sailer M, Fischl B, Salat D, et al. Focal thinning of the cerebral cortex in multiple sclerosis. Brain 2003;126:1734–1744. [DOI] [PubMed] [Google Scholar]
- 31.Howell OW, Reeves CA, Nicholas R, et al. Meningeal inflammation is widespread and linked to cortical pathology in multiple sclerosis. Brain 2011;134:2755–2771. [DOI] [PubMed] [Google Scholar]
- 32.Calabrese M, Magliozzi R, Ciccarelli O, Geurts JJ, Reynolds R, Martin R. Exploring the origins of grey matter damage in multiple sclerosis. Nat Rev Neurosci 2015;16:147–158. [DOI] [PubMed] [Google Scholar]
- 33.Saji E, Arakawa M, Yanagawa K, et al. Cognitive impairment and cortical degeneration in neuromyelitis optica. Ann Neurol 2013;73:65–76. [DOI] [PubMed] [Google Scholar]
- 34.Geurts JJ, Calabrese M, Fisher E, Rudick RA. Measurement and clinical effect of grey matter pathology in multiple sclerosis. Lancet Neurol 2012;11:1082–1092. [DOI] [PubMed] [Google Scholar]
- 35.Amiry-Moghaddam M, Ottersen OP. The molecular basis of water transport in the brain. Nat Rev Neurosci 2003;4:991–1001. [DOI] [PubMed] [Google Scholar]
- 36.Pittock SJ, Weinshenker BG, Lucchinetti CF, Wingerchuk DM, Corboy JR, Lennon VA. Neuromyelitis optica brain lesions localized at sites of high aquaporin 4 expression. Arch Neurol 2006;63:964–968. [DOI] [PubMed] [Google Scholar]
- 37.Wingerchuk DM, Weinshenker BG. Neuromyelitis optica (Devic's syndrome). Handb Clin Neurol 2014;122:581–599. [DOI] [PubMed] [Google Scholar]
- 38.Blanc F, Noblet V, Jung B, et al. White matter atrophy and cognitive dysfunctions in neuromyelitis optica. PLoS One 2012;7:e33878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chanson JB, Lamy J, Rousseau F, et al. White matter volume is decreased in the brain of patients with neuromyelitis optica. Eur J Neurol 2013;20:361–367. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Wang J, Daams M, et al. Differential patterns of spinal cord and brain atrophy in NMO and MS. Neurology 2015;84:1465–1472. [DOI] [PubMed] [Google Scholar]
- 41.Lucchinetti CF, Guo Y, Popescu BF, Fujihara K, Itoyama Y, Misu T. The pathology of an autoimmune astrocytopathy: lessons learned from neuromyelitis optica. Brain Pathol 2014;24:83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedman AM, van Haren NE, Schnack HG, Kahn RS, Hulshoff Pol HE. Human brain changes across the life span: a review of 56 longitudinal magnetic resonance imaging studies. Hum Brain Mapp 2012;33:1987–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato DK, Callegaro D, Lana-Peixoto MA, et al. Distinction between MOG antibody-positive and AQP4 antibody-positive NMO spectrum disorders. Neurology 2014;82:474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.