Although P-PIT was developed to detect diseases associated with known human polyomaviruses, the identification of a new polyomavirus in rats suggests that it may have utility as a broad-based screen for new, as well as known polyomaviruses. Our findings suggest that RatPyV2 may be a commensal infection of laboratory rats that can lead to disseminated disease in T cell immune-deficient rats. Infection of the X-SCID rats with RatPyV2 and Pneumocystis carinii is a potential model for coinfection pathogenesis and treatment options during transplant preclinical studies.
KEYWORDS: epitheliotropism, immune suppression, KIV, P-PIT, WUV, X-SCID, inclusion body, laboratory animals, polyomavirus, simian virus 40
ABSTRACT
Polyomaviruses (PyVs) are known to infect a wide range of vertebrates and invertebrates and are associated with a broad spectrum of diseases, including cancers, particularly in immune-suppressed hosts. A novel polyomavirus, designated rat polyomavirus 2 (RatPyV2), was identified from a breeding colony of rats having X-linked severe combined immunodeficiency. Using a human panpolyomavirus immunohistochemistry test (P-PIT), RatPyV2 was initially detected in the parotid salivary gland of a colony member. Rolling circle amplification using DNA from harderian and parotid glands identified a novel 5.1-kb polyomavirus genome closely related to human Washington University (WU) and Karolinska Institute (KI) and vole polyomaviruses but notably divergent from Rattus norvegicus PyV1 (RnorPyV1; also designated RatPyV1). Further screening showed RatPyV2 inclusion body infection in the lung epithelium and variably in other respiratory, reproductive, and glandular tissues of 12/12 (100%) rats.
IMPORTANCE Although P-PIT was developed to detect diseases associated with known human polyomaviruses, the identification of a new polyomavirus in rats suggests that it may have utility as a broad-based screen for new, as well as known polyomaviruses. Our findings suggest that RatPyV2 may be a commensal infection of laboratory rats that can lead to disseminated disease in T cell immune-deficient rats. Infection of the X-SCID rats with RatPyV2 and Pneumocystis carinii is a potential model for coinfection pathogenesis and treatment options during transplant preclinical studies.
INTRODUCTION
Polyomaviruses (PyVs) are a family of small DNA viruses known to infect a variety of mammals, birds, fish, and scorpions (1, 2). They are members of the family Polyomaviridae and are nonenveloped icosahedral viruses, with circular double-stranded DNA genomes of approximately 5,000 base pairs (bp) (3). An early viral gene region encodes multiple regulatory proteins, called tumor antigens (T-Ag), by alternative splicing, and a late gene region encodes structural capsid proteins, including VP1, VP2, and VP3 (4).
Most mammalian polyomaviruses cause subclinical infections with lifelong persistence in their natural hosts. According to serological studies, asymptomatic infection occurs with 12 of the 13 known polyomaviruses detected in humans (5–14). Polyomavirus-related diseases, which include nephritis (BK virus [BKV]) (15), encephalitis (JC virus [JCV]) (16), Merkel cell carcinoma (Merkel cell polyomavirus [MCV]) (17), skin dysplasia (trichodysplasia spinulosa-associated polyomavirus [TSV] and human polyomavirus 7 [HPyV7]) (18, 19), and pneumonitis (Washington University [WU] virus [WUV]) (20, 21), can occur among immune-suppressed individuals, including posttransplantation and AIDS patients. A panpolyomavirus immunohistochemistry test (P-PIT), comprised of three antibodies (Pab416, Xt7, and 2t2), recognizes well-conserved antigenic epitopes of polyomavirus early proteins and has been show to detect T antigens of all 13 human polyomaviruses (21). Toptan et al. found positive P-PIT staining for all currently known polyomavirus-related diseased human tissues.
To date, only seven distinct rodent PyVs have been fully sequenced: mouse PyV, mouse pneumotropic PyV, hamster PyV, Mastomys PyV, bank vole PyV, common vole PyV, and Rattus norvegicus PyV1 (RnorPyV1) (22–27). These polyomaviruses were largely found as commensal infections. RnorPyV1, closely related to mouse and hamster PyV, appears to persist with no signs of disease in feral Norway rats. However, in 1984, Ward et al. reported a widespread infection in a colony of athymic nude rats manifesting with parotid sialoadenitis, bronchitis, rhinitis, and harderian adenitis (28). Immunohistochemical (IHC) staining at the time with an anti-simian virus 40 (SV40) T antigen antibody was reactive with infected tissues; however, no viral sequences were obtained.
Here, we describe the identification of a new polyomavirus (RatPyV2) associated with disseminated viral inclusion body disease in X-linked severe combined immune deficiency (X-SCID) rats that have a genetically disrupted interleukin-2 receptor gamma gene (IL2RG) (29). The majority of these laboratory rats infected with RatPyV2 had decreased fecundity but were otherwise asymptomatic. However, some rats developed acute respiratory distress associated with Pneumocystis carinii and RatPyV2 coinfection, as well as chromodacryorrhea (red tear secretion from the harderian gland). Phylogenetic analyses based on large T (LT) sequences show that RatPyV2 belongs to the betapolyomavirus genus as proposed by the International Committee in Taxonomy of Viruses (ICTV) (30). Phylogenetic analysis with virus protein 1 (VP1), similar to LT analysis, shows RatPyV2 to be most closely related to human WU and Karolinska Institute (KI) polyomaviruses and more remotely related to RatPyV1.
RESULTS
Viral outbreak in X-SCID rat colony.
During quarterly diagnostic screening, serologic positivity for P. carinii was detected in an X-SCID rat breeding colony. Four (2 male and 2 female) rats exhibiting respiratory distress and chromodacryorrhea were euthanized, and lungs were collected for histopathology evaluation (Fig. 1; see Fig. S1 in the supplemental material). PCR testing confirmed infection with P. carinii (data not shown), and all rats in the colony began treatment with 250 mg/kg of body weight/day sulfamethoxazole (SMZ) pulse treatment for 2 weeks orally in water bottles. Rats were given 2 weeks off and a subsequent second round of SMZ. Follow-up serology performed at quarterly testing indicated no active infection for P. carinii or for other known rat pathogens, including cytomegalovirus and mouse adenoviruses 1 and 2 (Table 1).
FIG 1 .
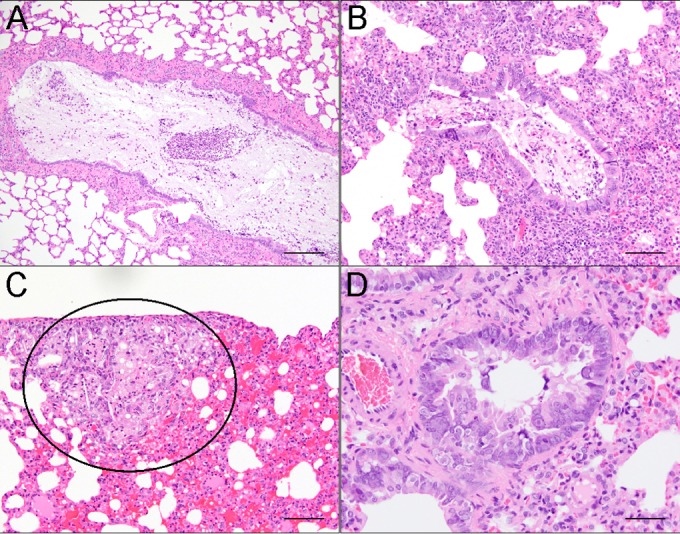
Lesions in hematoxylin-and-eosin (H&E)-stained lung sections of X-SCID rats infected with Pneumocystis carinii. (A) Bronchi were filled with neutrophils, mucous, foamy macrophages, and cell debris. Scale bar = 200 μm. (B) Bronchioles were filled with material similar to that in bronchi. Interstitial pneumonia was present in adjacent alveoli. Scale bar = 100 μm. (C) Subpleural foci consisted of alveoli filled with foamy macrophages and occasional cholesterol clefts (circled area), in addition to alveolar hemorrhage. Scale bar = 100 μm. (D) Bronchiolar hyperplasia was present, with intranuclear inclusion bodies within the bronchiolar epithelium. Scale bar = 50 μm.
TABLE 1 .
Pathogens included in quarterly (3 months) rodent surveillance testing
| Test type | Pathogen |
|---|---|
| Serology | Sendai virus (SEND) |
| Pneumonia virus of mice (PVM) | |
| Sialodacryoadenitis virus (SDAV) | |
| Kilham rat virus (KRV, RV) | |
| Toolan’s H1 virus (H-1) | |
| Rat parvovirus (RPV) | |
| Rat minute virus (RMV) | |
| Reovirus (REO-3) | |
| Rat theilovirus (RTV, TMEV, REV) | |
| Rat coronavirus (RCV) | |
| Pneumocystis carinii (previously thought to be rat respiratory virus [RRV]) | |
| Lymphocytic choriomeningitis (LCMV) | |
| Hantaan virus (HANT) | |
| Mouse adenovirus (MAV) | |
| Cilia-associated respiratory bacillus (CARB) | |
| Mycoplasma pulmonis (MPUL) | |
| Encephalitozoon cuniculi (ECUN) | |
| PCR | Pinworms of the genera Aspiculuris and Syphacia |
| Ectoparasites |
Silver staining of Pneumocystis carinii-coinfected lung samples in areas of interstitial pneumonia in H&E-stained tissue sections (left, top right) revealed black staining of cyst walls (bottom right). Left, ×20 magnification; right, ×100 magnification. Download Figure S1, PDF file, 0.9 MB (996.9KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Other than the 4 rats mentioned above, no rats within the colony displayed overt clinical symptoms. However, from a reproductive standpoint, female rats within this colony usually had 4 to 8 pups per litter, and the reproductive life span consisted of 2 litters per animal. Rarely, females had third litters with very poor survival rates, suggesting markedly reduced fecundity. Female rats were unable to maintain pregnancy after 6 months of age.
Although diagnostic screening excluded all known rat pathogens except P. carinii, histopathological changes, such as inclusion bodies, suggested a viral cause that could not be explained by P. carinii infection of the lung alone (Fig. 1). Therefore, a total of 8 additional rats (6 adults and 2 6-week-old weanlings) were examined via gross and microscopic pathology of all organs to look for pathological and immunohistochemical evidence of viral disease. Characteristic gross findings expected for the X-SCID strain (29) included severe thymic hypoplasia, unidentifiable lymph nodes, and hypoplastic spleens. In addition to gross findings indicative of pneumonia (see Fig. S2 and Table S2 in the supplemental material), we observed microscopic alterations, including intranuclear inclusions (Fig. 2), inflammation, and hyperplastic and dysplastic changes in the epithelia of multiple organs: nasal mucosa and lung (Fig. 1), parotid and submandibular salivary and harderian glands, reproductive organs (prostate and uterine epithelium), and kidney (see Table S3 in the supplemental material).
FIG 2 .
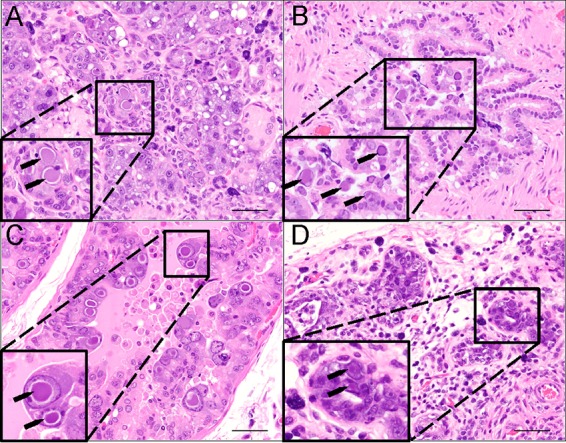
Hematoxylin-and-eosin (H&E) staining of multiple organs in X-SCID rats reveals epithelial hyperplasia with cytomegaly and intranuclear inclusion bodies (black arrows). (A) Parotid salivary gland acinar epithelium was hyperplastic, with regions of necrosis and loss. The interstitium was expanded by lymphocytes, plasma cells, and mast cells. (B) Bronchiolar epithelium was hyperplastic, with enlarged apical epithelial cells containing intranuclear inclusions. (C) Prostate glandular epithelium was severely hyperplastic and disorganized. Cytomegalic cells with extremely large intranuclear inclusions were a prominent feature. (D) There was severe loss of glands within the harderian gland. The remaining glands were often hyperplastic and disorganized, with areas of necrosis. Regions between the remaining glands contain fibrous connective tissue, lymphocytes, plasma cells, and mast cells. Scale bars = 50 μm.
(A) Gross lung lesions were present in polyomavirus-infected rats. Lungs were mottled red-pink with 1- to 4-mm white pleural foci. Scattered similarly sized red foci were present on the pleural surface and throughout the lungs. Cranioventral atelectasis (arrow) was also evident within the cranial lobe of the right lung. (B) Control, normal rat lungs. Download Figure S2, PDF file, 0.8 MB (806.7KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the 1984 report mentioned above, a similar condition associated with polyomavirus infection was described in athymic nude rats (28). We therefore applied a recently developed panpolyomavirus immunohistochemistry test (P-PIT) comprised of a cocktail of three antibodies that, when combined, can detect all known human PyV T antigens (21).
In all 12 rats, tissue sections showing intranuclear inclusion bodies by hematoxylin-and-eosin (H&E) examination also demonstrated correspondingly strong nuclear immunoreactivity by P-PIT (data not shown; representative examples are presented in Fig. 3A and 7). Tissues not affected by inclusion body changes, such as the heart, liver, spleen, and brain, were immunonegative (data not shown). Some degree of selectivity or tropism for distinct types of epithelia is discernible. For example, lesions in the salivary gland (Fig. 3A) were significantly more prominent in the serous portions of the gland than in the mucinous portions, where there were rare areas of necrosis, mixed infiltrates, and intranuclear inclusions. Follow-up immunostaining using each of the three antibodies comprising the P-PIT cocktail singly revealed strong PAb416 reactivity and weak Xt7 reactivity, while 2t2 antibody was unreactive on the diseased tissues (data not shown).
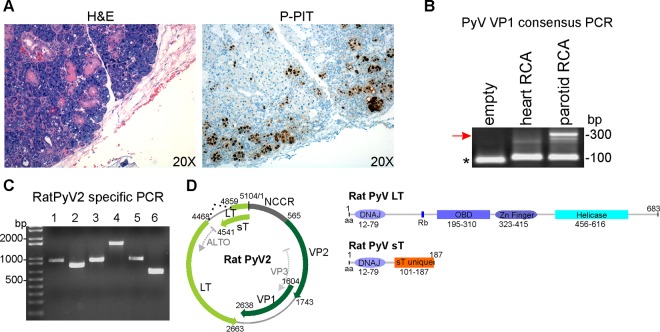
FIG 3 .
(A) Left, P-PIT staining of parotid gland. Right, H&E staining of the parotid salivary gland shows multiple viral inclusion bodies. Immunostaining with P-PIT antibody cocktail PAb416, xt7, and 2t2 was positive (brown) largely in the nucleus of infected cells. (B) Consensus PCR for PyV VP1. Rolling circle-amplified (RCA) DNA from FFPE heart and parotid tissues was subjected to nested PCR using consensus primers. Agarose gel electrophoresis shows an ~300-bp band (red arrow). Primer dimers are indicated by the asterisk. (C) Whole-genome amplification by RatPyV2-specific primers. Virus-specific primers (see Table S1 in the supplemental material) were designed and used to generate 6 overlapping PCR products. Lanes contain products amplified with the indicated primer pair: 1, F2-R3; 2, F4-R5; 3, F6-R7; 4, F8-R10; 5, F11-R12; 6, F13-R1. (D) Genome map of RatPyV2. The RatPyV2 genome comprises 5,104 base pairs (bp). The putative early region encodes large T (LT) and small T (sT) antigens (both light green). Intron splice sites for LT are indicated with dotted black lines. The putative late region comprises viral capsid proteins (VP1 and VP2; both dark green). The noncoding control region (NCCR; dark grey) contains the putative origin of replication and promoters. Examples of predicted open reading frames (ORFs) (ALTO-like ORF and VP3) are indicated by dotted light grey arrows in the early and late region, respectively. Schematic diagram on the right shows putative conserved domains of LT and sT. Amino acid (aa) positions for conserved DNAJ, pocket protein binding (Rb), origin binding (OBD), helicase, and unique sT domains are shown.
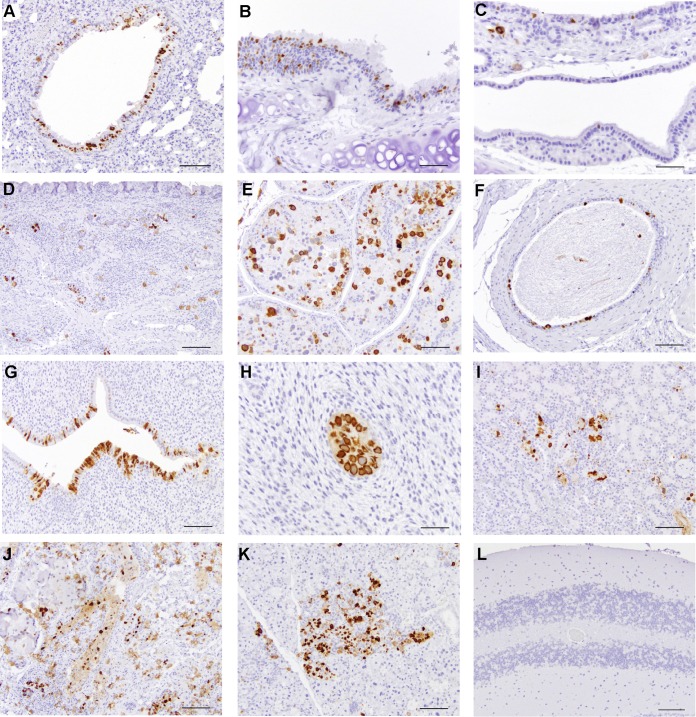
FIG 7 .
Representative images from immunohistochemistry performed on all organs of adult rats (female and male) with PAb416. Positively stained tissues included bronchiolar epithelium of the lung (A), tracheal epithelium and submucosa (B), laryngeal mucosa and submucosa (C), nasal mucosa and submucosa (D), prostate gland epithelium (E), vas deferens epithelium (F), uterine endometrial epithelium (G), uterine endometrial gland (H), focally damaged renal tubules within the kidney (I), harderian gland epithelium (J), salivary gland acini (K), and cerebellum (L). Scale bar = 100 µm (A, D, E, F, G, I, J, K, and L); scale bar = 50 µm (B, C, and H).
Primers used in this study. Download Table S1, PDF file, 0.1 MB (98.3KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gross findings present in RatPyV2-infected rats. N/A, not applicable because organ was not examined in this group. Download Table S2, PDF file, 0.1 MB (52.7KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Histopathologic lesions in RatPyV2-infected rats. *, interstitial pneumonia was characterized by mixed interstitial inflammatory infiltrates and type II pneumocyte hyperplasia; **, intranuclear inclusion bodies were present in epithelial cells of these organs in every group where tissue was examined; ***, lesion was only present in a single male examined; N/A, not applicable because organ was not examined in this group; —, lesion not present. Download Table S3, PDF file, 0.1 MB (77.8KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome characterization and phylogenetic analysis of RatPyV2.
Rolling circle amplification (RCA) products obtained using the DNA extracted from formalin-fixed, paraffin-embedded (FFPE) sections of parotid salivary gland and heart tissues were amplified by polyomavirus consensus PCR for conserved LT (data not shown) and VP1 sequences (Fig. 3B). In concert with the IHC staining, consensus PCR using degenerate primers and parotid tissue DNA resulted in ~300-bp products for VP1 and T-Ag. Based on the initial sequencing analysis, we generated virus-specific primers for genome walking (see Table S1 in the supplemental material). We obtained six overlapping PCR products using fresh tissue DNA isolated from the harderian and parotid glands of an infected rat (Fig. 3C) which were sequenced to 2×depth. The sequences from both tissues were identical. Assembly of overlapping PCR segments revealed a previously unknown polyomavirus, which we designated RatPyV2. The RatPyV2 genome (GenBank accession number KX574453) is 5,104 bp in length and has gene syntheny with other polyomaviruses. The genome encodes open reading frames for 683-amino-acid (aa) large T (LT), 187-aa small T (sT), 34- aa VP1, and 393-aa VP2 proteins (Fig. 3D). RatPyV2 LT splicing is predicted to occur, with a splice donor site between nucleotides 4853 and 4862 (CAAG/GTACAT) and an acceptor site between nucleotides 4467 and 4480 (TTTTTTTTAAAG/GT) within the large T sequence, which would generate a predicted protein of 683 aa (Fig. 3D). Motif analysis shows the putative LT protein to possess conserved PyV domains, including a DnaJ domain (aa 12 to 79, HPDKGG box), an Rb-binding site (aa 160 to 164, LHCDE), an origin binding domain (aa 195 to 310), a zinc finger domain (aa 323 to 415), and a helicase domain (aa 456 to 616) (Fig. 3D). The unspliced early region transcript is also predicted to encode an sT protein with conserved protein phosphatase 2A binding site residues. Overall, RatPyV2 shares 49.13% genome identity with RnorPyV1 (accession number KR075946.1).
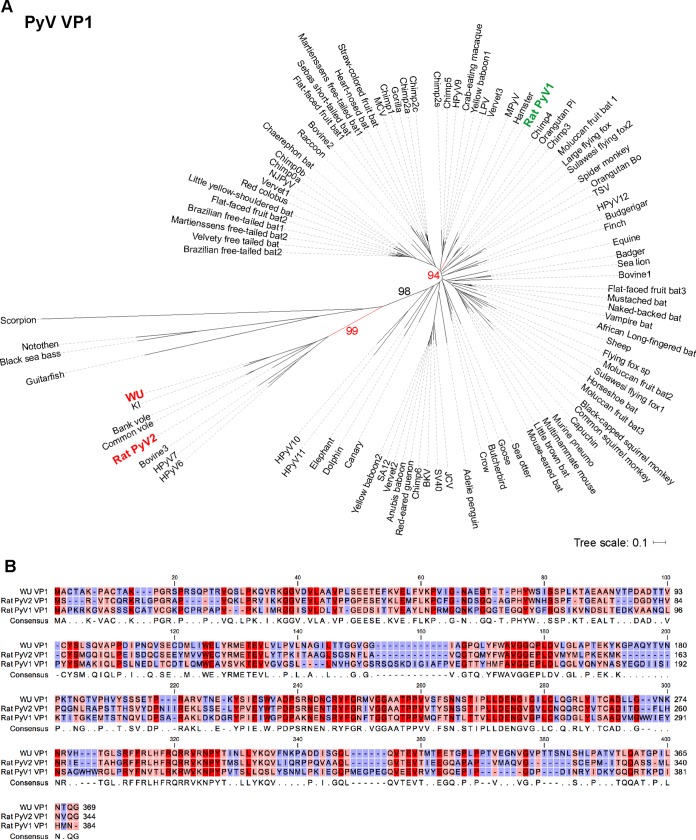
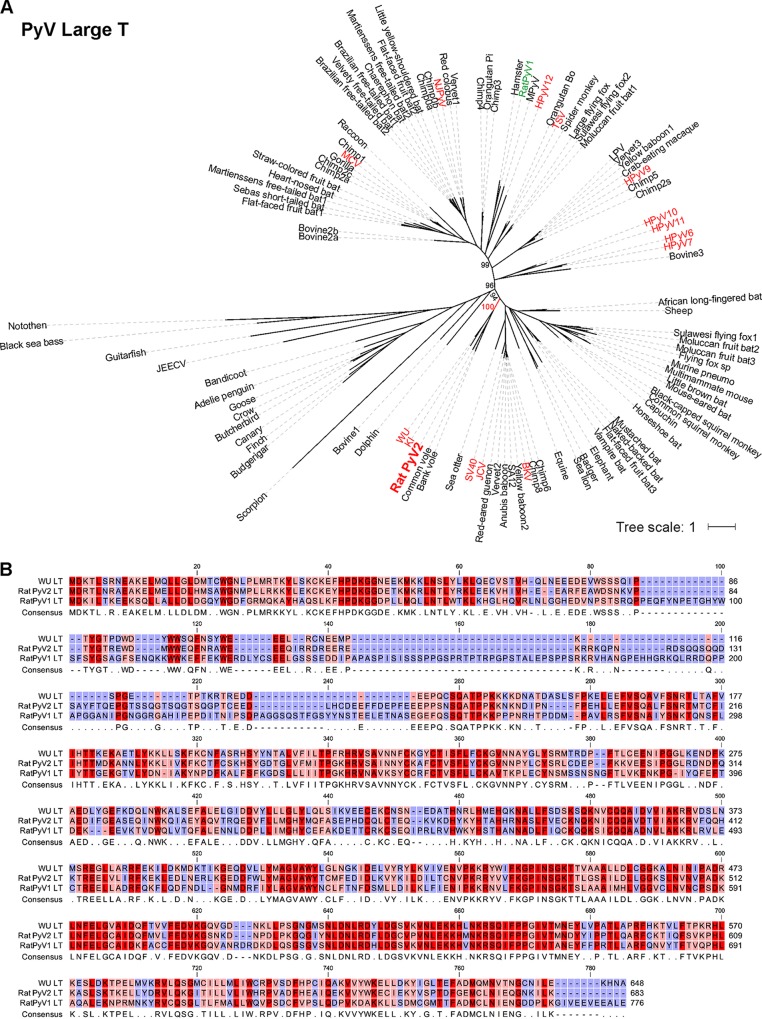
Phylogenetic analysis of VP1 and LT protein sequences revealed that RatPyV2 is most closely related to WUV, KIV, and vole PyVs (Fig. 4 and 5). Consistent with this close homology, IHC staining of parotid gland tissue with WUV VP1-specific antibody was strongly positive and correlated with PAb416 staining (Fig. 6).
FIG 4 .
Phylogenetic analysis of 100 full-length PyV VP1 protein sequences. (A) Consensus tree of 1,000 bootstrap replicates was generated by the neighbor-joining method using MEGA7. Percent bootstrap values are given for selected nodes. RatPyV2 (red, boldface) is closely related to WU (red, boldface) and highly divergent from RatPyV1 (green, boldface). The phylogenetic tree was visualized using iTOL. (B) VP1 amino acid sequence alignment of WU, RatPyV1 (RnorPyV1), and RatPyV2 sequences was generated using CLC Genomics Workbench software. Residue conservation ranges from red (conserved) to pink (mostly conserved) to blue (divergent). A minus sign indicates missing residue at the given position.
FIG 5 .
Phylogenetic analysis of 103 full-length PyV LT protein sequences. (A) Consensus tree of 1,000 bootstrap replicates was generated by the neighbor-joining method using MEGA7. Percent bootstrap values are given for selected nodes. All known P-PIT-reactive PyV T antigens are shown in red. RatPyV1 is shown in green. The phylogenetic tree was visualized using iTOL. (B) LT amino acid sequence alignment of WU, RatPyV1, and RatPyV2 sequences was generated using CLC Genomics Workbench software. Residue conservation ranges from red (conserved) to pink (mostly conserved) to blue (divergent). A minus sign indicates missing residue at the given position.
FIG 6 .
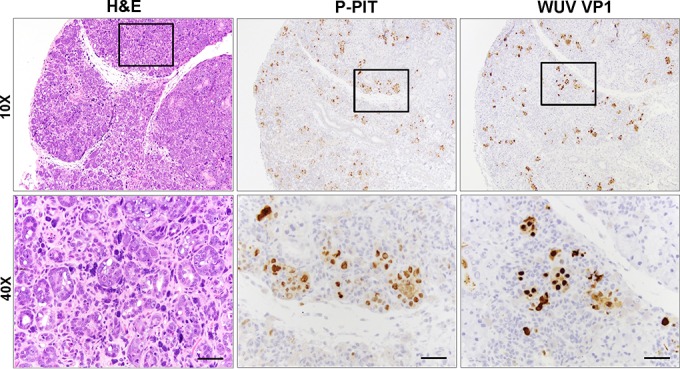
Immunohistochemistry analysis of parotid salivary gland tissue samples. H&E staining (left) showed glandular hyperplasia and inclusion bodies. P-PIT (middle) and WUV VP1 (right) staining correlated with H&E findings and revealed RatPyV2 T antigen and VP1 expression in these lesions. Scale bar = 100 μm. Images are shown at 10× and 40× the original magnification.
RatPyV2 is associated with disseminated inclusion body disease in X-SCID rats.
Immunohistochemistry was performed on organs of two adult rats with PAb416 antibody (Fig. 7). Positively staining tissues included bronchiolar epithelium of the lung, tracheal epithelium and submucosa, vas deferens epithelium, prostate gland epithelium, laryngeal mucosa and submucosa, nasal mucosa and submucosa, uterine endometrial epithelium, uterine endometrial glands, harderian gland epithelium, salivary gland acini, and focally damaged renal tubules in the kidney of one rat.
After the initial 12 animals were characterized, a female breeder was sacrificed postpartum for complications related to parturition (retained placenta and endometritis). In addition to the previously described respiratory and glandular lesions, prominent intranuclear inclusion bodies were present in the epithelium of active mammary glandular epithelium, and pAB416 staining was strongly positive in these areas (see Fig. S3 in the supplemental material). Although the pathogenesis of RatPyV2 is still unclear, transmission via milk (or colostrum) is a potential cause of spread to young animals within the colony.
The intranuclear inclusion bodies present in the epithelium of active mammary glandular epithelium are strongly positive (brown) for pAB416 staining. Images are at 4× (left) and 20× (right) the original magnification. Download Figure S3, PDF file, 2.8 MB (2.9MB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We further verified that RatPyV2 infection was disseminated throughout the colony by PCR and sequencing analysis. Using DNA extracted from FFPE lung tissues from all 12 rats (R1 to R12) and the female rat (Rx) mentioned above that presented with postpartum complications, we performed PCR using primers for the β-actin housekeeping gene and RatPyV2 VP1 (Fig. 8A). R12, from which we sequenced the complete RatPyV2 genome, was used as the positive control. As a negative control, we used lung tissue DNA from an immunocompetent Nagase strain rat that shared the same cubicle with the X-SCID colony. We detected β-actin PCR products from all samples except those from R1, indicating suboptimal extracted DNA integrity for PCR studies. VP1 PCR products of 103 bp (amplified using primer pair F7-R14; see Table S1 in the supplemental material for primers) were observed in all β-actin-positive samples except those from the Nagase strain rat used as a negative control. For sequence confirmation and analysis, a larger region of VP1 was amplified using a different primer pair (primer pair F14-R7; see Table S1). Although the DNA from two samples (R9 and R10) was not amplifiable over this larger genomic region, sequencing of the remaining 10 samples, spanning nucleotides 1885 to 2316, revealed two single nucleotide polymorphisms leading to changes in amino acids at nucleotide positions 2085 (A/G) and 2092 (A/C) in different rats (Fig. 8B).
FIG 8 .
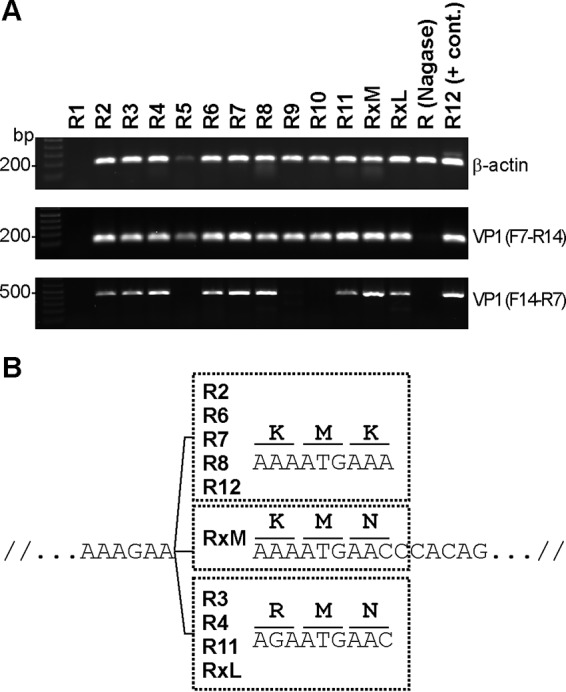
PCR detection of RatPyV2 infection in entire X-SCID rat colony. (A) DNA was extracted from FFPE lung tissue sections of all 12 X-SCID rats (R1 to R12), a female rat (Rx) with postpartum complications (mammary tissue, RxM, and lung tissue, RxL), and an immunocompetent Nagase strain rat [R (Nagase)]. β-Actin PCR products (191 bp) are amplifiable for all rat samples except those from R1. VP1 products (103 bp; primers F7-R14) were detected in all X-SCID rats (R2 to R12 and Rx) but not in the Nagase strain rat. A 432-bp fragment of VP1 was amplified using the F14-R7 primer pair and analyzed by sequencing. (B) Single nucleotide polymorphisms (SNP) detected in tissues from different rats are shown. For R3, R4, R11, and Rx (from both mammary and lung sites), an SNP at nucleotide position 2092 (A/C) leading to a K/N amino acid change was detected. R3, R4, R11, and RxL demonstrated an SNP at nucleotide position 2085 (A/G) leading to a K/R amino acid change.
DISCUSSION
In this study, we identified a novel rat polyomavirus infection in an X-SCID rat colony using the recently developed P-PIT assay that is designed to identify human polyomaviruses. This allowed us to isolate RatPyV2 sequences from immunoreactive tissues. Being immunocompromised, these rats are susceptible to a variety of opportunistic infections. The X-SCID rat colony was first suspected to harbor P. carinii as a cause for the respiratory symptoms. Following treatment of this colony for P. carinii, clinical symptoms subsided; however, evidence of viral disease was still present on histopathology despite negative tests for a comprehensive list of pathogens (Table 1), leading to identification of RatPyV2. P. carinii has long been known to cause pneumonia in immunosuppressed rats, but over the last several years, studies have also linked P. carinii to infectious interstitial pneumonia (IIP), a common, transient, usually mild disease of immunocompetent laboratory rats that had previously been attributed to an alleged virus referred to as rat respiratory virus (31, 32). Although RatPyV2 caused significant histopathologic lesions in multiple organs, including the lungs, clinical symptoms, such as respiratory distress and chromodacryorrhea, were only manifested by rats coinfected with P. carinii. However, in the majority of the colony, we observed reduced fecundity independent of P. carinii infection.
Ward et al. in 1984 described electron microscopic evidence for polyomavirus infection in 32 athymic nude (NIH-rnu) rats (28) that possessed spontaneous mutations in the FOXN1 gene (33). The pathology described by these authors included inclusion bodies in the salivary glands, harderian glands, lungs, and nasal glands, findings that are similar those in our X-SCID (IL2RG-SCID) rats infected with RatPyV2. Both FOXN1 and IL2RG mutations generate a T cell immunodeficient phenotype in experimental animals and in humans (29, 34–40). While it is not known whether RatPyV2 was responsible for the disease described in the NIH-rnu nude rats by Ward et al. (28), these findings are consistent with T cell surveillance being critical for host control of rat polyomavirus replication. The identification of this tractable virus-host model can allow experimental manipulation to assess the role of T cell immune surveillance in polyomavirus replication control.
In addition to IHC analysis performed on different tissues of the 12 rats, we further verified the extent of infection in this colony by PCR using RatPyV2-specific primers for regions in the VP1 locus. All 12 rats showed pulmonary infection by RatPyV2. However, the IHC and PCR assays used in this study do not address whether additional rat polyomaviruses are present.
Additional studies are needed to define the natural history of RatPyV2 infection. It is not known whether this is a common commensal infection of rats or if this is a rare and isolated infection. Based on the biology of other mammalian polyomaviruses, we suspect RatPyV2 to be a relatively common infection of rats that manifests as a disease in a setting of severe T cell immunodeficiency. If RatPyV2 is common, control measures might be devised (such as breeding RatPyV2-free rodents) to improve experimental animal health. Whether RatPyV2 causes disease in immunocompetent rats is also unknown, but our study reveals that this can be readily assayed with available antibodies and by PCR.
Polyomavirus-host evolutionary studies suggest coevolution of polyomaviruses with their hosts (41). Given the broad phylogenetic radiation of polyomaviruses among rodents, it is likely that additional rat polyomaviruses are present in Rattus populations. More importantly, both FOXN1 and IL2RG mutations are responsible for genetic T cell immunodeficiency syndromes in children (34). If T-cell immune surveillance is particularly critical for commensal polyomavirus control, our findings highlight the need to search for unrecognized polyomavirus infections as causes for disease in susceptible populations. Although P-PIT was not developed to discover nonhuman polyomaviruses, its identification of this agent in rats suggests that it may have utility as a broad-based screen for new, as well as known human polyomaviruses.
MATERIALS AND METHODS
Animals.
X-SCID rats were obtained from Kyoto University, Institute of Laboratory Animals, Graduate School of Medicine, Kyoto, Japan, on 2 June 2015 subsequent to a 3-month quarantine at Charles River, Inc. (Wilmington, MA). They were housed in numbers of 2 adults or fewer per static microisolator cage on sterilized hardwood chip bedding, fed irradiated Purina LabDiet rodent chow 5P76, and given water ad libitum. All rats were housed in a separate cubicle within the animal facility’s repository. The breeding scheme of this colony was pair-timed mating with weaning at 28 days.
All animal procedures conformed to NIH guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh (protocol number 16048182). The University of Pittsburgh’s Animal Care and Use Program (ACUP) (unit number 000496) is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Testing for rat infections.
For rodent health surveillance and disease detection, quarterly diagnostic screening, including serology (multiplexed fluorometric immunoassay [MFIA]), PCR, and parasitology, were performed by Charles River, Inc., Research Animal Diagnostic Services (Wilmington, MA) (Table 1). At each bedding change, soiled bedding from each of the cages was added to the sentinel (Sprague-Dawley rat) cage, thus exposing the sentinels to diseases carried by colony animals. For lung sections, Grocott’s methenamine silver (GMS) staining was performed on 5-µm-thick tissue sections for the detection of fungal elements.
Histology and immunohistochemical staining.
Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 5-µm thickness, and stained with hematoxylin and eosin for evaluation by light microscopy. Immunohistochemistry staining was performed according to a previously published protocol (21). Briefly, slides were deparaffinized in xylene and rehydrated through sequential ethanol solutions. Following incubation with 3% hydrogen peroxide for 15 min, epitope retrieval was performed using 1 mM EDTA buffer, pH 8.0 (125°C for 3 min and 90°C for 15 s), in an antigen retrieval chamber (Decloaking Chamber; Biocare Medical). Sections were incubated with serum-free protein block buffer (Dako), followed by primary antibodies diluted in antibody buffer (1% bovine serum albumin [BSA], 0.1% gelatin, 0.5% Triton X-100, 0.05% sodium azide in phosphate-buffered saline [PBS], pH 7.4) overnight at 4°C in a humidified chamber. The antibodies used include PAb416 (clone Ab-2, catalog number DP02, 1:200; Millipore), 2t2 (1:5 hybridoma supernatant, or approximately 8-μg/ml concentrated monoclonal antibody [MAb]), Xt7 (1:5 hybridoma supernatant, or approximately 8-μg/ml concentrated MAb), and mouse WUV VP1 MAb (NN-Ab06, 1:2,000; kind gift of David Wang [42]). Tissue sections were developed with mouse EnVision polymer (Dako) for 30 min at room temperature, reacted with diaminobenzidine (DAB; Dako), and counterstained with hematoxylin (Dako).
Genomic DNA extraction from FFPE or fresh tissue.
DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) sections of heart and parotid gland tissues using the QIamp DNA FFPE tissue kit according to the manufacturer’s instructions. For genomic DNA isolation from fresh tissues, harderian and parotid gland tissues were minced in 1 ml of PBS, transferred to microcentrifuge tubes, and centrifuged for 5 min at 6,000 × g. The tissue pellets were incubated in 0.5 ml of lysis buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 25 mM EDTA, pH 8.0, 1% SDS) with proteinase K (Promega) added to a 0.5-mg/ml final concentration overnight at 56°C with gentle shaking. This tissue solution was extracted with an equal amount of phenol chloroform-isoamyl alcohol (25:24:1; Amresco) for 10 min at room temperature and centrifuged for 5 min at 16,000 × g (Eppendorf 5424R). The aqueous layer was transferred into a new tube and mixed with 0.5 vol of 7.5 M ammonium acetate, 2 vol of 100% ethanol. After centrifugation for 5 min at 16,000 × g, the DNA pellet was rinsed with 70% ethanol, air dried, and subsequently dissolved in Tris-EDTA (TE) buffer.
Rolling circle amplification, PCR, and sequencing.
FFPE tissue DNA was subjected to rolling circle amplification (RCA) using phi29 DNA polymerase (TempliPhi; GE Healthcare). Consensus PCR was carried out using 1/10 of the RCA template, consensus primers (see Table S1 in the supplemental material), and 2.5 U of Taq polymerase (NEB) supplemented with Thermopol buffer. The cycling conditions for HPyV T-Ag primers or VP1-1 primers were initial denaturation for 5 min at 95°C, followed by 45 cycles each of 95°C for 30 s, 45°C for 1 min, and 68°C for 1 min and a final elongation at 68°C for 5 min. Nested PCR was performed with consensus VP1-2 primers, using 4 µl of the first PCR product as the template, at 95°C for 5 min, 45 cycles each of 95°C for 30 s, 53°C for 1 min, and 68°C for 30 s, and a final extension at 68°C for 5 min. PCR products were separated by agarose-gel electrophoresis, and DNA was purified using the QIAquick gel extraction kit (Qiagen) prior to sequencing (MCLAB). Based on the initial sequencing analysis, virus-specific contig primers (see Table S1 in the supplemental material) were synthesized (IDT) and used to amplify the entire genome with Q5 high-fidelity Taq polymerase (NEB). For genome-walking PCR, we used DNA isolated from fresh harderian and parotid gland tissues. The cycling conditions for overlapping contigs were 3 min at 98°C, followed by 35 cycles each of 98°C for 10 s, 60 to 66°C for 30 s, and 72°C for 1 min and a final elongation at 72°C for 2 min. PCR samples were separated on a 1% agarose gel, extracted (Qiagen), and sequenced (MCLAB). Sequence analysis and genome assembly were done using ContigExpress and Vector NTI software (Invitrogen). Analysis of the predicted protein sequences for family domains was performed with Motif Scan on the European Bioinformatics Institute (EBI) website (http://www.ebi.ac.uk/).
Phylogenetic analysis.
Currently, known polyomavirus annotated maps and protein sequences are given at http://home.ccr.cancer.gov/Lco/PyVE.asp. The full-length VP1 and LT protein sequences were aligned by ClustalW, and the phylogenetic trees were generated by the neighbor-joining method with 1,000 bootstrap replicates, implemented in the Molecular Evolutionary Genetics Analysis version 7 (MEGA7) software package (43). The phylogenetic trees were visualized using iTOL (http://itol.embl.de/) (44).
Accession number(s).
The RatPyV2 genome has been deposited in GenBank under accession number KX574453.
ACKNOWLEDGMENTS
We are grateful to Alejandro Soto-Gutierrez for sample contribution and to Cindy Stone, Megan Lambert, Robin Frederick, and Dave Bolette for technical support. We thank David Wang for kindly providing the mouse MAb WUV VP antibody. We also thank Meredith Monk and Collin Wolcott for help with the manuscript.
Y.C. and P.S.M. are supported by American Cancer Society Professorships and by support from the UPMC and Pittsburgh Foundations, respectively. This project used the University of Pittsburgh Cancer Institute Tissue and Research Pathology Services, Animal Facility and Li-COR Imaging Facility, which is supported in part by NIH grant number P30CA047904.
REFERENCES
- 1.Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC. 2011. Taxonomical developments in the family Polyomaviridae. Arch Virol 156:1627–1634. doi: 10.1007/s00705-011-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck CB, Van Doorslaer K, Peretti A, Geoghegan EM, Tisza MJ, An P, Katz JP, Pipas JM, McBride AA, Camus AC, McDermott AJ, Dill JA, Delwart E, Ng TFF, Farkas K, Austin C, Kraberger S, Davison W, Pastrana DV, Varsani A. 2016. The ancient evolutionary history of polyomaviruses. PLoS Pathog 12:e1005574. doi: 10.1371/journal.ppat.1005574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo Ma, McGeoch DJ, Pringle CR, Wickner RB. 2000. Virus taxonomy. Classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA. [Google Scholar]
- 4.DeCaprio JA, Imperiale MJ, Major EO. 2013. Polyomaviruses, p 1633–1661. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed). Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog 5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA. 2009. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst 101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazem S, van der Meijden E, Kooijman S, Rosenberg AS, Hughey LC, Browning JC, Sadler G, Busam K, Pope E, Benoit T, Fleckman P, de Vries E, Eekhof JA, Feltkamp MCW. 2012. Trichodysplasia spinulosa is characterized by active polyomavirus infection. J Clin Virol 53:225–230. doi: 10.1016/j.jcv.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 9.van der Meijden E, Kazem S, Burgers MM, Janssens R, Bouwes Bavinck JN, de Melker H, Feltkamp MCW. 2011. Seroprevalence of trichodysplasia spinulosa-associated polyomavirus. Emerg Infect Dis 17:1355–1363. doi: 10.3201/eid1708.110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicol JT, Touzé A, Robinot R, Arnold F, Mazzoni E, Tognon M, Coursaget P. 2012. Seroprevalence and cross-reactivity of human polyomavirus 9. Emerg Infect Dis 18:1329–1332. doi: 10.3201/eid1808.111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicol JTJ, Robinot R, Carpentier A, Carandina G, Mazzoni E, Tognon M, Touzé A, Coursaget P. 2013. Age-specific seroprevalences of Merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa-associated polyomavirus. Clin Vaccine Immunol 20:363–368. doi: 10.1128/CVI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 8:e58021. doi: 10.1371/journal.pone.0058021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim ES, Meinerz NM, Primi B, Wang D, Garcea RL. 2014. Common exposure to STL polyomavirus during childhood. Emerg Infect Dis 20:1559–1561. doi: 10.3201/eid2009.140561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicol JTJ, Leblond V, Arnold F, Guerra G, Mazzoni E, Tognon M, Coursaget P, Touzé A. 2014. Seroprevalence of human Malawi polyomavirus. J Clin Microbiol 52:321–323. doi: 10.1128/JCM.02730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner SD, Field AM, Coleman DV, Hulme B. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253–1257. doi: 10.1016/S0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 16.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257–1260. doi: 10.1016/S0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 17.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Meijden E, Janssens RWA, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MCW. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog 6:1–10. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho J, Jedrych JJ, Feng H, Natalie AA, Grandinetti L, Mirvish E, Crespo MM, Yadav D, Fasanella KE, Proksell S, Kuan SF, Pastrana DV, Buck CB, Shuda Y, Moore PS, Chang Y. 2015. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J Infect Dis 211:1560–1565. doi: 10.1093/infdis/jiu524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siebrasse EA, Nguyen NL, Willby MJ, Erdman DD, Menegus MA, Wang D. 2016. Multiorgan WU polyomavirus infection in bone marrow transplant recipient. Emerg Infect Dis 22:24–31. doi: 10.3201/eid2201.151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toptan T, Yousem SA, Ho J, Matsushima Y, Stabile LP, Fernández-Figueras MT, Bhargava R, Ryo A, Moore PS, Chang Y. 2016. Survey for human polyomaviruses in cancer. JCI Insight 1:1–26. doi: 10.1172/jci.insight.85562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross L. 1976. The fortuitous isolation and identification of the polyoma virus. Cancer Res 36:4195–4196. [PubMed] [Google Scholar]
- 23.Kilham L. 1952. Isolation in suckling mice of a virus from C3H mice harboring Bittner milk agent. Science 116:391–392. [DOI] [PubMed] [Google Scholar]
- 24.Graffi A, Schramm T, Graffi I, Bierwolf D, Bender E. 1968. Virus-associated skin tumors of the Syrian hamster: preliminary note. J Natl Cancer Inst 40:867–873. [PubMed] [Google Scholar]
- 25.Orba Y, Kobayashi S, Nakamura I, Ishii A, Hang’ombe BM, Mweene AS, Thomas Y, Kimura T, Sawa H. 2011. Detection and characterization of a novel polyomavirus in wild rodents. J Gen Virol 92:789–795. doi: 10.1099/vir.0.027854-0. [DOI] [PubMed] [Google Scholar]
- 26.Nainys J, Timinskas A, Schneider J, Ulrich RG, Gedvilaite A. 2015. Identification of two novel members of the tentative genus Wukipolyomavirus in wild rodents. PLoS One 10:e0140916. doi: 10.1371/journal.pone.0140916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehlers B, Richter D, Matuschka FR, Ulrich RG. 2015. Genome sequences of a rat polyomavirus related to murine polyomavirus, Rattus norvegicus polyomavirus 1. Genome Announc 3:e00997-15. doi: 10.1128/genomeA.00997-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward JM, Lock A, Collins MJ, Gonda MA, Reynolds CW. 1984. Papovaviral sialoadenitis in athymic nude rats. Lab Anim 18:84–89. doi: 10.1258/002367784780864884. [DOI] [PubMed] [Google Scholar]
- 29.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. 2010. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One 5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvignac-Spencer S, Feltkamp MCW, Daugherty MD, Moens U, Ramqvist T, Johne R, Ehlers B. 2016. A taxonomy update for the family Polyomaviridae. Arch Virol 161:1739–1750. doi: 10.1007/s00705-016-2794-y. [DOI] [PubMed] [Google Scholar]
- 31.Livingston RS, Besch-Williford CL, Myles MH, Franklin CL, Crim MJ, Riley LK. 2011. Pneumocystis carinii infection causes lung lesions historically attributed to rat respiratory virus. Comp Med 61:45–59. [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson KS, Dole V, Parker NJ, Momtsios P, Banu L, Brouillette R, Simon MA, Albers TM, Pritchett-Corning KR, Clifford CB, Shek WR. 2012. Pneumocystis carinii causes a distinctive interstitial pneumonia in immunocompetent laboratory rats that had been attributed to “rat respiratory virus.” Vet Pathol 49:440–452. doi: 10.1177/0300985811432351. [DOI] [PubMed] [Google Scholar]
- 33.Vos JG, Kreeftenberg JG, Kruijt BC, Kruizinga W, Steerenberg P. 1980. The athymic nude rat. II. Immunological characteristics. Clin Immunol Immunopathol 15:229–237. doi: 10.1016/0090-1229(80)90033-1. [DOI] [PubMed] [Google Scholar]
- 34.Palamaro L, Romano R, Fusco A, Giardino G, Gallo V, Pignata C. 2014. FOXN1 in organ development and human diseases. Int Rev Immunol 33:83–93. doi: 10.3109/08830185.2013.870171. [DOI] [PubMed] [Google Scholar]
- 35.Allenspach E, Rawlings DJ, Scharenberg AM 26 August 2003. X-linked severe combined Immunodeficiency. In Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, Stephens K (ed), GeneReviews. University of Washington, Seattle, WA: https://www.ncbi.nlm.nih.gov/books/NBK1410/. [PubMed] [Google Scholar]
- 36.Yee A, De Ravin SS, Elliott E, Ziegler JB, Contributors to the Australian Paediatric Surveillance Unit . 2008. Severe combined immunodeficiency: a national surveillance study. Pediatr Allergy Immunol 19:298–302. doi: 10.1111/j.1399-3038.2007.00646.x. [DOI] [PubMed] [Google Scholar]
- 37.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. 2008. Pillars article: Interleukin-2 receptor γ chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73: 147–157. J Immunol 181:5817–5827. [PubMed] [Google Scholar]
- 38.Leonard WJ. 1996. The molecular basis of X-linked severe combined immunodeficiency: defective cytokine receptor signaling. Annu Rev Med 47:229–239. doi: 10.1146/annurev.med.47.1.229. [DOI] [PubMed] [Google Scholar]
- 39.Fischer A, Cavazzana-Calvo M, De Saint Basile G, DeVillartay JP, Di Santo JP, Hivroz C, Rieux-Laucat F, Le Deist F. 1997. Naturally occurring primary deficiencies of the immune system. Annu Rev Immunol 15:93–124. doi: 10.1146/annurev.immunol.15.1.93. [DOI] [PubMed] [Google Scholar]
- 40.Leblond V, Autran B, Cesbron JY. 1997. The SCID mouse mutant: definition and potential use as a model for immune and hematological disorders. Hematol Cell Ther 39:213–221. doi: 10.1007/s00282-997-0213-2. [DOI] [PubMed] [Google Scholar]
- 41.Madinda NF, Ehlers B, Wertheim JO, Akoua-Koffi C, Bergl RA, Boesch C, Boji Mungu Akonkwa D, Eckardt W, Fruth B, Gillespie TR, Gray M, Hohmann G, Karhemere S, Kujirakwinja D, Langergraber K, Muyembe J-J, Nishuli R, Pauly M, Petrzelkova KJ, Robbins MM, Todd A, Schubert G, Stoinski TS, Wittig RM, Zuberbühler K, Peeters M, Leendertz FH, Calvignac-Spencer S. 2016. Assessing host-virus co-divergence for close relatives of Merkel cell polyomavirus infecting African great apes. J Virol 90:8531–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siebrasse EA, Pastrana DV, Nguyen NL, Wang A, Roth MJ, Holland SM, Freeman AF, Mcdyer J, Buck CB, Wang D. 2015. WU polyomavirus in respiratory epithelial cells from lung transplant patient with Job syndrome. Emerg Infect Dis 21:103–106. doi: 10.3201/eid2101.140855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Silver staining of Pneumocystis carinii-coinfected lung samples in areas of interstitial pneumonia in H&E-stained tissue sections (left, top right) revealed black staining of cyst walls (bottom right). Left, ×20 magnification; right, ×100 magnification. Download Figure S1, PDF file, 0.9 MB (996.9KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Gross lung lesions were present in polyomavirus-infected rats. Lungs were mottled red-pink with 1- to 4-mm white pleural foci. Scattered similarly sized red foci were present on the pleural surface and throughout the lungs. Cranioventral atelectasis (arrow) was also evident within the cranial lobe of the right lung. (B) Control, normal rat lungs. Download Figure S2, PDF file, 0.8 MB (806.7KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S1, PDF file, 0.1 MB (98.3KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gross findings present in RatPyV2-infected rats. N/A, not applicable because organ was not examined in this group. Download Table S2, PDF file, 0.1 MB (52.7KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Histopathologic lesions in RatPyV2-infected rats. *, interstitial pneumonia was characterized by mixed interstitial inflammatory infiltrates and type II pneumocyte hyperplasia; **, intranuclear inclusion bodies were present in epithelial cells of these organs in every group where tissue was examined; ***, lesion was only present in a single male examined; N/A, not applicable because organ was not examined in this group; —, lesion not present. Download Table S3, PDF file, 0.1 MB (77.8KB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The intranuclear inclusion bodies present in the epithelium of active mammary glandular epithelium are strongly positive (brown) for pAB416 staining. Images are at 4× (left) and 20× (right) the original magnification. Download Figure S3, PDF file, 2.8 MB (2.9MB, pdf) .
Copyright © 2016 Rigatti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.