Abstract
Since their discovery approximately three decades ago, sperm-borne RNAs, both large/small and coding/noncoding, have been reported in multiple organisms, and some have been implicated in spermatogenesis, early development, and epigenetic inheritance. Despite these advances, isolation, quantification, and annotation of sperm-borne RNAs remain nontrivial. The yields and subspecies of sperm-borne RNAs isolated from sperm can vary drastically depending on the methods used, and no cross-species analyses of sperm RNA contents have ever been conducted using a standardized sperm RNA isolation protocol. To address these issues, we developed a simple RNA isolation method that is applicable to sperm of various species, thus allowing for reliable interspecies comparisons. Based on RNA-Seq analyses, we established SpermBase ( www.spermbase.org), a database dedicated to sperm-borne RNA profiling of multiple species. Currently, SpermBase contains large and small RNA expression data for mouse, rat, rabbit, and human total sperm and sperm heads. By analyzing large and small RNAs for conserved features, we found that many sperm-borne RNA species were conserved across all four species analyzed, and among the conserved small RNAs, sperm-borne tRNA-derived small noncoding RNAs and miRNAs can target a large number of genes known to be critical for early development.
Keywords: early development, epigenetics, noncoding RNA, reprogramming, sperm
INTRODUCTION
Despite the cessation of transcription and shedding of the cytoplasm at the final stages of spermiogenesis, mature spermatozoa have been shown to possess diverse populations of both small and large RNAs [1–5]. These RNA populations, along with the paternal genome, can be delivered into the oocyte during fertilization, where they can persist beyond zygotic genome activation [5, 6]. Since their discovery in 1989, sperm-borne RNAs have been shown to function in spermatogenesis and during fertilization and early embryonic development, suggesting that they are not merely the remnants of sperm development [4, 7–10]. Sperm with aberrant miRNA contents display compromised fertilization rate and preimplantation embryonic development when injected into wild-type oocytes [5]. Additionally, sperm small RNAs have been found to play a role in epigenetic inheritance [11–17].
The study of sperm RNAs has been challenging, in part, because of the difficulties associated with sperm RNA isolation. Compared to other cell types, sperm contain fewer RNAs, and some RNAs appear to be localized to the nucleus and associated protamine-packed chromatin highly enriched in disulfide bonds resistant to lysis by detergents [4, 18, 19]. Therefore, it is no surprise that different RNA extraction procedures have been shown to lead to highly variable sperm-borne RNA contents; this issue is compounded by the inherent heterogeneity of RNA populations among individual sperm samples [20–24]. Additionally, a sperm RNA isolation protocol that works for one species does not necessarily work for another, because of interspecies differences in sperm morphology and chromatin condensation [25–27]. The aforementioned issues surrounding the study of sperm-borne RNAs render the comparative analyses of data obtained using different experimental approaches unreliable. Therefore, it is necessary that a broadly applicable methodology be established for studying the sperm RNA contents across species. To this end, we developed a simple and effective protocol for sperm RNA isolation, which can be applied to multiple species with only minor modifications. Based on sperm RNA-Seq data, we established SpermBase (www.spermbase.org), a database dedicated to sperm RNA expression profiling for various species. Currently, SpermBase contains expression data of both large and small RNAs in sperm of four mammalian species (rat, mouse, human, and rabbit), with plans to expand to more species in the future. To demonstrate the utility of SpermBase, we compared the sperm RNA-Seq data and identified highly conserved mammalian sperm-borne RNAs among the four mammalian species.
MATERIALS AND METHODS
Sperm Collection
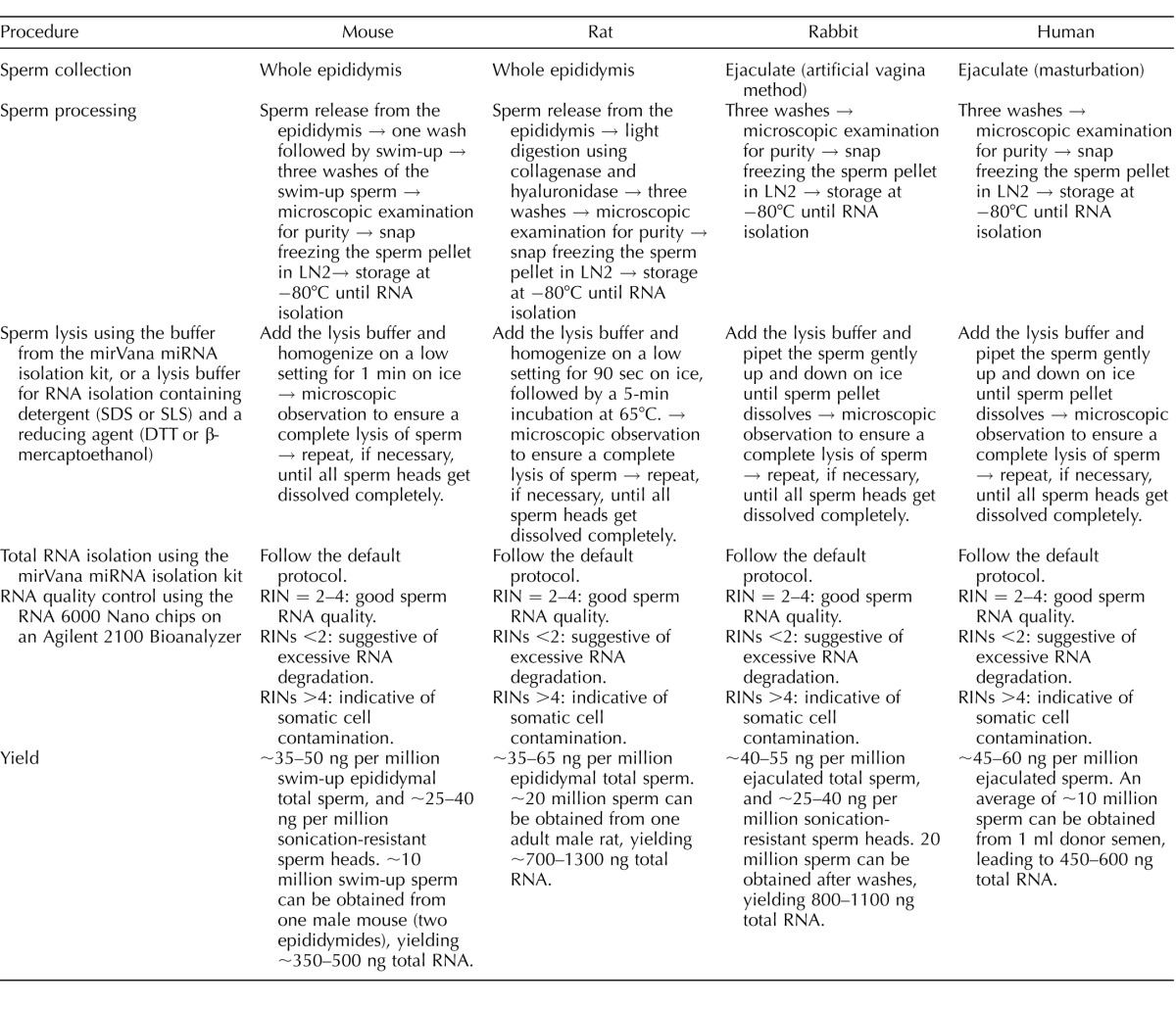
The Institutional Animal Care and Use Committee of the University of Nevada, Reno, approved the use of mice and rats (Protocol #00494), as well as rabbits (Protocol #00536) in this study. A summary of the sperm processing procedures is shown in Table 1.
TABLE 1.
Summary of sperm RNA isolation procedures.
Mouse (C57BL/6J; JAX) total sperm and sperm head collection were conducted as follows: Epididymides from a mouse were placed in 37°C HEPES-buffered human tubule fluid (HEPES-HTF) and dissected into smaller fragments. The sperm were allowed to escape from the epididymis during a 30-min incubation at 37°C. The sperm-containing supernatants were collected and centrifuged for 5 min at 700 × g. After removing the supernatants, fresh HEPES-HTF was added gently on top of the sperm pellet. During a 30-min incubation at 37°C, sperm were allowed to swim up from the pellet. The swim-up sperm were collected with the supernatant. After pelleting the sperm at 700 × g for 5 min, the sperm were washed three times with 1× Dulbecco PBS and centrifugation at the same speed. During the third wash, a small aliquot of sperm suspension was examined under a phase-contrast microscope to determine the purity. The purity was >98% in all samples used for this study. For total sperm isolation, we performed the procedure individually for each mouse, and the total sperm of three mice were pooled for total RNA isolation. Two biological replicates (each with pooled total sperm RNA from three mice) were used for large or small RNA sequencing. For sperm head isolation, total sperm from five mice were sonicated (Bioruptor UCD-200; Diagenode) in PBS for 3 min and subsequently pelleted at 700 × g for 5 min. The sonicated sperm (consisting now of separated sperm heads and tails) was then added to a 4.5-ml 83.5% sucrose cushion and centrifuged at 100 000 × g for 1 h (SW41Ti rotor; Beckman); afterwards, the sperm head pellet was collected. Because sonication broke all contaminating somatic cells, the sperm head purity was close to 100%. We used sperm heads purified from five mice for RNA isolation and the subsequent sequencing with two biological replicates.
Rat (Sprague Dawley; Charles River) total sperm were isolated as follows: Rat epididymides were dissected and minced in F12 culture medium containing 0.1% bovine serum albumin followed by a 30-min incubation at 37°C. The sperm-containing supernatants were collected and washed with PBS by centrifugation (800–1000 × g for 5 min). Sperm pellets were resuspended in 200 μl NIM medium (121.6 mM KCl, 7.8 mM Na2HPO4, 1.4 mM KH2PO4, 0.1% polyvinyl alcohol, and 10 mM EDTA), 100 μl collagenase (200 U/ml; Sigma), and 100 μl hyaluronidase (100 U/ml; Sigma) followed by an incubation at 37°C for 1 h, with occasional mixing. Rat total sperm were then washed three times using 500 μl NIM though centrifugation (4000 × g for 3 min) and the purity was >95%, as determined by phase-contrast microscopic observation. The rat sperm pellets were snap frozen in liquid nitrogen followed by storage at −80°C until RNA isolation. For total sperm isolation, we performed the procedure individually for each rat, and the total sperm of three rats was pooled for total RNA isolation and sequencing. Three biological replicates (each with pooled total sperm RNA from three rats) were used for large or small RNA sequencing.
Rabbit (New Zealand White; Charles River) ejaculates were collected using the artificial vagina method, as described [28]. Rabbit sperm were washed three times with HEPES-HTF medium and the purity was >98% based on phase-contrast microscopy. The rabbit sperm pellets were snap frozen in liquid nitrogen followed by storage at −80°C until RNA isolation. Three rabbits were used for collecting ejaculates, and sperm from the three were individually processed for RNA isolation, library construction, and sequencing.
Human sperm samples used in this study were deidentified human donor sperm samples purchased from California CryoBank Inc. Three donors were all healthy adult men (19–38 yr old) who underwent rigorous health screening and met all the criteria for a qualified sperm donor, as described in the company's website (https://cryobank.com/uploadedFiles/Cryobankcom/_forms/pdf/brochures/DonorPyramid.pdf). The use of purchased, deidentified human sperm for RNA isolation required no institutional review board approval, as determined by the University of Nevada, Reno (documents available upon request). The cryopreserved human sperm were thawed by incubation in a water bath at 37°C followed by three washes with HEPES-HTF, and the purity of the human sperm was >0.01% (fewer than 1 round cell per 10 000 sperm) based on phase-contrast microscopic observation. The sperm pellets were subjected to RNA isolation using the method described below. Three human sperm samples were processed and sequenced individually.
Sperm RNA Isolation
Total RNA was isolated using the mirVana miRNA Isolation Kit (Life Technologies) following the manufacturer's instructions with modifications at the lysis step. The same procedures were used for total sperm and sperm heads of each species. Differences in the lysis step of the sperm RNA isolation procedures for the four species are summarized in Table 1. For mouse sperm, after the addition of the lysis buffer containing guanidine thiocyanate and β-mercaptoethanol (a reducing agent required for a complete lysis of the sperm heads), the frozen sperm pellets were homogenized using a homogenizer (BenchMark) at low settings for 1 min on ice. For human and rabbit samples, after the addition of the lysis buffer, the frozen sperm pellets were pipetted up and down using a handheld pipette (Eppendorf) on ice, until the pellet dissolved. For rat samples, after the addition of the lysis buffer, the frozen sperm pellets were homogenized using a homogenizer (BenchMark) at low settings for 90 sec, followed by a 5-min incubation at 65°C. After lysis, a small aliquot (∼5–10 μl) was placed on a glass slide followed by covering with a coverslip for microscopic examination to ensure sperm nuclei were completely lysed. The completely lysed sperm samples were subjected to the remaining default protocol for RNA isolation. For RNA quality control, sperm RNA samples were analyzed using the RNA 6000 Nano chips on an Agilent 2100 Bioanalyzer (Agilent). The RNA integrity number (RIN) was measured to estimate the sperm RNA quality.
RNA-Seq
Library preparation was performed using the Ion Total RNA-Seq Kit v2 (Life Technologies). Large RNA libraries were prepared using the whole transcriptome protocol provided with the kit, and small RNA libraries were prepared using the small RNA protocol. Large and small RNA libraries were barcoded using Ion Xpress Barcode Adapters (Life Technologies). Quality control was performed using Agilent High Sensitivity chips (Agilent) and Experion DNA 1K kits (Bio-Rad). Libraries were loaded onto Ion PI chips via the Ion PI Template OT2 200 v3 (for small RNAs) or v2 (for large RNAs), and Ion PI Sequencing 200 v3 (for small RNAs) or v2 (for large RNAs) Kits, and sequenced on an Ion Proton Sequencer (Life Technologies).
Bioinformatics
RNA-Seq datasets have been deposited into the NCBI GEO database with the accession number of GSE81216. The large RNA-Seq data were annotated as follows: Reads were trimmed using fastx_trimmer and fastq_quality_trimmer (t = 30), and the resulting trimmed reads were mapped to the genome of each respective species (mouse, mm10; rat, rn5; human, hg19; rabbit, oryCun2) with TopHat (v2.09; default settings plus -b2-very-sensitive -r 200 and -mate-std-dev to 100) [29]. Illumina iGenome references (ENSEMBL) were used for rat, mouse, and human and generated via TopHat for rabbit (ENSEMBL, release 76) [29, 30]. The aligned reads were then assembled using Cufflinks (v2.1.1; default settings plus -frag-bias-correct, -max-bundle-length 1e7, and -multi-read-correct) using the same genome reference versions and mask GTF files containing all known RNA for each species (ENSEMBL) [30]. Expressed genes in the data with the “protein_coding” biotype (ENSEMBL) were extracted for further study and are housed on SpermBase. The percentage of exon coverage for each coding transcript identified in our RNA-Seq data, for each species and sample type, was calculated using Bedtools (bedtools coverage -s -a -b -sorted) and the results for each replicate were averaged [31]. general feature format (GFF) files (mm10, mouse; rn5, rat; oryCun2, rabbit; hg19, human) were obtained from UCSC [32].
The small noncoding (snc) RNA-Seq data were annotated as follows: reads <15 nt and >50 nt were discarded. The remaining reads were matched to known sncRNAs, consisting of, when available for each species, mature miRNAs (miRBase release 21), tRNAs (Genomic tRNA Database; mm10, mouse; rn5, rat; oryCun2, rabbit; hg19, human), Piwi-interacting RNAs (piRNAs) (piRNABank), rRNA (ENSEMBL, release 76), small nucleolar RNAs (snoRNAs) (ENSEMBL, release 76), small nuclear RNAs (snRNAs) (ENSEMBL, release 76), and mtRNA (ENSEMBL, release 76), using Sequery (zero to two mismatches allowed) [30, 33–36]. Unmatched reads were matched to mouse testis [37] and sperm endo-siRNA [5] with Sequery (zero mismatches allowed). The remaining unmatched reads were aligned to the genome of each respective species (human, hg19; mouse, mm10; rat, rn5; rabbit, oryCun2) via Bowtie (settings -n 2 -k 3 -best -S -al -q) [38]. Aligned reads were matched to the genomic coordinates of known mature miRNA, tRNA, rRNA, snRNA, snoRNA, and mtRNA, when available for each species (same databases used previously). Additional annotation of the rabbit sncRNA-Seq data was performed because of the incomplete annotation of the rabbit sncRNA transcriptome. Unmatched, aligned reads 18–25 nt in length were matched to known rat, human, and mouse mature miRNA (miRBase release 21) via Sequery (zero to two mismatches allowed) [33, 36]. Unmatched, aligned reads 26–32 nt in length were matched to known rat, human, and mouse piRNA (piRNABank) via Sequery (zero to two mismatches allowed) [35, 36]. Remaining unmatched, aligned reads 26–32 nt in length were analyzed by piRNA Predictor to detect novel rabbit piRNA [39]. The remaining unmatched, aligned reads 26–32 nt in length were then matched to the novel rabbit piRNA via Sequery (zero to two mismatches) [36]. Read counts were obtained by in-house Python scripts. Reads were normalized as reads per 100 000 aligned reads (RPK). Genes with fewer than 1 RPK were not included in the expression tables of SpermBase; when a gene had greater than 1 RPK in one sample type (e.g., total sperm), but not the other (e.g., sperm head), the expression of the gene was included for both sample types. The sncRNA-Seq reads that initially matched to tRNA genes were extracted and matched to the 5′ and 3′ halves of the full length tRNA (split at the 3′ end of the anticodon), via Sequery (zero to two mismatches allowed) [36]. Reads ≥27 nt and ≤26 nt were named halves and tRFs, respectively, and further classified as 5′ halves, tRF-5′s, 3′ halves, and tRF-3′s based on whether the read aligned to the 5′ or 3′ half of the tRNA.
The conserved sperm miRNA gene targets were predicted using RNAhybrid (settings -n 50 -m 50 000 -c -d xi,θ -p 0.05 -e -20) and miRanda (default settings; score ≥140 and energy ≤-20) against the 3′ untranslated region (UTR) of their respective species (ENSEMBL, release 76), discarding predictions not made by both programs [30, 40–42]. Human miRNAs were used to match against rabbit 3′ UTR, because of the lack of confirmed mature miRNA sequences for the conserved miRNA identified. The 5′ half gene targets were predicted using RNAhybrid (settings -n 50 -m 50 000 -c -d xi,θ -p 0.01 -e -20) against the 5′ UTR, coding DNA sequence (CDS), and 3′ UTR sequences of their respective species (ENSEMBL, release 76). The xi and θ values for each analysis were determined by RNAcalibrate (settings -n 50 -m 50 000 -s), using randomly selected mature miRNA (miRBase, release 21) and known 5′ half sequences (tRFDB, human and mouse) (Supplemental Table S1; Supplemental Data are available online at www.biolreprod.org) [33, 40, 43].
Gene ontology (GO) term enrichment analysis was performed using the g:Profiler suite [44, 45]. Input gene lists were ordered by the number of miRNA or 5′ halves that targeted each gene (highest to lowest), then by the average P value (lowest to highest). The top 2000 and 4000 genes of the miRNA and 5′ half ordered lists, respectively, were analyzed in g:GOST (settings: significant only, ordered query, no sorting, or hierarchical sorting—moderate, functional categories with 3–1000 terms only) [44, 45]. Word clouds were generated using Genes2WordCloud (http://www.maayanlab.net/G2W/) using the biological process (BP) terms found to be significantly enriched (P ≤ 0.05) using the above settings (no sorting), and the aforementioned ordered lists (miRNA and 5′ half putative gene targets) or the total sperm coding genes (reads per kilobase of transcript per million mapped reads [RPKM] ≥ 3) found to be conserved across species [46].
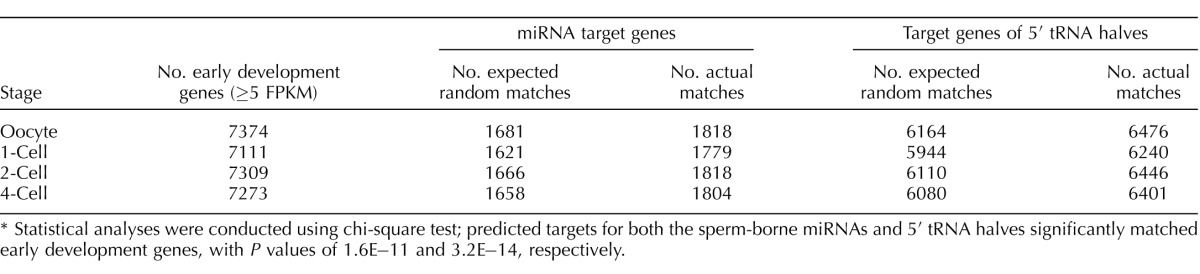
The significance of the number of observed matches between the predicted conserved miRNA and 5′ tRNA half gene targets and genes present in early development (≥5 fragments per kilobase of exons per million reads mapped) was determined using a chi-square test. Early development gene expression was obtained from the Database of Transcriptome in Mouse Early Embryos (DBTMEE) [47]. The observed number of matches was compared to the number of expected random matches, which was defined as the number of early development genes for each stage multiplied by the percentage of all known mouse coding genes that was targeted by the conserved miRNA or 5′ tRNA halves.
RESULTS
Complete Lysis of Sperm Nuclei is Key to Maximal Sperm RNA Yields
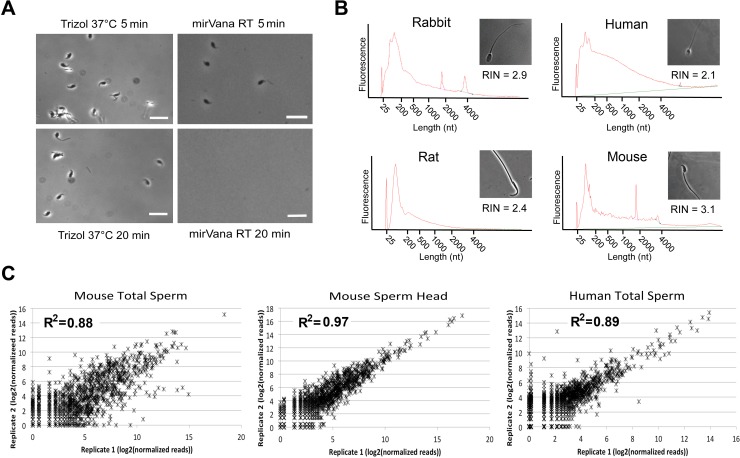
Trizol, one of the most commonly used lysis solutions for RNA isolation, was used in several sperm RNA profiling studies [26, 27, 48–51]. However, Trizol is optimal for RNA isolation from somatic cells, but does not work efficiently for sperm RNA isolation, as revealed in our tests (Fig. 1A). After incubation of the mouse sperm (at a concentration of 5000–10 000 sperm/μl) at 37°C for 20 min, the number of sperm decreased by ∼15%–20%. In contrast, mouse sperm in the mirVana lysis buffer were almost completely dissolved after 20 min of incubation at room temperature (Fig. 1A). This was not surprising given that Trizol solution contains neither detergents nor reducing agents, whereas the sperm chromatin is well known for the enriched disulfide bonds, which can only be broken down by both detergents (SDS or sodium lauryl sulfate [SLS]) and reducing agents (e.g., dithiothreitol [DTT] or β-mercaptoethanol). Given that a large number of sperm RNAs are embedded inside sperm nuclei [4, 18, 19], it is critical to lyse sperm heads completely so that sperm RNAs can be fully released. To this end, we attempted to establish a “modular” RNA isolation protocol so that it is applicable to RNA isolation from sperm of any species. We adopted the mirVana Total RNA Isolation Kit because it uses a lysis buffer containing both 0.5% N-lauroyl sarcosine (a detergent) and 0.1 M β-mercaptoethanol (a reducing agent), which are required for the complete lysis of the sperm heads. It should be noted that other similar kits with both detergents and reducing agents in their lysis solutions (e.g., the RNeasy Kit by Qiagen, as used previously [18]) can be as efficacious as the mirVana Kit for sperm RNA extraction. As outlined in Table 1, human and rabbit sperm heads could be easily lysed by pipetting up and down in the lysis buffer, whereas mouse sperm required mechanical homogenization and rat sperm even needed heating in addition to homogenization.
FIG. 1.
Quality and reproducibility of the sperm RNA isolation procedure reported. A) Comparison of the capability of Trizol and the mirVana lysis buffer to dissolve the mouse sperm heads. Bar = 10 μm. B) Representative electropherograms of total RNA isolated from mouse and rat epididymal sperm and rabbit and human ejaculated sperm along with their respective RINs, as determined using an Agilent Bioanalyzer 2100. C) Scatter plots showing the distribution of the number of normalized small RNA sequencing reads (log2 values) in two biological replicates of mouse and human total sperm and mouse sperm heads. R2 values represent Pearson correlation coefficients.
Electropherograms of the extracted sperm-borne RNAs were used to assess sperm RNA quality (Fig. 1). A lack of intact 18S and 28S rRNAs in sperm renders the traditional RIN conventions inappropriate for assessing sperm RNA quality [23, 52]. Instead of a RIN of ∼10 being ideal for other cell types, we observed that a RIN of ∼3 is indicative of high-quality sperm RNA, consistent with a previous report [53]. Lower RINs (<2) typically represent excessive RNA degradation, whereas higher RINs (>4) may indicate somatic cell RNA contamination. We also observed that, in addition to a RIN of 2–4, sperm RNA electrophoretic profiles across species are characterized by a large population of RNA <200 nt in length (Fig. 1), consistent with previous observation in mouse [50], human [52], horse [26], bull [54], and domestic swine [55].
Epididymal sperm collected from one adult male mouse routinely yielded ∼350–500 ng total RNA using our method (Table 1), which is sufficient for RNA-Seq analyses of both large and small RNAs. Similarly, a comparable amount of total RNA was obtained from sperm collected from one adult male rat, one ejaculate from one male rabbit, or a man (Table 1). Consistent with previous reports showing sperm nuclei/heads contain higher concentration of RNAs [4, 7, 18, 19, 56], our data suggest that sperm heads contain ∼60%–80% of all sperm-borne RNAs, at least in mice and rabbits (Table 1). Therefore, it is critical to completely lyse the sperm heads in order to obtain the maximal RNA yields. This may also explain why ∼70–80 mice were used to obtain enough RNA for constructing one library for RNA-Seq in an earlier study using Trizol [50].
The numbers of small RNA sequencing reads in two biological replicates of the mouse and human total sperm as well as mouse sperm heads were analyzed, and excellent correlations between replicates (R2 = 0.88–0.98) demonstrated a high degree of reproducibility of our sperm RNA isolation method (Fig. 1C). By comparing our mouse total sperm small RNA dataset with that of a previous report [50] where Trizol was used for sperm RNA isolation, we found that the alignment rate (i.e., percentage of sequencing reads aligned to the genome) of our data is much greater than that of their data (∼30% of ours vs. ∼13% of theirs) (Supplemental Fig. S1). Moreover, 24 303 unique small RNAs were identified from our dataset, whereas 18 915 were annotated from their dataset using exactly the same annotation pipeline (Supplemental Fig. S1). Although 13 318 small RNAs were common in both datasets, our dataset yielded more unique small RNAs (10 985 vs. 5597) compared to theirs (Supplemental Fig. S1). These data further support that our sperm RNA isolation method can lead to better sequencing results, which allows for identification of more sperm-borne RNA species.
Sperm-Borne mRNAs
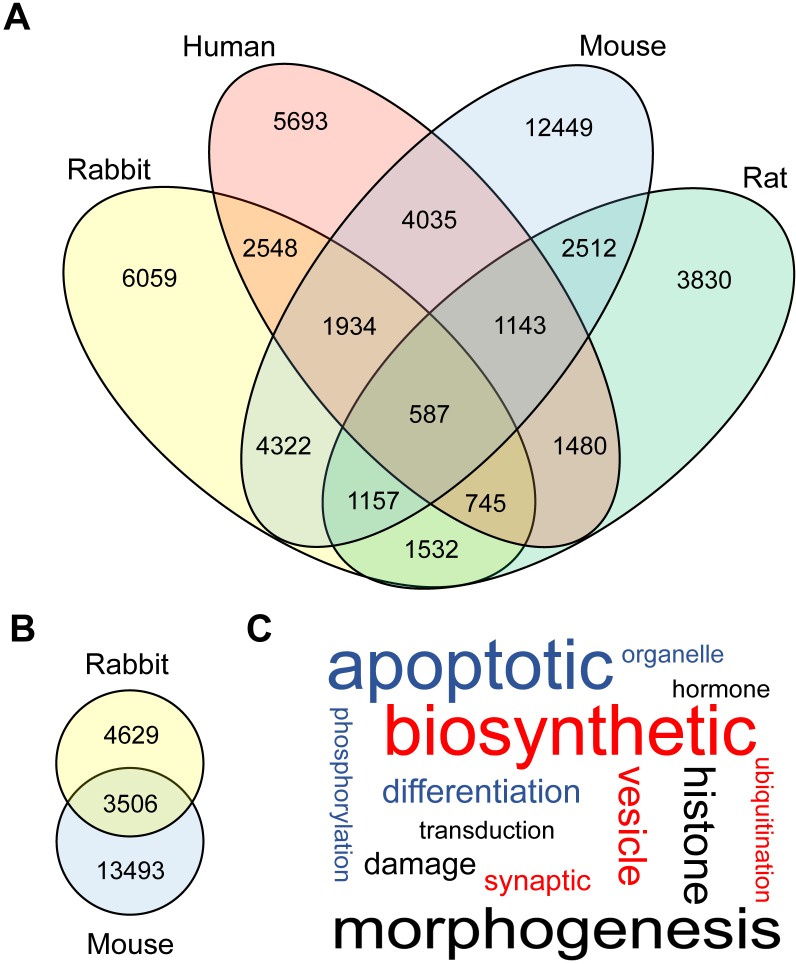
RNA-Seq was performed using total RNA isolated from both sperm heads (for rabbit and mouse) and total sperm (for rat, rabbit, mouse and human) to determine the expression of coding genes. Previous investigations into the mRNA content of mammalian sperm have found diverse populations of coding genes within both mouse and human sperm [2, 57]. Similarly, we observed several thousand coding genes present in the total sperm and sperm head samples (Fig. 2, A and B). The sperm coding transcriptome was previously shown to consist of many fragmented mRNAs [2]. We investigated the transcript integrity in each of our samples and found that the degree of “intactness” (i.e., the percentage of the mature transcript represented by sequencing reads in the data) varied considerably among the mRNA identified in our data (Supplemental Table S2). Next, we assessed the degree to which sperm-borne mRNAs are conserved across mammals. After removing any genes with expression levels below 3 RPKM from consideration, we identified 587 unique mRNAs present in the total sperm of all four species (Fig. 2A and Supplemental Table S3A). Using the same cutoff, we observed 3506 unique mRNAs that were commonly expressed in the mouse and rabbit sperm heads (Fig. 2B and Supplemental Table S3B). GO term enrichment analyses were performed on the conserved total sperm mRNAs identified (Fig. 2C and Supplemental Table S4). “Morphogenesis” and other development-related terms were the most prevalent amongst the significantly enriched BP terms (Fig. 2C and Supplemental Table S4A).
FIG. 2.
Sperm-borne mRNA fragments. A) Venn diagram showing the number of sperm-borne mRNA fragments conserved among four mammalian species (mouse, rat, rabbit, and human). B) Venn diagram showing conserved mRNA fragments between rabbit and mouse sperm heads. Only genes with common names were considered to be conserved ones. C) Word cloud showing the BP terms significantly enriched (P ≤ 0.05) among the conserved coding genes in total sperm of four species (mouse, rat, rabbit, and human) analyzed in this study. The size of each word is based on its frequency within the enriched BP terms, and the original data can be found in Supplemental Table S4A (no filtering).
Sperm-Borne Small RNAs
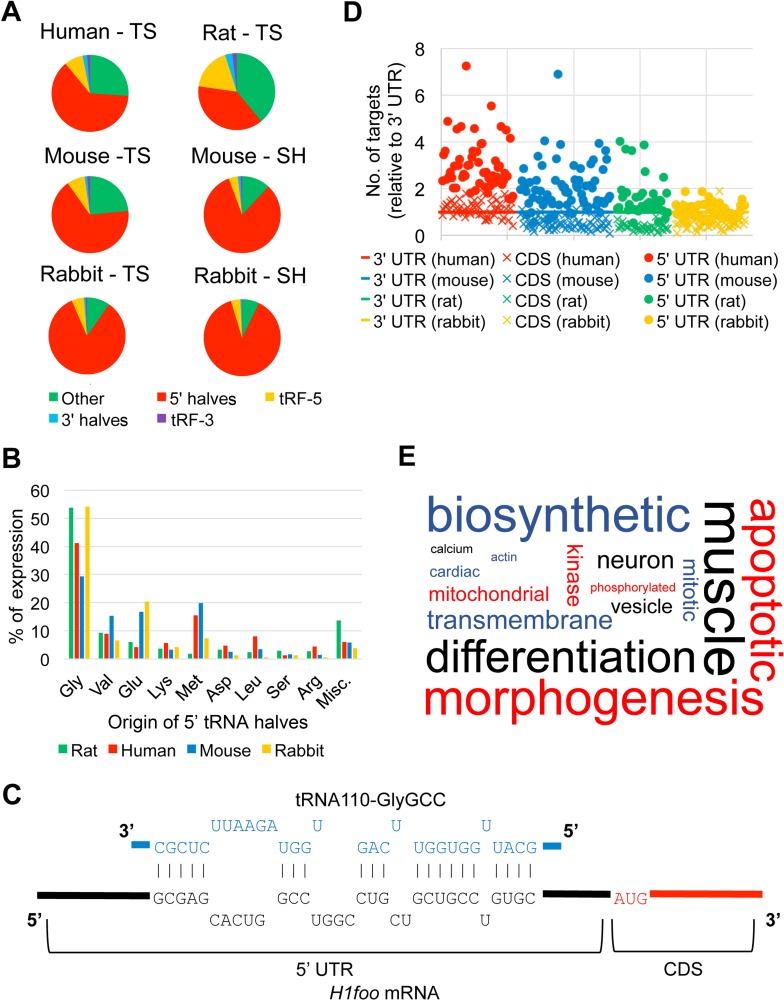
In addition to mRNAs, we sequenced sncRNA in human, rat, rabbit, and mouse total sperm, as well as rabbit and mouse sperm heads. Consistent with previous reports, we observed diverse sncRNA populations in the sperm of each species surveyed [3, 50, 51, 58]. The most abundant classes of sncRNA in sperm were miRNA, tRNA-derived sncRNA (tsRNA), piRNA, and mitochondrial genome-encoded small RNA (mitosRNA) (Fig. 3 and Supplemental Table S5). Interestingly, significant differences were observed in RNA contents between total sperm and sperm heads (Fig. 3). The tsRNAs were much more abundant in sperm heads than in total sperm, suggesting that tsRNAs are mainly localized to the sperm heads. Reduced proportions of mitosRNAs and miRNAs in sperm heads suggest that these two types of small RNAs, in addition to tsRNAs, are the major small RNA constituents in other compartments of the total sperm. Similarly, in rabbit sperm heads, tsRNAs were the predominant small RNA species, followed by mitosRNAs and miRNAs; mitosRNAs were much more abundant in the total sperm compared to sperm heads, suggesting that mitosRNAs are mainly localized to the nonhead regions of the total sperm. Considering their consistent abundance in both total sperm and sperm heads, we focused on four small RNA classes (piRNAs, mitosRNAs, miRNAs, and tsRNAs) while investigating the sncRNA conservation in mammalian sperm.
FIG. 3.
Sperm small RNA contents. Pie charts showing the proportional distribution of each of the eight sncRNA populations in total sperm (TS) or sperm heads (SH) based on sncRNA datasets currently available in SpermBase. The original data can be found in Supplemental Table S5.
piRNAs.
Previous work has demonstrated that piRNAs are highly expressed in male germ cells and play an essential role during spermatogenesis [59]. However, little is known about the purpose of their presence in mature sperm [1]. Unlike miRNAs, piRNAs are poorly conserved in their sequences across species [60]. Therefore, we investigated the general features of the piRNA populations in each species. In pachytene spermatocytes, there is a burst in piRNA expression—the piRNAs expressed in the meiotic phase of spermatogenesis are called “pachytene piRNAs,” whereas the piRNAs expressed in spermatogonia are referred to as the “prepachytene piRNAs” [59, 61]. Prepachytene piRNAs are typically 26–28 nt in length and have a strong preference towards uracil and adenine at their first and 10th nucleotide positions, respectively. In contrast, pachytene piRNAs are typically 30 nt in length and only possess a preference for adenine at their first nucleotide [62]. Based on these characteristics, we assessed whether the piRNAs found in total sperm samples were prepachytene or pachytene in origin. The majority of piRNAs in total sperm samples were 29–32 nt in length across species (Supplemental Fig. S2A), and our analyses of nucleotide preferences showed a strong bias towards uracil for the first nucleotide (Supplemental Fig. S2B) and only a slight preference towards adenine at the 10th nucleotide (Supplemental Fig. S2C). These data suggest that the majority of the rabbit, human, rat, and mouse sperm-borne piRNAs are pachytene piRNAs.
Mitochondrial genome-encoded small RNAs.
The majority of the sncRNA-Seq reads aligned to mitochondrial genome were much shorter than their matching full-length transcripts. Many of the sequences aligned to an mtRNA were a consistent length, suggesting that they are not derived from the degradation of intact transcripts, but rather are mitosRNAs that were purposefully produced, as reported previously [63] (Supplemental Figs. S3–S7). The lack of a consensus sequence length for mitosRNAs has been observed previously [63]. We also found that sperm-borne mitosRNAs varied in length depending on the origin of their mitochondrial genes, and this feature was conserved across all four species surveyed (Supplemental Figs. S4–S7). The mitosRNAs ranked second in terms of relative abundance among all four major sperm-borne small RNA species, i.e., tsRNAs > mitosRNAs > miRNAs > piRNAs (Fig. 3).
miRNAs.
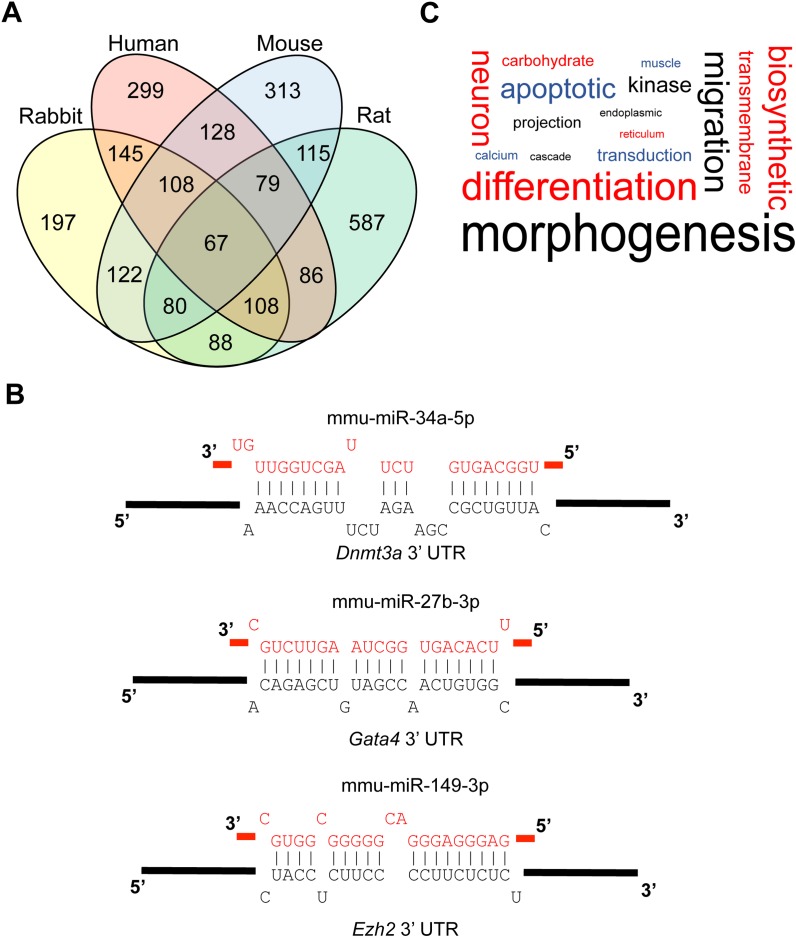
Several groups have previously investigated the miRNA contents in sperm, for a variety of organisms such as mouse [50, 64], human [2, 4], bull [58, 65–67], and pig [68]. Despite this cache of sperm miRNA expression information, cross-species comparisons using these existing data would be, as discussed previously, intrinsically unreliable because of differences in methodology [20, 21, 23–27]. Using the standardized miRNA expression data available in SpermBase, we were able to identify 67 miRNAs that were present in the total sperm samples of all four species (Fig. 4A and Supplemental Table S6A). These miRNAs accounted for the majority of all miRNA expression in every species surveyed (Supplemental Table S6B). Many of the conserved miRNAs were members of the same clusters (i.e., their genes were within 10 kb of one another); for example, one of these clusters contains the miR-34b/c family, which, along with miR-449a/b/c, is known to be involved in the regulation of spermatogenesis and male fertility in mice [10]. Three (miR-34c-3p, miR-19b-3p, and miR-148b-3p) of the seven miRNAs that were, in another study, found to be differentially expressed when comparing bulls of moderate and high fertility were also members of the 67 conserved miRNAs [65].
FIG. 4.
Sperm-borne miRNAs. A) Venn diagram showing sperm miRNAs conserved among four mammalian species (mouse, rat, rabbit, and human). B) Examples of critical early development genes predicted to be targeted by conserved sperm-borne miRNAs. C) Word cloud showing the BP terms significantly enriched (P ≤ 0.05) in the predicted targets of conserved sperm miRNAs among the four mammalian species analyzed in this study (mouse, rat, rabbit, and human). The size of each word is based on its frequency within the enriched BP terms and the original data can be found in Supplemental Table S8 (no filtering).
To assess whether the conservation of these miRNAs across species held any functional significance, we predicted the gene targets for all 67 miRNAs in silico using both RNAhybrid and miRanda to compare the miRNAs to 3′ UTR sequences, discarding the gene target predictions not made by both programs [40, 42]. The predicted gene targets of each conserved miRNA can be found in Supplemental Table S7. Several of these predicted targets, such as Dnmt3a, Ezh2, and Gata4, are known for their important role in early development (Fig. 4B) [69–76]. Many miRNAs are functionally redundant, a phenomenon that is commonly observed in single miRNA knockout mouse models, the majority of which lack any aberrant phenotype [9, 77]. Because of this, we used genes that were targeted by at least 2 of the 67 conserved miRNAs for our subsequent studies. In order to gauge whether the putative functions of these miRNAs are conserved, we performed a GO term enrichment analysis for every species on the top 2000 redundantly targeted genes, ranked by the number of matching conserved miRNAs, then by the average P value of each predicted gene target. The results of the GO term enrichment analysis are summarized in Supplemental Table S8. Interestingly, “morphogenesis” and development-related terms were the most prevalent terms for every species surveyed (Fig. 4C and Supplemental Table S8). Likewise, of the top 20 BP terms identified in our interspecies comparison, 6 were related to development (Supplemental Table S8G). These data suggest that these 67 miRNAs night have a regulatory role during early development, and this role is conserved across mammalian species.
To further assess whether the conserved sperm miRNAs are active during early development, we compared the list of redundantly targeted (i.e., targeted by at least two miRNAs) murine genes to genes known to be expressed in the very early stages of development (e.g., oocyte to four-cell). We found that the numbers of predicted gene targets that matched to early development genes were significantly higher than the expected number of random matches (Table 2), supporting the putative role of the 67 conserved sperm miRNAs as regulators of gene expression during early development.
TABLE 2.
Predicted targets of highly conserved sperm-borne miRNAs and 5′ tRNA halves compared to early development genes.*
Statistical analyses were conducted using chi-square test; predicted targets for both the sperm-borne miRNAs and 5′ tRNA halves significantly matched early development genes, with P values of 1.6E−11 and 3.2E−14, respectively.
tRNA-derived sncRNAs.
While annotating the sncRNA-Seq data for SpermBase, we observed that tRNAs accounted for more reads than any other sncRNAs, with the exception of human total sperm, in which mitosRNAs were more abundant (Fig. 3 and Supplemental Table S5). A closer look at these tRNA-aligned reads revealed that the majority of them were ∼30 nt, indicating that they actually represented tsRNAs and not intact mature tRNA species (Supplemental Fig. S8). These tsRNAs have previously been identified in a myriad of organisms and cell types, including sperm, where they were found to be highly abundant [51, 78]. Similar to piRNAs, tsRNAs are divided into further subgroups based on their length as well as their origins. The tsRNA species of 27 nt or longer were classified as 5′ or 3′ tRNA halves depending on whether they were derived from the 5′ or 3′ half of the mature tRNA (split at the anticodon), whereas those 19–26 nt in length were classified as tRF-5′s or tRF-3′s depending on their half preference [79]. We found that 5′ tRNA halves were the most abundant tsRNAs in every species and sample type surveyed, especially sperm heads, consistent with previous findings in mouse sperm (Fig. 5A) [51]. The predominance of 5′ halves in sperm-borne small RNA populations, in addition to speculation that they may play an important role in epigenetic inheritance, led us to focus our attention on this particular class of tsRNAs [16, 17, 51, 80–82].
FIG. 5.
Sperm-borne tsRNAs. A) Pie charts showing the proportional distribution of each of the five tsRNA subclasses in total sperm (TS) and sperm heads (SH) in the four mammalian species analyzed in this study. “Other” refers to tRNA-aligned reads that were not classified as tsRNAs. B) Histogram showing the origin of sperm-borne 5′ tRNA halves in the four mammalian species. The tRNA genes from which the 5′ tRNA halves were derived were grouped by amino acid (AA) and ranked. “Misc” refers to amino acids not shown on the table. C) Example of potential binding of the 5′ UTR of H1foo mRNA by a 5′ tRNA half. D) Scatter plots showing the relative number of targeting sites for the 5′ tRNA halves in different regions of the target genes (5′ UTR, CDS and 3′ UTR). For each species, the number of predicted gene targets for each 5′ tRNA half when matched against 5′ UTR, CDS, and 3′ UTR sequences was normalized to the number of 3′ UTR-based gene targets observed. E) Word cloud showing the BP terms significantly enriched (P ≤ 0.05) in the predicted targets of conserved sperm-borne 5′ tRNA halves among the four mammalian species analyzed in this study (mouse, rat, rabbit, and human). The size of each word is based on its frequency within the enriched BP terms and the original data can be found in Supplemental Table S10 (no filtering).
To assess the similarity of the 5′ tRNA halves between the species on SpermBase, we sorted the total sperm 5′ tRNA halves by the amino acid of their precursor tRNAs. In every organism, tRNAGly species accounted for the most 5′ tRNA halves, followed by tRNAGlu, tRNAVal, tRNAMet, and tRNALys (Fig. 5B). This finding is in agreement with previous studies on murine sperm-borne 5′ tRNA halves, which found that 5′ tRNA halves from tRNAGly and tRNAGlu were the most abundant tsRNAs in sperm [51]. The observation that the majority of the 5′ tRNA halves in each species surveyed are derived from the same group of tRNAs also indicates that the production and retention of these sncRNAs is conserved across mammalian sperm, similar to miRNAs (Fig. 4A).
Several studies have found that 5′ tRNA halves may possess the ability to act as posttranscriptional gene regulators, similar to miRNAs [83–86]. In order to evaluate whether the 5′ tRNA halves that we found in the SpermBase data were capable of complementary sequence-based gene regulation, we performed unbiased gene target prediction analyses, matching the 5′ halves against the 3′ UTR, as well as the 5′ UTR and CDS sequences available for each species. Instead of using both miRanda and RNAhybrid as we did for the conserved sperm miRNAs, we used solely RNAhybrid. This is because RNAhybrid accounts for the length of the target sequence, ensuring that the longer 3′ UTR and CDS sequences would not receive a higher number of random matches than the 5′ UTR [40]. The predicted gene targets for each species are provided in Supplemental Table S9. We observed that, on average, the 5′ UTR (Fig. 5C) was targeted the most, compared to the 3′ UTR and CDS sequences, and that this was true for all four species (Fig. 5D and Supplemental Table S9). The proportion of genes that were targeted by more than one 5′ tRNA half was also consistently higher on average for the 5′ UTR-based analyses (∼88% targeted by at least two 5′ tRNA halves) when compared to the CDS- and 3′ UTR-based analyses (∼68% and ∼78%, respectively), suggesting that there were fewer random target predictions when the 5′ UTR sequences were analyzed. Based on these findings, we utilized the 5′ UTR-based putative gene targets for our subsequent analyses.
From the 5′ UTR-based gene target predictions, we selected the top 4000 redundantly targeted genes, which were ranked according to the number of 5′ tRNA halves that targeted each gene, then by the average P value for each prediction, to perform a GO term enrichment analyses for each species. The results are summarized in Supplemental Table S10. Terms relevant to development and morphogenesis were common in the results for every species, similar to what was observed for the conserved sperm miRNAs (Figs. 4C and 5E and Supplemental Tables S7G and S9G). Of the top 20 BP terms for the 5′ tRNA halves, three were directly related to development (Supplemental Table S10G). Interestingly, the 5′ tRNA halves BP term ranked second on Supplemental Table S10G was “WNT signaling pathway,” a pathway vital for proper early embryonic development across the animal kingdom [87]. Additionally, “catabolic” and “metabolic” were prevalent in the enriched terms (Fig. 5E). This is not surprising, as sperm-borne 5′ tRNA halves have previously been linked to altered expression of metabolic genes in the early embryo [16]. Overall, it seems that, like the conserved sperm miRNAs, sperm-borne 5′ tRNA halves might also play a role in regulating gene expression during early development. To investigate this further, we compared the redundantly targeted murine genes (i.e., targeted by at least two 5′ tRNA halves) identified in the 5′ UTR-based analyses to genes known to be expressed throughout early development. As also seen with the putative miRNA targets, the number of predicted targets for 5′ tRNA halves that matched these early development genes was significantly higher than the number of anticipated random matches, providing additional evidence that 5′ tRNA halves may play a functional role after fertilization (Table 2). The 5′ tRNA halves were predicted to target a much larger percentage of the early development genes (∼88%) relative to the miRNAs (∼25%) (Table 2). This observation is consistent with another recent study, which determined that sperm-borne 5′ tRNA halves targeted ∼80% of the eight-cell embryo transcriptome [16].
SpermBase Is Easy to Use and Will Be Expanded to Cover More Species
To share sperm RNA profiling data, we established SpermBase, a publicly accessible database, which can be found at www.spermbase.org. The website is separated into five main pages: Home, Search, Method, Species, and FAQ & Contact. The homepage (Supplemental Fig. S9) provides an introduction to the database itself, touching on the topics discussed above in the introduction of this article. Links can be found throughout the introduction to other parts of the SpermBase website.
At the Search page, users can search SpermBase for expression data for their gene(s) of interest. Each RNA that was found in our data is categorized by its original or, if it was identified by the authors, its given name (“Gene”). As discussed below, the reads aligned to tRNA genes were classified as different subclasses (e.g., 5′ tRNA halves, tRF-5) of tsRNAs. The naming convention for these tsRNAs was to add the subclass of tsRNAs after the original tRNA name, e.g., the 5′ halves of “trna1000-PheGAA” would be named “trna1000-PheGAA-5halves.” Each RNA is also organized by the species it was identified in (“Species”), the “Sample” type (i.e., total sperm or sperm head), the class of large or small RNA, e.g., mRNA, piRNA (“Class”), and its expression (“Expression”). The expression values shown in SpermBase are the normalized read counts, given as RPK for sncRNA genes and RPKM for mRNAs. The sequences (5′ → 3′) of the tsRNAs and novel rabbit piRNAs we identified in this study are also provided under the “Sequence” column.
The Method page describes the methodology (discussed previously) used for sperm RNA isolation followed RNA-Seq analyses for building SpermBase. As SpermBase expands to include additional species, we will update this page with the lysis stage parameters used for the new species. Users who employ our modular RNA isolation protocol on sperm from animals not described in SpermBase are encouraged to send us information on the lysis parameters that they found to be most optimal for that species.
On the Species page, a list of each species currently available on SpermBase is provided, along with planned future additions to SpermBase. Users can obtain the expression data for each individual species on the Download page; these data are available in multiple formats (.txt, .csv, and Excel .xls). The files can be downloaded by either left clicking the file or right clicking to “Save link as…” if the user wishes to rename the file prior to downloading.
The FAQ & Contact page provides answers to frequently asked questions related to SpermBase and detailed contact information. Users who have questions not addressed in the FAQ are encouraged to email SpermBase. Troubleshooting queries and comments about the design and functionality of SpermBase are also welcome.
DISCUSSION
Currently, SpermBase contains sperm RNA-Seq data of four mammalian species (human, mouse, rat, and rabbit). To ensure the comparability of multispecies sperm RNA data in SpermBase, we conducted RNA-Seq on sperm RNA samples isolated using our standard sperm RNA isolation protocol. The key to high yield of high-quality sperm RNAs is to dissolve the sperm heads completely, which requires reducing agents, mechanical disruptions, and increased temperature during lysis, depending on the extent to which the sperm chromatins are compacted. In general, the more compact the sperm heads are, the more vigorous treatment is needed in the lysis step. In our protocol, the rat sperm requires longer homogenization (90 sec) plus heating (65°C for 5 min) for a complete lysis of the sperm chromatin, whereas human and rabbit sperm only need up and down pipetting. Interestingly, rat sperm appear to be much more compact than human and rabbit sperm.
Another interesting observation is the difference in RNA contents between total sperm and sperm heads. The fact that RNAs in sperm heads account for 60%–80% of the total sperm-borne RNA contents suggests that numerous RNAs are localized to the sperm nucleus and may be associated with sperm chromatin, as suggested in previous reports [51]. Because of the considerable difference in RNA contents between sperm heads and total sperm, it is critical to verify that all sperm heads are completely lysed through microscopic observation after the lysis step. It is our hope that, in addition to using the data on SpermBase, other groups will utilize our RNA extraction method and quality control metric (i.e., RIN of 2–4 as an indicator of ideal sperm RNA quality) for their own sperm RNA profiling studies. Although we are confident in the efficacy of our method, we do caution other groups to make sure that they initially test and, if necessary, optimize the lysis step for their own samples, to ensure the complete lysis of the entire sperm cells. In the future, we hope to develop an RNA extraction method that can be applied to sperm from any species, without making any alterations to the protocol. Other future improvements to SpermBase will include the addition of more sperm head fraction data, as well as the inclusion of other species, expanding the utility of SpermBase for the scientific community. Currently, we are working on adding large and small RNA expression data for the sperm of zebrafish, horse, monkey, and bull.
Using SpermBase data, we investigated the conserved general features of mammalian sperm mRNA and small RNA populations. We limited our analyses of the conservation of sperm-borne small RNAs to four classes, piRNAs, mitosRNAs, miRNAs, and tsRNAs, because they represent the most abundant small RNA species in both total sperm and sperm heads. The piRNA populations in the total sperm of all four mammals appeared to be pachytene in origin. As the sequences of individual piRNAs are poorly conserved across species, it is difficult at this time to say whether the piRNA present in sperm serve some biological purpose or are just the random remnants of the pachytene piRNA expression burst [59]. In every species (with the exception of human), mitosRNAs appear to be the second most abundant small RNA species in both total sperm and sperm heads; however, their physiological role remains unknown. A total of 67 conserved miRNAs account for the majority of all miRNAs expressed in each of the four species examined. The highly conserved sperm-borne miRNAs may suggest a potential role in fertilization and early development.
Our analyses indicate that tsRNAs are the most abundant sperm small RNAs in every species (with the exception of human), with 5′ tRNA halves as the most dominant subclass, consistent with a previous report [51]. In every species surveyed, the bulk of these 5′ tRNA halves originated from the same precursor mature tRNAs, namely tRNAGly, tRNAGlu, tRNAVal, tRNAMet, and tRNALys. Unlike miRNAs, which preferentially bind to the 3′ UTR of their target genes [88], sperm-borne 5′ tRNA halves preferentially target the 5′ UTR. Interestingly, the genes that are targeted by these 5′ tRNA halves are mostly related to early development, suggesting the sperm-borne tsRNAs may also have a role in regulating early development. Previous studies have demonstrated that 5′ tRNA halves have the potential to act as posttranscriptional regulators by binding to the 5′ UTR in a reverse complementary manner in cell lines [83–86]. In another study, 5′ tRNA halves inhibited translation via the displacement of eIF4G/eIF4A from the 5′ ends of mRNA [84]. In a recent report, Chen et al. determined that putative 5′ tRNA half target sites in the CDS were less frequent than the number observed in promoter regions (i.e., 2 kb upstream of the transcriptional start site) [16]. The data housed at SpermBase will undoubtedly be a boon to future investigations into the mechanism behind the proposed regulatory functions of sperm-borne 5′ halves.
Sperm-borne RNAs have been implicated as potential mediators of epigenetic inheritance [11–17, 89]. Of particular interest are the sperm-borne tsRNAs, as a methyltransferase involved with tsRNA production, Dnmt2, was found to be necessary for the transmission of two epigenetically inherited phenotypes [81]. Although the effects of Dnmt2 deficiency on the tsRNA populations in sperm have yet to be determined, in other cell types, the production of 5′ half species was found to increase in the absence of Dnmt2 [90]. It is therefore possible that changes in sperm-borne 5′ half levels can have an effect on the early embryo; this is in agreement with the results of our analyses of the predicted gene targets of the 5′ tRNA halves, suggesting that 5′ tRNA halves interact with genes related to development. Recently, in three separate epigenetic inheritance models, expression of sperm-borne 5′ tRNA halves was observed to be affected by an altered diet (i.e., high fat or low protein) or vinclozolin (a common agricultural fungicide) exposure [16, 17, 82]. Future investigations into other examples of epigenetic inheritance could potentially benefit from the tsRNA expression data and consensus sequences housed in SpermBase.
Since their discovery, research on sperm-borne RNAs has revealed that they are not simply the remnants of spermatogenesis—to the contrary, the likelihood that sperm RNAs play a functional role in many different contexts (e.g., posttranscriptional regulation during early development and epigenetic inheritance) has only increased in recent years. The difficulties associated with sperm RNA isolation and sperm RNA contents in various species have no doubt slowed the advance of efforts to elucidate additional functions and to assess the conservation of these RNAs across evolution. These issues will be alleviated, at least partially, by SpermBase, which provides public expression data as well as a simple and effective RNA extraction methodology, thereby making the study of sperm-borne RNA more accessible to other labs. We hope that, with this contribution, we will see an acceleration of our understanding of the physiological roles of sperm-borne RNAs.
Footnotes
Supported, in part, by grants from the NIH (HD060858, HD071736 and HD085506 to W.Y.) and the Templeton Foundation (PID: 50183 to W.Y.). Sequencing was conducted in the Single Cell Genomics Core supported, in part, by a NIH grant (1P30GM110767). RNA-Seq datasets have been deposited into the NCBI GEO database with the accession number of GSE81216.
These authors contributed equally to this work.
REFERENCES
- Casas E, Vavouri T. Sperm epigenomics: challenges and opportunities. Front Genet. 2014;5:330. doi: 10.3389/fgene.2014.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendler E, Johnson GD, Mao S, Goodrich RJ, Diamond MP, Hauser R, Krawetz SA. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013;41:4104–4117. doi: 10.1093/nar/gkt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP. A survey of small RNAs in human sperm. Hum Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update. 2013;19:604–624. doi: 10.1093/humupd/dmt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, Yan W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development. 2016;143:635–647. doi: 10.1242/dev.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- Pessot CA, Brito M, Figueroa J. Concha, II, Yanez A, Burzio LO. Presence of RNA in the sperm nucleus. Biochem Biophys Res Commun. 1989;158:272–278. doi: 10.1016/s0006-291x(89)80208-6. [DOI] [PubMed] [Google Scholar]
- Boerke A, Dieleman SJ, Gadella BM. A possible role for sperm RNA in early embryo development. Theriogenology. 2007;68(suppl 1):S147–S155. doi: 10.1016/j.theriogenology.2007.05.058. [DOI] [PubMed] [Google Scholar]
- Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Tang C, Zhang Y, Wu J, Bao J, Zheng H, Xu C. Yan W. mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol Open. 2015;4:212–223. doi: 10.1242/bio.201410959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Yuan S, Oliver D, Schuster A, Zheng H, Yan W. Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice. Sci Rep. 2015;5:9266. doi: 10.1038/srep09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A. 2015;112:13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich RJ, Anton E, Krawetz SA. Isolating mRNA and small noncoding RNAs from human sperm. Methods Mol Biol. 2013;927:385–396. doi: 10.1007/978-1-62703-038-0_33. [DOI] [PubMed] [Google Scholar]
- Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Arch Androl. 2007;53:161–167. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- Mao S, Goodrich RJ, Hauser R, Schrader SM, Chen Z, Krawetz SA. Evaluation of the effectiveness of semen storage and sperm purification methods for spermatozoa transcript profiling. Syst Biol Reprod Med. 2013;59:287–295. doi: 10.3109/19396368.2013.817626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Sendler E, Goodrich RJ, Hauser R, Krawetz SA. A comparison of sperm RNA-seq methods. Syst Biol Reprod Med. 2014;60:308–315. doi: 10.3109/19396368.2014.944318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan M, Martinez A, Llonch S, Pujol A, Vernaeve V, Vassena R. Effect of ribonucleic acid (RNA) isolation methods on putative reference genes messenger RNA abundance in human spermatozoa. Andrology. 2015;3:797–804. doi: 10.1111/andr.12053. [DOI] [PubMed] [Google Scholar]
- Cappallo-Obermann H, Schulze W, Jastrow H, Baukloh V, Spiess AN. Highly purified spermatozoal RNA obtained by a novel method indicates an unusual 28S/18S rRNA ratio and suggests impaired ribosome assembly. Mol Hum Reprod. 2011;17:669–678. doi: 10.1093/molehr/gar037. [DOI] [PubMed] [Google Scholar]
- Lalancette C, Platts AE, Johnson GD, Emery BR, Carrell DT, Krawetz SA. Identification of human sperm transcripts as candidate markers of male fertility. J Mol Med (Berl) 2009;87:735–748. doi: 10.1007/s00109-009-0485-9. [DOI] [PubMed] [Google Scholar]
- Johnson V. From a sperm's eye view—revisiting our perception of this intriguing cell In: Proceedings of the 53rd Annual Convention of the American Association of Equine Practitioners; 1–5 December 2007. Orlando, Florida 104–177. [Google Scholar]
- Das PJ, Paria N, Gustafson-Seabury A, Vishnoi M, Chaki SP, Love CC, Varner DD, Chowdhary BP, Raudsepp T. Total RNA isolation from stallion sperm and testis biopsies Theriogenology 2010. 74 1099 1106, 1106e1091–1106e1092. [DOI] [PubMed] [Google Scholar]
- Shafeeque CM, Singh RP, Sharma SK, Mohan J, Sastry KV, Kolluri G, Saxena VK, Tyagi JS, Kataria JM, Azeez PA. Development of a new method for sperm RNA purification in the chicken. Anim Reprod Sci. 2014;149:259–265. doi: 10.1016/j.anireprosci.2014.06.032. [DOI] [PubMed] [Google Scholar]
- Bredderman PJ, Foote RH, Yassen AM. An improved artificial vagina for collecting rabbit semen. J Reprod Fertil. 1964;7:401–403. doi: 10.1530/jrf.0.0070401. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A. Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortogero N, Hennig GW, Langille C, Ro S, McCarrey JR, Yan W. Computer-assisted annotation of murine Sertoli cell small RNA transcriptome. Biol Reprod. 2013;88:3. doi: 10.1095/biolreprod.112.102269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci U S A. 2011;108:13159–13164. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Kang L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics. 2011;27:771–776. doi: 10.1093/bioinformatics/btr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J. Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T. Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh C, Jenkins SL, Dannenfelser R, Ma'ayan A. Genes2WordCloud: a quick way to identify biological themes from gene lists and free text. Source Code Biol Med. 2011;6:15. doi: 10.1186/1751-0473-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Shirahige K, Ohsugi M, Nakai K. DBTMEE: a database of transcriptome in mouse early embryos. Nucleic Acids Res. 2015;43:D771–D776. doi: 10.1093/nar/gku1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthipan S, Selvaraju S, Somashekar L, Kolte AP, Arangasamy A, Ravindra JP. Spermatozoa input concentrations and RNA isolation methods on RNA yield and quality in bull (Bos taurus) Anal Biochem. 2015;482:32–39. doi: 10.1016/j.ab.2015.03.022. [DOI] [PubMed] [Google Scholar]
- Bissonnette N, Levesque-Sergerie JP, Thibault C, Boissonneault G. Spermatozoal transcriptome profiling for bull sperm motility: a potential tool to evaluate semen quality. Reproduction. 2009;138:65–80. doi: 10.1530/REP-08-0503. [DOI] [PubMed] [Google Scholar]
- Kawano M, Kawaji H, Grandjean V, Kiani J, Rassoulzadegan M. Novel small noncoding RNAs in mouse spermatozoa, zygotes and early embryos. PLoS One. 2012;7:e44542. doi: 10.1371/journal.pone.0044542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, Zhou Q, Chen Q, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Sendler E, Lalancette C, Hauser R, Diamond MP, Krawetz SA. Cleavage of rRNA ensures translational cessation in sperm at fertilization. Mol Hum Reprod. 2011;17:721–726. doi: 10.1093/molehr/gar054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert I, Bissonnette N, Boissonneault G, Vallee M, Robert C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction. 2007;133:1073–1086. doi: 10.1530/REP-06-0292. [DOI] [PubMed] [Google Scholar]
- Yang CC, Lin YS, Hsu CC, Wu SC, Lin EC, Cheng WT. Identification and sequencing of remnant messenger RNAs found in domestic swine (Sus scrofa) fresh ejaculated spermatozoa. Anim Reprod Sci. 2009;113:143–155. doi: 10.1016/j.anireprosci.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction. 2011;141:21–36. doi: 10.1530/REP-10-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Zeng P, Wang Z, Liu M, Xu W, Dai J, Zhao X, Zhang D, Liang D, Chen X, Shi S, Zhang M, et al. Estimated diversity of messenger RNAs in each murine spermatozoa and their potential function during early zygotic development. Biol Reprod. 2014;90:94. doi: 10.1095/biolreprod.114.117788. [DOI] [PubMed] [Google Scholar]
- Stowe HM, Calcatera SM, Dimmick MA, Andrae JG, Duckett SK, Pratt SL. The bull sperm microRNAome and the effect of fescue toxicosis on sperm microRNA expression. PLoS One. 2014;9:e113163. doi: 10.1371/journal.pone.0113163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y, Kang JY, Wang X, Li H, Hua MM, Zhao S, Hu SD, et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24:680–700. doi: 10.1038/cr.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SR, Juliano CE. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol Reprod Dev. 2013;80:632–664. doi: 10.1002/mrd.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikar O, Da Ros M, Korhonen H, Kotaja N. Chromatoid body and small RNAs in male germ cells. Reproduction. 2011;142:195–209. doi: 10.1530/REP-11-0057. [DOI] [PubMed] [Google Scholar]
- Ortogero N, Schuster AS, Oliver DK, Riordan CR, Hong AS, Hennig GW, Luong D, Bao J, Bhetwal BP, Ro S, McCarrey JR, Yan W. A novel class of somatic small RNAs similar to germ cell pachytene PIWI-interacting small RNAs. J Biol Chem. 2014;289:32824–32834. doi: 10.1074/jbc.M114.613232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Ma HY, Park C, Ortogero N, Song R, Hennig GW, Zheng H, Lin YM, Moro L, Hsieh JT, Yan W. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 2013;23:759–774. doi: 10.1038/cr.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez J, Alonso L, Cardenas DB, Artaza-Alvarez H. Hourcade Jde D, Martinez S, Brieno-Enriquez MA, Del Mazo J. Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA. 2015;21:946–962. doi: 10.1261/rna.048215.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlind M, Stalhammar H, Olsson B, Klinga-Levan K. Expression of miRNAs in bull spermatozoa correlates with fertility rates. Reprod Domest Anim. 2015;50:587–594. doi: 10.1111/rda.12531. [DOI] [PubMed] [Google Scholar]
- Du Y, Wang X, Wang B, Chen W, He R, Zhang L, Xing X, Su J, Wang Y, Zhang Y. Deep sequencing analysis of microRNAs in bovine sperm. Mol Reprod Dev. 2014;81:1042–1052. doi: 10.1002/mrd.22426. [DOI] [PubMed] [Google Scholar]
- Govindaraju A, Uzun A, Robertson L, Atli MO, Kaya A, Topper E, Crate EA, Padbury J, Perkins A, Memili E. Dynamics of microRNAs in bull spermatozoa. Reprod Biol Endocrinol. 2012;10:82. doi: 10.1186/1477-7827-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry E, Safranski TJ, Pratt SL. Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology. 2011;76:1532–1539. doi: 10.1016/j.theriogenology.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Hu YC, Okumura LM, Page DC. Gata4 is required for formation of the genital ridge in mice. PLoS Genet. 2013;9:e1003629. doi: 10.1371/journal.pgen.1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon N, Raiwet D, Viger RS, Silversides DW. Novel pre- and post-gastrulation expression of Gata4 within cells of the inner cell mass and migratory neural crest cells. Dev Dyn. 2008;237:1133–1143. doi: 10.1002/dvdy.21496. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XJ, Wang X, Ma X, Sun SC, Zhou X, Zhu C, Liu H. EZH2 is essential for development of mouse preimplantation embryos. Reprod Fertil Dev. 2014;26:1166–1175. doi: 10.1071/RD13169. [DOI] [PubMed] [Google Scholar]
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18:225–233. doi: 10.1038/ncb3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair G, Guibert S, Bender A, Weber M. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 2014;15:545. doi: 10.1186/s13059-014-0545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Olive V, Minella AC, He L. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci Signal. 2015 doi: 10.1126/scisignal.2005813. 8:re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megel C, Morelle G, Lalande S, Duchene AM, Small I, Marechal-Drouard L. Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci. 2015;16:1873–1893. doi: 10.3390/ijms16011873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani J, Rassoulzadegan M. A load of small RNAs in the sperm—how many bits of hereditary information? Cell Res. 2013;23:18–19. doi: 10.1038/cr.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A, Skinner MK, Yan W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet. 2016;2:1–10. doi: 10.1093/eep/dvw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21:368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Yan W. Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol Cell Endocrinol. 2014;398:24–30. doi: 10.1016/j.mce.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]