Abstract
Genetic studies show that Msx2 and Dlx5 homeodomain (HD) proteins support skeletal development, but null mutation of the closely related Dlx3 gene results in early embryonic lethality. Here we find that expression of Dlx3 in the mouse embryo is associated with new bone formation and regulation of osteoblast differentiation. Dlx3 is expressed in osteoblasts, and overexpression of Dlx3 in osteoprogenitor cells promotes, while specific knock-down of Dlx3 by RNA interference inhibits, induction of osteogenic markers. We characterized gene regulation by Dlx3 in relation to that of Msx2 and Dlx5 during osteoblast differentiation. Chromatin immunoprecipitation assays revealed a molecular switch in HD protein association with the bone-specific osteocalcin (OC) gene. The transcriptionally repressed OC gene was occupied by Msx2 in proliferating osteoblasts, while Dlx3, Dlx5, and Runx2 were recruited postproliferatively to initiate transcription. Dlx5 occupancy increased over Dlx3 in mature osteoblasts at the mineralization stage of differentiation, coincident with increased RNA polymerase II occupancy. Dlx3 protein-DNA interactions stimulated OC promoter activity, while Dlx3-Runx2 protein-protein interaction reduced Runx2-mediated transcription. Deletion analysis showed that the Dlx3 interacting domain of Runx2 is from amino acids 376 to 432, which also include the transcriptionally active subnuclear targeting sequence (376 to 432). Thus, we provide cellular and molecular evidence for Dlx3 in regulating osteoprogenitor cell differentiation and for both positive and negative regulation of gene transcription. We propose that multiple HD proteins in osteoblasts constitute a regulatory network that mediates development of the bone phenotype through the sequential association of distinct HD proteins with promoter regulatory elements.
Vertebrate development is orchestrated by hundreds of homeodomain (HD) proteins, which can be classified into subgroups based on their sequences and relationships of their homeobox motifs (7). The meshless (Msx) and distaless (Dlx) genes form two distinct but closely related subfamilies of homeobox genes that play essential roles during skeletal formation and in the development of the central nervous system (7, 14, 57, 76). Genetic, cellular, and biochemical evidence suggests that three Msx genes and at least six Dlx genes function during multiple phases of skeletal development, as exemplified by their expression patterns and actions during early, middle, and late stages of craniofacial, axial, and appendicular skeletal formation (7, 39, 42, 49, 57, 74). Initially, the differential expression patterns of Msx and Dlx genes confer spatial information on the mesenchyme of branchial arches and limbs. At later stages of embryonic development, HD proteins support the formation of more-defined skeletal structures, primarily by regulating epithelial-mesenchymal signaling.
Targeted gene disruption of Msx1 and especially Msx2 results in numerous developmental alterations that include defects in the calvarial bones of the skull, chondrogenic craniofacial bone abnormalities, defective skull ossification, and endochondral bone formation (42, 68). Dlx5 is involved in craniofacial development (1, 12) and limb initiation (20). Dlx5-deficient mice exhibit a mild delay in ossification of long bones, but there is no effect on expression of the Runx2 transcription factor, which is essential for osteogenesis (1). The double null of Dlx5/Dlx6 has a more severe phenotype, further supporting a role for these mammalian Dlx genes in specification of skeletal elements (15, 64). Dlx1 and Dlx2 pattern the dentition, and the null mice exhibit perinatal lethality and ectopic skull components (62, 81, 82). However, Dlx3 null mice die during early embryogenesis from placental failure; thus, a skeletal defect cannot be identified (53, 63).
Expression of Dlx3, -5, and -7 is bone morphogenetic protein 2 (BMP2) dependent in early gastrulation and during cellular differentiation of various phenotypes (44, 59, 69). Recent microarray analyses of osteogenic culture models have revealed that several HD proteins, including the Msx and Dlx families, are rapidly induced in response to BMP2-mediated osteoblast differentiation (4, 26, 27). Among the Dlx HD proteins identified in our studies, Dlx3 was induced by 1 h and peaked from 4 to 8 h after BMP2 treatment, coincident with the onset of commitment of C2C12 cells to the osteogenic lineage, as reflected by the induction of bone-related phenotypic genes beginning at 8 h (4). Although Dlx3 has been implicated in skeletal development, a direct role for Dlx3 in bone formation has not been identified. In humans, a 4-bp deletion in the Dlx3 gene is responsible for tricho-dento-osseous syndrome (60, 61, 85). In the mouse embryo, Dlx3 has been reported in multiple tissues, including the ectoplacental cone, the chorionic plate, placenta, branchial arches, and the developing hair follicle, as well as in differentiating ameloblasts, odontoblasts, and keratinocytes (53, 54, 63). Thus, we selected Dlx3 from our microarray to study its functional activity and contribution to osteoblastogenesis.
The expression profiles of Msx1, Msx2, and Dlx5 have been studied during chondrocyte and osteoblast differentiation, as have their regulatory roles in the transcription of bone-related genes (18, 21, 32, 40, 78, 84). Bone-related promoters, including osteocalcin (OC), osteopontin (OP), collagen type I, and bone sialoprotein (BSP), contain multiple HD binding motifs (6, 8, 17, 18, 28, 29, 32, 34, 65, 66, 75, 84). Msx2 represses transcription of OC (9, 32, 84) and collagen (18), while Dlx5 activates collagen I (78, 79). However, Dlx5 does not appear to activate other gene promoters, including OC (66) and BSP (5, 38, 75, 87). Yet, these genes are induced in response to forced expression of Dlx5 or osteogenic factors, like BMP2, which rapidly induces expression of these HD proteins. Other studies have shown that the HD regulatory sequences in the OC and collagen promoters play a critical role in retaining osteoblast-specific gene transcription (17, 28, 29, 31, 83, 86). Together, these findings raise compelling questions related to the involvement of different HD proteins in mechanisms mediating enhancer activity of tissue-specific genes essential for bone formation.
Transcriptional control of the prototypical bone-specific OC gene has been well characterized and serves as a model for examining regulation by HD proteins. The OC gene undergoes extensive chromatin remodeling to accommodate the transcriptional regulators that control its expression during osteoblast growth and differentiation (51, 52, 70, 71). The gene is suppressed in nonosseous and proliferating osteoprogenitor cells and then transcriptionally activated by Runx2/Cbfa1 in postproliferative osteoblasts. OC expression is further induced by C/EBP and the hormone 1,25(OH)2D3 to maximal levels of expression during the mineralization stage or earlier (25). A cis regulatory sequence conserved in the segment of the proximal promoter of all species, known as the OC box (−76 to −99 in rat), encompasses a core HD protein binding site (CAATTAGT) that restricts OC expression to osteoblasts (31, 32). Thus, the OC gene serves as a marker for determining in vivo mechanisms by which HD proteins regulate OC transcription during progression of osteoblast differentiation.
Here, we have studied the regulatory role of Dlx3 in bone-specific transcriptional control during development of the osteoblast phenotype, as well as its functional relationship to the Msx2 and Dlx5 proteins. Our findings show, for the first time, expression of Dlx3 in the skeleton, primarily in early-stage osteogenic lineage cells. Both overexpression and knock-down of Dlx3 support a function in promoting osteoblast differentiation. Chromatin immunoprecipitation (ChIP) studies demonstrate in vivo temporal recruitment of Dlx3 in relation to the Msx2 and Dlx5 HD proteins to the OC gene during osteoblast differentiation. We observe two molecular switches in HD protein-DNA interactions. Msx2 associates with OC chromatin when the gene is repressed, while Dlx3 and Dlx5 are recruited with Runx2, the tissue-specific activator of bone formation. A second switch coincides with the mineralization stage of osteoblast differentiation, when Dlx3 association decreases and Dlx5 recruitment increases. The temporal occupancy of Dlx3 followed by Dlx5 in the OC promoter correlates with increased transcription represented by the increased occupancy of RNA polymerase II (Pol II). Our studies demonstrate a role of Dlx3 in promoting osteogenic differentiation and a temporal recruitment of HD proteins to chromatin for regulation of osteogenic genes during development of the osteoblast phenotype.
(Components of this study were performed by Jeremy Karlin in partial fulfillment of the requirements for a B.S. degree from Worcester Polytechnical Institute, Worcester, Mass.)
MATERIALS AND METHODS
In situ hybridization.
Sagittal paraffin sections of mouse embryos were subjected to in situ radioactive hybridization as described by Mackem and Mahon (47). RNA probes corresponding to the sense and antisense strands of mouse Dlx3 partial cDNA (from 199 bp to TGA plus untranslated region) were prepared using T7 and T3 RNA polymerases and 33P-labeled UTP (Amersham Biosciences, Piscataway, N.J.).
Cell cultures.
Rat osteosarcoma (ROS 17/2.8) cells were cultured and maintained in F-12 medium (Gibco-BRL, Grand Island, N.Y.) supplemented with 5% fetal bovine serum (FBS).
Primary rat osteoblast (ROB) cells were isolated from calvaria of fetal rats at day 21 of gestation by three sequential digestions with collagenase P (2 mg/ml; Boehringer Mannheim, Indianapolis, Ind.) at 37°C and 0.25% trypsin (Gibco-BRL) treatment as detailed previously (2, 56). Cells from the third digestion were plated at a density of 4 × 105 cells/100-mm dish and fed every second day with minimal essential medium (MEM; Gibco-BRL) supplemented with 10% FBS, 50 μg of ascorbic acid/ml, and 10 mM β-glycerol phosphate to induce differentiation at confluency.
The mouse MC3T3-E1 osteoblastic cell line was maintained in α-MEM supplemented with 10% FBS and transfected at low density (50% confluency) using Fugene 6 (Roche Diagnostics Corp., Indianapolis, Ind.) according to the manufacturer's procedure with the indicated plasmids. After washing of reagents, cells were induced towards the osteogenic phenotype in medium containing 25 μg of ascorbic acid/ml and 10 mM β-glycerophosphate.
Antibodies.
The affinity-purified polyclonal antibody was raised against a 16-amino-acid synthetic peptide (amino acids 242 to 256) of the murine Dlx3 protein as previously described (10). Mouse monoclonal Msx(1 + 2) antibody (4G1) against bacterially expressed gallus Msx2 protein was obtained from Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa. Affinity-purified polyclonal Dlx5 antibodies (Y20 and C20) were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). Both antibodies were raised against a peptide mapping within an internal region (Y20) or the C terminus (C20) of human DLX5. Because the Msx and Dlx families share many common sequences in addition to their DNA binding domains, the specificities of the different antisera obtained from commercial sources used in these studies were verified with in vitro-transcribed and -translated HD proteins. The antibody to Dlx3 was previously reported to exhibit specificity (10). The Msx(1 + 2) antisera showed no cross-reactivity in Western blot studies and electrophoretic mobility shift assays (EMSAs) with Dlx5 or Dlx3. The Dlx5 (Y20) antibody showed no cross-reactivity with Msx2 or Dlx3. These findings are available at the website http://labs.umassmed.edu/steinlab/MCB2004.
Nuclear extracts, oligonucleotides, probes, and EMSA.
Nuclear extracts were prepared from 106 ROS 17/2.8 cells or day 4, 12, or 20 primary rat osteoblasts according to the Dignam method (16). Aliquots of supernatant enriched with nuclear proteins were quick-frozen in a dry ice-ethanol bath and stored at −80°C for use in Western blot analysis and gel mobility shift assays.
Oligonucleotides were synthesized representing the OC box wild type (OC-24) and mutants (mTT and mCC1) of the rat OC promoter sequence. The plus strand (10 pmol) was labeled with [γ-32P]ATP for 1 h at 37°C with T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.). Annealing with minus strand was performed by addition of a threefold excess amount (30 pmol) followed by boiling for 5 min and slow cooling to room temperature. The unincorporated nucleotides were removed using a quick-spin G-25 Sephadex column (Roche Molecular Biochemicals, Indianapolis, Ind.).
EMSA reaction mixtures were prepared using 10 fmol of radiolabeled probe and 2.5 to 5 μg of nuclear extract according to the procedure developed for optimal HD protein binding (30). Briefly, the DNA-protein binding reactions were carried out at room temperature for 10 min. Protein-DNA complexes were separated on a 6.5% (40:0.5) nondenaturing polyacrylamide-0.5× Tris-borate-EDTA gel. For antibody immunoshift analysis, approximately 100 to 200 ng of antibody against Msx(1 + 2), Dlx3, or Dlx5 was incubated with nuclear extract at 22°C for 0.5 h prior to the probe addition. The samples were electrophoresed at 200 V for 3 h. After running, the gels were dried and autoradiographed at −70°C or room temperature according to the signal intensity.
Expression plasmids and promoter regulation studies.
Rat osteosarcoma (ROS 17/2.8) cells were plated at a density of 0.5 × 106 per ml, 24 h prior to transfection. Cells were transfected at 60 to 70% confluency using ExGen 500 transfection reagent (MBI Fermentas, Hanover, Md.). One microgram of OC promoter construct (−208 CAT) or empty vector (PGEM CAT), 0.2 to 0.5 μg of Msx2, Dlx5, or Dlx3 expression vector, and 100 ng of Renilla luciferase plasmid were added to 50 μl of 150 mM NaCl. ExGen 500 (3.3 μl per μg of DNA) was added to 50 μl of 150 mM NaCl, vortexed immediately, and then mixed with DNA-NaCl solution and kept at room temperature for 10 min. After addition of 0.9 ml of complete medium, the mixture was applied to cells already washed once with phosphate-buffered saline (PBS). Plates were incubated at 37°C for 2 to 3 h, followed by a PBS wash and change to complete medium. Cells were harvested 24 h posttransfection. For chloramphenicol acetyltransferase (CAT) reporter assays, cells were lysed with 300 μl of reporter lysis buffer (Promega, Madison, Wis.) for 20 to 30 min at room temperature. Cell lysate (20 μl) was incubated with reaction mixture containing acetyl coenzyme A and radiolabeled chloramphenicol for 2 h at 37°C. The products were separated by thin-layer chromatography, and the amount of [14C]chloramphenicol incorporated was quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
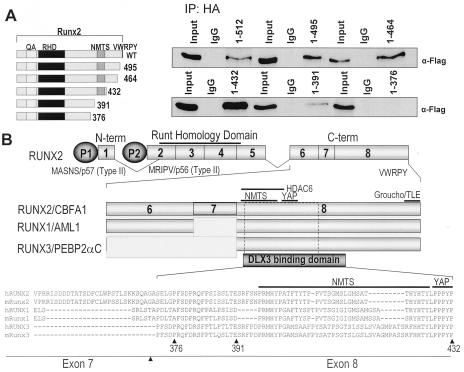
To study the Dlx3-interacting domain present in Runx2, different deletion mutants from the C terminus of Runx2 were used in the same transfection protocol (described above) using ROS 17/2.8 cells. Flag-tagged Dlx3 (5 μg) was cotransfected with 5 μg of hemagglutinin (HA)-tagged wild-type and different deletions of Runx2 (Δ495, Δ464, Δ432, Δ391, and Δ376) as described earlier (25, 73). Coimmunoprecipitations were performed using anti-HA polyclonal antibody (Santa Cruz Biotechnology) 24 h posttransfection. After four washings with 1× PBS, the immunocomplexes were separated in a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and immunoblotted with anti-Flag mouse monoclonal antibody (Sigma Aldrich, St. Louis, Mo.). The pull-down efficiency was also confirmed by Western blotting with rabbit polyclonal HA antibody (Santa Cruz Biotechnology).
Probes and Northern blot analysis.
The following cDNA probes were used to study the expression of HD proteins during osteoblast growth and differentiation. The BamHI-XhoI fragments corresponding to the cDNA sequence of Msx2 (807 bp), Dlx3 (864 bp), and Dlx5 (870 bp) were random-primed labeled and used for hybridization. We note that two Msx2 transcripts with an identical expression profile were observed (32); only the top one is shown in this study. The EcoRI-HindIII fragment for OC and the EcoRI fragment for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as probes for Northern blot analysis. Total cellular RNA was extracted with TRIzol reagent (Gibco-BRL) according to the manufacturer's instructions. Primary rat osteoblast cells were harvested at different days and resuspended in TRIzol solution (1 ml of TRIzol per 100 μl of pellet) to extract RNA by established procedures (Invitrogen Life Technologies, Carlsbad, Calif.). Total RNA (10 to 15 μg) was separated on a 1% formaldehyde agarose gel, transferred to nylon membrane (Amersham Biosciences, Piscataway, N.J.), and hybridized with [α-32P]dCTP-labeled Msx2, Dlx5, Dlx3, OC, and GAPDH probes as previously described (3). Blots were subjected to autoradiographic exposure overnight at −70°C.
RT-QPCR.
The DNase I-treated total cellular RNA was reverse transcribed using Invitrogen's Superscript first-strand synthesis system. A negative control was created in the absence of the reverse transcriptase enzyme. All cDNA sequences specific for bone phenotypic markers were analyzed using Primer Express software to predict optimum reverse transcription-PCR (RT-PCR) primer sets (Table 1), except for GAPDH primers, which were purchased from Applied Biosystems. The quantitative PCRs (QPCRs) were carried out in 50-μl volumes on 96-well plates using Applied Biosystem's ABI Prism 7000 sequence detection system and software according to Applied Biosystem's recommended protocols for either the Sybr Green dye detection method (mastermix purchased from Eurogentec) or the TaqMan 5′-nuclease probe method (mastermix and probes purchased from Applied Biosystems). The two-step PCRs were set up with a melting temperature of 95°C and an annealing-elongation temperature of 60°C for 40 cycles. Each set of PCRs for each gene was performed in duplicate simultaneously on the same plate from the same cDNA. Linear, three-point standard curves were also established in duplicate for each set of gene primers by using the threshold cycle, the point at which each set of reactions reached the logarithmic portion of the PCR curve. The 60°C dissociation protocol option was selected for reactions with the Sybr Green reagent. All transcript levels were normalized to that of GAPDH.
TABLE 1.
Primers for real-time PCR assays
| Genea | Species | Amplicon (bp) | Oligonucleotide |
|---|---|---|---|
| Collagen I* | Mouse | 51 | Forward primer: 5′-GTA TCT GCC ACA ATG GCA CG-3′ |
| Reverse primer: 5′-CTT CAT TGC ATT GCA CGT CAT-3′ | |||
| BSP* | Mouse | 51 | Forward primer: 5′-GCA CTC CAA CTG CCC AAG A-3′ |
| Reverse primer: 5′-TTT TGG AGC CCT GCT TTC TG-3′ | |||
| Osteopontin* | Mouse | 51 | Forward primer: 5′-TTT GCT TTT GCC TGT TTG C-3′ |
| Reverse primer: 5′-CAG TCA CTT TCA CCG GGA GG-3′ | |||
| AP | Mouse | 75 | Forward primer: 5′-TTG TGC CAG AGA AAG AGA GAG A-3′ |
| Reverse primer: 5′-GTT TCA GGG CAT TTT TCA AGG T-3′ | |||
| Taqman probe: 5′-TAC TGG CGA CAG CAA G-3′ | |||
| OC | Mouse | 59 | Forward primer: 5′-CTG ACA AAG CCT TCA TGT CCA A-3′ |
| Reverse primer: 5′-GCG GGC GAG TCT GTT CAC TA-3′ | |||
| Taqman probe: 5′-AGG AGG GCA ATA AG-3′ |
*, primer used for Sybr Green dye detection.
Western blot analysis.
Whole-cell lysates were prepared as previously described (25). ROB nuclear extract (20 μg) was added to SDS-polyacrylamide gel electrophoresis sample buffer, boiled for 10 min, and separated on SDS-10% polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes by using a semidry transblot apparatus for 30 min at 10 V. Blots were blocked with 5% nonfat milk in PBS-0.1% Tween 20 (PBST) buffer (1× PBS is 8.1 mM Na2HPO4, 1.9 mM NaH2PO4, 0.137 M NaCl, and 2.7 mM KCl [pH 7.4]) for 1 h at room temperature. Blots were probed with primary antibodies at a dilution of 1:2,000 against Msx(1 + 2), Dlx3, Dlx5, Runx2, CDK2, and α-actin in PBST containing 2% milk at room temperature for 1 to 2 h or overnight at 4°C. Bound specific antibody was detected using a secondary anti-rabbit or anti-mouse antibody coupled to horseradish peroxidase in a 1:5,000 dilution. The immunoreactive bands were visualized by autoradiography with chemiluminescence substrate (Perkin-Elmer Life Sciences, Boston, Mass.).
Coimmunoprecipitation.
Both untransfected and Msx2- and Dlx3-transfected ROS 17/2.8 cells, as well as untransfected ROB cells at three stages of differentiation, were used in these studies. Approximately 107 cells/immunoprecipitation were lysed in 800 μl of Nonidet P-40 (NP-40) lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40, 1× Complete protease inhibitor [Roche Molecular Biochemicals], 25 μM MG132 [Sigma Aldrich]) for 15 min at 4°C, followed by centrifugation at 16,000 × g for 15 min. The supernatant was transferred to a clean microcentrifuge tube and precleared with 40 μl of protein A/G plus agarose beads (Santa Cruz Biotechnology Inc.) at 4°C for 30 min. The beads were collected by centrifugation at 1,000 × g for 5 min at 4°C. Approximately 100 μg of nuclear extract from different rat osteoblast time points as indicated was also added to a final volume of 800 μl in lysis buffer and precleared as mentioned above. Msx(1 + 2), Dlx3 antibody, and normal immunoglobulin G (IgG) (3 μg each) were added to the precleared lysates following incubation at 4°C for 2 h. To precipitate immunocomplexes, 50 μl of protein A/G plus agarose beads was added and further incubated at 4°C with agitation for 1 h. Beads were washed three times with 1× PBS containing 1× protease inhibitors and 25 μM MG132, suspended in 20 to 30 μl of 2× SDS sample buffer, and analyzed by Western blotting.
RNA interference (RNAi) of Dlx3.
The mouse MC3T3-E1 osteoblastic cells at 30 to 50% confluency were transfected using Oligofectamine (Invitrogen Life Technologies) with small interfering RNA (siRNA) duplexes specific for murine Dlx3 obtained from QIAGEN Inc. (Stanford, Calif.) at different concentrations (50, 100, and 200 nM). The siRNA duplexes were r(CCC UGU GUU GCA AGU CGA A) dTdT and r(UUC GAC UUG CAA CAC AGG G) dAdG. The cells were also transfected with control siRNA duplexes specific for green fluorescent protein (GFP) using the same concentrations to check the transfection efficiency, or as a nonspecific control. Opti-MEM 1 (a reduced serum medium from Invitrogen) was used to dilute the siRNA duplexes and Oligofectamine and for transfection. After treating the cells with siRNA for 4 h, the cells were supplemented with α-MEM containing 30% FBS for a final concentration of 10% in the medium. The siRNA experiment was carried out for 72 h, at which time the cells were harvested for total protein and RNA to analyze the knock-down effect of Dlx3 siRNA on endogenous Dlx3 and its knock-down effect on other osteoblast-specific markers by real-time QPCR.
ChIP assays.
To cross-link protein with DNA, ROB cells were incubated for 10 min at room temperature in 1× PBS (3 ml/plate) containing 1% formaldehyde, 25 μM MG132 (Calbiochem/Sigma), and 1× protease inhibitor (Roche Molecular Biochemicals). A final concentration of 0.125 M glycine was added to the 1% formaldehyde-PBS solution for neutralization. Cells were collected in PBS after plates were washed twice with ice-cold PBS. The harvested cells were lysed in a lysis buffer containing 25 mM HEPES (pH 7.8), 1.5 mM MgCl2, 10 mM KCl, 0.1% NP-40, 1 mM dithiothreitol, 25 μM MG132, and 1× Complete protease inhibitor. To isolate the nuclei, cells were homogenized for 20 strokes in a Dounce homogenizer followed by centrifugation at 200 × g at 4°C. The nuclei pellet was resuspended in 300 μl (300 μl/100-mm plate) of sonication buffer (50 μM HEPES [pH 7.9], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 25 μM MG132, 1× Complete protease inhibitor). Samples were sonicated to reduce the DNA length to 0.2 to 0.6 kb. Cellular debris was removed by centrifugation at 14,000 rpm for 15 min at 4°C, and chromatin solutions were distributed into multiple 1-ml aliquots that were used as the starting material for all subsequent steps.
Chromatin aliquots were precleared with 100 μl of a 25% (vol/vol) suspension of 2 μg of single-stranded DNA-coated protein A/G and 1 mg of bovine serum albumin/ml. Samples were used directly for immunoprecipitation reactions with 2 μg of Msx(1 + 2), Dlx3, Dlx5, Pol II (Covance Inc.), or Runx2 (M-70; Santa Cruz Biotechnology) antibody and normal rabbit or mouse IgG as a control. ChIP reactions were allowed to proceed for 2 to 4 h at 4°C on a rotating wheel. Immune complexes were mixed with 100 μl of a 25% (vol/vol) precoated protein A/G agarose suspension followed by incubation for 1 h at 4°C on a rotating wheel. Beads were washed three times with low-salt, high-salt, and lithium salt buffers (70, 71). After a final wash with Tris-EDTA buffer, the beads were collected by brief centrifugation and the immunocomplexes were eluted twice by adding 150 μl of a freshly prepared solution of 1% SDS-0.1 M NaHCO3. The samples were adjusted to 0.2 M NaCl, and protein-DNA cross-linking was reversed by incubating at 68°C overnight. The samples were treated with 100 μg of proteinase K/ml followed by phenol-chloroform extraction and ethanol precipitation using 5 μg of glycogen as carrier. An aliquot (2 to 3 μl) of each sample was assayed for PCR to detect the presence of specific DNA fragments using appropriate oligos from the proximal OC promoter spanning bp −198 to −28. This region contains the OC box, C/EBP sites, and Runx2 site C. The primers were as follows: forward, 5′-GGC AGC CTC TGA TTG TGT CC-3′ (−198 to −179); reverse, 5′-TAT ATC CAC TGC CTG AGC GG-3′ (−47 to −28). PCR conditions were 28 cycles of 95°C for 60 s, 94°C for 50 s, 57°C for 50 s, and 68°C for 60 s, and then 68°C for 7 min.
RESULTS
Dlx3 is expressed in osteogenic lineage cells at sites of new bone formation.
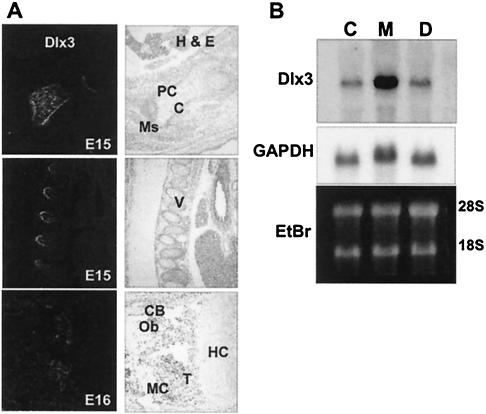
Studies in the mouse suggested Dlx3 is involved in craniofacial development (63), but Dlx3 had not been examined in cells of bone tissues or during osteoblast differentiation. In situ hybridization of whole embryo (embryonic day 15 [E15]) sagittal sections showed Dlx3 is expressed in perichondrium and mature chondrocytes of the developing limb (Fig. 1A, top panels) and vertebrae (Fig. 1A, middle panels), but not in the condensing mesenchyme. In more mature limbs (E16), Dlx3 expression was observed in periosteal cells, in cells surrounding the primary spongiosa trabeculae in the metaphysis (Fig. 1A, lower panel), and in endosteal osteoblasts of diaphyseal cortical bone. Dlx3 expression was not detected in the prehypertrophic and hypertrophic zones of the growth plate (Fig. 1A, lower panels). The predominant Dlx3 expression in the metaphysis and its absence in marrow were further confirmed by Northern blot analyses of postnatal bone (Fig. 1B). Trabecular bone expressed Dlx3 up to 10-fold more than intramembranous calvarial bone. Our results demonstrate for the first time that Dlx3 expression occurs in cells of the osteogenic lineage, in the putative osteoprogenitors that reside in the perichondrium-periosteum, as well as in active osteoblasts on the bone surfaces. These findings suggest Dlx3 may be related to osteoblast differentiation, formation of bone tissue, and regulation of phenotypic genes.
FIG. 1.
Dlx3 is expressed in osteogenic tissue in vivo. (A) In situ hybridization of mouse embryo sections. Left panels: paraffin sections were hybridized with a Dlx3-labeled probe (see Materials and Methods). Right panels: Dlx3-positive skeletal and cellular detail structures are identified by hematoxylin and eosin staining of the same section shown in the left panel. Top: developing limb bone in 15 day postcoitum embryos with Dlx3 expression in perichondrium (PC) and chondroblasts (C) and absence in the condensing mesenchyme (Ms). Middle: developing vertebral bodies (V) show Dlx3 in the bony trabeculae and surrounding periosteum and absence in the mesenchyme portions of the developing ribs. Lower panel: mature limb at E16, demonstrating Dlx3 in osteoblasts (Ob) associated with cortical bone (CB) and trabeculae (T) in the metaphysis. Note the lack of Dlx3 expression in marrow cavity (MC) and the hypertrophic cartilage zone of the growth plate (HC). (B) Expression of Dlx3 in mouse bone tissue, based on Northern analysis. Calvaria, metaphysis, and diaphysis of femurs from a 7-month-old mouse were dissected and washed free of adherent tissue and marrow prior to preparation of total cellular RNA. Total cellular RNA (10 μg per lane) was loaded on a 1% formaldehyde agarose gel and hybridized with Dlx3 probe (left panel). Ethidium bromide staining of the 28S and 18S rRNA bands showed equivalent loading.
Dlx3 is developmentally expressed during osteoblast growth and differentiation and promotes development of the osteoblast phenotype.
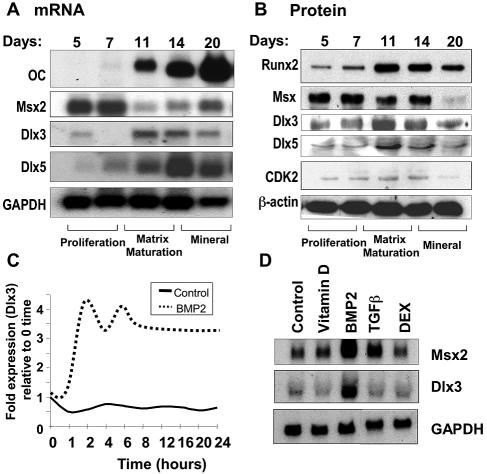
To address the physiological significance of Dlx3 expression during the early stage of osteoblast differentiation in vivo, we initially examined the cellular representation of Dlx3 relative to other HD proteins during the ex vivo maturation of primary rat calvaria-derived osteoblasts, a well-characterized model that produces a bone-like mineralized extracellular matrix (2, 56). We determined the levels for each of the HD proteins in relation to the expression of cell growth and phenotypic markers for the stages of osteoblast differentiation by both Northern and Western blot analyses (Fig. 2). Msx2 mRNA and protein were maximal in proliferating cells (days 5 to 7), although Msx2 protein remained detectable during formation of the multilayered bone nodules (matrix maturation stage). In contrast, Dlx3 mRNA and protein were expressed at the highest levels from days 11 to 14, when the mature osteoblast was induced throughout the culture, as reflected by OC expression (day 11). Dlx5 mRNA peaked later than Dlx3 (day 14); its expression was consistently sustained more robustly than that of Dlx3 in the most-differentiated mineralization stage in independent time courses. Both Dlx3 and Dlx5 protein levels exhibited an overlap in expression in relation to increased osteoblast differentiation, reflected by increased OC and Runx2 upregulation. The transient upregulation of Dlx3 during rat osteoblast differentiation was similar to its temporal expression during induction of the osteoblast phenotype in BMP2-treated premyogenic C2C12 cells (Fig. 2C [plotted from microarray data presented in reference 4]).
FIG. 2.
Temporal expression of Dlx3 during osteoblast growth and differentiation in relation to that of the Msx2 and Dlx5 HD proteins. Cell layers from the culture of calvaria-derived primary rat osteoblasts were harvested at the indicated days for either total cellular RNA by Northern blot analysis (A) or nuclear proteins for Western blot analysis (B). (A) Total cellular RNA as indicated was separated on a 1% formaldehyde-agarose gel, blotted, and hybridized with corresponding cDNA probes of HD proteins (described in Materials and Methods). OC mRNA is shown relative to the HD proteins as a marker of osteoblast maturation, while GAPDH mRNA levels served as a loading control. (B) Runx2 is shown as a marker of osteoblast differentiation. β-Actin is shown as a control for protein loading. (C) Dlx3 expression in BMP2-treated (300 ng/ml) C2C12 cells, plotted from the expression levels reported by gene microarray profiling (4). We normalized Dlx3 expression to GAPDH and plotted BMP2-treated and untreated control cell values as relative expression from zero to time points up to 24 h. (D) Dlx3 responsiveness to BMP2 compared to that of other osteogenic factors in primary calvaria-derived ROB cells. Northern blot analysis results of total cellular RNA from osteoblasts that were treated with 1,25(OH)2D3 (10 nM), TGF-β1 (250 ng/ml), BMP2 (100 ng/ml), and dexamethasone (10 nM) at day 11 of culture and harvested 24 h posttreatment are shown.
We next examined Dlx3 regulation by BMP2 in primary osteoblasts and compared it to other factors that modulate osteoblast differentiation and phenotypic genes in primary ROB cells (day 11) (Fig. 2D). After 24 h of treatment with BMP2, a 10-fold induction of Dlx3 was observed, while other mediators of skeletal development (transforming growth factor β1 [TGF-β1]) or regulators of osteoblast genes (vitamin D and the glucocorticoid dexamethasone) had little or no significant effect on Dlx3 expression. Msx2 was induced by both BMP2 and TGF-β1. This finding supports the hypothesis that Dlx3 is an early response gene of BMP2-induced bone formation. Taken together, these findings of a temporal profile of Msx2, Dlx3, and Dlx5 expression suggest a coordination of HD protein functional activities in regulating expression of genes involved in BMP2-mediated osteoblast differentiation.
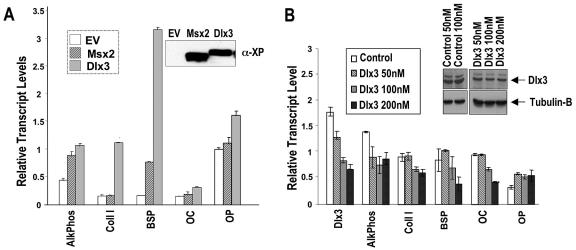
To directly address a functional role for Dlx3 in osteogenic differentiation, we determined the consequences of upregulated expression of Dlx3 compared to that of Msx2 (18) on osteoblast markers. We selected the mouse preosteoblastic MC3T3-E1 cell line, which does not require exogenous BMP2 for differentiation, for these studies. Cells were transiently transfected with Dlx3 or Msx2 and then allowed to differentiate. The data in Fig. 3 demonstrate that Dlx3 upregulated endogenous expression of all osteoblast marker genes, while Msx2 affected only three phenotypic genes. The Western blot analysis showed comparable Msx2 and Dlx3 expression levels (Fig. 3, insert). Notably, collagen type I and BSP were robustly induced only by Dlx3 (10- and 20-fold, respectively), while alkaline phosphatase (AP), OP, and OC showed a 1.5- to 2.2-fold response to Dlx3. Msx2 stimulated AP 1.5-fold, but BSP was stimulated 4.5-fold.
FIG. 3.
Osteogenic regulation and functional activities of Dlx3 compared to Msx2 on osteoblast gene expression. (A) Effects of overexpression of Msx2 and Dlx3 compared to empty vector (EV) on endogenous osteogenic genes in MC3T3-E1 cells are shown. Cells were transfected with 5 μg of cytomegalovirus promoter-driven Msx2 or Dlx3 expression plasmid per 100-mm plate and the HD proteins were detected by antibody to the Xpress tag. Levels of expression were similar, as shown in the insert Western blot. Phenotypic gene expression was determined 48 h posttransfection by real-time PCR analysis, using either Sybr Green dye (Eurogentec) or Taqman probes (Applied Biosystems). Primers and probes used in QPCR to analyze phenotypic genes are shown in Table 1. A typical experiment is shown with error bars that represent triplicate samples for real-time PCR. (B) RNAi of Dlx3 in MC3T3-E1 cells. After 72 h of treatment with either Dlx3-specific or control GFP-specific siRNA duplexes at the indicated doses, QPCR was performed on the RNA prepared from the harvested cells as described for panel A.
To further document the functional role of endogenous Dlx3 in promoting osteoblast differentiation and expression of phenotypic genes, we treated MC3T3-E1 cells with siRNA specific for Dlx3. The siRNA duplex inhibited Dlx3 expression in a dose-dependent manner to a maximum of approximately 55 to 60% at 100 nM, compared to the control levels of Dlx3 in cells treated with the same concentration of GFP RNAi (Fig. 3B). Complementary to the Dlx3 overexpression studies (Fig. 3A), we found inhibition of AP (by 30%), BSP (by 50%), and OC (by 50%). Collagen type I was decreased by 20%, and OP did not exhibit a change. Notably, the bone phenotypic genes that reflect osteoblast maturation, BSP and OC, were decreased in the same range as inhibition of Dlx3 expression. These results from both overexpression and RNAi suggest that Dlx3 is a mediator of osteogenic differentiation.
Dlx3 DNA binding activity reflects cellular protein levels and regulation of the OC gene.
Several osteoblast genes (collagen type I, BSP, OP, and OC) contain functional HD regulatory elements, and their expression was increased by Dlx3 (Fig. 3). However, the findings of these studies could not differentiate between a primary transcriptional response or an increase in osteogenic gene expression secondary to a more differentiated stage promoted by Dlx3 and/or the collagen matrix. The OC gene provides an example of a bone-specific marker that contains a conserved 24-nucleotide OC box sequence in the proximal promoter with a classic HD binding site as its core motif (Fig. 4). We therefore evaluated activity of Dlx3 for DNA-protein interactions at this regulatory motif by using nuclear extracts from osteoblasts at different stages of maturation. Distinct HD protein interactions were formed at the OC box as a function of osteoblast differentiation (Fig. 4A). The protein-DNA complexes from previously reported ROS17/2.8 osteosarcoma cells (lane 1) were compared to those from primary rat osteoblasts from the proliferation stage (day 4), the period of cellular multilayering and nodule formation (day 12), and the mineralization stage (day 20), when OC is expressed at peak levels (Fig. 4B). During ROB differentiation, four major HD complexes (competed by the homeobox consensus sequences) (Fig. 4C) were formed. The most prominent HD complex present in ROS 17/2.8 cells was barely detected in day 4 and day 12 ROB nuclear extracts, but it was present at significant levels in day 20 extracts (Fig. 4B). This differentiation-related HD complex correlated to high OC mRNA levels in both ROS and mature ROB cells.
FIG. 4.
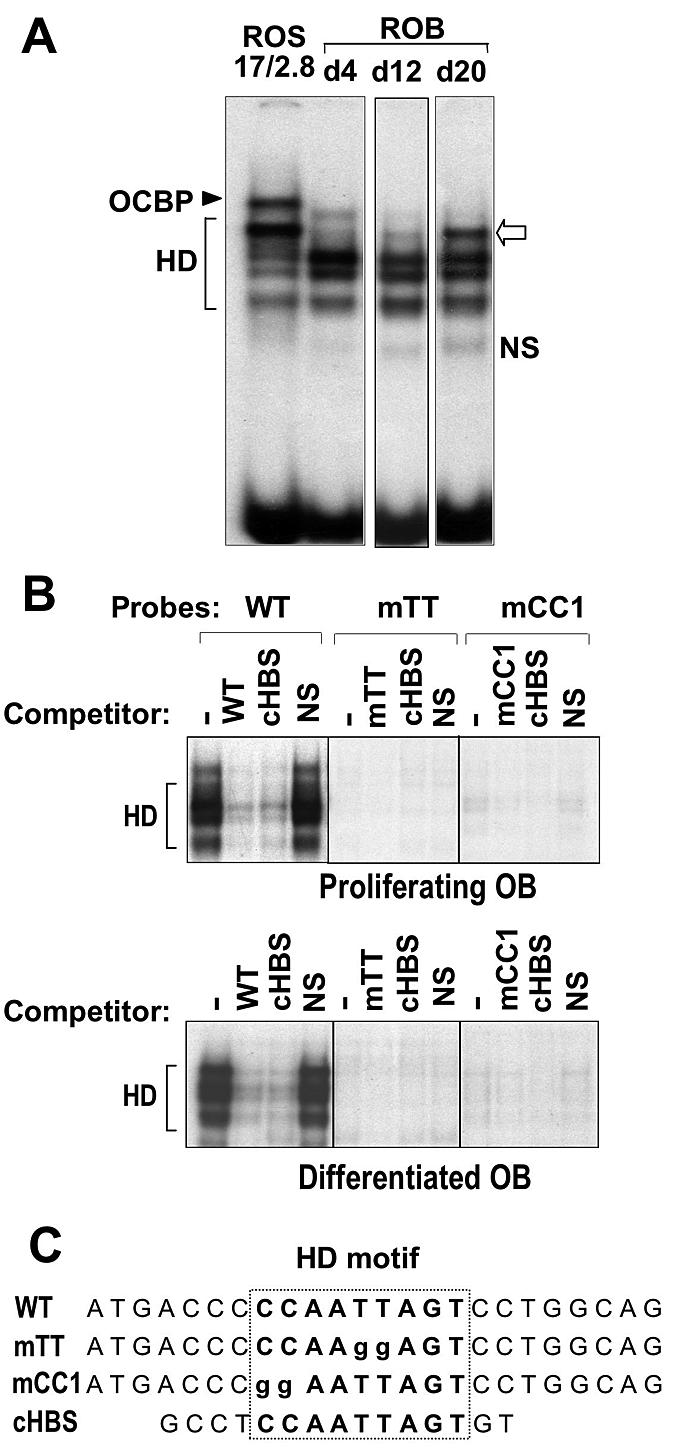
HD protein complexes formed at the OC box contribute to OC gene transcription. (A) Nuclear extracts from confluent ROS 17/2.8 cells (which constitutively express OC) (lane 1) and ROB cells were prepared at the indicated days (lanes marked d4, d12, and d20). The wild-type OC box 24-nucleotide oligonucleotide was 5′-end labeled with [γ-32P]ATP, and 10 fmol of the probe was incubated with 5 μg of nuclear extract. The protein-DNA complexes were separated under conditions which optimized for HD protein binding as described in Materials and Methods. Open arrow, the most prominent HD complex present in ROS 17/2.8 cells, barely detected in day 4 and day 12 ROB nuclear extracts but present at significant levels in day 20 extract. (B) Binding and competition studies using nuclear extracts from proliferating or differentiating primary rat osteoblasts. The labeled probes used in this study are described in Table 1. Specificity of the HD complexes can be seen by complete competition with wild-type oligo and the HD consensus sequence (cHBS). The mTT and mCC1 mutation inhibited the binding of all HD-related proteins, and neither probe formed a non-HD binding complex. (C) Sequences of the probes used in these studies.
The OC box binding protein (30, 31), a previously characterized non-HD protein complex in ROS 17/2.8 cells, was not observed in nuclear extracts from the normal ROB cells at any stage (Fig. 4A). Figure 4C further confirms this observation from binding studies using wild-type, mTT, and mCC1 oligonucleotides as probes with ROB cell nuclear extracts. The OC box binding protein complex, which should bind to the mCC1 probe (30, 31), was not formed from nuclear extracts of normal osteoblasts during the growth and differentiation stages. Rather, all complexes were HD protein related (Fig. 4B, competitor cHBS lane).
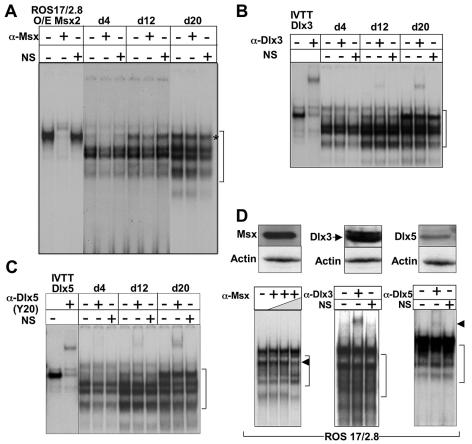
Antibody supershift assays were then used both to identify HD proteins in the complexes in primary rat calvarial osteoblast cells and to assess quantitative changes in HD proteins associated with complexes formed during stages of osteoblast differentiation (Fig. 5). The Msx1/2 antibody resulted in a block shift (Fig. 5A) in nuclear extracts from day 4 and day 12 samples, but no change was observed in the mature osteoblast nuclear extracts (day 20) (Fig. 5A), consistent with cellular protein levels. Addition of the Dlx3 antibody did not lead to a change in any of the bands in the day 4 nuclear extracts (Fig. 5B). In contrast, extracts from differentiated cells (day 12) showed a supershifted complex in the lanes containing Dlx3 antibody in day 12 and day 20 nuclear extracts. Although peak Dlx3 protein occurred on day 12, the binding of Msx to the OC probe on day 12 may compete with Dlx3 protein binding. The Dlx5 (Y20) antibody also resulted in an increasing supershift throughout the course of rat osteoblast differentiation (Fig. 5C) that reflected the cellular protein levels shown in Fig. 2B. Thus, both Dlx3 and Dlx5 have binding activity for the HD element in the postproliferative osteoblasts. The finding of Msx2 binding activity predominantly in proliferating cells, and Dlx3 and Dlx5 in differentiated cells, suggests a switch in HD-mediated protein binding and a change from a repressor protein-DNA (Msx2) complex to an activating complex containing either Dlx3 or Dlx5 as differentiation progresses. The OC gene was constitutively expressed in ROS 17/2.8 cells which abundantly expressed Dlx3 (Fig. 5D). In these nuclear extracts, a prominent supershift with the Dlx3 antibody demonstrated that Dlx3 contributes more to formation of the complex than Dlx5 or Msx proteins. In conclusion, these findings indicate selectivity for the binding of each of these HD proteins to the OC gene for regulation of its transcription between the proliferation and differentiation stages of the osteoblast. Importantly, Dlx3 and Dlx5 are associated with active transcription of OC in normal osteoblasts and the osteosarcoma cell lines.
FIG. 5.
Msx2, Dlx3, and Dlx5 selectively bind to the OC HD regulatory element at different stages of osteoblast differentiation. EMSAs are shown for nuclear extracts from calvaria-derived rat osteoblasts (A to C) and ROS 17/2.8 cells, and each panel represents supershift assays with a different antibody, as follows: (A) anti-Msx(1 + 2); (B) anti-Dlx3; (C) anti-Dlx5. Brackets indicate the multiple HD protein complexes, while the asterisk denotes the complex in primary ROB cells either blocked or supershifted by antibodies. Lane 1, probe only; lanes 2 and 3, mobility of overexpressed (O/E) Msx2 in ROS 17/2.8 cells or in vitro-transcribed and -translated (IVTT) Dlx3 and Dlx5 HD proteins as positive controls. Panel A also includes in lane 2 the Msx2 antibody to demonstrate the block shift property. The following lanes of each panel include nuclear extracts from the three stages of osteoblast differentiation at day 4, day 12, or day 20, with (+) or without (−) antibody. Panel B shows a supershift with Dlx3 antibody, which was present on day 12 and increased on day 20. Panel C is the supershift Dlx5 antibody, which showed Dlx5 was most represented on day 20. In each gel shift, 200 ng of antibody was added and a nonspecific antibody control lane included antisera against retinoblastoma protein as described in Materials and Methods. (D) Representation of HD proteins in nuclear extracts. EMSA showed representation of the HD protein in ROS 17/2.8 cells (top panel, Western blot of Msx 1 and -2, Dlx3, Dlx5, and actin) with antibodies against the complexes. A partial block shift was observed for Msx with increasing antibody, and a supershift was observed with Dlx3 and Dlx5 antibodies.
Dlx3 regulates OC transcription through protein-DNA and protein-protein interactions.
To directly address Dlx3 regulation of the OC gene, we performed a series of transfection studies with HD proteins using preosteoblast MC3T3 cells, in which we found osteogenic stimulation by Dlx3 (Fig. 3). Msx2 coexpression with OC-CAT resulted in dose-dependent repression of the OC promoter activity (Fig. 6A, left panels). In contrast, Dlx3 increased OC basal promoter activity at the lower expression levels and had a modest inhibitory effect (82% of control) at the higher dose. The observation that Dlx3 activated OC expression at low concentrations was consistent with our earlier findings with increased endogenous OC gene (Fig. 3), where we expressed Dlx3 at levels required to promote osteoblast differentiation. We also examined Dlx3 regulation of OC in the presence of stimulated promoter activity by Runx2 (Fig. 6A, right panels) and found a dose-dependent repression by Dlx3 at both low and high doses. To test the hypothesis that Dlx3 may attenuate Runx-mediated OC expression, Dlx3 activity on the OC promoter was examined in ROS 17/2.8 cells, representative of the mature osteoblast phenotype with high Runx2 and OC levels. In this cell, expression of either Msx2 or Dlx3 inhibits both OC promoter activity and endogenous OC gene expression (data not shown). Taken together, these studies demonstrated a direct role, but potential dual function, of Dlx3 in regulating OC transcription. In immature osteoblasts (MC3T3-E1 cells), Dlx3 can modestly stimulate OC gene expression. However, Dlx3 appears to repress OC at high cellular levels of Runx2 and Dlx3 in mature osteoblasts expressing constitutive OC.
FIG. 6.
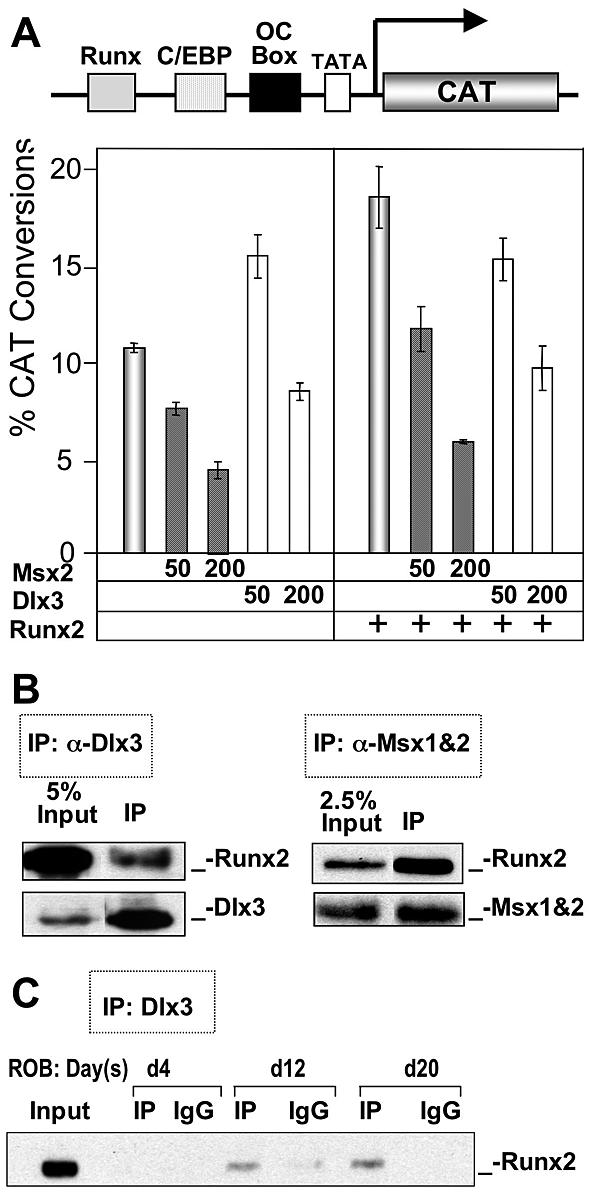
Dlx3 both activates and represses OC promoter activity through protein-DNA and protein-protein interactions, respectively. (A) The mouse preosteoblast cell line MC3T3-E1 was cotransfected with either empty vector or two concentrations of Msx2 or Dlx3 (50 and 200 ng/well on six-well plates) and the proximal OC promoter (−208 OC-CAT) using Fugene 6 (Roche Molecular Biologicals). Promoter activity was normalized with cotransfection with Renilla luciferase. The effects of Msx2 and Dlx3 at low dose (50 ng) and high dose (200 ng) on basal OC promoter activity (left) were compared to Runx2-induced conditions (right). (B) The protein-protein interaction between Runx2 and Dlx3 or Msx2 was demonstrated in coimmunoprecipitation studies. ROS 17/2.8 cells were cotransfected with Msx2 or Dlx3 and Runx2. Msx2 (4G1) and Dlx3 (10) antibodies were used to pull down the immunocomplexes. Runx2 mouse monoclonal antibody was then used for Western blotting to confirm the presence of Runx2 in the complex. (C) Endogenous nuclear proteins from the indicated stages of ROB differentiation were immunoprecipitated using Dlx3 antibody, and the presence of Runx2 in the precipitate was detected by Western blotting. The increase in the Dlx3-Runx2 complex was consistent with increased Runx2 cellular levels (Fig. 2). The coimmunoprecipitation assays included normal immunoglobulins (IgG) from either rabbit (Dlx3) or mouse (Runx2) which were used as controls. Input represents 10% of the sample of day 20 nuclear extracts.
To identify a mechanism for the Dlx3 inhibitory activity, we examined whether Dlx3 physically interacts with Runx2 in osteoblasts, as was reported for Runx2-Msx2 and Runx2-Dlx5 interactions (72). Figure 6B demonstrates that endogenous Runx2 can be coimmunoprecipitated by Dlx3 antibody in ROS 17/2.8 osteoblastic cells, similar to Msx2, which is consistent with the Msx2-Runx2 interaction previously reported in myogenic C2C12 cells (72). We next examined the Runx2-Dlx3 interaction during ROB differentiation and found an increase in the formation of an interacting complex from day 4 to day 20 (Fig. 6C). To further investigate the interacting domain on Runx2 for Dlx3 (Fig. 7A), coimmunoprecipitation studies were performed using anti-HA antibody to pull down mutant Runx2 followed by Western blotting with anti-Flag antibody (Dlx3 Flag tagged). The results indicated that the region between amino acids 391 and 432 exhibits a partial loss in interaction, whereas the region between amino acids 361 and 391 shows a complete loss of formation of the Runx2-Dlx3 complex. Thus, the Runx2 domain from amino acids 432 to 361 is required for Dlx3 interaction. Interestingly, this domain comprises the transcriptionally active nuclear matrix targeting sequence (amino acids 391 to 428) (Fig. 7B). Thus, Dlx3 is competent to stimulate OC expression through protein-DNA interactions in immature bone cells, but in a cellular environment with high Runx2 and Dlx3 proteins, as we observed in mature primary osteoblasts and ROS 17/2.8 cells, Dlx3 can interact directly with Runx2 to attenuate OC gene expression. The mechanism involves Dlx3 interaction in a Runx2 domain that has been described as an activating domain (23, 89).
FIG. 7.
Dlx3 interacts with a Runx2-specific domain in the C terminus. (A) The functional interacting domain of Runx2 was mapped by overexpression of HA-tagged Runx2 deletion mutants (shown on right) and Flag-tagged Dlx3 in ROS 17/2.8 cells. Coimmunoprecipitation was carried out using HA polyclonal antibody, and the interacting domain was characterized by Western blotting with anti-Flag antibody. The coimmunoprecipitation assays included normal immunoglobulins (IgG) from either rabbit (Dlx3) or mouse (Runx2), which were used as controls. Input represents 5% sample of the total cell lysate. (B) Illustration of the Runx2-Dlx3 interacting domain comparing the amino acid sequence among Runx family members of human and mouse origin. The top line shows the exon organization of mouse Runx2. The position of exons 6, 7, and 8 with respect to the interacting protein domain is also shown in the last line (arrows). The interacting domain (amino acids 376 to 432) includes the well-characterized nuclear matrix targeting signal and sequences essential for the Dlx3 interaction. The solid lines over Runx2 in the middle panel and over the sequence in the lower panel indicate the amino acids for each domain.
Dlx3 and Dlx5 are recruited to the OC promoter with induction of gene transcription.
Our functional studies suggested that Dlx3 binding to the OC homeobox sequence can activate or repress OC gene expression. We therefore performed ChIP assays to determine in vivo occupancy and functional regulation of the OC gene by Dlx3 compared to that of Msx2 and Dlx5 during osteoblast growth and differentiation (Fig. 8). We selectively amplified the proximal OC promoter containing the OC box, which supports tissue-specific expression (Fig. 8A). Our results demonstrated that Msx2 occupies the OC promoter on day 4, in the absence of Runx2 recruitment to the promoter in proliferating cells, consistent with its repressive function on OC gene expression in vivo (Fig. 8B). Similar to the EMSA results, Dlx3 and Dlx5 strongly associated with the OC promoter after the proliferation period (day 12, matrix maturation), in parallel with Runx2 interaction. At this time, Msx2 no longer associated with OC chromatin. In the mineralization stage, Dlx3 association with the promoter strikingly decreased, while Dlx5 interaction with OC chromatin increased. The reappearance of Msx2 at the OC promoter on day 20 may reflect the small percentage of the postmineralization apoptotic cells in which OC is downregulated, previously characterized in this cell model (45). We note that in Fig. 2 there was an increase in Msx2 mRNA on day 20; however, OC expression was maximal on day 20, representing the majority of the cell populations. We further addressed whether these molecular switches were also related to recruitment of RNA Pol II to favor OC transcription. In Fig. 8C, we demonstrate in a separate experiment that the increase in Dlx5 and Runx2 occupancy of the OC promoter was associated with an increase in RNA Pol II. In summary, the key findings of these ChIP data from multiple independent experiments are the switch in occupancy of the HD protein at the OC box from Msx2 in proliferating cells (day 4) to Dlx3/Dlx5, when OC is expressed in differentiated cells together with significant activation of the OC gene expression reflected by Runx2 occupancy of the promoter. A second switch occurs as cells progress from the matrix maturation stage to mineralization, with a decrease in Dlx3 occupancy and an increase in Dlx5, which is principally associated with OC chromatin in mature osteoblasts (schematically illustrated in Fig. 8D).
FIG. 8.
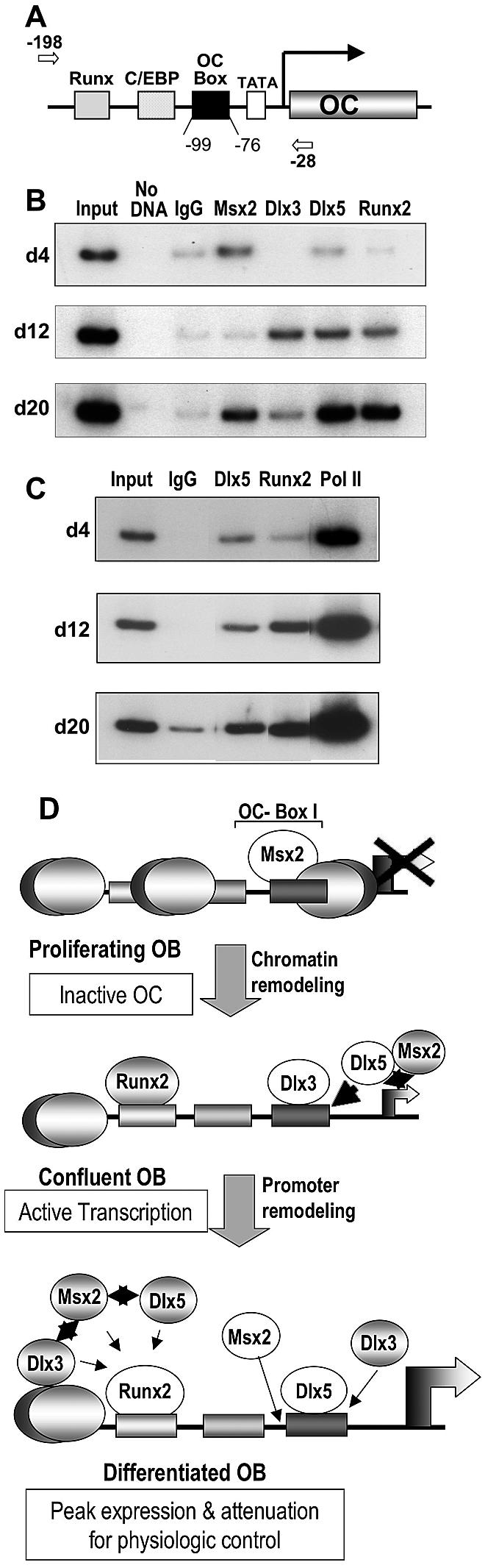
In vivo occupancy of HD proteins in the proximal OC promoter during osteoblast growth and differentiation. (A) The primers used to amplify the key regulatory elements present in the proximal OC promoter fragment in the ChIP assays are indicated in the top line. The arrows indicate the position of the forward (−190) and reverse (−28) primers. (B) Nuclei were prepared from rat calvaria-derived osteoblast cultures at the indicated stages of ROB differentiation (days 4, 12, and 20) for ChIP assays. Formaldehyde-cross-linked chromatin samples were used directly for immunoprecipitation reactions with 2 μg of each Msx (1 and 2), Dlx3, Dlx5 (Y20), PolII, and Runx2 antibody. Immunocomplexes were reversed at 68°C overnight, and the DNA fragments were purified and assayed by PCR. Normal IgG (2 μg) was used as a control for each time point. Input represents 0.2% of each chromatin fraction used for immunoprecipitation. The ChIP data presented are representative of four experiments in which all threetime points were derived from the same osteoblast preparation. (C) Nuclei were derived from calvarial osteoblasts as for panel B for ChIP assays, using the indicated antibodies to demonstrate increased transcription of OC with increased occupancy of Dlx5 and Runx2. (D) Schematic illustration of the ChIP results showing molecular switching in HD protein association with OC chromatin for negative and positive regulation of transcription. The proximal promoter regulatory elements during osteoblast growth and differentiation of the rat OC gene are represented with bound transcription factors at each stage of osteoblast differentiation. The stages are described on the left with the OC transcriptional status indicated. In proliferating cells where OC is not expressed and lacks DNase I hypersensitivity, Msx2 may be bound to linker DNA (between nucleosomes) or to nucleosomal DNA associated with histone deacetylases. Options for occupancy of the HD site by Dlx3 and Dlx5, as well as the potential for protein-protein interactions of Runx2 with Msx2, Dlx3, and Dlx5 to attenuate OC transcription, are shown. Msx2-Dlx5/Dlx3 heterodimerization may also contribute to physiological levels of OC transcription (see text for details). White circles indicate prominent occupancy of the factor at the indicated stages.
From in vitro binding data, in vivo ChIP studies, and functional assays, we conclude that HD proteins contribute to a regulatory network for promoting osteoblast differentiation and controlling the expression of phenotypic genes, such as the OC gene, through different stages of maturation. Occupancy of the OC promoter by the HD proteins is consistent with cellular protein levels (Fig. 2) and EMSA results (Fig. 4). The schematic presented in Fig. 8D represents the temporal regulation of the OC gene by protein-DNA interactions during osteoblast differentiation as indicated in the ChIP studies, as well as the data from our coimmunoprecipitation studies that demonstrated HD protein heterodimer formation with Runx2. Msx2 mainly contributed to maintaining repression of OC in the growth period (Fig. 8D, top panel). Dlx3 and Dlx5 occupied the promoter coincident with Runx2, chromatin remodeling, and OC transcription (Fig. 8D, middle panel), while Dlx5 occupancy was relatively favored when OC mRNA was at the peak level (Fig. 8D, lower panel). The lower panel illustrates several options for HD protein regulation of physiologic levels of OC by both promoting gene expression through direct protein-DNA interactions and by attenuating transcription through Runx2-HD complex formation, which increased during osteoblast differentiation for Runx2-Dlx3, as demonstrated in our coimmunoprecipitation studies (Fig. 6C).
DISCUSSION
Our primary finding is the expression of Dlx3 in osteogenic cells and a role for Dlx3 in the upregulation of bone-related genes to promote osteoblast differentiation. Using OC gene expression as a model that reflects stages of osteoblast maturation, a second key finding was the in vivo temporal association of Msx2, Dlx3, and Dlx5 with OC chromatin and a molecular switch from Msx2 occupancy in the repressed OC promoter in osteoprogenitors to Dlx3, Dlx5, and Runx2 occupancy in the activated OC gene in the postproliferative osteoblast. Finally, we have shown that Dlx3 can mediate increased transcription through protein-DNA interactions at HD response elements as well as repress Runx2-mediated activity through protein-protein interactions. Our findings suggest that these HD proteins function through multiple mechanisms in a regulatory network to support osteoblast differentiation.
Dlx3 is expressed in skeletal progenitor cells to promote osteoblast differentiation.
In vivo and ex vivo osteoblast expression studies together with functional data support the concept that Dlx3 is a regulator of skeletal development, osteoblast differentiation, and bone tissue-specific gene expression. In the embryo, restricted Dlx3 expression is observed in the skeleton, with the highest levels of Dlx3 concentrated in the perichondrium and developing cartilage, periosteal osteoprogenitor cells, and osteoblasts. The high level of Dlx3 mRNA observed in trabecular bone of the embryo and adult skeleton is consistent with Dlx3 expression in relation to new bone formation by the osteoblast. A role for Dlx3 in the early stages of osteoblastogenesis is further indicated by the ability of Dlx3 expression to promote upregulation of endogenous genes that reflect osteoblast differentiation in the preosteoblastic MC3T3-E1 cell line. However, upregulation of osteoblast genes is not observed in the nonosseous C3H10T1/2 cell line with Dlx3 nor in ROS 17/2.8 mature osteoblastic cells, again suggesting restricted requirements to a committed early-stage osteoblast for Dlx3 function in differentiation.
The overlapping, yet distinct expression profiles for Msx and Dlx proteins in early development (7) and in later organogenesis, e.g., in tooth morphogenesis (13, 46), suggest cell type-specific expression with some redundancy due to spatial and temporal expression of HD proteins. Our primary calvarial osteoblast studies reveal that Msx2, Dlx3, and Dlx5 exhibit overlapping expression patterns during stages of differentiation but associate with OC gene chromatin at defined windows of transcriptional activity. Thus, a hierarchy of Msx2, Dlx3, and Dlx5 distinct temporal functions during osteoblast differentiation is indicated. This coordination of HD protein activities is consistent with observations in other systems. For example, during keratinocyte differentiation, Msx1 and Msx2 are expressed in the undifferentiated basal keratinocyte, analogous to high Msx2 expression in proliferating preosteoblasts (40, 67). Dlx3 is then upregulated with the onset of keratinocyte differentiation (54, 67). Thus, our novel findings from studies with the osteoblast model and the in vivo analysis of HD protein regulation of the OC gene strengthen the concept that Dlx3 contributes to the onset of a cellular phenotype.
While null mutations of HD proteins support their functions in skeletal development, more direct evidence for the roles of HD proteins in promoting bone formation is derived from studies demonstrating the effects of HD protein expression in osteoblastic cells. Msx2 regulates cellular proliferation and differentiation of skeletal mesenchyme (35, 43, 58, 67) and has long been associated with phenotypic gene repression. It was the first HD protein implicated in repression of osteoblast, ameloblast, and chondrogenic differentiation (7, 32, 42, 68, 80, 84). However, a few recent reports have suggested that Msx2 may have some osteogenic-enhancing activity in specific cell types (11, 24, 88). Our studies showed that Msx2, relative to Dlx3, modestly increased only two osteogenic markers but repressed promoter activity of OC and other osteoblastic genes (18). Related to a potential Msx2 role in determining the osteoblast phenotype indirectly as a repressor protein in pluripotent cells is the observation that Msx2, like Dlx3 and Dlx5, is an immediate-early response gene to BMP2, an inductant of bone formation.
Dlx5 was the second HD protein well studied for its role in skeletogenesis. Dlx5 is activated in osteoblasts and is robustly expressed at mineralizing fronts and bone-forming surfaces (33). Studies have demonstrated that Dlx5 is also a positive regulator of chondrocyte differentiation (21) and can promote the differentiation of chick and mouse osteoblasts, as reflected in increased OC expression (19, 50, 78). The present studies now show that Dlx3 may also enhance osteoblastogenesis. Thus, multiple HD proteins are recruited for osteoblast differentiation, and the indication is that they function coordinately to support the different stages of osteoblast maturation. Notably, Dlx proteins exhibit coordinated activities in other biological systems. For example, both differential and overlapping expression patterns of Dlx2 and Dlx3 have been identified in craniofacial development (63). Also, sequential regulation of basal ganglia differentiation by a number of Dlx proteins has been documented (41). Furthermore, nested Dlx5 and Dlx6 expression in the branchial arch developmentally patterns the skull and jaw (15, 64).
We found that Dlx3 and Dlx5 expression overlap in part during the matrix maturation stage, with Dlx3 expression becoming lower in the later mineralization stage. ChIP assays showed both Dlx3 and Dlx5 occupancy of the OC gene at the onset of its activation. These findings are consistent with our identification of Dlx3 in contributing to osteoblast differentiation and a temporal and combinatorial requirement for Dlx3 and Dlx5 in the control of OC gene expression during osteoblast differentiation. Taken together, the findings suggest that the Dlx proteins may function in a complex regulatory pathway for mediating cellular differentiation.
OC gene regulation by HD proteins identifies multiple mechanisms of transcriptional control.
We and others previously showed that mutations of the HD element significantly decrease activity (30 to 40%) of the native OC promoter, which is expressed in vivo in a skeletal cell-specific manner (22, 32, 84). Furthermore, in the absence of the Runx2 sites, which are essential for activation of OC transcription and vitamin D enhancement (36, 73), some transcription of OC is still supported by the proximal promoter (29, 32). These findings have established that the OC box/HD core regulatory element is a key regulator of OC transcription. The studies presented herein show that multiple HD protein-DNA complexes form at the OC box. An osteoblast differentiation-related complex (Fig. 5) appears to change in composition from the preosteoblast to the mature bone cell. The gel shift assays suggest that this complex comprises the Dlx proteins. In proliferating cells, the complex consists of Msx2, although binding activity in vitro is relatively weak compared to cellular protein levels in the proliferation stage or in ROS 17/2.8 cells. However, Msx2 forms heterodimers with Dlx5 (9) and Dlx3 (data not shown), complexes with reduced DNA binding affinities (55, 90). Thus, in vitro DNA binding assays can be useful in determining binding properties that may not necessarily correlate to protein cellular levels or to in vivo chromatin occupancy of the factor. Results from using a short oligonucleotide should be interpreted with caution.
The ChIP assays demonstrated a clear molecular switch from Msx2 to Dlx3 and Dlx5 association with the OC promoter during progression of the osteoblast phenotype. Msx2 occupies the promoter in proliferating cells, where the OC gene is repressed. Msx2 can repress gene transcription through protein-protein interactions, such as with histone deacetylases (48) or by inhibiting transactivating factors C/EBP (91) and Runx2 (72). Dlx3 and Dlx5 associate with the OC gene at the onset of transcriptional activation, concomitant with Runx2 occupancy of the OC promoter. Here, we observed a significant increase in the recruitment of RNA Pol II, reflecting active transcription of the OC gene at this stage. Dlx3 association with OC chromatin peaked at this time. Although DNase I hypersensitivity has not been observed in proliferating osteoblasts (52), Msx2 occupancy of the OC gene promoter, as detected by ChIP assay, suggests that Msx2 may restrain expression of genes during proliferation that are subsequently expressed later in differentiation. The OC gene is not acetylated in proliferating cells (70). The occupancy of RNA Pol II on the OC promoter in the proliferating period suggests that the OC gene is poised for transcription (37, 77). Of interest, reciprocal expression of OC and Msx2 has been observed in situ in calvarial tissue cells (9), consistent with the direct mechanism our ChIP studies have defined by recruitment of Msx2 to the repressed OC gene.
In the mineralization stage, Dlx5 occupancy of the HD site is maximal, while Dlx3 interactions with OC chromatin decrease from the matrix maturation stage (day 12), suggesting that Dlx3 and Dlx5 may have coordinated molecular roles in the regulation of OC transcription. Thus, the ChIP studies identified a primary mechanism of OC transcriptional control during osteoblast differentiation resulting from the reciprocal occupancy of the OC HD element by Msx2 and the Dlx proteins during bone cell differentiation, as well as a temporal occupancy of Dlx3 and Dlx5 on the OC promoter. This finding suggests mutually exclusive protein-DNA interactions of Msx2 and Dlx3/Dlx5 at the OC box during the transition from proliferating cells (OC not expressed) to differentiated osteoblasts (OC gene on). The reappearance of Msx2 on the promoter at day 20 may reflect a minor population of apoptotic cells in mineralizing cultures in which OC is downregulated (45). However, the increase in RNA Pol II we observed with increasing Runx2 and Dlx5 on day 20 indicates that most cells are expressing OC at maximal levels. Thus, the promoter occupancy profile suggests that the Dlx proteins mediate enhancer function in osteoblasts directly as DNA binding proteins.
A second mechanism operative in the physiological control of OC transcription by Dlx3, like other HD proteins, is via protein-protein interactions, as heterodimers with other HD proteins (7, 10) or with regulatory factors at other elements (72, 91). An increase in the Dlx3-Runx2 complex from day 12 to day 20 occurs, while Dlx3 occupancy of the HD site in the promoter is decreased. Our results demonstrate that the protein-protein interactions between Dlx3 and Runx2, similar to formation of a Dlx5-Runx2 heterodimer complex (72), are functionally related to inhibition of Runx2-mediated gene transcription. Here, we have shown that the inhibition of Runx2 activity is the result of Dlx3 interaction at an essential and Runx2-specific regulatory domain (23, 89). Thus, we propose HD proteins may attenuate Runx activity for physiological control of OC transcription, which is particularly important in the late stages of osteoblast differentiation, when OC expression levels are rapidly increasing. In conclusion, multiple options and combinatorial mechanisms via protein-DNA and protein-protein interactions can be executed by HD proteins in a coordinated manner for either activation or repression of bone-related genes during osteoblast differentiation.
Acknowledgments
This work was supported by NIH grants DE12528, PO1 AR48818, and AR39588.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Acampora, D., G. R. Merlo, L. Paleari, B. Zerega, M. P. Postiglione, S. Mantero, E. Bober, O. Barbieri, A. Simeone, and G. Levi. 1999. Craniofacial, vestibular and bone defects in mice lacking the distal-less-related gene dlx5. Development 126:3795-3809. [DOI] [PubMed] [Google Scholar]
- 2.Aronow, M. A., L. C. Gerstenfeld, T. A. Owen, M. S. Tassinari, G. S. Stein, and J. B. Lian. 1990. Factors that promote progressive development of the osteoblast phenotype in cultured fetal rat calvaria cells. J. Cell. Physiol. 143:213-221. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2003. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Balint, E., D. Lapointe, H. Drissi, C. van der Meijden, D. W. Young, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2003. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89:401-426. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, G. L., T. T. Della, B. Sommer, M. F. Young, and L. C. Gerstenfeld. 2002. Transcriptional regulation restricting bone sialoprotein gene expression to both hypertrophic chondrocytes and osteoblasts. J. Cell. Biochem. 87:458-469. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, G. L., A. Javed, S. M. Waller, M. H. Kamal, K. E. Hebert, M. Q. Hassan, A. Bellahcene, A. J. van Wijnen, M. F. Young, J. B. Lian, G. S. Stein, and L. C. Gerstenfeld. 2003. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 63:2631-2637. [PubMed] [Google Scholar]
- 7.Bendall, A. J., and C. Abate-Shen. 2000. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene 247:17-31. [DOI] [PubMed] [Google Scholar]
- 8.Benson, M. D., J. L. Bargeon, G. Xiao, P. E. Thomas, A. Kim, Y. Cui, and R. T. Franceschi. 2000. Identification of a homeodomain binding element in the bone sialoprotein gene promoter that is required for its osteoblast-selective expression. J. Biol. Chem. 275:13907-13917. [DOI] [PubMed] [Google Scholar]
- 9.Bidder, M., T. Latifi, and D. A. Towler. 1998. Reciprocal temporospatial patterns of Msx2 and osteocalcin gene expression during murine odontogenesis. J. Bone Miner. Res. 13:609-619. [DOI] [PubMed] [Google Scholar]
- 10.Bryan, J. T., and M. I. Morasso. 2000. The Dlx3 protein harbors basic residues required for nuclear localization, transcriptional activity and binding to Msx1. J. Cell Sci. 113:4013-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, S. L., J. S. Shao, N. Charlton-Kachigian, A. P. Loewy, and D. A. Towler. 2003. Msx2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J. Biol. Chem. 278:45969-45977. [DOI] [PubMed] [Google Scholar]
- 12.Davideau, J. L., P. Demri, T. T. Gu, D. Simmons, C. Nessman, N. Forest, M. MacDougall, and A. Berdal. 1999. Expression of DLX5 during human embryonic craniofacial development. Mech. Dev. 81:183-186. [DOI] [PubMed] [Google Scholar]
- 13.Davideau, J. L., P. Demri, D. Hotton, T. T. Gu, M. MacDougall, P. Sharpe, N. Forest, and A. Berdal. 1999. Comparative study of MSX-2, DLX-5, and DLX-7 gene expression during early human tooth development. Pediatr. Res. 46:650-656. [DOI] [PubMed] [Google Scholar]
- 14.Davidson, D. 1995. The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 11:405-411. [DOI] [PubMed] [Google Scholar]
- 15.Depew, M. J., T. Lufkin, and J. L. Rubenstein. 2002. Specification of jaw subdivisions by Dlx genes. Science 298:381-385. [DOI] [PubMed] [Google Scholar]
- 16.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodig, M., M. S. Kronenberg, A. Bedalov, B. E. Kream, G. Gronowicz, S. H. Clark, K. Mack, Y. H. Liu, R. Maxon, Z. Z. Pan, W. B. Upholt, D. W. Rowe, and A. C. Lichtler. 1996. Identification of a TAAT-containing motif required for high level expression of the COL1A1 promoter in differentiated osteoblasts of transgenic mice. J. Biol. Chem. 271:16422-16429. [DOI] [PubMed] [Google Scholar]
- 18.Dodig, M., T. Tadic, M. S. Kronenberg, S. Dacic, Y. H. Liu, R. Maxson, D. W. Rowe, and A. C. Lichtler. 1999. Ectopic Msx2 overexpression inhibits and Msx2 antisense stimulates calvarial osteoblast differentiation. Dev. Biol. 209:298-307. [DOI] [PubMed] [Google Scholar]
- 19.Erceg, I., T. Tadic, M. S. Kronenberg, I. Marijanovic, and A. C. Lichtler. 2003. Dlx5 regulation of mouse osteoblast differentiation mediated by avian retrovirus vector. Croat. Med. J. 44:407-411. [PubMed] [Google Scholar]
- 20.Ferrari, D., A. Harrington, C. N. Dealy, and R. A. Kosher. 1999. Dlx-5 in limb initiation in the chick embryo. Dev. Dyn. 216:10-15. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari, D., and R. A. Kosher. 2002. Dlx5 is a positive regulator of chondrocyte differentiation during endochondral ossification. Dev. Biol. 252:257-270. [DOI] [PubMed] [Google Scholar]
- 22.Frenkel, B., C. Capparelli, M. van Auken, J. Bryan, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. Activity of the osteocalcin promoter in skeletal sites of transgenic mice and during osteoblast differentiation in bone marrow-derived stromal cell cultures: effects of age and sex. Endocrinology 138:2109-2116. [DOI] [PubMed] [Google Scholar]
- 23.Geoffroy, V., D. A. Corral, L. Zhou, B. Lee, and G. Karsenty. 1998. Genomic organization, expression of the human CBFA1 gene, and evidence for an alternative splicing event affecting protein function. Mamm. Genome 9:54-57. [DOI] [PubMed] [Google Scholar]
- 24.Gotoh, M., K. Notoya, Y. Ienaga, M. Kawase, and H. Makino. 2002. Enhancement of osteogenesis in vitro by a novel osteoblast differentiation-promoting compound, TAK-778, partly through the expression of Msx2. Eur. J. Pharmacol. 451:19-25. [DOI] [PubMed] [Google Scholar]
- 25.Gutierrez, S., A. Javed, D. Tennant, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 26.Harris, S. E., D. Guo, M. A. Harris, A. Krishnaswamy, and A. Lichtler. 2003. Transcriptional regulation of BMP-2 activated genes in osteoblasts using gene expression microarray analysis: role of Dlx2 and Dlx5 transcription factors. Front. Biosci. 8:S1249-S1265. [DOI] [PubMed] [Google Scholar]
- 27.Harris, S. E., and M. A. Harris. 2001. Gene expression profiling in osteoblast biology: bioinformatic tools. Mol. Biol. Rep. 28:139-156. [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs, A. A. J., C. Banerjee, R. Bortell, T. A. Owen, J. L. Stein, G. S. Stein, and J. B. Lian. 1993. Identification and characterization of two proximal elements in the rat osteocalcin gene promoter that may confer species-specific regulation. J. Cell. Biochem. 53:240-250. [DOI] [PubMed] [Google Scholar]
- 29.Heinrichs, A. A. J., R. Bortell, M. Bourke, J. B. Lian, G. S. Stein, and J. L. Stein. 1995. Proximal promoter binding protein contributes to developmental, tissue-restricted expression of the rat osteocalcin gene. J. Cell. Biochem. 57:90-100. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann, H., J. Green, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2000. Expression screening of factors binding to the osteocalcin bone-specific promoter element OC box I: isolation of a novel osteoblast differentiation-specific factor. J. Cell. Biochem. 80:156-168. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann, H. M., T. L. Beumer, S. Rahman, L. R. McCabe, C. Banerjee, F. Aslam, J. A. Tiro, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 1996. Bone tissue-specific transcription of the osteocalcin gene: role of an activator osteoblast-specific complex and suppressor hox proteins that bind the OC box. J. Cell. Biochem. 61:310-324. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann, H. M., K. M. Catron, A. J. van Wijnen, L. R. McCabe, J. B. Lian, G. S. Stein, and J. L. Stein. 1994. Transcriptional control of the tissue-specific, developmentally regulated osteocalcin gene requires a binding motif for the Msx family of homeodomain proteins. Proc. Natl. Acad. Sci. USA 91:12887-12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holleville, N., A. Quilhac, M. Bontoux, and A. H. Monsoro-Burq. 2003. BMP signals regulate Dlx5 during early avian skull development. Dev. Biol. 257:177-189. [DOI] [PubMed] [Google Scholar]
- 34.Hullinger, T. G., Q. Pan, H. L. Viswanathan, and M. J. Somerman. 2001. TGFβ and BMP-2 activation of the OPN promoter: roles of smad- and hox-binding elements. Exp. Cell. Res. 262:69-74. [DOI] [PubMed] [Google Scholar]
- 35.Ishii, M., A. E. Merrill, Y. S. Chan, I. Gitelman, D. P. Rice, H. M. Sucov, and R. E. Maxson, Jr. 2003. Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development 130:6131-6142. [DOI] [PubMed] [Google Scholar]
- 36.Javed, A., S. Gutierrez, M. Montecino, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 1999. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol. Cell. Biol. 19:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 23:6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiyoshima, T., M. Yamauchi, C. Wong, A. Jheon, B. Ganss, and J. Sodek. 2002. An L1 element disrupts human bone sialoprotein promoter: lack of tissue-specific regulation by distalless5 (Dlx5) and runt homeodomain protein2 (Runx2)/core binding factor a1 (Cbfa1) elements. Gene 299:205-217. [DOI] [PubMed] [Google Scholar]
- 39.Kraus, P., and T. Lufkin. 1999. Mammalian Dlx homeobox gene control of craniofacial and inner ear morphogenesis. J. Cell. Biochem. Suppl. 32-33:133-140. [DOI] [PubMed]
- 40.Lian, J. B., G. S. Stein, J. L. Stein, A. J. van Wijnen, L. McCabe, C. Banerjee, and H. Hoffmann. 1996. The osteocalcin gene promoter provides a molecular blueprint for regulatory mechanisms controlling bone tissue formation: role of transcription factors involved in development. Connect. Tissue Res. 35:15-21. [DOI] [PubMed] [Google Scholar]
- 41.Liu, J. K., I. Ghattas, S. Liu, S. Chen, and J. L. Rubenstein. 1997. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev. Dyn. 210:498-512. [DOI] [PubMed] [Google Scholar]
- 42.Liu, Y. H., R. Kundu, L. Wu, W. Luo, M. A. Ignelzi, Jr., M. L. Snead, and R. E. Maxson, Jr. 1995. Premature suture closure and ectopic cranial bone in mice expressing Msx2 transgenes in the developing skull. Proc. Natl. Acad. Sci. USA 92:6137-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Y. H., Z. Tang, R. K. Kundu, L. Wu, W. Luo, D. Zhu, F. Sangiorgi, M. L. Snead, and R. E. Maxson. 1999. Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: a possible mechanism for MSX2-mediated craniosynostosis in humans. Dev. Biol. 205:260-274. [DOI] [PubMed] [Google Scholar]
- 44.Luo, T., M. Matsuo-Takasaki, J. H. Lim, and T. D. Sargent. 2001. Differential regulation of Dlx gene expression by a BMP morphogenetic gradient. Int. J. Dev. Biol. 45:681-684. [PubMed] [Google Scholar]
- 45.Lynch, M., C. Capparelli, J. L. Stein, G. S. Stein, and J. B. Lian. 1998. Apoptosis during bone-like tissue development in vitro. J. Cell. Biochem. 68:31-49. [PubMed] [Google Scholar]
- 46.Maas, R., and M. Bei. 1997. The genetic control of early tooth development. Crit. Rev. Oral Biol. Med. 8:4-39. [DOI] [PubMed] [Google Scholar]
- 47.Mackem, S., and K. A. Mahon. 1991. Ghox 4.7: a chick homeobox gene expressed primarily in limb buds with limb-type differences in expression. Development 112:791-806. [DOI] [PubMed] [Google Scholar]
- 48.Mehra-Chaudhary, R., H. Matsui, and R. Raghow. 2001. Msx3 protein recruits histone deacetylase to down-regulate the Msx1 promoter. Biochem. J. 353:13-22. [PMC free article] [PubMed] [Google Scholar]
- 49.Merlo, G. R., B. Zerega, L. Paleari, S. Trombino, S. Mantero, and G. Levi. 2000. Multiple functions of Dlx genes. Int. J. Dev. Biol. 44:619-626. [PubMed] [Google Scholar]
- 50.Miyama, K., G. Yamada, T. S. Yamamoto, C. Takagi, K. Miyado, M. Sakai, N. Ueno, and H. Shibuya. 1999. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev. Biol. 208:123-133. [DOI] [PubMed] [Google Scholar]
- 51.Montecino, M., B. Frenkel, J. Lian, J. Stein, and G. Stein. 1996. Requirement of distal and proximal promoter sequences for chromatin organization of the osteocalcin gene in bone-derived cells. J. Cell. Biochem. 63:221-228. [DOI] [PubMed] [Google Scholar]
- 52.Montecino, M., J. Lian, G. Stein, and J. Stein. 1996. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry 35:5093-5102. [DOI] [PubMed] [Google Scholar]
- 53.Morasso, M. I., A. Grinberg, G. Robinson, T. D. Sargent, and K. A. Mahon. 1999. Placental failure in mice lacking the homeobox gene Dlx3. Proc. Natl. Acad. Sci. USA 96:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morasso, M. I., N. G. Markova, and T. D. Sargent. 1996. Regulation of epidermal differentiation by a Distal-less homeodomain gene. J. Cell Biol. 135:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newberry, E. P., T. Latifi, and D. A. Towler. 1998. Reciprocal regulation of osteocalcin transcription by the homeodomain proteins Msx2 and Dlx5. Biochemistry 37:16360-16368. [DOI] [PubMed] [Google Scholar]
- 56.Owen, T. A., M. Aronow, V. Shalhoub, L. M. Barone, L. Wilming, M. S. Tassinari, M. B. Kennedy, S. Pockwinse, J. B. Lian, and G. S. Stein. 1990. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell. Physiol. 143:420-430. [DOI] [PubMed] [Google Scholar]
- 57.Panganiban, G., and J. L. Rubenstein. 2002. Developmental functions of the Distal-less/Dlx homeobox genes. Development 129:4371-4386. [DOI] [PubMed] [Google Scholar]
- 58.Park, G. T., and M. I. Morasso. 1999. Regulation of the Dlx3 homeobox gene upon differentiation of mouse keratinocytes. J. Biol. Chem. 274:26599-26608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park, G. T., and M. I. Morasso. 2002. Bone morphogenetic protein-2 (BMP-2) transactivates Dlx3 through Smad1 and Smad4: alternative mode for Dlx3 induction in mouse keratinocytes. Nucleic Acids Res. 30:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price, J. A., D. W. Bowden, J. T. Wright, M. J. Pettenati, and T. C. Hart. 1998. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum. Mol. Genet. 7:563-569. [DOI] [PubMed] [Google Scholar]
- 61.Price, J. A., J. T. Wright, K. Kula, D. W. Bowden, and T. C. Hart. 1998. A common DLX3 gene mutation is responsible for tricho-dento-osseous syndrome in Virginia and North Carolina families. J. Med. Genet. 35:825-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu, M., A. Bulfone, I. Ghattas, J. J. Meneses, L. Christensen, P. T. Sharpe, R. Presley, R. A. Pedersen, and J. L. Rubenstein. 1997. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 185:165-184. [DOI] [PubMed] [Google Scholar]
- 63.Robinson, G. W., and K. A. Mahon. 1994. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mech. Dev. 48:199-215. [DOI] [PubMed] [Google Scholar]
- 64.Robledo, R. F., L. Rajan, X. Li, and T. Lufkin. 2002. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 16:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossi, P., and B. de Crombrugghe. 1987. Identification of a cell-specific transcriptional enhancer in the first intron of the mouse alpha 2 (type I) collagen gene. Proc. Natl. Acad. Sci. USA 84:5590-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryoo, H.-M., H. M. Hoffmann, T. L. Beumer, B. Frenkel, D. A. Towler, G. S. Stein, J. L. Stein, A. J. van Wijnen, and J. B. Lian. 1997. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol. Endocrinol. 11:1681-1694. [DOI] [PubMed] [Google Scholar]
- 67.Sargent, T. D., and M. I. Morasso. 1999. Differentiation of vertebrate epidermis, p. 553-567. In S. A. Moody (ed.), Cell lineage and fate determination. Academic Press, New York, N.Y.
- 68.Satokata, I., L. Ma, H. Ohshima, M. Bei, I. Woo, K. Nishizawa, T. Maeda, Y. Takano, M. Uchiyama, S. Heaney, H. Peters, Z. Tang, R. Maxson, and R. Maas. 2000. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 24:391-395. [DOI] [PubMed] [Google Scholar]
- 69.Shea, C. M., C. M. Edgar, T. A. Einhorn, and L. C. Gerstenfeld. 2003. BMP treatment of C3H10T1/2 mesenchymal stem cells induces both chondrogenesis and osteogenesis. J. Cell. Biochem. 90:1112-1127. [DOI] [PubMed] [Google Scholar]
- 70.Shen, J., H. Hovhannisyan, J. B. Lian, M. A. Montecino, G. S. Stein, J. L. Stein, and A. J. van Wijnen. 2003. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Mol. Endocrinol. 17:743-756. [DOI] [PubMed] [Google Scholar]
- 71.Shen, J., M. A. Montecino, J. B. Lian, G. S. Stein, A. J. van Wijnen, and J. L. Stein. 2002. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D dependent, bone tissue-specific transcription. J. Biol. Chem. 277:20284-20292. [DOI] [PubMed] [Google Scholar]
- 72.Shirakabe, K., K. Terasawa, K. Miyama, H. Shibuya, and E. Nishida. 2001. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells 6:851-856. [DOI] [PubMed] [Google Scholar]
- 73.Sierra, J., A. Villagra, R. Paredes, F. Cruzat, S. Gutierrez, G. Arriagada, J. Olate, M. Imschenetzky, A. J. van Wijnen, J. B. Lian, G. S. Stein, J. L. Stein, and M. Montecino. 2003. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyl transferase activity. Mol. Cell. Biol. 23:3339-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simeone, A., D. Acampora, M. Pannese, M. D'Esposito, A. Stornaiuolo, M. Gulisano, A. Mallamaci, K. Kastury, T. Druck, K. Huebner, and E. Boncinelli. 1994. Cloning and characterization of two members of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA 91:2250-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sodek, J., R. H. Kim, Y. Ogata, J. Li, M. Yamauchi, Q. Zhang, and L. P. Freedman. 1995. Regulation of bone sialoprotein gene transcription by steroid hormones. Connect. Tissue Res. 32:209-217. [DOI] [PubMed] [Google Scholar]
- 76.Stock, D. W., D. L. Ellies, Z. Zhao, M. Ekker, F. H. Ruddle, and K. M. Weiss. 1996. The evolution of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA 93:10858-10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Studitsky, V. M., W. Walter, M. Kireeva, M. Kashlev, and G. Felsenfeld. 2004. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 29:127-135. [DOI] [PubMed] [Google Scholar]
- 78.Tadic, T., M. Dodig, I. Erceg, I. Marijanovic, M. Mina, Z. Kalajzic, D. Velonis, M. S. Kronenberg, R. A. Kosher, D. Ferrari, and A. C. Lichtler. 2002. Overexpression of Dlx5 in chicken calvarial cells accelerates osteoblastic differentiation. J. Bone Miner. Res. 17:1008-1014. [DOI] [PubMed] [Google Scholar]
- 79.Tadic, T., I. Erceg, M. L. Stover, D. W. Rowe, and A. C. Lichtler. 2001. Dlx5 induces expression of COL1A1 promoter contained in a retrovirus vector. Croat. Med. J. 42:436-439. [PubMed] [Google Scholar]
- 80.Takahashi, K., G. H. Nuckolls, I. Takahashi, K. Nonaka, M. Nagata, T. Ikura, H. C. Slavkin, and L. Shum. 2001. Msx2 is a repressor of chondrogenic differentiation in migratory cranial neural crest cells. Dev. Dyn. 222:252-262. [DOI] [PubMed] [Google Scholar]
- 81.Thomas, B. L., M. H. Porteus, J. L. Rubenstein, and P. T. Sharpe. 1995. The spatial localization of Dlx-2 during tooth development. Conn. Tissue Res. 32:27-34. [DOI] [PubMed] [Google Scholar]
- 82.Thomas, B. L., A. S. Tucker, M. Qui, C. A. Ferguson, Z. Hardcastle, J. L. Rubenstein, and P. T. Sharpe. 1997. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development 124:4811-4818. [DOI] [PubMed] [Google Scholar]
- 83.Thomas, H. F., J. A. Feldman, A. Bedalov, C. O. Woody, S. H. Clark, K. Mack, and A. C. Lichtler. 1995. Identification of regulatory elements necessary for the expression of the COL1A1 promoter in murine odontoblasts. Connect. Tissue Res. 33:81-85. [DOI] [PubMed] [Google Scholar]
- 84.Towler, D. A., S. J. Rutledge, and G. A. Rodan. 1994. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol. Endocrinol. 8:1484-1493. [DOI] [PubMed] [Google Scholar]
- 85.Wright, J. T., K. Kula, K. Hall, J. H. Simmons, and T. C. Hart. 1997. Analysis of the tricho-dento-osseous syndrome genotype and phenotype. Am. J. Med. Genet. 72:197-204. [DOI] [PubMed] [Google Scholar]
- 86.Xu, S. C., M. A. Harris, J. L. Rubenstein, G. R. Mundy, and S. E. Harris. 2001. Bone morphogenetic protein-2 (BMP-2) signaling to the Col2α1 gene in chondroblasts requires the homeobox gene Dlx-2. DNA Cell Biol. 20:359-365. [DOI] [PubMed] [Google Scholar]
- 87.Yang, R., and L. C. Gerstenfeld. 1997. Structural analysis and characterization of tissue and hormonal responsive expression of the avian bone sialoprotein (BSP) gene. J. Cell. Biochem. 64:77-93. [DOI] [PubMed] [Google Scholar]
- 88.Yoshizawa, T., F. Takizawa, F. Iizawa, O. Ishibashi, H. Kawashima, A. Matsuda, N. Endo, and H. Kawashima. 2004. Homeobox protein MSX2 acts as a molecular defense mechanism for preventing ossification in ligament fibroblasts. Mol. Cell. Biol. 24:3460-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zaidi, S. K., A. Javed, J.-Y. Choi, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2001. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J. Cell Sci. 114:3093-3102. [DOI] [PubMed] [Google Scholar]
- 90.Zhang, H., G. Hu, H. Wang, P. Sciavolino, N. Iler, M. M. Shen, and C. Abate-Shen. 1997. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol. Cell. Biol. 17:2920-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou, Y. L., Y. Lei, and M. L. Snead. 2000. Functional antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J. Biol. Chem. 275:29066-29075. [DOI] [PubMed] [Google Scholar]