Abstract
Many proteins have been proposed to be involved in retinoblastoma protein (pRB)-mediated repression, but it is largely uncertain which cofactors are essential for pRB to repress endogenous E2F-regulated promoters. Here we have taken advantage of the stream-lined Drosophila dE2F/RBF pathway, which has only two E2Fs (dE2F1 and dE2F2), and two pRB family members (RBF1 and RBF2). With RNA interference (RNAi), we depleted potential corepressors and looked for the elevated expression of groups of E2F target genes that are known to be directly regulated by RBF1 and RBF2. Previous studies have implicated histone deacetylase (HDAC) and SWI/SNF chromatin-modifying complexes in pRB-mediated repression. However, our results fail to support the idea that the SWI/SNF proteins are required for RBF-mediated repression and suggest that a requirement for HDAC activities is likely to be limited to a subset of targets. We found that the chromatin assembly factor p55/dCAF-1 is essential for the repression of dE2F2-regulated targets. The removal of p55 deregulated the expression of E2F targets that are normally repressed by dE2F2/RBF1 and dE2F2/RBF2 complexes in a cell cycle-independent manner but had no effect on the expression of E2F targets that are normally coupled with cell proliferation. The results indicate that the mechanisms of RBF regulation at these two types of E2F targets are different and suggest that p55, and perhaps p55's mammalian orthologs RbAp46 and RbAp48, have a conserved function in repression by pRB-related proteins.
The E2F and the retinoblastoma protein (pRB) families of transcription factors are critical regulators of cell cycle progression and differentiation in metazoans (14). pRB family members interact with E2F proteins (7, 26, 36, 54) to form complexes that repress the transcription of genes required for cell cycle progression and DNA synthesis (22, 43). This repression is relieved during the G1 to S phase transition of the cell cycle, allowing the coordinated expression of a diverse collection of proliferation-related genes.
E2F is a heterodimer composed of one E2F and one DP subunit. Seven E2F and two DP genes have been characterized in mammalian cells (10, 12, 54). Aside from the recently identified E2F7 protein, all E2Fs require DP dimerization in order to bind DNA. Synchronized cell experiments show that the E2F family can be loosely divided into two subgroups based on their presence at E2F-regulated genes during either E2F transcriptional activation or repression (39, 42, 55, 64, 65). E2F1, E2F2, and E2F3 mediate activation, and E2F4, E2F5, E2F6, and E2F7 mediate repression. Ablation of activator E2Fs (E2F1, E2F2, and E2F3) result in lack of activation of E2F target genes, reduced S-phase entry, and inability to proliferate (66), whereas cells deficient for E2F4 and E2F5 are unable to respond to certain cell cycle arrest signals (19).
pRB is a member of a family of three closely related mammalian proteins that includes p107 and p130 (8). Together, these proteins are referred to as the pocket proteins, a name that alludes to their sequence similarity in a domain that mediates interactions with viral oncoproteins. Pocket proteins have similar functions, including their ability to interact and repress E2F-mediated transcription, and are all regulated by phosphorylation by cyclin-dependent kinases.
Pocket proteins can repress E2F-dependent promoters in several different ways. Initially, pRB was thought to act simply by masking the activation domain of E2F1 (20, 21). However, subsequent studies showed that, once recruited to a promoter, pRB can suppress the effects of other transcription factors, an effect frequently described as active repression. Active repression is thought to be due primarily to pRB's interactions with a variety of chromatin-remodeling and/or chromatin-modifying complexes. These interactions are generally dependent on an LXCXE binding cleft in the pocket domain of pRB, the binding site for viral oncoproteins, and have been proposed to be mediated by LXCXE-like motifs present in several different types of transcriptional repressors.
One of the first chromatin-remodeling proteins to be linked to pRB was Brahma-related gene 1 (BRG1), the ATPase subunit of the mammalian SWI/SNF complex (13). Other studies showed that BRG1 and a second mammalian Brahma homolog, hBRM, can cooperate with pRB to repress E2F transcriptional activation (58). Genetic screens for modifiers of an E2F-dependent phenotype and a separate screen for modifiers of a cycE hypomorphic allele suggested that brahma and other SWI/SNF components might act as regulators of Drosophila E2F activity (49, 52).
Some experiments have strongly suggested that pRB's role in repressing E2F transcription is, at least in part, facilitated by recruitment of histone deacetylase (HDAC) proteins. The HDACs, by virtue of their action on histones, are thought to condense chromatin structure (6, 15, 30, 31). Trichostatin A (TSA), an inhibitor of many HDACs, has been found to interfere with pRB-mediated repression at some but not all promoters (30, 51). This has prompted searches for chromatin-associated complexes that might mediate either the TSA-sensitive or TSA-insensitive effects of pRB. Suggested corepressors include the histone methyltransferase SUV39H1 and heterochromatin protein HP1 (38), the Polycomb group proteins (9), the C-terminal binding proteins E1A and CtIP/CtBP (32), DNA methyltransferase (DNMT1) (45), and the high-mobility-group protein HBP1 (56). Other studies have found that pRB can interfere with components of the basal transcriptional machinery, such as TAFII250, TFIIA, and TFIID, and suggested that this might underlie its repressive function (33, 46, 50).
While there are many attractive models to explain how pRB can repress transcription, there is surprisingly little information about which of these interactions are critical for endogenous pRB proteins to function at their natural targets. pRB repression has been studied primarily with overexpressed proteins and artificial reporter constructs. The action of the endogenous protein has been difficult to address, in part because much of the regulation of E2F-regulated genes is mediated by p107 and p130, and it is unclear exactly when, and where, pRB is the primary regulator of endogenous E2F-dependent promoters. In addition, until the recent development of short interfering RNA methods, it has been technically difficult to test the functional significance of putative corepressors by specifically removing that protein from the cell. Moreover, the large number of putative pRB corepressors and the diverse collection of E2F-regulated targets raise the possibility that the essential role of any individual pRB-associated protein may be limited and difficult to define.
In mammalian systems, the use of inhibitors such as TSA suggests that HDAC proteins are likely to be essential for repression at a subset of E2F targets (4, 30, 34, 68), and experiments with dominant-negative mutants indicate that BRG1 is required for the repression of the Cyclin A promoter (51). The elevated expression of several E2F-regulated genes (Cyclin E and Cyclin A) in mouse embryonic fibroblasts lacking Suv39H1 and Suv39H2 supports the idea that pRB-recruited methylases are involved in pRB-mediated repression (38), and a similar derepression of Cyclin D1 was also recently reported in differentiated cells treated with short interfering RNA to Suv39h (1).
The challenge of identifying the corepressors that mediate the repression of E2F-regulated promoters may be more easily addressed with Drosophila cells than mammalian cells. Drosophila contains two E2F genes (de2f1 and de2f2), one DP gene (dDP), and two pRB-related genes (rbf1 and rbf2), which all have properties similar to those of their mammalian counterparts. Thus, the Drosophila E2F/DP/RB pathway contains significantly fewer family members than mammalian cells, and hence the potential for functional redundancies would appear to be less. In addition, microarray analysis of RNA interference (RNAi)-treated cells has provided a detailed picture of the Drosophila E2F transcriptional program. This work has revealed several different classes of E2F-regulated promoters that depend on distinct combinations of dE2F and RBF proteins for their regulation (11). At some targets E2F regulation provides cell cycle-dependent oscillations in gene expression; at other targets dE2F/RBF proteins confer stable repression that is resistant to cell cycle progression. At both types of targets, repression requires RBF proteins and appears to be mediated through dE2F/dDP complexes.
Here we have taken advantage of this experimental system to investigate the role of a series of putative corepressors in the repression of two distinct classes of dE2F/RBF-regulated promoters. Given the high degree of functional conservation between the mammalian and Drosophila E2F/DP/pRB pathways, the detailed information about the Drosophila E2F transcriptional program, and the utility of RNAi in Drosophila tissue culture cells, this approach provides an interesting and direct test of whether individual proteins are required for the repression of endogenous dE2F/RBF regulated targets. Surprisingly, the results reveal no general requirement for HDAC proteins or SWI/SNF components in the repression of E2F-regulated genes. Instead we find that p55/dCAF-1 is essential for the repression of several E2F targets. Remarkably, p55 is specifically required for the repression of dE2F2-regulated promoters. Besides implicating p55 in RBF-mediated repression, these results strongly suggest that there is a fundamental difference in the way that RBF acts at E2F targets that are cell cycle regulated and the way that it acts at developmentally regulated dE2F2-repressed promoters.
MATERIALS AND METHODS
Cell culture and RNAi.
Drosophila melanogaster SL2 cells were cultured at 25°C in Schneider's insect medium (Sigma) supplemented with 10% fetal bovine serum. RNAi was performed as previously described with 50 μg of double-stranded RNA (dsRNA) (reference 53 and references therein). Samples for RNA isolation and Western blot analysis were collected 8 days after dsRNA administration. TSA (Sigma) and sodium butyrate (Sigma) were added to the culture medium in the concentrations described.
Western blot analysis and chromatin immunoprecipitation assays.
Immunoblotting was performed with standard techniques. Antibodies against the following proteins were used: dE2F1, dE2F2, dDP, RBF1, and RBF2 have been described (reference 53 and references therein); dRPD3 (gift from Pierre Spierer and David Wassarman); BRM (gift from John Tamkun); MOR (gift from C. Peter Verrijzer); OSA (gift from Jessica Treisman); SNR1 (gift from Andrew Dingwall); E(Z) (gift from Rick Jones); dimethyl histone-3-lysine-9 (Upstate Biotechnology); GRO (Developmental Studies Hybridoma Bank [DSHB]); dSIR2 (gift from Stefan Åström); p55 and p105 (gifts from Jessica Tyler); ESC and Su(Z)12 (gifts from Peter Harte); dSAP18 (gift from David Wassarman); ISWI (gift from Carl Wu); tubulin (E7 mouse ascites from DSHB). Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (11, 16). The following forward and reverse primers for CG8399 were used: 5′-TCCAATTTTCCAATTGTCAGCCGATG-3′ and 5′-GCAATTTTGATCTGCGCATTTCGG-3′, respectively.
RNA isolation, Northern blotting, and reverse transcription-PCR.
Total RNA was isolated from cells with Trizol (Invitrogen). Northern blotting was performed as previously described (16). Reverse transcription-PCR was achieved with the first-strand cDNA synthesis kit (Amersham) and performing reverse transcription-PCR for 25 cycles following standard procedures (47). Analysis of DNA was performed with a duplex PCR. The primer sets used were ISWI forward (5′-AGCTGTCGGAGATTCTGCGAG-3′) and reverse (5′-TGCATAAGGATGTTCTGCAGTCG-3′); PH-p forward (5′-AGCCAGCAACAGCAGCACTC-3′) and reverse (5′-GAAATGACGTTCATCGGTGAGAAG-3′); and RP49 forward (5′-CCAGCATACAGGCCCAAGATC-3′) and reverse (5′-AGGAACTTCTTGAATCCGGTGG-3′).
Fractionation and immunoprecipitation assays.
Nuclear extracts were prepared from dechorionated Drosophila embryos harvested 2 to 12 h after egg deposition as described previously (5); 250 μl of nuclear extract was applied to a 24-ml Superose 6 gel filtration column (Amersham-Pharmacia) equilibrated with 10 mM HEPES, pH 7.6, 300 mM KCl, 10% glycerol, and 0.5 mM phenylmethylsulfonyl fluoride (buffer EX300). The column was resolved with buffer EX300, and 500-μl fractions were collected. Even-numbered fractions were precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blot to determine the RBF1 elution profile; 250-μl aliquots of odd-numbered RBF1 peak fractions were immunoprecipitated with monoclonal RBF1 and c-Myc (control) antibodies with standard procedures.
Stable cell lines.
SL2 cell lines stably expressing Flag-tagged dRPD3, Flag-tagged RBF1, and Flag-tagged RBF2 were established and maintained as described previously (3).
Transient transfections.
Drosophila SL2 cells were RNAi-treated for 5 days with subsequent transfection with Cellfectin (Invitrogen) with 10 μg of plasmid DNA as described previously (53). Cells were harvested 46 h posttransfection and assayed for β-galactosidase and luciferase activities as described previously (16). In reporter assays, 2 μg each of reporter plasmid and pIE4-lacZ normalizing plasmid were transfected with increasing amounts of dE2F2. All reporter assays were performed in triplicate.
BrdU labeling.
Cells (2 × 106) were plated into a 60-mm dish. After 3 days, the HDAC inhibitor TSA or sodium butyrate was administered at the indicated concentrations, and samples were harvested 16 h post-inhibitor treatment. Samples were labeled with BrdU (Amersham). Samples were fixed, and BrdU was detected with a mouse anti-BrdU antibody (Becton Dickinson). Fluorescein-labeled anti-mouse immunoglobulin G (Vector Laboratories) was used as a secondary antibody. HDAC inhibitor-treated or RNAi-treated cells were labeled with BrdU and analyzed for BrdU incorporation with a Becton Dickinson FACScan and CellQuest program. The results generated were from multiple independent experiments.
RESULTS
dE2F- and RBF-mediated repression of E2F-regulated genes require different subsets of dE2F and RBF proteins.
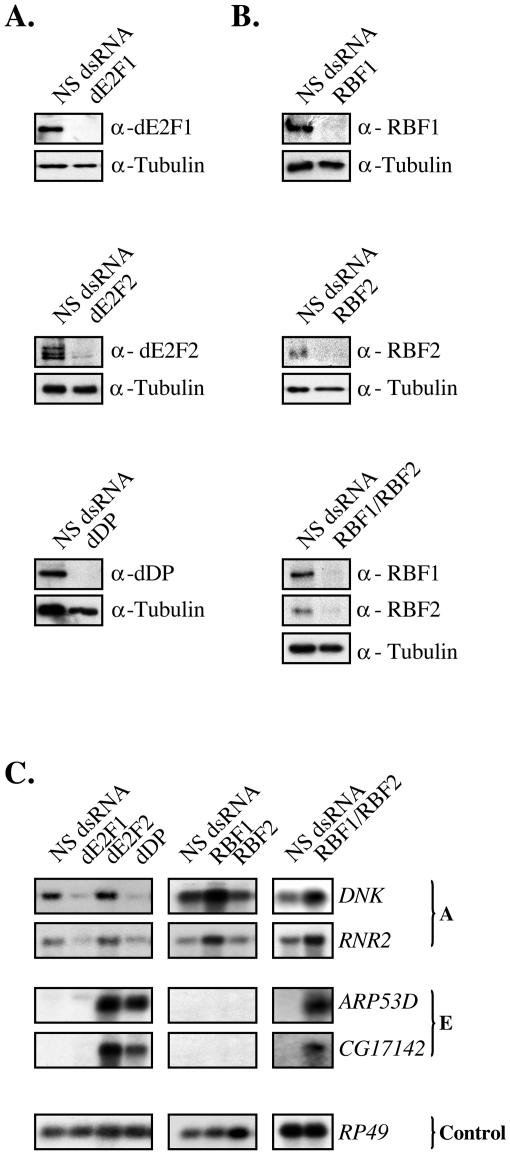
Dimova et al. subdivided Drosophila E2F-regulated genes into five different categories (groups A to E) based on the relative contributions of dE2F1/dDP to activation of gene expression or dE2F2/dDP to the repression of gene expression (11). In order to begin to examine the mechanisms underlying the RBF-mediated repression of these E2F targets, we first selected representatives of groups A and E, the two groups with clearly distinguishable forms of dE2F and RBF control, to use for further analysis (Fig. 1). In Fig. 1 and throughout this study, we monitored levels of tubulin to control for loading differences on the Western analyses and RP49 to control for the Northern blot experiments.
FIG. 1.
Representatives of group A and group E, two sets of differentially regulated dE2F target genes. (A) Western blot analysis of whole-cell extracts from dE2F RNAi-treated cells. Cells were incubated with dsRNA targeting dE2F1, dE2F2, or dDP for 8 days. A luciferase control was used as a nonspecific (NS) double-stranded RNA. Each blot was probed with two antibodies, one specific for the RNAi target protein and tubulin as a loading control. Note that three different forms of dE2F2 are recognized by multiple monoclonal and polyclonal antibodies to dE2F2. The reason for the varied mobility of dE2F2 is not known. (B) Same as panel A except cells were incubated with dsRNA targeting RBF1, RBF2, or both RBFs (RBF1/RBF2). (C) Northern blot analysis of total RNA extracted from RNAi-mediated depleted cells. Blots were probed for the expression of A genes (DNK and RNR2), E genes (ARP53D and CG17142), and RP49 (control).
DNK and RNR2 were chosen to represent group A. As illustrated in Fig. 1C (middle panel), these transcripts are normally expressed in asynchronously dividing SL2 cells and the depletion of RBF1 but not RBF2 increases gene expression. Hence these genes are repressed specifically by RBF1. ARP53D and CG17142 were chosen to represent group E. The expression of these genes is strongly repressed in cycling SL2 cells, however transcripts are readily detected when RNAi is used to remove dE2F2 or dDP (Fig. 1C, left panel). No expression of ARP53D and CG17142 was seen when RBF1 or RBF2 was removed individually, but they were expressed when both RBF1 and RBF2 were depleted (Fig. 1C, middle and right panels). Hence these genes appear to be constitutively, and redundantly, repressed in SL2 cells by dE2F2/dDP/RBF1 and dE2F2/dDP/RBF2 complexes. DNK, RNR2, ARP53D, and CG17142 were suited for this analysis because (i) their transcripts were readily detected on Northern blots, (ii) the changes seen in the RNAi depleted cells were robust, and (iii) the changes following RNAi were representative of the behavior of group A and group E genes in general. Earlier ChIP experiments have shown that these promoters are directly regulated by dE2F/RBF proteins (with dE2F1, dE2F2, dDP, and RBF1 being present at DNK and other A group gene promoters and dE2F2, dDP, RBF1, and RBF2 being present at ARP53D, CG17142, and other E group gene promoters) (11). Thus, A and E group genes are repressed by RBF proteins. However, both groups exhibit different types of dE2F regulation.
HDAC inhibition fails to derepress most A and E group E2F targets.
Several groups have proposed the repression of E2F targets is carried out through the recruitment of HDAC activities by pRB or pRB family members (6, 15, 24, 30, 31, 51). Thus, we began by examining the role of Drosophila HDACs in repression of the A and E group genes.
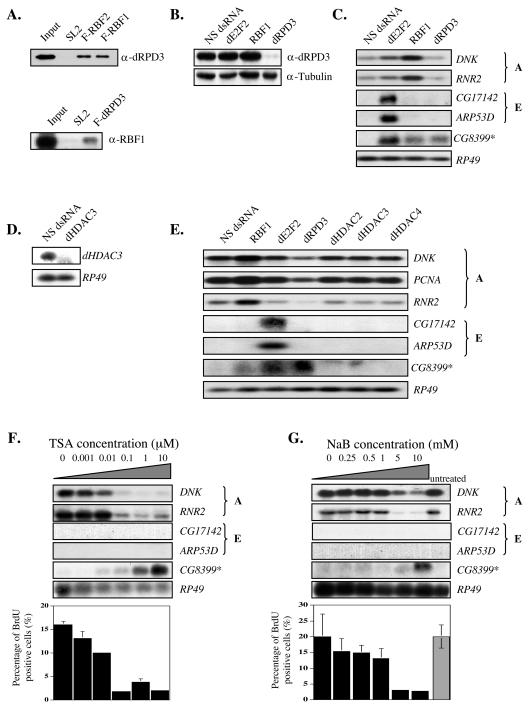
We first asked whether RBF1 interacts with dRPD3, the best-characterized Drosophila HDAC that has been shown to mediate the effects of other corepressor proteins. Immunoprecipitation and Western blot analysis with stable cell lines expressing Flag-tagged versions of RBF1, RBF2, or dRPD3 showed that dRPD3 coimmunoprecipitates with Flag-RBF1 and Flag-RBF2 and that RBF1 coimmunoprecipitates with Flag-dRPD3 (Fig. 2A). To test the significance of this physical interaction in E2F regulation, we used RNAi to deplete dRPD3 and examined the effects on E2F-regulated promoters. Depletion of dRPD3 failed to relieve repression of the E group genes and, surprisingly, gave a decrease in A group gene transcripts rather than the increase that would have been expected if the promoters were repressed by pRB/HDAC complexes (Fig. 2B and C). Since there are three additional Drosophila HDAC homologs (dHDAC2/HDAC6, dHDAC3, and dHDAC4) that might be alternative candidates for an RBF-recruited repressor, we treated cells with RNAi to each of these. Depletion of these dHDACs had no effect on the expression of either A or E group genes (Fig. 2D and E).
FIG. 2.
Inhibiting HDAC activity does not increase the expression of group A or group E dE2F targets. (A) RBFs associate with the dRPD3 histone deacetylase. The upper panel shows extracts from control SL2 cells and SL2 cells stably expressing Flag-tagged RBF1 (F-RBF1) and RBF2 (F-RBF2). Extracts were subjected to affinity purification with anti-Flag-coupled beads. After binding and washing, complexes were then eluted with Flag peptide and subjected to SDS-PAGE and Western blot analysis with a dRPD3 antibody. The lower panel shows extracts from control SL2 cells and SL2 cells stably expressing Flag-tagged dRPD3 (F-dRPD3). Extracts were treated as in the upper panel except an RBF1 antibody was used. (B) Western blot analysis of whole-cell extracts from dRPD3 RNAi-treated cells. Cells were incubated with NS, dE2F2, RBF1, or dRPD3 dsRNA. The blot was probed with antibodies to dRPD3 and tubulin as a control. (C) Northern blot analysis of cell extracts used in panel B. Blots were probed for the same E2F target genes as in Fig. 1C in addition to an atypical D group gene, CG8399. CG8399 served as an internal positive control for the effectiveness of the dRPD3 RNAi and is labeled with an asterisk (*) throughout the paper to denote the fact that its properties are unusual and do not appear to be representative of E2F targets in general. (D) Northern blot analysis of dHDAC3 RNAi-treated cells. Cells were incubated with NS or dHDAC3 dsRNA. Blots were probed for dHDAC3 or RP49 as a control. RNAi of dHDAC2 and dHDAC4 could not be verified via Western blot or Northern blot analyses due to lack of antibody and detection sensitivity, respectively. Reverse transcription-PCRrevealed that Drosophila HDAC2 and dHDAC4 were slightly expressed despite RNAi treatment targeting each of these homologs. (E) Northern blot analysis of cell extracts from Drosophila HDAC RNAi-treated cells. Cells were incubated with dHDAC2, dHDAC3, or dHDAC4 in addition to NS, dE2F2, RBF1, or dRPD3 dsRNA as controls. Blots were probed for the same E2F target genes as in panel C in addition to PCNA (an A group gene). (F) Cells were treated with TSA for 18 h at the indicated concentrations (0 to 10 μM) with subsequent Northern blot analysis probing for the same genes as in panel C. The bar graph shows the percentage of BrdU-positive cells. BrdU incorporation experiments were performed in triplicate; no standard deviation is shown when it was less than 1% of the data points. (G) Same as in panel F but with 0 to 10 mM sodium butyrate (NaB). Untreated cells were included to serve as an additional control to the mock-treated control (0 mM).
Plausibly, the lack of derepression of RBF-repressed genes in the RNAi-treated cells could be due to functional redundancy between dHDACs. As an alternative approach, we used the well-characterized HDAC inhibitors TSA and sodium butyrate to provide a more general knockdown of HDAC activity. Titration of TSA and sodium butyrate had no effect on the repression of the group E genes ARP53D and CG17142. In a manner that was reminiscent of the effects of dRPD3 RNAi, the addition of TSA and sodium butyrate caused a decrease in A group gene expression (Fig. 2F and G, upper panels). This decrease correlated with a decrease in the number of S-phase cells and may be a secondary effect of changes in the cell cycle distribution of the RNAi-treated cells (Fig. 2F and G, lower panels).
During the course of these experiments, we examined the expression of CG8399, an atypical member of group D genes that has the unusual characteristic of being derepressed in cells depleted of RBF1 alone. Interestingly, expression of CG8399 is strongly increased in cells depleted of dRPD3 or in cells treated with TSA or sodium butyrate. We have examined additional A and E group genes, but CG8399 is the only E2F target gene that we have found, thus far, that shows this effect. These changes confirm that the inhibition of HDAC activity resulting from the TSA, sodium butyrate, and RNAi treatments is sufficient to give clear changes in gene expression, but these treatments do not result in the derepression of any of A or E group genes tested. We cannot formally exclude the possibility that HDACs repress A group genes in S phase and that a decrease in the percentage of S-phase cells might mask an increase in gene expression. Nevertheless, these results indicate that although RBF proteins associate with HDAC proteins, there is no general requirement for HDAC activity for the repression of either of these types of E2F-controlled promoters.
p55 is required specifically for the repression of E group genes.
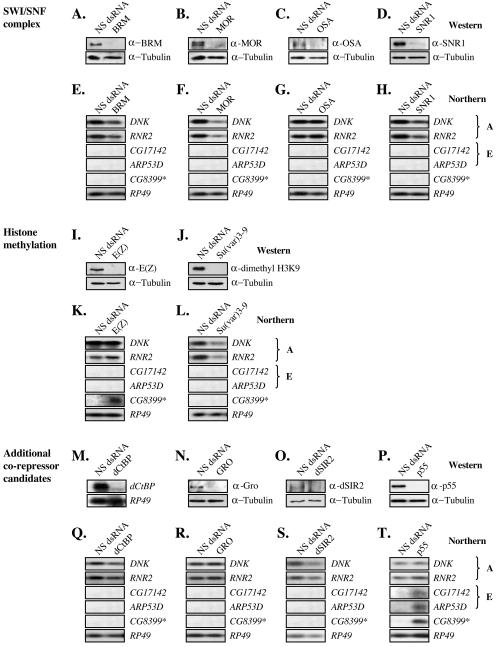
Since Drosophila HDACs appeared to be unimportant for the repression of E2F target genes, we expanded the study to examine the role of other potential coregulators. We compiled a list of candidates that included components of the SWI/SNF complex, methyltransferases, and other potential corepressor proteins and used Northern blot analysis of RNAi-treated cells to ask whether these proteins were required for repression of the selected A and E group E2F targets. In the majority of cases we saw no change in the expression of E2F targets in the RNAi-treated cells (Fig. 3). The limitation of this approach, as with any high-throughput RNAi approach, is that it was not always possible to confirm the depletion of the relevant protein. In some cases antibodies have not been described, in other cases antibodies exist but lacked the required specificity or sensitivity. Shown in Fig. 3 are the examples where we were able to confirm that the protein or mRNA levels were substantially decreased by the RNAi treatment, including BRM, MOR, OSA, SNR1, E(Z), Su(var)3-9, dCtBP, GRO, dSIR2, and p55. In addition, no derepression was seen in cells treated with RNAi to SKI, SMOX, SMR, Bx42, DNMT2, or BAP60 or following treatment with 5-azacytidine, a DNA methyltransferase inhibitor (data not shown).
FIG. 3.
Influence of candidate Drosophila pRB corepressors on dE2F target gene expression. (A to D) Western blot analysis of whole-cell extracts from cells incubated with dsRNA for SWI/SNF subunits (A) BRM, (B) MOR, (C) OSA, or (D) SNR1. The blots were probed with the appropriate antibodies to BRM, MOR, OSA, and SNR1, respectively. Tubulin served as a control. (E to H) Corresponding Northern blot analysis of extracts used in panels A to D. Extracts were probed with E2F target genes as used in Fig. 2C. (I and J) Western blot analysis of whole-cell extracts from cells incubated with dsRNA for histone methyltransferases (I) E(Z) and (J) Su(var)3-9. Blots were probed with antibodies to (I) E(Z) and (J) dimethyl histone-3-lysine-9 to serve as an indirect measurement of Su(var)3-9 depletion. (K and L) Corresponding Northern blot analysis of the extracts used in panels I and J probed with the E2F target genes used in Fig. 2C. (N to P) Western blot analysis of whole-cell extracts from cells incubated with dsRNA for additional corepressor candidates (N) GRO, (O) dSIR2, or (P) p55. Blots were probed with the appropriate antibodies to GRO, dSIR2, p55, and tubulin as a control. (N) In the case of dCtBP, depletion of dCtBP was detected by Northern blot analysis. (Q to T) The extracts used in panels M to P were probed with E2F target genes as used in Fig. 2C.
SWI/SNF complexes have previously been proposed to play an important role in pRB-mediated repression. However, we saw no increase in the expression of A or E group genes in cells depleted of BRM, MOR, OSA, or SNR1 (Fig. 3A to H). Indeed, transcripts from A group genes were decreased following RNAi-mediated depletion of BRM, MOR, and SNR1. It is unclear whether this decrease occurs because SWI/SNF complexes are needed for activation of gene expression or as an indirect effect of a reduction in S-phase population in the RNAi treated cultures (data not shown). Nevertheless these results provide no evidence to support the idea that SWI/SNF complexes have an essential role in RBF-mediated repression.
This initial survey revealed two proteins that were required for the repression of E2F-regulated targets. The depletion E(Z) resulted in the upregulation of CG8399 but had no effect on the E group genes, or on A group genes tested (Fig. 3I and 3K). In addition, the depletion of p55 resulted in the derepression of CG8399 and both E group genes tested, but had no effect on A group genes (Fig. 3P and 3T). The expression of other E group (CG3505, CG3105) and D group (CG8316) genes was also increased following depletion of p55, suggesting that the changes are likely to be common features of both D and E group E2F targets (data not shown). We conclude that D and E group genes are repressed by a mechanism that is different from A group genes. These results raise the possibility that dE2F2/RBF-regulated genes are controlled by a p55-dependent process.
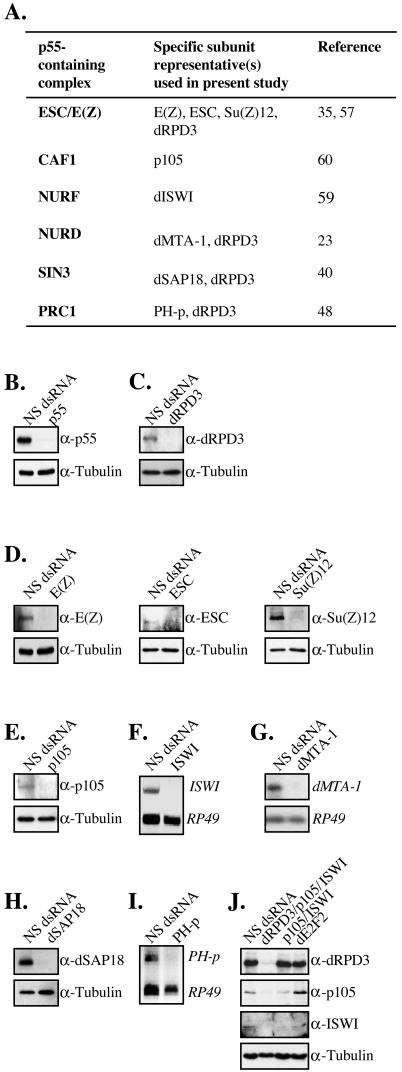
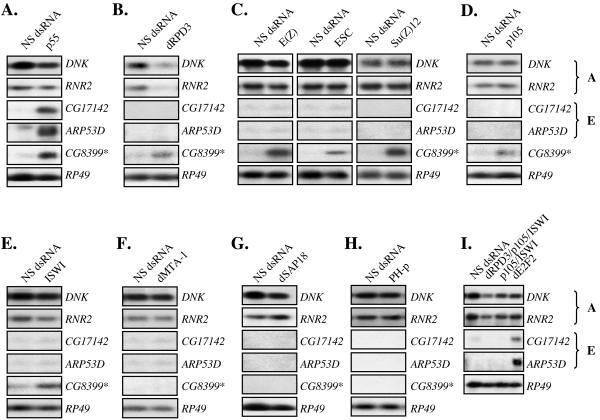
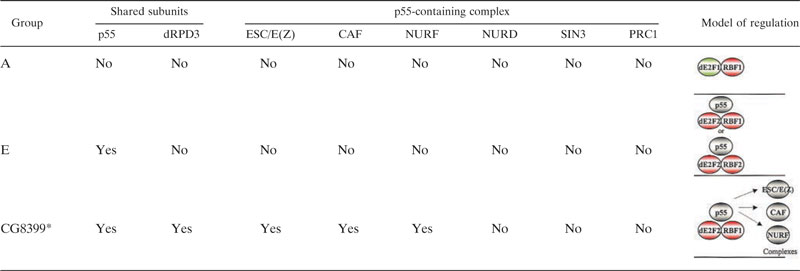
p55 is a histone-binding protein (61). The human ortholog was initially isolated as an RB-binding protein (RbAp46/48), but it subsequently has been shown to be a component of several chromatin-associated complexes (28, 41, 44). Therefore, to extend our observations, we examined several p55-containing complexes, including ESC/E(Z), CAF, NURF, NURD, SIN3, and PRC1, by performing additional rounds of RNAi treatment to deplete previously described p55-associated proteins (Fig. 4). The expression of CG8399 was derepressed following RNAi treatment to five different components of the ESC/E(Z) complex, E(Z), ESC, Su(Z)12, p55, and dRPD3, supporting the idea that this complex is required for repression of CG8399 (Fig. 5A to 5C). However, CG8399 expression was also detected following RNAi depletion of the CAF subunit p105 and the ISWI subunit of the NURF complex, indicating that several different p55-containing complexes affect the transcription of this gene (Fig. 5D and 5E).
FIG.4.
Specific depletion of p55-associated subunits by RNAi. (A) Drosophila p55-containing complexes examined in the present study, listing representatives used for subsequent RNAi. (B to E, H, and J) Western blot analysis of components of p55-containing complexes depleted by RNAi. Antibodies to p55, dRPD3, E(Z), ESC, Su(Z)12, p105, dSAP18, and ISWI were used to monitor the depletion of the corresponding target, and tubulin was used as a control. (F and I) SL2 cells were incubated with double-stranded RNA for ISWI or PH-p. Total RNA was extracted and reverse transcribed into cDNA. Specific depletions were confirmed by subjecting the cDNA to linear PCR amplification with a duplex reaction comprising specific primers for ISWI or PH-p, with RP49 as a control. (G) Northern blot analysis of dMTA-1 RNAi-treated cells. Blots were probed for dMTA-1 or RP49 as a control.
FIG. 5.
dE2F2/RBF target (group E) genes are not regulated by the p55-containing complexes examined. (A to I) Northern blot analysis of the RNAi-treated cells corresponding to the extracts shown in Fig. 4. The blots were probed sequentially with the same E2F target genes used in Fig. 2C.
In contrast to CG8399, the repression of CG17142 and ARP53D was unchanged by these treatments. Notably, repression of CG17142 and ARP53D was maintained in cells treated with RNAi to components of the ESC/E(Z) complex [E(Z), ESC, and Su(Z)12], the CAF complex (p105, p180), the NURF complex [ISWI, NURF38, and E(bx)], the NURD complex (dMTA1-like, dMBD2/3, and dMi2), the SIN3 complex (dSin3, dSAP18, and CG4756/dSAP30), or PRC1 complex (PH-p) (Fig. 5, summarized in Table 1, and data not shown). Moreover, E group genes remained repressed in cells treated with multiple dsRNAs that led to the depletion of CAF p105 and ISWI, or the depletion of dRPD3, CAF p105, and ISWI (Fig. 4J and 5I).
TABLE 1.
Divergent requirement for p55 between A and E group genesa
Derepression of A and E group genes directed by p55, dRPD3, or various p55-containing complexes, ESC/E(Z), CAF, NURF, NURD, SIN3, and PRC1. Derepression is denoted by yes and lack of derepression by no. See Fig. 2 legend for explanation of the asterisk.
These results suggest three general possibilities to explain the specific requirement for p55. Either the repression of CG17142 and ARP53D E group promoters is mediated specifically by p55, or it may require a p55-containing complex that we have not yet identified, or it may be mediated by two or more redundantly acting complexes that have a common dependency on p55.
Links between p55 and dE2F/RBF.
Four additional lines of evidence support the idea that p55 is involved in the repression of E group genes by dE2F/RBF proteins.
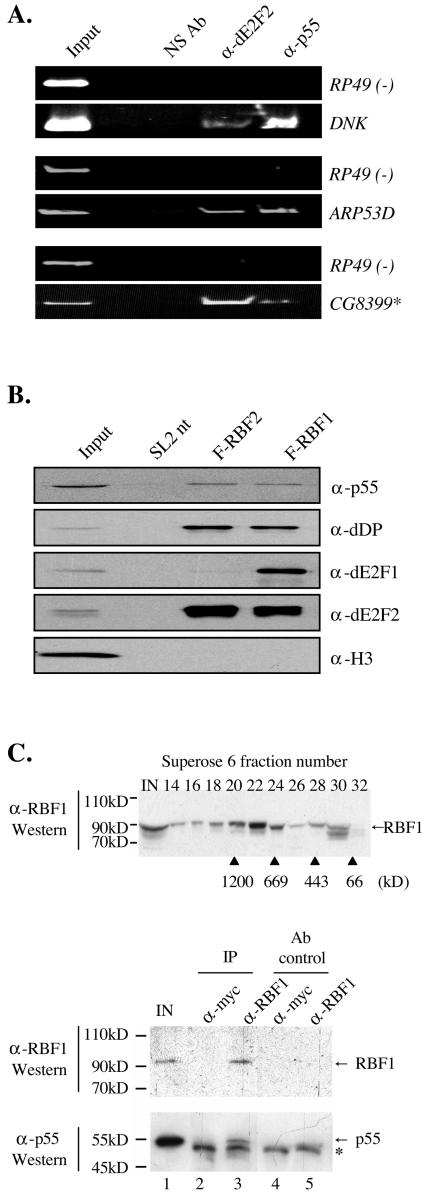
First, ChIP experiments showed that p55 is present at D and E group promoters (Fig. 6A). Immunoprecipitates prepared with antisera to both dE2F2 and p55 showed a strong enrichment of ARP53D and CG8399 promoter sequences over a control sequence (RP49). Interestingly, ChIP experiments showed that p55 is also present at the A group promoter DNK. This result mirrors previous ChIP data showing that dE2F2 is present at all five groups of E2F targets (A to E) but is only rate limiting for gene expression at a subset of targets (C to E) (11). Hence p55, like dE2F2, has the opportunity to act at multiple E2F-regulated promoters.
FIG. 6.
p55 associates with dE2F and RBF proteins. (A) p55 binds to the E gene promoter ARP53D. Antibodies against dE2F2, p55, and rabbit anti-mouse serum (NS Ab) were used to enrich for the DNK, ARP53D, and CG8399 promoter regions. (B) p55 immunoprecipitates with both RBF1 and RBF2. Extracts from control SL2 cells and SL2 cells stably expressing Flag-tagged RBF1 (F-RBF1) and RBF2 (F-RBF2) were subjected to affinity purification with anti-Flag-coupled beads. After binding and washing, complexes were then eluted withFlag peptide and subjected to SDS-PAGE and Western blot analysis with a p55 antibody. dDP, dE2F1, and dE2F2 immunoprecipitates served as positive controls and histone 3 (H3) as a negative control. (C) RBF1 exists in multisubunit complexes. (Upper panel) Drosophila embryo nuclear extracts were fractionated over a Superose 6 sizing column. Fractions were precipitated with trichloroacetic acid and then probed for RBF1 by Western blot analysis. Molecular size standards are shown at the bottom of the upper panel. (Lower two panels) Aliquots of fraction 21 (1 MDa) were immunoprecipitated with anti-RBF1 antibody (α-RBF1) or an irrelevant antibody (α-Myc). Immunoprecipitates were analyzed by Western blot analysis for the presence of RBF1 (∼90-kDa band in upper panel, lanes 1 and 3) and p55 (55-kDa band in lower panel, lanes 1 and 3) showing RBF1 complexes contain p55. The controls include fraction 21 immunoprecipitated with anti-Myc (lane 2), and aliquots of fractionless immunoprecipitation buffer immunoprecipitated with anti-RBF1 (lane 4) or anti-Myc (lane 5). The presence of RBF1 and p55 in the RBF1 immunoprecipitate (lane 3, upper panel) and their absence in Myc and control antibody immunoprecipitates (lanes 2, 4, and 5) indicate the specific association of RBF1 and p55. Lane 1 refers to the input (IN), and the asterisk (*) refers to the immunoglobulin heavy chain. Molecular size standards are shown to the left of all three panels.
Second, biochemical studies show p55 associates with the RBFs. p55 coimmunoprecipitates specifically with RBF1 and RBF2 from Drosophila SL2 cells stably expressing Flag-tagged proteins (Fig. 6B). In addition, a crude fractionation of Drosophila embryo nuclear extracts revealed the existence of large (approximately 1 MDa) RBF-containing complexes (Fig. 6C), from which p55 was found to coimmunoprecipitate with RBF1.
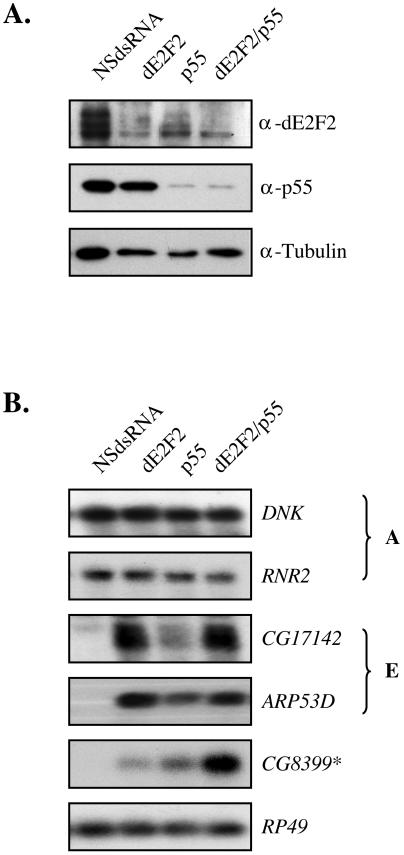
Third, to investigate whether dE2F2 and p55 act together or independently at E group promoters we examined the effects of codepleting both proteins. RNAi to both p55 and dE2F2 gave no additive derepression of the ARP53D and CG17142 promoters than targeting either protein alone, consistent with the idea that they act in the same complex (Fig. 7B). In contrast, removing both proteins had a strongly synergistic effect on the derepression of CG8399, consistent with the idea that p55 is a component of several different repressor complexes that act on this promoter.
FIG. 7.
Promoter-specific synergy between p55 and dE2F2. (A) Western blot analysis of whole-cell extracts from dE2F2 and p55 coRNAi-treated cells. Cells were incubated with dsRNA targeting dE2F2, p55, or both proteins. Each blot was probed with antibodies to dE2F2, p55, and tubulin as a loading control. (B) Corresponding Northern blot analysis to the samples used in panel A. Blots were probed with the same target genes as in Fig. 2C.
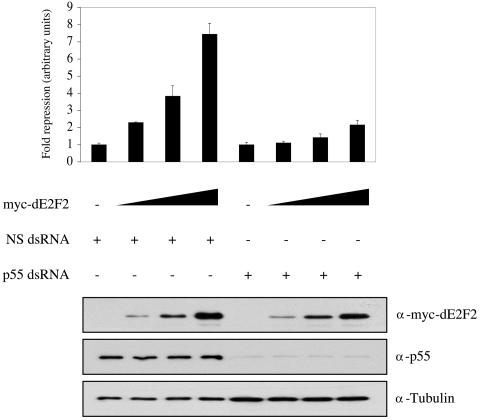
Fourth, to determine whether dE2F2 repression requires p55 we depleted p55 by RNAi and examined the ability of dE2F2 to repress transcription from an E2F reporter construct in transient transfection assays. Although the removal of dE2F2 has only a minimal effect on the expression of PCNA in rapidly dividing SL2 cells (Fig. 2) (11, 16), previous studies have shown that dE2F2 is present at the endogenous PCNA promoter and ectopic expression of dE2F2 is able to repress transcription from PCNA reporter constructs in SL2 cells (16). dE2F2-mediated repression in this assay depends on the E2F-binding sites in the reporter, can be titrated, is reversed by dE2F1, and requires RBF proteins (16, 17, 53). We note that this type of assay cannot be done with E group promoters because E group genes are fully repressed in SL2 cells by the endogenous dE2F2 proteins; the reporters that we have generated containing upstream sequences from E group genes do not give any measurable activity in SL2 cells to repress (E.-J. Kwon and N. Dyson, unpublished data). With the PCNA reporter as an artificial assay for dE2F2-mediated repression, we cotransfected it with increasing amounts of dE2F2 expression plasmid into cells treated with either white (control) or p55 RNAi. As shown in Fig. 8, the repressor activity of dE2F2 was severely compromised in cells lacking p55, suggesting that most of the repressive effect of dE2F2 is mediated via p55 or p55-dependent complexes.
FIG. 8.
dE2F2-mediated repression is severely compromised in the absence of p55. SL2 cells were treated with white (NS) or p55 double-stranded RNA and transfected with the PCNA reporter along with increasing amounts of pIE4-mycdE2F2 (0, 50, 150, and 450 ng). The level of downregulation of the PCNA reporter construct in the different conditions tested is expressed as the decrease compared to the activity of the PCNA reporter in the absence of exogenous dE2F2 (repression). Ectopic expression of dE2F2 represses a dE2F/RBF-responsive target gene (PCNA, group A) in a dose-dependent manner. dE2F2-mediated repression is increased up to sevenfold but impaired to twofold in the absence of p55. Western blot analysis with anti-Myc and anti-p55 antibodies confirm the dose-dependent delivery of dE2F2 and the depletion of p55, respectively.
DISCUSSION
The role and regulation of the E2F/pRB pathway is conserved between mammals and Drosophila, and it seems likely, therefore, that pRB-related proteins use similar mechanisms to repress transcription in different species. Attempts to investigate this subject in mammalian cells have faced two major obstacles. First, there is a great deal of uncertainty over the identity of the mammalian promoters that are directly, and specifically, controlled by pRB-mediated repression. While E2F is the best-known target of pRB regulation, there is disagreement in the literature over whether pRB is present at most E2F-regulated promoters, and it is uncertain precisely when the repression of E2F-regulated genes can be attributed specifically to pRB as opposed to p107 or p130. Second, until recently it has not been possible to easily inhibit gene function in mammalian cells. Consequently, most studies of pRB-linked repressors have relied on overexpression experiments, artificial promoter constructs, and/or fusion proteins that allow pRB to repress unnatural targets to implicate pRB-associated proteins in transcriptional repression. While several different mechanisms of pRB-mediated repression clearly can occur, it is largely uncertain which of these processes are required for endogenous pRB proteins to repress the expression of endogenous genes. Indeed, given the myriad of possibilities and the likelihood that these operate redundantly, it was quite conceivable that no single protein might prove to be generally required for RB-mediated repression.
Here we have taken advantage of the recent identification of sets of Drosophila promoters that are directly regulated by dE2F and RBF proteins and the emergence of RNAi-based approaches to investigate this issue. Of the candidates tested, the most important player in RBF-mediated repression appears to be p55. The results show that p55 is essential for the repression of E (and D) group genes, E2F-regulated genes that are stably repressed by dE2F2/RBF complexes in cycling cells. Surprisingly, the depletion of p55 had no effect in the expression of A group genes even though both sets of E2F targets are known to be directly regulated by RBF proteins (see Table 1). This distinction strongly suggests that the mechanism of RBF-mediated repression is different at the two types of E2F target genes. One simple model, based on the fact that the two sets of genes are primarily dependent on different dE2Fs, is that A group genes are predominantly controlled by dE2F1-mediated activation and the inhibition of activation by RBF1, whereas E group genes are controlled by active repression through dE2F2/dDP/RBF1 or dE2F2/dDP/RBF2 complexes that is mediated by p55.
p55 is a Drosophila ortholog of RbAp46/48, two closely related proteins that were originally identified through their ability to associate with pRB in vitro (41, 61). p55, like RbAp46/48, physically associates with RBF proteins, and ChIP experiments confirm that it is indeed present at E group promoters. p55 and RbAp46/48 show functional similarity, interacting with histones and chromatin, but they are not known to possess any intrinsic enzymatic activity. We cannot formally exclude the possibility that p55 acts alone as an RBF-recruited repressor, but p55 and RbAp46/48 are best known as components of large chromatin-associated complexes and most likely act in conjunction with other proteins to modify histones and/or remodel nucleosomes (2, 28, 44, 63, 67). In keeping with this, we were able to coimmunoprecipitate p55 and RBF1 from size-separated fractions containing complexes of approximately 1 MDa.
We have used RNAi to deplete components of several well-studied p55-containing complexes, including the CAF, NURF, NURD, SIN3, PRC1, and ESC/E(Z) complexes, but none of these gave the full changes in expression that we found by removing p55. This may indicate that E group genes are repressed by multiple, redundantly acting complexes, each containing p55 as an essential component. Alternatively, repression of E group genes may be carried out by a novel p55-containing complex. Interestingly, the expression of E genes was higher in dE2F2-depleted cells than in p55-depleted cells (Fig. 7B). This could be explained in two ways. One explanation is that dE2F2 is more important for repression than p55. This situation might arise if dE2F2 is part of multiple repressor complexes, only some of which contain p55 as an essential component. An alternative explanation is that this simply reflects differences in the extent of functional inactivation achieved by RNAi.
Further insight into the mechanism of repression issue will likely require the biochemical purification of endogenous dE2F/RBF complexes. It is intriguing that a heterologous experimental system, in which human pRB was found to repress transcription in Saccharomyces cerevisiae revealed a requirement for the p55 ortholog msi1 (24). Furthermore, microinjection experiments have suggested that RbAp48 may be required for the repression of an E2F-driven reporter (37). In addition, several of the complexes that have been suggested to be involved in pRB-mediated repression in mammalian cells are known to contain RbAp46/48 (25, 37, 62). The Caenorhabditis elegans homolog of p55, lin-53 (rba-1), is a member of the same class of synthetic multivulval mutants as lin35 (the worm pRB homolog), suggesting that their products may act in association with one another (29). Thus, it seems quite likely that RB-related proteins may share a generalized and conserved requirement for p55/RbAp46/48-related proteins in active repression.
HDACs and SWI/SNF proteins have been proposed previously to have critical roles in RB-mediated repression, but the importance of these activities was not evident in this study. Although it is difficult to draw strong conclusions from negative data, we were surprised by the consistent lack of derepression of E2F-regulated genes in multiple experiments that targeted HDAC and SWI/SNF activity. The simplest interpretation is that these proteins are not generally required for RBF-mediated repression. Possibly, the roles of SWI/SNF and HDAC complexes may be redundant with other repressors and therefore are not apparent in this specific type of loss-of-function experiment. The derepression of CG8399 in several different RNAi-treated cells illustrates the potential for multiple repressor complexes, although, intriguingly, none of the complexes implicated in the repression of this promoter appear to be able to compensate fully for one another.
Another possibility is that these cells lack a signal needed to activate certain repressor complexes. For example, SWI/SNF proteins have recently been reported to be specifically recruited to E2F-regulated promoters following DNA damage (27). It is a formal possibility that none of the treatments described here to target HDAC and SWI/SNF complexes were sufficient to fully inactivate the endogenous functions. We targeted these complexes in multiple ways in order to minimize this possibility. Since the RNAi depletions or pharmacological agents were sufficient to affect the expression of other genes and to interfere with cell cycle progression, we think that this is unlikely to be the correct explanation for the lack of derepression of E2F targets. As RNAi-mediated depletion of dRPD3, MOR, or BRM reduced the number of S-phase cells, it is conceivable that the changes in cell cycle distribution were sufficient to mask some degree of derepression at A group genes. Finally, we note that group E genes seem to be permanently repressed by dE2F2/RBF complexes in SL2 cells. These RNAi assays may be best suited to identifying proteins that are important for the maintenance of repression rather than the proteins responsible for establishing the repressed state. If HDAC and SWI/SNF act early in the repression of E group genes but are not needed to maintain this effect, then the inhibition of these activities might not give a dramatic change.
This initial study explored only a sample set of dE2F/RBF targets, and future genome-wide studies will be needed to determine how the distinctions between group A and group E genes segregate across the full range of dE2F/RBF-regulated genes. Nevertheless, these results clearly show that different types of dE2F/RBF target genes have different requirements for corepressor proteins. Hence, the question of what coregulators are needed for RBF proteins to repress transcription will have several different answers, and these will vary depending on the type of E2F control that is operative at a given promoter. Moreover, the differences between CG8399 (atypical D group gene) and ARP53D/CG17142 (E group genes) in the RNAi-treated cells highlight the fact that even within a general subtype of E2F targets (dE2F2-repressed genes) individual dE2F-regulated promoters may well depend on different combinations of corepressor complexes.
Despite this complexity, there are two positive conclusions that emerge from this study. The first is that, although redundancy may exist between dE2F/RBF-recruited corepressors, some individual components are required for RBF-mediated repression. Hence, this is a tractable problem, and with RNAi, it is possible to determine which of the many proteins that have been proposed to be important for pRB-mediated repression are indeed required for it to function at its endogenous targets. The second conclusion is that clearly definable themes will emerge from these approaches; that is, groups of dE2F/RBF targets that have a similar pattern of regulation will likely share a common dependency on a certain type of corepressor. The fact that repression of D and E group genes requires p55 is one example, and one wonders what other proteins are required for RBF to act at each of the various subgroups of E2F targets. Several subclasses of E2F-regulated genes have been identified in Drosophila melanogaster and further examination of this issue may be more tractable in Drosophila cells than mammalian cells, where the sheer size of the E2F/DP/pRB families may make it difficult to distinguish and classify different types of E2F control.
Acknowledgments
We thank Dessislava Dimova, Anders Näär, and Michael Pazin for critical reading of the manuscript. We are grateful to Lori Pile and David Wassarman for sharing the RNAi protocol and Stefan Åström, Andrew Dingwall, Rick Jones, Alexander Mazo, Peter Harte, John Tamkun, Pierre Spierer, Jessica Treisman, Jessica Tyler, David Wassarman, Carl Wu, and C. Peter Verrijzer for generously providing reagents. We thank Olivier Stevaux, Marie Classon, Fred Dick, Simon Boulton, Jeremy Mason, Louis Mahadevan, and members of the Dyson, Näär, Hariharan, and Harlow laboratories for advice and fruitful discussions. Technical assistance with the RNAi and chromatin immunoprecipitation assays was kindly provided by O.S. and D.D., respectively.
This work was supported by NIH grant GM53203 to N.D.
REFERENCES
- 1.Ait-Si-Ali, S., V. Guasconi, L. Fritsch, H. Yahi, R. Sekhri, I. Naguibneva, P. Robin, F. Cabon, A. Polesskaya, and A. Harel-Bellan. 2004. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 23:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 3.Bouazoune, K., A. Mitterweger, G. Langst, A. Imhof, A. Akhtar, P. B. Becker, and A. Brehm. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21:2430-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutillier, A. L., E. Trinh, and J. P. Loeffler. 2002. Constitutive repression of E2F1 transcriptional activity through HDAC proteins is essential for neuronal survival. Ann. N. Y. Acad. Sci. 973:438-442. [DOI] [PubMed] [Google Scholar]
- 5.Brehm, A., G. Langst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 7.Chellappan, S. P., S. Hiebert, M. Mudryj, J. M. Horowitz, and J. R. Nevins. 1991. The E2F transcription factor is a cellular target for the RB protein. Cell 65:1053-1061. [DOI] [PubMed] [Google Scholar]
- 8.Classon, M., and N. Dyson. 2001. p107 and p130: versatile proteins with interesting pockets. Exp. Cell Res. 264:135-147. [DOI] [PubMed] [Google Scholar]
- 9.Dahiya, A., S. Wong, S. Gonzalo, M. Gavin, and D. C. Dean. 2001. Linking the Rb and polycomb pathways. Mol. Cell 8:557-569. [DOI] [PubMed] [Google Scholar]
- 10.de Bruin, A., B. Maiti, L. Jakoi, C. Timmers, R. Buerki, and G. Leone. 2003. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278:42041-42049. [DOI] [PubMed] [Google Scholar]
- 11.Dimova, D. K., O. Stevaux, M. V. Frolov, and N. J. Dyson. 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17:2308-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Stefano, L., M. R. Jensen, and K. Helin. 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22:6289-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 14.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira, R., L. Magnaghi-Jaulin, P. Robin, A. Harel-Bellan, and D. Trouche. 1998. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. USA 95:10493-10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frolov, M. V., D. S. Huen, O. Stevaux, D. Dimova, K. Balczarek-Strang, M. Elsdon, and N. J. Dyson. 2001. Functional antagonism between E2F family members. Genes Dev. 15:2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolov, M. V., O. Stevaux, N. S. Moon, D. Dimova, E. J. Kwon, E. J. Morris, and N. J. Dyson. 2003. G1 cyclin-dependent kinases are insufficient to reverse dE2F2-mediated repression. Genes Dev. 17:723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuyama, T., F. Tie, and P. J. Harte. 2003. Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis 35:114-124. [DOI] [PubMed] [Google Scholar]
- 19.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 20.Helin, K., E. Harlow, and A. Fattaey. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 13:6501-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiebert, S. W. 1993. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRB-mediated growth suppression. Mol. Cell. Biol. 13:3384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, B. K., O. W. Liu, F. A. Dick, N. Dyson, E. Harlow, and M. Vidal. 2001. Histone deacetylase-dependent transcriptional repression by pRB in yeast occurs independently of interaction through the LXCXE binding cleft. Proc. Natl. Acad. Sci. USA 98:8720-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21:2918-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam, E. W., and N. B. La Thangue. 1994. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr. Opin. Cell Biol. 6:859-866. [DOI] [PubMed] [Google Scholar]
- 27.Liu, K., Y. Luo, F. T. Lin, and W. C. Lin. 2004. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRB-independent and E2F1-specific control for cell survival. Genes Dev. 18:673-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677:3-11. [DOI] [PubMed] [Google Scholar]
- 29.Lu, X., and H. R. Horvitz. 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95:981-991. [DOI] [PubMed] [Google Scholar]
- 30.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 31.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 32.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, A. J., C. Sardet, and R. E. Herrera. 2002. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol. Cell. Biol. 22:856-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 36.Nevins, J. R. 1992. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258:424-429. [DOI] [PubMed] [Google Scholar]
- 37.Nicolas, E., S. Ait-Si-Ali, and D. Trouche. 2001. The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein. Nucleic Acids Res. 29:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 40.Pile, L. A., and D. A. Wassarman. 2000. Chromosomal localization links the SIN3-RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 19:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian, Y. W., Y. C. Wang, R. E. Hollingsworth, Jr., D. Jones, N. Ling, and E. Y. Lee. 1993. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature 364:648-652. [DOI] [PubMed] [Google Scholar]
- 42.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridgway, P., and G. Almouzni. 2000. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci. 113:2647-2658. [DOI] [PubMed] [Google Scholar]
- 45.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 46.Ross, J. F., X. Liu, and B. D. Dynlacht. 1999. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol. Cell 3:195-205. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 49.Secombe, J., J. Pispa, R. Saint, and H. Richardson. 1998. Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for cyclin E in cell proliferation control during eye imaginal disc development. Genetics 149:1867-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao, Z., S. Ruppert, and P. D. Robbins. 1995. The retinoblastoma-susceptibility gene product binds directly to the human TATA-binding protein-associated factor TAFII250. Proc. Natl. Acad. Sci. USA 92:3115-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siddiqui, H., D. A. Solomon, R. W. Gunawardena, Y. Wang, and E. S. Knudsen. 2003. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol. Cell. Biol. 23:7719-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staehling-Hampton, K., P. J. Ciampa, A. Brook, and N. Dyson. 1999. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics 153:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevaux, O., D. Dimova, M. V. Frolov, B. Taylor-Harding, E. Morris, and N. Dyson. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21:4927-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevaux, O., and N. J. Dyson. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14:684-691. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 56.Tevosian, S. G., H. H. Shih, K. G. Mendelson, K. A. Sheppard, K. E. Paulson, and A. S. Yee. 1997. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 11:383-396. [DOI] [PubMed] [Google Scholar]
- 57.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 58.Trouche, D., C. Le Chalony, C. Muchardt, M. Yaniv, and T. Kouzarides. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. USA 94:11268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 60.Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger, P. J. Harte, R. Kobayashi, and J. T. Kadonaga. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyler, J. K., M. Bulger, R. T. Kamakaka, R. Kobayashi, and J. T. Kadonaga. 1996. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 16:6149-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaute, O., E. Nicolas, L. Vandel, and D. Trouche. 2002. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 30:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 64.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 67.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]