Abstract
Phosphorylation is important for p53 protein stabilization and activation after DNA damage. Serine 389 of p53 is specifically phosphorylated after UV irradiation, whereas gamma radiation activates p53 through a different pathway. To study the in vivo significance of p53 phosphorylation at serine 389, we generated a physiological mouse model in which p53 phosphorylation at serine 389 is abolished by alanine substitution. Homozygous mutant p53.S389A mice are viable and have an apparently normal phenotype. However, cells isolated from these mice are partly compromised in transcriptional activation of p53 target genes and apoptosis after UV irradiation, whereas gamma radiation-induced responses are not affected. Moreover, p53.S389A mice show increased sensitivity to UV-induced skin tumor development, signifying the importance of serine 389 phosphorylation for the tumor-suppressive function of p53.
The p53 gene is the most frequently mutated tumor suppressor gene in human cancers (18), and it has been extensively shown that p53 plays an important role in cellular responses to a variety of stresses. In unstressed cells, p53 is present in a latent form and is maintained at low levels through rapid targeted degradation by MDM2 (17, 19, 27). However, in response to damaged DNA, nucleotide depletion, hypoxia, or several other genotoxic stresses, signaling pathways that transiently stabilize the p53 protein cause rapid accumulation of activated p53 in the nucleus (reviewed in reference 29). Stabilization, and thus accumulation, of the p53 protein is thought to result primarily from disruption of the p53-MDM2 interaction, which targets the p53 protein for ubiquitin-mediated degradation (39). Activation of the p53 protein occurs via protein modifications, such as phosphorylation, acetylation, and sumoylation, through multiple, potentially interacting yet distinct pathways (1, 2). The activated p53 protein functions predominantly as a tetrameric transcription factor in various cellular processes to prevent tumor development. The repertoire of p53 functions includes regulation of the cell cycle, induction of apoptosis, senescence, facilitating DNA repair, and antagonizing angiogenesis (1, 12, 39). Through these functions, predominantly mediated by transcriptional activation of target genes (12), p53 prevents cells with damaged DNA from dividing and becoming cancerous. Hence, complete loss of p53 function in mice leads to rapid tumor development, as is the case in p53 knockout mice (4).
Posttranslational p53 modifications predominantly occur at N- or C-terminal amino acids, and several protein kinases that phosphorylate p53 have been identified (reviewed in references 1 and 2). Modifications in the N-terminal domain are thought to hinder the p53-MDM2 interaction, whereas modifications in the C-terminal domain are thought to disrupt nonspecific DNA binding or induce conformational changes that prevent interactions between the C terminus and the core DNA-binding domain needed for activating and stabilizing the p53 protein.
Ser389 is one of the target residues phosphorylated in p53. Kinases that target this site in vitro are casein kinase II (26, 33), the double-stranded-RNA-activated protein kinase (PKR) (9), and p38 MAP kinase (20, 25). However, the responsible kinase in vivo has not been identified.
The kinetics of modification of individual p53 residues appear to be very specific and highly dependent on the nature of the stress-inducing agent (1). Notably, UV, but not gamma, radiation triggers mouse Ser389 (equivalent to human Ser392) phosphorylation (24, 32). This provides evidence for the existence of a distinct pathway responsible for activation of p53 after exposure to UV versus gamma radiation. In several in vitro assays using p53 phosphorylation site mutants transfected into cell lines, phosphorylation of Ser389 enhanced the DNA-binding capacity of p53; however, these data are contradictory. That is, p53 proteins with mutations at residue 389 displayed wild-type transactivation properties, but other phenotypes, such as inefficient tetramer formation, were also observed (13, 15, 16, 34, 40), demonstrating the ongoing controversy over the functions and effects of Ser389 phosphorylation. Altogether, it is still unclear whether Ser389 phosphorylation is involved in the major function of p53, namely, tumor suppression.
Obviously, understanding the involvement of p53 in carcinogenesis is of great importance for the development of new medical treatments. Several different (mutant) p53 mouse models have been generated for this purpose. Most of these models, however, are neither comparable to the human tumor situation nor physiologically relevant, due to overexpression of mutant p53 protein or complete knockout of the p53 allele. It is therefore important to study p53 using mouse models with more subtle modifications in this gene, thus mimicking the human situation and allowing studies to be done in a more physiologically relevant way (without overexpression). The first studies beginning to analyze the importance of p53 phosphorylation in such an in vivo system have recently been reported (7, 43, 45). Embryonic stem (ES) cells were generated with a p53.S23A mutation (Ser23 is the murine analog to human Ser20). Phosphorylation of p53 at Ser23 prevents MDM2 binding and thus degradation of p53 (42). ES cells, mouse embryonic fibroblasts (MEFs), and thymocytes with the p53.S23A homozygous mutation did not show defects in p53 stability, induction of p53 target genes, or induction of apoptosis following DNA damage (45), suggesting that phosphorylation of p53 at Ser23 alone is not sufficient to activate p53. In contrast, analysis of ES cells with a p53.S18A mutation (Ser18 is the murine analog to human Ser15) shows that Ser18 phosphorylation is necessary for p53 stabilization and its function as a transcriptional factor (8). Furthermore, cells isolated from mice with an S18A mutation display impaired transcriptional and apoptotic responses (7). However, in an additional study, the presence of the S18A mutation did not alter the proliferation rate and the p53-mediated G1 arrest induced by DNA damage in MEFs. Furthermore, the mice developed normally and were not susceptible to spontaneous tumorigenesis, nor did they rescue the embryonic-lethal phenotype of MDM2-null mice (43). This clearly demonstrates the difference in the requirement for each individual phosphorylation site in p53 for its activation and its separate cellular functions. The effects of these phosphorylation mutants on tumor development after exposure to genotoxic stress have not been reported.
To enable in vivo functional analysis of the Ser389 phosphorylation site in p53 in a well-defined system, we generated mice with a single point mutation in the p53 gene, resulting in a serine-to-alanine substitution at amino acid residue 389. This substitution blocks phosphorylation at this site. p53.S389A mice were generated by site-directed mutagenesis of relevant p53 exon sequences, subsequent homologous recombination in ES cells, and Cre-mediated excision of selectable marker genes. The hypothesis is that this single-mutation approach will disturb only a specific subset of p53 protein functions rather than all and will leave the expression levels of mutant p53 genes unchanged. Here, we describe the effects of the p53.S389A mutation in mice on both spontaneous and carcinogen-induced tumor development. Moreover, the effects of the p53.S389A mutation on several known p53-dependent processes, such as transcriptional activation of target genes, apoptosis, and cell cycle arrest, are analyzed in cells isolated from the mice.
MATERIALS AND METHODS
p53.S389A mutant targeting vector.
A previously described murine p53 mutant targeting vector (10) was used. Here, a single-base-pair mutation was introduced in exon 11, leading to a serine-to-alanine substitution at codon 389. The primers used in combination with the QuikChange site-directed mutagenesis kit (Stratagene) were as follows (mutated nucleotides are in boldface): p53.S389A#1, 5′ GTG GGG CCT GAC GCA GAC TGA CTG CC 3′; p53.S389A#2, 5′ GGC AGT CAG TCT GCG TCA GGC CCC AC 3′. Approximately 0.5 kb downstream of the p53 gene, a pGK-neo marker cassette flanked by loxP sites was incorporated, and a pGK-DTA selection marker was cloned at the 3′ end of the vector for negative selection (both plasmids were kindly provided by F. Gertler, Massachusetts Institute of Technology, Cambridge, Mass.). The p53 sequences of the targeting vector were sequenced to exclude additional (undesired) mutations.
Homologous-recombination experiments in ES cells and generation of mice.
A NotI-linearized targeting vector was electroporated into J1 ES cells using standard procedures (21). G418-resistant ES cell clones were analyzed for homologous integration of the targeting vector by Southern blot analysis of EcoRI-digested DNA using probe I (10) (Fig. 1A), and the presence of the mutation was verified by a PCR- and digestion-based assay with the following primers: p53in10#2, 5′ GGT GGT GAC AGT TGT GAT AAT AAT 3′ (intron 10), and p53ex11#2, 5′ TTG CAG AAT GGA AGG AAA GTA 3′ (exon 11).
FIG. 1.
Introduction of a p53.S389A point mutation into ES cells and mice. (A) Scheme for targeting one allele of the murine p53 gene in ES cells with a targeting vector containing the p53.S389A mutation (*), a pGK-neo selectable marker cassette flanked by loxP sites, and a pGK-DTA selectable marker cassette for negative selection. Homologous integration of the vector results in an additional EcoRI site (in the pGK-neo promoter), which is used for identification by Southern blot analysis of neo-resistant ES cell clones using probe I. Deletion of the pGK-neo marker cassette was accomplished by crossing mice with the p53.S389A mutation with Deleter mice expressing Cre in their germ line and was verified by Southern blot analysis using a BamHI digest and probe II. The resulting mutant p53.S389A allele differs from the wild-type allele, in addition to having the desired p53.S389A mutation in p53 exon 11, only in the presence of one loxP site downstream of the p53 gene. Expression of the p53.S389A mutation was verified by RT-PCR, followed by sequencing of the entire p53 coding sequence using RNA isolated from several tissues of p53.S389A mice. (B, BamHI; R, EcoRI; H, HindIII; Xho, XhoI; X, XbaI). (B) Presence of the S389A mutation was determined by PCR amplification of exon 11, followed by digestion with HinfI. The S389A mutation results in the loss of one HinfI restriction site. −, negative control.
The resulting 674-bp wild-type PCR product contains three HinfI restriction sites, one of which is absent in the mutant S389A PCR product. Digestion of the PCR product with HinfI results in the following fragment sizes: wild type, 282, 242, 87, and 63 bp; p53S389A/+, 524, 282, 242, 87, and 63 bp; and p53S389A/S389A, 524, 87, and 63 bp (Fig. 1B). Correctly targeted ES cell clones with the S389A heterozygous mutation (p53S389A/+) were injected into blastocysts. The resulting chimeras were mated with C57BL/6 mice. One clone gave rise to germ line transmission of the S389A mutated allele. These mice were subsequently bred to Cre transgenic deleter mice (kindly provided by K. Rajewski, University of Cologne, Cologne, Germany). The absence of the neo marker cassette in offspring mice was confirmed by analyzing BamHI-digested tail DNA by Southern blotting, using probe II (Fig. 1A). Routinely, mice were genotyped by PCR, followed by HinfI digestion.
Western blot analysis.
MEFs were expanded and plated at 1.2 × 106/10-cm-diameter plate. Twenty-four hours later, the cells were washed with phosphate-buffered saline (PBS) and exposed to 20-J/m2 UV-C or 5 Gy of gamma radiation. At several time points after the treatment, the MEFs were rinsed with PBS and collected in 200 μl of ice-cold lysis buffer (Giordano buffer [50 mM Tris HCl {pH 7.4}, 250 mM NaCl, 5 mM EDTA {pH 8.0}, 0.1% Triton X-100]-12% glycerol, supplemented with complete mini-protease inhibitor cocktail tablets [Roche] and phosphatase inhibitor cocktails 1 and 2 [Sigma]). The cells were lysed on ice for 20 to 40 min, followed by 15 min of centrifugation at 4°C to remove cellular debris. Total protein concentrations were determined using a BCA Protein Assay kit (Pierce).
To detect p53, immunoprecipitation was applied. One microgram of anti-p53 mouse monoclonal antibody (Ab-4; Oncogene Research Products) was cross-linked to 20 μl of protein G-protein A-agarose beads (Oncogene Research Products) with the cross-linker dimethylpilimidate dihydrochloride (Sigma). The beads with cross-linked antibody were incubated overnight with ∼250 μg of cells/protein extract at 4°C, washed with Giordano buffer, and eluted in sample buffer. The concentrated samples were separated on sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels (NuPAGE; Invitrogen) and transferred to Hybond polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). Forty-five micrograms of original lysates was also separated and transferred as described above. The membrane was blocked for 1 h with Western buffer (PBS-0.2% Tween 20-5% nonfat dry milk [Nutricia]), followed by three washes for 15 min each time with PBS-0.2% Tween 20. The membranes were incubated for at least 12 h at 4°C with either anti-p53 mouse monoclonal antibody (Ab-1; Oncogene Research Products) or anti-phospho-p53 rabbit polyclonal antibody (Ser392; Cell Signaling). Incubation for 1 h at room temperature with horseradish peroxidase (HRP)-linked antiactin affinity-purified goat polyclonal antibody (I-19-HRP; Santa Cruz) was done on the membrane with remnants from the immunoprecipitation. Primary antibodies were detected by incubating the membrane for 1 h at room temperature with HRP-linked sheep anti-mouse immunoglobulin G or HRP-linked donkey anti-rabbit immunoglobulin G (Amersham Pharmacia Biotech), and staining was done using ECL-plus reagent (Amersham Pharmacia Biotech). The membranes were scanned using a PhosphorImager (Storm 860; Molecular Dynamics), and ratios were determined by Totallab version 2.00 (Nonlinear Dynamics).
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed using the p53 NuShift Plus kit (Active Motif). p53 antibody 421 (Ab-1) at 1 μg/μl was used in place of the supplied polyclonal anti-p53 antibody. MEFs were plated at 106 per 10-cm-diameter plate; 24 h later, the MEFs were washed with PBS and exposed to UV-C light (20 J/m2). After 6 h, the MEFs were rinsed with PBS, scraped from the plate (combining five plates), and pelleted. The cells were lysed in 150 μl of cell lysis buffer (100 mM Tris-HCl [pH 8.0],100 mM NaCl, 1% Triton X-100, 10% glycerol, supplemented with inhibitors as used for Western blot analysis) by rocking them for 30 min at 4°C, followed by 15 min of centrifugation at 4°C to remove cellular debris.
Total protein concentrations were determined using a BCA Protein Assay kit, and 50 μg of lysate was used per binding reaction. The supplied probe was used for end labeling with T4 polynucleotide kinase and [γ-32P]ATP. The sequence of the supplied probe used was 5′ AGC TGG ACA TGC CCG GGC ATG TCC 3′, homologous to the p53 binding consensus sequence, 5′ PuPuPuC(A/T)(A/T)GPyPyPy 3′, as described earlier (14, 30). Binding reactions were run on a native 4% polyacrylamide gel (200 V for 1.5 h) with 0.25× Tris-borate-EDTA as a running buffer. The gels were dried for 1 h at 80°C. The amount of complex formed in each sample was quantitated using Totallab version 2.00, and the values were normalized for the amount of p53 protein present in the sample (as detected by Western blotting).
RT-PCR.
MEFs were plated at 106 per 10-cm-diameter plate; 24 h later (at ∼80% confluence), the cells were washed with PBS and exposed to UV-C light (20 J/m2). The cells were collected either 6 or 16 h after treatment. RNAs were harvested using an RNeasy kit (Qiagen), and reverse-transcriptase reactions were performed using the First-Strand cDNA Synthesis kit (Amersham Bioscience). Real-time PCR (RT-PCR) (Applied Biosystems) was performed according to the manufacturer's specifications.
The primers used were as follows: p21, 5′ CCT GAC AGA TTT CTA TCA CTC CA 3′ and 5′ AGG CAG CGT ATA TCA GGA G 3′; mdm2, 5′ CTC TGG ACT CGG AAG ATT ACA GCC 3′ and 5′ CCT GTC TGA TAG ACT GTC ACC CG 3′; bax, 5′ GTT TCA TCC AGG ATC GAG CAG 3′ and 5′ CCC CAG TTG AAG TTG CCA TC 3′; noxa, 5′ TCG CAA AAG AGC AGG ATG AG 3′ and 5′ CAC TTT GTC TCC AAT CCT CCG 3′; GAPDH, 5′ TCA CCA CCA TGG AGA AGG C 3′ and 5′ GCT AAG CAG TTG GTG GTG CA 3′.
Primer sequences were selected using the Primer Express program and were checked for gene specificity by BLASTN. The relative mRNA expression levels were normalized to the value of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each reaction.
Analysis of apoptosis and cell cycle arrest in MEFs.
Primary MEFs were prepared and cultured as described previously (6). Experiments were performed with early-passage MEFs (prior to passage 6). For UV-induced apoptosis, the MEFs were expanded and plated onto 24-well plates (3 × 104/well). After 24 h, the cells were washed with PBS and exposed to UV-C radiation at 12 or 20 J/m2. For trypan blue exclusion assays, both floating and adherent cells were harvested at several time points after exposure and resuspended in 0.4% trypan blue solution (Gibco BRL). The ratio of blue (apoptotic) to white (viable) cells was subsequently determined (>100 cells per sample). For the cell death detection enzyme-linked immunosorbent assay (ELISA) kit (Roche Molecular Biochemicals), adherent cells were lysed 16 h after the UV-C treatment. Apoptotic levels in each well (based on the enrichment of mono- and oligonucleosomes in the cytoplasm of the apoptotic cells) were subsequently determined according to the manufacturer's instructions. Retroviral infection of MEFs with adenovirus oncogene E1A and subsequent assessment of apoptosis after exposure to 0.2 μg of doxorubicin (Sigma)/ml was done as described previously (5). For all assays, each time point was measured in duplicate, and several independent MEF cell lines were tested. The values of exposed samples were corrected for background apoptosis levels found in untreated cultures of the same cell line.
For cell cycle analysis, MEFs were expanded and plated at 8 × 105/10-cm-diameter plate; 24 h later, the cells were exposed to 5 Gy of gamma radiation. After 18 h, the MEFs were collected and prepared, and fluorescence-activated cell sorter (FACS) analyses were performed as described previously (5). After FACS analysis, fractions of cells in each phase of the cell cycle were quantified using CellQuest software (Becton Dickinson).
CD4/CD8 staining of thymocytes.
Mice (6 to 9 weeks old) were exposed to a single 5-Gy dose of gamma radiation using a gamma cell irradiator with a Cs source. Twenty-four or 48 h later, the thymus was isolated, and single-cell suspensions of thymocytes were generated. The thymocytes (106) were incubated for 5 min at room temperature with red cell removal buffer (160 mM NH4Cl, 0.1 mM EDTA, 12 mM NaHCO3), washed with 3 ml of 2% PBS-fetal calf serum (FCS), and resuspended in 100 μl of 2% PBS-FCS with anti-CD4 and -CD8 antibodies (1:100; BD Biosciences Pharmingen). The cells were stained for 30 min on ice, washed with 2% PBS-FCS, resuspended in 500 μl of 0.1% PBS-propidium iodide (PI), and analyzed by FACS analysis. After FACS analysis, fractions of CD4/CD8-positive thymocytes were quantified using CellQuest software (Becton Dickinson).
Analysis of apoptosis in mouse thymocytes.
For in vitro analysis of apoptosis, thymocytes were isolated from mice (6 to 9 weeks old), treated with several doses of doxorubicin (adriamycin) or dexamethasone (Sigma), and stained with fluorescein isothiocyanate-labeled annexin V antibody (Pharmingen) and PI (Sigma) as described previously (10, 31). Relative amounts of apoptotic cells were determined by binding of annexin V and subsequent FACS analysis.
Analysis of spontaneous and UV-B-induced tumor development.
For spontaneous tumor development, mice were crossed with C57BL/6 mice (F3 generation) and followed as they aged. Sick animals were sacrificed, and tissues were collected. For UV radiation experiments, a total of 14 wild-type, 13 p53S389A/+, 22 p53S389A/S389A, and 14 p53+/− mice, all consisting of 50% males and 50% females and bred into a hairless background (HR7:SKH; F2 generation), were exposed to a daily UV-B dose of 600 or 300 J/m2 using Philips TL12 lamps. All mice were checked weekly for development of tumors. Tumors and tissues were processed for histopathological analysis in formaldehyde and embedded in paraffin. Sections were cut at 5 μm and stained with hematoxylin and eosin. In vivo experiments with mice were performed in compliance with the International Guiding Principles for Biomedical Research Involving Animals (developed by the Council for International Organization of Medical Sciences), and the National Institute of Public Health and Environment (RIVM) Institutional Animal Care and Use Committee evaluated the study protocols.
Statistical analysis.
Statistical analyses were carried out using Kaplan Meyer analysis for tumorigenesis studies and the two-tailed Student t test for in vitro assays, with P values of <0.05 considered statistically significant.
RESULTS
Generation of mice with a serine-to-alanine mutation at p53 codon 389.
To study the significance of p53 phosphorylation at residue Ser389 in vivo in a physiologically relevant manner, we generated a knockin mouse model using gene targeting. In this model, codon 389 of the p53 gene is mutated, changing the TCA (serine) codon into a GCA (alanine) codon. This p53.S389A mutation should abolish phosphorylation of the p53 protein at amino acid residue 389. A targeting vector harboring the p53.S389A mutation was successfully created by site-directed mutagenesis and introduced into the mouse genome by homologous recombination in ES cells. Homologous recombinant clones were identified by Southern blot analysis of ES cell DNA (Fig. 1A and results not shown). The neo marker cassette, flanked by loxP sites, was subsequently removed by in vivo Cre-mediated excision using Deleter mice (41), and homozygous mutant mice were obtained after the mice were bred. In these mice, expression of the p53.S389A mutated allele was verified by RT-PCR analysis. Homozygous p53.S389A mice (p53S389A/S389A) have a unique point mutation only in the p53 coding sequence and a single residual loxP site downstream of the p53 gene. The genotypes of the mice and cells used in the different experiments were verified through a PCR-digestion-based assay (Fig. 1B). Homozygous p53.S389A mice are born at Mendelian ratios, are viable and fertile, and have no obvious phenotypic abnormalities.
p53.Ser389 phosphorylation and p53 protein levels after UV or gamma irradiation.
We verified whether the p53.S389A mutation indeed abolishes phosphorylation of the p53 protein at amino acid residue 389. For this, we exposed MEFs isolated from p53S389A/S389A and wild-type mice to 20-J/m2 UV radiation and analyzed them by Western blotting for phosphorylation at Ser389 using phosphospecific antibodies. As a negative control, MEFs were exposed to a single 5-Gy dose of gamma radiation. As expected, in wild-type MEFs, increasing levels of Ser389-phosphorylated p53 were observed in a time-dependent manner after UV irradiation (Fig. 2A). In MEFs isolated from p53S389A/S389A mice, no phosphorylation of serine 389 could be detected after UV irradiation (Fig. 2A). This demonstrates the successful inactivation of the Ser389 phosphorylation site. As expected, exposure to gamma radiation did not result in phosphorylation of Ser389 in wild-type or S389A mutant MEFs (Fig. 2B).
FIG. 2.
Phosphorylation of p53.S389 and total p53 protein levels in wild-type and p53.S389A MEFs after exposure to UV or gamma radiation. Phosphorylation of Ser389 and induction of p53 protein levels in wild-type (+/+) and p53S389A/S389A (SA/SA) MEFs after treatment with 20 J of UV-C/m2 (A) or 5 Gy of gamma radiation (B). Protein extracts were prepared from MEFs at different time points after the treatment, and phosphorylation of Ser389 and total p53 protein levels were visualized by Western blotting after immunoprecipitation. The remnant was moved to an additional Western blot, which was incubated with actin antibody to control for loading.
In untreated cells, p53 is present at low levels due to rapid turnover. In response to DNA damage, active p53 protein levels increase significantly, primarily due to increased protein stability. Modification by phosphorylation is considered an important regulator of p53 protein stability, so we investigated whether the p53.S389A mutation influences p53 protein levels after DNA damage. p53+/+, p53S389A/S389A, and, as a negative control, p53−/− MEFs (not shown) were exposed to either 20- J/m2 UV or 5 Gy of gamma radiation, and p53 protein levels were determined at several time points after treatment. Total p53 protein levels accumulate rapidly in wild-type MEFs after exposure to both UV and gamma radiation (Fig. 2). Comparison of p53 protein levels in UV-exposed wild-type and p53S389A/S389A MEFs revealed slightly reduced levels in the p53S389A/S389A MEFs at 3 and 6 h after UV irradiation. This phenomenon proved to be a recurring event in several independent experiments, but differences in p53 levels encountered in wild-type and mutant MEFs are small. After gamma irradiation, the levels of induction of p53 protein in p53S389A/S389A MEFs were indistinguishable from the levels encountered in wild-type MEFs. In conclusion, mutating the serine 389 codon of the p53 protein into that for alanine abolishes phosphorylation at this site and has a slight effect on total p53 protein levels after UV irradiation.
DNA binding by p53 protein after UV irradiation.
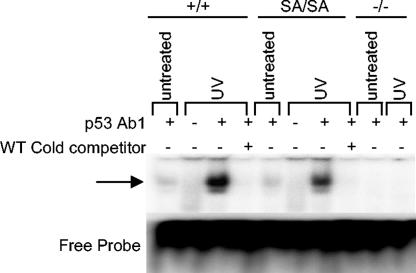
An important function of p53 is the transcriptional activation of downstream target genes. For this purpose, a tetramer of activated p53 proteins binds a p53-specific DNA promoter. Using EMSA, we tested the effect of the p53.S389A mutation on p53-specific DNA binding after UV irradiation. Adding a cold competitor that completely eliminated p53 DNA binding, as well as the use of p53 knockout MEFs, proved the binding specificity for the target sequence (Fig. 3).
FIG. 3.
DNA binding by p53 protein after UV irradiation. EMSAs were performed using a p53-specific oligonucleotide probe, in wild-type (+/+), p53S389A/S389A (SA/SA), and p53−/− (−/−) MEFs. All samples were isolated 6 h after irradiation with 20-J/m2 UV-C and were incubated with the labeled probe in the presence or absence of p53-specific antibody and wild-type (WT) cold competitor.
In untreated wild-type and p53S389A/S389A MEFs, comparable amounts of p53 tetramer bound to DNA (Fig. 3). Apparently, binding of p53 to its target sequence is not dependent on phosphorylation at Ser389 in untreated cycling MEFs. In response to UV radiation, levels of p53 complex binding DNA dramatically increased in wild-type MEFs (Fig. 3). Interestingly, although in p53S389A/S389A MEFs, levels of DNA-binding p53 tetramer also increased compared to untreated mutant MEFs, the relative quantity of the complex, corrected for the amount of p53 protein present in the lysate, was clearly reduced compared to that observed in wild-type MEFs. Therefore, a lack of phosphorylation at Ser389 results in reduced DNA-binding activity of the p53 tetramer after UV irradiation.
Expression of p53 target genes after UV radiation.
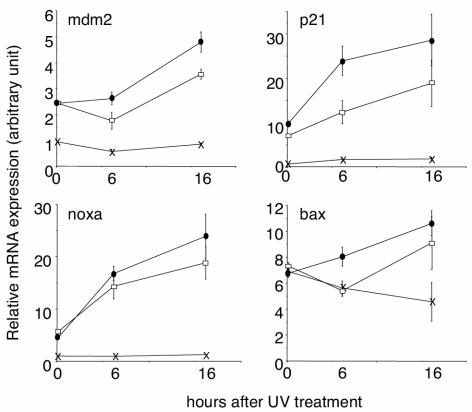
A key function of p53 is the transcriptional activation of target genes involved in cellular processes, such as apoptosis and cell cycle arrest. Since DNA binding of the p53 complex after UV irradiation is inhibited by the S389A mutation, we analyzed the expression levels of several well-known p53 target genes in cells exposed to UV radiation. For this purpose, we analyzed by RT-PCR the gene expression levels of mdm2, the negative regulator of p53 activity (27); p21, associated with the cell cycle arrest function of p53 (6); and noxa and bax, both implicated in p53-dependent apoptosis (35-37).
Endogenous expression levels of all tested genes, except for bax, were significantly lower in p53−/− MEFs, and after exposure to UV radiation, a lack of transcriptional induction was observed in p53−/− MEFs, demonstrating the p53 dependence of the regulation of these genes in our cellular assay. In contrast, the gene expression levels of all tested genes were equal in untreated wild-type and p53S389A/S389A MEFs (Fig. 4). After UV irradiation, both wild-type and p53S389A/S389A MEFs showed an induction of the relative gene expression levels of all tested genes. However, the induction levels in p53S389A/S389A MEFs of all four genes are clearly reduced or delayed compared to that in wild-type MEFs. At 6 h, these data were significantly different for mdm2, p21, and bax, but this was not the case at 16 h. Comparable results were obtained for expression levels of the cyclin G gene, also shown to be implicated in p53-dependent apoptosis (38) (results not shown). In conclusion, MEFs with a homozygous p53.S389A mutation are compromised in transcriptional activation of well-known p53 target genes after exposure to UV radiation, whereas endogenous levels in nonexposed cells are not affected.
FIG. 4.
Expression of p53 target genes in MEFs. Wild-type (•), p53S389A/S389A (□), and p53−/− (×) MEFs were exposed to 20-J/m2 UV-C radiation. At 6 and 16 h after treatment, RNA was isolated and RT-PCR was performed on different p53 target genes. The relative gene expression was normalized to the gene expression of GAPDH in the same sample, and average values with standard deviations from triplicate reactions are shown. Note that a different graduation scale is applied to each graph.
Apoptosis and G1 cell cycle arrest after UV irradiation.
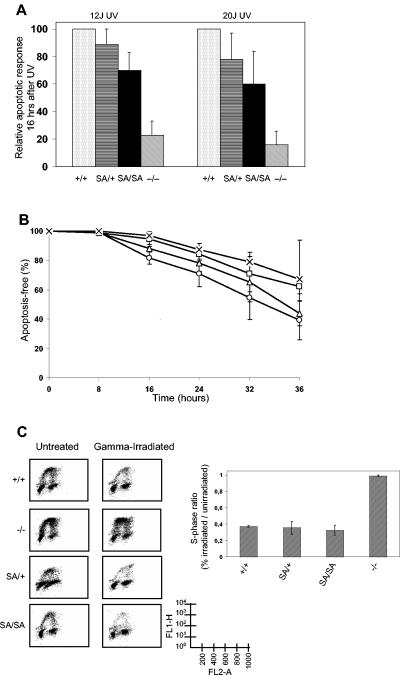
After exposure to DNA damage, p53 is able either to induce G1 cell cycle arrest (G1 arrest), presumably to allow the cells to remove DNA damage, or to induce apoptosis, seemingly when the cells have high levels of sustained DNA damage. Also, after exposure to the DNA-damaging agent UV radiation, p53 residue 389 becomes phosphorylated (Fig. 2A). With the p53.S389A mouse model, it is now possible to analyze whether this particular phosphorylation plays a role in these distinct cellular processes. We exposed MEFs of several genotypes to 12- or 20-J/m2 UV radiation and analyzed apoptotic responses using two different assay systems at several time points after treatment (Fig. 5). Cell death detection ELISA (Fig. 5A) shows that wild-type MEFs display extensive apoptosis, whereas p53−/− MEFs are highly resistant (response relative to that of the wild type, <20%), as was described previously (3). p53S389A/S389A MEFs clearly showed an apoptotic response (relative apoptotic responses of ∼70 and ∼60%) between those of wild-type and p53−/− MEFs after exposure to 12- and 20-J/m2 UV radiation. p53S389A/+ MEFs showed an apoptotic response (relative apoptotic responses of ∼89 and ∼78%) between those of wild-type and p53S389A/S389A MEFs, which is likely indicative of a gene dose effect of the mutation on UV-induced apoptosis.
FIG. 5.
UV-induced apoptosis and gamma radiation-induced cell cycle arrest in MEFs. (A) Wild-type (+/+), p53S389A/+ (SA/+), p53S389A/S389A (SA/SA), and p53−/− (−/−) MEFs were exposed to 12- or 20-J/m2 UV-C radiation, and the apoptotic response was measured 16 h later with a cell death ELISA detection assay. The apoptotic response in UV-irradiated samples is normalized to the apoptotic response in untreated cultures from the same MEF cell line. The apoptotic response of each genotype is subsequently depicted relative to the response in wild-type cells. For each data point, at least three independent MEF cell lines of each genotype were analyzed in independent experiments. (B) Wild-type (○), p53S389A/+ (▵), p53S389A/S389A (□), and p53−/− (×) MEFs were exposed to 20-J/m2 UV-C radiation, and the apoptotic response was measured 8, 16, 24, 32, and 36 h later with a trypan blue exclusion apoptosis assay. The apoptotic response in UV-irradiated MEFs is normalized similarly to that in Fig. 5A. For each data point, at least two independent MEF cell lines of each genotype were analyzed in independent experiments. (C) Wild-type (+/+), p53S389A/+ (SA/+), p53S389A/S389A (SA/SA), and p53−/− (−/−) MEFs were exposed to 5 Gy of gamma radiation, and cell cycle arrest was determined 18 h later by FACS analysis. Left, representative examples of FACS analysis. Right, quantitation of arrest responses of MEFs to gamma radiation. The degree of G1 arrest is indicated by a ratio of the S-phase fraction of cells after gamma exposure to the S-phase fraction of untreated cells. Each bar represents the average and standard deviation of multiple experiments. FL1-H, BrdU incorporation; FL2-A, DNA content.
The reduced apoptotic response observed in p53S389A/S389A MEFs compared to wild-type MEFs was confirmed using a different, commonly used apoptotic assay (trypan blue exclusion, as described by Attardi et al. [5]). For this purpose, cells were exposed to 20-J/m2 UV radiation, and the ratio of viable to dead cells was calculated at several time points after the treatment (Fig. 5B). Again, the same order of increasing resistance to UV-induced apoptosis was observed, from wild-type to p53S389A/+ to p53S389A/S389A to p53−/− MEFs. Differences between the apoptotic responses of wild-type and S389A mutant cells were statistically significant at all time points except 8 h after UV exposure.
Whether the affected apoptosis is specific for UV radiation or is attributable to a more general effect on apoptosis was analyzed by investigating another p53-dependent apoptotic response, i.e., that induced by expression of activated oncogenes (5). p53S389A/S389A and wild-type MEFs were transfected with activated adenovirus oncogene E1A, and apoptosis was measured after the exposure of the transfected cells to doxorubicin (0.2 μg/ml). Wild-type MEFs expressing E1A rapidly undergo apoptosis after this treatment, whereas p53−/− cells are resistant (5). p53S389A/S389A MEFs responded like wild-type cells in this assay (data not shown), indicating that p53 residue Ser389 phosphorylation is not necessary for oncogene-mediated cell death but is specifically used for UV-induced apoptosis.
To confirm the specificity of the role of S389 phosphorylation in response to UV radiation, we tested the cellular response in p53.S389A MEFs after exposure to gamma radiation. Gamma radiation is known to induce p53-dependent G1 arrest in MEFs (6). Wild-type MEFs efficiently undergo G1 arrest upon gamma irradiation, resulting in a decreased percentage of cells in S phase, whereas p53−/− MEFs do not (Fig. 5C), confirming observations made by others (6). Both p53S389A/S389A and p53S389A/+ MEFs undergo G1 arrest as efficiently as wild-type MEFs. Thus, the p53.S389A mutation seems to influence the UV-induced apoptotic response of MEFs but not oncogene-induced apoptosis or gamma radiation-induced G1 arrest.
p53-dependent and -independent apoptosis after gamma radiation and doxorubicin.
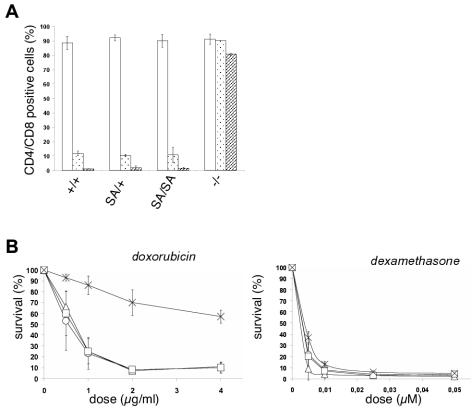
Thymocytes undergo p53-dependent apoptosis after exposure in vivo or in vitro to gamma radiation or to compounds known to induce DNA double-strand breaks (e.g., doxorubicin) (31). To test the involvement of p53.Ser389 phosphorylation in this apoptotic response, we exposed p53S389A/S389A mice to 5 Gy of gamma radiation. Thymocytes were isolated 24 or 48 h after treatment and were stained with anti-CD4 and anti-CD8 antibodies. Approximately 90% of untreated thymocytes from wild-type mice are CD4/CD8 double positive, which is similar to the rate for thymocytes of p53.S389A mutant mice (Fig. 6A). Twenty-four hours after exposure to gamma radiation, only 12% of CD4/CD8-double-positive thymocytes can still be detected in the thymuses of wild-type mice, and after 48 h just 1% can still be detected, whereas at the same time points, 90 and 80% of the thymocytes in p53−/− mice survive the treatment. This clearly shows the p53 dependence of the apoptotic process related to this treatment. After both 24 and 48 h, the apoptotic responses of p53S389A/+ and p53S389A/S389A thymocytes are similar to that of wild-type thymocytes. These findings indicate that phosphorylation of p53 residue Ser389 does not play a crucial role in activating p53 for this cellular response.
FIG. 6.
In vivo and in vitro apoptosis in thymocytes from p53.S389A mice. (A) For analysis of in vivo-induced apoptosis, mice of the indicated genotypes (+/+, wild type; SA/+, p53S389A/+; SA/SA, p53S389A/S389A; and −/−, p53−/−) were exposed to a single dose of gamma radiation (5 Gy; whole body). Twenty-four and 48 h later, thymocytes were isolated, and the percentage of CD4/CD8-double-positive cells was determined by FACS analysis. The bars represent three independent experiments (i.e., mice). Open bars, untreated thymocytes; speckled bars, 24 h after gamma irradiation; hatched bars, 48 h after gamma irradiation. (B) For analysis of in vitro-induced apoptosis, thymocytes were isolated from mice of different genotypes and propagated in cell culture. The cells were either left untreated or exposed to increasing doses (as indicated) of doxorubicin (p53-dependent apoptosis) or dexamethasone (p53-independent apoptosis). After 24 h, the thymocytes were stained with PI and annexin V, and the viable cells were counted by FACS analysis. The relative percentage of viable cells (negative for both PI and annexin V) for each sample is shown. All values are normalized to the number of cells remaining viable in untreated cultures derived from the same animal and stained simultaneously. The data represent at least two independent experiments (i.e., mice). ○, wild-type; ▵, p53S389A/+; □, p53S389A/S389A; ×, p53−/−.
To determine whether this unaffected apoptotic response in thymocytes depended on the damaging agent applied, thymocytes of the same genotypes were treated in vitro with increasing doses of a different double-strand break-inducing compound, doxorubicin. Wild-type cells undergo apoptosis after this treatment, whereas p53−/− cells are highly resistant (Fig. 6B). Here, p53S389A/S389A and p53S389A/+ thymocytes also behaved like wild-type thymocytes, underscoring the observation made previously that p53 responses initiated by DNA double-strand breaks are not affected in cells carrying a p53.S389A mutation. As a control, thymocytes were exposed to dexamethasone, a compound inducing p53-independent apoptosis, and as expected, cells from all tested genotypes underwent apoptosis efficiently (Fig. 6B).
Survival and spontaneous tumor development in p53.S389A mice.
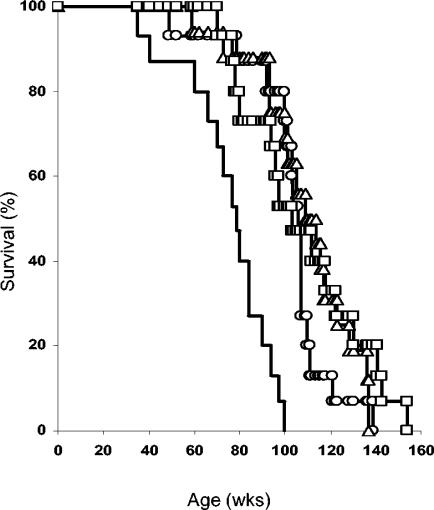
It is well known that loss of functional p53 protein predisposes both mice and humans to the development of tumors (4, 11, 18). To analyze whether phosphorylation of p53 residue Ser389 plays a role in the ability of p53 to suppress tumorigenesis, we monitored wild-type, p53S389A/+, p53S389A/S389A (all littermates), and, as a control, p53+/− mice as they aged (>2 years) for spontaneous tumor development or other abnormalities. Mice with a homozygous or heterozygous p53.S389A mutation have survival curves similar to those for wild-type mice (Fig. 7) (P = 0.29). For all these genotypes, the 50% survival time is ∼110 weeks. p53+/− mice showed reduced survival (50% survival time, ∼75 weeks) compared to wild-type mice, confirming previous observations (4, 11). Taken together, these observations indicate that the presence of the p53.S389A mutation in mice has no obvious effect on their survival.
FIG. 7.
Survival of p53.S389A mice. Mice of the indicated genotypes were monitored as they aged for development of tumors or other aberrations to determine survival. Moribund animals were killed, and their tissues were analyzed by histopathology. Genotypes: wild-type, ○; p53S389A/+, ▵; p53S389A/S389A, □; p53+/−, no symbol. The number of animals analyzed per genotype is indicated in Table 1. Each group consisted of approximately equal numbers of males and females. wks, weeks.
The majority of animals developed tumors, and the frequency of tumor-bearing animals (71 to 75%) appeared not to be influenced by the presence of the p53.S389A mutation (Table 1). p53+/− mice in this study had a slightly higher incidence of tumors (87%), again comparable to observations made by others (4, 11, 21). Among the tumors, lymphomas (follicular center cell; lymphoblastic), along with mesenchymal tumors (sarcomas) of various kinds and epithelial tumors in various tissues, were frequently observed (Table 1). Heterozygous mutant p53.S389A mice displayed a tumor spectrum similar to the spectrum observed in wild-type mice; however, homozygous mutant mice seemed to develop more lymphomas and fewer epithelial tumors than wild-type mice. In p53+/− mice, mainly sarcomas were found (60%), and to a lesser extent lymphomas (30%), a phenomenon described before for these mice (4, 11, 21).
TABLE 1.
Spontaneous tumor development in aged p53.S389A mutant mice
| Genotype | No. of mice analyzed | Tumor-bearing mice [no. (%)] | % Tumors
|
|||
|---|---|---|---|---|---|---|
| Sarcomaa | Epithelial tumorab | Lymphomaa | Othersa | |||
| Wild-type | 14 | 10 (71) | 20 | 46 | 27 | 7 |
| p53S389A/+ | 15 | 11 (73) | 28 | 44 | 28 | 0 |
| p53S389A/S389A | 16 | 12 (75) | 29 | 24 | 47 | 0 |
| p53+/− | 15 | 13 (87) | 60 | 5 | 30 | 5 |
Tumor types presented as percentage of total number of tumors found per genotype.
Adenocarcinoma, hepatocellular adenoma or carcinoma, bronchioloalveolar adenoma or carcinoma, pars distalis adenoma, or squamous cell carcinoma.
In conclusion, abolishing phosphorylation at residue Ser389 does not seem to affect the tumor-suppressive function of p53 in spontaneous tumor development but does seem to affect the tumor spectrum.
UV-induced skin tumor development in p53.S389A mice.
Since Ser389 is specifically phosphorylated after UV irradiation and the presence of the p53.S389A mutation appeared to compromise UV-induced responses in MEFs, the tumor suppressor function of p53 after exposure to UV radiation was studied. To analyze this, skin tumor development in response to UV-B, the main UV radiation inducer of skin cancer, was determined. To facilitate UV-B irradiation, wild-type, p53S389A/S389A, p53S389A/+, and p53+/− mice were bred into a hairless background (SKH:HR7). The minimal erythemal dose for p53S389A/S389A, p53S389A/+, and p53+/− mice was identical to that for wild-type mice (data not shown), namely, 900 J/m2. The presence of a S389A mutation apparently does not result in an altered acute sensitivity of the skin to UV-B radiation.
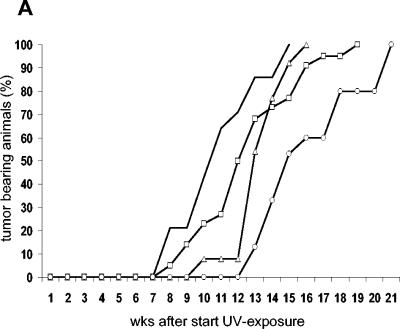
In the chronic UV-B radiation experiment, mice (all littermates) were exposed to a daily dose of 300 or 600 J/m2. p53S389A/S389A and p53+/− mice developed skin tumors significantly earlier(P = 0.001 and 0.0001, respectively) than their wild-type littermates after daily exposure to 600 J/m2 (Fig. 8A). Within 8 weeks after the start of UV-B irradiation, the first p53S389A/S389A and p53+/− mice developed skin tumors, whereas the first skin tumor on a wild-type mouse was encountered 5 weeks later. p53S389A/+ mice showed a skin tumor response intermediate between the responses of homozygous mutants and wild-type animals. The development of skin tumors, however, was significantly earlier than that of wild-type animals (P = 0.021). There was also a significant difference in tumor development between p53+/− and p53S389A/S389A mice (P = 0.0459), indicating that loss of one p53 allele renders mice more sensitive to UV-induced tumor development than abolishing phosphorylation of Ser389. Also, exposure to 300 J/m2 resulted in a significantly decreased latency time for skin tumor development in p53S389A/S389A and p53+/− mice compared to wild-type mice, but at that dose, heterozygous mutant p53S389A/+ mice develop tumors at the same rate as wild-type mice (data not shown).
FIG. 8.
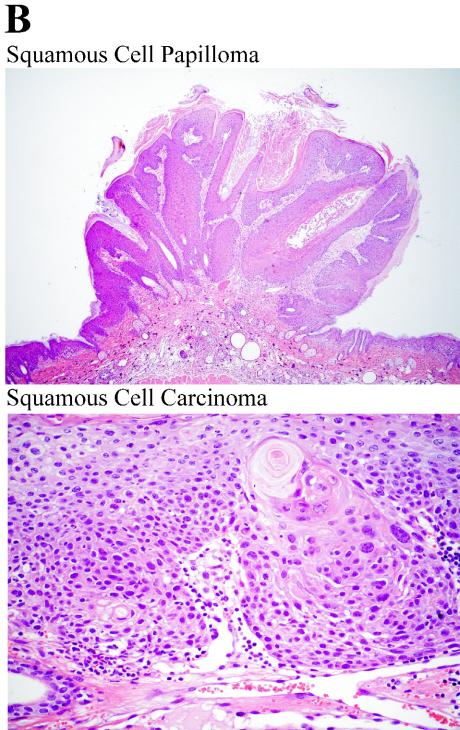
UV-B-induced skin tumor development in p53.S389A mice. (A) Mice were bred into a hairless background (SKH:HR7) and exposed to a daily UV-B radiation dose of 600 J/m2. Wild-type, ○; p53S389A/+, ▵; p53S389A/S389A, □; p53+/−, no symbol; wks, weeks. (B) Representative examples of squamous cell carcinoma and squamous cell papilloma, both isolated from UV-B-irradiated p53S389A/S389A mice.
The UV radiation regimen applied eventually resulted in 100% tumor-bearing mice; however, the tumor yield per animal was slightly increased in p53S389A/S389A mice compared to that in wild-type mice (an average of 10.3 tumors per UV-B-exposed skin of p53S389A/S389A mice versus 7.2 in wild-type mice). The skin tumor types that p53+/−, p53S389A/S389A, and p53S389A/+ mice developed were identical to those detected on the wild-type animals, namely, papillomas and squamous cell carcinomas (Fig. 8B shows representative examples). In conclusion, abolishing the phosphorylation of serine 389 in mice resulted in increased UV-B-induced skin tumor development, the first clear in vivo demonstration of the requirement for phosphorylation of p53 for its tumor suppressive action.
DISCUSSION
p53 activation through phosphorylation is an important posttranslational event, and several functional phosphorylation sites have been assigned throughout the protein (1). Though they are recognized as important, the molecular roles of the specific modifications have not yet been elucidated in vivo. Several studies suggest that phosphorylation of specific residues in p53 is necessary for the protein to become stabilized and active and thus to fulfill its biochemical and/or cellular functions, such as DNA binding, induction of apoptosis, and suppression of cellular transformation (reviewed in reference 1). However, the vast majority of data supporting an important role for phosphorylation are obtained through in vitro transformation assays employing transfected p53-encoding plasmids, raising the important question of whether these observations accurately reflect the in vivo situation.
Ser389 is phosphorylated in response to specific types of DNA damage, such as those induced by UV radiation (24, 32), but remains unphosphorylated after exposure to gamma radiation. This could provide an explanation for the different cellular processes that p53 induces after exposure of cells to these different DNA-damaging agents. For example, gamma irradiation of MEFs results exclusively in p53-dependent cell cycle arrest, whereas UV irradiation of MEFs results mainly in apoptosis (3). It is tempting to postulate that, depending on the cellular context, different types of DNA damage induce different p53 modifications that in turn provide signals directing a specific p53 response.
Phosphorylation of p53 at residue 389 has been shown to enhance tetramer formation, a clear indication of the involvement of this phosphorylation in p53 activation (40). Transfection studies to determine the precise role of p53 Ser389 phosphorylation by blocking it through mutating the residue into alanine resulted in ambiguous observations. Some studies showed that p53 tetramer formation, DNA-binding capacity, transcription regulation activity, and growth suppression activity were all abolished (16, 34), whereas other studies showed these functions to be unaltered (13, 15). A straightforward explanation for this apparent discrepancy might be the use of different cell lines and unequal expression levels of transfected p53 protein in the studies. Even so, the results would imply that phosphorylation of Ser389 is not always, or not absolutely, required for p53 activation.
To determine the involvement of Ser389 phosphorylation in p53 functions in a physiologically relevant manner and to enable the in vivo analysis of the requirement for this phosphorylation, we generated mice with a single point mutation in the p53 gene, resulting in a serine-to-alanine substitution at residue 389. Homozygous mutant mice are viable and show no obvious abnormalities. The S389A mutation did not influence the latency time for spontaneous tumor development, and moreover, the number of tumor-bearing animals was identical to that in wild-type controls. Only the tumor spectrum in homozygous p53.S389A mice appeared altered compared to wild-type and heterozygous mutant mice, since more lymphomas and fewer epithelial tumors were found. Intriguingly, homozygous p53 knockout mice also predominantly develop lymphomas spontaneously (4, 11). However, given the small numbers of mice that were analyzed in the study described here and the mixed genetic background (F3 generation; strains 129SV and C57BL/6) of the mice, the significance of this finding needs to be confirmed and will therefore be the subject of future studies. However, if this shift in tumor types is indeed corroborated, it might provide new insights into the specific roles of p53 in tumor development in different cell types.
The observation that heterozygous and homozygous p53 knockout mice are highly prone to spontaneous tumor development clearly demonstrated the tumor-suppressive function of p53 (4, 11). Therefore, our results with the p53.S389A mice indicate that without genotoxic stress, Ser389 phosphorylation alone is not absolutely required for p53 tumor-suppressive action. Recently, Chao et al. and Sluss et al. also described the lack of spontaneous tumor development in mice harboring a serine-to-alanine substitution at an N-terminal phosphorylation site (residue Ser18) (7, 43). Taken together, these findings are not entirely unexpected, since phosphorylation of p53 is mainly induced upon exposure to specific genotoxic compounds.
In contrast, an adverse effect on tumor suppression would be expected when p53.S389A mice are challenged with a specific DNA-damaging agent. Indeed, chronic exposure of these mutant mice to UV radiation resulted in a significantly reduced latency time for skin tumor development compared to their wild-type littermates. Moreover, a slightly increased tumor yield was observed. Even heterozygous S389A mice showed an altered response compared to wild-type mice, suggesting a gene dosage effect of p53 alleles consistent with an analogous observation in heterozygous p53 knockout mice (44). Although the observed adverse effects are modest, our data clearly demonstrate for the first time that without proper Ser389 phosphorylation, p53 partly loses its capacity for preventing tumorigenesis when cells are exposed to specific genotoxic stresses. However, a comparison of UV-induced tumor development in p53.S389A mice with that observed in p53+/− mice reveals different effects of a lack of Ser389 phosphorylation and the complete absence of one allele. The latter, as also described by others (22), results in rapid development of UV-induced skin tumors. Here, we show that p53+/− mice are more sensitive to UV radiation than p53.S389A mice. Apparently, in S389A mutant cells, p53 still becomes active, albeit to a lesser extent, and is able to fulfill at least part of its function. Preliminary results show that short exposures (i.e., 2 days) to UV-B in vivo indeed result in induction of p53 protein in the skin of p53.S389A mutant mice, as analyzed by immunostaining of irradiated skin sections with CM5 antibody. The number of p53-positive cells was reduced compared to wild-type mice; however, differences between the two genotypes were again small. A reasonable explanation could be that activation of p53 through phosphorylation is redundant and that even though lack of phosphorylation at one site results in decreased functional activity of p53, multiple sites need to be mutated to completely inactivate the protein. In line with this assumption, Kapoor et al. have shown in vitro that cooperative phosphorylation at multiple sites is required to completely activate p53 after UV irradiation (23). Moreover, targeted mutation of other p53 phosphorylation sites in ES cells and thymocytes has also been shown to result in only partial defects, again implying redundancy in p53 modifications (7).
Our findings after chronic UV exposure in vivo are supported by in vitro studies with MEFs isolated from the S389A mice, bearing in mind the differences in the UV sources and cell types analyzed. The S389A mutant MEFs show several defects in their responses to UV radiation. First, total p53 protein levels are diminished after UV irradiation in p53.S389A MEFs, whereas p53 levels are comparable to those in wild-type MEFs after gamma irradiation. In addition, DNA binding of the p53 complex to its target sequence is reduced in our mutant MEFs upon UV irradiation. As a consequence, an impaired capacity to activate downstream target genes can be expected, and indeed, the gene expression levels of several well-known p53 targets are reduced. Altogether, it is reasonable to assume that these observed defects in the p53.S389A MEFs are responsible for the adverse effects on cellular response in p53.S389A mutant mice and cells, since the apoptotic response in S389A MEFs is impaired after UV irradiation. In line with the in vivo UV-induced tumor development, apoptotic responses of the p53.S389A mutant cells are intermediate between those observed in wild-type and homozygous p53 knockout cells. As stated before, redundancy is likely to be responsible for this.
Taken together, our results clearly show that Ser389 phosphorylation is involved in some, but not all, functions of p53. We hypothesize that, upon the induction of specific DNA damage, lack of phosphorylation of Ser389 hampers proper DNA binding of p53 to promoter sequences of its target genes, either directly or through inefficient formation of tetrameric complexes. As a consequence, the activity of downstream target genes, implicated in p53-dependent cellular functions aimed at eliminating damaged cells, is reduced and/or delayed. Eventually, this may result in an increased chance of fixation of mutations in critical genes, such as oncogenes and/or tumor suppressor genes, leading to cancer development. In line with phosphorylation of Ser389, this response appears to be DNA-damaging agent specific. Supporting this, our preliminary results indeed show that tumor development in p53.S389A mutant mice after gamma irradiation is minimal and indistinguishable from that in wild-type mice, whereas p53+/− mice are more sensitive (E. M. Hoogervorst, W. Bruins, C. T. M. van Oostrom, G. J. van den Aardweg, R. B. Beems, J. van den Berg, T. Jacks, H. van Steeg, and A. de Vries, unpublished data).
The increased sensitivity to UV-induced skin tumors of the p53.S389A mice shows a clear correlation between a biochemical modification of p53—phosphorylation of Ser389—and an important cellular function, i.e., tumor suppression after exposure to a genotoxic agent. Therefore, the applied strategy of a physiologically relevant single-mutation mouse model appears to be valuable for analyzing the importance of specific phosphorylation and/or other modification events in p53 for its functions in vivo. Future studies of Ser389 and other p53 modification residues, using a variety of DNA-damaging agents, will likely reveal their roles in p53 functions. Moreover, since the ways in which different modifications are interrelated remain to be elucidated, it will be worthwhile to generate and analyze mice with combinations of modification mutations.
Acknowledgments
We are grateful to A. G. Jochemsen and R. Stad for their assistance with the p53 Western blot analyses, J. de Wit for assistance with gamma radiation experiments, the Central Animal Facility Laboratory (RIVM-CDF) for their biotechnological support, J. Robinson for assistance with histopathological analyses, M. Luijten and S. Banus for help with statistical analyses, and S. Wijnhoven and T. M. Breit for critical reading of the manuscript.
This work was supported by the Dutch Cancer Society (KWF) grant RIVM 2000-2352 and NIH/NIEHS (Comparative Mouse Genomics Centers Consortium) grant 1UO1 ES11044-02.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2000. Signaling to p53: breaking the posttranslational modification code. Pathol. Biol. 48:227-245. [PubMed] [Google Scholar]
- 2.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 3.Attardi, L. D., A. de Vries, and T. Jacks. 2004. Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene 23:973-980. [DOI] [PubMed] [Google Scholar]
- 4.Attardi, L. D., and T. Jacks. 1999. The role of p53 in tumour suppression: lessons from mouse models. Cell. Mol. Life Sci. 55:48-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attardi, L. D., E. E. Reczek, C. Cosmas, E. G. Demicco, M. E. McCurrach, S. W. Lowe, and T. Jacks. 2000. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev . 14:704-718. [PMC free article] [PubMed] [Google Scholar]
- 6.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 7.Chao, C., M. Hergenhahn, M. D. Kaeser, Z. Wu, S. Saito, R. Iggo, M. Hollstein, E. Appella, and Y. Xu. 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 278:41028-41033. [DOI] [PubMed] [Google Scholar]
- 8.Chao, C., S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2000. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl. Acad. Sci. USA 97:11936-11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuddihy, A. R., A. H. Wong, N. W. Tam, S. Li, and A. E. Koromilas. 1999. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene 18:2690-2702. [DOI] [PubMed] [Google Scholar]
- 10.de Vries, A., E. R. Flores, B. Miranda, H. M. Hsieh, C. T. van Oostrom, J. Sage, and T. Jacks. 2002. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc. Natl. Acad. Sci. USA 99:2948-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donehower, L. A. 1996. The p53-deficient mouse: a model for basic and applied cancer studies. Semin. Cancer Biol. 7:269-278. [DOI] [PubMed] [Google Scholar]
- 12.El Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 13.Fiscella, M., N. Zambrano, S. J. Ullrich, T. Unger, D. Lin, B. Cho, W. E. Mercer, C. W. Anderson, and E. Appella. 1994. The carboxy-terminal serine 392 phosphorylation site of human p53 is not required for wild-type activities. Oncogene 9:3249-3257. [PubMed] [Google Scholar]
- 14.Funk, W. D., D. T. Pak, R. H. Karas, W. E. Wright, and J. W. Shay. 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol. 12:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, S. R., L. E. Campbell, and D. W. Meek. 1996. Phosphorylation of p53 at the casein kinase II site selectively regulates p53-dependent transcriptional repression but not transactivation. Nucleic Acids Res. 24:1119-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao, M., A. M. Lowy, M. Kapoor, A. Deffie, G. Liu, and G. Lozano. 1996. Mutation of phosphoserine 389 affects p53 function in vivo. J. Biol. Chem. 271:29380-29385. [DOI] [PubMed] [Google Scholar]
- 17.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 19.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C., W. Y. Ma, A. Maxiner, Y. Sun, and Z. Dong. 1999. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J. Biol. Chem. 274:12229-12235. [DOI] [PubMed] [Google Scholar]
- 21.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, W., H. N. Ananthaswamy, H. K. Muller, and M. L. Kripke. 1999. p53 protects against skin cancer induction by UV-B radiation. Oncogene 18:4247-4253. [DOI] [PubMed] [Google Scholar]
- 23.Kapoor, M., R. Hamm, W. Yan, Y. Taya, and G. Lozano. 2000. Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene 19:358-364. [DOI] [PubMed] [Google Scholar]
- 24.Kapoor, M., and G. Lozano. 1998. Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc. Natl. Acad. Sci. USA 95:2834-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller, D., X. Zeng, X. Li, M. Kapoor, M. S. Iordanov, Y. Taya, G. Lozano, B. Magun, and H. Lu. 1999. The p38MAPK inhibitor SB203580 alleviates ultraviolet-induced phosphorylation at serine 389 but not serine 15 and activation of p53. Biochem. Biophys. Res. Commun. 261:464-471. [DOI] [PubMed] [Google Scholar]
- 26.Keller, D. M., X. Zeng, Y. Wang, Q. H. Zhang, M. Kapoor, H. Shu, R. Goodman, G. Lozano, Y. Zhao, and H. Lu. 2001. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7:283-292. [DOI] [PubMed] [Google Scholar]
- 27.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 28.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 29.Ljungman, M. 2000. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia 2:208-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo, P. K., J. Y. Chen, P. P. Tang, J. Lin, C. H. Lin, L. T. Su, C. H. Wu, T. L. Chen, Y. Yang, and F. F. Wang. 2001. Identification of a mouse thiamine transporter gene as a direct transcriptional target for p53. J. Biol. Chem. 276:37186-37193. [DOI] [PubMed] [Google Scholar]
- 31.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 32.Lu, H., Y. Taya, M. Ikeda, and A. J. Levine. 1998. Ultraviolet radiation, but not gamma radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc. Natl. Acad. Sci. USA 95:6399-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meek, D. W., S. Simon, U. Kikkawa, and W. Eckhart. 1990. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 9:3253-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne, D. M., R. H. Palmer, and D. W. Meek. 1992. Mutation of the casein kinase II phosphorylation site abolishes the anti-proliferative activity of p53. Nucleic Acids Res. 20:5565-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyashita, T., S. Krajewski, M. Krajewska, H. G. Wang, H. K. Lin, D. A. Liebermann, B. Hoffman, and J. C. Reed. 1994. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9:1799-1805. [PubMed] [Google Scholar]
- 36.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 37.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, K., and D. Beach. 1994. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 13:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 40.Sakaguchi, K., H. Sakamoto, D. Xie, J. W. Erickson, M. S. Lewis, C. W. Anderson, and E. Appella. 1997. Effect of phosphorylation on tetramerization of the tumor suppressor protein p53. J. Protein Chem. 16:553-556. [DOI] [PubMed] [Google Scholar]
- 41.Schwenk, F., U. Baron, and K. Rajewsky. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shieh, S. Y., Y. Taya, and C. Prives. 1999. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 18:1815-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sluss, H. K., H. Armata, J. Gallant, and S. N. Jones. 2004. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell. Biol. 24:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatachalam, S., Y. P. Shi, S. N. Jones, H. Vogel, A. Bradley, D. Pinkel, and L. A. Donehower. 1998. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 17:4657-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, Z., J. Earle, S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2002. Mutation of mouse p53 Ser23 and the response to DNA damage. Mol. Cell. Biol. 22:2441-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]