The growth factor independence 1 (Gfi-1) zinc finger (ZF) protein and its homologs have been implicated in processes as diverse as oncogenesis, apoptosis, proliferation, cell fate specification, and differentiation. Studies with vertebrates have focused mostly on hematopoietic lineages whereas research with Caenorhabditis elegans and Drosophila has focused on the nervous system. However, recent reports have shown that Gfi-1 is also expressed in neurons in mice and zebra fish. In this review, we attempt to summarize the available data and elaborate on the similarities and differences between expression patterns, loss- and gain-of-function phenotypes, and modes of action of these genes. We draw parallels between several in vivo observations in which Gfi-1 and its homologs seem to cooperate with basic helix-loop-helix (bHLH) transcription factors in different tissues. These observations suggest new hypotheses for the roles and interactions of Gfi/Pag-3/Senseless (GPS) proteins in different organisms. Finally, we present data which suggest that GPS proteins may play a critical role in determination or early differentiation of many cell types.
STRUCTURAL AND FUNCTIONAL SIMILARITIES AND DIFFERENCES BETWEEN THE GPS PROTEINS
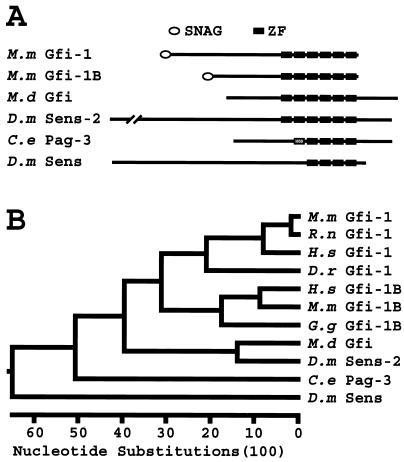
In 1993 Gilks and colleagues reported the isolation and preliminary characterization of the Gfi-1 gene. Upon culturing Moloney murine leukemia virus-induced rat T-cell lymphoma lines in interleukin-2 (IL-2)-free medium, they observed that new proviral insertions gave rise to IL-2-independent cell lines (35). The Gfi-1 locus was one of the insertion sites responsible for this phenotype. Since then, several groups have identified and studied orthologs of Gfi-1 in mice (37, 108), humans (78, 84), and zebra fish (23), as well as its closest homologs, i.e., Gfi-1B in vertebrates, senseless in Drosophila, M.d Gfi in Musca, and pattern of gene expression 3 (pag-3) in C. elegans (13, 29, 32, 43, 45, 49, 55, 56, 71, 96). The Drosophila gene CG31632, which we named senseless-2, is also a Gfi-1 homolog. GPS genes encode transcription factors with C2H2-type ZFs, which are the only motifs conserved among all of these proteins (Fig. 1-A). Gfi-1, Gfi-1B, M.d Gfi, and Senseless-2 contain six, Pag-3 contains five, and Senseless contains four ZFs. The phylogenetic analysis of the full-length GPS amino acid sequences suggests that Gfi-1 and Gfi-1B might have arisen from the duplication of an ancestral senseless-2/M.d Gfi-like gene (Fig. 1B). This implies that senseless and pag-3 were probably the first GPS genes to diverge from the other extant genes in this group, assuming that Gfi-1 and Gfi-1B are the only functional vertebrate GPS genes (96, 101).
FIG. 1.
Structural similarities and differences between the GPS proteins. (A) Domain structures of GPS proteins. Since Gfi-1 and Gfi-1B have similar lengths in different vertebrate species, a representative is shown for each. The first ZF of Pag-3 is shaded because of low homology with corresponding ZFs in other GPS proteins (see Fig. 2). The proteins are aligned relative to their ZFs. The drawings are to scale except for D. melanogaster Sens-2, which contains 200 extra amino acids at its N terminus. (B) Phylogenetic analysis of the GPS proteins. The evolutionary tree of the full-length GPS proteins was drawn based on the ClustalW algorithm. Note that Senseless behaves as an outlier. The MegAlign software of the DNASTAR (Madison, Wis.) package was used for this analysis. H.s, Homo sapiens; M.m, Mus musculus; R.n, Rattus norvegicus; D.r, Danio rerio; G.g, Gallus gallus; M.d, Musca domestica; D.m, Drosophila melanogaster; C.e, Caenorhabditis elegans.
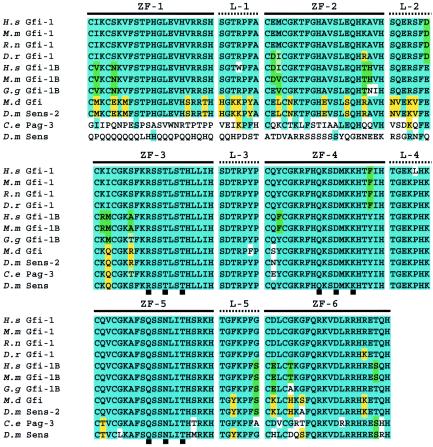
As shown in Fig. 2, the highest degree of homology exists between ZFs 3 to 5 of Gfi-1, Gfi-1B, M.d Gfi, and Senseless-2, which correspond to ZFs 2 to 4 of Pag-3 and to ZFs 1 to 3 of Senseless. ZFs 3 to 5 of Gfi-1 have been shown to be necessary and sufficient for binding its consensus recognition element, taAATCac(a/t)gca (uppercase indicates the core sequence) (109). As these three ZFs are conserved among all GPS proteins, it is conceivable that they all have similar binding sites. Indeed, the consensus binding site of Gfi-1B has been shown to be virtually identical to that of Gfi-1 (96), and Senseless and Pag-3 bind this consensus sequence with high affinity (1, 45). Interestingly, the degree of similarity between the ZFs of the GPS proteins seems to be beyond what is needed for similar DNA-binding properties. Not only are the amino acids in ZFs 3 to 5 predicted to contact DNA identical in all of these proteins (Fig. 2) (76, 77), but these three ZFs also exhibit a high level of conservation in amino acids that do not bind DNA and in the domains that link the ZFs. Finally, the ZFs that are dispensable for DNA binding also show a high level of conservation. These observations strongly suggest that the ZF motifs might endow the GPS proteins with functional similarities other than binding a similar DNA element, including interactions with other proteins or RNA elements.
FIG. 2.
Extensive sequence identity between GPS proteins in the ZFs involved in DNA binding (ZF-3 to -5) and their linker motifs (L-3 and L-4). Alignment of the ZFs of GPS proteins is shown. Blue boxes show the most conserved residues. The second most conserved residues are marked green (predominantly vertebrate) or yellow (predominantly invertebrate). In the case of equal numbers, the amino acids belonging to Gfi-1 are arbitrarily highlighted in blue. Black boxes underneath the sequences indicate amino acids predicted to contact DNA. H.s, Homo sapiens; M.m, Mus musculus; R.n, Rattus norvegicus; D.r, Danio rerio; G.g, Gallus gallus; M.d, Musca domestica; D.m, Drosophila melanogaster; C.e, Caenorhabditis elegans.
Vertebrate GPS proteins can act as transcriptional repressors (36, 96). They contain a Snail/Gfi-1 (SNAG) domain, which corresponds to the N-terminal 20 amino acids and has been shown to mediate the transcriptional repressor activity of Gfi-1 and Gfi-1B (36). However, there is one report suggesting that Gfi-1 functions as a transcriptional repressor even in the absence of the SNAG domain, albeit more weakly than the full-length protein (66). The latter function seems to require protein-protein interactions between Gfi-1, histone deacetylases, and a corepressor called ETO (for eight-twenty-one), which was first identified as a part of the fusion protein formed in the (8;21) translocation associated with acute myelogenous leukemia (25). ETO interacts with the ZFs of Gfi-1, and its Drosophila homolog is called nervy because of its expression in the central nervous system (CNS) and peripheral nervous system (PNS) (26). Interestingly, although Senseless, Senseless-2, Pag-3, and M.d Gfi do not have the SNAG domain, some data suggest that they also act as transcriptional repressors. Indeed, in the absence of pag-3 function, touch neuron-specific genes are ectopically expressed in other neurons (49); M.d Gfi might repress a housefly cytochrome P450 gene, CYP6D1 (56), and Senseless has been implicated in repressing proneural gene expression in cells other than sensory organ precursors (SOPs) (45). Also, loss of senseless is associated with upregulation of the Rough protein and the pointed gene in presumptive R8 photoreceptor cells (28, 29). Finally, Chandrasekaran and Beckendorf reported an increase in the expression of the proapoptotic genes reaper and hid in senseless mutant embryonic salivary glands (14). These findings strongly suggest that the invertebrate GPS proteins also repress the transcription of several target genes, possibly via recruiting corepressors. In addition, both Senseless and Pag-3 have motifs very similar to the consensus binding motif for the C-terminal binding protein (CtBP), a context-dependent transcriptional cofactor that can act as a corepressor in many contexts (16, 79). The interaction between Senseless and CtBP has been confirmed in vitro and in vivo (our unpublished data). It is therefore possible that Senseless and Pag-3 compensate for the lack of a SNAG domain via interactions with additional corepressors. It is worth mentioning that Gfi-1 and Gfi-1B do not have a consensus CtBP-binding motif. Finally, M.d Gfi and Senseless-2 contain alanine-rich domains, which have been shown in some cases to be involved in transcriptional repression (46, 65).
Although in most cases Gfi-1 and Gfi-1B act as DNA-binding transcriptional repressors, there might be exceptions to this rule. Specifically, Gfi-1B and Gfi-1 can activate the transcription of a promoter containing four copies of their consensus binding site in an erythroid cell line (74). Interestingly, the socs-1 promoter activity was repressed in the same cell line, as expected from a previous report (48, 74). This suggests that Gfi-1 and Gfi-1B can affect target promoters differentially according to the cellular and promoter contexts. Also, deletion of the Gfi-1-binding element in the promoter of the β1 soluble guanylyl cyclase gene results in significantly decreased luciferase activity in transient-transfection assays with a human neuroblastoma cell line, suggesting that Gfi-1 might function as an activator of this gene in specific cells (92).
The study of Senseless in Drosophila has also provided evidence for a dual role as a transcriptional repressor and activator. Specifically, low levels of Senseless can repress the transcription of the proneural bHLH gene achaete in a DNA-binding-dependent manner. However, higher levels of Senseless can synergize with proneural proteins on the achaete promoter and strongly activate transcription, even after mutation of the Senseless-binding site in the achaete promoter (45). One can envisage that the activator function of Senseless on the achaete promoter might be entirely mediated through repression of a negative regulator of bHLH proteins or genes (see below). However, specific point mutations that abolish DNA binding and repressor function of Senseless are still able to synergistically activate achaete transcription in cell culture assays (our unpublished data). Moreover, a recent report also suggests the dual role for Senseless. While Senseless function is necessary for sensory organ-promoting effects of a gene called phyllopod, Senseless-binding sites in the phyllopod enhancer play a negative role in the regulation of phyllopod activity (80). Of note, there are glutamine-rich domains in Senseless and Senseless-2 that might mediate transcriptional activation (46). Hence, the present data suggest that Senseless and probably other GPS proteins might act as both activators and repressors. However, it is possible that the activator functions may not require DNA binding.
GPS PROTEINS PREVENT APOPTOSIS
Apoptosis plays a central role in animal development and tumor formation (67). One of the biological processes that GPS proteins regulate is apoptosis. Gfi-1 was originally identified as a proviral integration site that rendered a T-cell lymphoma line independent of IL-2 (35). Gfi-1 overexpression downregulates the expression of proapoptotic genes and inhibits apoptosis both in immortalized IL-2-dependent T cells and in explanted primary thymocytes (37). Also, Gfi-1 overexpression renders Jurkat T cells insensitive to activity-induced cell death, an apoptotic response initiated by a strong T-cell receptor stimulation in peripheral T lymphocytes (54). These observations are in agreement with loss-of-function data indicating a severely decreased thymic cellularity in Gfi-1-deficient mice, which is thought to be in part due to Gfi-1's role in preventing apoptosis in early T-cell precursors (43, 105). Gfi-1's involvement in survival might be crucial for its role as an oncoprotein and its cooperation with Myc. It has been suggested that cells that are forced into a proliferative mode become sensitive to the induction of apoptosis, perhaps to decrease the risk of neoplasm formation (67). Interestingly, some oncoproteins, like Myc, are potent inducers of events that can lead to the activation of the apoptotic cascade (51, 52). Therefore, preventing apoptosis and promoting proliferation (see below) is probably part of the mechanism by which overexpression of Gfi-1 can result in full malignant transformation of malignancies promoted by Myc and Pim oncoproteins (91).
Gfi-1 is also involved in preventing apoptosis in the cochlear hair cells of the inner ear (101). In Gfi-1 mutant mice, hair cells are initially specified but fail to differentiate properly and are disorganized. Later on, the hair cells go through apoptosis in a basal to apical order along the cochlea. Interestingly, the vestibular hair cells do not undergo apoptosis in the absence of Gfi-1, although they are also abnormal and disorganized. This suggests that preventing apoptosis is certainly not the only role Gfi-1 plays in the inner ear, nor is Gfi-1 the only protein that promotes hair cell survival. Nevertheless, Gfi-1 seems to be involved in sensing and/or implementing the survival signals that the cochlear hair cells need to receive in order to continue their differentiation.
Senseless also prevents programmed cell death in Drosophila embryos. In senseless null mutant embryos, SOPs form and divide, only to go through apoptosis during PNS organ differentiation (71). The antiapoptotic role of Senseless has been studied in great detail in embryonic salivary glands (14). In salivary glands, Senseless is not involved in cell fate specification. However, it is specifically required for survival, as loss of senseless function can be rescued by a concomitant loss of proapoptotic genes or by overexpression of the antiapoptotic protein P35 (41). As mentioned, Senseless seems to regulate the transcription of the proapoptotic genes reaper and possibly hid. It is interesting that blocking apoptosis fails to rescue the senseless mutant phenotype in the embryonic PNS, suggesting roles other than preventing cell death in the embryo (14). A similar observation has been made for the developing eye, where unrecruited, undifferentiated cells are normally eliminated by apoptosis. Inhibition of apoptosis, however, failed to rescue the senseless loss-of-function phenotype in the eye (30).
Involvement in apoptosis extends to another GPS protein, Pag-3. In pag-3 mutants there are extra cell corpses along the ventral nerve cord because an increased number of cells undergo programmed cell death. Interestingly, in some lineages, cells that normally die via apoptosis survive in pag-3 mutants (13). It is therefore not obvious how Pag-3 regulates apoptosis in worms, especially considering the fact that at least part of the apoptosis-related phenotypes observed in pag-3 mutants might be secondary to cell fate determination abnormalities. It remains to be seen whether Pag-3 regulates the expression of proapoptotic genes in C. elegans.
GPS PROTEINS AND PROLIFERATION
Both loss- and gain-of-function studies suggest a role for Gfi-1 in proliferation. During T-cell activation, the interaction between IL-2 and its receptor is essential for antigen-mediated proliferation and an efficient immune response (35). Ligation of T-cell receptors on peripheral T cells results in activation and a transient increase in Gfi-1 expression. Transgenic expression of Gfi-1 in either T-cell lines or primary T cells increases activation- (19, 54, 85) and cytokine-induced proliferation (19). IL-6-mediated signaling through STAT3 is involved in the proliferation of T cells upon antigenic stimulation, and the observation that Gfi-1 physically interacts with the STAT-3 inhibitor PIAS3 might offer an explanation for Gfi-1's positive role in proliferation (85). The differentiation of CD4+ T cells into Th2 lymphocytes involves a large amount of cellular proliferation, and IL-4 is key to this process. IL-4 induces Gfi-1 expression in Th2 lymphocytes, and prolonged Gfi-1 expression results in the expansion of these cells (107). Overexpression of Gfi-1 renders lymphoma cell lines independent of IL-2 for their growth, suggesting a role in cell proliferation (35). As mentioned above, loss of Gfi-1 function results in reduced thymic cellularity because of increased cell death and lack of proliferation (105).
Albeit at a much lower frequency than Gfi-1, Gfi-1B has also been implicated in accelerating lymphomagenesis by cooperating with other oncoproteins (96). Forced expression of Gfi-1B in human primary CD34+ hematopoietic progenitors resulted in a dramatic expansion of erythroblasts (74), suggesting a role for Gfi-1B in proliferation. These erythroblasts failed to proceed to mature red blood cells and underwent massive apoptosis. However, loss-of-function studies did not point to a specific role for Gfi-1B in proliferation. Specifically, Gfi-1B mutant hematopoietic progenitors could generate rapidly proliferating colonies of arrested erythroid precursors in response to growth factors (88). The same phenomenon is observed in Gfi-1B mutant megakaryocytic precursors in response to growth factors. These data suggest that the developmental block in the erythroid/megakaryocytic pathways generated by loss of Gfi-1B function is less likely to be because of a failure of these cells to respond to growth-promoting cues from the environment.
The evidence for a role of the invertebrate GPS proteins in cell proliferation is not very obvious. In pag-3 mutants, some of the neuroblasts undergo extra rounds of division, suggesting that instead of exiting the cell cycle and proceeding with neuronal differentiation, some of the progeny of the pag-3 mutant neuroblasts adopt a neuroblast fate themselves (13). These data suggest that Pag-3 might link the neuroblast fate determination to terminal neuronal differentiation, in part via controlling the cell cycle.
Senseless might also play a role in controlling the cell cycle and proliferation. In sensLyra mutants, ectopic expression of intermediate levels of Senseless in the third-instar wing pouch results in wing margin tissue loss in the adult flies (70). The loss of tissue is not due to an increase in cell death (2). Instead, there is a severe downregulation of the mitosis-inducing phosphatase string (cdc25), suggesting that lack of proliferation might in part account for the senseless gain-of-function phenotype. This would imply that specific levels of Senseless may inhibit the proliferation of some cells, besides being able to turn many other epidermal cells into neurons (71). However, it is not yet clear if and how Senseless regulates the cell cycle in the SOPs and their progeny.
In summary, Gfi-1 has a well established role in cell cycle regulation and proliferation. Although Gfi-1B can act as an oncoprotein, it is not obvious whether it promotes cell proliferation when expressed normally. Finally, while there are reasons to assume that invertebrate GPS proteins might regulate cell cycle progression, future studies will have to establish the exact role that each GPS protein plays in this biological process.
THE LINK BETWEEN BHLH AND GPS PROTEINS
bHLH transcriptional activators are key regulators of precursor determination and differentiation in numerous developmental processes, including myogenesis, neurogenesis, hematopoiesis, and the development of the heart, limb, and pancreas (9, 15, 27, 40, 68, 72, 93). They have also been implicated in tumor formation (17). Among the best-studied groups of the bHLH superfamily are the proneural proteins. These proteins are necessary and sufficient to generate neural precursors from ectodermal primordia (38, 47, 63, 99). They form heterodimers with class I bHLH activators such as Daughterless and E12/E47 to bind specific DNA sequences called E-boxes and to activate the transcription of their target genes (12, 64, 98). To prevent overproduction of neural tissue, proneural proteins induce inhibitory cell-cell interactions mediated by Notch signaling, which ultimately prevent proneural protein upregulation in most cells of the proneural field (4, 42, 60, 94). Notch signaling induces transcription of the genes of the Enhancer of Split complex [E(spl)-C], a group of bHLH transcription factors that repress proneural gene expression by several mechanisms (18, 34, 45, 59). Proneural proteins themselves are direct transcriptional activators of E(spl)-C gene expression (94). These observations strongly suggest that in order to be upregulated and to specify the SOPs, proneural proteins should overcome the negative regulation of Notch signaling/E(spl) proteins. Several lines of evidence suggest that Senseless is one of the main factors that confers to the proneural proteins the ability to escape this inhibition: senseless transcription is directly activated by proneural proteins; Senseless synergizes transcriptionally with proneural proteins such as Achaete and Daughterless to upregulate proneural gene expression; Senseless synergizes genetically with proneural proteins in forming sensory organs, as comisexpression of Senseless and proneural proteins greatly increases the number of sensory organs formed (45, 71); and sensory bristles formed upon Senseless misexpression are very closely spaced, suggesting inefficient Notch signaling (80). It is interesting that in many tissues, low levels of Senseless in cells around the presumptive SOP repress proneural gene expression and thereby antagonize neuronal fate. This has led to the notion that Senseless might act as a binary switch during adult peripheral neurogenesis.
Senseless interacts with bHLH activators in several other contexts in flies and cooperates with bHLH proteins to enforce the fate they have initiated. In eye imaginal discs, Senseless is activated by the bHLH protein Atonal to suppress Rough, which is an inhibitor of the R8 photoreceptor fate normally promoted by Atonal (29). In embryonic SOPs, senseless expression depends on proneural proteins. Senseless in turn is necessary for the proper differentiation of the SOP progeny in the embryo (71). Finally, in embryonic salivary glands, senseless expression is initiated by Fork head (Fkh) and maintained by Sage, a salivary gland-specific bHLH activator that promotes the survival of salivary cells. Hence, in all tissues where Senseless is expressed, we observe that its expression is initiated and/or maintained via bHLH proteins. The only known exception is the wing margin, where the broad, low-level senseless expression seems to be under the control of Wingless (45, 75).
Gfi-1 is expressed in both the CNS and a variety of sensory organs. It has been detected in olfactory epithelia, Merkel cells of the skin, the dorsal epithelium of the tongue where taste papilla develop, optic epithelia, and the otic vesicle (23, 101). In the inner ear, Gfi-1 mRNA is initially expressed in a broad domain. Later on, this expression becomes confined to the precursors of the inner ear hair cells. This pattern is very similar to the expression pattern of Math1, one of the two mouse homologs of the Drosophila proneural gene atonal, which is necessary for the generation of the inner ear hair cells (8). Direct regulation of senseless by proneural proteins combined with evidence for similar expression patterns of Gfi-1 and bHLH proteins suggests that Gfi-1 expression might also be regulated by one or more bHLH proteins. Although the early expression of Gfi-1 does not seem to depend on Math1 (101), Math1 is a good candidate for being upstream of Gfi-1 to control its hair cell-specific expression, as Gfi-1 expression is abolished in the presumptive hair cells of Math1 mutants.
The functional relationship between Gfi-1 and bHLH proteins in the inner ear is also quite reminiscent of the Senseless/proneural relationship in flies. Math1 and atonal are highly conserved: both genes specify cells with similar functions (inner ear hair cells and fly mechanoreceptors) and are functionally interchangeable, as each gene can substitute for the function of the other (7, 103). Interestingly, the role of Gfi-1 in inner ear hair cell development is also similar to the role of Senseless in Drosophila embryonic PNS (see above). In Gfi-1 mutant mice, inner ear hair cells are specified but fail to differentiate properly, and the mutant mice are deaf and ataxic (101). A similar relationship might also exist between Gfi-1 and Math5, the other mouse Atonal homolog involved in retinal development (11, 102). Loss of Math5 blocks differentiation of most retinal ganglion cells and results in severely reduced Gfi-1 expression in these cells (104). This observation suggests that Gfi-1 might be activated by Math5 to ensure the proper differentiation or survival of the retinal ganglion cells. These data are in line with the hypothesis that GPS proteins might be involved in consolidating the fate specified by bHLH transcriptional activators.
More recently, Gfi-1 was found to be necessary for the differentiation of pulmonary neuroendocrine cells. Importantly, it was shown to be expressed in neuroendocrine lung cancer cell lines, especially in small-cell lung carcinomas (SCLCs) (58a). It should be noted that Mash1, a mouse homolog of the Drosophila proneural genes achaete and scute, is necessary for pulmonary neuroendocrine cell formation and is highly expressed in SCLCs (6, 10). Hence, this is yet another example where proneural proteins may activate Gfi-1. Gfi-1 in turn seems to be involved in enforcing the neuroendocrine fate induced by a proneural-type protein. Similar to the SOP formation paradigm, Notch signaling opposes the neuroendocrine phenotype conferred by Hash-1 (the human homolog of Mash-1) to cultured SCLC cells (95). However, Gfi-1 promotes the neuroendocrine phenotype and probably helps the bHLH protein to escape the negative regulation of Notch signaling.
T-cell maturation is another process in which Gfi-1 and bHLH activators promote a similar outcome. Specifically, similar to E47 mutant mice, Gfi-1 mutant mice develop significantly more CD8+ T cells and show an accelerated positive selection of double-positive thymocytes (5, 105). The observation that Id1 (for inhibitor of DNA binding) and Id2, which are negative regulators of the E47 protein, are upregulated in Gfi-1 mutant thymocytes suggests that Gfi-1 might repress Id1 and Id2 expression and thereby be required together with E47 for a correct CD4/CD8 lineage decision (5, 105). It would be interesting to test whether Gfi-1 expression in thymocytes depends on E47 activity. Also, Id1 and Id2 are homologs of the Drosophila protein Extra macrochaetae (Emc), one of the negative regulators of proneural protein function (24, 33). Therefore, repression of emc transcription might be one of the mechanisms by which Senseless upregulates proneural proteins.
Finally, lymphomagenesis is another setting in which GPS proteins cooperate with bHLH activators. The Myc family of oncoproteins are involved in the formation of a variety of human cancers (17). Like other transcriptional activators containing bHLH domains, dimers of the Myc proteins bind E-box DNA elements and activate the transcription of their target genes (64). Interestingly, the low oncogenic potential of Gfi-1 increases dramatically when it is coexpressed with Myc and another oncoprotein, Pim-1 (90, 108). Similarly, the presence of Gfi-1 clearly accelerates the process of lymphoma formation initiated by Myc and Pim-1. Indeed, Gfi-1 has been identified as a frequent target of Moloney proviral insertions in the lymphomas of rats, mice, and Myc- and Pim-1-transgenic mice (3, 35, 62, 89, 91, 96).
Altogether, GPS proteins seem to cooperate with bHLH activators in many cellular and developmental contexts. GPS expression seems to be regulated by bHLH proteins in both vertebrates and invertebrates, and there is ample evidence that this regulation might be direct in flies. Although Senseless is required for proneural protein upregulation and maintenance in adult SOPs, a similar role for Gfi-1 or Gfi-1B has not been observed yet (88, 101). Indeed, a recent report suggests that in vertebrates, another ZF protein called MyT1 synergizes with Neurogenin1 and antagonizes Notch signaling to induce neurogenesis, a role that is played by Senseless in fly SOP formation (81). Finally, the GPS-bHLH cooperation is of a synergistic nature in some contexts both in flies and in vertebrates. Accordingly, inhibition of Gfi-1 function might be a fruitful approach in treating malignancies such as SCLC, in which bHLH and GPS proteins are coexpressed (58a). Even more exciting is the idea of using GPS proteins to synergize with bHLH proteins in generating cell types that are lost to disease or trauma. For example, the generation of inner ear hair cells by exogenous Math1 delivery in rat cochlear and utricular explants and in adult guinea pigs (58, 106) may be enhanced by codelivery of Gfi-1 with Math1.
SIMILAR FACTORS OTHER THAN BHLH PROTEINS MIGHT CONTROL GPS EXPRESSION
Similarities between the expression and function of senseless and Gfi-1 in sensory organs of flies and mice suggest that similar proteins might regulate the transcription of these genes. Although bHLH proteins regulate senseless expression in many contexts, senseless expression does not depend solely on proneural proteins. The low-level expression of senseless in the wing margin and the initiation of senseless expression in the embryonic salivary glands is independent of bHLH proteins (14, 45). One of the senseless enhancers identified so far corresponds to a 200-bp fragment which is sufficient for expression in almost all of the embryonic SOPs and a large number of pupal SOPs (45) (Fig. 3). Although the single E-box in this enhancer has a major role in gene expression, a residual β-galactosidase expression in some embryonic and pupal precursors is observed when this E-box is mutated (45), suggesting that proteins other than proneural proteins regulate this enhancer. Sequence analysis with the MatInspector software (82) predicted several putative binding sites for other transcription factors in this 200-bp enhancer (Fig. 3). Alignment of the senseless genomic regions of Drosophila melanogaster and Drosophila pseudoobscura showed that some of the binding sites predicted by MatInspector are highly conserved between these two species, which diverged around 25 to 30 million years ago (87). Specifically, the 200-bp senseless enhancer contains conserved putative binding sites for Pax-2 and the GATA protein Pannier, both of which are important for PNS development (31, 39, 57). Also, the winged helix transcription factor Fkh initiates senseless expression in embryonic salivary gland placodes prior to its becoming dependent on the heterodimer of Sage and Daughterless bHLH proteins (14). Interestingly, homologs of all of these proteins are known to be involved in inner ear hair cell development: Pax-2 has been shown to be a major regulator of inner ear patterning, Fkh10 mutant mice exhibit vestibular and hearing impairments, and GATA-3 is involved in the morphogenesis of the inner ear (44, 53, 61, 97). These observations suggest that a similar battery of transcription factors might regulate PNS-specific expression of senseless and Gfi-1.
FIG. 3.
Evolutionarily conserved putative binding sites for several transcription factors are present in a senseless enhancer. Comparison of a 200-bp D. melanogaster (D.m) senseless enhancer with the corresponding enhancer of D. pseudoobscura (D.p) is shown. Boxes indicate identical nucleotides. The predicted binding sites are marked by the horizontal lines above the sequences.
Although the expression of senseless and pag-3 has been identified only in the nervous system (with the exception of Drosophila embryonic salivary gland), Gfi-1 and Gfi-1B are expressed in many other tissues. Gfi-1 expression has been detected in the thymus, spleen, testis, bone marrow, lung, and gut (35, 96, 101). Gfi-1B is expressed in the spleen, bone marrow, fetal liver, fetal thymus, and testis (19, 86, 96). Is there a reason to think that similarities between transcriptional regulators of senseless and its vertebrate homologs might go beyond the nervous system? We believe that there is some evidence in favor of this speculation. There is indeed precedence for similar gene regulation mechanisms between the fly PNS and the vertebrate hematopoietic system. GATA proteins regulate multiple aspects of hematopoietic development (73). An oligomeric complex containing the hematopoietic bHLH protein TAL-1/SCL and GATA-1 has been implicated in regulating transcription in erythroid cells (100). In Drosophila, a similar complex has been shown to directly regulate proneural gene expression in vivo (83). Careful inspection of the above-mentioned 200-bp senseless enhancer indicates the proximity of the conserved E-box and GATA-binding sites (Fig. 3). Together, these observations suggest that Gfi-1 and Gfi-1B expression might also be regulated by a GATA-bHLH complex.
Another common mode of GPS regulation might be negative autoregulation. This mechanism was first discovered by Jia and colleagues in C. elegans, where they showed that pag-3 message was upregulated in pag-3 loss-of-function mutants (50). Recently, it was reported that Gfi-1 can bind the Gfi-1 promoter and repress its own expression in T cells (20).
In summary, Gfi-1 is expressed in precursors of organs involved in almost all types of senses, including hearing, balance, vision, touch, smell, and taste (101), and senseless is expressed in precursors of all PNS organs in Drosophila. Given the fact that homologs of several transcription factors predicted to regulate senseless expression play important roles in vertebrate PNS development, it is only natural to speculate that they might be involved in Gfi-1 regulation too. Moreover, similar transcriptional regulatory complexes have been shown to function in both vertebrate hematopoietic and fly nervous systems (83, 100). This should encourage efforts to study the transcriptional regulation of senseless, watching for possible parallels with vertebrate hematopoiesis.
GPS PROTEINS PLAY IMPORTANT ROLES IN CELL FATE DETERMINATION AND DIFFERENTIATION
Studies of the invertebrate GPS proteins implicate their involvement in both specification and differentiation. Senseless, for example, plays a variety of roles in different cell types, ranging from SOP fate specification in adult external sensory organs to differentiation of R8 photoreceptors in the eye to a pure antiapoptotic role in embryonic salivary glands (14, 29, 71). However, vertebrate GPS proteins so far have been found to function mostly after lineage commitments have been made. These observations suggest that playing a role in early lineage decisions might be one of the differences between vertebrate and invertebrate GPS proteins, perhaps parallel to the divergent roles of their corresponding bHLH proteins (40). However, we cannot exclude the possibility that redundancy between Gfi-1 and Gfi-1B functions might belie their role in precursor selection, at least in some contexts.
Pag-3 has also been shown to function in cell fate determination and differentiation. In pag-3 mutants, some of the neuroblast progeny seem to adopt the fate of their mother cells (13). Specifically, instead of producing a terminally differentiated neuron, one of the Pn.aaa progeny behaves like a neuroblast in the absence of pag-3 function. Pag-3 is also involved in differentiation of several neuronal lineages in C. elegans (13). Therefore, Pag-3 shares with Senseless the ability to function at various stages during the development of different cell lineages.
Loss-of-function studies have established that Gfi-1B plays an essential role in the generation of red blood cells and platelets (88). Gfi-1B mutant embryos form immature primitive erythrocytes that are able to perform only a limited degree of oxygenation. Definitive erythropoiesis, however, is disrupted, as there are no adult red blood cells in these embryos, leading to death by E15. Based on marker analysis, it was concluded that in the absence of Gfi-1B, hematopoietic progenitors commit to the erythroid lineage but fail to mature. Gfi-1B has a similar role in platelet formation; there is a block in megakaryopoiesis in Gfi-1B mutant embryos after cells are committed to the megakaryocytic or perhaps erythoid/megakaryocytic lineage (88). Although Gfi-1B is expressed in a myeloid cell line, loss of Gfi-1B function does not result in a gross abnormality in the shape or number of myeloid cells.
Gfi-1 also functions in a lineage-specific fashion. Although lack of Gfi-1 is compatible with the formation of mature T and B cells, Gfi-1 plays an important role in the early differentiation steps of lymphocytes (see above) (43, 105). Moreover, loss of Gfi-1 function is not compatible with the formation of normal neutrophils beyond the promyelocyte stage, as Gfi-1 regulates neutrophil development in a cell-autonomous fashion (43, 55). Instead of normal granulocytes, Gfi-1 mutant mice accumulate “atypical myeloid” cells with characteristics of both granulocyte and macrophage lineages in their blood as they age. In fact, coexpression of monocytic and granulocytic lineage markers in these cells suggests that Gfi-1 might also oppose the adoption of macrophage fate by myeloid precursors. Some mutant animals survive up to a year without antibiotics, in agreement with the finding that the atypical myeloid cells preserve some of the capacities needed to defend against pathogens, such as phagocytosis and oxidative burst activity. However, these cells do not seem to provide an efficient defense mechanism, as the mutant animals are highly susceptible to infection and abscess formation by gram-positive bacteria (43). Also, the Gfi-1-deficient macrophages tend to produce increased levels of inflammatory cytokines in response to the bacterial lipopolysaccharide, which can kill the mutant mice even at low doses (55). Interestingly, heterozygous mutations in Gfi-1 have been found in patients with neutropenia (21, 78). These mutations are able to act in a dominant negative fashion in transient-transfection assays, and it has been suggested, based on several lines of evidence, that a derepression of the gene Ela2 (encoding neutrophil elastase) might underlie the hematopoietic problems in the affected patients. A similar increase in Ela2 transcript level has also been reported for the bone marrow cells of the Gfi-1 mutant mice (43).
Together, these data indicate that both Gfi-1 and Gfi-1B are required for the differentiation of specific cell types after the lineage commitment has occurred, unlike Senseless and Pag-3, for which earlier roles have also been established. However, circumstantial evidence suggests that Gfi-1 and Gfi-1B might also regulate earlier steps in hematopoietic development. First, many of the known “lineage-restricted” hematopoietic transcription factors are also expressed at lower levels in uncommitted progenitors (73). Indeed, expression of both Gfi-1 and Gfi-1B has been detected in hematopoietic stem cells (74), suggesting an earlier role before the lineage commitments are made. Second, chromatin immunoprecipitation assays have shown that Gfi-1 is recruited to the Gfi-1B promoter in both myeloid and lymphoid cell lines (22), which is compatible with the idea that one protein might regulate the expression of the other. Furthermore, Gfi-1B is able to function as a direct transcriptional repressor of Gfi-1 in both a T-cell line and primary thymocytes (20). Taken together, these observations suggest the possibility that the effects of loss of one protein might be in part compensated for by the presence of the other, either at wild-type levels or, if there is indeed mutual repression, at higher-than-wild-type levels. This means that the earliest phenotype that can be observed from the loss of one gene would be at a stage in which each of the two has assumed its lineage-specific role. It is important to note that there is data in the literature suggesting that Gfi-1 and Gfi-1B might not be interchangeable in all cellular and developmental contexts: Gfi-1 is found much more frequently than Gfi-1B as a proviral insertion site in T-cell malignancies (3), and, unlike the case for Gfi-1, Gfi-1B overexpression results in defective T-cell activation (19). Nevertheless, given their structural similarities, it is reasonable to think that in some contexts the two proteins might perform redundant functions. In summary, the versatility of the vertebrate GPS proteins in regulating several cellular processes and the involvement of their invertebrate counterparts in determination and differentiation of some of the early neural precursors should encourage the study of their role in survival and maintenance of stem cells.
EPILOGUE
Although different GPS proteins have mostly been studied in different biological contexts, the picture emerging from the recent reports suggests that the parallels between the functions of these proteins may be more significant than originally thought. It is therefore natural to speculate on what those parallel functions might be and to design experiments to validate the speculations. The evidence for similarities between the regulation and function of this small group of ZF proteins both encourages further studies to identify the mechanism of their currently known functions and holds the promise for the discovery of novel aspects of their biological roles. Several important questions need to be answered. What are the phenotypes of Gfi-1 Gfi-1B double-knockout mice? What is the functional importance of Gfi-1 expression in sensory organs other than the inner ear? What is the loss-of-function phenotype of Gfi-1 in the CNS? Is Gfi-1B expressed anywhere outside the hematopoietic system? Does Senseless or Senseless-2 play a role in Drosophila hemocyte development? Is the expression of pag-3 regulated by a bHLH protein?
This all brings to mind the old “elephant in the dark” story beautifully told by the 13th century mystic poet Rumi to metaphorically describe one of the challenges faced by the seekers of truth (69). Briefly, an elephant was brought for exhibition to a city where no one had seen an elephant before and put in a dark room. Wise men were asked to touch the animal and report their experience to others. The person who touched the ear assumed that the beast was fan-shaped, the one who felt the leg reported the animal to be like a pillar, and the one who laid his hand on its back said that it was similar to a throne. Rumi suggests that the difference would have gone out of their words had each of them carried a candle in his hands. We anticipate that the collective efforts of researchers to study the regulation and function of GPS proteins in different contexts will eventually shed light on the conserved functions of these proteins.
Acknowledgments
We thank Melih Acar, Hillary Andrews, Vafa Bayat, Lee Grimes, Haluk Lacin, Clayton Morrison, Elaine Seto, Hiroshi Tsuda, Koen Venken, and Huda Zoghbi for comments on the manuscript and Cheng-Ting Chien and Lee Grimes for sharing unpublished data.
H.J.-N. is supported by NIH Medical Genetics Research Fellowship Program grant T32-GMO7526. This work was supported by a grant from NASA. H.J.-N. is an associate and H.J.B. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aamodt, E., L. Shen, M. Marra, J. Schein, B. Rose, and J. B. McDermott. 2000. Conservation of sequence and function of the pag-3 genes from C. elegans and C. briggsae. Gene 243:67-74. [DOI] [PubMed] [Google Scholar]
- 2.Abbott, L. A. 1986. Restoration of wing margins in Lyra mutants of Drosophila melanogaster. Dev. Biol. 115:233-248. [Google Scholar]
- 3.Akagi, K., T. Suzuki, R. M. Stephens, N. A. Jenkins, and N. G. Copeland. 2004. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32:D523-D527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 5.Bain, G., M. W. Quong, R. S. Soloff, S. M. Hedrick, and C. Murre. 1999. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J. Exp. Med. 190:1605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball, D. W., C. G. Azzoli, S. B. Baylin, D. Chi, S. Dou, H. Donis-Keller, A. Cumaraswamy, M. Borges, and B. D. Nelkin. 1993. Identification of a human achaete-scute homolog highly expressed in neuroendocrine tumors. Proc. Natl. Acad. Sci. USA 90:5648-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Arie, N., B. A. Hassan, N. A. Bermingham, D. M. Malicki, D. Armstrong, M. Matzuk, H. J. Bellen, and H. Y. Zoghbi. 2000. Functional conservation of atonal and Math1 in the CNS and PNS. Development 127:1039-1048. [DOI] [PubMed] [Google Scholar]
- 8.Bermingham, N. A., B. A. Hassan, S. D. Price, M. A. Vollrath, N. Ben-Arie, R. A. Eatock, H. J. Bellen, A. Lysakowski, and H. Y. Zoghbi. 1999. Math1: an essential gene for the generation of inner ear hair cells. Science 284:1837-1841. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand, N., D. S. Castro, and F. Guillemot. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3:517-530. [DOI] [PubMed] [Google Scholar]
- 10.Borges, M., R. I. Linnoila, H. J. van de Velde, H. Chen, B. D. Nelkin, M. Mabry, S. B. Baylin, and D. W. Ball. 1997. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386:852-855. [DOI] [PubMed] [Google Scholar]
- 11.Brown, N. L., S. Patel, J. Brzezinski, and T. Glaser. 2001. Math5 is required for retinal ganglion cell and optic nerve formation. Development 128:2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabrera, C. V., and M. C. Alonso. 1991. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 10:2965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cameron, S., S. G. Clark, J. B. McDermott, E. Aamodt, and H. R. Horvitz. 2002. PAG-3, a Zn-finger transcription factor, determines neuroblast fate in C. elegans. Development 129:1763-1774. [DOI] [PubMed] [Google Scholar]
- 14.Chandrasekaran, V., and S. K. Beckendorf. 2003. senseless is necessary for the survival of embryonic salivary glands in Drosophila. Development 130:4719-4728. [DOI] [PubMed] [Google Scholar]
- 15.Charite, J., D. G. McFadden, and E. N. Olson. 2000. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development 127:2461-2470. [DOI] [PubMed] [Google Scholar]
- 16.Chinnadurai, G. 2003. CtBP family proteins: more than transcriptional corepressors. Bioessays 25:9-12. [DOI] [PubMed] [Google Scholar]
- 17.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 18.Delidakis, C., and S. Artavanis-Tsakonas. 1992. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 89:8731-8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doan, L. L., M. K. Kitay, Q. Yu, A. Singer, S. Herblot, T. Hoang, S. E. Bear, H. C. Morse III, P. N. Tsichlis, and H. L. Grimes. 2003. Growth factor independence-1B expression leads to defects in T cell activation, IL-7 receptor alpha expression, and T cell lineage commitment. J. Immunol. 170:2356-2366. [DOI] [PubMed] [Google Scholar]
- 20.Doan, L. L., S. D. Porter, Z. Duan, M. M. Flubacher, D. Montoya, P. N. Tsichlis, M. Horwitz, C. B. Gilks, and H. L. Grimes. 2004. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 32:2508-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan, Z., and M. Horwitz. 2003. Gfi-1 oncoproteins in hematopoiesis. Hematology 8:339-344. [DOI] [PubMed] [Google Scholar]
- 22.Duan, Z., and M. Horwitz. 2003. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl. Acad. Sci. USA 100:5932-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dufourcq, P., S. Rastegar, U. Strahle, and P. Blader. 2004. Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr. Patterns 4:53-57. [DOI] [PubMed] [Google Scholar]
- 24.Ellis, H. M., D. R. Spann, and J. W. Posakony. 1990. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell 61:27-38. [DOI] [PubMed] [Google Scholar]
- 25.Erickson, P. F., M. Robinson, G. Owens, and H. A. Drabkin. 1994. The ETO portion of acute myeloid leukemia t(8;21) fusion transcript encodes a highly evolutionarily conserved, putative transcription factor. Cancer Res. 54:1782-1786. [PubMed] [Google Scholar]
- 26.Feinstein, P. G., K. Kornfeld, D. S. Hogness, and R. S. Mann. 1995. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics 140:573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firulli, A. B., D. G. McFadden, Q. Lin, D. Srivastava, and E. N. Olson. 1998. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 18:266-270. [DOI] [PubMed] [Google Scholar]
- 28.Frankfort, B. J., and G. Mardon. 2004. Senseless represses nuclear transduction of Egfr pathway activation. Development 131:563-570. [DOI] [PubMed] [Google Scholar]
- 29.Frankfort, B. J., R. Nolo, Z. Zhang, H. Bellen, and G. Mardon. 2001. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 32:403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankfort, B. J., K. L. Pepple, M. Mamlouk, M. F. Rose, and G. Mardon. 2004. Senseless is required for pupal retinal development in Drosophila. Genesis 38:182-194. [DOI] [PubMed] [Google Scholar]
- 31.Fu, W., H. Duan, E. Frei, and M. Noll. 1998. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development 125:2943-2950. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs, B., T. Wagner, N. Rossel, M. Antoine, H. Beug, and J. Niessing. 1997. Structure and erythroid cell-restricted expression of a chicken cDNA encoding a novel zinc finger protein of the Cys + His class. Gene 195:277-284. [DOI] [PubMed] [Google Scholar]
- 33.Garrell, J., and J. Modolell. 1990. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell 61:39-48. [DOI] [PubMed] [Google Scholar]
- 34.Giagtzoglou, N., P. Alifragis, K. A. Koumbanakis, and C. Delidakis. 2003. Two modes of recruitment of E(spl) repressors onto target genes. Development 130:259-270. [DOI] [PubMed] [Google Scholar]
- 35.Gilks, C. B., S. E. Bear, H. L. Grimes, and P. N. Tsichlis. 1993. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 13:1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimes, H. L., T. O. Chan, P. A. Zweidler-McKay, B. Tong, and P. N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimes, H. L., C. B. Gilks, T. O. Chan, S. Porter, and P. N. Tsichlis. 1996. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc. Natl. Acad. Sci. USA 93:14569-14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guillemot, F., L. C. Lo, J. E. Johnson, A. Auerbach, D. J. Anderson, and A. L. Joyner. 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75:463-476. [DOI] [PubMed] [Google Scholar]
- 39.Haenlin, M., Y. Cubadda, F. Blondeau, P. Heitzler, Y. Lutz, P. Simpson, and P. Ramain. 1997. Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11:3096-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan, B. A., and H. J. Bellen. 2000. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 14:1852-1865. [PubMed] [Google Scholar]
- 41.Hay, B. A., T. Wolff, and G. M. Rubin. 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120:2121-2129. [DOI] [PubMed] [Google Scholar]
- 42.Heitzler, P., M. Bourouis, L. Ruel, C. Carteret, and P. Simpson. 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122:161-171. [DOI] [PubMed] [Google Scholar]
- 43.Hock, H., M. J. Hamblen, H. M. Rooke, D. Traver, R. T. Bronson, S. Cameron, and S. H. Orkin. 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18:109-120. [DOI] [PubMed] [Google Scholar]
- 44.Hulander, M., W. Wurst, P. Carlsson, and S. Enerback. 1998. The winged helix transcription factor Fkh10 is required for normal development of the inner ear. Nat. Genet. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 45.Jafar-Nejad, H., M. Acar, R. Nolo, H. Lacin, H. Pan, S. M. Parkhurst, and H. J. Bellen. 2003. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 17:2966-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janody, F., R. Sturny, V. Schaeffer, Y. Azou, and N. Dostatni. 2001. Two distinct domains of Bicoid mediate its transcriptional downregulation by the Torso pathway. Development 128:2281-2290. [DOI] [PubMed] [Google Scholar]
- 47.Jarman, A. P., Y. Grau, L. Y. Jan, and Y. N. Jan. 1993. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73:1307-1321. [DOI] [PubMed] [Google Scholar]
- 48.Jegalian, A. G., and H. Wu. 2002. Regulation of Socs gene expression by the proto-oncoprotein GFI-1B: two routes for STAT5 target gene induction by erythropoietin. J. Biol. Chem. 277:2345-2352. [DOI] [PubMed] [Google Scholar]
- 49.Jia, Y., G. Xie, and E. Aamodt. 1996. pag-3, a Caenorhabditis elegans gene involved in touch neuron gene expression and coordinated movement. Genetics 142:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia, Y., G. Xie, J. B. McDermott, and E. Aamodt. 1997. The C. elegans gene pag-3 is homologous to the zinc finger proto-oncogene gfi-1. Development 124:2063-2073. [DOI] [PubMed] [Google Scholar]
- 51.Juin, P., A. O. Hueber, T. Littlewood, and G. Evan. 1999. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 13:1367-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Juin, P., A. Hunt, T. Littlewood, B. Griffiths, L. B. Swigart, S. Korsmeyer, and G. Evan. 2002. c-Myc functionally cooperates with Bax to induce apoptosis. Mol. Cell. Biol. 22:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karis, A., I. Pata, J. H. van Doorninck, F. Grosveld, C. I. de Zeeuw, D. de Caprona, and B. Fritzsch. 2001. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J. Comp. Neurol. 429:615-630. [DOI] [PubMed] [Google Scholar]
- 54.Karsunky, H., I. Mende, T. Schmidt, and T. Moroy. 2002. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene 21:1571-1579. [DOI] [PubMed] [Google Scholar]
- 55.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K. W. Schmid, U. Duhrsen, and T. Moroy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295-300. [DOI] [PubMed] [Google Scholar]
- 56.Kasai, S., and J. G. Scott. 2001. A house fly gene homologous to the zinc finger proto-oncogene Gfi-1. Biochem. Biophys. Res. Commun. 283:644-647. [DOI] [PubMed] [Google Scholar]
- 57.Kavaler, J., W. Fu, H. Duan, M. Noll, and J. W. Posakony. 1999. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development 126:2261-2272. [DOI] [PubMed] [Google Scholar]
- 58.Kawamoto, K., S. Ishimoto, R. Minoda, D. E. Brough, and Y. Raphael. 2003. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J. Neurosci. 23:4395-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58a.Kazanjian, A., D. Wallis-Schultz, N. Au, R. Nigam, K. J. T. Venken, P. T. Cagle, B. F. Dickey, H. J. Bellen, C. B. Gilks, and H. L. Grimes. Growth factor independence-1 (GFI1) is expressed in primary human neuroendocrine lung carcinomas and mediates differentiation of pulmonary neuroendocrine cells. Cancer Res., in press. [DOI] [PubMed]
- 59.Knust, E., H. Schrons, F. Grawe, and J. A. Campos-Ortega. 1992. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 132:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunisch, M., M. Haenlin, and J. A. Campos-Ortega. 1994. Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc. Natl. Acad. Sci. USA 91:10139-10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawoko-Kerali, G., M. N. Rivolta, and M. Holley. 2002. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J. Comp. Neurol. 442:378-391. [DOI] [PubMed] [Google Scholar]
- 62.Liao, X., Y. Tang, S. K. Chattopadhyay, J. W. Hartley, and H. C. Morse III. 1997. Upregulation of Gfi-1, a gene involved in IL-2-independent growth of T cells, in a murine retrovirus-induced immunodeficiency syndrome. In Vivo 11:9-12. [PubMed] [Google Scholar]
- 63.Ma, Q., Z. Chen, I. del Barco Barrantes, J. L. de la Pompa, and D. J. Anderson. 1998. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20:469-482. [DOI] [PubMed] [Google Scholar]
- 64.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maurer, P., F. T'Sas, L. Coutte, N. Callens, C. Brenner, C. Van Lint, Y. de Launoit, and J. L. Baert. 2003. FEV acts as a transcriptional repressor through its DNA-binding ETS domain and alanine-rich domain. Oncogene 22:3319-3329. [DOI] [PubMed] [Google Scholar]
- 66.McGhee, L., J. Bryan, L. Elliott, H. L. Grimes, A. Kazanjian, J. N. Davis, and S. Meyers. 2003. Gfi-1 attaches to the nuclear matrix, associates with ETO (MTG8) and histone deacetylase proteins, and represses transcription using a TSA-sensitive mechanism. J. Cell Biochem. 89:1005-1018. [DOI] [PubMed] [Google Scholar]
- 67.Meier, P., A. Finch, and G. Evan. 2000. Apoptosis in development. Nature 407:796-801. [DOI] [PubMed] [Google Scholar]
- 68.Naya, F. J., H. P. Huang, Y. Qiu, H. Mutoh, F. J. DeMayo, A. B. Leiter, and M. J. Tsai. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11:2323-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nicholson, R. A. 1930. The Mathnawi of Jalalu' ddin Rumi, edited with critical notes, translation, and commentary, vol. IV. Gibb Memorial New Series IV. Gibb Memorial Trust, London, United Kingdom.
- 70.Nolo, R., L. A. Abbott, and H. J. Bellen. 2001. Drosophila Lyra mutations are gain-of-function mutations of senseless. Genetics 157:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nolo, R., L. A. Abbott, and H. J. Bellen. 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102:349-362. [DOI] [PubMed] [Google Scholar]
- 72.Olson, E. N., and W. H. Klein. 1994. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 73.Orkin, S. H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57-64. [DOI] [PubMed] [Google Scholar]
- 74.Osawa, M., T. Yamaguchi, Y. Nakamura, S. Kaneko, M. Onodera, K. Sawada, A. Jegalian, H. Wu, H. Nakauchi, and A. Iwama. 2002. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood 100:2769-2777. [DOI] [PubMed] [Google Scholar]
- 75.Parker, D. S., J. Jemison, and K. M. Cadigan. 2002. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 129:2565-2576. [DOI] [PubMed] [Google Scholar]
- 76.Pavletich, N. P., and C. O. Pabo. 1993. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science 261:1701-1707. [DOI] [PubMed] [Google Scholar]
- 77.Pavletich, N. P., and C. O. Pabo. 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 252:809-817. [DOI] [PubMed] [Google Scholar]
- 78.Person, R. E., F. Q. Li, Z. Duan, K. F. Benson, J. Wechsler, H. A. Papadaki, G. Eliopoulos, C. Kaufman, S. J. Bertolone, B. Nakamoto, T. Papayannopoulou, H. L. Grimes, and M. Horwitz. 2003. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 34:308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phippen, T. M., A. L. Sweigart, M. Moniwa, A. Krumm, J. R. Davie, and S. M. Parkhurst. 2000. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J. Biol. Chem. 275:37628-37637. [DOI] [PubMed] [Google Scholar]
- 80.Pi, H., S. K. Huang, C. Y. Tang, Y. H. Sun, and C. T. Chien. 2004. phyllopod is a target gene of proneural proteins in Drosophila external sensory organ development. Proc. Natl. Acad. Sci. USA 22:8378-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quan, X. J., T. Denayer, J. Yan, H. Jafar-Nejad, A. Philippi, O. Lichtarge, K. Vleminckx, and B. A. Hassan. 2004. Evolution of neural precursor selection: functional divergence of proneural proteins. Development 131:1679-1689. [DOI] [PubMed] [Google Scholar]
- 82.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramain, P., R. Khechumian, K. Khechumian, N. Arbogast, C. Ackermann, and P. Heitzler. 2000. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol. Cell 6:781-790. [DOI] [PubMed] [Google Scholar]
- 84.Roberts, T., and J. K. Cowell. 1997. Cloning of the human Gfi-1 gene and its mapping to chromosome region 1p22. Oncogene 14:1003-1005. [DOI] [PubMed] [Google Scholar]
- 85.Rodel, B., K. Tavassoli, H. Karsunky, T. Schmidt, M. Bachmann, F. Schaper, P. Heinrich, K. Shuai, H. P. Elsasser, and T. Moroy. 2000. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 19:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodel, B., T. Wagner, M. Zornig, J. Niessing, and T. Moroy. 1998. The human homologue (GFI1B) of the chicken GFI gene maps to chromosome 9q34.13-A locus frequently altered in hematopoietic diseases. Genomics 54:580-582. [DOI] [PubMed] [Google Scholar]
- 87.Russo, C. A., N. Takezaki, and M. Nei. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12:391-404. [DOI] [PubMed] [Google Scholar]
- 88.Saleque, S., S. Cameron, and S. H. Orkin. 2002. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16:301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scheijen, B., J. Jonkers, D. Acton, and A. Berns. 1997. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J. Virol. 71:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmidt, T., H. Karsunky, E. Gau, B. Zevnik, H. P. Elsasser, and T. Moroy. 1998. Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene 17:2661-2667. [DOI] [PubMed] [Google Scholar]
- 91.Schmidt, T., M. Zornig, R. Beneke, and T. Moroy. 1996. MoMuLV proviral integrations identified by Sup-F selection in tumors from infected myc/pim bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 24:2528-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sharina, I. G., E. Martin, A. Thomas, K. L. Uray, and F. Murad. 2003. CCAAT-binding factor regulates expression of the beta1 subunit of soluble guanylyl cyclase gene in the BE2 human neuroblastoma cell line. Proc. Natl. Acad. Sci. USA 100:11523-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shivdasani, R. A., E. L. Mayer, and S. H. Orkin. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373:432-434. [DOI] [PubMed] [Google Scholar]
- 94.Singson, A., M. W. Leviten, A. G. Bang, X. H. Hua, and J. W. Posakony. 1994. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 8:2058-2071. [DOI] [PubMed] [Google Scholar]
- 95.Sriuranpong, V., M. W. Borges, R. K. Ravi, D. R. Arnold, B. D. Nelkin, S. B. Baylin, and D. W. Ball. 2001. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 61:3200-3205. [PubMed] [Google Scholar]
- 96.Tong, B., H. L. Grimes, T. Y. Yang, S. E. Bear, Z. Qin, K. Du, W. S. El-Deiry, and P. N. Tsichlis. 1998. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol. Cell. Biol. 18:2462-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Torres, M., E. Gomez-Pardo, and P. Gruss. 1996. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development 122:3381-3391. [DOI] [PubMed] [Google Scholar]
- 98.Vaessin, H., M. Brand, L. Y. Jan, and Y. N. Jan. 1994. daughterless is essential for neuronal precursor differentiation but not for initiation of neuronal precursor formation in Drosophila embryo. Development 120:935-945. [DOI] [PubMed] [Google Scholar]
- 99.Villares, R., and C. V. Cabrera. 1987. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell 50:415-424. [DOI] [PubMed] [Google Scholar]
- 100.Wadman, I. A., H. Osada, G. G. Grutz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wallis, D., M. Hamblen, Y. Zhou, K. J. Venken, A. Schumacher, H. L. Grimes, H. Y. Zoghbi, S. H. Orkin, and H. J. Bellen. 2003. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130:221-232. [DOI] [PubMed] [Google Scholar]
- 102.Wang, S. W., B. S. Kim, K. Ding, H. Wang, D. Sun, R. L. Johnson, W. H. Klein, and L. Gan. 2001. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang, V. Y., B. A. Hassan, H. J. Bellen, and H. Y. Zoghbi. 2002. Drosophila atonal fully rescues the phenotype of Math1 null mice: new functions evolve in new cellular contexts. Curr. Biol. 12:1611-1616. [DOI] [PubMed] [Google Scholar]
- 104.Yang, Z., K. Ding, L. Pan, M. Deng, and L. Gan. 2003. Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol. 264:240-254. [DOI] [PubMed] [Google Scholar]
- 105.Yucel, R., H. Karsunky, L. Klein-Hitpass, and T. Moroy. 2003. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 197:831-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng, J. L., and W. Q. Gao. 2000. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3:580-586. [DOI] [PubMed] [Google Scholar]
- 107.Zhu, J., L. Guo, B. Min, C. J. Watson, J. Hu-Li, H. A. Young, P. N. Tsichlis, and W. E. Paul. 2002. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity 16:733-744. [DOI] [PubMed] [Google Scholar]
- 108.Zornig, M., T. Schmidt, H. Karsunky, A. Grzeschiczek, and T. Moroy. 1996. Zinc finger protein GFI-1 cooperates with myc and pim-1 in T-cell lymphomagenesis by reducing the requirements for IL-2. Oncogene 12:1789-1801. [PubMed] [Google Scholar]
- 109.Zweidler-McKay, P. A., H. L. Grimes, M. M. Flubacher, and P. N. Tsichlis. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]