Abstract
The paired box 6 (Pax6) gene encodes a transcription factor essential for eye development in a wide range of animal lineages. Here we describe the cloning and characterization of Pax6 gene from the blind hydrothermal vent tubeworm Ridgeia piscesae (RpPax6). The deduced RpPax6 protein shares extensive sequence identity with Pax6 proteins from other species and contains both the paired domain and a complete homeodomain. Phylogenetic analysis indicates that it clusters with the corresponding sequence from the closely related species Platynereis dumerilii (P. dumerilii) of Annelida. Luciferase reporter assay indicate that RpPax6 protein suppresses the transcription of sine oculis (so) in D. melanogaster, interfering with the C-terminal of RpPax6. Taking advantage of Drosophila model, we show that RpPax6 expression is not able to rescue small eye phenotype of ey2 mutant, only to cause a more severe headless phenotype. In addition, RpPax6 expression induced apoptosis and inhibition of apoptosis can partially rescue RpPax6-induced headless phenotype. We provide evidence RpPax6 plays at least two roles: it blocks the expression of later-acting transcription factors in the eye development cascade, and it promotes cell apoptosis. Our results indicate alternation of the Pax6 function may be one of the possible causes that lead the eye absence in vestimentiferan tubeworms.
Introduction
Members of the paired box (Pax) gene family encode transcription factors that are characterized by a DNA-binding paired domain of 128 amino acids located at the amino terminal end [1]. Additionally, some Pax proteins contain a partial or complete paired-type homeodomain and/or an octapeptide motif. Members of the Pax gene family are involved in the regulation of wide range of developmental processes, including segmentation and organogenesis [2, 3]. Pax6 is one of the well-characterized Pax genes which plays critical role in eye development in diverse animal lineages. It contains both a paired and a complete homeodomain. The genomic organization, domain sequences, and function are highly conserved [4, 5].
Mutations in the Pax6 gene cause aniridia (a panocular disorder primarily characterized by complete or partial absence of iris tissue) in human and small eye (sey) phenotype (microphthalmia and small body size) in mouse [6, 7]. Mutations in the Drosophila Pax6 homolog eyeless (ey) cause partial to complete loss of the compound eye as well as surrounding sensory bristles. Moreover, misexpression of Pax6 genes from many divergent species including Drosophila [8], ribbon worm [9], squid [10] and ascidian [11] induce ectopic eyes in Drosophila. In frogs, Pax6 misexpression can also induce ectopic eyes [12]. The conservation of regulatory cascade required for eye morphogenesis is further supported by the fact that eya-Eya, so-Six, and atonal-Ath5 gene families all act immediately downstream of Pax6 in both fruit fly and mouse eye development [13–16], indicating the Pax6 dependent eye developmental pathway can be traced at least in the last common ancestor of protostome and deuterostome (urbilateria) and has been adapted to the control of development of different visual systems found in both clades.
The comparative analyses of bilaterian eyes have revealed that the simplest morphology comprises one photorceptor cell and one pigment cell [17]. In Annelida, two distinct types of pigmented cerebral eyes were characterized: larval and adult eyes. Adult annelid eyes are multicellular, whereas larval annelid eyes are comparatively simple, usually only comprising of two cells: one rhabdomeric photoreceptor cell and one pigment cell. Larval-type eyes match the prototypical two-celled eye and are present in larvae of species of many protostomian lineages [18, 19, 20] as well as deuterostomian lineages including hemichordates [21] and cephalochordates [22]. The larval eyes are usually replaced by the eyes of the adults during later development [23–26]. The adult multicellular eyes show a very characteristic structure with photoreceptor cell processes traversing the pigment cell layer. This type of eyes is found widespread in various protostomian lineages including polychaetes [23, 27], molluscs [28], sipunculans [29] and onychophorans [30]. Both larval and adult eyes are molecularly characterized in the marine annelid Platynereis dumerilii. Pax6 was found to be expressed in the larval but not adult eyes of P. dumerilii [23].
Based the resemblance to the fossilized tubes, modern vestimentifera tubes old as 430 million years form a derived clade of vestimentifera within the annelid radiation.” [31]. Vestimentiferans are important members of deep-sea chemosynthetic communities, which include hydrothermal vents, cold seeps, whale falls and reduced sediments. Due to their deep-sea habitats the first member of vestimentifera Siboglinum weberi was not reported until early in the 20th century [32]. More than 100 species of vestimentiferans, have been described. Like many deep-sea organisms as well as cave-dwelling animals, the vestimentiferans tubeworms lack eyes. They rely on microbial endosymbionts for their energetic needs. Vestimentiferan tubeworms have been extensively studied though research has focused primarily on phylogeny and bacterial symbionts [33–35]. Far fewer studies have explored the molecular machinery used in the regulation of development and innate immune system.
To further our understanding of the biomolecular mechanisms underlying the eye absence in vestimentifera lineage, we isolated Pax6 gene of vestimentiferan Ridgeia piscesae. R. piscesae is the foundation specie in many Juan de Fuca Ridge hydrothermal vent communities [36, 37]. In order to investigate the function of the RpPax6, the model organism D. melanogaster was applied for genetic analysis. D. melanogaster is accessible for a broad range of genetic and molecular techniques and has been widely used for the elucidating the function of Pax6 from various species. RpPax6 protein is found to be able to suppresses the transcription of sine oculis (so) which can interfere with the activation of eye development network. In addition RpPax6 is associated with the elevated level of apoptosis. Our results suggest that functional alternation of RpPax6 may be involved in the eye absence in vestimentiferan tubeworms.
Materials and Methods
cDNA cloning of R. piscesae Pax6
The cDNA library of R. piscesae was previously constructed [38]. Sequences of cDNA clones were analyzed and a clone bearing putatively entire ORF of Pax6 was identified. To obtain the full length cDNAs of Pax6 gene, the 3’ and 5’ ends were obtained by rapid amplification of cDNA ends (RACE) approaches using 3’ -Full RACE Core Set with PrimeScript™ RTase and 5’ -Full RACE Kit with TAP (TaKaRa, Japan) following the manufacturer’s instructions. The PCR products were ligated into pMD-19T vector (TaKaRa, Japan) and transformed into the competent E. coli TOP10 cells. Positive clones with the expected-size inserts were determined by colony PCR and DNA sequencing.
Phylogenetic analysis
Accession numbers for sequences included in the analyses are: Doryteuthis opalescens Pax6 (DoPax6): AAB40616; Euprymna scolopes Pax6 (EsPax6): AAM74161; Crassostrea gigas Pax6 (CgPax6): XP_011433289; Idiosepius paradoxus Pax6 (IpPax6): BAM74253; Ambystoma mexicanum Pax6 (AmPax6): AAD50903; Anolis carolinensis Pax6 (AcPax6): XP_008104750; Cavia porcellus Pax6 (CpPax6): XP_003464531; Aotus nancymaae Pax6 (AnPax6): XP_012307699; Xenopus laevis Pax6 (XlPax6): AF154555; Homo sapiens Pax6 (HsPax6): NP_000271; Platynereis dumerilii Pax6 (PdPax6): CAJ40659; Terebratalia transversa Pax6 (TtPax6): ADZ24784; Lottia gigantea Pax6 (LgPax6): XP_009066032; Cupiennius salei Pax6 (CsPax6): CEH19758; Saccoglossus kowalevskii Pax6 (SkPax6): NP_001158383; Limulus polyphemus Pax6 (LpPax6): XP_013778820; Drosophila melanogaster eyeless (DmEy): AAF59318; Mus musculus Pax2 (MmPax2): CAA39302.1. The amino acid sequences of Pax proteins were aligned and a phylogenetic tree was generated using the Mega 3 [39]. The neighbor joining method with 1000 bootstrap replications was used.
Fly stocks
The following fly stocks were used:
y w;+/+;ey-GAL4/TM6B,Tb
y w;+/+;GMR-GAL4
y w;+/+;dpp-GAL4/TM6B,Tb
y w;+/+;da-GAL4
y w;+/+;da-GAL4,tub-GAL80ts
y w;+/+;UAS-RpPax6
y w;+/+;UAS-ey
y w;UAS-P35/cyo,y+;FRT-82B
y w;+/cyo
y w;+/+;+/TM6B,Tb
w t;ey2
y w;+/+;UAS-mPax6/Tm3;ey2
Construction of plasmids
We constructed a set of chimeric molecules in which individual segments of ey were deleted and replaced with the corresponding region of RpPax6. The eyN+RpPax6C chimera was created by replacing the N terminal of RpPax6 with amino acids 1–471 of ey. The eyPD+RpPax6HD chimera was created by replacing the PD of RpPax6 with amino acids 36–164 of ey. Similarly, we generated a set of chimeric molecules in which individual segments of RpPax6 were deleted and replaced with the corresponding region of ey. The RpPax6PD+eyHD chimera was created by replacing the PD of ey with amino acids 17–143 of RpPax6. The RpPax6N+eyC chimera was generated by replacing the N terminal segment of ey with amino acids 1–296 of RpPax6.
The firefly luciferase reporter plasmid (pGL3-so and pGL3-eya) were constructed by inserting the 428 bp of so10 and a 398 bp of so5 fragments [13] and -499 to +100 fragment eya gene promoter region PCR amplified from the Drosophila genome in pGL3-basic vector (Promega, America) at Kpn I and Xho I sites. The plasmid pUAST-RpPax6 was generated by subcloning the RpPax6 cDNA into the Drosophila transformation vector pUAST using EcoR I and Xho I. The transgenes encoding wild-type and mutated RpPax6 and ey proteins were constructed as follows: pCMV-myc-RpPax6, pCMV-myc-ey, pCMV-myc-RpPax6N+eyC, pCMV-myc-eyN+RpPax6C, pCMV-myc-eyPD+RpPax6HD, pCMV-myc-RpPax6PD+eyHD were prepared by subcloning the inserts from the corresponding pEASY-T5 vectors into vector pCMV-myc plasmid (Clontech, Japan) using EcoR I and Xho I. Several transgenic lines of each construct were obtained by P-element-mediated germline transformation according to standard procedures. All of the constructs were confirmed by DNA sequencing.
Temperature-shift experiments
Conditional expression of RpPax6 was carried out by using y w;+/+;UAS-RpPax6/da-GAL4, tub-GAL80ts line. GAL80ts is a temperature sensitive form of the GAL80 repressor. At 25°C degrees, the GAL80ts represses the activity of GAL4; however, when the temperature is shifted to 29°C degrees, the GAL80ts is inactivated, allowing for the GAL4 transcription factor to bind to the UAS binding sites and express the gene of interest [40]. Larvae were raised at 18°C (GAL80ts permissive temperature) for 120 h then shift to 29°C (GAL80ts restrictive temperature) for 24 h. Twenty third instar larva were collected. Larger discs (including the wing and eye-antenna discs) were dissected and incubated for 5 minutes in 5 g/mL acridine orange in phosphate-buffered saline (PBS) [41]. The organs were then placed in fresh PBS and analyzed immediately for nuclear staining on a Leica M165FC fluorescence stereomicroscope (Leica, Wetzlar, Germany). Independent triplicate experiments were performed.
Luciferase assays
Transient transfections of 293T cells were performed in 24-well plates with the transfection reagent Lipofectamine 2000 (Invitrogen, America). For each well, cells were transfected with 100 ng of pSV-β-galactosidase (Promega, America) as transfection efficiency control, 200 ng reporter vector (pGL3-so) and 200 ng expression vector (pCMV-myc-RpPax6, pCMV-myc-ey, pCMV-myc-RpPax6N+eyC, pCMV-myc-eyN+RpPax6C, pCMV-myc-eyPD+RpPax6HD, pCMV-myc-RpPax6PD+eyHD or pCMV-myc). pCMV-myc vector was used as a negative control. At 6 h post-transfection the medium was replaced. Cells were harvested 30 h after transfection, rinsed with PBS, resuspended in reporter cell lysis buffer (Promega, America), and incubated for 10 minutes at room temperature. The lysate was centrifuged at 12,000 × g for 5 min to pellet the cell debris. The supernatants were transferred to a fresh tube. A 10 μL-aliquot of the extract was added to 25 μL of the luciferase assay substrate (Promega, America) and the luminescence of the samples were read immediately on a Glomax™ 20/20 Luminometer (Promega, America). Each transfection was performed in triplicate.
β-Galactosidase activity was used for normalizing the transfection efficiency and protein input. A 10 μL-aliquot of the cell extract was mixed with 290 μL of O-nitrophenyl-β-D-Galactopyranoside (ONPG) solution (880 μg/ml ONPG, 67 mM Na3PO4, 1 mM MgCl2, 45 mM β-mercaptoethanol, pH 7.5) (Sangon Biotech, Shanghai). The absorbance of the mixture was determined at 420 nm after 30 min of incubation at 37°C. Each transfection was performed in triplicate. Results are expressed as means of the ratio between firefly luciferase activity and β-Galactosidase activity whereas control is 100%.
Results
Cloning and phylogenetic analysis RpPax6
Analyses of the cDNA library from long-skinny Ridgeia piscesae identified a putative Pax6 gene. The cloned full length Pax6 cDNA contained 312 bp of 5’ untranslated region (UTR), 1410 bp of open reading frame (ORF) encoding 470 amino acids, and 650 bp of 3’ UTR. The predicted protein product contains the paired domain and a complete homeodomain, but not an octapeptide, exhibiting the structural features characteristic of the Pax6 protein [5]. Therefore, we designated this gene RpPax6 (R. piscesae Pax6). The nucleotide and amino acid sequences have been assigned to the GenBank Accession Number KT380855.
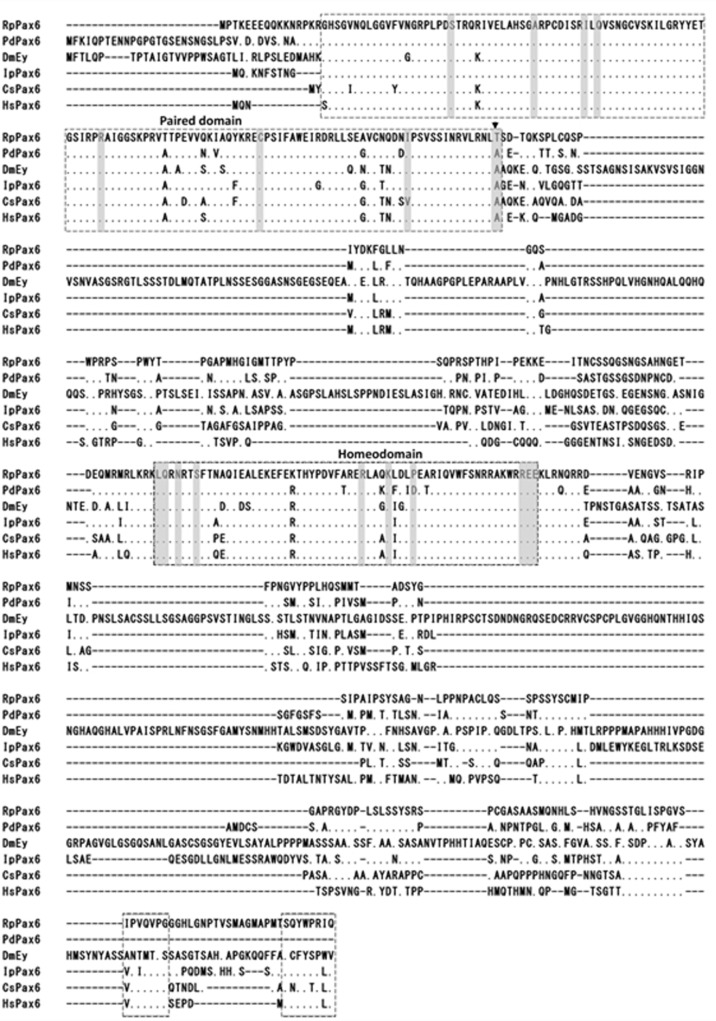
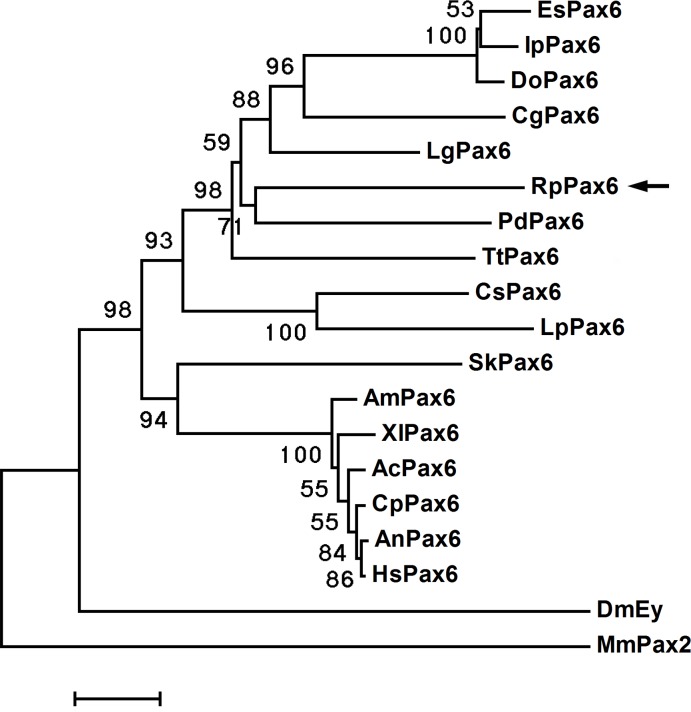
Amino acids sequence alignment of RpPax6 and the pax6 from different species reveal that the paired domains and homeodomains are highly conserved (Fig 1). Positions of Pax6 specific amino acids in the conserved domain [5] are conserved in RpPax6 except one amino acid change. Typical of most paired domains found in Pax6 genes have an asparagine at position 128. However, RpPax6 encode a threonine at this position. The C-terminal region (C) comprises 152 aa, rich in serine (18%), proline (13%) and asparagine (6%), and contains conserved termination motif. Phylogenetic tree based on full-length Pax6 amino acid sequences was constructed with neighbor joining method (Fig 2). DmEy from D. melanogaster branched out at the base of the tree other Pax6 proteins were divided into two clades, corresponding protostome and deuterostome respectively. The clade of protostomian lineage comprises two subclades. Pax6 from lophotrochozoan species form a monophyletic subclade. Among this group the identified RpPax6 clusters with the corresponding sequence from the closely related species P. dumerilii of Annelida. The sequences from arthropoda species form the sister group of lophotrochozoan subclade. In the deuterostome clade all vertebrate members of Pax6 gathered together whereas Pax6 from hemichordate S. kowalevskii (SkPax6) branch out independently at the basal place.
Fig 1. Comparison of amino acid sequences of Pax6 genes.
Identical amino acids are indicated by dots. The introduced gaps are indicated by dashes. The paired domain, homeodomain and conserved C-terminal motif are boxed. Pax6-specific amino acids [5] are shaded. Arrowhead indicates a single amino acid change of the Pax6-specific amino acids.
Fig 2. Phylogenetic position of RpPax6.
A neighbor-joining tree based on a comparison of the deduced amino acid sequences of full-length clones of Pax6 with mouse Pax2 included as outgroup. Numbers at nodes indicate the levels of bootstrap support based on data for 1,000 replicates; only values greater than 50% are shown. Bar 5% estimated sequenced divergence.
RpPax6 has repressive effect on the promoter activity of so
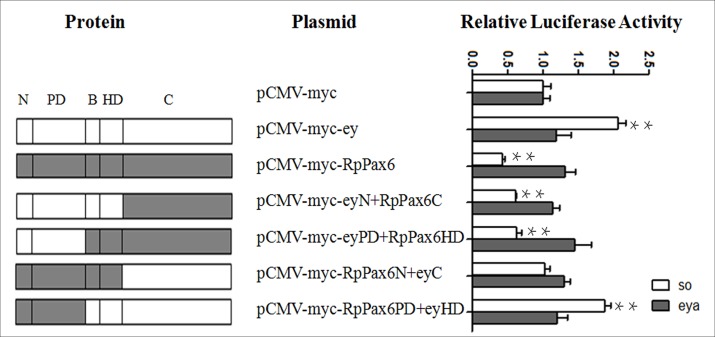
RpPax6 shares 91.4% and 88.3% identity at the paired domain and homeodomain with the ortholog of Drosophila ey at the amino acid level. Drosophila ey protein has been known as a critical regulator of early retinal development and sine oculis (so) is the direct target of ey [42]. In addition the eye-specific enhancer of eya is known to be induced in response to ectopic expression of ey [43]. To test if RpPax6 is involved in the transcriptional regulation of ey downstream target, we examined the effect of RpPax6 and ey proteins with or without mutation on the transcriptional activity of so and eya. Reporter vectors (pGL3-so and pGL3-eya) were constructed by cloning so and eya promoters upstream of a luciferase reporter. 293T cells were transfected with luciferase reporter vector and an expression vector bearing chimeric ey and RpPax6 constructs. As demonstrated in Fig 3, neither ey, RpPax6 or chimeric fusion proteins has significant effect on the promoter activity of pGL3-eya, suggesting that ey acts through other factors to regulate eya. Meanwhile, cotransfection of the expression vector pCMV-myc-ey and the pCMV-myc-RpPax6PD+eyHD with the report construct pGL3-so resulted in two-fold increase in the luciferase activity as compared with the level observed when cotransfecting with control pCMV-myc plasmid, indicating that so promoter was activated by ey and RpPax6PD+eyHD. In contrast the expression of RpPax6, eyN+RpPax6C and eyPD+RpPax6HD resulted in significant suppression of luciferase activity, indicating RpPax6 and eyN+RpPax6C eyPD+RpPax6HD have repressive effect on the promoter activity of so. Compared with the full-length RpPax6, RpPax6 with paired domain swapped with the corresponding portion of ey (eyN+RpPax6C and eyPD+RpPax6HD) had a much lesser effect on so promoter. In the case of RpPax6 with C region replaced by the counterpart of ey, the repressive effect of RpPax6 was not detected. These results indicate that RpPax6 protein suppresses the transcription of so and the C region of RpPax6 plays crucial role in the repressive activity.
Fig 3. Structural-functional analysis of ey and RpPax6.
Schematic summaries of the original and chimeric constructs are listed in the left column. The right column shows the effect of ey and RpPax6 proteins with or without mutation on promoter activity of pGL3-so and pGL3-eya. The assay was carried out in 293T cells as described in Materials and Methods. Luciferase activities are shown relative to those of pCMV-myc. pSV-β-galactosidase was included in transfection as an internal control of the transfection efficiency. The values are the means from three independent experiments ±SE. **P < 0.01.
Ectopic expression of RpPax6 in D. melanogaster
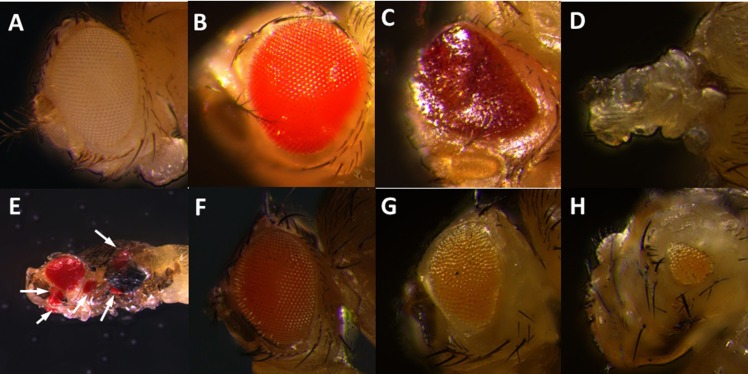
The GAL4/UAS binary system was used to drive the expression of RpPax6 in various Drosophila tissues. Four GAL4 drivers including eyeless-GAL4 (ey-GAL4), GMR-GAL4, decapentaplegic-GAL4 (dpp-GAL4), and daughterless-GAL4 (da-GAL4) were employed (Table 1). The ey-GAL4 driver line expresses GAL4 ubiquitously in the eye-antenna discs throughout early larval development when all the cells are proliferating. During the third instar larval stage, the expression is restricted to the cycling cells anterior to the morphogenetic furrow [8, 44].GMR-GAL4 expresses behind the morphogenetic furrow in the eye disc. dpp-GAL4 directs GAL4 expression in all of the imaginal discs throughout development, and in only a limited portion of each disc [45]. da-GAL4 is used to express GAL4 ubiquitously. We observed early lethality in flies expressing RpPax6 under the control of all four GAL4 driver lines. Global expression of the RpPax6 using da-GAL4 leads to embryonic lethality. Expression of RpPax6 using dpp-GAL4 caused lethality during larval and pupal stages. Dissection of the pupae showed the animals could not start metamorphosis of the eyes, wings and legs. Expression of RpPax6 using ey-GAL4 exhibits pupal lethality. Examination of these dead pharate adults in the pupal case revealed that most head structures and both eyes were missing (Fig 4D). Dissection of the third instar larvae showed abnormal morphology of eye-antennal discs (Fig 5C). Expression of RpPax6 using GMR-GAL4 results pupal lethality with eclosion rate of 7.9%. The pharates and adults show rough eye phenotype with a relatively normal eye size and severe ommatidia loss (Fig 4C).
Table 1. Phenotypes associated with ectopic expression of ey and RpPax6.
| ey-GAL4 | GMR-GAL4 | dpp-GAL4 | da-GAL4 | |
|---|---|---|---|---|
| Expression | Eye | Behind the morphogenetic furrow in the eye disc. | Limited portion of in all of the imaginal discs | Ubiquitously |
| UAS-ey | Pink eyes with non-uniformly reduced size(Fig 4G and 4H) | Pupal lethal (100%) Ectopic eyes on wings, Legs and head(Fig 4E). | ||
| UAS-RpPax6 | Pupal lethal (100%) Abnormal eye-antennal discs in third instar stage (Fig 5C). Headless phenotype in dead pharate (Fig4D). | Pupal lethality (92.1%) rough eye and severe ommatidia loss (Fig 4C). | Larval and pupal lethal (100%) Under development in the eyes, wings and legs. | Embryonic lethal (100%) |
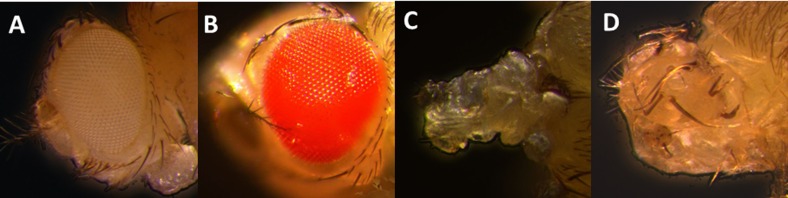
Fig 4. Eye phenotypes caused by misexpression of RpPax6 and ey genes.
(A, B) head of y w and UAS-RpPax6 pharate. (C) Rough eye in a UAS-RpPax6/GMR-GAL4 pharate. (D) UAS-RpPax6/ey-GAL4 pharate with most head structures and both eyes absent. (E) Ectopic eyes in UAS-ey/dpp-GAL4 fly (arrows). (F) Head of UAS-ey pharate. (G, H) Right and left eye of UAS-ey/ey-GAL4 pharate. The left eye was significantly reduced in size and the right eye was slightly reduced.
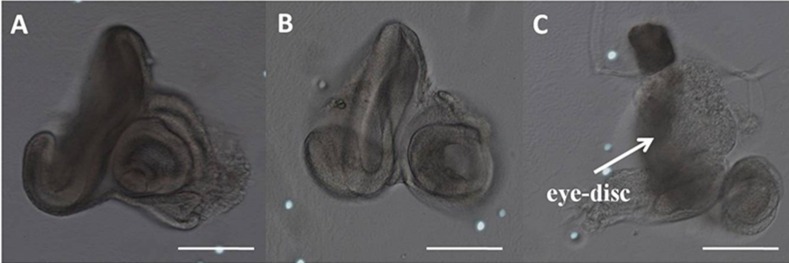
Fig 5. Third instar eye-antenna discs.
(A): y w (B): UAS-RpPax6 (C): UAS-RpPax6/ey-GAL4 flies. The boxed areas show growth defects in the eye disc of UAS-RpPax6/ey-GAL4 larvae. Scale bars = 100 μm.
To delineate the function of RpPax6 the Drosophila Pax6 homologue gene ey was expressed using dpp-GAL4 and ey-GAL4. The result show that expression of ey with dpp-GAL4 leads to significant pupal lethality. Flies dissected from the pupal case show the formation of ectopic eyes on wings, legs and head. Expression of ey with ey-GAL4 produced pink eyes with non-uniformly reduced size. As shown in Fig 4G and 4H, the left eye of the fly was significantly reduced in size whereas the right eye was slightly reduced.
RpPax6 expression induced apoptosis
Expression of RpPax6 using ey-GAL4 cause abnormal eye-antennal disc, defects in the head region, and death during the pupal stage. The reduced size of the fly head could be due to cell death induced by RpPax6. This is supported by the observation that throughout expression of RpPax6 with da-GAL4 caused earlier and more severe lethality than restricted expression with ey-GAL4, GMR-GAL4 and dpp-GAL4 (Table 1). To investigate the effects of RpPax6 expression to the cell death, y w;+/+;UAS-RpPax6 stock was crossed to the line y w;+/+;da-GAL4,tub-GAL80ts. The cross was reared at 18°C for 120 h to ensure tight suppression of RpPax6 expression until third instar stage. And then RpPax6 was induced by 29°C temperature shift to inactivate GAL80 and subsequently activate Gal4 activity. The third instar larval wing and eye-antenna discs were examined by using acridine orange staining, a vital dye that preferentially labels apoptotic cells [41]. Larva with temperature shift-up had higher level of dying cells in both discs than those raised continuously reared at 18°C (Fig 6), suggesting RpPax6 expression causes cell death.
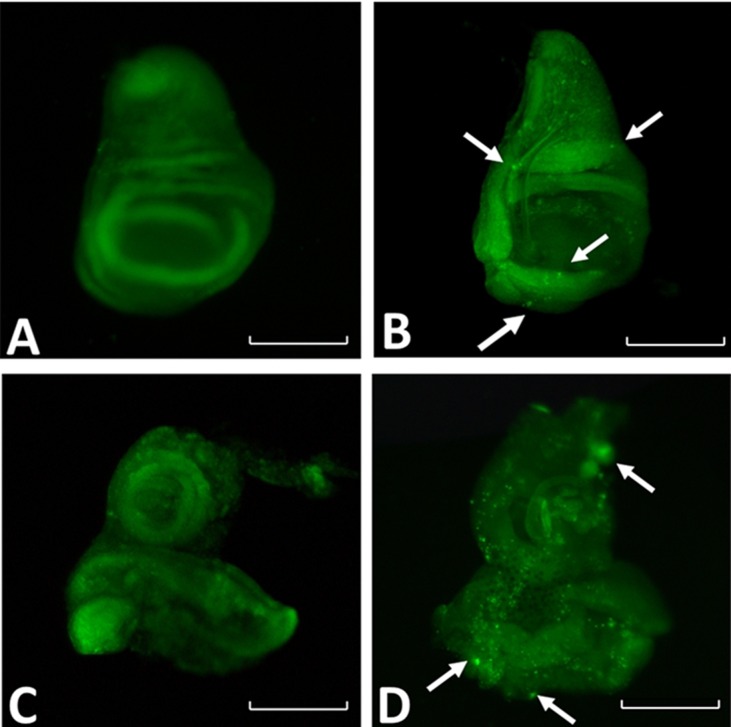
Fig 6. Expression of RpPax6 promotes apoptotic cells were identified by acridine orange staining.
Arrow points to a cluster of apoptotic cells. All discs are reproduced at the same magnification. Scale bars = 100 μm. (A) Third instar wing disc of UAS-RpPax6/da-GAL4,tub-GAL80ts reared continuously at 18°C exhibit no cell death as indicated by a lack of nuclear acridine orange staining. (B) Expression of RpPax6 for 24 h (UAS-RpPax6/da-GAL4,tub-GAL80ts, shifted to 29°C) lead to cell death in third instar wing disc as indicated by nuclear acridine orange staining (green spots). (C) Third instar of eye-antenna discs of UAS-RpPax6/da-GAL4,tub-GAL80ts reared continuously at 18°C show a few cell death. (D) Expression of RpPax6 for 24 h (UAS-RpPax6/da-GAL4,tub-GAL80ts, shifted to 29°C) lead to high frequency of apoptotic cells in third instar eye-antenna discs.
RpPax6 can not rescue ey2 mutant
Rescue experiments were set up to assess the importance of the RpPax6 in vivo. To allow eye-specific expression we used ey-Gal4 to drive UAS-ey, UAS-mPax6 (mouse Pax6) and UAS- RpPax6 genes in ey2 mutant background. ey mutation ey2 is amorphic for ey function in the eye disc. Flies homozygous for ey2 have reduced eyes. Expression of RpPax6 induced by ey-GAL4 in an ey2 mutant background caused pupal lethality. The dead pharates exhibited headless phenotype and the third instar larvae showed abnormal morphology of eye-antennal discs (data not shown), as the condition in UAS-RpPax6/ey-GAL4 flies. In addition, in the control cross, in which both full-length ey and mPax6 proteins were misexpressed in the same genetic background, the ey2 mutant was fully rescued. The eyes were morphologically normal and often of normal size (Fig 7). The rescue results suggest that RpPax6 expression can not induce eye development like ey and mPax6 do.
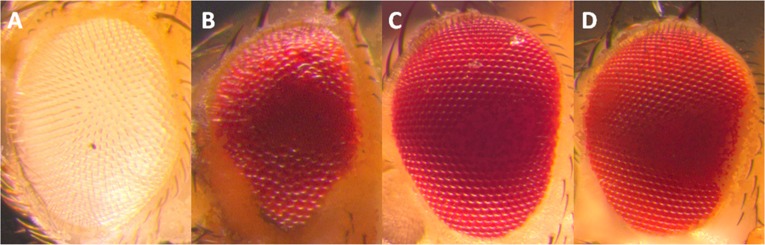
Fig 7. Mutant rescue by targeted expression of ey and mPax6.
(A) eye of y w fly. The ey2 mutant phenotype (small eye, see Fig B) can be completely rescued by targeted expression of ey (C) and mPax6 (D) driven by ey-GAL4.
Inhibition of apoptosis partially rescued RpPax6-induced headless phenotype
Our results revealed that flies expressing RpPax6 with ey-GAL4 die as pharates showing a headless phenotype(Fig 4D). If the defects are caused by apoptosis, it might be possible to rescue the headless phenotype by inhibition of apoptosis. To test this possibility the effect of a well-characterized anti-apoptotic viral protein P35 that inhibits downstream effector caspases was assessed. Coexpression of RpPax6 together with P35 with the use of ey-GAL4 resulted in the suppression of the RpPax6 induced defects. Although the rescued flies cannot eclose, almost all pharate adults exhibit an eyeless phenotype with most head structures recovered (Fig 8D). Reduction in the severity of the defect strongly supports a role for caspase-dependent apoptotic cell death in RpPax6 induced abnormalities.
Fig 8.
Eye phenotype caused by expression of RpPax6 is partially rescued by inhibition of apoptosis (A, B) head of y w and UAS-RpPax6 pharate. (C) UAS-RpPax6/ey-GAL4 pharate with most head structures and both eyes absent. (D) UAS-RpPax6/ey-GAL4 pharate headless phenotype can be largely rescued by coexpression of p35, an inhibitor of apoptosis.
Discussion
Covering nearly two-thirds of the Earth’s surface, deep-sea regions are known to harbor complex communities with impressively high numbers of species, showing remarkable morphological and physiological adaptations [46–48]. Due to the difficulties in the sampling and preserving of individual specimens, molecular studies of deep-sea organisms are still rare. R. piscesae is the foundation species in many Juan de Fuca Ridge hydrothermal vent communities [36, 37] within depths ranging from 1570–3250 m [35, 49–51]. R. piscesaeis presumed to have lost eyes during evolution because eyes are widespread throughout many annelid taxa [23, 52–54]. Likewise absence of eyes is observed in many species living in ocean depths beyond the penetration of daylight. These blind species might have dispensed with vision due to the lack of selective advantage in a dark environment [55, 56].
Pax6 play a central role in the core gene regulatory network that governs the development of the eye, a function that is well conserved in both invertebrates and vertebrates. The eye regulatory genes including ey, toy, so, eya and dac control early eye development in Drosophila [57]. Toy acts upstream to induce ey expression [58, 59]. Ey activates expression of so, eya and dac [13, 43, 60]. The latter three are part of a positive feedback loop controlling ey expression [61–63] and further regulate more peripheral genes in the eye development network such as atonal (Ato) [56, 64]. The gene networks have meanwhile also been extended to the respective mammalian proteins [65–67]. Thus Pax6 proteins (ey and toy) function upstream in eye gene hierarchies, orchestrating all downstream events. To test if Pax6 is also involved in eye absence in deep sea organisms, we identified and characterized Pax6 of R. piscesae. Comparison of the deduced amino acid sequence with Pax6 proteins from other species revealed that two characteristic domains (PD and HD) are highly conserved, indicating selection pressure was exerted due to functional constraints.
Drosophila Pax6 homolog ey has been shown to bind directly to the so promoter and activate so expression [13]. However, our functional analysis by dual luciferase assay revealed that RpPax6 can directly target on so, not as activator but as repressor. As DNA binding transcription factor, Pax genes regulate the development of multiple organs through the utilization of PD and HD combination for target gene promoter recognition [68]. The results (Fig 3) non-conserved C region moiety plays crucial role in the suppression of so promoter activity. Previous study has revealed that transactivation domain in ey as well as Pax6 of quail are both located in the c-terminus of the protein [69, 70]. It is possible that the C-terminal moiety of RpPax6 achieved repression ability either directly or by activation of a repressor. In addition ey has been found to be a putative transcriptional repressor. The repressive activity lies in the linker region between paired domain and homeodomain [69]. Our result show RpPax6 with N-terminal moiety swapped with the corresponding portion of ey (eyN+RpPax6C and eyPD+RpPax6HD) had little impact on the activity of the so promoter (Fig 3), indicating the presence of repressive activity within the N-terminal moiety of RpPax6.
The ability to induce ectopic eyes through Pax6 misexpression has been demonstrated in Drosophila and vertebrates [12]. Pax6 genes of various species including C. elegans [71] mouse, Drosophila [72], sea squirt [11], squid [10], and lancelet [71] are capable to induce supernumerary eyes upon targeted ectopic expression by means of the GAL4-system in D. Melanogaster [8, 10]. While the reciprocal experiment, expression of Drosophila ey and its paralog twin of eyeless (toy) genes in Xenopus embryos, induces the development of vertebrate eye structures [73]. Our experiments show flies expression of UAS-ey under the control of dpp-GAL4 generated many ectopic eye structures on wings, legs and head, in line with previous research [8]. By contrast, none of the UAS-RpPax6 transgenic lines was able to induce ectopic eye morphogenesis under the same conditions. It is conceivable that RpPax6 expression cause repression of so and lead to the failure to trigger the eye formation process.
To elucidate the role of RpPax6 in eye developmental cascade, we misexpressed ey, mPax6 and RpPax6 in an ey mutant background. Rescuing the ey2 mutant by ey and mPax6 leads to full recovery of eye size. However, expression of RpPax6 in the same background is lethal with a lethal phase during the pupal stage, in agreement with the lethal phase of the severe ey mutant eyD. eyD mutants has been described to develop into fully formed headless adults that fail to eclose from their pupal cases (pharate adults) [59]. Interestingly, similar headless phenotypes was also produced in pharates of RpPax6 expressed ey2 mutant. The EyD protein not only lacks the entire homeodomain, but also 660 amino acids in the C-terminus [74] show that overexpression of ey can rescue the lethality of homozygous eyD mutants and also suppress the eyD phenotype and partially restore head development. It has been suggested that the remaining paired domain is be able to bind to the cluster of binding sites within the eye enhancer of so, but the modifications in the C-terminus might interfere with normal transcriptional activation. Similarly, our Luciferase reporter assay revealed that RpPax6 protein suppresses the transcription of sine oculis (so) and the C-terminal of RpPax6 plays crucial role in the repressive activity. We postulate that RpPax6 expression induced by ey-GAL4 might interfere with the developmental pathway during eye-antenna disc formation and lead to the headless phenotype.
In D. melanogaster, the compound eyes and most head structures (head capsule, antenna, ocelli) develop entirely from the eye-antennal discs [75], in which process ey gene is involved in cell proliferation, differentiation and migration/adhesion [76, 77]. The headless phenotype characterized by the lack of structures derived from eye-antennal discs suggests massive cell death during development [56, 78]. As evident from acridine orange staining of RpPax6 expressed larva and rescue of headless by the expression of the baculovirus P35 protein in eye-antennal discs (Fig 8D), the headless phenotype of UAS-RpPax6/ey-GAL4 pharates results from the induction of apoptosis by expression of RpPax6. Paradoxically, authentic Pax genes are associated with differentiation, proliferation and anti-apoptosis during development process [76, 77]. Inhibition of ey [58, 79] as well as other Pax genes including Pax2, Pax8 [80], Pax3 and Pax7 [81] induce apoptosis. Interestingly, Pax6 is involved in the eye regression of a blind cave form of the teleost Astyanax mexicanus. Pax6 sequence is found to be identical in the eyeless cave form (cavefish) and eyed surface form (surface fish) [82]. However, during embryonic development in cavefish increased expression of sonic hedgehog (Shh) suppresses Pax6 and increases expression of Shh-regulated genes, which further results in lens apoptosis and eye degeneration. The apoptosis and Pax6 down regulation are common to several independently derived cavefish populations, suggesting the importance in the evolution of eye degeneration [83–86]. The results of our study indicate that alternation of Pax6 function may cause the activation of apoptosis and further contribute the blindness in vestimentiferan tubeworms. Fly models provide powerful genetic systems to dissect the role of transcription factor change in developmental evolution. In the present study we have used this tool to provide some initial molecular insights into the mechanisms of transcriptional regulation of RpPax6. Further in vivo investigations are necessary to validate the function in the normal context.
Acknowledgments
We are grateful to Renjie Jiao and the Bloomington Stock Center for fly stocks.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Scientific Research Foundation of Third Institute of Oceanography (SOA, grant: 2015001), the China Ocean Mineral Resources R & D Association (COMRA, grants: DY125-22-QY-05 and DY-125-15-T-08), the Natural Science Foundation of China (grants: 31271567 and 31272684), the Fujian Marine Hi-tech Industry Program (grant: 2014-05), and the Aquatic Sanxin Engineering Project of Jiangsu Province (grant: D2015-11-5). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blake JA, Ziman MR. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014; 141(4):737–751. 10.1242/dev.091785 [DOI] [PubMed] [Google Scholar]
- 2.Relaix F. Pax genes: Master regulators of development and tissue homeostasis. Semin Cell DevBiolI. 2015; 44:62–63. [DOI] [PubMed] [Google Scholar]
- 3.Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997; 19(9):755–765. 10.1002/bies.950190905 [DOI] [PubMed] [Google Scholar]
- 4.Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999; 15(9):371–377. [DOI] [PubMed] [Google Scholar]
- 5.Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu Rev Neuro Sci. 1997; 20(1):483–532. [DOI] [PubMed] [Google Scholar]
- 6.Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991; 354:522–525. 10.1038/354522a0 [DOI] [PubMed] [Google Scholar]
- 7.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991; 113(4):1435–1449. [DOI] [PubMed] [Google Scholar]
- 8.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995; 267:1788–1792. [DOI] [PubMed] [Google Scholar]
- 9.Loosli F, Kmita-Cunisse M, Gehring WJ. Isolation of a Pax-6 homolog from the ribbonworm Lineus sanguineus. P Nat Acad Sci USA. 1996; 93(7):2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring WJ, et al. Squid Pax-6 and eye development. P Nat Acad Sci USA. 1997; 94(6):2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glardon S, Callaerts P, Halder G, Gehring WJ. Conservation of Pax-6 in a lower chordate, the ascidian Phallusia mammilata. Development. 1997; 124(4):817–825. [DOI] [PubMed] [Google Scholar]
- 12.Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999; 126(19):4213–22. [DOI] [PubMed] [Google Scholar]
- 13.Niimi T, Seimiya M, Kloter U. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999; 126(10):2253–2260. [DOI] [PubMed] [Google Scholar]
- 14.Xu PX, Woo I, Her H. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997; 124(1):219–231. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, Ranade S, Cai CQ. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006; 133(24):4881–4889. 10.1242/dev.02669 [DOI] [PubMed] [Google Scholar]
- 16.Riesenberg AN, Le TT, Willardsen MI. Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis. 2009; 47(3):175–187. 10.1002/dvg.20479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Land MF, Nilsson DE. Animal Eyes. New York: Oxford University Press; 2002. [Google Scholar]
- 18.Leys SP, Degnan BM. Cytological basis of photoresponsive behavior in a sponge larva. Biol Bull US. 2001; 201(3):323–338. [DOI] [PubMed] [Google Scholar]
- 19.Nordström K, Seymour J, Nilsson D. A simple visual system without neurons in jellyfish larvae. P Roy Soc Lond B Bio. 2003; 270(1531): 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.vonDöhren J, Bartolomaeus T. Ultrastructure and development of the rhabdomeric eyes in Lineus viridis (Heteronemertea, Nemertea). Zoology. 2007; 110(5):430–438. 10.1016/j.zool.2007.07.006 [DOI] [PubMed] [Google Scholar]
- 21.Brandenburger J, Woolacott R, Eakin R. Fine structure of eyespots in tornarian larvae (Phylum: Hemichordata). Cell Tissue Res. 1973; 142(1):89–102. [DOI] [PubMed] [Google Scholar]
- 22.Lacalli TC. Frontal eye circuitry, rostral sensory pathways and brain organization in amphioxus larvae: evidence from 3D reconstructions. P Roy Soc Lond B Bio. 1996; 351(1377):243–63. [Google Scholar]
- 23.Arendt D, Tessmar K, de Campos-Baptista MI, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereisdumerilii and evolutionary conservation of larval eyes in bilateria. Development. 2002; 129(5):1143–1154. [DOI] [PubMed] [Google Scholar]
- 24.Rhode B. Development and differentiation of the eye in Platynereisdumerilii (Annelida, Polychaeta). J Morphol. 1992; 212(1):71–85. [DOI] [PubMed] [Google Scholar]
- 25.Rhode B. Larval and adult eyes in Capitella sp. I (Annelida, Polychaeta). J Morphol. 1993; 217(3): 327–335. [DOI] [PubMed] [Google Scholar]
- 26.Bartolomaeus T. Different photoreceptors in juvenile Ophelia rathkei (Annelida, Opheliida). Microfauna Marina. 1993; 8:99–114. [Google Scholar]
- 27.Hermans C, Eakin R. Fine structure of the eyes of an alciopidpolychaete, Vanadistagensis. Z. Morphol. J Morphol. 1974; 79(4):245–267. [Google Scholar]
- 28.Yamamoto T, Tasaki K, Sugawara Y. Fine structure of the octopus retina. J Cell Biol. 1965; 25(2): 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermans CO, Eakin RM. Fine structure of the cerebral ocelli of a sipunculid, Phascoloso maagassizii. Zeitschriftfür Zellforschung und mikroskopische Anatomie. Cell Tissue Res. 1969; 100 (3):325–339. [DOI] [PubMed] [Google Scholar]
- 30.Eakin RM, Westfall JA. Fine structure in the eye of Peripatus (Onychophora). Cell Tissue Res. 1965; 68(2):278–300. [DOI] [PubMed] [Google Scholar]
- 31.Little CTS, Herrington RJ, Maslennikov VV, Morris NJ, Zaykov VV. Silurian hydrothermal vent community from the southern Urals, Russia. Nature. 1997; 385:146–148. [Google Scholar]
- 32.Caullery M. Surles Siboglinidae, type nouveau dinvertkbres receuillis par l’expedition du Siboga. Cr Acad Bulg Sci. 1914; 158:2014–2017. [Google Scholar]
- 33.McMullin ER, Hourdez S, Schaeffer SW. Phylogeny and biogeography of deep sea vestimentiferan tubeworms and their bacterial symbionts. Symbiosis. 2003; 34(1):1–41. [Google Scholar]
- 34.Kojima S, Hashimoto T, Hasegawa M, Murata S, Ohta S, Seki H, et al. Close phylogenetic relationship between vestimentifera (tube worms) and annelida revealed by the amino acid sequence of elongation factor-lα. J Mol Evol. 1993; 37(1):66–70. [DOI] [PubMed] [Google Scholar]
- 35.Schulze A. Phylogeny of Vestimentifera (Siboglinidae, Annelida) inferred from morphology. Zool Scr. 2003; 32(4):321–342. [Google Scholar]
- 36.Tunnicliffe V. The biology of hydrothermal vents: ecology and evolution. Oceanogr Mar Biol. 1991; 29:319–407. [Google Scholar]
- 37.Urcuyo IA, Massoth GJ, Julian D, Fisher CR. Habitat, growth and physiological ecology of a basaltic community of Ridgeia piscesae from the Juan de Fuca Ridge. Deep-Sea Res Pt I. 2003; 50(6):763–780. [Google Scholar]
- 38.Ruan L, Bian X, Wang X. Molecular characteristics of the tubeworm, Ridgeia piscesae, from the deep-sea hydrothermal vent. Extremophiles. 2008; 12(5):735–739. 10.1007/s00792-008-0172-8 [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bionform. 2004; 5(2):150–163. [DOI] [PubMed] [Google Scholar]
- 40.Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, et al. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. P Nat Acad Sci USA. 2004; 101(44):15811–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993; 117(1):29–43. [DOI] [PubMed] [Google Scholar]
- 42.Halder G, Callaerts P, Flister S. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998; 125(12): 2181–2191. [DOI] [PubMed] [Google Scholar]
- 43.Zimmerman JE, Bui QT, Liu H, et al. Molecular genetic analysis of Drosophila eyes absent mutants reveals an eye enhancer element. Genetics. 2000; 154(1):237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauck B, Gehring WJ, Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. P Nat Acad Sci USA. 1999; 96(2):564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karim FD, Rubin GM. Ectopic expression of activated ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998; 125(1):1–9. [DOI] [PubMed] [Google Scholar]
- 46.Gage JD, Tyler PA. Deep-sea biology: a natural history of organisms at the deep-sea floor Cambridge: Cambridge University Press; 1991. [Google Scholar]
- 47.Herring P. The biology of the deep ocean Oxford: Oxford University Press; 2002. [Google Scholar]
- 48.Rex MA. Community structure in the deep-sea benthos. Annu Rev Ecol Syst. 1981; 12:331–353. [Google Scholar]
- 49.Jones ML. On the Vestimentifera, new phylum: six new species, and other taxa, from hydrothermal vents and elsewhere. P Biol Soc Wash. 1985; 6:117–185. [Google Scholar]
- 50.Tunnicliffe V, McArthur AG, McHugh D. A biogeographical perspective of the deep-sea hydrothermal vent fauna. Adv Mar Biol. 1998; 34:353–442. [Google Scholar]
- 51.Nyholm SV, Robidart J, Girguis PR. Coupling metabolite flux to transcriptomics: insights into the molecular mechanisms underlying primary productivity by the hydrothermal vent tubeworm Ridgeia piscesae. Biol Bull US. 2008; 214(3):255–265. [DOI] [PubMed] [Google Scholar]
- 52.Purschke G, Arendt D, Hausen H, Müller MC. Photoreceptor cells and eyes in Annelida. Arthropod Struct Dev. 2006; 35(4):211–230. 10.1016/j.asd.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 53.Suschenko D, Purschke G. Ultrastructure of pigmented adult eyes in errant polychaetes (Annelida): Implications for annelid evolution. Zoomorphology. 2009; 128(1):75–96. [Google Scholar]
- 54.Purschke G, Nowak KH. Ultrastructure of pigmented eyes in Dorvilleidae (Annelida, Errantia, Eunicida) and their importance for understanding the evolution of eyes in polychaetes. Acta Zool Stockholm. 2015; 96(1):67–81. [Google Scholar]
- 55.Raupach MJ, Mayer C, Malyutina M. Multiple origins of deep-sea Asellota (Crustacea: Isopoda) from shallow waters revealed by molecular data. P RoySoc B Biol Sci. 2009; 276 (1658):799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warrant E. Vision in the dimmest habitats on earth. J Comp Physiol A. 2004; 190(10):765–789. [DOI] [PubMed] [Google Scholar]
- 57.Friedrich M. Ancient mechanisms of visual sense organ development based on comparison of the gene networks controlling larval eye, ocellus, and compound eye specification in Drosophila. Arthropod Struct Dev. 2006; 35(4):357–378. 10.1016/j.asd.2006.08.010 [DOI] [PubMed] [Google Scholar]
- 58.Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999; 3(3):297–307. [DOI] [PubMed] [Google Scholar]
- 59.Kronhamn J, Frei E, Daube M. Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless. Development. 2002; 129(4):1015–1026. [DOI] [PubMed] [Google Scholar]
- 60.Punzo C, Kurata S, Gehring WJ. The eyeless homeodomain is dispensable for eye development in Drosophila. Gene Dev. 2001; 15(13):1716–1723. 10.1101/gad.196401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997; 91(7):893–903. [DOI] [PubMed] [Google Scholar]
- 62.Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997; 91(7):881–891. [DOI] [PubMed] [Google Scholar]
- 63.Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development. 2005; 132(12):2771–2782. 10.1242/dev.01841 [DOI] [PubMed] [Google Scholar]
- 64.Lynch VJ, Wagner GP. Revisiting a classic example of transcription factor functional equivalence: are Eyeless and Pax6 functionally equivalent or divergent? J Eep Zool Part B. 2011; 316(2):93–98. [DOI] [PubMed] [Google Scholar]
- 65.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Gene Dev. 1999; 13(24):3231–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002; 22(19):6759–6766. 10.1128/MCB.22.19.6759-6766.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, et al. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999; 19(10):6815–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Bi. 2007; 23(1):645–673. [DOI] [PubMed] [Google Scholar]
- 69.Weasner BM, Weasner B, DeYoung SM, Michaels SD, Kumar JP. Transcriptional activities of the Pax6 gene eyeless regulate tissue specificity of ectopic eye formation in Drosophila. Dev Biol. 2009; 334(2):492–502. 10.1016/j.ydbio.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005; 96(3):171–184. 10.1093/jhered/esi027 [DOI] [PubMed] [Google Scholar]
- 71.Carriere C, Plaza S, Caboche J, Dozier C, Bailly M, Martin P. Nuclear localization signals, DNA binding, and transactivation properties of quail Pax-6 (Pax-QNR) isoforms. Cell Growth Differ. 1995; 6(12): 1531–1540. [PubMed] [Google Scholar]
- 72.Lionakis MS; Lewis RE; May GS; Wiederhold NP; Albert ND; Halder G; Kontoyiannis DP. Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis. 2005; 191(7):1188–95. 10.1086/428587 [DOI] [PubMed] [Google Scholar]
- 73.Onuma Y, Takahashi S, asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. P Nat Acad Sci USA. 2002; 99(4):2020–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacobsson L, Kronhamn J, Rasmuson-Lestander Å. The Drosophila Pax6 paralogs have different functions in head development but can partially substitute for each other. Mol Genet Genomics. 2009; 282(3):217–231. 10.1007/s00438-009-0458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jordanova A, Irobi J, Thomas FP. Disrupted function and axonal distribution of mutant tyrosyl-Trna synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006; 38(2):197–202. 10.1038/ng1727 [DOI] [PubMed] [Google Scholar]
- 76.van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002; 11(10):1161–1167. [DOI] [PubMed] [Google Scholar]
- 77.Morante J, Erclik T, Desplan C. Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development. 2011; 138(4):687–693. 10.1242/dev.056069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Punzo C, Plaza S, Seimiya M, Schnupf P, Kurata S, Jaeger J, et al. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development. 2004; 131:3943–3953. 10.1242/dev.01278 [DOI] [PubMed] [Google Scholar]
- 79.Halder G, Callaerts P, Flister S. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998; 125(12): 2181–2191. [DOI] [PubMed] [Google Scholar]
- 80.Bouchard M, Souabni A, Mandler M, Neubüser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002; 16(22):2958–2970. 10.1101/gad.240102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Relaix F, Rocancourt D, Mansouri A. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005; 435(7044): 948–953. 10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- 82.Behrens M, Langecker TG, Wilkens H, Schmale H. Comparative analysis of Pax-6 sequence and expression in the eye development of the blind cave fish Astyanax fasciatus and its epigean conspecific. Mol Biol Evol. 1997; 14(3):299–308. [DOI] [PubMed] [Google Scholar]
- 83.Jeffery WR, Martasian DP. Evolution of eye regression in the cavefish Astyanax: apoptosis and the Pax-6 gene. Am Zool. 1998; 38(4):685–696. [Google Scholar]
- 84.Jeffery WR, Strickler AG, Yamamoto Y. To see or not to see: Evolution of eye degeneration in Mexican blind cavefish. Integr Cpmp Biol. 2003; 43(4):531–541. [DOI] [PubMed] [Google Scholar]
- 85.Jeffery WR. Adaptive evolution of eye degeneration in the Mexican blind cavefish. J Hered. 2005; 96(3):185–196. 10.1093/jhered/esi028 [DOI] [PubMed] [Google Scholar]
- 86.Yamamoto Y, Jeffery WR. Probing teleost eye development by lens transplantation. Methods. 2002; 28(4):420–426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.