Abstract
Transcriptional regulation, a multiple-step process, is still poorly understood in the important pig pathogen Mycoplasma hyopneumoniae. Basic motifs like promoters and terminators have already been described, but no other cis-regulatory elements have been found. DNA repeat sequences have been shown to be an interesting potential source of cis-regulatory elements. In this work, a genome-wide search for tandem and palindromic repetitive elements was performed in the intergenic regions of all coding sequences from M. hyopneumoniae strain 7448. Computational analysis demonstrated the presence of 144 tandem repeats and 1,171 palindromic elements. The DNA repeat sequences were distributed within the 5’ upstream regions of 86% of transcriptional units of M. hyopneumoniae strain 7448. Comparative analysis between distinct repetitive sequences found in related mycoplasma genomes demonstrated different percentages of conservation among pathogenic and nonpathogenic strains. qPCR assays revealed differential expression among genes showing variable numbers of repetitive elements. In addition, repeats found in 206 genes already described to be differentially regulated under different culture conditions of M. hyopneumoniae strain 232 showed almost 80% conservation in relation to M. hyopneumoniae strain 7448 repeats. Altogether, these findings suggest a potential regulatory role of tandem and palindromic DNA repeats in the M. hyopneumoniae transcriptional profile.
Introduction
Mycoplasma hyopneumoniae is a diminutive bacterium, characterized by a small genome (0.92 Mb) with a low GC content [1]. It is commonly associated with mycoplasmal pneumonia in pigs [2] and infected animals are affected by a sporadic, dry and non-productive cough, retarded growth rate and inefficient utilization of feed [3]. Until now, the genomes of six strains of M. hyopneumoniae have been sequenced [1, 4–7] and the availability of their sequences has enabled whole comparative genome analyses [8]. Transcription, a multi-step mechanism, is finely regulated in all forms of life and is still poorly understood in M. hyopneumoniae. The occurrence of transcription units [4, 9, 10], promoters [11, 12] and terminators [13] has already been described in this species, but the existence of other regulatory sequences remains to be elucidated.
DNA repeats, widespread and characterized in eukaryote genomes, can also play an important role in prokaryote genomic regulation [14]. It is usually hypothesized that repeats arise by successive duplications and several causal mechanisms, like homologous recombination, slipped-strand mispairing of DNA polymerase or by events of genomic transposition [15]. Bacterial repeats are commonly classified as low-complexity repeats and longer repeats. Low-complexity repeats can be composed of simple oligonucleotides (typically ranging from mononucleotide to pentanucleotide in size) repeated several times in a head-to-tail configuration (tandem), while longer repeats may include complex and spaced repeats, such as palindromes, transposable elements, inverted sequences, minisatellites and large tandem repeats spread throughout the genome [16].
DNA repeats can be found within coding-sequences (CDS), in intergenic regions, or in transposable elements, reflecting both regulatory and structural requirements for the bacterial chromosome [17, 18]. Phase variation, an important mechanism of evolution and adaptation of prokaryotes, is capable of switches in an ON/OFF way the expression of important genes. The presence of repetitive elements, especially simple sequence repeats (SSR) is the major source of variability in these cases. Mutations in SSRs within the coding region can be related to the shifting of the reading frame of proteins. Mutations in noncoding or promoter-located SSRs generate more subtle variations in protein expression levels due to alterations in the relative spacing and positional orientation of promoter elements [18, 19]. The presence of repetitive elements within intergenic regions has already been related to pathogenicity through phase variation in Neisseria species, Haemophilus parainfluenzae, Moraxella catarrhalis [20] and Mycoplasma hyorhinis [18]. Many other phenotypes associated with repetitive genotypes were reviewed by Belkum et al. [14] involving microbial evolution, pathogenesis and molecular epidemiology. In Mycoplasma spp., the presence of some repetitive elements has been described in Mycoplasma genitalium [21–24], Mycoplasma gallisepticum [23, 25, 26], Mycoplasma bovis [27], M. hyorhinis [28], Mycoplasma fermentans [29], Mycoplasma synoviae [23, 30], Mycoplasma pneumoniae [23, 24, 31], Mycoplasma hominis [32], Mycoplasma penetrans [23], Mycoplasma pulmonis [23, 24, 33], Mycoplasma mobile [23] and Mycoplasma mycoides [23, 34] genomes. In M. hyopneumoniae the presence of SSR elements [23] and some specific palindromic repeats [35] have already been reported, however a genome-wide approach correlating DNA repeats with their possible genomic regulatory consequences is not yet available. Therefore, in this work, an in silico prediction associated with experimental validation was performed, aiming to verify and associate the presence of tandem and palindromic repeats as transcriptional regulatory sites.
Materials and Methods
DNA repeats in silico analysis
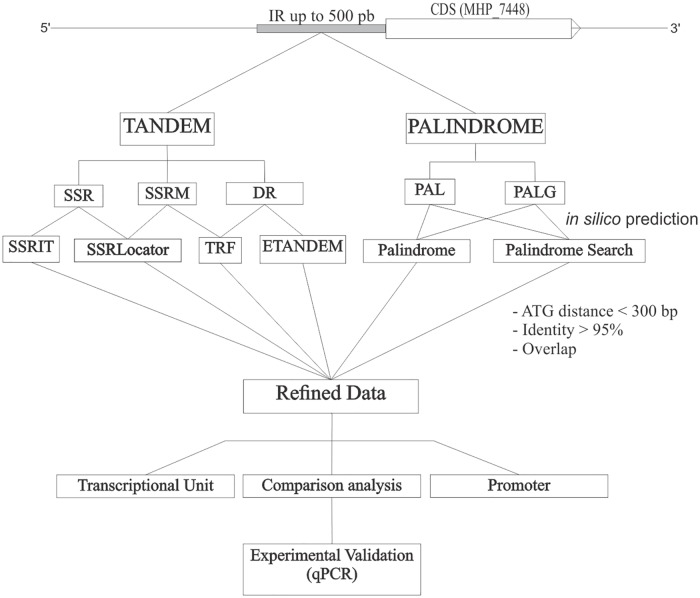
Based on the most common repeats already described in prokaryote genomes, especially in Mycoplasma spp. [36], two types of DNA repeats were selected to be investigated in this work (tandem and palindromic elements). Tandem sequences were divided into the following: i) Simple Sequence Repeats (SSR); ii) Simple Sequence Repeats of Mononucleotides (SSRM) and iii) Direct Repeats (DR). Palindromic elements were searched using two approaches: allowing gaps (PALG) or not allowing gaps (PAL). In-house PERL scripts were used to extract intergenic regions up to 500 bp upstream from the start codon of all M. hyopneumoniae strain 7448 (NSDC AE017244.1) CDSs which were used as input for the software’s prediction. The annotation of the genome was manually cured, with confident translational start codons. For each type of repeat sequence, two independent software packages were used, as detailed below.
In the present work, SSR is defined as a sequence containing 2 to 10 repeated nucleotides as described by Huang et al. [36]. The algorithms used to search for SSR were SSRLocator [37] and Simple Sequence Repeat Identification Tool (SSRIT) [38].
SSRM is defined as a mononucleotide repeated at least 8 times in a head-to-tail orientation. The software packages used to predict SSRM were Tandem Repeat Finder (TRF) [39] and SSRIT. Only perfect repeats of SSR and SSRM were analysed, once mismatches were not allowed in this approach.
DR is defined as an element with 11 to 50 nucleotides repeated at least twice in a tandem way. TRF and ETANDEM from The European Molecular Biology Open Software Suite (EMBOSS) [40] were used to predict this type of repeat. In this case, output data from TRF and ETANDEM were filtered to include only DR repeats that had identity of ≥95% within the repeated copies.
PALG is an inverted repeat of 9 to 20 nucleotides containing additional gaps ranging from 5 to 15 bases. Palindrome from EMBOSS and Palindrome Search [41] were the algorithms used in this approach.
PAL is an inverted repeat of 9 to 15 bases with no gaps. The same software was used to predict PAL and PALG, and only elements that had the maximum of 2 mismatches between the inverted repeat were considered.
Just PALG and PAL elements that were located up to 300 bp distant from the ATG were analyzed, the remaining data was excluded. After the individual search of each element, the results were grouped and all overlaps were eliminated according to the following hierarchy: PAL, SSRM, DR, SSR and PALG.
All repetitive elements found in noncoding regions of M. hyopneumoniae strain 7448 were classified based on the downstream CDS, ATG distance, association with promoter sequences and transcription unit distribution.
A search for common motifs among repeat classes was performed through the web server MEME SUITE [42], using default parameters. A complete workflow of the search strategy was represented in Fig 1.
Fig 1. Pipeline of repeat search strategy.
Up to 500 bp of 5’ intergenic regions (IR) of all M. hyopneumoniae strain 7448 (MHP_7448) CDSs were used for in silico prediction of tandem and palindromic DNA repeats.
Comparative analysis
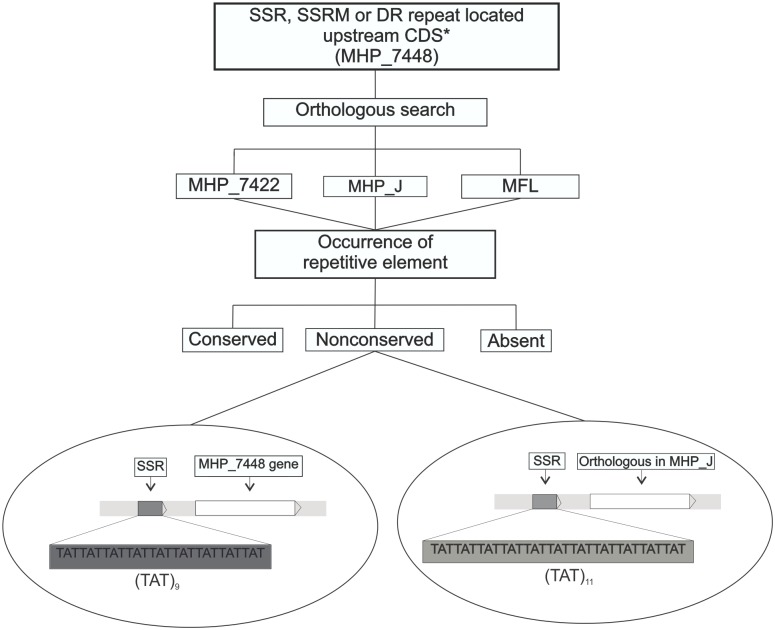
To validate the relevance of repetitive elements found, comparison analysis was performed using three different approaches. Initially, all tandem repeats (SSR, SSRM and DR) found upstream of the first gene of M. hyopneumoniae strain 7448 transcription units [9, 10] were compared against three other mycoplasma genomes: M. hyopneumoniae strain 7422 –pathogenic; M. hyopneumoniae strain J–nonpathogenic; and Mycoplasma flocculare ATCC 27716 –phylogenetically related to M. hyopneumoniae and nonpathogenic. Therefore, a BLAST [43] search was performed with the first CDS of each transcription unit against the genomes of the three organisms mentioned above. The corresponding tandem motif was manually localized in the 5´upstream region of each orthologous gene. One of three classifications could be assigned to each repeat: conserved (C)–the repetitive element was exactly the same in sequence and number; nonconserved (NC)–the copy number of the tandem element was different; and absent (A)–the repetitive element was not found in the orthologous gene. A schematic pipeline of the tandem comparison can be seen in Fig 2.
Fig 2. Pipeline of tandem repeat comparison analysis among mycoplasma genomes.
An example of nonconserved SSR between M. hyopneumoniae strain 7448 (MHP_7448), and M. hyopneumoniae strain J (MHP_J) can be observed above. A (TAT)9 repeat found in the MHP_7448 gene intergenic region was classified as nonconserved, as a (TAT)11 repeat was localized in the respective orthologous intergenic region in MHP_J. Abbreviations: M. hyopneumoniae strain 7422 (MHP_7422); M. flocculare (MFL). *First CDS of the transcription unit.
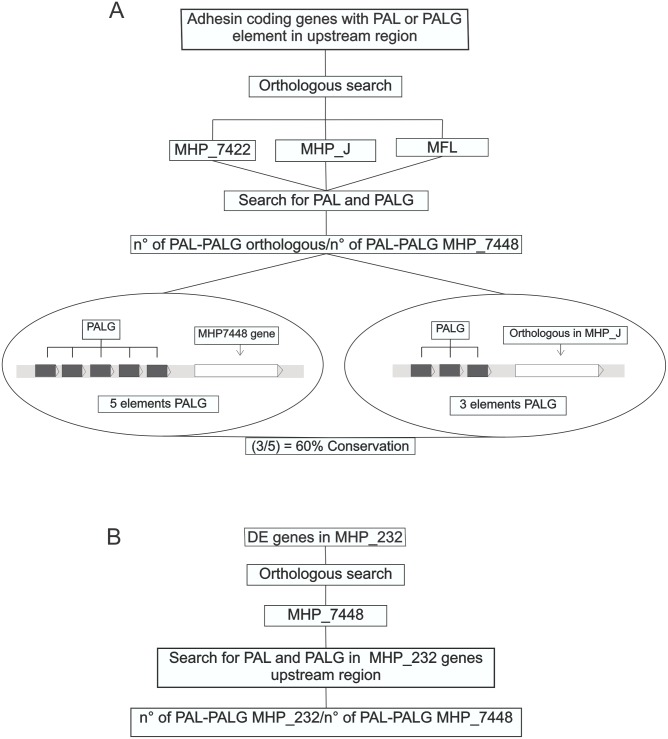
The second comparison analysis was performed in genes coding for putative adhesins [44]. Each palindromic repeat (PAL or PALG) already found upstream of the start codon of adhesin-coding genes in M. hyopneumoniae strain 7448 were compared among the same mycoplasma genomes previously described. The repeat search was conducted directly upstream of the region of the orthologous gene or in the upstream region of the corresponding transcription unit. The comparison of PAL and PALG among mycoplasmas genomes were performed by Pattern Locator software [45]. Differently from the tandem repeat strategy, just two classifications could be assigned to the PALG or PAL elements: 1) conserved–the inverted repeat that composed the palindromic element is similar (maximum of 3 mismatches) to that found in M. hyopneumoniae strain 7448 or 2) absent–the repetitive element PALG or PAL was not identified. The level of conservation (%) of palindromic repeats between mycoplasma genomes was calculated by dividing the number of conserved repeats found in the noncoding region of orthologue gene by the total number of repeats found in the respective M. hyopneumoniae strain 7448 gene (Fig 3A).
Fig 3. Pipeline of repeat conservation in putative adhesin-coding genes and differentially expressed genes from strains of M. hyopneumoniae.
(A) Comparison analysis of adhesins, with an example of relative conservation between MHP_7448 genes that have 5 PALG repeats and the respective orthologues in MHP_J that have only 3. The percentage (%) conservation between MHP_7448 and MHP_J repeats was obtained by dividing the number of repeat elements. (B) Repeats present in differentially expressed (DE) genes found in M. hyopneumoniae strain 232 (MHP_232) were compared with those found in MHP_7448 in the same way as in (A). Abbreviations as in Fig 2.
The third approach was based on previous studies of M. hyopneumoniae strain 232, in which microarray assays have identified genes that are differentially expressed in some growing adverse situations, such as exposure to norepinephrine [46] or hydrogen peroxide [47], in vitro infection [48], iron depletion [49] and heat shock [50]. Using BLAST search [43] orthologous of the differentially expressed genes from M. hyopneumoniae strain 232 were found in M. hyopneumoniae strain 7448 genome. A comparison analysis of palindromic repeats that were predicted in M. hyopneumoniae strain 7448 against M. hyopneumoniae strain 232 was performed as described for the adhesin genes search strategy (Fig 3B). As palindromic elements can form secondary structure, the ΔG was evaluated by QuickFold algorithm, [51] using default parameters.
qPCR experiments
Culture conditions, RNA isolation and cDNA synthesis by Reverse Transcription (RT) were performed as described in Siqueira et al. [10]. Target and primer descriptions for M. hyopneumoniae strain 7448, M. hyopneumoniae strain J and M. flocculare ATCC 27716 reactions are available in S1 Table. Primers were designed using Vector NTI Advance 10 (Invitrogen, USA).
Quantitative PCR (qPCR) assay was performed using 1:50 cDNA as template and Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, USA) on 7500 Real-Time PCR Systems (Applied Biosystems, USA). The qPCR reactions were carried out at 90°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A melting curve analysis for each primer pair was done to verify primer efficiency.
Relative expression of each gene was evaluated in M. hyopneumoniae strain 7448, M. hyopneumoniae strain J and M. flocculare ATCC 27716 RNAs. Relative expression of mRNA was calculated by the 2-ΔCt method [52]. The threshold cycle (CT) values were normalized to the reference gene lon (MHP7448_0524). Reference gene was determined in a specific assay where expression profiles of several genes were evaluated in the three mycoplasma RNAs tested (M. hyopneumoniae strain 7448, M. hyopneumoniae strain J and M. flocculare ATCC 27716). The gene that showed no differential expression in all mycoplasma genomes tested was used in relative expression calculations. Three technical and two biological replicates were done for each target evaluated. Statistical analyses were performed using GraphPad Prism 6 software by One-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
Results
In silico prediction
Computational analysis predicted the presence of 340 tandem repeats and 1,879 palindromic elements in the genome of M. hyopneumoniae strain 7448. Among the tandem repeats, 272 SSRM, 55 SSR and 13 DR repeats were found. Palindromic elements were divided into 689 PAL and 1,190 PALG (Table 1). These results were further analysed and all overlapping sequences were excluded. We could observed that PAL elements, in most of the cases, overlapped partially with PALG (Table 1) and therefore PAL overlapping elements were excluded from further analysis. The decision to exclude was based on the capability of the selected PALG elements to form stronger secondary structure, representing an interesting physical modification in the DNA molecule [53]. As a result, 59% of previously located repeats were maintained in the M. hyopneumoniae strain 7448 genome, distributed as 144 tandem (111 SSRM, 29 SSR and 4 DR repeats) and 1,171 palindromic repeats (73 PAL and 1,098 PALG) (Table 1; S2 Table). The presence of the tandem repeats and palindromic elements was confirmed by utilization of two independent software packages. The best results were found with the SSR repeats wherein almost 90% of elements found were predicted by both algorithms tested (Table 1).
Table 1. Computational analysis of tandem and palindromic repeats in the M. hyopneumoniae strain 7448 genome.
| Repeats | Software | General Data | Refined Data | ||
|---|---|---|---|---|---|
| N° repeats | N° CDS* | N° repeats | N° CDS* | ||
| SSRM | TRF | 1 | 1 | 0 | 0 |
| SSRLocator | 229 | 165 | 82 | 68 | |
| TRF x SSRLocatora | 42 | 41 | 29 | 29 | |
| Total | 272 | 192 | 111 | 89 | |
| SSR | SSRIT | 5 | 4 | 2 | 2 |
| SSRLocator | 3 | 3 | 1 | 1 | |
| SSRIT x SSRLocatorb | 47 | 44 | 26 | 25 | |
| Total | 55 | 48 | 29 | 28 | |
| DR | TFR | 4 | 4 | 2 | 2 |
| Etandem | 8 | 8 | 1 | 1 | |
| TRF x etandemc | 1 | 1 | 1 | 1 | |
| Total | 13 | 13 | 4 | 4 | |
| PAL | Palindrome | 226 | 169 | 26 | 25 |
| Palindrome Search | 244 | 174 | 27 | 26 | |
| Palindrome x Palindrome Searchd | 219 | 169 | 20 | 19 | |
| Total | 689 | 314 | 73 | 62 | |
| PALG | Palindrome | 843 | 336 | 779 | 321 |
| Palindrome Search | 168 | 134 | 149 | 119 | |
| Palindrome x Palindrome Searche | 179 | 138 | 170 | 132 | |
| Total | 1190 | 393 | 1098 | 373 | |
General data represent independent software prediction. Refined data show filtered sequences, without overlaps.
*Number of CDS found downstream the predicted repeats.
aSSRM repeats that were predicted by the both software (TRF and SSRLocator).
bSSR repeats that were predicted by the both software (SSRIT and SSRLocator).
cDR repeats that were predicted by the both software (TRF and etandem)
dPAL and ePALG predicted by the both software (Palindrome and Palindrome Search).
The distribution of repetitive elements among 5´upstream regions of M. hyopneumoniae strain 7448 CDSs demonstrated an average of 3 PALG, 1 PAL, 1 SSRM, 1 SSR and 1 DR repeat per 5’ upstream region of a unique CDS (Table 1). Moreover, different combinations of palindrome and tandem repeats could be observed in a single intergenic region (S2 Table). Comparative analysis of nucleotide sequences in each class of repetitive element was unable to determine a common motif for tandem repeat or palindromic sequence, as they were diverse in nucleotide composition and length (S2 Table).
Repeat Classification
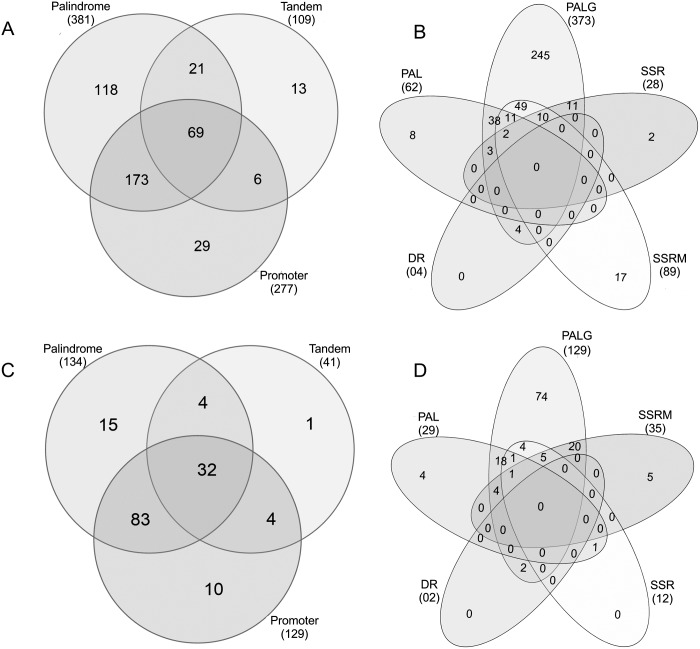
All 730 genes and 162 transcriptional units (polycistronic and monocistronic) [10] of M. hyopneumoniae strain 7448 were classified according to the cis-elements found in the upstream region of the respective start codon. Analyses of all genes revealed that 429 of them (59%) had putative promoter sequences (according to Weber et al. [12]) or repetitive element (present analysis) in the 5’ upstream region. Among these 429 genes, the most representative element in the 5’ upstream region were palindromic repeats plus promoter sequences (40%), followed by individual palindromic repeats (28%) and the combination of all putative sequences (tandem, palindromic and promoter) representing 16% (Fig 4A).
Fig 4. Repeat distribution in the intergenic regions of M. hyopneumoniae strain 7448.
(A) All intergenic regions of the genes were analysed in relation to the presence of tandem repeats, palindromic sequences or putative promoter motifs. (B) Tandem repeats and palindromic sequences were distributed in SSR, SSRM, DR, PAL and PALG throughout genes intergenic regions. The same was done for the 5’ upstream region of the first gene of each transcription unit described in genome (C) and (D).
The distribution of tandem and palindromic elements showed that individually, PALG were present in the majority of the genes 5’ upstream regions investigated (57%). The combination of PALG plus SSRM repeats was seen in 11% of genes 5’ upstream regions. PALG plus PAL elements were seen in 9% of genes 5’ upstream regions. None of the genes had exclusively DR repeats or a combination of all repeat types in a single 5’ upstream region (Fig 4B).
Considering all 162 transcriptional units of M. hyopneumoniae strain 7448, 149 (92%) of them contained a putative regulatory element (repeat or promoter) in the respective 5’ upstream region from the start gene of the transcription unit. Among the 149 transcription units, 56% contained palindromic elements associated with putative promoters; 21% showed the presence of the three elements (tandem, palindrome and promoter), and 10% contained only palindromic sequences. The three situations mentioned above represented 87% of all possible cases of elements in the regulatory regions of the transcriptional units (Fig 4C). The classification of palindromic sequences and tandem repeats in the 5’ region of the transcription units resulted in 50% of transcription units with PALG, 13% with PALG plus SSRM and 12% having PAL plus PALG. None of the transcription unit 5’ upstream regions showed the presence of an exclusive DR or SSR element, and finally, none possessed the five elements together (Fig 4D).
Comparative analysis: conservation of repeat elements in orthologous CDSs
Aiming to understand the prevalence of distribution of the repeat elements identified in M. hyopneumoniae strain 7448, a comparative analysis was performed with other swine Mycoplasma species. In this work, the presence of repeat sequence in the 5’ upstream regions of orthologous genes in M. hyopneumoniae strain 7422 (pathogenic) and M. hyopneumoniae strain J (nonpathogenic), and also with the phylogenetically related nonpathogenic mycoplasma species, M. flocculare, was explored (see Fig 2 for the pipeline analysis).
A total of 45 genes (S3 Table) that presented tandem repeats (SSR, SSRM or DR) in their 5’ upstream regions and were positioned as the first gene of a transcriptional unit were selected for comparison analysis. Concerning the 45 M. hyopneumoniae strain 7448 genes investigated by the BLAST approach, 97%, 90% and 67% of them displayed orthologous genes in the genome of M. hyopneumoniae strain 7422, M. hyopneumoniae strain J and M. flocculare, respectively. Repeat comparison analysis was performed only among the orthologous genes found in all genomes and localized in the upstream region of the first gene in each transcription unit. Therefore, 4 DR repeats, 13 SSR and 44 SSRM elements were selected for further comparative studies.
As expected, detailed analysis revealed that conservation in repeat sequences among orthologous genes was higher between strains of the same species (M. hyopneumoniae strain 7422 and M. hyopneumoniae strain J) than between different species (M. hyopneumoniae and M. flocculare) (Fig 5A; S3 Table). However, differences were found between M. hyopneumoniae strain 7448 versus M. hyopneumoniae strain 7422 and M. hyopneumoniae strain 7448 versus M. hyopneumoniae strain J when each repetitive element was analysed. Greater differences could be seen in SSRM elements, which are 60% conserved in M. hyopneumoniae strain 7422, 30% in M. hyopneumoniae strain J and had no conserved elements in M. flocculare. Interestingly, although the number of repetitive elements classified as “nonconserved” maintained the same pattern previously observed for strains of M. hyopneumoniae, it increased during analysis between M. hyopneumoniae strain 7448 and M. flocculare orthologous genes. DR and SSR showed lower values of divergence in copy number (nonconserved) compared with SSRM (see Fig 5B). Also, the number of orthologues that did not share repetitive elements with M. hyopneumoniae strain 7448 was higher in the M. flocculare genome, mainly within the DR repeat class, which was completely absent. Lower absence values were seen in SSR repeats compared with DR repeats and none SSRM were absent in the tested situation (see Fig 5C).
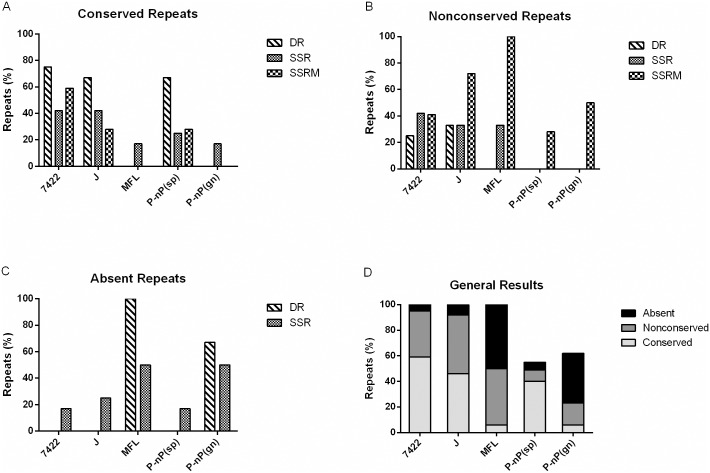
Fig 5. Conservation of repetitive elements in Mycoplasma genomes.
Graphics represent percent of relative conservation (%) of conserved (A), nonconserved (B) and absent (C) SSR, SSRM, and DR repeats found in M. hyopneumoniae strain 7448 intergenic regions in relation to strains 7422, J and M. flocculare (MFL) orthologues. A pathogenic versus nonpathogenic comparison was done and defined as P-nP. Repeats conserved between M. hyopneumoniae strain 7448 and M. hyopneumoniae strain 7422 were investigated in relation to MHP_J (P-nP (sp)) and MFL (P-nP(gn)). (D) Graphic represents the average of all tandem repeats found in each situation tested.
To understand the relation between the presence of repetitive elements and pathogenicity, a further comparative analysis was performed considering the following: repeats must be identical in M. hyopneumoniae strain 7448 and M. hyopneumoniae strain 7422 orthologous genes and only those elements that fitted this feature were analysed in relation to M. hyopneumoniae strain J or M. flocculare orthologues. These comparisons were named “Pathogenic-non-Pathogenic species” (P-nP(sp)) and “Pathogenic-non-Pathogenic genera” (P-nP(gn)), respectively. Among M. hyopneumoniae strains, two DR repeats types, 12 SSR and 39 SSRM could be analysed. Higher conservation could be observed in P-nP(sp), and a reversed scenario was seen in P-nP(gn), where nonconserved and absence of tandem repeats were found (Fig 5; S3 Table). These results demonstrated that although the number of DR repeats is almost the same in the orthologous genes of M. hyopneumoniae strains, the number of SSR and SSRM decreases. Moreover, the numbers of orthologues that share repeat conservation among the pathogenic M. hyopneumoniae strains and M. flocculare orthologues drastically decrease (see Fig 5).
In summary, a comparison of M. hyopneumoniae strain 7448 orthologous genes revealed that 58% of repeats were conserved, 36% were nonconserved and 6% were absent in M. hyopneumoniae strain 7422 orthologous. Considering M. hyopneumoniae strain J, the values change to 46% of repeat conservation, 46% of nonconservation and 8% of absence in the orthologous genes. M. flocculare orthologues demonstrated only 6% of repeat conservation, 44% of nonconservation and 50% of absence. The comparison among the orthologous genes relating to pathogenicity P-nP(sp) showed 40% of repeat conservation and 9% and 6% of nonconservation and absence, respectively. The P-nP(gn) analysis revealed that only 6% of repeats were conserved and 56% were nonconserved or absent among orthologous genes (Fig 5D; S3 Table).
Repetitive elements in the upstream regions of the putative adhesin-coding genes from the three M. hyopneumoniae strains and M. flocculare were searched using the same approach as described above. Sixteen M. hyopneumoniae putative adhesin-coding genes [44] were selected for comparison analysis. In these genes, palindrome repeats (PAL and PALG) were found with an average of 5 repeat elements per gene in M. hyopneumoniae strain 7448. In the majority of the genes coding for adhesins (12 genes), palindromic elements were located directly in the CDS 5’ upstream region. However, this situation was not applied to MHP7448_0005, MHP7448_0006, MHP7448_0361 and MHP7448_0362 CDSs. Therefore, a search for repetitive elements was performed in the upstream region of Transcriptional Unit (TU)_01 (containing MHP7448_0005 and MHP7448_0006) and TU_63 (containing MHP7448_361 and MHP7448_362). Using this approach, PAL and PALG elements could be found. Comparative analysis revealed that all M. hyopneumoniae strain 7422 and M. hyopneumoniae strain J adhesin orthologous genes were present in the respective genome and a palindromic repeat conservation level of 87% and 69%, respectively, were established. Comparison between M. hyopneumoniae strain 7448 and M. flocculare demonstrated only 27% of conserved elements in 5’ upstream region of the orthologue genes (Table 2; S4 Table).
Table 2. The presence of repetitive elements in upstream regions of putative adhesin-coding genes.
| MHP_7448 | N° ortholog element /N° MHP_7448 elements | |||
|---|---|---|---|---|
| Gene ID | Product | MHP_7422 | MHP_J | MFL |
| MHP7448_361 | P29 | 5/5 (100%) | 5/5 (100%) | 3/5 (60%) |
| MHP7448_362 | P69 | |||
| MHP7448_497 | P76 | 5/5 (100%) | 3/5 (60%) | 1/5 (20%) |
| MHP7448_198 | P97 | 6/6 (100%) | 3/6 (50%) | - |
| MHP7448_108 | P97 | 3/3 (100%) | 2/3 (67%) | 1/3 (33%) |
| MHP7448_272 | P97 | 5/6 (83%) | 5/6 (83%) | 4/6 (67%) |
| MHP7448_199 | P102 | 4/5 (80%) | 5/5 (100%) | 1/5 (20%) |
| MHP7448_107 | P102 | 1/1 (100%) | 1/1 (100%) | 0/1 (0%) |
| MHP7448_271 | P102 | 1/1 (100%) | 0/1 (0%) | 0/1 (0%) |
| MHP7448_105 | P102 | 3/3 (100%) | 1/3 (33%) | 3/3 (100%) |
| MHP7448_663 | P146 | 7/7 (100%) | 6/7 (86%) | 4/7 (57%) |
| MHP7448_496 | P216 | 2/2 (100%) | 1/2 (50%) | 1/2 (50%) |
| MHP7448_373 | LppS | 1/2 (50%) | 0/2 (0%) | 1/2 (50%) |
| MHP7448_372 | LppT | 1/5 (20%) | 2/5 (40%) | 2/5 (40%) |
| MHP7448_006 | MgPa | 5/5(100%) | 5/5(100%) | 3/5(60%) |
| MHP7448_005 | Mgpa | |||
| Total | 49/56 (87%) | 39/56 (69%) | 24/56 (27%) | |
No MHP7448_198 orthologous gene was found in M. flocculare (MFL) genome using BLAST. Abbreviations: M. hyopneumoniae strain 7448 (MHP_7448), M. hyopneumoniae strain 7422(MHP_7422), and M. hyopneumoniae strain J (MHP_J).
Experimental validation
Based on in silico comparison analysis, 12 CDSs with different tandem repeat compositions in 5’ upstream regions were selected to perform the experimental procedures (3 CDSs with DR repeats, 4 with SSR repeats, and 5 with SSRM repeats). Some of these CDSs encode important proteins to bacterial survival, like transporters or signalling molecules, and others encode hypothetical proteins. Nine genes with palindromic sequences at the upstream region of putative adhesins proteins were also selected (Table 3).
Table 3. Influence of repeat composition on mycoplasma gene expression.
| MHP_7448 | MHP_J | MFL | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Repeat Analyzed | Gene ID | Product | Conservation | Expression | p-value | Conservation | Expression | p-value | |
| DR_01_TE | rpsP | 30S ribosomal protein S16 | C | up | *** | A | - | ns | |
| DR_01_T | MHP7448_0397 | hypothetical protein | C | up | ** | A | - | ns | |
| DR_06_E | MHP7448_0197 | hypothetical protein | NC | - | ns | A | - | ns | |
| SSR_49_LI | MHP7448_0485 | hypothetical protein | NC | - | ns | NC | up | ** | |
| SSR_30_LI | MHP7448_0484 | hypothetical protein | NC | - | ns | NC | up | *** | |
| SSR_42_LI | MHP7448_0623 | ABC transporter ATP-binding—Pr1 | C | - | ns | C | - | ns | |
| SSR_05_LI | MHP7448_0087 | GTP-binding protein | A | up | **** | A | up | * | |
| SSRM_07_TL | sipS | signal peptidase I | NC | do | *** | NC | do | **** | |
| SSRM_69_L | glyA | glycine hydroxymethyltransferase | NC | up | * | NC | up | * | |
| SSRM_15_TL | MHP7448_0272 | P97-like | NC | up | **** | NC | up | *** | |
| SSRM_10_L | MHP7448_0108 | P97-like copy 2 | NC | - | ns | NC | up | * | |
| SSRM_195_L | MHP7448_0505 | lipoprotein | C | - | ns | NC | up | *** | |
| PALG_E_472a | |||||||||
| PALG_E_473a | |||||||||
| PALG_E_474a | MHP7448_0361 | P29 | 100% | up | *** | 60% | - | ns | |
| PALG_E_975a | MHP7448_0362 | P69 | up | ** | - | ns | |||
| PALG_ES_134a | |||||||||
| PALG_E_1055 | MHP7448_0497 | P76 | 60% | up | *** | 20% | - | ns | |
| PALG_E_1056 | |||||||||
| PALG_E_572 | |||||||||
| PALG_E_573 | |||||||||
| PALG_ES_183 | |||||||||
| PALG_E_291 | MHP7448_0108 | P97 | 67% | - | ns | 33% | up | * | |
| PALG_E_859 | |||||||||
| PALG_S_1184 | |||||||||
| PALG_E_381 | MHP7448_0272 | P97 | 83% | up | **** | 67% | up | *** | |
| PALG_E_382 | |||||||||
| PALG_E_383 | |||||||||
| PALG_E_385 | |||||||||
| PALG_E_925 | |||||||||
| PALG_E_926 | |||||||||
| PALG_E_863 | MHP7448_0107 | P102 | 100% | NT | NT | 0% | up | * | |
| PALG_E_924 | MHP7448_0271 | P102 | 0% | up | ** | 0% | up | ** | |
| PALG_E_237b | |||||||||
| PALG_E_238b | |||||||||
| PALG_E_239b | MHP7448_0006 | MgPa | 100% | - | ns | 60% | up | *** | |
| PALG_E_240b | MHP7448_0005 | Mgpa | - | ns | up | * | |||
| PAL_ES_24b | |||||||||
Asterisks indicate statistically significant differences in levels of expression, *0.01 < P < 0.05, **0.001 < P <0.01***, P < 0.001****, ns: non-significant. Genes with DNA repeats in the respective upstream region were classified as Conserved (C), Nonconserved (NC) and Absent (A). These genes were analysed and changes in the expression level of M. hyopneumoniae strain J (MHP_J) and M. flocculare (MFL) in relation to M. hyopneumoniae strain 7448 (MHP_7448) through qPCR assays were evaluated. Upregulation (up), downregulation (do) or no significantly differences in expression (-) could be observed among tested genes.
(a) PALG elements found in the 5’ upstream region of the first gene of the transcriptional unit containing MHP7448_0361 and MHP7448_0362 CDS.
(b) PALG elements found in the 5’ upstream region of the first gene of the transcriptional unit containing MHP7448_0005 and MHP7448_0006 CDS.
NT = not tested
Comparative transcription analysis were performed considering the conservation level of SSR, SSRM and DR elements in upstream regions of genes from M. hyopneumoniae strain J and M. flocculare, which are orthologous to M. hyopneumoniae strain 7448. Results revealed that, when conserved repeat (SSR_42_LI in both mycoplasmas analysed and SSRM_195_L in M. hyopneumoniae strain J), were found in upstream region of the gene, nonsignificant differences in the basal transcription level were observed. In the same way, in genes with nonconserved SSR repeats in intergenic region (SSR_49_LI and SSR_30_LI in M. hyopneumoniae strain J) nonsignificant differences in gene expression were seen. However, when nonconserved SSR repeats were analysed in M. flocculare, differences in gene expression were observed. The absence of SSR element (SSR_05_LI) in M. hyopneumoniae strain J and M. flocculare upstream region of orthologous demonstrated different transcription profile (Table 3). Genes with nonconserved SSRM elements (SSRM_07_TL, SSRM_69_L and SSRM_15_TL) in upstream region of M. hyopneumoniae strain J and M. flocculare demonstrated changes in level of transcripts. The exception was SSRM_10_L which only M. flocculare showed differences in gene expression (Table 3). Interestingly, genes with DR repeats in the upstream region displayed a different expression profile in relation to those with SSR and SSRM repeats, as only conserved elements (DR_01_TE; DR_01_T) demonstrated distinct expression profiles between M. hyopneumoniae strain 7448 and J (Table 3).
Detailed analyses of conserved SSR elements found in the 5’ upstream region of MHP7448_0623 orthologues from M. hyopneumoniae strain J and M. flocculare showed no significant differences in basal gene transcription levels in relation to M. hyopneumoniae strain 7448 (S1 Fig). However, among the mycoplasma orthologues, the glyA gene displays a nonconserved SSRM in the 5’ upstream region and exhibited differential expression in the tested condition (S1 Fig). The SSR element absence in MHP7448_0087 orthologues from M. hyopneumoniae strain J and M. flocculare also seems to influence in transcriptional, whereas differences in gene expressions were observed (S1 Fig).
As demonstrated in tandem repeats, adhesins coding genes revealed similar profile when conservation of palindromic elements was related to transcriptional levels. Conserved (100%) PAL and PALG present in MHP7448_0005 and MHP7448_0006 orthologues from M. hyopneumoniae strain J did not affect gene expression. However, when relative conservation dropped to 60% in orthologues of M. flocculare, basal transcript level varied significantly. The absence of palindromic repeats in MHP7448_0271 orthologues from M. hyopneumoniae strain J and M. flocculare resulted in significant differences in basal gene transcription level. Furthermore, even when few elements were lost in MHP7448_0272 orthologues from M. hyopneumoniae strain J and M. flocculare, distinct transcripts level were observed (Table 3 and S2 Fig). A detailed comparison of palindromic elements in adhesin-coding genes that were experimentally analysed is described in S4 Table.
In summary, the presence of nonconserved repetitive DNA elements (SSR and SSRM) among the 5’ upstream regions of M. flocculare orthologous had gene expression variations under the tested conditions. In M. hyopneumoniae strain J only the classified absent repeats (SSR) demonstrate some potential bias in transcription. In relation to PAL and PALG the loss of some elements (lower conservation) could be related to the observed differences in gene expression. Orthologous of MHP7448_497, MHP7448_272 and MHP7448_272 in M. hyopneumoniae strain J and MHP7448_108, MHP7448_107, MHP7448_271 and MHP7448_272 could support this data (Table 3). All these findings suggest a putative regulatory influence on gene expression when tandem or palindromic repeats were present in 5’ upstream region of the gene.
Repeat presence in differentially regulated CDS
In order to understand the possible role of repetitive elements in transcriptional regulation, a search for the presence of palindromic elements was performed in genes with differential expression profiles, previously reported [46–50]. Since the molecular pathways that could modulate the regulation of these genes are unknown, the presence of conserved palindromic repeats in intergenic region could suggest a probably source of cis-regulatory elements. A total of 243 differentially expressed genes in M. hyopneumoniae strain 232 were compared against the M. hyopneumoniae strain 7448 genome (S5 and S6 Tables). Orthologous could be assigned (using BLAST approach) to 206 out of 243 (85%) of the differentially expressed genes, in the M. hyopneumoniae strain 7448 genome. An average of 4 palindromic elements was present in intergenic regions of these 206 orthologous genes present in M. hyopneumoniae strain 7448 (S6 Table). Comparison with orthologous genes from M. hyopneumoniae strain 232 showed that 103 (61%) genes had exactly the same element found in M. hyopneumoniae strain 7448 and only 17 (10%) did not have any corresponding elements (S5 and S6 Tables). In conclusion, almost 80% of the palindromic elements were conserved among the differentially expressed genes analysed. The ΔG values of PAL and PALG repeats found were evaluated and in general, demonstrated satisfactory potential to form secondary structures (S5 and S6 Tables).
Discussion
Prokaryote genomes are extremely diverse in terms of nucleotide composition and the presence of distinct patterns of repeat sequences that could affect the physical properties of DNA molecules [36]. In this work, a global analysis of tandem and palindromic repetitive elements found in noncoding sequences of M. hyopneumoniae strain 7448 was reported. In silico analysis revealed that the majority of the repeat sequences found were classified as palindromic elements (1,171 elements) compared with tandem repeats (144 elements). Similar results were also reported by Huang et al. [36] who investigated tandem and palindromic repeats in protein-coding sequences and intergenic regions through a global analysis of more than 1,000 genomes, including Mycoplasma agalactiae, M. bovis, M. fermentans and M. mycoides. Further analysis of the repetitive elements identified in our work demonstrated that common motifs between tandem or palindromic repetitive elements could not be established. The pattern frequently observed was the presence of AT-rich sequences in all elements investigated. In SSRM, for example, mononucleotides containing only Adenine (A) or Thymine (T) repeated 8 to 25 times were observed, in accordance with the AT-rich genome of M. hyopneumoniae strain 7448 [1].
Combinations of palindromic elements, tandem repeats and promoter sequences could be detected in 92% of all M. hyopneumoniae strain 7448 transcriptional units demonstrating that different mechanisms of regulation can be considered (see S2 Table). Results showed that 83% of SSR and SSRM repeats were located proximal to putative promoter sequences and 76% of palindromic elements revealed similar profile. The totality of tandem repeat class was associated with promoters (S2 Table). Previous work demonstrated that copy number of tandem repeats next to putative promoter sequences can modulate RNA polymerase action, by spacing the promoter region in distinct way, affecting gene expression [54]. Moreover, palindromes can form cruciform structures and mediate promoter sequence availability or create physical barriers that could be broken in a regulatory way [53]. DNA repeats have already been described as being involved in phase variation resulting in diversity of pathogenic phenotypes and other bacterial biological processes [19]. Therefore, aiming to establish the role of the repetitive elements found in M. hyopneumoniae strain 7448, a comparative investigation of tandem elements was performed in relation to mycoplasma genomes. Although subtle differences were observed, in general, the level of repeat conservation was higher between the two pathogenic M. hyopneumoniae strains (7448 and 7422), compared with nonpathogenic mycoplasmas (M. hyopneumoniae strain 7448 versus M. hyopneumoniae strain J and M. hyopneumoniae strain 7448 versus M. flocculare) as shown in Fig 5. Adhesin proteins are known to be essential to mycoplasma host infection and for the establishment of the disease [44]. To evaluate the correlation of the palindromic repeats associated with these genes, another comparison analysis was done. A comparative analysis of the PAL and PALG elements among the orthologue adhesin genes in the three M. hyopneumoniae strains and M. flocculare was performed with similar results as previous demonstrated for the tandem repeats. Adhesin-coding genes show greater differences in the number of palindromes in non-pathogenic strains (M. hyopneumoniae strain J and M. flocculare) than the two M. hyopneumoniae pathogenic strains (7448 and 7422). An example was seen in MHP7448_0107, MHP7448_0271 and MHP7448_497 genes, which showed 100% conservation in M. hyopneumoniae strain 7422 and a marked reduction in palindromic elements in M. hyopneumoniae strain J and M. flocculare. In this work tandem repeats (SSR and SSRM) were also reported in 3 of the 16 putative adhesin coding genes analysed (MHP7448_0108, MHP7448_0272 and MHP7448_0373). Previous authors have reported that tandem repeats, more precisely SSR, were able to influence adhesin gene expression [21, 25] but differences in palindromic repeats had not yet been reported.
To know the role of the repetitive elements found among the 5’ upstream regions in different CDSs, an experimental analysis was performed with orthologous genes from two M. hyopneumoniae strains (7448 and J) and M. flocculare. Detailed analysis of comparative studies of the presence of tandem repeats (SSR, SSRM and DR) in different genes and palindromic elements found in putative adhesin-coding genes were the basis of the experimental investigation. In general, the results of qPCR assay revealed that conserved elements among distinct mycoplasmas seem to have no influence on gene expression, as observed in the MHP7448_0623 gene (Table 3). Whereas when nonconserved or absent elements were investigated, a relation between differences in repetitive elements found in 5’ upstream regions and gene expression could be suggested (Table 3). Interestingly, a SSR found in the 5’ upstream region of the MHP7448_0087 (GTP-binding protein) gene was conserved in all pathogenic strains of mycoplasma (7448 and 7422) and absent in nonpathogenic mycoplasmas (M. hyopneumoniae strain J and M. flocculare). The same pattern was present in SSRM found in the 5’ upstream region of the glyA (glycine hydroxymethyltransferase) and sipS (signal peptidase I) genes, which are conserved in pathogenic mycoplasmas strains and nonconserved in nonpathogenic mycoplasmas (S3 Table). Differences in gene expression were observed among tested mycoplasma RNAs of M. hyopneumoniae strain J and M. flocculare in relation to M. hyopneumoniae strain 7448 in MHP7448_0087, glyA and sipS genes (Table 3), suggesting that some DNA repeats analysed may be involved in mycoplasma pathogenicity. GTP-binding protein, glycine hydroxymethyltransferase enzyme and signal peptidase I coding genes are important to cell viability and in some cases have already been related to mycoplasma pathogenicity process [55–57]. Promoter sequences associated with tandem repeats were present in all regulatory sequences of experimentally tested genes (S2 Table), reinforcing the hypothesis that repetitive elements may be interfering in the promoter region and consequently the transcription of related genes [20, 54].
The adhesin-coding genes are important virulence factors and seem to be regulated depending on the presence or absence of palindromic repeats. Therefore, it is possible to suggest a relation between the differences in gene transcriptional level and the divergence of DNA repeats found in the 5’ upstream regions among mycoplasma orthologous genes, as observed in genes coding for adhesin proteins P102 (MHP7448_0271) and P97 (MHP7448_0272) and genes coding for proteins in the MgPa operon (MHP7448_0005 and MHP7448_0006). Similar results were previously demonstrated for genes coding adhesin in M. genitalium (MgPa adhesin proteins) [21] and in M. gallisepticum (M9/pMGA adhesin proteins) [25], whereas repetitive DNA could influence transcriptional regulation.
The repeats found in the M. genitalium genome represent hypervariable sites in the MgPa operon, which encodes two adhesin proteins that represent surface proteins required for the development of the terminal organelle structure and attachment of the organism to host epithelial cells. Such adhesin variation may allow this organism to evade the host immune response and to adapt to diverse host microenvironments, thus establishing persistent infection [21]. In M. gallisepticum, the GAA trinucleotide repeat region regulates M9/pMGA gene expression (which encodes adhesin(s) associated with haemagglutination). Depending on the copy number of GAA in the intergenic regions, gene expression can be inhibited [25]. In M. bovis, repetitive elements were involved in a family of phase- and size-variable membrane surface lipoprotein antigens [27].
Besides transcriptional regulation, DNA repeats could also be involved in translation regulation. Identification of transcription start site (TSS) was performed for 23 genes of M. hyopneumoniae strain 7448 [12]. The analysis of these 23 genes in relation to the presence of DNA repeats revealed that 15 of them have at least one repetitive element located in the 5’ UTR region (S7 Table). This data demonstrates another potential mechanism of regulation by DNA repeats as they could influence in the process of mRNA translation into its protein product.
Previous studies have demonstrated differential expression of genes from M. hyopneumoniae 232 growing under several specific culture conditions [46–50], so a detailed analysis was performed to better understand the differences in transcriptional level of genes that diverge in the presence of repetitive elements in the 5’ upstream region. All M. hyopneumoniae strain 7448 orthologues from differentially expressed genes of the M. hyopneumoniae strain 232 were selected and analysed for repeat conservation. This study revealed that almost all differentially expressed genes had palindromic elements in their 5’ upstream regions. Orthologous gene comparisons between the two M. hyopneumoniae strains (7448 and 232) showed that the PAL and PALG repeats were approximately 80% conserved in M. hyopneumoniae strain 232, reinforcing the putative regulatory role of these repetitive elements since temperature, pH and others factors can influence the formation of secondary structures, as well as could perturb the stability of the nucleotide bindings [58].
In this work, the presence of SSR, SSRM, DR, PAL and PALG DNA repeats found in the 5’ upstream regions of M. hyopneumoniae strain 7448 CDSs was described. Relevance in transcriptional regulation of the DNA repeats found could be established through comparison analysis, demonstrating that repeats could be perpetuated among related mycoplasmas, and some of them could be involved in pathogenicity. Experimental assays revealed differential expression of genes differing in repetitive elements in the 5’ upstream region, reinforcing the putative regulatory role of palindromic and tandem repeats. Previously described differentially expressed genes show palindromic elements in the upstream region of the start codon and are conserved between two strains of M. hyopneumoniae. All of these findings suggest the importance of repetitive DNA elements in M. hyopneumoniae, and contribute to expand the source of regulatory sequences that can modulate gene expression in the important pig pathogen M. hyopneumoniae.
Supporting Information
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(XLSX)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by Coordenação de Apoio de Pessoal de Nível Superior – CAPES. CAPES-Biologia Computacional (Process number: 23038.010043/2013-02) and MCTI/CNPq/ Universal (Process number: 445228/2014-8). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vasconcelos AT, Ferreira HB, Bizarro CV, Bonatto SL, Carvalho MO, Pinto PM, et al. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J Bacteriol. 2005;5568–77. 10.1128/JB.187.16.5568-5577.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thacker EL. Diagnosis of Mycoplasma hyopneumoniae. Anim Health Res Rev. 2004;5(2):317–20. [DOI] [PubMed] [Google Scholar]
- 3.Kuhnert P, Overesch G. Molecular epidemiology of Mycoplasma hyopneumoniae from outbreaks of enzootic pneumonia in domestic pig and the role of wild boar. Vet Microbiol. 2014;174(1–2):261–6. 10.1016/j.vetmic.2014.08.022 [DOI] [PubMed] [Google Scholar]
- 4.Siqueira FM, Thompson CE, Virginio VG, Gonchoroski T, Reolon L, Almeida LG, et al. New insights on the biology of swine respiratory tract mycoplasmas from a comparative genome analysis. BMC Genomics. 2013;14:175 10.1186/1471-2164-14-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minion FC, Lefkowitz EJ, Madsen ML, Cleary BJ, Swartzell SM, Mahairas GG. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J Bacteriol. 2004;7123–33. 10.1128/JB.186.21.7123-7133.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Feng Z, Fang L, Zhou Z, Li Q, Li S, et al. Complete genome sequence of Mycoplasma hyopneumoniae strain 168. J Bacteriol. 2011;193(4):1016–7. 10.1128/JB.01305-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Xiao S, Li M, Guo S, Li S, Luo R, et al. Comparative genomic analyses of Mycoplasma hyopneumoniae pathogenic 168 strain and its high-passaged attenuated strain. BMC Genomics. 2013;14:80 10.1186/1471-2164-14-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Fang L, Li M, Li S, Guo S, Luo R, et al. Comparative genomics of Mycoplasma: analysis of conserved essential genes and diversity of the pan-genome. PLoS One. 2012;7(4):e35698 10.1371/journal.pone.0035698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siqueira FM, Gerber AL, Guedes RL, Almeida LG, Schrank IS, Vasconcelos AT, et al. Unravelling the transcriptome profile of the Swine respiratory tract mycoplasmas. PLoS One. 2014;9(10):e110327 10.1371/journal.pone.0110327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siqueira FM, Schrank A, Schrank IS. Mycoplasma hyopneumoniae transcription unit organization: genome survey and prediction. DNA Res. 2011;18(6):413–22. 10.1093/dnares/dsr028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siqueira FM, de Souto Weber S, Cattani AM, Schrank IS. Genome organization in Mycoplasma hyopneumoniae: identification of promoter-like sequences. Mol Biol Rep. 2014;41(8):5395–402. 10.1007/s11033-014-3411-3 [DOI] [PubMed] [Google Scholar]
- 12.Weber Sde S, Sant'Anna FH, Schrank IS. Unveiling Mycoplasma hyopneumoniae promoters: sequence definition and genomic distribution. DNA Res. 2012;19(2):103–15. 10.1093/dnares/dsr045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritsch TE, Siqueira FM, Schrank IS. Intrinsic terminators in Mycoplasma hyopneumoniae transcription. BMC Genomics. 2015;16:273 10.1186/s12864-015-1468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62(2):275–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treangen TJ, Abraham AL, Touchon M, Rocha EP. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiol Rev. 2009;33(3):539–71. [DOI] [PubMed] [Google Scholar]
- 16.Achaz G, Rocha EP, Netter P, Coissac E. Origin and fate of repeats in bacteria. Nucleic Acids Res. 2002;30(13):2987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocha EP, Danchin A, Viari A. Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol Biol Evol. 1999;16(9):1219–30. [DOI] [PubMed] [Google Scholar]
- 18.Citti C, Nouvel LX, Baranowski E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 2010;5(7):1073–85. 10.2217/fmb.10.71 [DOI] [PubMed] [Google Scholar]
- 19.Bayliss CD, Palmer ME. Evolution of simple sequence repeat-mediated phase variation in bacterial genomes. Ann N Y Acad Sci. 2012;1267:39–44. 10.1111/j.1749-6632.2012.06584.x [DOI] [PubMed] [Google Scholar]
- 20.Peak IR, Jennings MP, Hood DW, Bisercic M, Moxon ER. Tetrameric repeat units associated with virulence factor phase variation in Haemophilus also occur in Neisseria spp. and Moraxella catarrhalis. FEMS Microbiol Lett. 1996;137(1):109–14. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Jensen JS, Mancuso M, Hamasuna R, Jia Q, McGowin CL, et al. Genetic variation in the complete MgPa operon and its repetitive chromosomal elements in clinical strains of Mycoplasma genitalium. PLoS One. 2010;5(12):e15660 10.1371/journal.pone.0015660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Jensen JS, Mancuso M, Hamasuna R, Jia Q, McGowin CL, et al. Variability of trinucleotide tandem repeats in the MgPa operon and its repetitive chromosomal elements in Mycoplasma genitalium. J Med Microbiol. 2012;61(2):191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mrazek J. Analysis of distribution indicates diverse functions of simple sequence repeats in Mycoplasma genomes. Mol Biol Evol. 2006;23(7):1370–85. 10.1093/molbev/msk023 [DOI] [PubMed] [Google Scholar]
- 24.Rocha EP, Blanchard A. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 2002;30(9):2031–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Dybvig K, Panangala VS, van Santen VL, French CT. GAA trinucleotide repeat region regulates M9/pMGA gene expression in Mycoplasma gallisepticum. Infect Immun. 2000;68(2):871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Panangala VS, Dybvig K. Trinucleotide GAA repeats dictate pMGA gene expression in Mycoplasma gallisepticum by affecting spacing between flanking regions. J Bacteriol. 2002;184(5):1335–9. 10.1128/JB.184.5.1335-1339.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178(18):5395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yogev D, Rosengarten R, Watson-McKown R, Wise KS. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5' regulatory sequences. Embo j. 1991;10(13):4069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theiss P, Wise KS. Localized frameshift mutation generates selective, high-frequency phase variation of a surface lipoprotein encoded by a mycoplasma ABC transporter operon. J Bacteriol. 1997;179(12):4013–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noormohammadi AH, Markham PF, Whithear KG, Walker ID, Gurevich VA, Ley DH, et al. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect Immun. 1997;65(7):2542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musatovova O, Kannan TR, Baseman JB. Mycoplasma pneumoniae large DNA repetitive elements RepMP1 show type specific organization among strains. PLoS One. 2012;7(10):e47625 10.1371/journal.pone.0047625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Wise KS. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64(7):2737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons WL, Denison AM, Dybvig K. Resistance of Mycoplasma pulmonis to complement lysis is dependent on the number of Vsa tandem repeats: shield hypothesis. Infect Immun. 2004;72(12):6846–51. 10.1128/IAI.72.12.6846-6851.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gosney F, Corro M, Iob L, McAuliffe L, Nicholas RA. Variable number tandem repeat (VNTR) typing of strains of Mycoplasma mycoides subspecies mycoides small colony isolated from the north-eastern regions of Italy between 1990 and 1993. Vet Microbiol. 2011;147(1–2):220–2. 10.1016/j.vetmic.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 35.Chang LJ, Chen WH, Minion FC, Shiuan D. Mycoplasmas regulate the expression of heat-shock protein genes through CIRCE-HrcA interactions. Biochem Biophys Res Commun. 2008;367(1):213–8. 10.1016/j.bbrc.2007.12.124 [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, Mrazek J. Assessing diversity of DNA structure-related sequence features in prokaryotic genomes. DNA Res. 2014;21(3):285–97. 10.1093/dnares/dst057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maia LC, Palmieri DA, de Souza VQ, Kopp MM, de Carvalho FI, Costa de Oliveira A. SSR Locator: Tool for Simple Sequence Repeat Discovery Integrated with Primer Design and PCR Simulation. Int J Plant Genomics. 2008;2008:412696 10.1155/2008/412696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11(8):1441–52. 10.1101/gr.184001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–7. [DOI] [PubMed] [Google Scholar]
- 41.Freund T, Engel N. Finding approximate palindromes in genomic sequences. 2013.
- 42.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 44.Ferreira HB, Castro LA. A preliminary survey of M. hyopneumoniae virulence factors based on comparative genomic analysis. Genet Mol Biol. 2007;30(1):245–55. [Google Scholar]
- 45.Mrazek J, Xie S. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics. 2006;22(24):3099–100. 10.1093/bioinformatics/btl551 [DOI] [PubMed] [Google Scholar]
- 46.Oneal MJ, Schafer ER, Madsen ML, Minion FC. Global transcriptional analysis of Mycoplasma hyopneumoniae following exposure to norepinephrine. Microbiology. 2008;154(Pt 9):2581–8. 10.1099/mic.0.2008/020230-0 [DOI] [PubMed] [Google Scholar]
- 47.Schafer ER, Oneal MJ, Madsen ML, Minion FC. Global transcriptional analysis of Mycoplasma hyopneumoniae following exposure to hydrogen peroxide. Microbiology. 2007;153(11):3785–90. [DOI] [PubMed] [Google Scholar]
- 48.Madsen ML, Puttamreddy S, Thacker EL, Carruthers MD, Minion FC. Transcriptome changes in Mycoplasma hyopneumoniae during infection. Infect Immun. 2008;76(2):658–63. 10.1128/IAI.01291-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madsen ML, Nettleton D, Thacker EL, Minion FC. Transcriptional profiling of Mycoplasma hyopneumoniae during iron depletion using microarrays. Microbiology. 2006;152(4):937–44. [DOI] [PubMed] [Google Scholar]
- 50.Madsen ML, Nettleton D, Thacker EL, Edwards R, Minion FC. Transcriptional profiling of Mycoplasma hyopneumoniae during heat shock using microarrays. Infect Immun. 2006;74(1):160–6. 10.1128/IAI.74.1.160-166.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 53.Brazda V, Laister RC, Jagelska EB, Arrowsmith C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol Biol. 2011;12:33 10.1186/1471-2199-12-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gemayel R, Cho J, Boeynaems S, Verstrepen KJ. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes (Basel). 2012;3(3):461–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borges CL, Parente JA, Pereira M, Soares CM. Identification of the GTPase superfamily in Mycoplasma synoviae and Mycoplasma hyopneumoniae. Genet Mol Biol. 2007;30(1):212–8. [Google Scholar]
- 56.Moitinho-Silva L, Heineck BL, Reolon LA, Paes JA, Klein CS, Rebelatto R, et al. Mycoplasma hyopneumoniae type I signal peptidase: expression and evaluation of its diagnostic potential. Vet Microbiol. 2012;154(3–4):282–91. 10.1016/j.vetmic.2011.07.009 [DOI] [PubMed] [Google Scholar]
- 57.Arraes FBM, Carvalho MJ, Maranhão AQ, et al. Differential metabolism of Mycoplasma species as revealed by their genomes. Genet Mol Biol. 2007;30(1):182–9. [Google Scholar]
- 58.Chen C, Wang W, Ge J, Zhao XS. Kinetics and thermodynamics of DNA hybridization on gold nanoparticles. Nucleic Acids Res. 2009;37(11):3756–65. 10.1093/nar/gkp230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.