Abstract
The Ets family transcription factor PU.1 and the interferon regulatory factor (IRF)4 and IRF8 regulate gene expression by binding to composite DNA sequences known as Ets/interferon consensus elements (EICE). Although all three factors are expressed from the onset of B cell development, single deficiency of these factors in B cell progenitors only mildly impacts on bone marrow B-lymphopoiesis. Here we tested whether PU.1 cooperates with IRF factors in regulating early B cell development. Lack of PU.1 and IRF4 resulted in a partial block in development the pre-B cell stage. The combined deletion of PU.1 and IRF8 reduced recirculating B cell numbers. Strikingly, all PU.1/IRF4 and approximately 50% of PU.1/IRF8 double deficient mice developed pre-B cell acute lymphoblastic leukemia (B-ALL) associated with reduced expression of the established B-lineage tumor suppressor genes, Ikaros and Spi-B. These genes are directly regulated by PU.1/IRF4/IRF8, and restoration of Ikaros or Spi-B expression inhibited leukemic cell growth. In summary, we demonstrate that PU.1, IRF4 and IRF8 cooperate to regulate early B cell development and to prevent pre-B-ALL formation.
Keywords: PU.1, IRF4, IRF8, pre-B cells, acute lymphoblastic leukemia, Spi-B, Ikaros
INTRODUCTION
The Ets transcription factor PU.1 is a key regulator of hematopoiesis. Biochemical 1–6 and genome-wide analyses of DNA binding sites 7, 8 have demonstrated that PU.1 can bind to DNA alone at canonical Ets sites, or together with hematopoietic restricted factors IRF4 and IRF8 at composite Ets-interferon consensus elements (EICE) sites. The functional importance of these biochemical interactions has not been examined.
Germline or conditional deficiency of PU.1 (encoded by Spi1) in hematopoietic stem cells (HSC) results in the loss of macrophages, granulocytes, dendritic cells, lymphocytes and functional HSC activity 9–11. In contrast, conditional ablation of PU.1 expression in B cell progenitors only mildly affects B cell development 11, 12. Recent evidence suggests that the loss of PU.1 function in B cell progenitors is compensated for by the related Ets family member Spi-B, as combined deletion of PU.1 and Spi-B in the B cell lineage impairs B cell development in the bone marrow (BM) 13. Similar to the redundant function of PU.1 and Spi-B, deletion of either interferon regulatory factor (IRF)4 or IRF8 only mildly affects early B cell development while double deficiency results in a block at the pre-B cell stage 14–17.
PU.1 and IRF8 are both established tumor suppressors in the myeloid lineage, with their loss promoting acute myeloid leukemia (AML) and chronic myeloid leukemia (CML) respectively 18–24. However their roles in suppressing B-lineage acute lymphoblastic leukemia (B-ALL) are less clear. Rare mutations in SPI1 (PU.1) and IRF8 have been found in human pre-B-ALL 25, 26 and diffuse large B cell lymphoma (DLBCL)27, while SPIB expression is reduced in pre-B-ALL carrying the t(12;21) ETV6-RUNX1 translocation 28. IRF4 has been implicated in several B cell malignancies, including chronic lymphocytic leukemia 29 and multiple myeloma 30, and it was recently reported that IRF4 is 2-fold overexpressed in pediatric pre-B-ALL compared to unfractionated healthy BM 31. Irf4−/− mice do not develop pre-B-ALL, but IRF4 deficiency cooperates with oncogenes such as BCR-Abl 32 and c-Myc 33 to promote leukemogenesis in mouse models. Mice lacking both IRF4 and IRF8 initially show more accelerated myelopoiesis, a characteristic of the early stages of CML, but ultimately succumb to pre-B-ALL 34. Most recently, Spi-B and PU.1 have also been shown to act synergistically in repressing pre-B-ALL in mice 13, 35, further emphasizing the complex interplay within the Ets and IRF families in leukemia suppression.
Despite biochemical evidence that PU.1 interacts with IRF4/IRF8 in developing B cells, it remains unclear whether they functionally collaborate during B lymphopoiesis. To address this question, we generated mice that lack either PU.1 and IRF4 or PU.1 and IRF8 in the B cell lineage. PU.1/IRF4 double-deficient mice show a partial developmental block beyond the pre-B cell stage, while PU.1/IRF8 deficient mice have relatively normal early B cell development but impaired late B cell maturation in the BM. We recently showed that the loss of PU.1 and IRF8 negatively regulate immunoglobulin class-switching and plasma cell differentiation 36. Strikingly, mice deficient for PU.1/IRF4 or PU.1/IRF8 developed pre-B-ALL at high frequency. Characterization of these pre-B-ALL showed that the Ets-IRF complexes directly regulate the expression of a number of known tumor suppressors including Spi-B, Ikaros and Blnk. We therefore conclude that Ets factor, PU.1, cooperate with IRF4 and IRF8 to suppress the formation of acute leukemia. Significantly, IRF4 and SPIB are commonly also down-regulated in human B-ALL suggesting that the tumor suppressor activity of the ETS/IRF complex is also present in human pre-B cells.
MATERIALS AND METHODS
Experimental animals
PU.1fl/flCd19cre/+ 11, 37, Irf8−/− 19, Irf4−/− 38 and Mb1cre/+ 39 mouse strains have been previously reported. All experimental mice were maintained on a C57BL/6 background and used between 6–12 weeks of age, unless monitored for tumor formation. The mice were bred and maintained at the Walter and Eliza Hall Institute under Animal Ethics Committee guidelines.
Cell culture
Cells were cultured in alpha-modified Eagle medium with 2 mM L-glutamine, 1 mM, Na-pyruvate, 0.1 mM non-essential amino acids, 10 mM HEPES, pH 7.4, 100 μg/ml of streptomycin, 100 U/ml of penicillin, 50 μM β-mercaptoethanol (all supplements were from Sigma-Aldrich), 10% heat inactivated FCS and 2% IL-7 supernatant.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (GraphPad Software, San Diego, California, USA). A Student’s t-test with two-tailed distributions for two samples with equal variance was used.
Please also see the supplementary information section for a more detailed Materials and Methods description and animal numbers used for each experiment.
RESULTS
Overlapping of binding sites between PU.1, IRF4 and IRF8
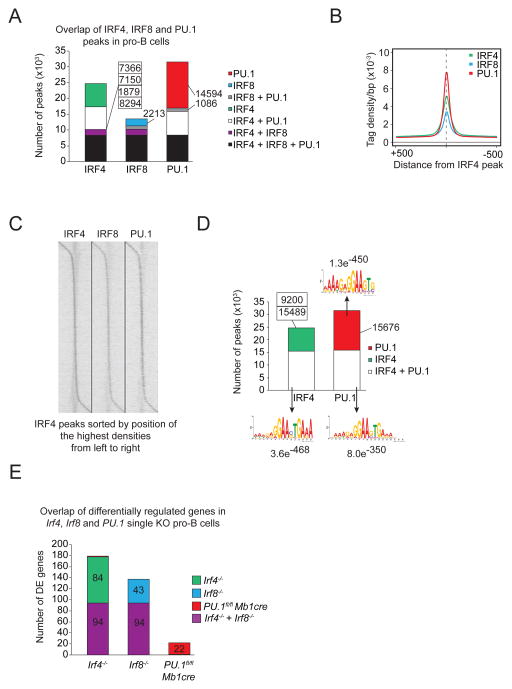
PU.1 can either bind to canonical Ets sites or in complex with IRF4 and IRF8 at composite Ets-interferon consensus elements (EICE) sites. To determine the global binding site pattern for PU.1, IRF4 and IRF8 we performed ChIP-sequencing experiments for all three transcription factors in pro-B cells. Approximately half of the PU.1 binding was detected at EICE sites together with either Irf4 or Irf8 and, as expected, the other half of binding sites being canonical ETS sites (Figure 1A–D). Additionally we compared the transcriptome of wild type pro-B cells to PU.1, IRF4 and IRF8 deficient pro-B cells. Interestingly, only few genes were found to be differentially expressed in the single deficient B cell progenitors, indicating redundant function of these three factors in early B cell development (Figure 1E). In summary, this data provides strong evidence for the relevance of the cooperative binding of PU.1/Irf to EICE motifs.
Figure 1.
Overlapping binding sites between PU.1, IRF4 and IRF8. A) Co-localization of IRF4, IRF8 and PU.1 peaks in pro-B cells were determined by multiple overlap analysis of the respective ChIP-seq data. The number of binding sites for each overlap is shown. B) The average density of IRF4− (green), IRF8− (blue) and PU.1-binding (red) in pro-B cells are shown for a 5 kb region centered on the summits of the IRF4 peaks. C) The densities of IRF4−, IRF8− and PU.1-binding in pro-B cells are displayed as a heat map, sorted by position of the highest IRF4 peak density, from upstream to downstream. D) Overlap of IRF4− and PU.1-binding in pro-B cells. The number of binding sites is indicated together with the top-ranked motif identified with the indicated E-value. For IRF4-only binding sites no significant motif could be found. E) Multiple overlap of differentially regulated genes of Irf4, Irf8 and PU.1 single KO pro-B cells. The number of differentially regulated genes is shown for the respective overlap.
PU.1 and IRF8 cooperate to regulate early B cell development
To test a functional collaborative role of PU.1 and IRF8 proteins during B cell development, we crossed mice with specific inactivation of PU.1 in the B cell lineage with Irf8 mutant mice (PU.1fl/flCd19creIrf8−/−). For simplicity, we will refer to PU.1fl/flCd19cre mice as PU.1 cKO and PU.1fl/flCd19creIrf8−/− as PU.1/IRF8 DKO. As reported previously 11, 13, B cell-specific inactivation of PU.1 resulted in a 2-fold increase in early B cell progenitor numbers and a reduction of recirculating mature B cells in the BM (Supplementary Figure 1). Similar results were obtained in PU.1fl/flMb1cre mice, in which PU.1 is deleted at a slightly earlier stage compared to PU.1fl/flCd19cre (39 and data not shown). IRF8 deficiency also led to a mild increase in pro/pre-B cell numbers and a 2-fold reduction in recirculating B cells (Supplementary Figure 1B, D–G). Strikingly, the combined loss of PU.1/IRF8 resulted in a further reduction in transitional and recirculating B cells compared to that observed in single mutant mice (Supplementary Figure 1B, F, G).
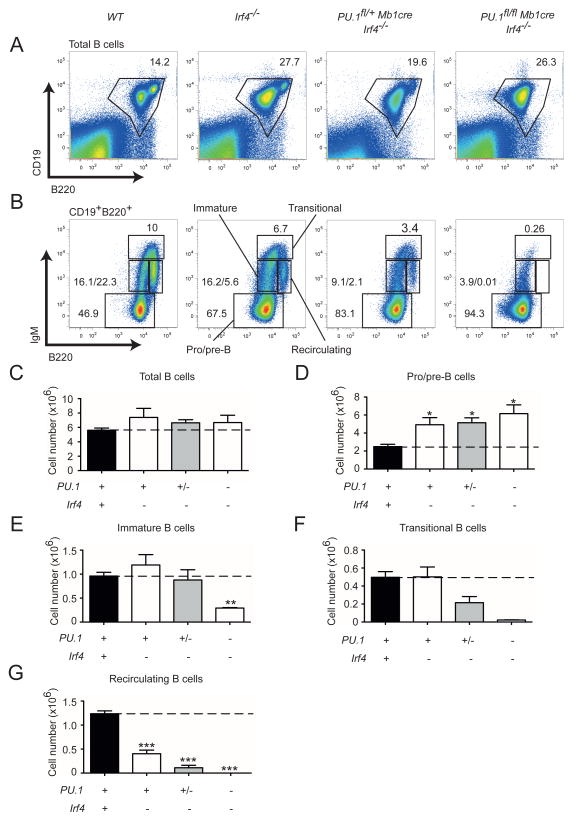
PU.1 and IRF4 regulate B cell development in a dose dependent manner
To test if PU.1 also cooperates with IRF4 during B cell development PU.1fl/flMb1cre mice were crossed to Irf4−/− mice to generate PU.1/IRF4 DKO mice, which lack both proteins only in the B cell compartment. Similar to IRF8 deficient mice, IRF4 loss resulted in a moderate increase in pro-/pre-B cells and a 2-fold decrease in recirculating B cells (Figure 2). Like PU.1/IRF8 deficiency, a severe reduction of recirculating B cells was observed in PU.1/IRF4 DKO mice (Figure 2B, G). Analysis of Irf4−/− mice lacking a single copy of PU.1 (PU.1fl/+Mb1creIrf4−/−) demonstrated a dose dependency of this Ets-IRF complex as the loss of transitional and recirculating B cells was more pronounced than in Irf4−/− mice (Figure 2F, G). Taken together, these results identify an important collaboration between PU.1 and IRF4 or IRF8 during BM B cell development and maturation.
Figure 2.
Analysis of B cell development in the absence of PU.1 and IRF4. (A–B) BM cells were isolated from mice of the indicated genotypes and analyzed for CD19, B220 and IgM expression. Representative flow cytometric plots show the gates used to calculate the number of (A) total CD19+B220+ B cells and (B) CD19+ BM compartments based on B220 and IgM expression used to define pro/pre-B cells immature, transitional and recirculating B cells. (C–G) Total cell number of each cell population were calculated from the gating shown in (A) and (B). A simplified genotype nomenclature is shown below the graphs with symbols representing the existence of two (+), one (+/−) or no (−) functional alleles for the indicated genes. The genotypes are presented in the same order as in (A). Data are mean ± SD from 3 to 13 mice per genotype. p values compare the indicated groups using a paired t test (two tailed). * p <0.05, ** p<0.005, *** p<0.0005.
PU.1, IRF4 and IRF8 regulate early B cell differentiation
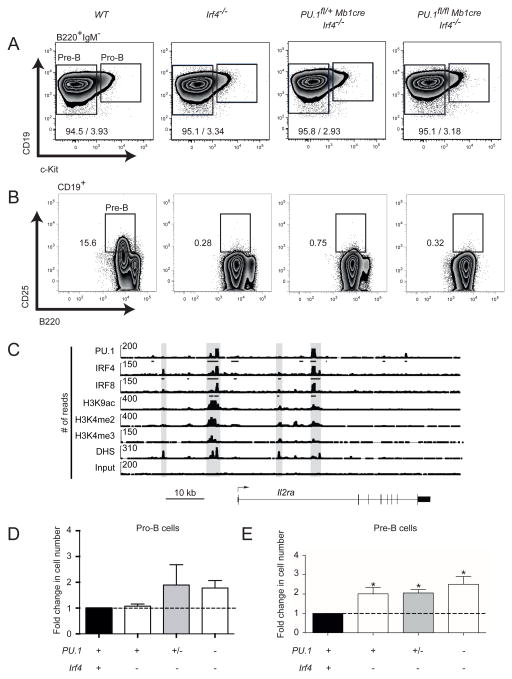
To investigate the pro/pre-B cell compartment in more detail, BM derived CD19+B220+ B cells of all mutant mice were analyzed for c-Kit and CD25 (Interleukin-2 receptor alpha (encoded by Il2ra)) expression. Pro-B cells are commonly identified as CD19+B220+IgM−cKit+ and pre-B cells as CD19+B220+CD25+ (Figure 3A–B, Supplementary Figure 2A–B). Pro-B cells were not affected by the loss of either IRF4 and slightly increased, although not statistically significantly, in the absence of IRF8 expression (Figure 3D and Supplementary Figure 2C). The number of pre-B cells remained similar in PU.1/IRF8 DKO (Supplementary Figure 2C). In our analysis of the pre-B cell compartment, we noted that IRF4 loss lead to the absence of CD25 expression (Figure 3B). ChIP-sequencing revealed that IRF4 directly binds the promoter and an enhancer region between exon 1 and 2 of the Il2ra gene in pro-B cells, suggesting that IRF4 directly regulates the expression of CD25 in pre-B cells (Figure 3C). Pre-B cells were therefore subsequently identified as B220+CD19+cKit−IgM− (Figure 3A). The presence of pre-B cells was independently confirmed by analyzing the expression of CD43 (Supplementary Figure 3). Pre-B cell numbers were significantly increased in the absence of PU.1 and IRF4 when compared to wt pre-B cells (Figure 3E).
Figure 3.
Analysis of the pro- and pre-B cell compartments in the absence of PU.1 and IRF4. BM cells were isolated from mice of the indicated genotypes were analyzed for the frequency of (A) CD19+B220+IgM−c-Kit+ pro-B and CD19+B220+IgM−c-Kit− pre-B cells. (B) Representative flow cytometric plots of CD25 expression on CD19+ cells. Box indicates the location of pre-B cells. (C) ChIP-seq mapping of IRF4, IRF8 and PU.1 binding plus the indicated histone modifications and DNase I hypersensitive sites (DHS) at the regulatory regions of Il2ra (encoding CD25) in pro-B cells. Gray boxes highlight the IRF4 binding peaks. Arrow shows the direction of Il2ra transcription. Bars below the ChIP-seq tracks indicate transcription factor-binding regions identified by MACS peak calling. (D–E) Fold change (normalized to the wild-type value set as 1) in the total number of each cell population from each genotype were quantified from the gating shown in (A–B). A simplified genotype nomenclature is shown below the graphs with symbols representing the existence of two (+), one (+/−) or no (−) functional alleles for the indicated genes. The full genotypes are presented in the same order as in (A). The data are mean ± SD from 3 to 13 mice per genotype. p values compare the indicated groups using a paired t test (two tailed). * p <0.05.
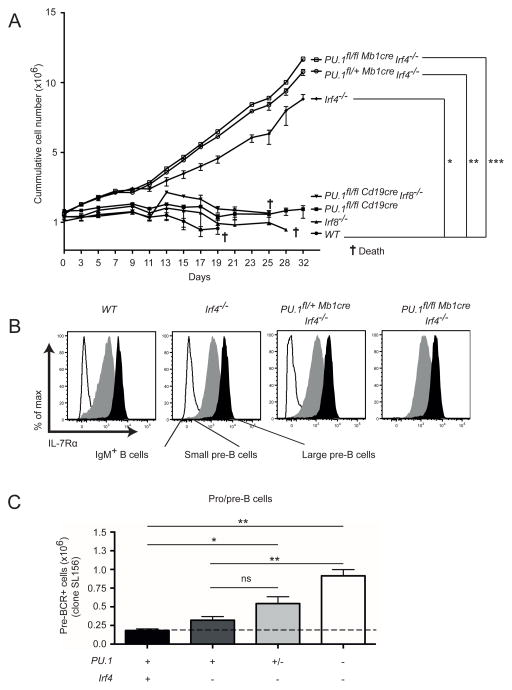
PU.1 and IRF4 mutant B cells are hyper-responsive to IL-7
Increased IL-7 dependent proliferation in vitro was previously reported for IRF4/IRF8 double-deficient pre-B cells 15. To test whether altered responsiveness to IL-7 is involved in the expansion of B cell progenitor cells observed in the absence of PU.1, IRF4 and IRF8, B220+ cells (predominantly pre-B cells) were isolated from the BM of mutant and wild-type mice, and cultivated in the presence of IL-7. Wild-type pre-B cells underwent a small transient proliferative response (Figure 4A). Interestingly, cells lacking IRF4, with or without PU.1, had a higher proliferative response to IL-7 and continued to proliferate strongly throughout the experiment (32 days). In contrast, cells lacking PU.1, IRF8 or both factors proliferated similarly to wild-type cells in this assay (Figure 4A). Although PU.1 has been implicated in regulating the Il7r gene 43, no differences in surface IL-7Rα expression was observed on small and large pre-B cells from mice lacking IRF4, with or without PU.1 (Figure 4B). This suggests that changes in the expression of IL-7Rα did not account for the hyper-responsiveness to IL-7.
Figure 4.
Enhanced IL-7 dependent proliferation and pre-BCR expression in the absence of PU.1, IRF4 and IRF8. (A) The BM cells of the indicated genotypes were isolated and enriched for B220+ cells. The cells were cultured in the presence of IL-7, and reseeded at the point when the cells of each indicated genotype were confluent. The cumulative frequencies of cells of each genotype were calculated based on the total number of cells on that day and the dilution factor used. (B) Expression levels of IL-7Rα on CD19+B220+IgM+ B cells, and small pre-B and large pre-B cells gated as in (Figure 3A). (C) BM cells of each genotype were isolated and analysed on the pro/pre-B cells (gated as in Figure 3A) for pre-BCR expression levels using an antibody that detects a conformational epitope of the pre-BCR but not with surrogate light chain, λ5, or VpreB in the absence of IgH. A simplified genotype nomenclature is shown below the graphs with symbols representing the existence of two (+), one (+/−) or no (−) functional alleles for the indicated genes. The full genotypes are presented in the same order as in Figure 2A. The data in (A) and (C) are mean ± SD between 3 to 8 mice per genotype. p values compare the indicated groups using a paired t test (two tailed). * p <0.05, ** p<0.005, *** p<0.0005. ns, not significant. The cross symbol indicates the death of the cells growing in culture.
The pre-B cell receptor (pre-BCR) is usually only transiently detectable on the surface of wild-type pre-B cells. However, double deficiency in IRF4/IRF8 resulted in increased surface pre-BCR expression 15. To examine how the combined loss of PU.1 and IRF4 impacts on this process, we examined pre-BCR expression on BM pre-B cells. Although no difference was observed in the number of pre-BCR+ cells between wild-type, PU.1, and IRF4 singularly deficient mice, the number of pre-BCR+ cells was increased significantly with the loss of one or both PU.1 alleles in the context of IRF4 deficiency (Figure 4C). Taken together, these data demonstrate that the complex of PU.1 and IRF4 is an important regulator of IL-7 responsiveness and pre-BCR signaling, the major pathways that are active in pre-B cells.
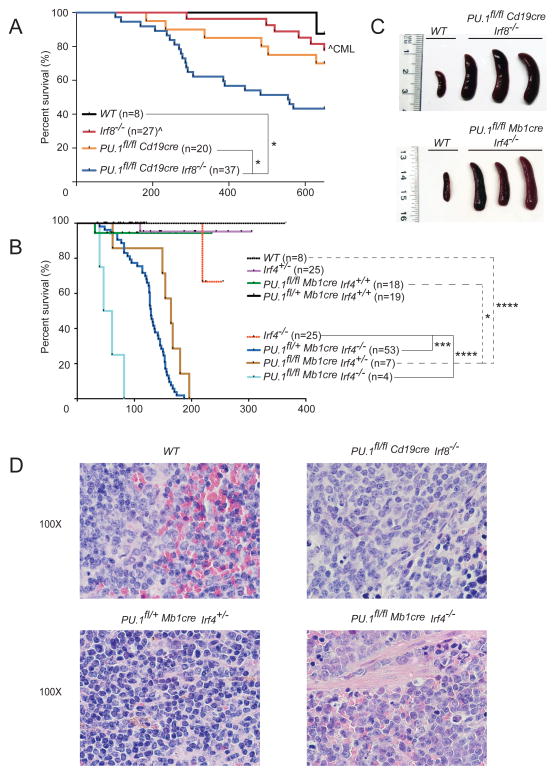
Combined loss of PU.1 and either IRF4 or IRF8 in B cells results in the development of pre-B cell acute lymphoblastic leukemia
To assess possible pathological consequences of the altered BM B cell differentiation observed in the various PU.1, IRF4 and IRF8 mutants, cohorts of single and double mutant mice were monitored for disease for up to 600 days (Figure 5A–B). 30% of PU.1 cKO and 56% of PU.1/IRF8 DKO mice required euthanasia during the monitoring period because of labored breathing and difficulty in locomotion. The illness was characterized by enlarged spleen, occasionally accompanied by enlarged lymph nodes (Figure 5C, data not shown). Leukemic cells could as well be detected in other organs such as bone marrow, peritoneum and peripheral blood at variable frequencies (data not shown). Histological analysis of the enlarged spleens revealed extensive infiltration of blast-like tumor cells disturbing the architectural integrity of the organ (Figure 5D, Supplementary Figure 4). The disease course was markedly accelerated in the absence of IRF4 with the median survival of PU.1/IRF4 DKO, as well as PU.1fl/+Irf4−/− and PU.1fl/flIrf4+/− mutant mice being 50, 130 and 180 days, respectively (Figure 5B). In contrast, the disease incidence over 600 days was much lower for wild-type or Irf8−/− mice (Figure 5A), with the 6 Irf8−/− mice succumbing to CML as previously reported 19. All tested PU.1/IRF8 and PU.1/IRF4 DKO tumors initiated leukemia after transfer of a limiting number of leukemic cells into non-irradiated syngeneic mice (Supplementary Table 1). In keeping with the literature 32, we did not observe any leukemia in mice lacking IRF4 alone, although some individuals had to be euthanized due to manifestations of the known immune deficiencies present in these mice (Figure 5B).
Figure 5.
Development of leukemia in mice lacking PU.1, IRF4 and/or IRF8. (A–B) Kaplan-Meier survival plot for aging mice of the indicated genotypes. n, indicates the number of mice recorded for each genotype. (C) Spleens from moribund mice of the indicated genotypes. (D) Histological section of the spleens from a moribund mouse of indicated genotype. Enlarged spleens showed infiltration of lymphocytes into the red pulp and destruction of the original organ architecture, when compared to the WT control. Magnification is indicated. Lower magnification images are shown in Supplementary Figure 4.
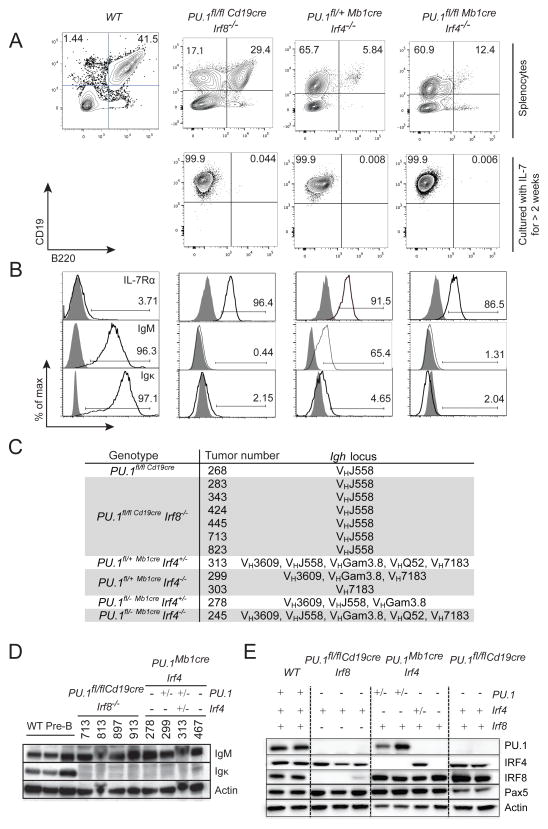
Flow cytometric analysis showed that the majority of splenic B cells in clinically ill mice consisted of CD19+B220− B cells, although in some cases, we also observed CD19+B220+ cells (Figure 6A). The splenic cells could be cultured with IL-7 for more than 2 weeks to enrich for CD19+ leukemic cells (Figure 6A, lower panel). The lack of B220 expression in PU.1-deficient cells was consistent with a previous report that PU.1 is required to activate Ptprc (Cd45), which codes for B220 in B cells 11, 12, 44. However, it is noteworthy, that the vast majority of PU.1-deficient pre-leukemic B cells expressed B220 (Figure 2, Supplementary Figure 1). The reason for this discrepancy is currently not known, but it is consistent with our previous report that CD45 expression was also lost on PU.1-deficient AML cells 20. Tumor cells from all genotypes expressed IL-7Rα and could be continuously grown in vitro in medium supplemented with IL-7 (Figure 6A and data not shown). Although wild-type splenic B cells expressed high levels of IgM, indicative of mature B cells, PU.1/IRF4 and PU.1/IRF8 DKO tumor cells expressed low amounts of IgM and even less Igκ on their surface suggestive of a pre-B cell phenotype (Figure 6B). PCR analysis for VDJ rearrangements of the Igh loci showed that all leukemic samples from PU.1/IRF8 DKO mice were monoclonal, and rearranged specifically the common VHJ558 variable region. In keeping with very rapid onset PU.1/IRF4 DKO leukemias, these exhibited predominantly oligoclonal Igh rearrangements (Figure 6C). Western blot analysis confirmed that the leukemic cells expressed IgM protein but not Igκ (Figure 6D). Taken together, these results suggested that PU.1/IRF-deficient tumors display a pre-B-ALL phenotype.
Figure 6.
Development of pre-B acute lymphoblastic leukemia in PU.1, IRF4 or IRF8 compound mutant mice. (A) Representative plots of CD19 and B220 expression from isolated splenocytes of moribund mice (upper panel), and cultured leukemic cells (lower panel) of the indicated genotypes. Data are compared to normal control wild-type splenocytes (WT). (B) Expression of IL-7Rα, IgM and Igκ on the gated CD19+B220− leukemic cells and CD19+B220+ WT B cells from (A). Gray histograms show unstained controls. (C) DNA was extracted from leukemias of indicated genotype and subjected to PCR for Igh VDJ rearrangements with sets of primers recognizing various VH gene families. The amplified samples were run on an agarose gel and the alleles assessed and shown. (D) Leukemic cells of the indicated genotypes were lysed and subjected to western blotting for IgM and Igκ. Actin serves as a control for equal protein loading. WT pre-B cells served as a positive control. Note the Igκ in WT pre-B cells is intracellular. Mouse tumor numbers are indicated. (E) Cultured leukemic cells of the indicated genotypes were lysed and subjected to western blotting for PU.1, IRF4, IRF8 and Pax5. Actin serves as a control for equal protein loading. Symbols in (D) and (E) represent the existence of two (+), one (+/−) or no (−) functional alleles for the indicated genes.
Loss of a single copy of PU.1 or IRF4 is sufficient to induce leukemia formation
Notably, mice lacking a single copy of PU.1 or Irf4 on a background of complete loss of their binding partner (IRF4 or PU.1, respectively) rapidly developed leukemia (Figure 5B). To test if leukemogenesis was due to haploinsufficiency or consequence of loss of heterozygosity (LOH) at the second PU.1 or Irf4 allele, western blotting was performed on leukemic cell lysates. PU.1 and IRF4 protein could still be detected in heterozygous cells, suggesting that leukemia development did not select for LOH of either gene (Figure 6E). Similarly, there was no evidence of cross-regulation of PU.1, Irf4 or Irf8 expression by any of these factors suggesting that the reduced gene dose of PU.1 or Irf4, and the resulting reduction in protein level is sufficient to allow leukemia progression.
The transcription factor PAX5 is mutated in approximately 30% of human pre-BALL 45, and the B cell specific enhancer of mouse Pax5 is known to contain in vivo occupied binding sites for PU.1, IRF4 and IRF8 46. Loss of Pax5 expression does not however appear to contribute to PU.1/IRF4 or PU.1/IRF8 DKO leukemogenesis, as Pax5 expression was unchanged in the singular or combined absence of PU.1, IRF4 and IRF8 (Figure 6E).
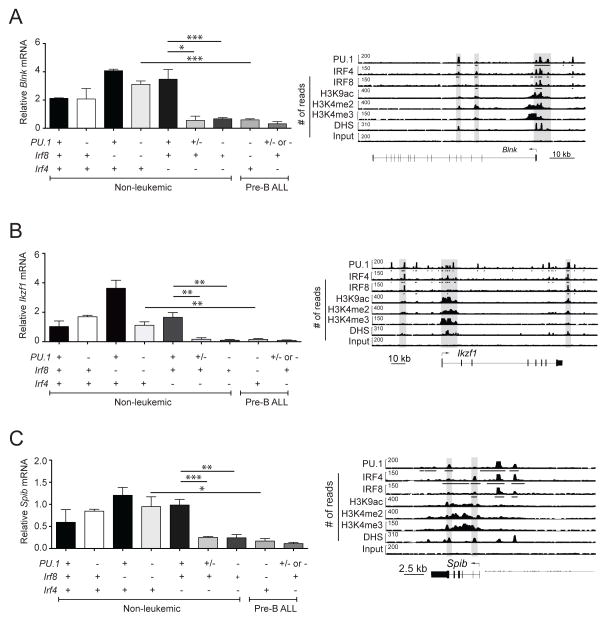
Blnk, Spi-B and Ikaros expression is reduced in PU.1/IRF-deficient leukemia
Blnk is an intracellular adapter protein that plays an important role in pre-BCR signaling and acts as a tumor suppressor of pre-B-ALL in mice and humans 47–49,26, 45, 50. Recently, Blnk was reported to be activated by PU.1 and Spi-B 13, 35. The transcription factor Ikaros, encoded by the Ikzf1 gene, plays a crucial role in early B-cell development 16, 51, 52 and pre-BCR signaling 53–55 and its gene is mutated at a high frequency in pediatric pre-B-ALL 45, 56, 57. Determination of PU.1, IRF4 and IRF8 binding sites in pro-B cells using ChIP-sequencing showed that all three transcription factors bound to the promoter and/or enhancer regions of Blnk, Spib and Ikzf1, indicating that PU.1/IRF complexes potentially regulate the expression of these genes (Figure 7A–C right). Indeed, the expression of all three transcripts was significantly reduced in PU.1/IRF-deficient pre-B-ALL samples. Importantly, Blnk, Spib and Ikzf1 were already expressed at reduced levels in non-leukemic PU.1/IRF4 DKO pre-B cells when compared to wild-type pre B cells, indicating that PU.1, together with IRF4, directly regulates the expression of these genes (Figure 7). The loss of one allele of PU.1 in Irf4−/− mutant mice was sufficient to reduce the expression of all three target genes (Figure 7). In contrast there was no difference in the expression levels of Blnk, Spib and Ikzf1 in non-leukemic PU.1/IRF8 DKO pre-B cells when compared to wild-type pre B cells (Figure 7). These results suggest that PU.1, IRF8 and IRF4 directly activate the transcription of Blnk, Spib and Ikzf1. Given that these genes are known tumor suppressor genes in mouse and human pre-B-ALL, it is likely that reduced expression of these genes contributes to the formation of pre-B-ALL in PU.1/IRF-deficient mice.
Figure 7.
Spib and Ikzf1 are transcriptionally activated by PU.1 and IRF proteins. (A–C) RNA samples from healthy pre-B cells (identified as B220+CD19+cKit−IgM−) and leukemias of the indicated genotypes were subjected to RT-qPCR to measure the relative expression of steady-state mRNA transcripts for Blnk (A), Ikzf1 (B) and Spib (C). Transcript frequencies were normalized to Hprt transcript levels. The data are mean ± SD between 2 to 10 samples per genotype. p values compare the indicated groups using a paired t test (two tailed). Lower panels, ChIP-seq analysis of PU.1, IRF4, and IRF8 binding and the indicated histone modifications and DNase I hypersensitive sites (DHS) at the regulatory regions of Blnk (A), Ikzf1 (B) and Spib (C) in short-term cultured pro-B cells. Gray boxes highlight the PU.1-IRF binding peaks. Arrows indicate the direction of gene transcription. Bars below the ChIP-seq tracks indicate transcription factor-binding regions identified by MACS peak calling. Symbols in (A–C) represent the existence of two (+), one (+/−) or no (−) functional alleles for the indicated genes. p values compare the indicated groups using a paired t test (two tailed). * p <0.05, ** p<0.005, *** p<0.0005.
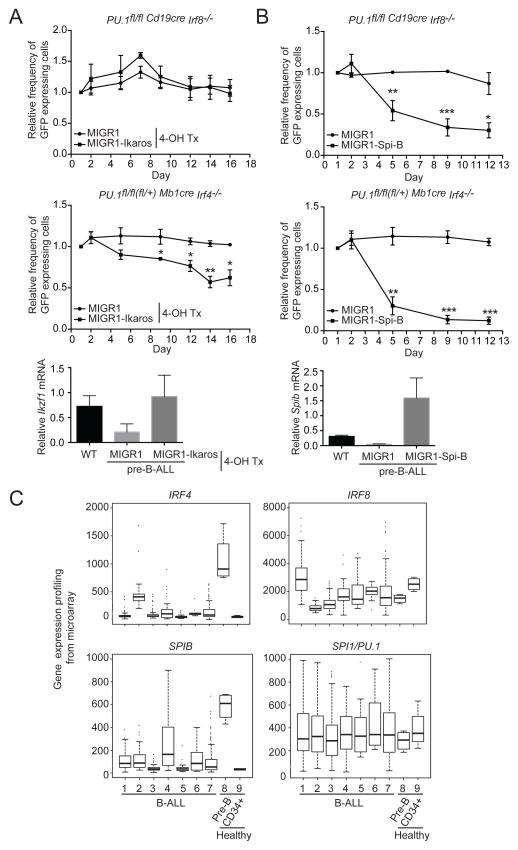
Ectopic expression of Ikaros and Spi-B reduces the growth of pre-B-ALL in culture
We next tested whether restoration of Ikaros, Spi-B or Blnk expression in the leukemic cells negatively influenced their in vitro growth and/or survival. PU.1/IRF4 and PU.1/IRF8 DKO leukemic pre-B cell lines were transduced with a retroviral construct coding for Ikaros, Spi-B or Blnk and GFP or only GFP. The percentage of GFP expressing cells was determined by flow cytometry over a period of 12–18 days of in vitro cultivation. Restoration of Ikaros expression had a negative effect on in vitro proliferation and/or survival in PU.1/IRF4-deficient cells but had no effect on the growth of PU.1/IRF8 DKO leukemic cells (Figure 8A). Restoration of Spi-B expression led to a more pronounced block in cell growth in both PU.1/IRF4 and PU.1/IRF8 DKO cells (Figure 8B), while surprisingly, Blnk re-expression did not inhibit the leukemic cell proliferation in this system (Supplementary Figure 5A–B) Collectively, these complementation studies show that reduced expression of Spi-B and to a lesser extent Ikaros are important factors in contributing to the pre-B-ALL phenotype.
Figure 8.
Ectopic expression of Spi-B and Ikaros oppose IL-7 dependent proliferation of pre-B-ALL. Leukemic cells from mice of the indicated genotypes were infected with Ikaros (A) or Spi-B (B)-expressing, or control MIGR1 (GFP) retroviral vectors. The estrogen analogue 4-hydroxytamoxifen (4-OH Tx) (0.5μM) was added at the start of infection to activate the expressed Ikaros-ERT2 fusion protein. Flow cytometry was used to determine the percentage of GFP+ cells over the indicated time course. Lower panel indicates the relative expression of mRNA transcript for Ikzf1 and Spib from infected cells from A and B. Data are the mean ± SD of 3 to 4 independent B-ALL samples per genotype and were normalized for relative frequency of GFP expression at day 1 (set as 1). p values compare the indicated data point to the MIGR1 control using a paired t test (two tailed* p <0.05, ** p<0.005, *** p<0.0005. (C) Gene expression data for IRF4, IRF8, SPIB and SPI1 (PU.1) from B-ALL patient samples and control BM. B-ALL cohorts: 1. Hyperdiploid (n=112); 2. TCF3/PBX1 t(1;19) (n=40); 3. ETV6/RUNX1 t(12;21) (n=100); 4. MLL (n=29), 5. BCR/ABL t(9;22) (n=22); 6. Hypodiploid (n=9); 7. Other B-ALL (n=174); 8. Normal CD19+CD10+ BM pre-B cells (n=4) and 9. Normal CD34+ hematopoietic progenitors (n=4). Note that IRF4 and SPIB are not expressed in early hematopoietic CD34+ progenitors in contrast to CD19+CD10+ pre-B cells of healthy individuals.
IRF4 and SPI-B expression are reduced in human B-ALL
Direct sequence mutations in IRF4, IRF8, SPIB or PU.1 (SPI1) in human B-ALL are rare 25, 26. To examine the expression of these factors in human B-ALL we have interrogated a public microarray data set derived from a large sample cohort (n=486 26), representing multiple clinical subtypes, and compared this to the expression of these factors in CD10+CD19+ pre-B cells and CD34+ hematopoietic progenitors derived from healthy donors (Figure 8C). While the expression of IRF8 and PU.1 (SPI1) was showed high variability across all samples, the expression levels of both IRF4 and SPIB were reduced in all B-ALL subtypes compared to normal pre-B cells (Figure 8C). This data is consistent with the tumor suppressor function of IRF4 and the reduced Spib expression we describe here in mouse B-ALL (Figure 7) and suggests a conserved role of the Ets-IRF binding activity in preventing B-ALL in both species.
DISCUSSION
Although the Ets-IRF DNA binding complex that recognizes an EICE consensus sequence was first described in B cells 1, 4, its importance relative to isolated Ets and IRF binding sites has not been clearly deciphered. Here we demonstrate that around half of the PU.1 binding is at EICE site indicating an important role of PU.1/IRF complexes in early B cell development. Despite a requirement for PU.1 in generating B cells from early hematopoietic progenitors 9, 43, 58, the function of this factor after commitment to the B cell lineage remains elusive. PU.1 is expressed at a uniform low level in B cells 37 and conditional mutagenesis demonstrated that B cell development is largely normal in its absence 11, 12. PU.1 may play a role in the divergence of B1 B cells from the conventional (B2) B cell lineage, as PU.1 deficient or hypomorphic B2 cells adopted a B1 like phenotype over time 12, 59.
One possible explanation for the minimal impact of PU.1 loss in B cells is the co-expression in B cells of the most related Ets protein in the mammalian genome, Spi-B. This hypothesis is strongly supported by the recent finding that PU.1/Spi-B DKO mice show a differentiation block at the pre-B cell stage and develop pre-B-ALL at high frequency 13. Strikingly, we show in this study that Spi-B expression was also markedly reduced in the absence of PU.1 and IRF4 or IRF8, and reintroduction of Spi-B inhibited the proliferation of leukemic cell lines.
IRF proteins have also been implicated in B cell development. Both IRF4 and IRF8 are expressed from the pro-B cell stage of differentiation, where they are under the control of Pax5 60. There is clear evidence for a synergy between IRF4 and IRF8 in B cells as DKO B cells show a developmental arrest at the pre-B cell stage due to a failure to rearrange Ig light chain genes 14–17. We found that PU.1/IRF4 DKO B cells showed a partial developmental block at the transition from pre-B cells to immature B cells. In contrast, the combined absence of PU.1 and IRF8 allowed a normal transition to the IgM+ immature B cell stage. PU.1/IRF8 DKO mice did, however, show a strong decrease of recirculating mature B cells in the BM and a 2-fold reduction in follicular B cells (data not shown). Care must be taken in interpreting these findings as the PU.1/IRF4 DKO model utilized Mb1-Cre to delete the floxed PU.1 allele in B cells, while the PU.1/IRF8 DKO strain used Cd19-Cre. As Mb1-Cre fully deletes at the pro-B cell stage 39, 61, while we and others have shown that Cd19-Cre deletion is partial at the pre-B cell stage (~80% deletion of PU.1 11, 13) and only deletes fully in B220+IgMint immature B cells. Nevertheless, single deficiency of PU.1 using either Cd19-Cre 11 or Mb1-Cre (data not shown) allows normal BM B cell development.
Intriguingly, pre-B cells from all IRF4 mutant mice were highly proliferative in response to IL-7, regardless of the presence of PU.1. This recapitulated the results obtained from studies by Lu et al. (2003) using IRF4/IRF8 DKO mice 15. While this hyperproliferation may be an important component of the tumor suppressor activity of IRF4, it is not sufficient to explain the leukemic potential of the cells, as IRF4 deficiency alone does not result in pre-B-ALL. IRF4 loss instead requires cooperating oncogenic events, such as BCR-ABL or constitutive c-Myc expression 32, 33. Lu et al. (2003) also found that IRF4/IRF8 DKO pre-B cells expressed a higher amount of the pre-BCR 15. Interestingly, while the pre-BCR was expressed on the cell surface at a similar frequency in wild-type and Irf4−/− B cells, we observed a striking increase in the level of pre-BCR in developing pre-B cells in PU.1/IRF4 DKO and PU.1−/+Irf4−/− pre-B cells, which may contribute to the large increase leukemogenesis when both PU.1 and Irf4 are mutated (see below).
The most striking aspect of this study is that all PU.1/IRF DKO mice develop pre-B-ALL that appears similar to the leukemia reported from PU.1/Spi-B DKO mice 13. The different genetic models that we have utilized varied in that the incidence of leukemia was greatly accelerated in PU.1/IRF4 DKO mice, compared to the PU.1/IRF8 DKO or single PU.1 KO strains. In keeping with this, PU.1/IRF4 DKO tumors were oligoclonal, while PU.1/IRF8 DKO tumors were monoclonal. Interestingly, the early onset of morbidity rate correlates with the loss of single PU.1 or Irf4 alleles, suggesting a dose dependent effect. This effect appeared to be caused by genuine haploinsufficiency as we have not detected any evidence of LOH in PU.1 or Irf4 heterozygous mice.
Our molecular studies did not reveal any evidence for transcriptional cross-regulation of PU.1, IRF4 and IRF8. However, a candidate approach based on qPCR and ChIPseq analyses revealed that the tumor suppressor genes, Blnk, Spib and Ikzf1, were direct targets of all three factors. Blnk and Ikzf1 are well known to promote early B cell differentiation by controlling pre-BCR signaling 40, 49, 53–55 and to suppress pre-B-ALL in both mice and humans 26, 45, 49, 50, 56, 57, although re-expression of Blnk in the leukemic cells in vitro did not alter the cell growth rates. Little is known about the role of SPIB in human acute leukemia, recent studies using PU.1/Spi-B DKO mice showed a high incidence of pre-B-ALL 13. In keeping with this, overexpression of Spi-B, and to a lesser extent Ikaros, inhibited the growth of our leukemic cell lines in vitro, thus acting as classical tumor suppressors.
Given the complex genetic interactions described here in mice that have lost expression of several tumor suppressors, it might be predicted that PU.1, IRF4 and IRF8 are not likely candidates to singularly act as drivers of human leukemogenesis. In keeping with this prediction there have only been sporadic reports of mutations in PU.1 (SPI1) and IRF8 in B-ALL 25, 26. Our analysis of the gene expression data from a very large cohort of B-ALL samples revealed that low IRF4 and SPIB expression was frequent, while IRF8 and PU.1 expression showed high variability and no clear directional trend. SPIB has also recently been reported to be down-regulated in the ETV6-RUNX1 subset of B-ALL 28, which is a finding consistent with our data. These data raise the possibility that activity of the various Ets/IRF complexes may also be compromised in human BALL.
Supplementary Material
Acknowledgments
We thank M. Reth and T. Mak for mice, J. Leahy for animal husbandry, and the institute flow cytometry facility for excellent technical assistance. We thank Markus Jaritz for bioinformatic analysis. This work was supported by program and project grants (APP1054925 to S.L.N., 637345 to S.C.) and fellowships (APP1058238 to S.L.N) from the National Health and Medical Research Council (NHRMC) of Australia. S.H.M.P. was supported by the Leukaemia Foundation of Australia and S.C. by an NHMRC Career Development Fellowship. Research of the Busslinger group was supported by Boehringer Ingelheim and an ERC Advanced Grant (291740-LymphoControl). This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIIS.
Footnotes
Conflict of interest disclosure: The authors declare no competing financial interests.
Supplementary information is available at Leukemia’s website.
References
- 1.Pongubala JM, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanno Y, Levi B-Z, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- 3.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes & Development. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 4.Eisenbeis CF, Singh H, Storb U. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2–4 enhancer. Mol Cell Biol. 1993;13:6452–6461. doi: 10.1128/mcb.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. Embo J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes & Development. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 7.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 10.Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 11.Polli M, Dakic A, Light A, Wu L, Tarlinton D, Nutt S. The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood. 2005;106:2083–2090. doi: 10.1182/blood-2005-01-0283. [DOI] [PubMed] [Google Scholar]
- 12.Ye M, Ermakova O, Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med. 2005;202:1411–1422. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokalski KM, Li SKH, Welch I, Cadieux-Pitre H-AT, Gruca MR, Dekoter RP. Deletion of genes encoding PU.1 and Spi-B in B cells impairs differentiation and induces pre-B cell acute lymphoblastic leukemia. Blood. 2011:1–33. doi: 10.1182/blood-2011-02-335539. [DOI] [PubMed] [Google Scholar]
- 14.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes & Development. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood. 2008;111:1396–1403. doi: 10.1182/blood-2007-08-110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma S, Turetsky A, Trinh L, Lu R. IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 18.Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL, et al. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood. 2004;104:3437–3444. doi: 10.1182/blood-2004-06-2234. [DOI] [PubMed] [Google Scholar]
- 19.Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 20.Metcalf D, Dakic A, Mifsud S, Di Rago L, Wu L, Nutt S. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:1486–1491. doi: 10.1073/pnas.0510616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenbauer F, Koschmieder S, Steidl U, Tenen DG. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106:1519–1524. doi: 10.1182/blood-2005-02-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 23.Scheller M, Schonheit J, Zimmermann K, Leser U, Rosenbauer F, Leutz A. Cross talk between Wnt/beta-catenin and Irf8 in leukemia progression and drug resistance. J Exp Med. 2013;210:2239–2256. doi: 10.1084/jem.20130706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117:2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouamar H, Abbas S, Lin AP, Wang L, Jiang D, Holder KN, et al. A capture-sequencing strategy identifies IRF8, EBF1, and APRIL as novel IGH fusion partners in B-cell lymphoma. Blood. 2013;122:726–733. doi: 10.1182/blood-2013-04-495804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niebuhr B, Kriebitzsch N, Fischer M, Behrens K, Gunther T, Alawi M, et al. Runx1 is essential at two stages of early murine B-cell development. Blood. 2013;122:413–423. doi: 10.1182/blood-2013-01-480244. [DOI] [PubMed] [Google Scholar]
- 29.Shukla V, Ma S, Hardy RR, Joshi SS, Lu R. A role for IRF4 in the development of CLL. Blood. 2013;122:2848–2855. doi: 10.1182/blood-2013-03-492769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer AL, Emre NC, Romesser PB, Staudt LM. IRF4: Immunity. Malignancy! Therapy? Clin Cancer Res. 2009;15:2954–2961. doi: 10.1158/1078-0432.CCR-08-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamaki M, Lambrou GI, Athanasiadou A, Tzanoudaki M, Vlahopoulos S, Moschovi M. Implication of IRF4 aberrant gene expression in the acute leukemias of childhood. PLoS One. 2013;8:e72326. doi: 10.1371/journal.pone.0072326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acquaviva J, Chen X, Ren R. IRF-4 functions as a tumor suppressor in early B-cell development. Blood. 2008;112:3798–3806. doi: 10.1182/blood-2007-10-117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathak S, Ma S, Trinh L, Eudy J, Wagner KU, Joshi SS, et al. IRF4 is a suppressor of c-Myc induced B cell leukemia. PLoS One. 2011;6:e22628. doi: 10.1371/journal.pone.0022628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo SH, Schatz JH, Acquaviva J, Singh H, Ren R. Cooperation between deficiencies of IRF-4 and IRF-8 promotes both myeloid and lymphoid tumorigenesis. Blood. 2010;116:2759–2767. doi: 10.1182/blood-2009-07-234559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu LS, Sokalski KM, Hotke K, Christie DA, Zarnett O, Piskorz J, et al. Regulation of B cell linker protein transcription by PU.1 and Spi-B in murine B cell acute lymphoblastic leukemia. J Immunol. 2012;189:3347–3354. doi: 10.4049/jimmunol.1201267. [DOI] [PubMed] [Google Scholar]
- 36.Carotta S, Willis SN, Hasbold J, Inouye M, Pang SHM, Emslie D, et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014:2169–2181. doi: 10.1084/jem.20140425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittrücker HW, Matsuyama T, Grossman A, Kündig TM, Potter J, Shahinian A, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. [Google Scholar]
- 39.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreiros-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–1782. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- 41.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 42.Schlissel MS, Corcoran LM, Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J Exp Med. 1991;173:711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 44.Anderson KL, Nelson SL, Perkin HB, Smith KA, Klemsz MJ, Torbett BE. PU.1 is a lineage-specific regulator of tyrosine phosphatase CD45. J Biol Chem. 2001;276:7637–7642. doi: 10.1074/jbc.M009133200. [DOI] [PubMed] [Google Scholar]
- 45.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 46.Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Minegishi Y, Rohrer J, Coustan-Smith E, Lederman HM, Pappu R, Campana D, et al. An essential role for BLNK in human B cell development. Science (New York, NY) 1999;286:1954–1957. doi: 10.1126/science.286.5446.1954. [DOI] [PubMed] [Google Scholar]
- 48.Pappu R, Cheng AM, Li B, Gong Q, Chiu C, Griffin N, et al. Requirement for B cell linker protein (BLNK) in B cell development. Science (New York, NY) 1999;286:1949–1954. doi: 10.1126/science.286.5446.1949. [DOI] [PubMed] [Google Scholar]
- 49.Jumaa H, Wollscheid B, Mitterer M, Wienands J, Reth M, Nielsen PJ. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity. 1999;11:547–554. doi: 10.1016/s1074-7613(00)80130-2. [DOI] [PubMed] [Google Scholar]
- 50.Jumaa H, Bossaller L, Portugal K, Storch B, Lotz M, Flemming A, et al. Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature. 2003;423:452–456. doi: 10.1038/nature01608. [DOI] [PubMed] [Google Scholar]
- 51.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 52.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010:4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heizmann B, Kastner P, Chan S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med. 2013;210:2823–2832. doi: 10.1084/jem.20131735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, et al. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol. 2014;15:294–304. doi: 10.1038/ni.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mullighan C, Downing J. Ikaros and acute leukemia. Leuk Lymphoma. 2008;49:847–849. doi: 10.1080/10428190801947500. [DOI] [PubMed] [Google Scholar]
- 57.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeKoter R, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 59.Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 60.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 61.Greig KT, de Graaf CA, Murphy JM, Carpinelli MR, Pang SHM, Frampton J, et al. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010;115:2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christie DA, Xu LS, Turkistany SA, Solomon LA, Li SK, Yim E, et al. PU.1 Opposes IL-7-Dependent Proliferation of Developing B Cells with Involvement of the Direct Target Gene Bruton Tyrosine Kinase. J Immunol. 2015;194:595–605. doi: 10.4049/jimmunol.1401569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.