Abstract
Lotus japonicus is a well-characterized model legume widely used in the study of plant-microbe interactions. However, datasets from various Lotus studies are poorly integrated and lack interoperability. We recognize the need for a comprehensive repository that allows comprehensive and dynamic exploration of Lotus genomic and transcriptomic data. Equally important are user-friendly in-browser tools designed for data visualization and interpretation. Here, we present Lotus Base, which opens to the research community a large, established LORE1 insertion mutant population containing an excess of 120,000 lines, and serves the end-user tightly integrated data from Lotus, such as the reference genome, annotated proteins, and expression profiling data. We report the integration of expression data from the L. japonicus gene expression atlas project, and the development of tools to cluster and export such data, allowing users to construct, visualize, and annotate co-expression gene networks. Lotus Base takes advantage of modern advances in browser technology to deliver powerful data interpretation for biologists. Its modular construction and publicly available application programming interface enable developers to tap into the wealth of integrated Lotus data. Lotus Base is freely accessible at: https://lotus.au.dk.
Lotus japonicus is a popular, well-characterized model legume1, widely used to study plant-microbe interactions due to its ability to establish a range of different types of relationship with microorganisms along the symbiosis–pathogenesis spectrum—ranging from biological nitrogen fixation2 and arbuscular mycorrhizal symbiosis3, to bacterial4 and fungal5 pathogenesis. The establishment of the LORE1 mutant population6,7,8 and the annotated sequence of the Lotus japonicus genome9 necessitated a centralized and freely available online resource for researchers working with this model legume. From its original incarnation as a LORE1 resource site to allow handling and processing of LORE1 mutant seeds orders, Lotus Base has grown to incorporate additional resources and toolkits tailored for the general needs of the research community. Although various Lotus databases have been made available to the public through different providers—such as the v3.0 genome through the Kazusa DNA Research Institute9; and the L. japonicus Gene Expression Atlas10, there is hitherto no publicly accessible repository to integrate all these data in a coherent manner. Due to the lack of a central information portal for Lotus japonicus—in spite of its popularity and utility as a model plant organism11,12,13, and its role in the study of biological nitrogen fixation14—we believe that Lotus Base is poised to benefit a large research community that does not traditionally have convenient access to such data.
Lotus Base is designed to be a user-friendly browser-based application that is operating system (OS)-agnostic and publicly accessible. In order to improve the workflow of researchers, Lotus Base provides functionalities that enable users to (1) search and retrieve sequence information; (2) identify functions and co-expression of Lotus gene(s) of interest; (3) view and order LORE1 lines that contain insertions in candidate gene(s); (4) visualize and annotate co-expression networks in Lotus; and (5) view and investigate gene structures and annotations of the latest Lotus genome version. In order to present a unified workflow, Lotus Base is designed with deep linking in mind where various toolkits can exchange information with each other. The secure and ethical design behind Lotus Base ensures that user information and credentials are properly stored and cryptographically encrypted during transmission, and that users have free access to, and retain ownership of, the data they have generated.
As a wide spectrum of technological competencies exist across the board within the research community, compounded by differential access to various computing technologies among researchers, Lotus Base was built from ground up with a focus on making tools simple to use, yet sufficiently verbose, for a common end-user. In short, Lotus Base ensures secure yet convenient access to Lotus genomics and expression data made coherent by deep linking by leveraging the latest browser technologies, avoiding the need for tedious software updates for related dependencies and/or plugins.
Methods & Data
Technologies
Lotus Base adopts a clean, minimal design principle for the front-end design, allowing users who are accustomed to normal browser use to acquaint themselves with the resource easily. Lotus Base is designed to be used by modern standards-compliant browsers, and is powered by Apache running on a CentOS7 server behind a load balancer. All communications between the end-user and our front-facing load balancer are SSL encrypted, while the load balancer communicates with our web server by normal HTTP protocols. Our database adopts an atomic design and is powered by either MySQL or PostgreSQL, depending on the needs of individual applications. In addition, a Python and R stack known as Anaconda15, powers some Lotus Base functionalities.
On the client end, we are serving pages via PHP 5.6, using HTML5 and CSS3, with user interactions assisted and enhanced with asynchronous JavaScript and XML (AJAX) and jQuery. We have implemented HMAC-SHA256 encryption16 for generation and verification of RFC 7519-compliant JSON web tokens (JWT)17 for user and API key authentication. Server-based sessions are frequently cycled to avoid session hijacking. All user login credentials are individually salted and cryptographically hashed, and are never stored or transmitted in plain text format.
Lotus Base is built using Grunt18, while the developer blog and application programming interface (API) documentation are generated by Jekyll19 and Slate20 respectively. Source control is done via git. The resource is designed to be extensible and modular, with the code base made open source through a GitHub repository (https://github.com/lotusbase/lotus.au.dk). Other features of Lotus Base are powered by various open-source projects, which together bring about a friendly, dynamic, and coherent user experience.
Overview of data provisioned by Lotus Base
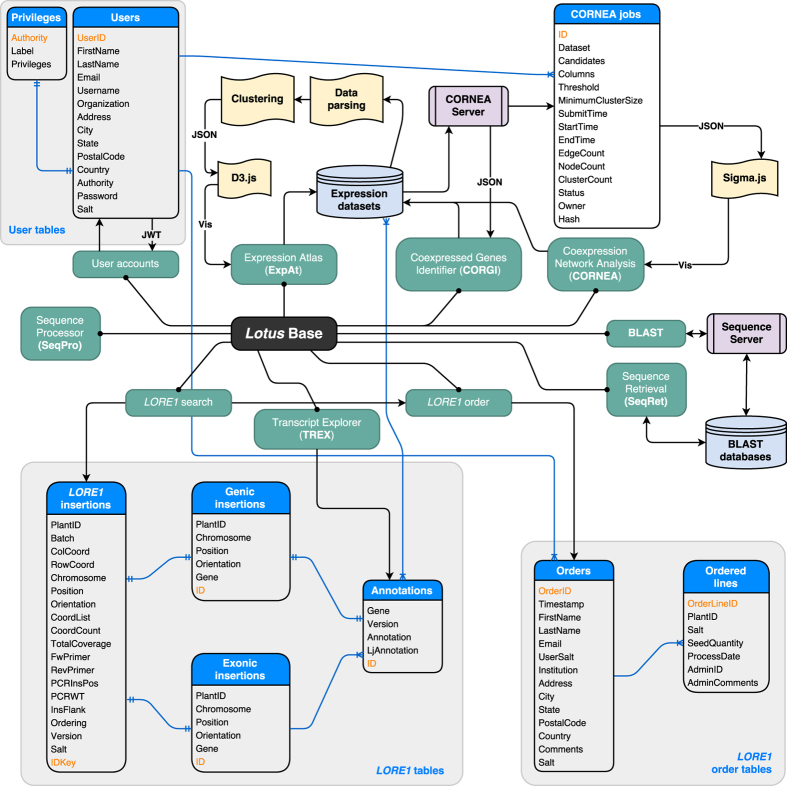
In the backend, Lotus Base constitutes various deeply integrated toolkits, which provide a coherent and simple workflow (Fig. 1). Lotus data are made publicly available (v2.5 of L. japonicus genome and proteins; and v3.0 of the following L. japonicus databases: genome, proteins, cDNA, and coding sequences). All available Lotus data that have been integrated to Lotus Base are outlined in Table 1.
Figure 1. The server-side design behind Lotus Base.
The resource consists of several tools deeply integrated with each other—LORE1 search, LORE1 order, Sequence Retrieval (SeqRet), BLAST, CORx toolkit (CORGI and CORNEA) and Expression Atlas (ExpAt). MySQL tables are indicated in blue entity boxes with column names listed. Highlighted column names, in orange, are used as primary indexes. Tables are grouped by the function they serve, in relation to individual tools. Due to space restraints, expression datasets are described in further detail in Fig. 4. An overview of all integrated datasets on Lotus Base is available in Table 1.
Table 1. List of datasets available through Lotus Base.
| Dataset | Toolkits |
|||||
|---|---|---|---|---|---|---|
| Lotus BLAST | LORE1 search | Genome browser | CORx toolkit | ExpAt | Gene page | |
| Genome | ||||||
| L. japonicus MG20 v2.5 genome | + | + | + | − | − | − |
| L. japonicus MG20 v3.0 genome | + | + | + | − | − | + |
| L. japonicus MG20 v3.0 cDNA | + | − | − | − | − | + |
| L. japonicus MG20 v3.0 CDS | + | – | − | − | − | + |
| Transcriptome | ||||||
| Lotus responses to draught stress33 | − | − | − | + | + | + |
| Lotus responses during arbuscular mycorrhizal symbiosis34 | − | − | − | + | + | + |
| Responses in wildtype and symbiotic mutants of Lotus during legume-rhizobium symbiosis35 | − | − | − | + | + | + |
| Salt acclimatization in Lotus japonicus Gifu36 | − | − | − | + | + | + |
| Salt acclimatization among Lotus ecotypes37 | − | − | − | + | + | + |
| Early Lotus root responses to germinating spore extract5 | − | − | − | −a | + | + |
| Predicted proteins | ||||||
| L. japonicus proteins v2.5 | − | + | + | − | − | − |
| L. japonicus proteins v3.0 | + | + | + | + | + | + |
| LORE1 resource | ||||||
| Danish collection: DK01–03, 05, 07–168 | − | + | + | − | − | + |
| Japanese collection: JPA, JPL, and JPP8 | − | + | + | − | − | + |
Datasets without any references represent original datasets generated in this work.
aExcluded from co-expression analysis due to low number of treatments/conditions available (3 only).
Genomic data
Lotus Base currently hosts the two latest versions of the L. japonicus genome assembly, versions 2.5 and 3.0 respectively. Both versions of the genome comprise six chromosomes and a single artificial chromosome 0 containing unassembled contigs interspersed with N spacers, with version 3.0 containing an additional mitochondrion genome. Version 2.5 of the genome includes sequence information from transformation-competent/bacterial artificial chromosome (TAC/BAC) clone Sanger-sequencing data, amounting to a total genome size of 397 Mb. Meanwhile, version 3.0 was assembled by integrating sequencing data from both TAC/BAC clone Sanger-sequencing and Illumina shotgun sequencing of up to 40× coverage, amounting to a total genome size of 448 Mb.
Genes and predicted proteins
Gene features such as mRNA, alternatively spliced transcripts (also known as isoforms), exons, and coding sequences were made available both in the form of (1) a GFF3 file, used in a customized JBrowse21 implementation; and (2) individual BLAST databases. Gene and protein predictions were based on Augustus22, Cufflinks23, Genemark24, and Glimmer25. Lotus Base currently hosts gene and protein predictions for two versions of the genome assembly—19,713 predicted genes and 38,482 transcripts for v2.5; 44,483 predicted genes and 98,302 transcripts for v3.0.
LORE1 resource
Lotus Base is an integrated, one-stop platform for the LORE1 insertional mutagenesis population, hitherto the largest plant mutagenesis population established (Table 2). The LORE1 insertional mutagenesis population and its accompanying data (Table 3), collectively known as the LORE1 resource, have been previously described8. Lotus Base hosts 121,531 mutant lines containing 629,631 unique insertions, sourced from 14 Danish (DK01–03, 05, 07–16; 108,133 lines) and 3 Japanese (JPA, JPL, and JPP; 13,398 lines) batches26. All LORE1 lines have been sequenced and the ± 1000 bp flanking sequences were used for automated primer design using Primer327. In addition, all LORE1 associated data can be downloaded at https://lotus.au.dk/data/lore1. The LORE1 resource on Lotus Base has so far delivered more than 185,000 seeds from 3,800 unique mutant lines, shipped to 21 countries worldwide. The resource has also seen its use in several reverse genetics studies28,29,30,31,32.
Table 2. A non-exhaustive overview of currently available mutageneis populations for model plants.
| Organism | Agent | Type | Method | Classification | Population | Reference |
|---|---|---|---|---|---|---|
| A. thaliana | T-DNA | Insertional | Agrobacterium transfection | Transgenic | 48,830 | 79 |
| A. thaliana | Ac/Ds | Insertional | Transposon | Transgenic | 559 | 80 |
| A. thaliana | Tto1 | Insertional | Retrotransposon | Transgenic | 255* | 81 |
| A. thaliana | Tnt1 | Insertional | Retrotransposon | Transgenic | ca. 400* | 82 |
| L. japonicus | LORE1 | Insertional | Retrotransposon | Non-transgenic | 121,531 | 8 |
| L. japonicus | Tnt1 | Insertional | Retrotransposon | Transgenic | 51* | 83 |
| M. truncatula | Tnt1 | Insertional | Retrotransposon | Transgenic | ca. 12,000 | 84 |
| M. truncatula | Tnt1 | Insertional | Retrotransposon | Transgenic | 2* | 85 |
| Lettuce | Tnt1 | Insertional | Retrotransposon | Transgenic | 10* | 86 |
| Rice | Tos17 | Insertional | Retrotransposon | Non-transgenic | 47,196 | 87 |
| Soybean | Tnt1 | Insertional | Retrotransposon | Transgenic | 27* | 88 |
| A. thaliana | EMS | Allelic SNP | TILLING | Non-transgenic | 6,764 | 89, 90 |
| L. japonicus | EMS | Allelic SNP | TILLING | Non-transgenic | 8,556 | 91 |
| M. truncatula | EMS | Allelic SNP | TILLING | Non-transgenic | 3,162 | 92 |
| Rice | EMS, NaN3, NMU | Allelic SNP | TILLING | Non-transgenic | 5,120 | 93, 94 |
| Soybean | EMS | Allelic SNP | TILLING | Non-transgenic | 116 | 95 |
| Tomato | EMS, NMU | Allelic SNP | TILLING | Non-transgenic | 5,508 | 96 |
| Wheat | EMS | Allelic SNP | TILLING | Non-transgenic | 1,536 | 97 |
Mutagenesis methods that have so far only established starter lines without generating a large-scale mutagenis population are marked with an asterisk (*). Abbreviations: EMS, ethyl methanesulfonate; NMU, N-nitroso-N-methylurea, SNP, single nucleotide polymorphism; TILLING, Targeting Induced Local Lesions in Genomes.
Table 3. A table describing all LORE1-associated data available through Lotus Base.
| Field | Description | Example value |
|---|---|---|
| PlantID | A seven-digit mutant plant identifier, in the format of 3xxxxxxx. | 30000001 |
| Batch | The sources of the LORE1 line, indicating its Danish (DKn) or Japanese origins (JPx). | DK01 |
| ColCoord | Column coordinate of the plant for FSTpoolit. This value can be used to resolve lines with identical LORE1 inserts. | C_1 |
| RowCoord | Row coordinate of the plant for FSTpoolit. This value can be used to resolve lines with identical LORE1 inserts. | R_1 |
| Chromosome | The chromosome which the LORE1 insert is mapped to. This value may differ among genome assembly versions. | chr0 |
| Position | Position of the LORE1 insert. | 146086605 |
| CoordList | Pool coordinate details of all lines containing a particular LORE1 insert. Used for resolving lines with identical inserts.Note: In the example value, mutant plants containing the same insertion are observed to originate from two coordinates, C_1#R_1 and C_49#R_43. | C_1#C_49#R_1#R_43 |
| CoordCount | Absolute counts of the number of reads associated with each pool coordinate.Note: In the example value, C_1 and R_1 have the highest column and row counts (89 and 215 respectively), therefore the mutant line with coordinates C_1#R_1 is likely the actual mutant line containing this particular insert. | 89#27#215#9 |
| TotalCoverage | Sequencing coverage at the LORE1 insert. | 340 |
| FwPrimer | Forward primer designed using Primer3, based on the ±1000 flanking sequence. | TGCCAGCACCTGCAAATGAGAATCA |
| RevPrimer | Reverse primer designed using Primer3, based on the ±1000 flanking sequence. | TGTCCAGGTCTTGCTGCCAAATCA |
| PCRInsPos | Size of PCR product if there is a positively identified LORE1 insert. | 516 |
| PCRWT | Size of PCR product if there is no LORE1 insert. | 507 |
| InsFlank | The ±1000 bp flanking sequence (2 kb int total) of the LORE1 insert. | TGTTTTCACCTTATATCTCT |
| Ordering* | A Boolean value indicating if the line is orderable or not. | 1 |
| Version | Lotus genome assembly version against which the LORE1 line is mapped. | 3.0 |
| Salt* | A unique 32-character hexadecimal identifier of a LORE1 insert. | 7cd0a4c8e9f10c4096d1782702267c59 |
| IDKey* | An auto-incremental, unique entry identifier for indexing purposes. | 131071 |
Rows that are not made available through data export are marked with an asterisk (*).
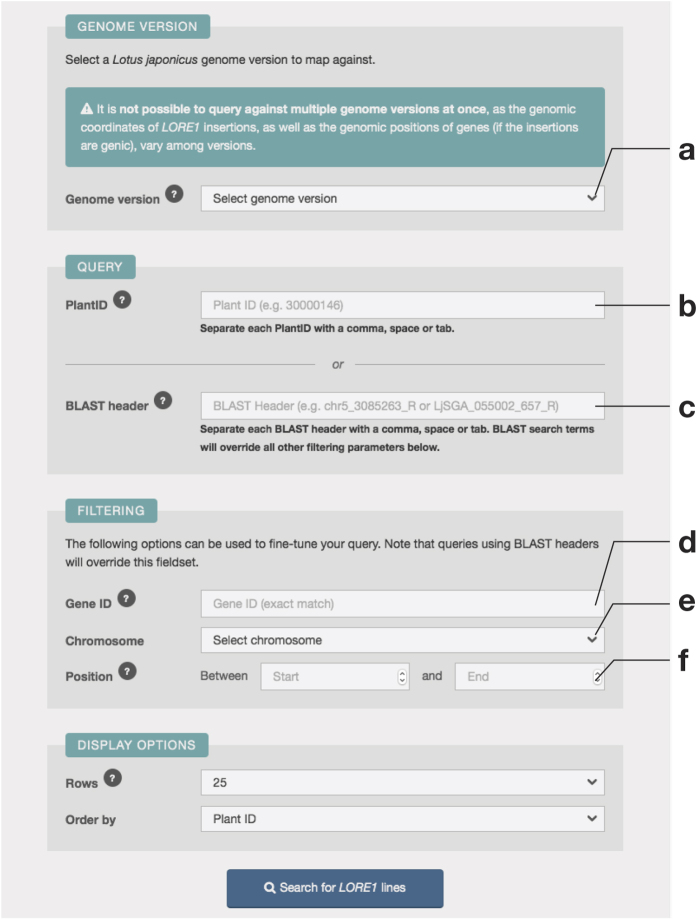
With such a large volume of data available, the LORE1 search form is designed to be intuitive and easy to use, allowing users to search for LORE1 lines of interest using a variety of user-defined criteria (Fig. 2). Users may search for LORE1 insertions based on: (1) a LORE1 mutant line identifier; (2) an insertion identifier, also known as a BLAST header, which is an underscore delimited string containing the chromosome, position and orientation of a LORE1 insert); (3) or the gene(s), if any, that the insertion is located in. Due to spatial constraints, data from all fields are not displayed on the search results page, although data export options are available on all pages.
Figure 2. The LORE1 search form.
Field A is compulsory, while fields B and C are mutually exclusive. Fields D, E and F are optional fields that allows user to further filter their results if desired. (a) A dropdown menu for the L. japonicus genome version—currently v2.5 and v3.0 are publicly available. (b) The unique eight-digit identifier of a LORE1 mutant line. One mutant line may have multiple BLAST headers. (c) A BLAST header, which is a unique LORE1 insertion identifier, is an underscore-delimited string of chromosome, position and orientation of the insert. Each BLAST header should uniquely map back to a single LORE1 insertion. (d) Filtering for LORE1 inserts that are inserted in a gene of interest. The gene identifier differs among L. japonicus genome versions. (e) The chromosome where the LORE1 insert is located in. (f) The genomic interval (inclusive on both ends), where the LORE1 insert must be located in. If only one value is provided (be it in the “start” or “end” field), then a specific genomic coordinate is enforced.
Orders can be placed on all Danish LORE1 lines (108,133; 89% of listed lines) for which seed stocks are available. Listed Japanese lines (13,398; 11% of listed lines) are included in our database but are not available for ordering—users are instead directed to LegumeBase (https://www.legumebase.brc.miyazaki-u.ac.jp/lore1BrowseAction.do) for ordering said lines.
Expression data
Lotus Base also offers Lotus-related expression data sourced from various studies. The first dataset was derived from the L. japonicus gene expression atlas (LjGEA) project10, which combined expression data from additional studies33,34,35,36,37. The whole LjGEA dataset consists of 81 conditions sourced from 6 independent published studies10, such as the investigation of draught responses33, effect of mycorrhizal and symbionts inoculation34,35, transcriptome changes in symbiosis defective mutants35, effect of salt and nitrate treatment35,36,37, and transcriptome regulation in various plant organs10. We have mapped probe identifiers from the LjGEA dataset against the annotated proteins of L. japonicus genome v3.0 by performing BLAST alignments of LjGEA probe set against the predicted transcripts from L. japonicus genome v3.0 and selecting for hits with the lowest E-value(s). In addition to the LjGEA dataset, we have also integrated expression data from Lotus roots in response to germinating spore exudates from arbuscular mycorrhiza5, containing 3 conditions.
Genome browser
The Lotus genome browser is powered by a customized version of JBrowse v1.12.021, with the following tracks publicly available: L. japonicus MG20 reference genome v3.0; predicted protein tracks; LORE1 insertions; genome gaps; repeat masks; and L. japonicus Gifu and MG20 RNAseq reads.
Lotus BLAST and SeqRet, an improved NCBI BLAST and sequence retrieval tool
SequenceServer v1.0.438 was modified according to our needs and serves as the backbone for Lotus Base Basic Local Alignment Search Tool (BLAST). Lotus BLAST currently runs using NCBI BLAST+ v2.2.31 executables39, allowing users to execute the total suite of BLAST algorithms—blastn, blastx, tblastn, tblastx, and blastp. Various toolkits on our site are integrated with an in-house developed Sequence Retrieval (SeqRet) tool, which allows real time retrieval of accession/identifier-based sequence information across all locally hosted Lotus BLAST databases. Users are presented with the option to view retrieved sequences in a modal box, or to download them as FASTA files for storage and/or further processing.
Sequence Processor (SeqPro)
The traditional wwwblast package from NCBI still outputs BLAST results in a monospaced, plain text format that can be problematic to parse for the end user. Users carrying such data from other sites may encounter difficulty in extracting useful sequence identifiers. Sequence Processor (SeqPro) tool is designed as a regular-expression based parser to handle wwwblast output and provide a tabular output. In addition, SeqPro also helps to remove line breaks and number lines from plain text FASTA outputs, which improved readability of sequences if users simply want to store the nucleotide/amino acid sequences without any accompanying metadata such as row counts, nucleotide position numbers, and unnecessary line breaks.
Transcript Mapper (TRAM)
As each Lotus genome assembly comes with a unique combination of predicted gene/transcript nomenclature and populations, we have designed a simple tool to aid users in mapping v2.5 to v3.0 transcripts and vice versa. A mapping table has a many-to-many relationship is precomputed by performing BLAST alignments between transcripts from both versions, and storing the highest confidence hits for all transcripts.
Transcript Explorer (TREX)
For users to glean quick information about their genes or transcripts of interest, we have designed the Transcript Explorer (TREX) tool, which is simply a full-text search engine that allows users to pull integrated information related to their search candidates. The search result is tabulated and summarized to display the working name (if any), and the function of the candidate gene/transcript, its position in the Lotus genome and any LORE1 lines with exonic insertions in the gene. Further information and deep links to other toolkits on the site, such as to ExpAt, LORE1 search, individual gene pages, are available in a dropdown menu for each candidate.
Expression Atlas (ExpAt)
We have developed a data-driven, web-based visualization tool for L. japonicus expression data. Visualization in the L. japonicus Expression Atlas (ExpAt) tool is powered by jQuery and d3.js40. The use of client-side JavaScript enables intuitive and dynamic customization, on-the-fly asynchronous clustering, and vector graphics export options—all of which are features unavailable in currently available expression data visualization tools for Lotus.
Search functionalities
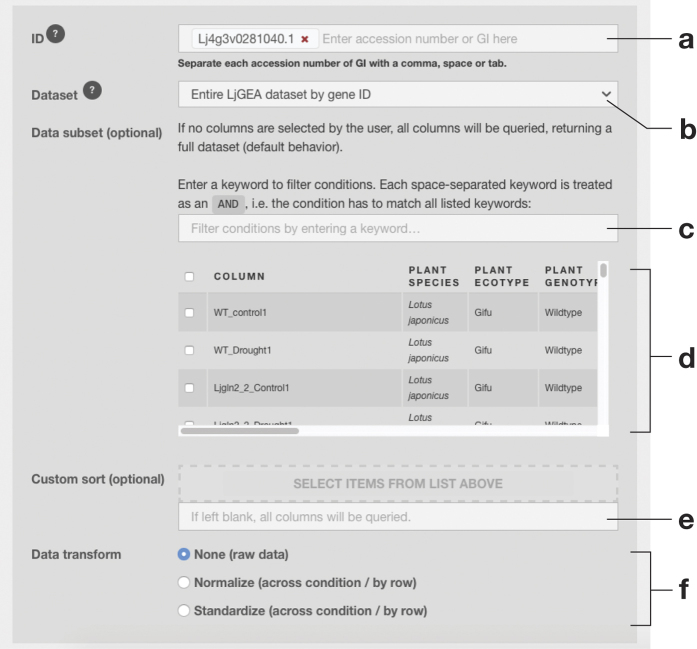
ExpAt features a simple search form to query the expression levels of candidates (genes or probes, depending on the dataset selected) against a list of published datasets (Fig. 3). The user can subset a dataset by checking individual conditions, which can also be filtered by user-defined keyword(s) using an in-browser full-text search engine implemented using Lunr.js41.
Figure 3. The design of ExpAt search form.
(a) The query for the expression level of candidate(s) of interest—gene, transcript, or probe identifiers are accepted. (b) A dropdown selection menu for an ExpAt dataset to base the query upon. When a dataset is selected, the metadata table in (d) will be asynchronously updated with the related metadata from the selected dataset. (c) A text field to perform full-text search, using user defined keywords, in order to filter the columns in the metadata table. (d) The metadata table containing column/condition-associated data. (e) A text field that accepts a comma-separated string of columns generated from a previous ExpAt search, if a certain sorting order of columns is desired. Users may also drag to reorder checked columns from the metadata table. (f) An option to transform the expression levels.
Table design
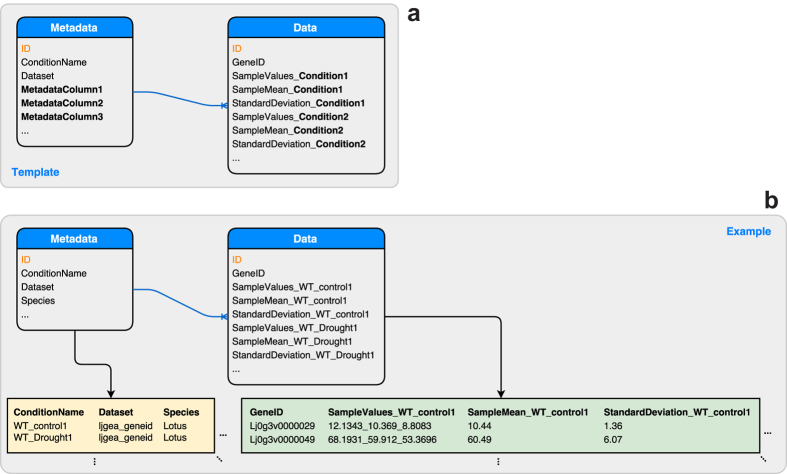
As expression datasets are multidimensional, we have devised a simple, two-table-based system to accommodate the data (Fig. 4). The “metadata” table contains all metadata associated with each column, such as the age of the plant, the treatment type and/or inoculation pressure. Contents of these metadata fields is fed into Lunr.js41 for in-browser full-text search. The “data” table contains all the expression data of each dataset. Each row in the “data” table presents a unique gene or probe. Each row is tagged with a unique identifier in the first column, followed by three sets of columns representing the raw data: the “sample values” column, where raw expression levels are delimited with an underscore; the “sample mean” column, where the arithmetic average of raw expression levels is stored; and the “standard deviation” column, where the sample standard deviation of raw expression levels is stored. There is therefore a one-to-three relationship between the “metadata” and “data” tables, as each condition maps to three independent data columns.
Figure 4. The organization of multi-dimensional expression data in the Expression Atlas (ExpAt) tool.
A two-table system is used—the “metadata” table is used to store metadata associated with each condition. The “data” table is used to store expression levels associated with each row identifier. Highlighted column names, in orange, are used as primary indexes. (a) A standard template used for all ExpAt datasets. (b) An example of what an ExpAt dataset may look like, featuring some data extracted from the LjGEA dataset.
Data transformation
For easing quick visual comparison across genes with significantly different levels of absolute expression, measured by either (1) reads per kilobase of transcript (RPKM) for RNAseq datasets, or (2) arbitrary Affymetrix units for Affymetrix MicroArray datasets, we included two possibilities to transform the expression levels, by normalization or standardization. Data normalization is simply the rescaling of expression values to fit the domain [0, 1], by subtracting the log-transformed sample expression levels, xs, with the lowest log-transformed expression level, (log10 x)min, followed by the division of the difference between the log-transformed maximum and minimum expression levels, as defined in equation (1). In order to allow comparison for extreme values, expression values are log10-transformed prior to normalization.
Meanwhile, data standardization10 serves to rescale the expression levels on a per row basis, across conditions, to have a mean of zero and a standard deviation of one. This is performed by subtracting the sample expression levels (xs) by the average expression level (μ) across all samples, and dividing the difference with the sample standard deviation computed across all samples (σ), as defined in equation (2).
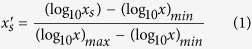 |
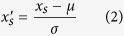 |
Clustering
Depending on the size of the matrix, we implemented either k-means clustering (for 1-by-n or n-by-1 matrices), or hierarchical agglomerative clustering (for matrices the size of, or larger than, 2-by-2). The clustering is performed asynchronously on the server-side using SciPy42. As clustering is based on heuristics and therefore non-deterministic in nature, users are encouraged to export the sorted order of either, or both axes, should they want to preserve the exact clustering order.
For k-means clustering, the default number of starting clusters is set to the square root of the number of conditions queried, rounded up to the nearest integer. For hierarchical agglomerative clustering, the cluster cutoff is set to 0.25 of the maximum cluster distance for both axes, and is allowed to vary between 0 and 1. Complete linkage is used by default, with the option of switching to single, centroid, median, ward, or weighted methods. The default linkage metric used is Euclidean, with other options available: Braycurtis, Canberra, Chebyshev, city block (Manhattan), correlation, cosine, standard Euclidean, squared Euclidean, normalized Hamming, Jaccard, or Minkowski.
CORNEA and CORGI: co-expression gene network visualization and co-expressed gene list retrieval
The co-expression (CORx) toolkit comprises the Co-Expression Network Analysis (CORNEA) and Co-expressed Gene Identifier (CORGI) tools. ExpAt and CORx toolkit share the same expression datasets. Co-expression gene networks in CORNEA are generated on the fly by a dedicated virtual server, which returns JSON-formatted data used for asynchronous network visualization with Sigma.js43 in the web browser. CORGI performs a similar function to CORNEA, but instead of generation a two-dimensional co-expression network, simply retrieves a one-dimensional slice by calling a unique gene or probe identifier, which in return generates a list of highly co-expressed entities with the gene or probe of interest.
Generating and displaying network jobs
All CORNEA and CORGI requests are handled by a central co-expression network threaded server setup implemented using Remote Python Call (RPyC)44. Both client and server-side logic will check for the validity of the job request, before submitting it to the server. An entry in a MySQL table is created per job for the purpose of storing user settings and metadata of the specific network. This information is freely accessible to the user and can be exported, if the user intends to recreate the network in the future, or to reuse similar settings for network generation using alternative datasets. The submission of a valid job will trigger a redirection to a job-specific URL, which will poll the server for the job status at a set interval until completion. Once the job is completed, the user will receive an email notification if they have indicated as such prior to job submission, containing links to view their live network in the CORNEA application, and to download all data associated with their network, contained in a gzipped JSON-formatted file. The file contains all the necessary information to display a co-expression network, and within it also stores network metadata such as correlation threshold, minimum cluster size, and job runtime.
Users may also visualize networks generated by previous jobs by uploading the JSON file, gzipped or decompressed, using a drag-and-drop interface implemented in CORNEA itself. Using client-side JavaScript, the browser will unzip—if the file is gzipped—and parse the JSON file, which is handed off to SigmaJS to handle the construction of the co-expression network.
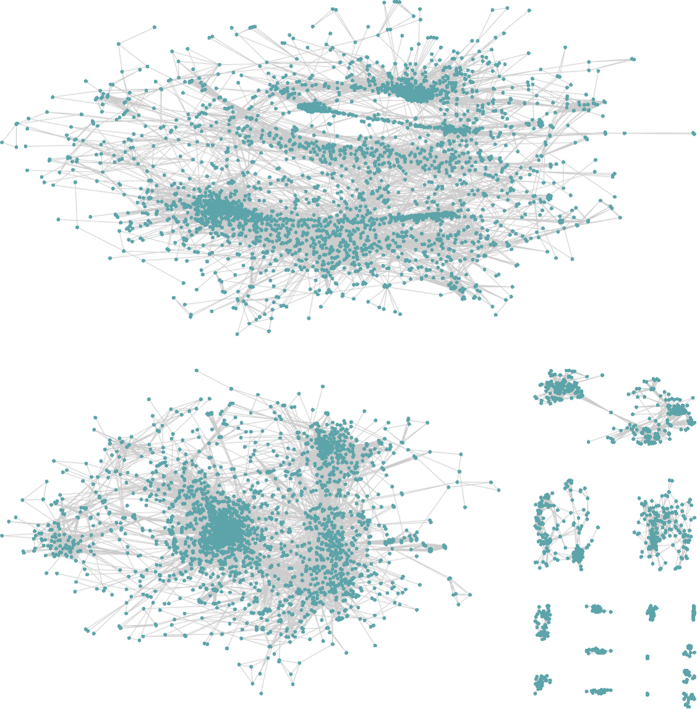
We anticipate that several basic co-expression network parameters may be heavily utilized, and in order to reduce the load on the server on generating identical or highly similar networks, we have therefore generated static networks that users can utilize for preliminary exploration. An example of a static network is one that was generated from expression data from the LjGEA dataset with an R2 threshold of 0.85, and a minimum cluster size of 15. The resulting network was produced in 4 minutes and 48 seconds, with a total of 7,839 nodes—connected by 273,018 edges and found in 17 mutually exclusive clusters (Fig. 5).
Figure 5. An example of a standard co-expression network generated by CORNEA, using the following parameters: LjGEA dataset with a minimum R2 value of 0.85 and a cluster size of 15 or larger.
The resulting network has 7,839 nodes connected by 273,018 edges and represented in 17 distinct clusters. The network took 4 minutes and 48 seconds to generate.
As CORNEA relies heavily on client-side JavaScript on parsing and displaying the co-expression network, the use of a modern, standards-compliant browser with an optimized, efficient JavaScript engine is strongly recommended.
Computation of co-expression relationships
Prior to pairwise calculation of correlation scores among genes or probes (collectively termed “candidates” hereon), the raw dataset is filtered in order to exclude candidates with highly similar expression pattern across conditions. For a dataset containing N number of candidates with a gene expression profile of ci, the candidate will be removed from analysis if its pattern falls below a dissimilarity threshold compared to another gene expression profile cj as seen in equation (3), while making exceptions for highly similar patterns with obvious peaks as defined in equation (4).
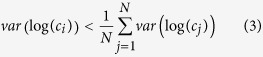 |
 |
The degree of co-expression of genes is calculated as the squared Pearson’s correlation coefficient (R2) between gene and/or probe pairs across conditions. Prior to submission of a CORNEA network generation job, the user is provided with an option to subset their conditions of interest from a list of all conditions available for a given dataset.
Node highlighting
To allow easy identification of the node(s) of interest, we implemented a highlight feature which allows the end-user to filter the displayed nodes in the network by (1) searching for a specific node, using an appropriate identifier depending on the type of dataset used, such as a gene identifier for the LjGEA dataset; or by (2) highlighting an array of nodes using a CSV file. The CSV file should contain no headers, and two columns—the first column containing the appropriate identifier for the queried dataset, and the second (optional) column containing arbitrary grouping (see supplementary, “File format for advanced node highlighting in CORNEA”). Additional columns in the CSV file will not be parsed, but can be used to store additional metadata.
Public API
To allow other developers to benefit from the scope of our Lotus data, we have developed a public API using Slim framework45, a PHP Standard Recommendation (PSR) 7-compliant46 representational state transfer conformant (REST) service. All API calls are to be authenticated with a secure and cryptographically generated JWT known as an API access token. API access tokens are freely available to developers who have signed up for an account with Lotus Base. Due to the possibility to forge HTTP referral headers, we do not enforce domain-based restrictions on API access tokens. However, any API access token can be revoked at the liberty of developers who have created them, in the event of suspicious use by unauthorized third parties.
Lotus Base API uses a versioning system in order to maintain compatibility with developers using various versions of the API, to account for the possibility of major updates and changes. The Lotus Base API is currently at version 1, and is accessible at https://lotus.au.dk/api/v1. Complete documentation of the Lotus Base API v1 is available at https://lotus.au.dk/docs/api/v1.
User accounts
Users may opt to sign up for a new account with Lotus Base for a more personalized experience. We have integrated several popular OAuth 2.0 identitiy providers—LinkedIn, GitHub, and Google—so that users can use alternative online services acting as identity providers to sign in, without the need to sign up manually. Existing users may also opt to integrate their Lotus Base user accounts with the aforementioned identity providers. Lotus Base adopts an ethical design principle giving users control over their own data and accounts. Private information of users is never shared with unaffiliated third parties, and their login credentials cryptographically salted and encrypted.
Usage and Application
As a proof-of-concept use of Lotus Base for a typical end user, we will choose to work with LjFls2, the Lotus ortholog of Arabidopsis FLS2 (AtFLS2). AtFLS2 encodes a bacterial flagellin receptor and is an important component in the induction of an evolutionarily conserved, first line defense responses in plants against pathogens47. The functionality of the Lotus ortholog, LjFls2, has also been previously confirmed48.
Identification and BLAST search for a Lotus ortholog of AtFLS2
The amino acid sequence of AtFLS2 (AT5G46330) was obtained from Araport49, and searched against the L. japonicus MG20 v3.0 protein database in Lotus BLAST. The top candidate was Lj4g3v0281040.1 with an E-value of 0 and a matching length of 1157. There were no other candidates with this degree of similarity, and a reverse BLASTp performed using the amino acid sequence of Lj4g3v0281040.1, retrieved using the SeqRet tool, against the Arabidopsis TAIR10 protein database revealed AtFLS2 as the single, high-confidence match. Therefore, Lj4g3v0281040.1 is tentatively named LjFls2 and referred to as such hereon.
LjFls2 is strongly expressed in Lotus roots
Next, we checked the expression of LjFls2 and compared it against the closest Lotus homologs of a handpicked subset of genes with distinct expression patterns in plant development using ExpAt (Table 4). We selected homologs of AtEIR150; AtSUC2, AtCOB, AtRHD351; and members of the cellulose synthase family, CesA family52, for their root-restricted expression. We also selected members of the SEPALATA family for their role in flower development53; AtZIFL1 and AtZIFL2 for their upregulated expression under draught conditions54; and members of the alpha-galactosidase family for their role in seed development in Arabidopsis55 and tomato56.
Table 4. List of handpicked genes for visualization in ExpAt and CORNEA, based on their expression in developmental stages and organs in Arabidopsis.
| Lotus organ |
Arabidopsis gene |
Putative Lotusorthologs |
||
|---|---|---|---|---|
| ID | Name | ID | Name | |
| Root | AT5G46330 | AtFLS2 | Lj4g3v0281040 | LjFls2 |
| Root | AT5G57090 | AtEIR1 | Lj4g3v2139970 | LjEir1 |
| Root | AT1G22710 | AtSUC2 | Lj2g3v0205600 | LjSuc2 |
| Root | AT5G60920 | AtCOB | Lj1g3v0414750 | LjCob |
| Root | AT4G32410 | AtCESA1 | Lj0g3v0249089 | LjCesA1 |
| Root | AT4G39350 | AtCESA2 | Lj4g3v2775550 | LjCesA2 |
| Root | AT5G05170 | AtESA3 | Lj0g3v0245539 | LjCesA3 |
| Root | AT3G13870 | AtRHD3 | Lj3g3v2693010 | LjRhd3 |
| Flower development | AT1G24260 | AtSEP3 | Lj4g3v2573630 | LjSep3 |
| Flower development | AT5G15800 | AtSEP1 | Lj2g3v1105370 | LjSep1 |
| Flower development | AT3G02310 | AtSEP2 | Lj4g3v1736080 | LjSep2 |
| Draught tolerance | AT5G13750 | AtZIFL1 | Lj1g3v2975920 | LjZifl1 |
| Draught tolerance | AT3G43790 | AtZIFL2 | Lj6g3v1052420 | LjZifl2 |
Genes were separated into three groups: root, flower development and draught tolerance. Lotus orthologs are discovered by performing a BLASTp search of the corresponding Arabidopsis genes against L. japonicus MG20 proteins v3.0 database, and by selecting the candidate of the highest confidence selected. Lotus orthologs inherit the name of their Arabidopsis counterparts, with the standard gene nomenclature used for Lotus. Abbreviations: CesA, cellulose synthase family; Cob, COBRA-like extracellular glycosyl-phosphatidyl inositol-anchored protein family; Eir1, ethylene-insensitive root 1; Fls2, flagellin-sensing 2; Rhd3, root hair defective 3; Sep, SEPALATA family; Suc2, sucrose-proton symporter 2; Zifl, zinc-finger-like protein family.
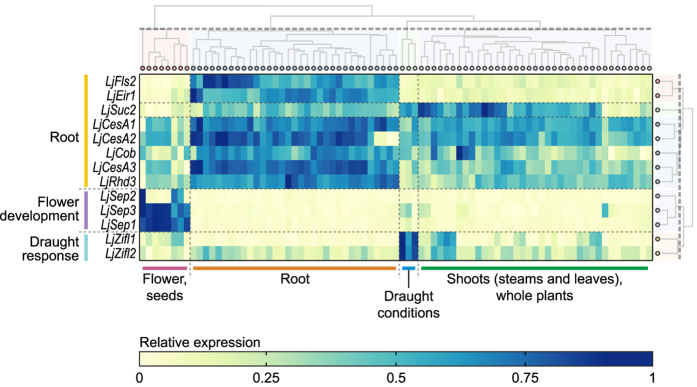
We discovered that LjFls2 has an expression pattern that strongly mirrors that of LjEir1, LjSuc2, LjCob, LjRhd3, and the CesA family members that show root expression in Arabidopsis, but not those of genes involved in other developmental stages and/or organs (Fig. 6). Hierarchical clustering was performed in ExpAt, using a Euclidean distance matrix over complete linkage based on squared Pearson’s correlation values (R2). This revealed distinct clusters of genes and conditions, with genes clustering into groups demarcated by developmental stage and organ in Arabidopsis, and conditions clustering into groups defined by organs and treatment conditions (Fig. 6).
Figure 6. The expression heatmap generated by ExpAt for our candidate gene, LjFls2 (Lj4g3v08201040), and other selected gene with distinct expression patterns.
Expression levels, expressed as arbitrary Affymetrix units in the vertical axis, are normalized across conditions (horizontal axis). Hierarchical agglomerative clustering was performed to generate a clustered heatmap, using complete linkage over a Euclidean distance matrix, with a clustering cutoff set to 0.4.
LjFls2 is located in the same co-expression cluster as genes with root-based expression
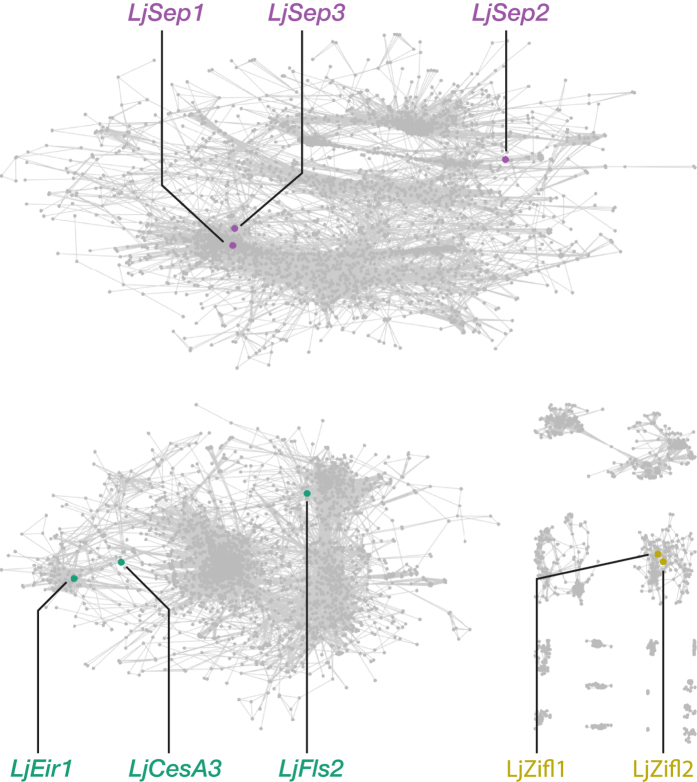
In order to visualize the co-expression network around LjFls2, we loaded the standard network generated from the LjGEA dataset in CORNEA, and highlighted network nodes using the gene list in Table 4 (Fig. 7; see supplementary “Node highlighting in CORNEA with selected genes”). Even when genes strongly expressed in the roots do not show highly correlated expression pattern (R2 ≤ 0.85) with LjFls2, they are still found in the same mega cluster, suggesting overall similarities in expression patterns. More importantly, flower development genes SEPALATA are found in another distinct mega cluster, and so are those involved in draught responses, LjZifl1 and LjZifl2.
Figure 7. The highlighted nodes of LjFls2 and other selected genes (see Table 4) in a standard co-expressed genes network map generated from the LjGEA dataset, using an R2 threshold of 0.85 and a minimum cluster size of 25.
Some root-based genes—LjCob, LjRhd3, LjSuc2, LjCesA1, and LjCesA2—were not found in the network, due to their expression patterns not meeting the minimum threshold on the squared Pearson’s correlation score (R2). Abbreviations: CesA, cellulose synthase family; Cob, COBRA-like extracellular glycosyl-phosphatidyl inositol-anchored protein family; Fls2, flagellin-sensing 2; Rhd3, root hair defective 3; Suc2, sucrose-proton symporter 2.
Taken together, this suggests that both ExpAt and CORNEA are reliable tools in not only differentiating, but correctly clustering, distinct gene expression patterns in Lotus. Moreover, both tools complement each other by providing a different perspective on the relationship of the expression patterns between candidate genes—ExpAt allows inference of relationship(s) among user-defined candidates, while CORNEA provides spatial information on how user-defined candidates fit into the overall expression network generated from a dataset.
Genes that are strongly co-expressed with LjFls2 have been functionally validated
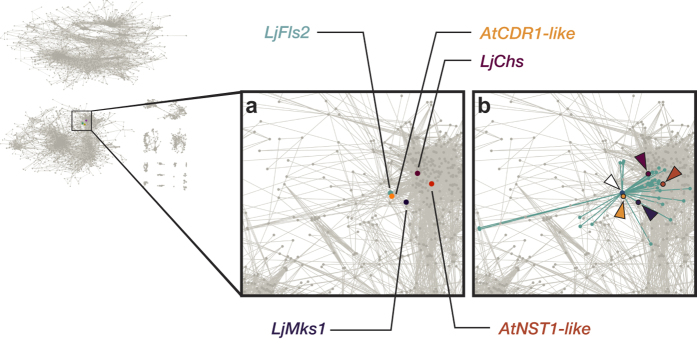
CORGI was used to generate a list of the top 25 highly co-expressed genes of LjFls2 (Table 5), and putative Lotus orthologs of four candidates whose expression patterns have been verified by published literature to be correlated with, or induced by, flagellin exposure—AtCDR1-like, AtNST1-like, MtCHS1-like, and AtMKS1-like. These genes were found not only in the same co-expression megacluster, but also directly connected to LjFLS2 in the network (Fig. 8).
Table 5. The top 25 highly co-expressed genes of LjFls2, generated by CORGI.
| Gene ID | Name/Description | R2 |
|---|---|---|
| Lj6g3v1880370 | PREDICTED: basic 7S globulin-like [Glycine max] gi|356557887|ref|XP_003547241.1| | 0.93336 |
| Lj4g3v2603590 | PREDICTED: stress response protein NST1-like [Cicer arietinum] gi|502140841|ref|XP_004504356.1| | 0.91121 |
| Lj0g3v0320039 | PREDICTED: probable receptor-like protein kinase At5g20050-like [Glycine max] gi|356563053|ref|XP_003549780.1| | 0.90673 |
| Lj4g3v2574990 | chalcone synthase CHS4 [Glycine max] gi|34148079|gb|AAQ62588.1| | 0.89932 |
| Lj2g3v1155180 | Protein MKS1 [Medicago truncatula] gi|357444747|ref|XP_003592651.1| | 0.89682 |
| Lj0g3v0173689 | PREDICTED: wall-associated receptor kinase 5-like [Glycine max] gi|356551203|ref|XP_003543967.1| | 0.89649 |
| Lj3g3v0602640 | n.a. | 0.88933 |
| Lj3g3v0602630 | phenylalanine ammonia-lyase [Lotus japonicus] gi|118142392|dbj|BAF36971.1| | 0.88933 |
| Lj3g3v0602620 | phenylalanine ammonia-lyase [Lotus japonicus] gi|118142392|dbj|BAF36971.1| | 0.88933 |
| Lj1g3v4590760 | phenylalanine ammonia-lyase [Lotus japonicus] gi|118142392|dbj|BAF36971.1| | 0.88901 |
| Lj2g3v1369250 | PREDICTED: zinc finger protein 5-like [Cicer arietinum] gi|502141274|ref|XP_004504507.1| | 0.88272 |
| Lj6g3v1418060 | PREDICTED: zinc finger protein 5-like [Cicer arietinum] gi|502141274|ref|XP_004504507.1| | 0.88272 |
| Lj0g3v0245779 | n.a. | 0.87638 |
| Lj0g3v0245769 | n.a. | 0.87638 |
| Lj0g3v0305019 | Uncharacterized protein TCM_040942 [Theobroma cacao] gi|508785660|gb|EOY32916.1| | 0.87633 |
| Lj4g3v2578250 | Rhg4-like receptor kinase II [Glycine max] gi|90655934|gb|ABD96566.1| | 0.87422 |
| Lj1g3v4590840 | phenylalanine ammonia-lyase [Lotus japonicus] gi|118142384|dbj|BAF36967.1| | 0.87387 |
| Lj3g3v1421800 | PREDICTED: U-box domain-containing protein 16-like [Cicer arietinum] gi|502146392|ref|XP_004506434.1| | 0.87052 |
| Lj3g3v1421810 | PREDICTED: U-box domain-containing protein 16-like [Cicer arietinum] gi|502146392|ref|XP_004506434.1| | 0.87052 |
| Lj0g3v0343089 | n.a. | 0.86410 |
| Lj6g3v0958320 | Uncharacterized protein TCM_040942 [Theobroma cacao] gi|508785660|gb|EOY32916.1| | 0.86281 |
| Lj2g3v2051190 | PREDICTED: 1-aminocyclopropane-1-carboxylate synthase-like [Glycine max] gi|356539620|ref|XP_003538294.1| | 0.86203 |
| Lj1g3v3716560 | GntR family transcriptional regulator [Cupriavidus basilensis] gi|493149530|ref|WP_006161625.1| | 0.86103 |
| Lj1g3v3716780 | n.a. | 0.86103 |
| Lj1g3v0129110 | n.a. | 0.86103 |
The candidates were pulled from a one-dimensional slice across the co-expression matrix generated by CORNEA, ranked by the squared Pearson’s correlation coefficient (R2) in descending order. CORGI returns 25 rows by default, but may be configured to return up to 100 candidates.
Figure 8. The location of four strongly co-expressed genes, relative to LjFls2, in the standard gene co-expression network generated from the LjGEA dataset.
Insets depict (a) the highlighted nodes of the three genes respectively, and (b) the immediate network around LjFls2, which contains co-expressed genes that meet the minimum threshold of R2 ≥ 0.85. Among them are AtCDR1-like (orange), AtNST1-like (red), MtCHS1-like (maroon), and AtMKS1-like (black). The white-filled triangle indicates the root candidate gene, LjFls2, which is partially occluded by an overlying node in (b). Abbreviations: CDR1, constitutive disease resistance 1; Chs, chalcone synthase; Fls2, flagellin-sensing 2; MKS1, mitogen-activated protein kinase substrate 1; NST1, no apical meristem (NAC) secondary wall thickening promoting factor 1.
Lj6g3v1880370 (1st, R2 = 0.933) is highly similar to a gene encoding for an aspartyl protease-like protein in Arabidopsis. A gene encoding an apoplastic aspartyl protease, AtCDR1, is found to play an important role in conferring salicylic acid-dependent resistance against Pseudomonas syringae in Arabidopsis57. Although the role of proteases in defense responses are yet to be clearly elucidated, it is hypothesized that they either aid in the processing of R proteins, or through enzymatic action generate ligands that are recognized by R proteins58,59,60.
Lj4g3v2603590 (2nd, R2 = 0.911) encodes a NST1-like protein, a member of a family of genes involved in the regulation of secondary cell wall thickening in Arabidopsis61 due to its role in lignin biosynthesis62. Lignification of plant cell walls may be induced by mechanical, environmental and disease stresses63,64, and treatment with bacterial flagellin has shown to induce lignin biosynthesis in plants65,66,67.
Lj4g3v2574990 (4th, R2 = 0.899) is a chalcone synthase (CHS) found in both alfalfa (Medicago truncatula) and Mexican lime (Citrus aurantifolia L.), and its expression is upregulated upon exposure to flagellin of their respective pathogens, Aphanomyces euteiches68 and Candidatus Phytoplasma aurantifolia69.
Lj2g3v1155180 (5th, R2 = 0.897) is the closest homolog of the Arabidopsis MKS1 (At3G18690), which encodes a protein that is substrate of AtMPK470, a kinase involved in the regulation of defense responses in plants71. More poignantly, AtMPK4 is activated by exposure to flagellin purified from P. syringae, an adapted pathogen of Arabidopsis, and results in phosphorylation of AtMKS1.
Multiple LORE1 lines with exonic insertions in LjFls2
Next, we retrieved LORE1 mutant lines that contain exonic insertions in the LjFls2 gene using the TREX tool. Out of the 40 LORE1 lines that contain insertions in LjFls2, 31 are exonic, of which 29 originate from the Danish collection and are therefore orderable through Lotus Base (Table 6). These 29 lines can be propagated (as F0 plants) and allowed to self-fertilize in order to generate F1 homozygous mutant lines, whose progenies (F2) will be useful for further phenotyping studies, if desired.
Table 6. All LORE1 lines containing a LORE1 insert in the LjFls2 gene, found across 16 batches of LORE1 populations.
| LORE1 ID | Batch | Chromosome | Position | Orientation | Insertion type |
|---|---|---|---|---|---|
| 30003492 | DK01 | chr4 | 3287060 | F | Exonic |
| 30012416 | DK03 | chr4 | 3286059 | R | Exonic |
| 30030341 | DK05 | chr4 | 3288262 | F | Intronic |
| 30030425 | DK05 | chr4 | 3286258 | F | Exonic |
| 30033827 | DK05 | chr4 | 3286209 | F | Exonic |
| 30034607 | DK05 | chr4 | 3287742 | F | Exonic |
| 30035947 | DK05 | chr4 | 3287150 | F | Exonic |
| 30056942 | DK07 | chr4 | 3287104 | F | Exonic |
| 30057743 | DK07 | chr4 | 3284745 | R | Exonic |
| 30057897 | DK07 | chr4 | 3285808 | F | Exonic |
| 30060694 | DK08 | chr4 | 3285199 | F | Exonic |
| 30003492 | DK01 | chr4 | 3287060 | F | Exonic |
| 30012416 | DK03 | chr4 | 3286059 | R | Exonic |
| 30061005 | DK08 | chr4 | 3285639 | R | Exonic |
| 30070461 | DK09 | chr4 | 3288029 | R | Intronic |
| 30071709 | DK09 | chr4 | 3288029 | R | Intronic |
| 30072232 | DK09 | chr4 | 3286020 | R | Exonic |
| 30072618 | DK09 | chr4 | 3287504 | R | Exonic |
| 30074013 | DK09 | chr4 | 3288029 | R | Intronic |
| 30075601 | DK09 | chr4 | 3286721 | F | Exonic |
| 30080413 | DK10 | chr4 | 3285894 | F | Exonic |
| 30083670 | DK10 | chr4 | 3285699 | R | Exonic |
| 30084653 | DK10 | chr4 | 3287937 | R | Intronic |
| 30088736 | DK11 | chr4 | 3284973 | R | Exonic |
| 30092255 | DK11 | chr4 | 3285297 | R | Exonic |
| 30095950 | DK12 | chr4 | 3285585 | F | Exonic |
| 30100097 | DK12 | chr4 | 3288274 | R | Intronic |
| 30100269 | DK12 | chr4 | 3286310 | F | Exonic |
| 30108970 | DK13 | chr4 | 3286450 | R | Exonic |
| 30109089 | DK13 | chr4 | 3287032 | R | Exonic |
| 30109124 | DK13 | chr4 | 3288292 | F | Intronic |
| 30109659 | DK13 | chr4 | 3284933 | R | Exonic |
| 30115606 | DK14 | chr4 | 3284648 | R | Exonic |
| 30117374 | DK14 | chr4 | 3285311 | F | Exonic |
| 30119789 | DK14 | chr4 | 3286700 | R | Exonic |
| 30119801 | DK14 | chr4 | 3286700 | R | Exonic |
| 30124389 | DK15 | chr4 | 3286500 | F | Exonic |
| 30138429 | DK16 | chr4 | 3285596 | F | Exonic |
| A04405 | JPA | chr4 | 3287360 | R | Exonic |
| L0530 | JPL | chr4 | 3288070 | R | Intronic |
| L4758 | JPL | chr4 | 3286032 | F | Exonic |
| P1585 | JPP | chr4 | 3288189 | R | Intronic |
Abbreviations: chr, chromosome; F, forward; R, reverse.
Discussion
In this paper, we introduced Lotus Base, an integrated information portal for genomic and expression data for the model legume L. japonicus. With the utilization of modern browser technology and cryptographically secure information transmission, Lotus Base poises itself to be at the forefront of accessibility, security, privacy and usability of large-scale scientific data without sacrificing usability. The lack of a central database for Lotus resources has been a strong driving force behind the creation of Lotus Base. This places Lotus japonicus on par with other popular model plants, such as A. thaliana, G. max, and M. truncatula, all of which have dedicated online platforms that serve integrated data, namely Araport49, the Arabidopsis Information Resource72, SoyBase73 and the Medicago truncatula Genome Database74.
Lotus Base distinguishes itself from other cross-species integration platform such as Legume Information System (LIS)75,76, PlantGDB77, and Phytozome78, by offering comprehensive species-specific data. In addition, Lotus genomic data is available on LIS75,76, PlantGDB77, Phytozome78, and through the Kazusa DNA Research Institute website9; and Lotus expression data on LjGEA10. However, there are hitherto neither LORE1 mutant population nor Lotus expression data integrated with the sequenced and annotated genome of Lotus. Yet, similar to the motivation behind Araport49 and LIS, Lotus Base is designed in response to a fragmented landscape of Lotus data available across various platforms, by bridging data sourced from various studies. The integration of various resources, such as the search and order system of 120,000+ LORE1 lines, the assimilation and deep linking of publicly available expression datasets, makes Lotus Base a convenient and feature-rich one-stop repository for Lotus resources. While other legume resources offer similar datasets, many features are non-standards compliant, rely on dated web technologies, lack user friendliness, or do not offer integrated data for easy data mining (Table 7). Moreover, the web-based implementation of Lotus Base aims to improve data availability and exchange among the Lotus research community, unimpeded by computer hardware and operation systems, or technological know-how of the end user.
Table 7. Comparison of features available on extant legume resources and Lotus Base.
| Legume Information System1 75 | Lotus japonicus2 or Medicago truncatula3 Gene Expression Atlas (Lj/MtGEA)10,98,99 | Kazusa DNA Research Institute4 9 | Medicago truncatula Genome Project5 74 | SoyBase6 73 | Lotus Base7 (this work) | |
|---|---|---|---|---|---|---|
| Species | 21 | 1; L. japonicus or M. truncatula | 1; L. japonicus | 1; M. truncatula | 1; G. max | 1; L. japonicus |
| Genome browser | Yes; GBrowse100 and JBrowse12 | Yes; for MtGEA only (available as external link) | Yes; GBrowse | Yes; JBrowse | Yes; GBrowse | Yes; JBrowse |
| Genetic map | No | No | Yes | Yes | Yes | No |
| Gene ontology | Yes | No | Yes | Yes | Yes | No |
| Data mining | Partial (only 3 species supported) | No | Yes | Yes | Yes | Yes |
| - Implementation | Intermine101 | No | Custom-designed solution | Intermine | Intermine | Custom-designed solution |
| Large-scale mutagenesis population & data | No | No | No | No | No | Yes; 121,531 LORE1 lines containing 629631 unique insertions |
| - Predicted gene model overlay | No | No | No | No | No | Yes |
| - Ordering and dispatching | No | No | No | No | No | Yes |
| BLAST | Yes; NCBI BLAST39 | Yes; NCBI BLAST | Yes; NCBI BLAST | Yes; NCBI BLAST | Yes; NCBI BLAST | Yes; SequenceServer38 powered by NCBI BLAST |
| - Programs | ||||||
| - - blastn | Yes | Yes | Yes | Yes | Yes | Yes |
| - - blastx | Yes | No | Yes | Yes | Yes | Yes |
| - - tblastn | Yes | No | Yes | Yes | Yes | Yes |
| - - tblastx | No | No | Yes | Yes | Yes | Yes |
| - - blastp | Yes | No | Yes | Yes | Yes | Yes |
| - Datasets | ||||||
| - - Genome | Yes | No | Yes | Yes | Yes | Yes |
| - - cDNA/mRNA | No | No | Yes | Yes | Yes | Yes |
| - - CDS | Yes | No | Yes | Yes | Yes | Yes |
| - - Proteins | No | No | Yes | Yes | Yes | Yes |
| - - Misc | – | Microarray chip target, sequences, and probes | – | Unspliced transcripts and BAC ends | – | LjGEA microarray chip probes |
| Gene expression | No | Yes | No | No | Yes | Yes |
| - Data transformation | No | Yes; normalization | No | No | Yes; normalization | Yes; normalization or standardization |
| - Data export | No | Yes; values are available as replica readouts or arithmetic means | No | No | Yes; only single values available in CSV format | Yes; values are available as replica readouts, arithmetic means, and pre-computed standard deviations |
| - Visualization | No | Yes; reliance on Adobe Flash, requires multiple windows to be opened | No | No | Yes; rudimentary and tabular based | Yes; using web-based data-driven approach, highly customizable |
| - Analysis | No | No | No | No | Yes; hierarchical clustering | Yes; asynchronous clustering—k-means or hierarchical—dependent on matrix size |
| - Co-expression analysis | No | Yes; single dimensional co-expression relationships | No | No | No | Yes; single and two-dimensional co-expression relationships, spatial network construction, and data-driven presentation |
| Public API | No | No | No | No | No | Yes |
| User documentation & help | Yes | Yes | No | Yes | Yes | Yes |
Abbreviations: BLAST, basic local alignment search tool; CDS, coding sequence; GEA: gene expression atlas.
The modular construction and open-source model of Lotus Base ensure continuity and encourage expansion and inclusion of additional dataset with relative ease in the future. In addition, the public API of Lotus Base aims to benefit a larger community by making Lotus data available to developers who are deploying applications that pull integrated data from our databases.
The introduction of Lotus BLAST allows deep integration of Lotus BLAST databases with other toolkits specifically designed to tackle data visualization and analysis. The implementation of various toolkits such as ExpAt, CORNEA and CORGI can be extrapolated to datasets unrelated to Lotus, or even scientific research in general. We demonstrated that ExpAt offers users a powerful way of visualizing co-expression relationships on a subset of user-defined candidates by leveraging on k-means or hierarchical clustering, while CORNEA presents users a two-dimensional, spatial chart of co-expression relationships among all genes from selected datasets. The use of data-driven documents in these toolkits reveal their prowess in the ability to visualize large volumes of data with ease, by combining the computational power of server-side technologies and the efficiency of client-side JavaScript interpreters. Many features on Lotus Base can therefore be adapted by the community as novel ways to represent, investigate, analyze, and visualize biological data. We believe that Lotus Base will not only make comprehensive Lotus data accessible to researchers easily, but also empower them to perform computationally intensive and complex analysis and visualization without the need for extensive technological skills. Taken in all, Lotus Base will benefit the legume research community and beyond, by providing a framework for a coherent scientific workflow and powerful tools for raw data interpretation.
Additional Information
How to cite this article: Mun, T. et al. Lotus Base: An integrated information portal for the model legume Lotus japonicus. Sci. Rep. 6, 39447; doi: 10.1038/srep39447 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Danish National Research Foundation grant DNRF79.
Footnotes
Author Contributions T.M. wrote the manuscript, created the figures, designed the front-end of Lotus Base, implemented client-side codes for majority of site functionalities and ExpAt visualization, set up the server and performed most of the back-end integration with various open source packages. A.B. wrote the server-side code for co-expression network server powering CORNEA and CORGI, and implemented client-side code for co-expression network visualization. V.G. generated all GFF files used for the JBrowse tracks. S.U.A. conceptualized site features and, along with J.S., coordinated and commented on the manuscript. All authors read and approved the final manuscript.
References
- Handberg K. & Stougaard J. Lotus japonicus, an Autogamous, Diploid Legume Species for Classical and Molecular-Genetics. Plant Journal 2, 487–496, doi: 10.1111/j.1365-313X.1992.00487.x (1992). [DOI] [Google Scholar]
- Radutoiu S. et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592, doi: 10.1038/nature02039 (2003). [DOI] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K. & Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827, doi: 10.1038/nature03608 (2005). [DOI] [PubMed] [Google Scholar]
- Bordenave C. D. et al. Defense responses in two ecotypes of Lotus japonicus against non-pathogenic Pseudomonas syringae. Plos One 8, e83199, doi: 10.1371/journal.pone.0083199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M., Mari A., Novero M. & Bonfante P. Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front Plant Sci 6, 480, doi: 10.3389/fpls.2015.00480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai E. et al. Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. The Plant Journal 69, 720–730, doi: 10.1111/j.1365-313X.2011.04826.x (2012). [DOI] [PubMed] [Google Scholar]
- Urbański D. F., Małolepszy A., Stougaard J. & Andersen S. U. Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. The Plant Journal 69, 731–741, doi: 10.1111/j.1365-313X.2011.04827.x (2012). [DOI] [PubMed] [Google Scholar]
- Małolepszy A. et al. The LORE1 insertion mutant resource. The Plant journal: for cell and molecular biology, doi: 10.1111/tpj.13243 (2016). [DOI] [PubMed] [Google Scholar]
- Sato S. et al. Genome structure of the legume. Lotus japonicus. DNA Res 15, 227–239, doi: 10.1093/dnares/dsn008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J. et al. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. The Plant journal: for cell and molecular biology 74, 351–362, doi: 10.1111/tpj.12119 (2013). [DOI] [PubMed] [Google Scholar]
- Pajuelo E. & Stougaard J. In Lotus japonicus Handbook (ed. Márquez Antonio J.) 3–24 (Springer Netherlands, 2005). [Google Scholar]
- Sato S. & Tabata S. Lotus japonicus as a platform for legume research. Current Opinion in Plant Biology 9, 128–132, doi: 10.1016/j.pbi.2006.01.008 (2006). [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Tabata S., Parniske M. & Stougaard J. Lotus japonicus: legume research in the fast lane. Trends Plant Sci 10, 222–228, doi: 10.1016/j.tplants.2005.03.008 (2005). [DOI] [PubMed] [Google Scholar]
- Madsen L. H. et al. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1, 10, doi: 10.1038/ncomms1009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaconda Software Distribution. Anaconda: Leading Open Data Science Platform Powered by Python v. 2.4.0. Continuum Analytics, Austin, USA. URL https://www.continuum.io/ (2015).
- Bider D. & Baushke M. SHA-2 Data Integrity Verification for the Secure Shell (SSH) Transport Layer Protocol, https://tools.ietf.org/html/rfc6668 (2012).
- Jones M., Bradley J. & Sakimura N. JSON Web Token (JWT), https://tools.ietf.org/html/rfc7519 (2015).
- Alman B. et al. Grunt: The JavaScript Task Runner v. 0.4.5. GitHub, San Franciso, USA. URL https://github.com/gruntjs/grunt (2014).
- Moore P., Bedwell J. & Rogers M. Jekyll: Simple, blog-aware, static sites v. 3.1.6. GitHub, San Franciso, USA. URL https://github.com/jekyll/jekyll/ (2008).
- Lord R. Slate: API docs generator v. 1.3.3. GitHub, San Franciso, USA. URL https://github.com/lord/slate (2013).
- Buels R. et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17, 66, doi: 10.1186/s13059-016-0924-1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M. & Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 Suppl 2, ii215–225 (2003). [DOI] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515, doi: 10.1038/nbt.1621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin A. V. & Borodovsky M. GeneMark. hmm: new solutions for gene finding. Nucleic Acids Res 26, 1107–1115 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Bratke K. A., Powers E. C. & Salzberg S. L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679, doi: 10.1093/bioinformatics/btm009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai E. et al. Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. The Plant journal: for cell and molecular biology 69, 720–730, doi: 10.1111/j.1365-313X.2011.04826.x (2012). [DOI] [PubMed] [Google Scholar]
- Untergasser A. et al. Primer3–new capabilities and interfaces. Nucleic Acids Res 40, e115, doi: 10.1093/nar/gks596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małolepszy A. et al. The deubiquitinating enzyme AMSH1 is required for rhizobial infection and nodule organogenesis in Lotus japonicus. Plant Journal 83, 719–731, doi: 10.1111/tpj.12922 (2015). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. Lotus japonicus Clathrin Heavy Chain1 Is Associated with Rho-Like GTPase ROP6 and Involved in Nodule Formation. Plant Physiology 167, 1497–1510, doi: 10.1104/pp.114.256107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L. et al. Network of GRAS Transcription Factors Involved in the Control of Arbuscule Development in Lotus japonicus. Plant Physiology 167, 854-+, doi: 10.1104/pp.114.255430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. R. et al. Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci Rep-Uk 6, doi: ARTN 2973310.1038/srep29733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D. E., Heckmann A. B., Novak O., Kelly S. & Stougaard J. CYTOKININ OXIDASE/DEHYDROGENASE3 Maintains Cytokinin Homeostasis during Root and Nodule Development in Lotus japonicus. Plant Physiology 170, 1060–1074, doi: 10.1104/pp.15.00650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz P. et al. Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol 188, 1001–1013, doi: 10.1111/j.1469-8137.2010.03440.x (2010). [DOI] [PubMed] [Google Scholar]
- Guether M. et al. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182, 200–212, doi: 10.1111/j.1469-8137.2008.02725.x (2009). [DOI] [PubMed] [Google Scholar]
- Høgslund N. et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. Plos One 4, e6556, doi: 10.1371/journal.pone.0006556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D. H. et al. Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. The Plant journal: for cell and molecular biology 53, 973–987, doi: 10.1111/j.1365-313X.2007.03381.x (2008). [DOI] [PubMed] [Google Scholar]
- Sanchez D. H. et al. Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. Plos One 6, e17094, doi: 10.1371/journal.pone.0017094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyam A. et al. Sequenceserver: a modern graphical user interface for custom BLAST databases. bioRxiv (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421, doi: 10.1186/1471-2105-10-421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock M. D3: Data-Driven Documents v. 4.1.0. GitHub, San Franciso, USA. URL https://github.com/d3/d3 (2010).
- Author lunr.js: Simple full-text search in your browser v. 0.7.1. GitHub, San Franciso, USA. URL https://github.com/olivernn/lunr.js (2011).
- Jones E., Oliphant E. & Peterson P. SciPy: Open Source Scientific Tools for Python v. 0.18.0. SciPy, Austin, USA. URL http://www.scipy.org/ (2001).
- Jacomy A. & Plique G. Sigma, a JavaScript library dedicated to graph drawing v. 1.1.0. GitHub, San Franciso, USA. URL https://github.com/jacomyal/sigma.js (2012).
- Filiba T. RPyC—Transparent, Symmetric Distributed Computing v. 3.3.0. Python Software Foundation, Wilmington, USA. URL http://rpyc.readthedocs.org (2014).
- Lockhart J., Smith A., Allen R. & Manricks G. Slim, a micro framework for PHP v. 3.0. GitHub, San Franciso, USA. URL https://github.com/slimphp/Slim (2011).
- O’Phinney M. W. PSR-7: HTTP message interfaces, http://www.php-fig.org/psr/psr-7/ (2015).
- Gomez-Gomez L., Felix G. & Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. The Plant journal: for cell and molecular biology 18, 277–284 (1999). [DOI] [PubMed] [Google Scholar]
- Lopez-Gomez M., Sandal N., Stougaard J. & Boller T. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J Exp Bot 63, 393–401, doi: 10.1093/jxb/err291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V. et al. Araport: the Arabidopsis information portal. Nucleic Acids Res 43, D1003–1009, doi: 10.1093/nar/gku1200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R. A., Grisafi P. & Fink G. R. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12, 2175–2187 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960, doi: 10.1126/science.1090022 (2003). [DOI] [PubMed] [Google Scholar]
- Burn J. E., Hocart C. H., Birch R. J., Cork A. C. & Williamson R. E. Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol 129, 797–807, doi: 10.1104/pp.010931 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E. & Yanofsky M. F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203, doi: 10.1038/35012103 (2000). [DOI] [PubMed] [Google Scholar]
- Haydon M. J. & Cobbett C. S. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiology 143, 1705–1719, doi: 10.1104/pp.106.092015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L. et al. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124, 1595–1604 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado J. A., Banik M. & Bewley J. D. The cloning and characterization of alpha-galactosidase present during and following germination of tomato (Lycopersicon esculentum Mill.) seed. J Exp Bot 52, 1239–1249 (2001). [PubMed] [Google Scholar]
- Xia Y. et al. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. Embo J 23, 980–988, doi: 10.1038/sj.emboj.7600086 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo A., Monteiro F. & Sebastiana M. Subtilisin-like proteases in plant-pathogen recognition and immune priming: a perspective. Front Plant Sci 5, 739, doi: 10.3389/fpls.2014.00739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F., Merritt P. M., Bao Z., Innes R. W. & Dixon J. E. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109, 575–588 (2002). [DOI] [PubMed] [Google Scholar]
- Xia Y. Proteases in pathogenesis and plant defence. Cell Microbiol 6, 905–913, doi: 10.1111/j.1462-5822.2004.00438.x (2004). [DOI] [PubMed] [Google Scholar]
- Mitsuda N. et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280, doi: 10.1105/tpc.106.047043 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q. & Dixon R. A. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16, 227–233, doi: 10.1016/j.tplants.2010.12.005 (2011). [DOI] [PubMed] [Google Scholar]
- Malinovsky F. G., Fangel J. U. & Willats W. G. The role of the cell wall in plant immunity. Front Plant Sci 5, 178, doi: 10.3389/fpls.2014.00178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E., Vanholme R., Boerjan W. & Molina A. The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5, 358, doi: 10.3389/fpls.2014.00358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M. et al. Expression patterns of flagellin sensing 2 map to bacterial entry sites in plant shoots and roots. J Exp Bot 65, 6487–6498, doi: 10.1093/jxb/eru366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenke D., Böttcher C. & Scheel D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant, Cell & Environment 34, 1849–1864, doi: 10.1111/j.1365-3040.2011.02381.x (2011). [DOI] [PubMed] [Google Scholar]
- Takakura Y. et al. Expression of a bacterial flagellin gene triggers plant immune responses and confers disease resistance in transgenic rice plants. Mol Plant Pathol 9, 525–529, doi: 10.1111/j.1364-3703.2008.00477.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapphoff T., Beutner C., Niehaus K. & Colditz F. Induction of distinct defense-associated protein patterns in Aphanomyces euteiches (Oomycota)-elicited and -inoculated Medicago truncatula cell-suspension cultures: a proteome and phosphoproteome approach. Mol Plant Microbe Interact 22, 421–436, doi: 10.1094/MPMI-22-4-0421 (2009). [DOI] [PubMed] [Google Scholar]
- Mardi M., Karimi Farsad L., Gharechahi J. & Salekdeh G. H. In-Depth Transcriptome Sequencing of Mexican Lime Trees Infected with Candidatus Phytoplasma aurantifolia. Plos One 10, e0130425, doi: 10.1371/journal.pone.0130425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E. et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. Embo J 24, 2579–2589, doi: 10.1038/sj.emboj.7600737 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M. et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120 (2000). [DOI] [PubMed] [Google Scholar]
- Huala E. et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29, 102–105 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D., Nelson R. T., Cannon S. B. & Shoemaker R. C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38, D843–846, doi: 10.1093/nar/gkp798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V. et al. MTGD: The Medicago truncatula genome database. Plant Cell Physiol 56, e1, doi: 10.1093/pcp/pcu179 (2015). [DOI] [PubMed] [Google Scholar]
- Dash S. et al. Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Res 44, D1181–1188, doi: 10.1093/nar/gkv1159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M. D. et al. The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res 33, D660–665, doi: 10.1093/nar/gki128 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick J. et al. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res 36, D959–965, doi: 10.1093/nar/gkm1041 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40, D1178–1186, doi: 10.1093/nar/gkr944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657, doi: 10.1126/science.1086391 (2003). [DOI] [PubMed] [Google Scholar]
- Altmann T. et al. Ac/Ds transposon mutagenesis in Arabidopsis thaliana: mutant spectrum and frequency of Ds insertion mutants. Mol Gen Genet 247, 646–652 (1995). [DOI] [PubMed] [Google Scholar]
- Okamoto H. & Hirochika H. Efficient insertion mutagenesis of Arabidopsis by tissue culture-induced activation of the tobacco retrotransposon Tto1. The Plant journal: for cell and molecular biology 23, 291–304 (2000). [DOI] [PubMed] [Google Scholar]
- Courtial B. et al. Tnt1 transposition events are induced by in vitro transformation of Arabidopsis thaliana, and transposed copies integrate into genes. Mol Genet Genomics 265, 32–42 (2001). [DOI] [PubMed] [Google Scholar]
- Iantcheva A. et al. Tnt1 retrotransposon as an efficient tool for development of an insertional mutant collection of Lotus japonicus. In Vitro Cell Dev-Pl 52, 338–347, doi: 10.1007/s11627-016-9768-3 (2016). [DOI] [Google Scholar]
- Tadege M. et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant Journal 54, 335–347, doi: 10.1111/j.1365-313X.2008.03418.x (2008). [DOI] [PubMed] [Google Scholar]
- d’Erfurth I. et al. Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. The Plant journal: for cell and molecular biology 34, 95–106 (2003). [DOI] [PubMed] [Google Scholar]
- Mazier M. et al. Successful gene tagging in lettuce using the Tnt1 retrotransposon from tobacco. Plant Physiology 144, 18–31, doi: 10.1104/pp.106.090365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A. et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15, 1771–1780, doi: 10.1105/tpc.012559 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y. Y. et al. Tnt1 Retrotransposon Mutagenesis: A Tool for Soybean Functional Genomics. Plant Physiology 161, 36–47, doi: 10.1104/pp.112.205369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C. M., Comai L., Greene E. A. & Henikoff S. Targeted screening for induced mutations. Nature Biotechnology 18, 455–457 (2000). [DOI] [PubMed] [Google Scholar]
- Henikoff S., Till B. J. & Comai L. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiology 135, 630–636, doi: 10.1104/pp.104.041061 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. et al. TILLING in Lotus japonicus Identified Large Allelic Series for Symbiosis Genes and Revealed a Bias in Functionally Defective Ethyl Methanesulfonate Alleles toward Glycine Replacements. Plant Physiology 151, 1281–1291, doi: 10.1104/pp.109.142190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C. et al. Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol J 7, 430–441, doi: 10.1111/j.1467-7652.2009.00410.x (2009). [DOI] [PubMed] [Google Scholar]
- Till B. J. et al. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol 7, 19, doi: 10.1186/1471-2229-7-19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. L., Henikoff S., Comai L. & Till B. J. In Rice Protocols (ed. Yinong Yang) 39–56 (Humana Press, 2013). [Google Scholar]
- Cooper J. L. et al. TILLING to detect induced mutations in soybean. BMC Plant Biol 8, 9, doi: 10.1186/1471-2229-8-9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia S. et al. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res Notes 3, 69, doi: 10.1186/1756-0500-3-69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C. et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol 9, 115, doi: 10.1186/1471-2229-9-115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito V. A. et al. A gene expression atlas of the model legume Medicago truncatula. The Plant journal: for cell and molecular biology 55, 504–513, doi: 10.1111/j.1365-313X.2008.03519.x (2008). [DOI] [PubMed] [Google Scholar]
- He J. et al. The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10, 441, doi: 10.1186/1471-2105-10-441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin M. J. In Current Protocols in Bioinformatics (John Wiley & Sons, Inc., 2002). [Google Scholar]
- Kalderimis A. et al. InterMine: extensive web services for modern biology. Nucleic Acids Res 42, W468–472, doi: 10.1093/nar/gku301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.