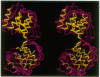
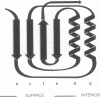
Abstract
5-enol-Pyruvylshikimate-3-phosphate synthase (EPSP synthase; phosphoenolpyruvate:3-phosphoshikimate 1-carboxyvinyltransferase, EC 2.5.1.19) is an enzyme on the pathway toward the synthesis of aromatic amino acids in plants, fungi, and bacteria and is the target of the broad-spectrum herbicide glyphosate. The three-dimensional structure of the enzyme from Escherichia coli has been determined by crystallographic techniques. The polypeptide backbone chain was traced by examination of an electron density map calculated at 3-A resolution. The two-domain structure has a distinctive fold and appears to be formed by 6-fold replication of a protein folding unit comprising two parallel helices and a four-stranded sheet. Each domain is formed from three of these units, which are related by an approximate threefold symmetry axis; in each domain three of the helices are completely buried by a surface formed from the three beta-sheets and solvent-accessible faces of the other three helices. The domains are related by an approximate dyad, but in the present crystals the molecule does not display pseudo-symmetry related to the symmetry of point group 32 because its approximate threefold axes are almost normal. A possible relation between the three-dimensional structure of the protein and the linear sequence of its gene will be described. The topological threefold symmetry and orientation of each of the two observed globular domains may direct the binding of substrates and inhibitors by a helix macrodipole effect and implies that the active site is located near the interdomain crossover segments. The structure also suggests a rationale for the glyphosate tolerance conferred by sequence alterations.
Full text
PDF


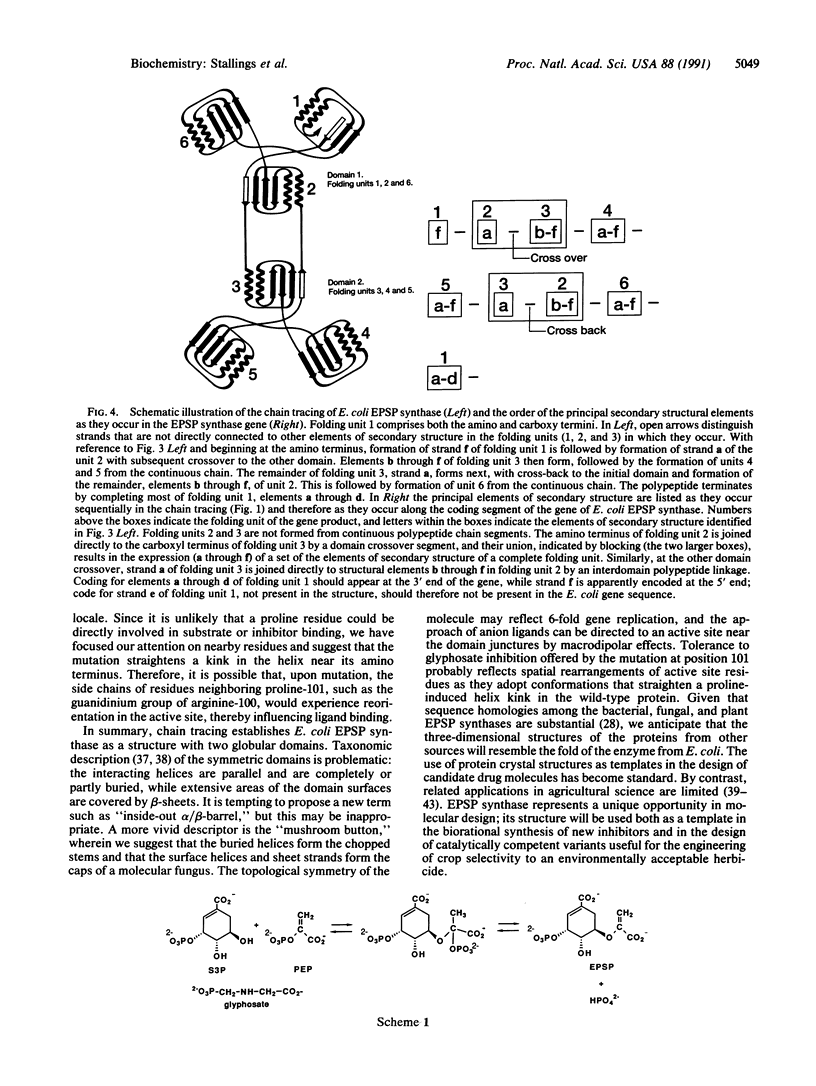
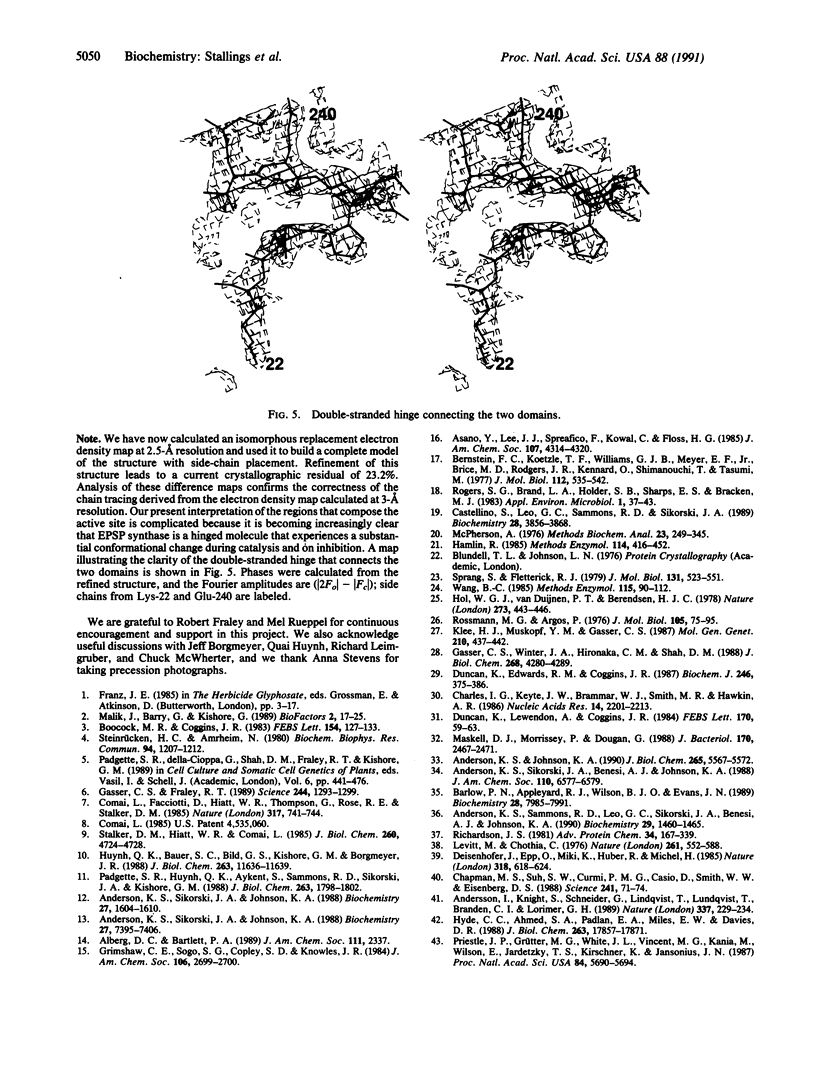
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson K. S., Johnson K. A. "Kinetic competence" of the 5-enolpyruvoylshikimate-3-phosphate synthase tetrahedral intermediate. J Biol Chem. 1990 Apr 5;265(10):5567–5572. [PubMed] [Google Scholar]
- Anderson K. S., Sammons R. D., Leo G. C., Sikorski J. A., Benesi A. J., Johnson K. A. Observation by 13C NMR of the EPSP synthase tetrahedral intermediate bound to the enzyme active site. Biochemistry. 1990 Feb 13;29(6):1460–1465. doi: 10.1021/bi00458a017. [DOI] [PubMed] [Google Scholar]
- Anderson K. S., Sikorski J. A., Johnson K. A. A tetrahedral intermediate in the EPSP synthase reaction observed by rapid quench kinetics. Biochemistry. 1988 Sep 20;27(19):7395–7406. doi: 10.1021/bi00419a034. [DOI] [PubMed] [Google Scholar]
- Anderson K. S., Sikorski J. A., Johnson K. A. Evaluation of 5-enolpyruvoylshikimate-3-phosphate synthase substrate and inhibitor binding by stopped-flow and equilibrium fluorescence measurements. Biochemistry. 1988 Mar 8;27(5):1604–1610. doi: 10.1021/bi00405a032. [DOI] [PubMed] [Google Scholar]
- Barlow P. N., Appleyard R. J., Wilson J. O., Evans J. N. Direct observation of the enzyme-intermediate complex of 5-enolpyruvylshikimate-3-phosphate synthase by 13C NMR spectroscopy. Biochemistry. 1989 Oct 3;28(20):7985–7991. doi: 10.1021/bi00446a003. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Boocock M. R., Coggins J. R. Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 1983 Apr 5;154(1):127–133. doi: 10.1016/0014-5793(83)80888-6. [DOI] [PubMed] [Google Scholar]
- Chapman M. S., Suh S. W., Curmi P. M., Cascio D., Smith W. W., Eisenberg D. S. Tertiary structure of plant RuBisCO: domains and their contacts. Science. 1988 Jul 1;241(4861):71–74. doi: 10.1126/science.3133767. [DOI] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Smith M., Hawkins A. R. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res. 1986 Mar 11;14(5):2201–2213. doi: 10.1093/nar/14.5.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Edwards R. M., Coggins J. R. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem J. 1987 Sep 1;246(2):375–386. doi: 10.1042/bj2460375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser C. S., Fraley R. T. Genetically engineering plants for crop improvement. Science. 1989 Jun 16;244(4910):1293–1299. doi: 10.1126/science.244.4910.1293. [DOI] [PubMed] [Google Scholar]
- Gasser C. S., Winter J. A., Hironaka C. M., Shah D. M. Structure, expression, and evolution of the 5-enolpyruvylshikimate-3-phosphate synthase genes of petunia and tomato. J Biol Chem. 1988 Mar 25;263(9):4280–4287. [PubMed] [Google Scholar]
- Hamlin R. Multiwire area X-ray diffractometers. Methods Enzymol. 1985;114:416–452. doi: 10.1016/0076-6879(85)14029-2. [DOI] [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Huynh Q. K., Bauer S. C., Bild G. S., Kishore G. M., Borgmeyer J. R. Site-directed mutagenesis of Petunia hybrida 5-enolpyruvylshikimate-3-phosphate synthase: Lys-23 is essential for substrate binding. J Biol Chem. 1988 Aug 25;263(24):11636–11639. [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Klee H. J., Muskopf Y. M., Gasser C. S. Cloning of an Arabidopsis thaliana gene encoding 5-enolpyruvylshikimate-3-phosphate synthase: sequence analysis and manipulation to obtain glyphosate-tolerant plants. Mol Gen Genet. 1987 Dec;210(3):437–442. doi: 10.1007/BF00327194. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Malik J., Barry G., Kishore G. The herbicide glyphosate. Biofactors. 1989 Mar;2(1):17–25. [PubMed] [Google Scholar]
- Maskell D. J., Morrissey P., Dougan G. Cloning and nucleotide sequence of the aroA gene of Bordetella pertussis. J Bacteriol. 1988 Jun;170(6):2467–2471. doi: 10.1128/jb.170.6.2467-2471.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A., Jr The growth and preliminary investigation of protein and nucleic acid crystals for X-ray diffraction analysis. Methods Biochem Anal. 1976;23(0):249–345. doi: 10.1002/9780470110430.ch4. [DOI] [PubMed] [Google Scholar]
- Padgette S. R., Huynh Q. K., Aykent S., Sammons R. D., Sikorski J. A., Kishore G. M. Identification of the reactive cysteines of Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase and their nonessentiality for enzymatic catalysis. J Biol Chem. 1988 Feb 5;263(4):1798–1802. [PubMed] [Google Scholar]
- Posnett D. N., Marboe C. C., Knowles D. M., 2nd, Jaffe E. A., Kunkel H. G. A membrane antigen (HC1) selectively present on hairy cell leukemia cells, endothelial cells, and epidermal basal cells. J Immunol. 1984 Jun;132(6):2700–2702. [PubMed] [Google Scholar]
- Priestle J. P., Grütter M. G., White J. L., Vincent M. G., Kania M., Wilson E., Jardetzky T. S., Kirschner K., Jansonius J. N. Three-dimensional structure of the bifunctional enzyme N-(5'-phosphoribosyl)anthranilate isomerase-indole-3-glycerol-phosphate synthase from Escherichia coli. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5690–5694. doi: 10.1073/pnas.84.16.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Rogers S. G., Brand L. A., Holder S. B., Sharps E. S., Brackin M. J. Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl Environ Microbiol. 1983 Jul;46(1):37–43. doi: 10.1128/aem.46.1.37-43.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Argos P. Exploring structural homology of proteins. J Mol Biol. 1976 Jul 25;105(1):75–95. doi: 10.1016/0022-2836(76)90195-9. [DOI] [PubMed] [Google Scholar]
- Sprang S., Fletterick R. J. The structure of glycogen phosphorylase alpha at 2.5 A resolution. J Mol Biol. 1979 Jul 5;131(3):523–551. doi: 10.1016/0022-2836(79)90006-8. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Hiatt W. R., Comai L. A single amino acid substitution in the enzyme 5-enolpyruvylshikimate-3-phosphate synthase confers resistance to the herbicide glyphosate. J Biol Chem. 1985 Apr 25;260(8):4724–4728. [PubMed] [Google Scholar]
- Steinrücken H. C., Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun. 1980 Jun 30;94(4):1207–1212. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]