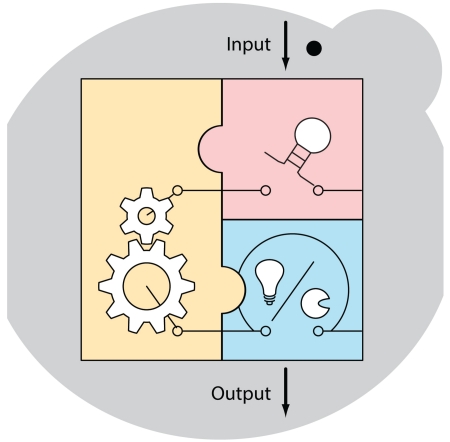
Summary
For synthetic biology applications, protein-based transcriptional genetic controllers are limited in terms of orthogonality, modularity, and portability. Although ribozyme-based switches can address these issues, their current two-stage architectures and limited dynamic range hinder their broader incorporation into systems-level genetic controllers. Here, we address these challenges by implementing an RNA-protein hybrid controller with a three-stage architecture that introduces a transcription-based amplifier between an RNA sensor and a protein actuator. To facilitate the construction of these more complex circuits, we use a model-guided strategy to efficiently match the activities of stages. The presence of the amplifier enabled the three-stage controller to have up to 200-fold higher gene expression than its two-stage counterpart and made it possible to implement higher-order controllers, such as multilayer Boolean logic and feedback systems. The modularity inherent in the three-stage architecture along with the sensing flexibility of RNA devices presents a generalizable framework for designing and building sophisticated genetic control systems.
Graphical abstract
Introduction
Engineered RNA switches can be used for various systems-level applications such as controlling virus replication (Bell et al., 2015) or guiding cell fates (Galloway et al., 2013). They exhibit compositional modularity (Townshend et al., 2015; Win and Smolke, 2007, 2008), sensing orthogonality (McKeague et al., 2015), and cross-species portability (Wei et al., 2013) and can provide a preferable alternative to protein-based transcriptional controllers, which are limited in these characteristics. One class of engineered RNA switches, consisting of a ligand-binding aptamer and a self-cleaving ribozyme, controls gene expression in response to a small molecule ligand by regulating mRNA levels through co- or post-transcriptional mechanisms in vivo (Liang et al., 2011). Typically, these ribozyme switch-based genetic controllers are implemented in a simple two-stage architecture: the RNA switch sensor directly transduces external signals to a protein actuator to perform desired functions (Figure 1A). To obtain a functional genetic controller, the activity range of the protein actuators must be matched to the dynamic range of the RNA switch. Strategies for such “level matching” include dynamic tuning (Galloway et al., 2013) or library-based screening of individual components (Liang et al., 2012).
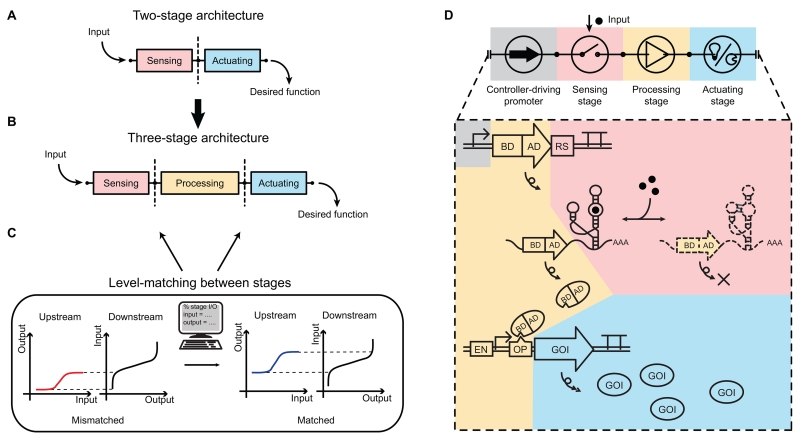
Figure 1. Three-stage controller architecture with model-facilitated level matching and the design of an RNA-based gene activation controller.
(A) Two-stage controller architecture with a sensing stage (pink) and actuating stage (blue).
(B) Three-stage controller architecture with a processing stage (yellow) between sensing and actuating stages.
(C) Level matching between stages. Mismatched levels (left, red and black curves) between stages versus computationally matched levels between stages (right, blue and black curves).
(D) Circuit diagram (top) and biological implementation (bottom) of a gene activation controller. Pink: sensor, small molecule-responsive RNA switches. Yellow: processor, transcription-based amplifiers. Blue: actuator, gene products. Biological elements: DNA (double lines), RNA (single lines), and proteins (ovals).
Abbreviations: AD: activation domain; BD: DNA-binding domain; RS: RNA switch; TA: transcription-based amplifier; EN: epigenetic enhancer; OP: operator site; GOI: gene-of-interest.
Although the ribozyme switch-based, two-stage architecture is straightforward for implementing inducible genetic control systems, it restricts the design of systems for more sophisticated applications. The first challenge is the relatively limited dynamic range of ribozyme switch-based controllers (Beisel et al., 2008; Wang et al., 2013). The use of inducible protein-based transcriptional controllers (e.g., Tet-ON device) as an alternative to RNA switches might allow for increased dynamic range; however, the engineering of these protein-based transcriptional controllers to exhibit different sensing or regulatory properties can be very challenging (Golynskiy et al., 2011). The second challenge is the lack of design freedom, as researchers must compromise between optimization for sensing function to the intracellular levels of the input signals and the activity of its gene regulatory function to the downstream actuating stage. Increasing the number of system stages can potentially relax this design constraint; however, the trade-off lies in the increased complexity to obtain matched activity levels between each stage.
Here, we propose a design framework that incorporates a ribozyme switch-based, three-stage genetic controller architecture with a model-guided approach to facilitate the level matching between stages (Figure 1B). We insert a transcription-based amplifier as the processing stage between the sensing and actuating stages to increase the overall system dynamic range and reduce the need to compromise between optimizing the sensor or the actuator. We begin with implementing a gene activation controller and use a data-driven approach to derive a predictive in silico model (Figure 1C), from which we obtain matched activity levels between stages in the controller with a large dynamic range that can drive sufficient expression to switch a cellular live-dead phenotype. We show that the designed activation controller possesses plug-and-play modularity at each stage, and can be extended to more complicated control systems. Our framework is further used to implement systems-level feedback controllers through an enzymatic loop. This system architecture provides design flexibility allowing alteration of the biological components at each stage, providing a generalizable approach to implement sophisticated control functions.
Results
Three-stage design framework for a gene activation controller
We started with the design of a gene activation controller – a fundamental regulatory system that responds to external signals and initiates gene expression (i.e., inducible gene-expression system). This framework comprises three stages: sensing, processing, and actuating (circuit symbol in Figure 1D). Small molecule-responsive RNA switches were selected for the sensing stage as they respond to a variety of inputs (Townshend et al., 2015). The processing stage performs genetic signal amplification, and was implemented with a transcription-based amplifier (or “amplifier”) to enable a broad range of gene expression in the activation controller. A protein actuator was used for the actuating stage to support diverse outputs, such as signal reporting, cellular phenotype, and enzyme activity. We implemented the proposed activation controller with two genetic cassettes: 1) a cassette comprising a promoter (referred to as “controller-driving promoter”; grey in Figure 1D) to express the trans element of the amplifier (yellow in Figure 1D) and an RNA switch (pink in Figure 1D) inserted in the 3′ UTR (untranslated region); 2) a cassette comprising the cis hybrid promoter of the amplifier to drive the expression of the protein actuator (blue in Figure 1D). In the absence of the input, the RNA switch adopts a cleavage-active conformation that increases the transcript degradation rate. The reduced levels of the trans element of the amplifier, encoded in the RNA switch-regulated transcript, results in reduced expression of the downstream protein actuator. Binding of the input to the RNA switch stabilizes the transcript, leading to higher levels of the trans element of the amplifier and increased levels of the protein actuator.
A processing stage expands the system dynamic range
We first engineered the transcription-based amplifier in the processing stage of the framework. Ideal amplifiers exhibit a large dynamic range, amplification gain (Figure S1A), and are orthogonal to the target inputs. We implemented a two-element transcription-based amplifier (Ajo-Franklin et al., 2007; Khalil et al., 2012): a trans synthetic transcription factor comprising a DNA-binding domain (BD; implemented with a bacterial protein LexA) fused to an activation domain (AD; implemented with a viral activator VP16) and nuclear localization signal (NLS); a cis hybrid promoter comprising operator (OP; implemented with LexAO) sites for binding and a minimal promoter (miniP; implemented with miniPGAL1) for transcription initiation (Figure 2A; see “Pre-optimized transcription-based amplifier design” in Document S1). Our initial amplifier exhibits no amplification activity (gain of ~0.4; defined as the expression ratio in the presence vs. absence of the amplifier) and a low expression level with large variability, necessitating the design of new amplifiers. We hypothesized that increasing the gene expression efficiency could improve the amplifier gain, specifically through expressing a more efficient trans element at low levels with a less-regulated cis hybrid promoter. We built an improved trans element by codon optimizing both BDLexA and ADVP16 for expression in our host (Saccharomyces cerevisiae). In addition, the BDLexA and ADVP16 were connected through a peptide linker to enhance protein folding (George and Heringa, 2002) and the NLS was moved from the C- to N-terminus for better nuclear transport (Kalderon et al., 1984). We built an improved cis element by interspacing the two LexAO sites with nucleosome-disfavoring sequences (NDS; “TTTTT”) (Raveh-Sadka et al., 2012), incorporating an upstream enhancer (EN) to reduce epigenetic silencing (Gaszner and Felsenfeld, 2006), and mutating the Mig1 inhibitory sites in the miniPGAL1 (Lundin et al., 1994) (boxes in Figure 2A). A reporter cassette was built using the redesigned cis hybrid promoter (PLexAO) to express an EGFP reporter, and integrated into the yeast chromosome (his3) to decrease expression variability.
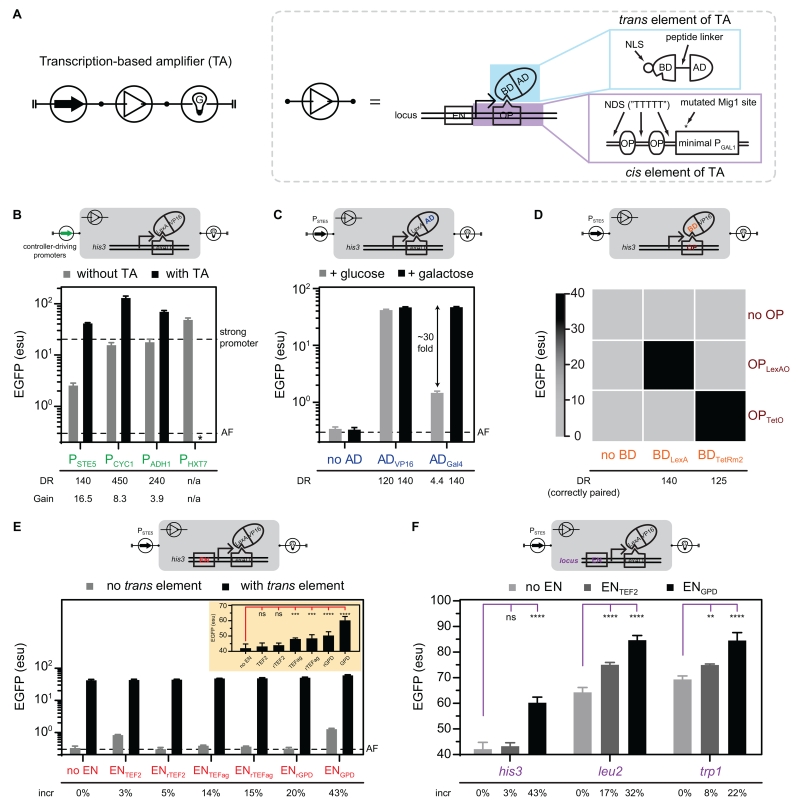
Figure 2. Design and optimization of the transcription-based amplifier.
(A) Circuit symbol (left) and compositional domains of a transcription-based amplifier (dash box). The trans element of the amplifier has (blue box) a nuclear localization signal (NLS), a DNA-binding domain (BD), a peptide linker, and an activation domain (AD). The cis element of the amplifier has (purple box) an upstream epigenetic enhancer (EN), two operator (OP) sites interspaced with nucleosome-disfavoring sequences (NDSs), and a minimal promoter from PGAL1 with mutated Mig1 site.
(B) Gain and dynamic range of the transcription-based amplifier under different controller-driving promoters. Both “with amplifier” (amplifier regulating EGFP) and “without amplifier” (controller-driving promoter directly regulating EGFP) were characterized. Bars represent mean values and error bars represent ± 1 s.d. of three biological replicates. The amplification gain (Gain) and dynamic range (DR) are displayed. Two dashed lines represent the expression levels of PTEF1 integrated at his3, and empty strain as the autofluorescence level (“AF”; CSY3). *No viable cells were observed when transforming the PHXT7-driven amplifier into yeast, which can be due to growth inhibition under excessive expression of the trans element of the amplifier. See also Figure S1A.
(C) Examination of the AD modularity by extending the BDLexA-based transcription-based amplifier to different ADs (ADVP16 or ADGal4). The constructs were characterized as Figure 2B, except two carbon sources (glucose or galactose) were supplied in the media. Bars represent mean values and error bars represent ± 1 s.d. of three biological replicates, and the dynamic range is displayed. See also Figure S1C.
(D) Examination of the BD modularity and orthogonality by extending the transcription-based amplifier to two trans elements (BDLexA and BDTetRm2) and their cognate cis elements. The constructs were characterized as Figure 2B. Mean values of three biological replicates for each construct are represented in the heat map, and the dynamic range is displayed only for the construct on the diagonal (correctly paired). See also Figures S1D and S2.
(E) The impact of EN on gene expression levels. Three ENs (ENGPD, ENTEF2, and ENTEFag) were inserted upstream of the cis element (CSY1101) of the BDLexA-based amplifier, in either native or reverse orientations. Either an empty plasmid (no trans element) or a trans element-harboring plasmid (with trans element) was transformed into the strains with EN-modulated cis reporters, and characterized as Figure 2B. Bars represent mean values and error bars represent ± 1 s.d. of three biological replicates, and the percentage increase in EGFP levels (“incr”; setting no EN to 0%) is displayed. The yellow inset illustrates the result of “with trans element” in linear scale for better visualization. ANOVA test was performed with an ad hoc Dunnett’s test. See also Figure S1E.
(F) Examination of EN modularity at distinct genomic loci. The EN-modulated cis reporters were integrated into three loci (his3, leu2, and trp1), and were characterized as Figure 2B. The y-axis is in linear scale for better visualization. Bars represent mean values and error bars represent ± 1 s.d. of three biological replicates, and the percentage increase in EGFP levels (“incr”; setting no EN in each locus to 0%) is displayed. ANOVA test was performed with an ad hoc Dunnett’s test.
Significance summary: p > 0.05 (ns), p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***), and p ≤ 0.0001 (****).
We established a standard unit (“esu”, external standard unit) to enable quantitative comparison of expression activities across experiments (Kelly et al., 2009). In each experiment, a reference construct (PTEF1 expressing EGFP on a low-copy plasmid; CSY3 + pCS2212) was measured in parallel and its fluorescence scaled to 100 esu. We expressed the new trans element (BDLexA-ADVP16) under four controller-driving promoters and measured their EGFP levels via flow cytometry (Figure 2B). The re-designed amplifier exhibits a large dynamic range (~450-fold) with an OFF state at the autofluorescence detection limit and an ON state that is 6-fold stronger than a strong promoter PTEF1 (integrated at his3; CSY1110). Moreover, the gain of the re-designed amplifier was larger than 1 (i.e., amplifying) under most controller-driving promoters. The largest gain was observed when using the weakest promoter (PSTE5), confirming our hypothesis that amplification efficiency affects gains.
Design modularity and orthogonality expands and improves amplifier performance
We demonstrated the modularity of the amplifier design by interchanging three compositional domains (AD, BD, EN). We started with AD modularity and substituted the ADVP16 with an AD from a yeast transcription factor Gal4 (ADGal4) (Traven et al., 2006). We tested the AD-replaced amplifier (BDLexA-ADGal4) and observed comparable ON states between the two ADs (Figure 2C). The ADVP16-based amplifier shows no significant dependency on the carbon sources examined; while the ADGal4-based amplifier exhibits a 30-fold reduction in the absence of galactose. This galactose-dependency may result from the repressor (Gal80p) binding domain at the C-terminus of ADGal4, in which the repressor is released in the presence of galactose (Lavy et al., 2012; Melcher and Xu, 2001).
We confirmed the BD modularity by replacing the BD in the trans element and the OP sites in the cis element. We selected the TetR protein as a candidate BD due to its high binding affinity to its OP sites (TetO) (Gossen and Bujard, 1992); however, TetR is responsive to tetracycline and thus introduces undesirable input-dependent gene-regulatory interference in our amplifier. We employed site-directed mutagenesis and DNA shuffling (Zhao and Arnold, 1997) to evolve a TetO-binding TetR variant (TetRm2) that is unresponsive to tetracycline (Figure S2), and used TetRm2 to construct a new amplifier (BDTetRm2-ADVP16). The results indicate that the BDTetRm2-based amplifier exhibits similar dynamic range and AD modularity as the BDLexA-based amplifier (Figure S1C). To verify the potential of using these amplifiers in a single genetic control system, we tested their orthogonality by pairing the BDs with each OP site. We observed strong amplification specificity between the cognate BD and OP, and no activity when the BD and OP were mispaired (Figures 2D and S1D).
We increased the dynamic range of the transcription-based amplifier by using modular enhancers to reduce epigenetic silencing. Three epigenetic enhancer sequences (ENGPD, ENTEF2, ENTEFag) (Bi and Broach, 1999; Bitter and Egan, 1984) were identified and placed in native or reverse orientation ~60 bp upstream of the first operator site in the cis hybrid promoter. Two enhancer sequences (ENGPD, ENTEFag) increased gene expression (14% to 40%) regardless of their orientation (p < 0.001), which is a typical characteristic of epigenetic regulators (Gaszner and Felsenfeld, 2006) (Figure 2E). The enhancers by themselves possess no or very limited activity in initiating transcription (p > 0.05), suggesting that the increase in expression is not due to tandem promoter repeats (Blazeck et al., 2012). We further examined the modularity of the enhancers (ENGPD, ENTEF2) by integrating the reporter cassettes at different genomic loci (his3, leu2, trp1). ENGPD was observed to be functional (p < 0.0001) in each integration locus, whereas ENTEF2 was functional only at leu2 and trp1 (Figure 2F).
Finally, we explored the effect of the distance between EN and its cis hybrid promoter on promoter strength. We designed randomized spacer sequences with %GC matched to the yeast genome (Zhu et al., 2014), and inserted them between ENGPD and PLexAO before integrating the construct into the trp1 locus. Four strains (with 0, 100, 200, 300 bp spacers, making the final distance 60, 160, 260, 360 bp) were characterized in the presence and absence of the trans element. The results indicate that an EN-promoter distance of 160 bp (the 100-bp spacer; CSY1141) can increase the expression by 6.6-fold (Figure S1E), while maintaining a low expression level in the absence of the trans element. Through this optimization process, we were able to obtain an improved amplifier design with increased amplification gains (from 0.4 to 190) and significantly reduced variability (Figure S1B).
Model-guided level matching to connect individual stages into an activation controller
We continued to demonstrate the construction of a three-stage activation controller with EGFP as a reporter (i.e., an EGFP-activator) (Figure 3A). One major challenge for assembling such an activation controller is to efficiently obtain matched activity levels between stages for optimal performance, commonly referring as “level matching” (defined as “the process of matching the input and output levels between stages in a system, such that the output level from the upstream stage is sufficient as an input to its downstream stage” (Wang et al., 2013)). To address this, we developed a modeling approach to facilitate the level-matching process. The modeling procedure involves characterizing individual components in vivo to derive data-driven models. Then, an in silico system is assembled from the models of individual components, together with a Monte Carlo algorithm to generate level-matching results at various conditions (see STAR Method).
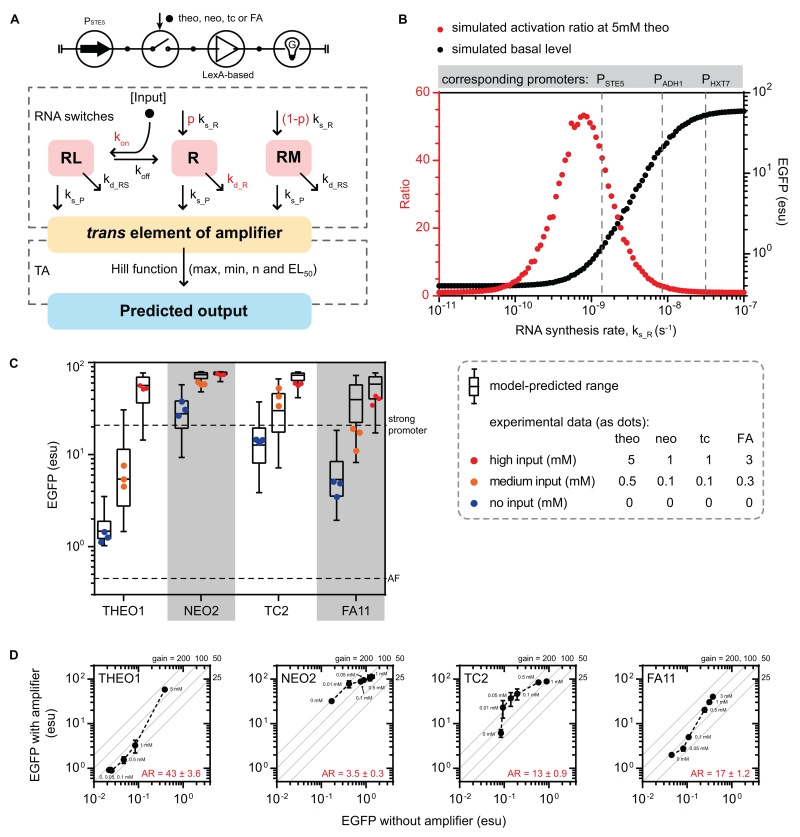
Figure 3. A model-guided approach to facilitate the design of the EGFP-activator.
(A) Circuit symbol (top) and in silico model (bottom) of the EGFP-activator. The model of the EGFP-activator comprises of an RNA switch model (first dashed box) with a transcription-based amplifier model (second dashed box). See also STAR Methods for modeling details.
(B) Applying the modeling procedure to facilitate the level-matching process between stages. The performance metrics of the modeled controller (THEO1-BDLexA-based EGFP-activator) was evaluated under a range of RNA synthesis rate (ks_R from 10−11 to 10−7 Ms−1). The median values (Monte Carlo, N = 1000) of the simulated activation ratio (red) and basal level (black) are plotted. Three in vivo characterized conditions (PSTE5, PADH1, and PHXT7) are displayed as dashed lines. See also Figure S3A.
(C) Comparison of the predicted and experimentally measured performance of the EGFP-activators. The modeling procedure (Monte Carlo, N = 5000; ks_R = 10−9 s−1) was applied to four EGFP-activators (with RNA switches THEO1, NEO2, TC2, and FA11). The predicted range of EGFP is represented as a box-and-whisker plot (bars representing: min, first quartile, median, third quartile, and max). EGFP level was measured after growing cells with added inputs for 6 hours, and is plotted against the predicted expression range (dots as individual replicates).
(D) Amplification gains exhibited by the EGFP-activators. For each EGFP-activator (“with amplifier”), a corresponding construct was built using the RNA switch to directly regulate EGFP expression (without amplifier). The constructs were characterized as Figure 3C. Each dot represents an input level (with the concentration labeled alongside), and error bars represent ± 1 s.d. of three biological replicates. Diagonal lines represent the levels of constant gain. See also Figure S3B.
Abbreviations: AR: activation ratio. In the RNA switch model - R, RL and RM: cleavable, input-bound non-cleavable, and misfolded non-cleavable RNA switch states in an mRNA; ks_R, and ks_P: synthesis rates of mRNA and protein; kd_R, kd_RS, and kd_P: degradation rates cleavable RNA, non-cleavable RNA, and protein; p: mRNA folding partition coefficient; kon and koff: input-aptamer binding constants. In the amplifier model - max: saturation level; min: leakage level; n: cooperativity; EL50: half-maximal response.
We hypothesized that the strength of the promoter plays a critical role in matched activity level between the RNA switch and the amplifier, from which the EGFP-activator can exhibit desired properties (e.g., low basal level and high activation ratio). We built an in silico EGFP-activator system using the models of THEO1 switch and the BDLexA-based amplifier, and simulated its performance at a range of promoter strengths (by varying the mRNA synthesis rate, ks_R). The simulation results indicate that there is an optimal range of ks_R (0.5–2×10−9 s−1) for an EGFP-activator to exhibit a high activation ratio while keeping a low basal level (Figure 3B). We experimentally verified the model predictions by constructing the EGFP-activator with three controller-driving promoters (PSTE5, PADH1, PHXT7) with relative expression strengths of 1:7:20 (Figure 2B). Each EGFP-activator was built by placing the THEO1 switch in the 3′ UTR of the trans element of the amplifier on a low-copy plasmid, which was transformed into an EGFP reporter-integrated strain (CSY1113). We characterized these EGFP-activators at three input levels (0, 0.5, 5 mM theophylline), and the results indicate that only the PSTE5-driven EGFP-activator exhibits a high input-controlled activation (activation ratio of ~40; Figures 3C and S3A).
We then leveraged the modularity of the activation controller framework to replace the sensing stage with other RNA switches (TC2, NEO2, FA11). We updated the parameters and applied the modeling procedure to predict the performance of each EGFP-activator. The data indicate that substitution of the RNA switch in the framework results in a new EGFP-activator that responds to different inputs (Figure 3C). In addition, the predicted activity ranges of each EGFP-activator matched the experimental data, suggesting the modeling procedure can be extended to other RNA switches. Nearly all of the designed EGFP-activators retain strong ON states, and typical system activation ratios ranged from 10- to 40-fold (Figures 3D and S3B). Although most EGFP-activators exhibit high gains (~50 to 200), each ribozyme switch-based activator exhibits different amplification characteristics (Figure 3D). For example, the gain of the FA11-based activator is relatively constant and input-independent (between 50 to 100); while the gain of the NEO2-based activator exhibits a lowpass-like characteristic that amplifies selectively at low-to-medium input levels (~200 compared to ~80).
Using the gene activation controller to regulate cellular phenotypes
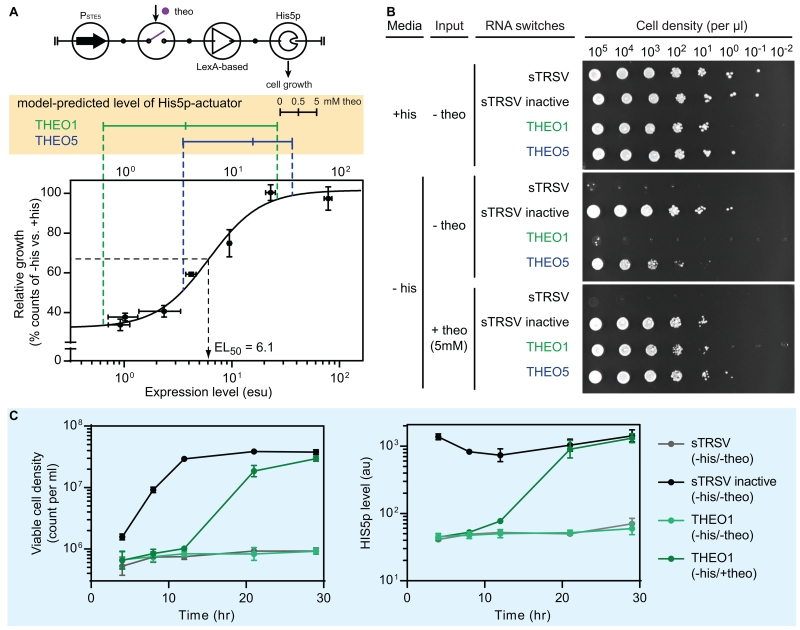
We tested the ability to modify the actuating stage of the activation controller framework to regulate phenotypes such as cell viability. We used an enzyme His5p (from Schizosaccharomyces pombe) as the actuator, whose expression can rescue the histidine auxotrophy of our strain and sustain cellular growth (Gueldener et al., 2002). To identify the growth-sustaining threshold level of His5p in yeast, His5p was fused to an N-terminal EGFP to enable quantitative measurement. We then developed a model describing cellular growth under different His5p levels (see STAR Method). The results indicate a clear transition from reduced cell growth (~30%) to resumed growth (~100%) across a two orders of magnitude range of His5p expression levels (Figure 4A). We used theophylline as our input with two previously tested activation controllers (THEO1, THEO5), and replaced the EGFP reporter with the EGFP-His5p fusion actuator. We first simulated the performance of these in silico His5p-activators (Figure S4A), and the results predict that the THEO1-based His5p-activator exhibits a His5p expression range that can shift the cells from reduced growth to nearly recovered growth (~30% to 95%; Figure 4A). In contrast, the THEO5-based His5p-activator exhibits a His5p expression range corresponding to higher basal growth at no input (50% to 98%; Figure 4A).
Figure 4. Application of the gene activation controller to control a cellular phenotype.
(A) Circuit symbol of the His5p-activator (top) and model-predicted His5p-activator performance. The His5p-dependent cell growth model (bottom) was obtained by fitting the cellular growth data (dots in the plot with error bars as ± 1 s.d.) with a Hill function. The protein levels of two His5p-activators (THEO1 in green and THEO5 in blue) were simulated (Monte Carlo, N = 5000) at three input levels (0, 0.5, and 5 mM of theophylline). The median values of the simulation results are displayed (yellow box). See also Figure S4A and STAR Methods for modeling details.
(B) A serial dilution plating experiment to assess input-controlled cellular growth. Yeast strains harboring the His5p-activators (sTRSV, sTRSV inactive, THEO1, and THEO5) were grown stationary phase, and diluted into a series (from 105 to 10−2 viable cells per μL). Two μL of each dilution were plated under three conditions: presence of the auxotrophic nutrient (+his, −theo), absence of nutrient and input (−his, −theo), and presence of input (−his, +theo). The assay was conducted in three biological replicates. See also Figure S4B.
(C) Time course cellular growth assays with His5p-activators. Strains harboring three His5p-activators (sTRSV, sTRSV inactive, and THEO1) were grown under two conditions: absence of nutrient and input (-his, -theo) and presence of input (-his, +theo). Viable cell counts and protein levels were measured over the course of experiment. Data are plotted as the mean values and error bars represent ± 1 s.d. of three biological replicates.
Abbreviations: his: histidine; theo: theophylline; sTRSV: small satellite RNA of tobacco ringspot virus (a self-cleavable ribozyme); sTRSV inactive: a non-cleavable mutant of sTRSV.
We validated the predictions by conducting a serial dilution experiment to show the viability of yeast under three conditions (histidine-only, theophylline-only, neither). We included two controls by replacing the RNA switch with either sTRSV (a self-cleaving ribozyme) or sTRSV inactive (a non-cleaving mutant ribozyme). In the histidine-only (+his, −theo) condition, we observed normal cell growth independent of the harbored His5p-activator; whereas in the −his, −theo condition, only cells harboring the sTRSV inactive-based His5p-activator produced sufficient histidine to sustain normal growth (Figures 4B and S4B). The cells harboring the THEO1-based His5p-activator exhibit a strong shift from no to full viability in the presence of input signal (theophylline), while the cells harboring the THEO5-based His5p-activator exhibit slightly improved viability, confirming the predictions of the model. To further demonstrate the advantages of a three-stage architecture, we implemented a functionally equivalent two-stage controller by placing the THEO1 switch directly in the 3′ UTR of the His5p actuator. The two-stage His5p-activator was found to exhibit a reduced device performance; specifically a lower dynamic range and higher basal level of growth (see “Two-stage device controls” in Document S1).
We then demonstrated that the optimal THEO1-based His5p-activator could shift yeast cells from minimal growth (i.e., sTRSV-like) to fully resumed growth (i.e., sTRSV inactive-like) upon induction. We performed a time course experiment in liquid media (-his) and measured the growth of the cells in the absence and presence of the input. The results indicate a strong input-dependent growth phenotype, and the density of cells harboring the THEO1-based His5p-activator was ~80% of that of cells with constant His5p expression (i.e., harboring sTRSV inactive) (left plot in Figure 4C). Moreover, the expression level of the phenotypic actuator (His5p) was increased upon induction, suggesting the resumed growth was a result of the His5p-activator function (right plot in Figure 4C).
Extending the RNA-based framework to open-loop controllers
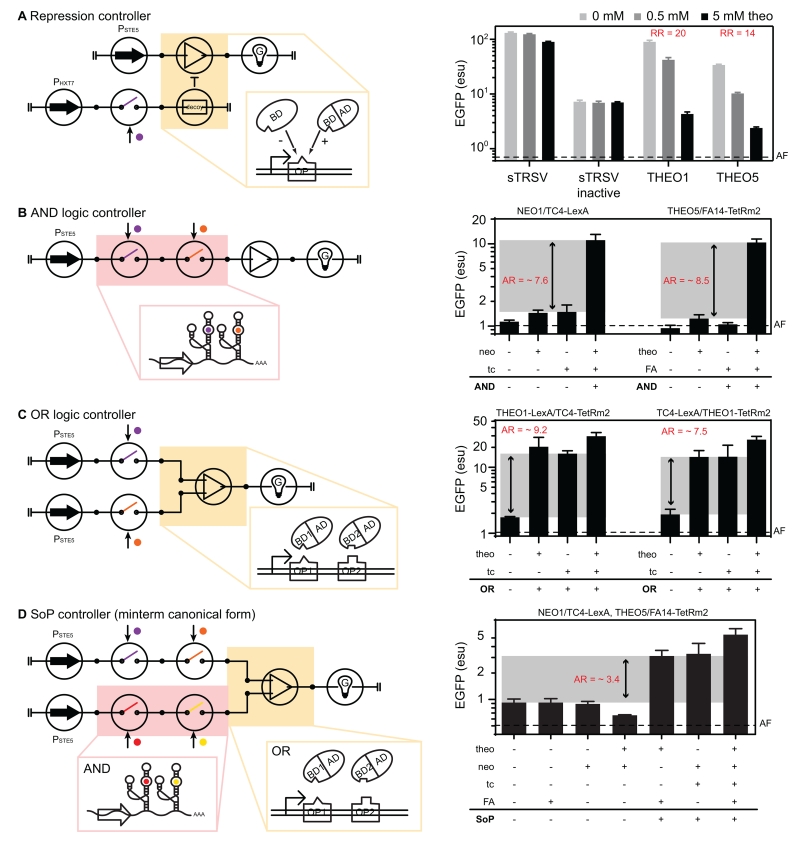
The gene activation controller framework can be extended to achieve more sophisticated control systems such as a repression or multi-input logic controller. We first examined extension of the processing stage of the framework to reverse regulatory mechanisms, specifically a repression controller that responds to external input to inhibit gene expression. We introduced a BD-only decoy factor in the processing stage, whose binding to the hybrid promoter inhibits the binding of the designed transcription factor for activation (Figure 5A). We used an RNA switch to regulate the decoy factor while keeping the transcription factor constantly expressed. As the input increases, more decoys are expressed to compete with the functional transcription factor for binding the OP sites in the cis hybrid promoter (i.e., competitive binding mechanism), leading to a stronger repression of gene expression. We applied these modifications to a BDLexA-based EGFP-activator (CSY1113 + pCS3442) and inserted two RNA switches (THEO1, THEO5) and two controls (sTRSV, sTRSV inactive) into the 3′ UTR of the decoy factor (BDLexA). The results demonstrate that the EGFP levels are different (~20-fold; p < 0.05 under paired t-test) between the presence of BDLexA (sTRSV inactive) and the absence of BDLexA (sTRSV) (Figure 5A), which confirmed the competitive binding mechanism used in the repression controller. The THEO1- and THEO5-based repressors exhibit strong input-controlled inhibition (20- and 14-fold reduction ratio; see definition in Document S1), which is comparable to the activation ratio of the designed activators.
Figure 5. Design of higher-order genetic control systems through expanding the framework.
(A) Circuit symbol of the repression controller (left). Four EGFP repressor-harboring strains (sTRSV, sTRSV inactive, THEO1, THEO5) were characterized as Figure 3C, except that EGFP was measured after 12 hours (right). Bars represent the mean values and error bars represent ± 1 s.d. of three biological replicates, and the RR is displayed.
(B-D) Boolean controller designs. Circuit symbol of each controller is displayed on the left: (B) AND controller, (C) OR controller, and (D) Sum-of-Product (SoP) controller. Different types of Boolean controllers were designed – two AND controllers (NEO1/TC4-BDLexA-based AND and THEO5/FA14-BDTetRm2-based AND), two OR controllers (THEO1-BDLexA/TC4-BDTetRm2-based OR and TC4-BDLexA/THEO1-BDTetRm2-based OR), and one SoP controller (NEO1/TC4-BDLexA/THEO5/FA14-BDTetRm2-based SoP). The constructs were characterized as Figure 3C, except that EGFP was measured after 24 hours (right). Bars represent the mean values and error bars represent ± 1 s.d. of three biological replicates. The Boolean AR (defined as the worst-case AR) is indicated.
Abbreviations: AR: activation ratio; RR: reduction ratio.
Next, we extended the framework to support the design of controllers that execute single-function Boolean logic (such as AND, OR) or combinatorial logic. We expanded the sensing stage to create the AND function by connecting distinct input-responsive RNA switches in series, such that the transcript is only stabilized in the presence of both inputs (Figure 5B) (Win and Smolke, 2008). The OR function was implemented at the processing stage by inserting different OP sites in the cis hybrid promoter, which becomes responsive to either transcription factor (Figure 5C). Finally, we combined the AND and OR controllers to implement the “Sum-of-Products” (SoP) canonical form of Boolean logic (e.g., “(X AND Y) OR Z”), into which any Boolean functions can be simplified and expressed (Figure 5D)
We first constructed two ANDs (NEO1/TC4-LexA executing “neo AND tc”, and THEO5/FA14-TetRm2 executing “theo AND FA”) and two ORs (THEO1-LexA/TC4-TetRm2 and TC4-LexA/THEO1-TetRm2, both executing “theo OR tc”) with EGFP as the reporter. Both AND and OR controllers were characterized by growing each device-harboring strain in media without input signals and then back-diluted into combinations of inputs. These Boolean controllers were found to have an OFF state close to the autofluorescence detection limit (Figure 5B and 5C). In addition, the designed controllers exhibit Boolean activation ratios (see definition in Document S1) between 7.5- and 9.2-fold between their ON and OFF states. In contrast, functionally equivalent two-stage implementation of AND controllers exhibit reduced Boolean activation ratios (1.6- and 1.8-fold) and significantly higher basal expression levels (see ”Two-stage device controls” in Document S1). We then combined the ANDs and ORs into the SoP controller to execute the function “(neo AND tc) OR (theo AND FA)”, and characterized this controller using a similar approach. Compared to the single-logic controllers, the SoP controller exhibits a reduced Boolean activation ratio (3.4 fold) and OFF states higher than the autofluorescence detection limit (Figure 5D). However, the SoP controller exhibits ON state expression levels only in the presence of the correct input combinations (e.g., theo and FA), confirming its function as a Boolean controller to respond to logic-formulated combinatorial input signals.
Achieving systems-level closed-loop control
Finally, we demonstrated the use of this framework to build closed-loop systems with an enzyme at the actuating stage to enable feedback adjustment of the input level. We designed two fundamental closed-loop topologies (positive or negative feedback system) by substituting the EGFP reporter with an input-producing enzyme, acting as a “feedback wire” to influence the level of input. We replaced the EGFP reporter in the activation controller with an input-producing enzyme to achieve positive feedback (PFB). When the input reaches above a critical level in the PFB system, the activation controller is turned on and leads to increasing expression of the input-producing enzyme, which further drives the input level to its maximum limit (i.e., all-or-none effect). On the other hand, replacing the EGFP reporter in the repression controller with an input-producing enzyme results in a negative feedback (NFB) system. The presence of input in the NFB system turns on the repression controller to inhibit the enzyme expression, and reduces the input level with faster dynamics compared to the open-loop configuration (e.g., a simple repression controller) (Rosenfeld et al., 2002).
We implemented the PFB system with theophylline as the input and a theophylline-producing enzyme (yCDM8) (Michener and Smolke, 2012) as the actuator in the activation controller framework. To verify the enzymatic output activity, an open-loop yCDM8-activator was built by decoupling the enzymatic output and input, by using a tetracycline-responsive RNA switch (TC1) to replace the theophylline switch. Cells harboring the open-loop yCDM8-activator were fed the substrate (caffeine) and titrated with different input (tetracycline) levels, and theophylline levels in the media were measured. The results indicate that cells harboring the yCDM8-activator exhibit increased production of theophylline as tetracycline increased, which confirmed the activity of the yCDM8-activator as an open-loop inducible activation controller (Figure S5A). We next examined the yCDM8-activator in its closed-loop configuration (i.e., PFB system) by using theophylline-responsive RNA switches as our sensing stage and titrating theophylline. Isotope-labeled substrate (13C-caffeine) was used to distinguish the enzymatic product (13C-theophylline) from the fed input (12C-theophylline). Similar to the open-loop controller, we observed a positive input-output relationship between the enzymatic product and the fed input in the PFB system-harboring cells (Figure S5B). We applied the same approach to build a NFB system (i.e, a yCDM8-repressor with theophylline as input), and tested its function with 13C-caffeine as a substrate at different levels of fed 12C-theophylline. The results demonstrate that the 13C-theophylline decreased when the 12C-theophylline increased, suggesting a negatively correlated relationship between the enzymatic output and the fed input in the NFB system-harboring cells (Figure S5C).
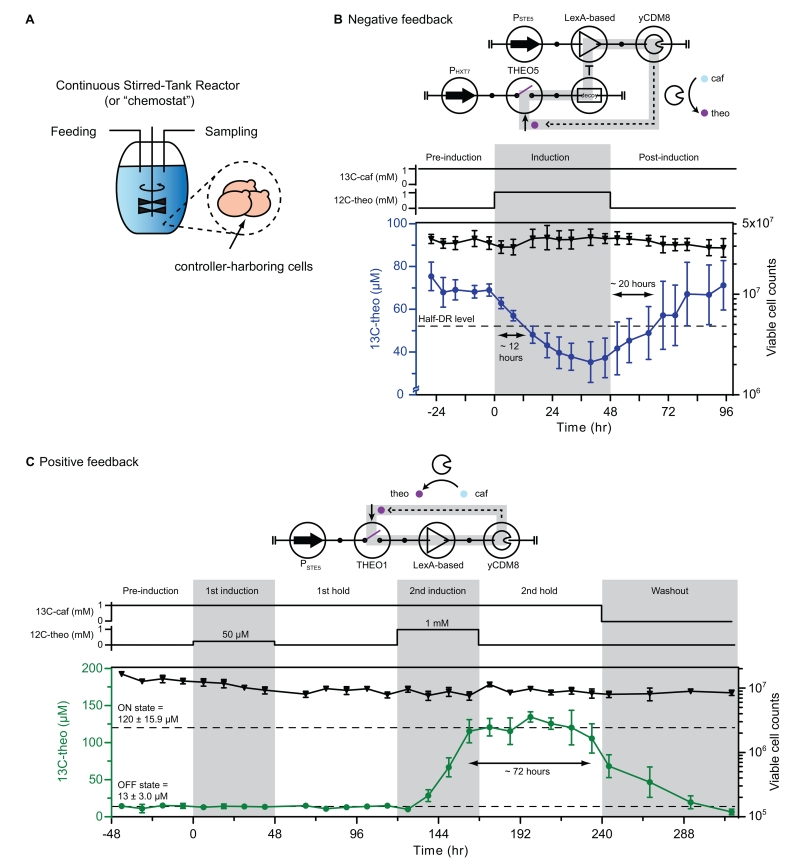
To better demonstrate the dynamic behavior of the PFB and NFB systems, we built a multi-chamber continuous-stirred tank reactor (“chemostat”) to assay our designed systems (Miller et al., 2013). The chemostat enables a steady-state cellular growth profile by continuously feeding fresh media and sampling each chamber (Figure 6A), upon which system feedback effects can be better examined. In a conventional closed-batch culture, a growth-dependent gene expression profile overrides designed feedback effects (Figure S5D). We first validated the temporal gene expression profile of the key components in the feedback systems to ensure their robustness in the long duration culture condition. A time course experiment was conducted on the transcription-based amplifier with EGFP as the reporter, and results indicate a strong and stable expression profile over time (Figure S5E). We performed a 2-day chemostat experiment, where cells harboring an RNA switch-regulated EGFP construct were grown with or without input. We observed a steady density of viable cells in the chemostat, suggesting that the cells are growing continuously in this environment (Figure S6A). In addition, the RNA switches exhibit an activation ratio of ~18, confirming the activity of RNA switches in the chemostat environment.
Figure 6. Dynamic characterization of positive and negative feedback systems.
(A) Schematic of continuous stirred-tank reactor (chemostat) (left). The chemostat was fed 13C-caf continuously and 12C-theo was pulsed during the induction period (right). The dilution rate was set to 5% per hour (~ 1 mL of a total 20 mL volume). Samples were collected every 6 hours during a 6-day experiment (for NFB) or every 12 hours during a 15-day experiment (for PFB). 13C-caf, 13C-theo, and 12C-theo in the media were quantified using LC-MS and viable cell counts were measured using flow cytometry.
(B) Circuit symbol of a negative feedback system built with THEO5-based yCDM8-repressor (left). The negative feedback (grey loop) is established through the antagonizing activity between actuator expression (yCDM8) and the signal level (theophylline). The construct was characterized as Figure 6A. Each data point represents the mean value and error bars represent ± 1 s.d. of four biological replicates. The dash line represents the half dynamic range (half-DR) level (i.e., 52.5 μM). The double arrows indicate the half-DR fall time (first) and rise time (second) of the characterized system. See also Figures S5C and S6B.
(C) Circuit symbol of a positive feedback system built with THEO1-based yCDM8-activator (left). The positive feedback (grey loop) is established through the synergistic activity between actuator expression (yCDM8) and the signal level (theophylline), resulting in bistable production states. The construct was characterized as Figure 6A. Each data point represents the mean value and error bars represent ± 1 s.d. of three biological replicates. The double arrows indicate the hold time for ON state of the characterized system. See also Figures S5A, S5B, and S6C.
Abbreviations: caf: caffeine; theo: theophylline.
The chemostat experiment with the NFB system was performed over 6 days with constant 13C-caffeine feeding and modulated 12C-theophylline input levels with three phases: pre-induction, induction, and post-induction (Figure 6B). THEO5-based NFB was selected for its low OFF state and characterized in the chemostat with a culture sampling rate of once every ~6 hours. The viable cells in each chamber were measured and results confirmed a steady-state growth environment during the course of the chemostat experiment. Cells harboring the NFB system were found to establish an initial steady production level (~70 μM; pre-induction). The production level began to decrease (at a rate of ~30 μM/day) when supplying external theophylline (induction), and relaxed back to its initial state (at a rate of ~25 μM/day) upon removal of the external input (post-induction). The response times of this closed-loop NFB system were 12 and 24 hours (fall time and rise time, respectively; see definitions in Document S1), which were faster than those in an open-loop configuration (24 and 72 hours, respectively; Figure S6B). The increase in response dynamics supports the closed-loop behavior with our THEO5-based NFB system.
The PFB system (THEO1-based) was characterized using the same chemostat setup. To fully explore the system characteristics, the experimental duration was extended to 15 days and two induction cycles (50 μM, 1 mM) of 12C-theophylline input levels were applied (Figure 6C). Cells harboring the PFB system exhibit an OFF state of 13 ± 3 μM and an ON state of 120 ± 16 μM when fully induced (1 mM; 2nd induction). Moreover, the PFB system maintained its ON state during the period of post-induction (2nd hold), indicating a molecular hysteresis effect in which the system produces enough theophylline to self-sustain even after removing the external induction. In contrast, at 50 μM of input (1st induction) the PFB system exhibits neither an increase in or sustained production level (p > 0.05 with standard one-way ANOVA, tested against the measured OFF state average (13 ± 3 μM)), which suggests that the activation threshold for this bistability is above 50 μM. To further substantiate that the observed feedback performance was mediated by the two molecules (13C-caffeine, 12C-theophylline), we added a final washout phase. The results demonstrate that the sustained ON state is disrupted when removing the caffeine substrate (Washout). In addition, we characterized a functionally equivalent two-stage PFB system in the same chemostat setup. The ON state of this two-stage PFB was not detected upon induction and was not maintained in the post-induction hold (i.e., no bistability; Figure S6C).
Discussion
We have addressed the challenges associated with the use of the conventional two-stage ribozyme switch-based architecture by expanding it to a three-stage architecture (resembling the layout of a standard, universal controller unit). The three-stage architecture was expanded to controllers with diverse functions through the incorporation of a decoy factor (repression controller), serial RNA switches (AND), multiple OP sites (OR), and phenotypic (live-death) and enzymatic (feedback) actuators. Our bottom-up approach to construct the controllers yields strong plug-and-play modularity that enables expansion to higher-order systems without significant component reorganization (Cameron et al., 2014). We achieved substitution of diverse components at the sensing (18 RNA switches), processing (2 BD-OP pairs, 2 ADs, 6 ENs), and actuating stages (EGFP, His5p, yCDM8).
Our use of RNA switches to compose synthetic input-controlled amplifiers (i.e., the activation controllers) increases the device performance beyond typical dynamic ranges, enabling system-scale applications (e.g., feedback) that have been largely implemented with inducible promoters (Wang et al., 2013). Our systems exhibit a two orders of magnitude gain, which is comparable to or higher than recently reported amplification gains (~15-20) (Bonnet et al., 2013; O’Shaughnessy et al., 2011; Wang et al., 2014). To the best of our knowledge, Wang and co-workers were the first to implement genetic amplifiers similarly with protein-based transcriptional controllers. However, these earlier designs were tightly coupled to prokaryotic expression mechanisms. The RNA-based sensor used in the framework has the advantage of leveraging the capacity of systematically generating de novo RNA aptamers (McKeague and Derosa, 2012), and incorporating them into RNA switches for engineering diverse sensing functions (Win et al., 2009) for different organisms (Wei et al., 2013). Design improvements, such as tuning optimal EN-promoter distance (Figure S2E) or cascading multiple transcription-based amplifiers (Hooshangi et al., 2005), are made feasible by the modularity and orthogonality of our framework.
The linearity of amplifiers (i.e., gain independent from the input levels) is another key characteristic, as linear amplifiers can serve as genetic detectors for their proportional input/output relationship; while nonlinear amplifiers can serve as signal filters or devices to increase cooperativity (Lu et al., 2009; Roquet and Lu, 2014). Although we achieved both linear and nonlinear amplifiers in our design, the exact mechanism of this linear/nonlinear property is not fully elucidated. We hypothesize the operating range of the RNA switch on the transition curve of a transcription-based amplifier (i.e., three regimes in a Hill function: non-amplifying, linear, saturation) can affect the linearity of amplifiers. Thus, our designed activation controllers can behave as a linear amplifier if the RNA switching range lies within the linear regime of the amplifier, and become nonlinear if the RNA switching range begins to cross multiple regimes. For example, a low pass filter-like activation controller can have its RNA switching cross between linear to saturation regimes, which corresponds to a constant gain (linear) at low input and small-to-no gains (saturation) at high input.
One drawback of increasing the number of stages in a genetic control system is the need for additional level matching between stages, which can require cycles of post hoc tuning to obtain a functionally active system. We addressed this challenge with a model-guided approach, in which we assembled an in silico controller and screened different genetic components to identify desired device performance. However, our model-guided approach was not successful in capturing all the dynamics of some genetic controllers (e.g., the FA11-based EGFP-activator), and the limitation may result from inaccurately used parameter distributions in the Monte Carlo simulations (i.e., assumed to be log-uniform for all randomized parameters in the model). The actual distributions of these biochemical or cellular parameters are less uniform, and may be related to growth (e.g., kd_R and kd_RS) (Milo and Phillips, 2015) or intrinsic to stochastic biochemical processes (e.g., kon and koff) (Kaern et al., 2005). Further studies to pinpoint the fundamental mechanisms of these cellular and biochemical processes may determine the most appropriate simulation algorithms, and thus will help increase the predictability of the model-guided level-matching approach, especially as system complexity increases.
We were able to expand our framework to the design of layered Boolean controllers with four orthogonal input signals. Although several studies have demonstrated the use of multilayer frameworks to design logic controllers, the focus has been largely on prokaryotic systems (Green et al., 2014; Moon et al., 2012). In some cases, physical separation of individual device-harboring cells is needed to enable cascading of each logic gate (Tamsir et al., 2011). Our framework presents an approach to implement layered logic controllers within a single cellular environment. However, we observed a gradual decrease of activation ratios in the designed controllers when the numbers and types of input-responsive RNA switches increase (i.e., activation ratios of ~20, 8, and 3.4 in the single-input activation controllers, double-input AND/OR controller, and quadruple-input SoP controller, respectively). This drop in activation ratios can potentially limit the total number of input signals that can be used in a synthetic control system. While the underlying causes are not entirely clear, one explanation is that the cells may be substantially more stressed in the presence of multiple inputs (Kaloriti et al., 2012). Efforts to engineer RNA switches with increased sensitivities (i.e., lower KD) may reduce the concentrations of input required, thereby reducing cell stress.
Closed-loop feedback topologies are one of the most commonly observed features in natural genetic regulation (Alon, 2007). We synthetically recapitulated this natural phenomenon using a small molecule input (theophylline) with its cognate RNA sensor/enzyme pair. Through establishing a continuous growth environment, we successfully decoupled the controllers from the natural growth-dependent feedback (Klumpp et al., 2009) and demonstrated a synthetic positive and negative feedback scheme. The dynamics of our feedback systems (days) are comparable to or slower than other reported small molecule-mediated feedback systems (e.g., days in a malonyl CoA-mediated system (Xu et al., 2014) and hours in a fatty acid-mediated system (Zhang et al., 2012); both in bacterial systems). The slower dynamics may be a result of a longer timescale associated with eukaryotic gene expression (Milo and Phillips, 2015), and the nutrient limitations required to maintain a constant growth rate in the chemostat. The system dynamics can be increased by supplying extra nutrients in the chemostat to achieve a higher growth rate (Ziv et al., 2013) or adding device accelerators to the controllers (e.g., incoherent feedforward loops (Mangan et al., 2006)).
Future applications will leverage this design framework to support systems biology research; e.g., studying small molecule-mediated feedback motifs in a gene network and examining their stability and stringency through tuning device characteristics with a spectrum of enzyme actuators. Our input-controlled RNA-based framework can be used to sense and re-direct metabolic fluxes to optimize production in reconstructed biosynthesis pathways. In addition, the design framework can offer a streamlined process to build phenotypic controllers that regulate complex cellular behaviors, in particular for devices that need a large response range (e.g., cellular memory (Ajo-Franklin et al., 2007)) or have a sensitive activity range (e.g., mating type differentiation (Galloway et al., 2013)). Finally, the RNA-based sensing stage in our framework can be expanded to other RNA-based regulatory mechanisms or different input types (Chang et al., 2012). These unique features of RNA-based devices and the modularity inherent in the three-stage architecture offer a generalizable framework to advance our capabilities to design increasingly sophisticated biological systems.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
• Small Molecules As Input Signals
Four small molecules were used as target binding inputs for in vitro RNA aptamer characterization experiments and in vivo feeding experiments: theophylline (≥ 99%; LOT# 091M0214V) was purchased from Sigma-Aldrich, neomycin (as fradiomycin sulfate, ≥ 97%; LOT# SCFUG-KI) was purchased from TCI America (Portland, OR), tetracycline (≥ 98%; LOT# BCBK1604V) was purchased from Sigma-Aldrich, and (6R)-folinic acid (LOT#154) was purchased from Schircks Laboratories.
• S. Cerevisiae Strains and Growth Media
The Saccharomyces cerevisiae strain W303α (leu2-3, 112; trp1-1; can1-100; ura3-1; ade2-1; his3-11, 15) was used as a host organism to harbor designed genetic controllers in all in vivo experiments. Yeast strains without any auxotrophic selection were grown in standard yeast extract peptone dextrose (YPD) media. Yeast strains with auxotrophic selection markers either from gene expression cassettes on the plasmids or amplified gene expression cassette DNA for genome-integration (either LEU, URA, or URA/TRP; see “Plasmids” in Supplemental Data 1) were grown in standard yeast nitrogen base (YNB) defined media (Becton, Dickinson and Company (BD)) and supplemented with the proper amino acid drop out solution (Clontech) to maintain selection. The YNB media was supplied with either 2% dextrose or galactose as the carbon source and its pH was adjusted to 5.8 using 5 M sodium hydroxide. Yeast strains with the antibiotic selection marker KanMX (only for genome-integration) were grown in YPD media with G418. Corresponding agar plates were also made for each selection condition.
METHOD DETAILS
• In Vitro Aptamer Characterization Assay
The SPR-based aptamer characterization assay was performed as previously described (Chang et al., 2014). A Biacore X100 instrument (GE Healthcare) at 25 °C with a CM5 sensor chip (GE Healthcare) was equilibrated with HBS-N buffer. The CM5 chip was conjugated with a DNA linker strand (5′-AmMC6-TTTTTTTTTTTTTTTTTTTTTTTT; ordered from Integrated DNA Technologies (IDT)) as follows. First, the carboxymethylated dextran surface of the CM5 chip was activated with a 1:1 volume ratio of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (GE Healthcare) and 0.1 M N-hydroxysuccinimide (GE Healthcare) for 7 min at a flow rate of 10 μL/min. A DNA: hexadecyltrimethylammonium bromide (Sigma-Aldrich) mixture was prepared in 10 mM HEPES buffer (Sigma-Aldrich) to a final concentration of 20 μM and 0.6 mM and injected over the activated surface for 10 min at a flow rate of 5 μL/min. Excess activated groups were blocked with 1 M ethanolamine (GE Healthcare), pH 8.5, for 7 min at a flow rate of 10 μL/min. The immobilization reaction was performed on both flow cells (FC1, FC2) and yielded approximately 3,000 RU of the DNA linker on each flow cell.
DNA templates of the aptamer sequences were purchased from IDT and included a 3′ 24-mer poly(A) sequence and a 5′ T7 promoter. DNA templates were amplified by PCR using forward and reverse primers Biacore_fwd and Biacore_rev. RNA aptamers were transcribed using the MEGAshortscript T7 Kit (Life Technologies) followed by purification with the RNA Clean & Concentrator kit (Zymo Research). In all experiments the HBS-N running buffer (10 mM HEPES, 150 mM NaCl, pH 7.4, purchased from GE Healthcare) was supplemented with 0.5 mM MgCl2 (Life Technologies). A 500 μM stock of each molecule was prepared fresh by dissolving in running buffer and filtered through a 0.2 μm membrane (Pall Corporation, Port Washington, NY). A nine-to ten-point dilution series was prepared for each molecule spanning these ranges: neomycin 3.2765 nM to 5 μM; tetracycline 8.192 nM to 50 μM; theophylline 26.2144 nM to 100 μM.
The Biacore X100 was primed three times with running buffer followed by three startup cycles. For each startup cycle, the aptamer (~70-100 ng/μL) was captured onto the sample flow cell (FC2) for 40 s at a flow rate of 5 μL/min, yielding ~2,500–4,000 RU. The surface was regenerated with 25 mM NaOH (GE Healthcare) and 1 M NaCl (Sigma-Aldrich) for 30 s at a flow rate of 30 μL/min over both flow cells. The aptamer was captured onto the sample flow cell (FC2) for 40 s at a flow rate of 5 μL/min, the target solutions (as prepared above) were injected over both flow cells at a flow rate of 30 μL/min, followed by injection of running buffer over both flow cells at a flow rate of 30 μL/min. Aptamer and target were removed from the sensor surface by injecting 25 mM NaOH, 1 M NaCl for 30 s at a flow rate of 30 μL/min over both flow cells.
SPR data processing and analysis were performed using Biacore X100 Evaluation Software version 2.0 (GE Healthcare). The double-referencing method was performed to process all datasets. Data from the sample flow cell (FC2) were referenced first by subtracting data from the reference flow cell (FC1) to correct for bulk refractive index changes, nonspecific binding, injection noise, matrix effects, and baseline drift. Reference-subtracted data (FC2 – FC1) were double-referenced with a blank injection of run buffer to account for any systematic drift over the course of the injection. Double-referenced data were fit to a steady-state affinity model for thermodynamic analysis or a 1:1 binding model for kinetic analysis when possible. Reported values are the mean and standard deviation of three experiments.
• In Silico Model for Stage Level Matching
We developed a modeling procedure to facilitate the level matching process when designing a multi-stage system. The procedure begins with building a computational model to describe the general dynamics of a class of genetic devices; for example, an input-binding kinetic model for RNA switches (see “Modeling RNA switch-controlled gene regulation” in Document S1), or a Hill function for TAs (see “Modeling transcription-based amplifiers” in Document S1). In these general models, one or more key parameters are set as unknowns and further determined in a device-by-device manner to best capture its dynamics. As shown in Figure 3A, each genetic device is characterized in vivo, and the data are then used to estimate the unknown parameter(s) to obtain a specific model for this particular device. After obtaining all the models for the devices used in a system, we proceed to assemble these device models into an in silico system. Finally, Monte Carlo simulation algorithms are applied to predict the activities of this in silico system. For example, an in silico system can be used to simulate varying parameter ranges or different compositional devices (e.g., controller-driving promoter strengths, or different RNA switches) to obtain an understanding of the optimal conditions for a matched activity level between device stages.
• Monte Carlo Simulation for Prediction
We used a standard Monte Carlo sampling technique to predict protein level based on a given computational model (i.e., model for RNA switch, TA, or an EGPF-activator). In order to capture the cell-to-cell variability of gene expression regulated by RNA switches in vivo, certain parameters in the model were randomized during simulation (see Supplemental Data 2). In each cycle of the Monte Carlo sampling, these parameters are randomly generated assuming a log uniform probability distribution, and the predicted protein level is evaluated based on solving the model, similar to using the pseudo-code described below with a total random sampling size of N = 1000 to 5000 (as specified).
Specify non-randomized parameters
Iterate the following cycle for N times
Generate randomized parameters
Solve the given model and calculate predicted protein level
End
This algorithm was implemented using MATLAB (Windows ver R2013b, MathWorks).
• Construction of Gene Expression Cassettes
The plasmid-based genetic expression cassettes were constructed in yeast centromere-containing shuttle vector (pRS314/TRP selection and pRS316/URA selection) (Sikorski and Hieter, 1989), with multiple restriction cloning sites. Each genetic component in an expression cassette was inserted into the plasmid in the following order using standard cloning methods: ClaI-promoter-HindIII-BglII-coding region-AvrII-XhoI-terminator-KpnI, as previously constructed and reported (McKeague et al., 2015). On the other hand, the genetic expression cassettes for genomic integration were constructed in the yeast non-replicable plasmids (pUG6/KanMX selection and pUG73/LEU selection) (Gueldener et al., 2002), with the following order: HindII-EN-spacer-SphI-promoter-AvrII-coding region-XmaI-terminator-SalI.
Each genetic component was made from either PCR amplifying from the genomes or directly synthesizing as double strand DNA fragments, with further DNA assembling using the Gibson method as needed (Gibson et al., 2009). Codon optimization to increase expression in S. cerevisiae was performed using the GeneArt GeneOptimizer program (Life Technologies). Double-stranded DNA fragments were synthetized as gblocks from IDT, and DNA oligonucleotides were synthesized either by IDT (> 80 nucleotides) or Stanford Protein and Nucleic Acid Facility (< 80 nucleotides). Pfu Hotstart polymerase or PfuUltra II (Agilent Technologies) was used for PCR reactions for DNA fragments shorter or longer than 2 kb, respectively. The restriction enzymes and ligases (T4 and Taq) used in the cloning were purchased from New England Biolabs (NEB), and reactions were performed according to manufacturer’s instructions. All cloned plasmid candidates were transformed into TOP10 Escherichia coli (Thermo Fisher Scientific), and plated on LB agar plates (EMD Millipore) with ampicillin (50 mg/L) for overnight growth in 37°C. The next day single colonies on the plates were first PCR screened with primers that bind 200~500 base pairs upstream and downstream of the inserts and further verified by sequencing. Sequencing to verify each candidate colony was performed by Elim Biopharmaceuticals Inc, by sending either purified plasmids or bacterial colonies. Plasmids containing the correct genetic expression cassette were prepared by growing 5 mL of overnight bacterial culture in LB liquid media (EMD Millipore) with ampicillin (50 mg/L). The plasmid DNA in each bacterial culture was collected using Econospin columns (Epoch Life Science) according to manufacturer’s instructions.
• Cloning of RNA Switches
To insert an RNA switch or ribozyme (see “RNA switches and ribozyme” in Supplemental Data 1) into the 3′ UTR of the coding region in the expression cassette of a plasmid, their DNA sequences were PCR amplified with a proper pair of Gibson primers (see “Primers” in Supplemental Data 1). These amplified Gibson inserts were assembled with target double-digested backbone vectors (AvrII/XhoI) using Gibson one-pot, isothermal DNA assembly (Gibson et al., 2009). The resultant plasmids have an RNA switch or ribozyme inserted between the coding region and its terminator (TADH1). To insert a second copy of an RNA switch, we PCR amplified its DNA sequence together with the TADH1 from a RNA switch-cloned plasmid with a pair of ligation primers (see “Primers” in Supplemental Data 1). The target vector was double-digested (XhoI/KpnI) and mixed with cut RNA switch insert (SalI/KpnI) for a T4 ligation reaction. The XhoI restriction site generates a sticky end that is compatible with that of SalI, but a non-digestible scar will occur after ligation.
• Construction of Yeast Strains
The genetic cassettes were functionally expressed in yeast through two approaches: 1) integrating genetic expression cassettes into the yeast genome, and 2) transforming plasmids containing genetic expression cassettes into yeast. Genomic integration was performed using a previously reported method (Thodey et al., 2014), in which the target expression cassette was amplified from the non-replicable plasmid with primers that provide ~100 base pairs of homology to the target integration site in the yeast genome. The PCR products were transformed into yeast cells using standard lithium acetate transformation protocol (Gietz and Schiestl, 2007). Integration events were selected by growth on G418 or leucine-dropout agar plates in 30°C incubator for 36 to 48 hours, and further confirmed by colony PCR screening at both ends of integration and by sequencing. As for plasmid-based expression, plasmids were directly transformed into yeast cells using the same lithium acetate protocol. To keep plasmids in the yeast, proper selection pressure was maintained during growth of the cells.
• Yeast Culture Conditions
Yeast cells were grown in three culture conditions: 1) closed-batch culture in which cells were inoculated into a closed environment for growth and harvested for characterization after a period of time; 2) density-controlled closed-batch culture in which cells were first grown to stationary phase, and harvested and then re-suspended in fresh media to a constant cell density with external input induction for characterization after a period of time; and 3) chemostat culture in which cells were inoculated into a continuous feeding and sampling environment. Details of each culture conditions are described as in the following sections. Four different input signals were fed into the cell cultures at three distinct levels: no, medium and high correspond to 0, 0.5, and 5 mM for theophylline; 0, 0.1, and 1 mM for neomycin; 0, 0.1, and 1 mM for tetracycline; 0, 0.3, 3 mM for (6R)-folinic acid.
• Closed-Batch Culture Condition
Closed-batch experiments were conducted to characterize our designed controllers in S. cerevisiae. Yeast cells were grown at two different scales, based on how much total volume was needed for the experiment: 1) 200 μL in 2 mL deep well 96-well plates (USA Scientific) covered by an AeraSeal film (Excel Scientific), 2) 2 mL in a long glass tube. The 1:10 ratio was kept between the culture and container volumes for good aeration. Device-harboring strains were picked as single colonies from YNB agar plates in triplicates, inoculated into YNB media (mostly with 2% glucose, but 2% galactose also used for AD modularity experiments) under proper dropout selections, and grown overnight at 30°C. Culture plates were placed in a Kuhner LT-X plate shaker (Adolf Kühner), shaking at 480 RPM with 80% humidity; while tubes were placed in an incubator (Forma Orbital Shaker; Thermo Fisher Scientific), shaking at 240 RPM. The next day, each stationary-phase culture was back-diluted (1:50) into same volume of fresh YNB media with different fed inputs for additional growth. Three feeding concentrations were used in the in vivo experiments (in the order of no, medium, and high levels): 0, 0.5, and 5 mM for theo; 0, 0.1, and 1 mM for tc; 0, 0.1, and 1 mM for neo; 0, 0.3, and 3 mM for FA. The yeast cultures were harvested after different durations of additional growth: 6 hours for transcription-based amplifier and EGFP-activator assays, 12 hours for EGFP-repressor assays, and 24 hours for Boolean controller assays. The cells and media were properly prepared for further experimental characterization.
For time-course closed-batch experiment, yeast cells were first grown in 2 mL culture media in glass tubes. Device-harboring strains were picked as single colonies from YNB agar plates in triplicates, inoculated into YNB media (2% glucose) under proper dropout selection for overnight growth. The next day, each culture was back-diluted (1:50) in 10 mL of fresh YNB media in 50 mL-flasks with different fed inputs. 100 μL of yeast cultures were sampled in each time point, and the cells and media were prepared for further experimental characterization.
• Density-Controlled Culture Condition
A density-controlled closed-batch culture condition was modified based on a reported two-phase fermentation process (Trenchard and Smolke, 2015) for our ribozyme switch-based yCDM8-activator experiments. Device-harboring yeast strains were first grown in YNB under proper dropout selection overnight to stationary phase, without the input signal or the substrate. 100 μL of each overnight culture was sampled and viable cellular density was measured using flow cytometry (see section “Flow cytometry and data analysis for fluorescence level measurements”). Then all cells were spun down at 4000 RPM (10 min, 4°C) and re-suspended in fresh YNB growth media (5 × 107 viable cell per mL), supplied with 1 mM of caffeine (Sigma-Aldrich) as the substrate for the yCDM8, and with or without the input signals (depending on which RNA switch is used). To keep the cells alive yet minimize further growth, galactose (2%) was used as carbon source in the post-growth phase instead of glucose (2%). Yeast cultures were harvested after another 24 hours of production, and the cells and media were properly prepared for further experimental characterization.
• Yeast Chemostat Culture Condition
To enable growing yeast cells in a continuous culture condition for a longer duration, we adapted a previously reported small-scale multi-chamber chemostat setup (Miller et al., 2013). Our chemostat was implemented using a multiplexed peristaltic pump (Manostat Pumps; Cole-Parmer) to enable yeast growth in glass tubes, with air pumping into each tube to generate back pressure for yeast culture sampling. This chemostat setup was composed of five major modules: 1) 12 glass tubes as chemostat chambers for culturing yeast, 2) a media tank used to supply fresh media to the chemostat chambers (through an influx needle), which influx flow rate controlled by a peristaltic pump, 3) a heat block set to maintain temperature of yeast culture at 30°C, 4) an air pump to supply humidified, filtered air (through an air needle) into each chemostat chamber, and also to keep yeast cells from precipitating at the bottom of the tube, and 5) 12 collectors connected to each chemostat chamber (through an efflux needle) for sample connection. Three sizes of silicone tubes were used to connect each module: 3/32″ × 7/32″, 1/4″ × 3/8″ and 1/2′ × 5/8′ (VWR International). All modules were connected together first and autoclaved before use.
Two media tanks were made: 1) a media tank without theophylline using 2× YNB (2% glucose) with proper dropout selection (used in both pre- and post-induction phases), and 2) a media tank without 1 mM theophylline using the same 2× YNB and dropout selection (used only in induction phase). For the feedback experiments, extra 1 mM of 13C-labeled caffeine (caffeine-(trimedthyl-13C3) from Sigma-Aldrich) was fed into the media as the substrate to allow distinguishing between the produced and fed theophylline. Device-harboring yeast strains (in three or four biological replicates) were first grown in YNB under proper dropout selections overnight to stationary phase. Fresh media were pumped in to the chemostat first (without inoculating cells) with gradually increasing peristaltic pump speeds (from 5, 10, to 20 RPM) until reaching its steady state. The working volume of chemostat chamber was adjusted to 20 mL (by adjusting the depth of the efflux needle in the chamber, as described in (Miller et al., 2013)). Then, the peristaltic pumped was turn off (i.e., no fresh media supplying to the chamber). 1 mL of each overnight yeast culture was inoculated into each chamber and grown overnight back to its stationary phase (as the closed-batch culture condition). The peristaltic pumped was turn back on to 20 RPM the next day, and the cell culture in each chamber was grown overnight into a new steady-state (with fresh media supplying to the chamber). The dilution rate of the chemostat was set to be at 5% per hour (i.e., 1 mL per hour). The status and quality of the cell culture in each chamber (cell density, cellular viability, and existence of contamination) were carefully monitored during this phase.
We began the chemostat experiment once all cell cultures in the chamber had reached the steady state (i.e., no significant changes in the cell density in each chamber for 12 hours). The culture volume in each chemostat chamber was maintained at 20 mL throughout the experiment. In the RNA switching experiment, two growth phases were established (pre-induction and induction) by switching the media tank at Time 0 hour (Figure S6A). In the feedback experiment, different growth phases were established (pre-induction, induction, post-induction/hold, and washout) by switching the media tank at denoted times. When changing the condition from the pre-induction to induction phase, we injected 0.8 mL of concentrated theophylline (80 mM made in 1:1 H20 and ethanol) into each chemostat chamber to boost up the theophylline concentration closer to our target 1 mM. Yeast cultures were collected continuously from each efflux, and each collection was harvested every 6 or 12 hours with a sampled volume between 5 to 7 mL (6 hours) or 11 to 14 mL (12 hours). The cells and media in the harvested sample were properly prepared for further experimental characterization.
• Yeast Cellular Growth Characterization
Two types of experiments (liquid- and plate-based) were conducted to characterize the live-death phenotype in His5p-activator harboring cells. In the liquid-based experiments, yeast cells were grown under the closed-batch culturing condition, in either 96-well plates, or long glass tubes with YNB media (2% glucose) under histidine dropout and the input signal (i.e., +his/0 mM theo, −his/0 mM theo, and −his/5mM theo) on top of the existing selection. 10 mM of 3-AT (3-Amino-1,2,4-triazole; purchased from Sigma-Aldrich) was added to the media to increase the cellular dependency on histidine (Win and Smolke, 2007). The yeast cultures were either harvested after 6 hours of back-dilution and re-growth, or sampled at different time points (i.e., time course experiments). The cells and media were properly prepared for further experimental characterization.
In the plate-based experiments, yeast cells were first grown in liquid cultures overnight to stationary phase, and their viable cell counts were measured before generating a serial dilution (from 105 to 10−2 viable cells per μL). Three types of plates were made with YNB media (2% glucose) under histidine dropout and the input signal (i.e., +his/0 mM theo, −his/0 mM theo, and −his/5mM theo) on top of the existing selection. 10 mM of 3-AT was also added to the plate. These plates were dried for 20 min in a chemical fume hood to reduce backdrop liquid from the cap during growth. 2 μL from each dilution were carefully spotted on the center of the plates, with proper spacing between each dilution. The plates were first placed in the incubator facing up with their cap slightly open to dry up the liquid culture for 2 hours, and then flipped back to the conventional orientation (facing down). The plates were grown in the 30°C incubator for 2 days, and imaged by G:BOX Chemi XT4 Imager (Syngene, Frederick, MD) using the “Upper White Light” mode.
• Evolution of TetR Mutants
We employed site-directed mutagenesis in combination with DNA shuffling (Zhao and Arnold, 1997) to evolve a TetO-binding TetR variant (TetRm2) that is not responsive to tetracycline. The binding mechanism between TetR and tetracycline involves the formation of a tetracycline-Mg2+ complex before binding to a pocket in TetR, which then represses TetR from binding to TetO (Ramos et al., 2005). The amino acid residues in the binding pocket of TetR that interact with tetracycline-Mg2+ include, but are not limited to: H64, N82, S85, F86, H100, T103, P105, Q109, Q116, and D178 (Hecht et al., 1993; Zhou et al., 2007).
We hypothesized that among a combination of these reported mutations, we could find a TetR mutant that keeps TetO-binding capability but becomes not responsive to tetracycline. Two parent TetR variants (the wild-type and the mutant TetRm0 with all 10 identified mutations) were synthesized by IDT as parents for DNA shuffling. A standard DNA shuffling method was used (Zhao and Arnold, 1997) to generate a library of variants that consist of combinatorial mutations. Two parent DNA fragments were first PCR amplified and then digested using DNase I (NEB) at 15°C for 7 min to achieve an average digested fragment length between 50 to 100 base pairs. These two digested fragments were purified by QIAquick PCR Purification Kit (Qiagen), and were combined into one pool for shuffling and reassembly into full size fragments with a high fidelity DNA polymerase (pfu) in a thermocycler (without primers). The assembled products were purified using gel extraction and further PCR amplified with primers before cloning into the plasmid to construct a library of expression cassettes.
Twenty TetR candidates were sequenced (Elim Biopharmaceuticals Inc.) and seven unique mutant combinations were found (TetRm1 to m7 in Figure S2A). The library was transformed into a yeast strain harboring the PTetO cis reporter regulating EGFP for screening. A titration experiment was set up to grow the strain at the closed-batch culture condition (see section “Continuous-growth (chemostat) culturing condition for S. cerevisiae”) and screen for a TetR mutant that is capable of activating EGFP gene expression at a relatively constant level under different levels of tetracycline (i.e., not responsive to tetracycline).
• Expansion of the Design Framework
The repression controller was built by introducing a BD-only decoy factor to compete with the synthetic transcription factor (i.e., BD-AD) for the binding of the OP site (grey box in Figure 5A). Input-controlled inhibition was achieved by using an RNA switch to regulate the level of the decoy factor. Specifically, we built the EGFP-repressor by modifying an EGFP-activator construct (CSY1113 + pCS3442 series). We first moved the BDLexA-ADVP16 (removing the RNA switch regulation) into the cis reporter harboring yeast (CSY1113) at genomic locus (Ieu2) to create strain CSY1116, and introduced an RNA-switch regulated decoy factor (BDLexA) on a plasmid (pCS3455 series). Device-harboring strains were built by transforming the BD-only plasmid into the EGFP-yeast cells (CSY1116 + pCS3455), and characterized in closed-batch culture with back-dilution (1:50) into three input levels. EGFP levels were assayed using flow cytometry after 6 hour of growth.
We also expanded the framework to construct Boolean controllers: an AND controller was built by inserting two RNA switches (responsive to different inputs) to regulate the trans element of the transcription-based amplifier (pink in Figure 5B), so that the transcript of the trans element of the amplifier is only stable in the presence of both input signals. An OR controller was built by merging two orthogonal OP sites in a single cis hybrid promoter (OPLexAO-OPTetO-miniPGAL1; yellow in Figure 5C) and placing two orthogonal trans elements under the regulation of two input-responsive RNA switches. In the presence of either input signal, one or both of the trans elements are expressed and activate the expression of the cis hybrid promoter-regulated gene. An SoP controller was built by combining these two single-layer controllers (AND and OR), with the mechanism of using two RNA switches regulating the first trans element of OR, and another two RNA switches to the second trans element of OR (as indicated in pink or yellow box, respectively). Therefore, as long as one of the transcripts coding for either the trans element of the amplifier is stabilized by the presence of two corresponding input signals, the Boolean controller is turned on for gene expression. Strains harboring each Boolean controller were characterized in close-batched culture condition with combinations of inputs (“-“ represents no input; “+” represents either 5 mM theophylline, 1 mM neomycin, 1 mM tetracycline, or 3 mM (6R)-folinic acid) as indicated. EGFP levels were assayed via flow cytometry after overnight growth.
QUANTIFICATION AND STATISTICAL ANALYSIS
• Flow Cytometry and Its Data Analysis
The fluorescent protein expression level in cells was quantified using flow cytometry. S. cerevisiae cultures harboring designed genetic controllers were sampled and spun down at 4500 RPM (5 min, 4°C). The culture supernatant was discarded, and the cells were re-suspended in 200 μL of 1× phosphate buffered saline (PBS) with 1% bovine serum albumin (BSA Fraction V, EMD Millipore), and 1 mg/L DAPI (Life Technologies) was added as viable cell stain. The cells were filtered with a nylon 6/6 woven mesh sheet (30 μm in mesh size and 40 μm in thread diameter; part # B000FMYHXO from Amazon) to remove large cell debris. The fluorescence of yeast cells was measured with a three-laser MACSQuant VYB flow cytometer (Miltenyi Biotec Inc.). EGFP was excited at 488 nm and captured by a 525/50 bandpass filter (channel B1 with PMT voltage 312). DAPI was excited at 405 nm and captured by a 450/50 bandpass filter (channel V1 with PMT voltage 303).
Fluorescence data processing was performed using the standard software package FlowJo (Mac ver. X). The machine raw data were first converted into FlowJo-compatible format (FCS3.1) and viable cells were isolated through three consecutive gates: 1) scatter gate: side scatter (log) vs. forward scatter (linear); 2) singlet gate: height vs. area of forward scatter (linear); 3) viability gate: side scatter (log) vs. negative DAPI region (log). The median and CV values of measured fluorescence level for the isolated group of viable cells was exported into an excel sheet for further processing and analysis.
Fluorescence data analysis and figure plotting were performed using Prism (Windows ver. 6; GraphPad). Two post-processing procedures were performed on the exported data. First, the color of tetracycline, which was observed in the EGFP fluorescence channel in a feeding experiment, was compensated by manually subtracting out the additional fluorescence. The compensation level was determined by comparing the measured EGFP fluorescence in a no fluorescence construct (CSY3 + pCS8) under the presence and absence of tetracycline. Second, a standard expression unit (“esu”, external standard unit) was established to enable quantitative comparison among experiments in this study. We chose a constitutively expressed genetic cassette (CSY3 + pCS2212) as the external standard reference, and this reference construct was grown side-by-side with other experimental constructs. The measured fluorescence levels of all experimental constructs were scaled accordingly by setting the fluorescence level of this standard construct to 100 esu.
• Small Molecule Concentration Analysis
The molecule concentrations in the media in the feedback experiments were determined using liquid chromatography-mass spectrometry (LC-MS). 1 mL of device-harboring cell cultures were sampled and spun down at max speed (10 min, 4°C) to ensure no cells or large cellular debris in the media. The supernatant was carefully sampled without disturbing the cell pellet and analyzed by LC–MS/MS using an Agilent 1260 infinity binary pump HPLC and Agilent 6420 triple quadrupole mass spectrometer with an electrospray ionization source. Chromatography was performed with a Zorbax Eclipse Plus C18 column (2.1 × 50 mm, 1.8 μm; Agilent Technologies). Mobile phase A was water with 0.1% formic acid, phase B was acetonitrile with 0.1% formic acid, and the flow rate was 0.4 mL/min at 40 °C. 2 μL samples were injected and separated with the following method: 10% B for 0.5 min, 10–50% B gradient for 3 min, 95% B for 1 min, and re-equilibrate for 2 min. The eluent was directed to the MS for 1–4 min with ESI source gas temperature at 350°C, gas flow of 11L/min, nebulizer pressure of 40 PSI. The quantification of molecules was based on the integrated peak area of the MRM chromatograms. The MRM transitions used were: 12C-theophylline (181.1 to 124.0 and 181.1 to 69.2), 13C-theophylline (183.1 to 125 and 183.1 to 70.2) and 13C-caffeine (198.1 to 140 and 198.2 to 112.1), each of which was determined using the MassHunter Optimizer software with appropriate standards. MRM peak areas were compared to a calibration curve of external standard peak areas to determine concentration.
• Statistical Analysis
Standard statistical analyses were conducted using Prism (Windows ver. 6; GraphPad). Three types of analyses were used in this study: 1) Paired t-test for data presented in Figure 5A, 2) standards one-way ANOVA for data presented in Figure 6C, and 3) standard two-way ANOVA for data presented in Figures 2E, 2F, S1E, and S2A; standard one-way ANOVA. The results and corresponding statistics (e.g. p values; significant levels) can be found in Supplemental Data 3.
DATA AND SOFTWARE AVAILABILITY
• Software
Please see Key Resource Table for information on the software used in the study.
• Data Resources
Please see Supplemental Data 2 as the MATLAB files used for simulation and associated model simulation raw data.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Theophylline | Sigma-Aldrich | LOT# 091M0214V |
| Neomycin (as fradiomycin sulfate) | TCI America | LOT# SCFUG-KI |
| Tetracycline | Sigma-Aldrich | LOT# BCBK1604V |
| (6R)-folinic acid | Schircks Laboratories | LOT# 154 |
| Caffeine | Sigma-Aldrich | C0750 |
| Caffeine-(trimedthyl-13C3) | Sigma-Aldrich | 485365 |
| Critical Commercial Assays | ||
| QIAquick PCR Purification kit | QIAGEN | 28106 |
| MEGAshortscript T7 kit | Life Technologies | AM1354 |
| RNA Clean & Concentrator kit | Zymo Research | R1019 |
| DNA Clean & Concentrator kit | Zymo Research | D4014 |
| EconoSpin Mini Spin Column | Epoch Life Science | 1910-050/250 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: Strain background W303 | ATCC | ATTC: 208353 |
| S. cerevisiae: Device-harboring strains | This paper |
Table S1 and Supplemental Data 8 |
| E. coli: TOP10 | Thermo Fisher Scientific |
C404010 |
| Recombinant DNA | ||
| Plasmids | This paper | Supplemental Data 8 |
| Sequence-Based Reagents | ||
| DNA primers | This paper | Supplemental Data 8 |
| Ribozyme-based RNA switches, and ribozymes | This paper | Supplemental Data 8 |
| DNA and protein parts | This paper | Supplemental Data 8 |
| Software and Algorithms | ||
| MATLAB (Windows ver R2013b) | MATLAB | www.mathworks.com/products/matlab |
| Prism (Windows ver. 6) | GraphPad | www.graphpad.com |
| Convex optimization package CVX | Grant and Boyd, 2013 | cvxr.com/cvx |
| GeneArt GeneOptimizer program | Thermo Fisher | www.thermofisher.com/ |
| MassHunter Optimizer software | Agilent | www.agilent.com |
Highlights.
Developed a 3-stage architecture for designing ribozyme-based genetic controllers
Designed amplifiers with a gain of up to 200-fold over unamplified controllers
Established a model-guided strategy to aid the level-matching process between stages
Extended the framework to the higher-order controller designs and feedback systems
Acknowledgements
We thank A. Cravens, M. Mathur, P. McLean, C. Schmidt, K. Thodey, K. Wei, J. Xiang for valuable comments on the manuscript; C. Torng for assistance with protein evolution; S. Galanie, K. Thodey, K. Wei, for valuable discussion and suggestions on experimental design. This work was supported by the National Institutes of Health, National Science Foundation, Defense Advanced Research Projects Agency, Bill & Melinda Gates Foundation (grants to C.D.S.), Stanford Bio-X Institute, Ministry of Education of Taiwan, Siebel Scholars Foundation (graduate student fellowships to Y.-H.W.), and Natural Sciences and Engineering Research Council of Canada (postdoctoral fellowship to M.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Y.-H.W. and C.D.S. conceived of this project and designed experiments. Y.-H.W., M.M., and T.M.H conducted experiments. Y.-H.W. designed computational models. All authors analyzed data, discussed results, and wrote the manuscript.
Conflict of Interest
The authors declare no competing financial interests.
References
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA. Rational design of memory in eukaryotic cells. Genes & development. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Beisel C, Bayer T, Hoff K, Smolke C. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol Syst Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes & development. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter GA, Egan KM. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene. 1984;32:263–274. doi: 10.1016/0378-1119(84)90002-7. [DOI] [PubMed] [Google Scholar]
- Blazeck J, Garg R, Reed B, Alper HS. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol Bioeng. 2012;109:2884–2895. doi: 10.1002/bit.24552. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- Chang AL, McKeague M, Liang JC, Smolke CD. Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform. Analytical chemistry. 2014;86:3273–3278. doi: 10.1021/ac5001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol. 2012;23:679–688. doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway KE, Franco E, Smolke CD. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341:1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- George RA, Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein engineering. 2002;15:871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Golynskiy MV, Koay MS, Vinkenborg JL, Merkx M. Engineering protein switches: sensors, regulators, and spare parts for biology and biotechnology. Chembiochem. 2011;12:353–361. doi: 10.1002/cbic.201000642. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B, Muller G, Hillen W. Noninducible Tet repressor mutations map from the operator binding motif to the C terminus. Journal of bacteriology. 1993;175:1206–1210. doi: 10.1128/jb.175.4.1206-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci U S A. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kaloriti D, Tillmann A, Cook E, Jacobsen M, You T, Lenardon M, Ames L, Barahona M, Chandrasekaran K, Coghill G, et al. Combinatorial stresses kill pathogenic Candida species. Medical mycology. 2012;50:699–709. doi: 10.3109/13693786.2012.672770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of biological engineering. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy T, Kumar PR, He H, Joshua-Tor L. The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation. Genes & development. 2012;26:294–303. doi: 10.1101/gad.182691.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell. 2011;43:915–926. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JC, Chang AL, Kennedy AB, Smolke CD. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M, Nehlin JO, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. Journal of molecular biology. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. Journal of nucleic acids. 2012;2012:748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeague M, Wang YH, Smolke CD. In vitro screening and in silico modeling of RNA-based gene expression control. Acs Chem Biol. 2015;10:2463–2467. doi: 10.1021/acschembio.5b00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Xu HE. Gal80-Gal80 interaction on adjacent Gal4p binding sites is required for complete GAL gene repression. The EMBO journal. 2001;20:841–851. doi: 10.1093/emboj/20.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metabolic engineering. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Miller AW, Befort C, Kerr EO, Dunham MJ. Design and use of multiplexed chemostat arrays. Journal of visualized experiments: JoVE. 2013:e50262. doi: 10.3791/50262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Phillips R. Rates and Duration In Cell Biology by the Numbers. Garland Science; New York, NY: 2015. pp. 233–306. [Google Scholar]
- Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–131. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiology and molecular biology reviews: MMBR. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nature genetics. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- Roquet N, Lu TK. Digital and analog gene circuits for biotechnology. Biotechnol J. 2014;9:597–608. doi: 10.1002/biot.201300258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. Journal of molecular biology. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townshend B, Kennedy AB, Xiang JS, Smolke CD. High-throughput cellular RNA device engineering. Nat Methods. 2015 doi: 10.1038/nmeth.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO reports. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenchard IJ, Smolke CD. Engineering strategies for the fermentative production of plant alkaloids in yeast. Metabolic engineering. 2015;30:96–104. doi: 10.1016/j.ymben.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Barahona M, Buck M. Engineering modular and tunable genetic amplifiers for scaling transcriptional signals in cascaded gene networks. Nucleic Acids Res. 2014;42:9484–9492. doi: 10.1093/nar/gku593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Wei KY, Smolke CD. Synthetic biology: advancing the design of diverse genetic systems. Annu Rev Chem Biomol. 2013;4:69–102. doi: 10.1146/annurev-chembioeng-061312-103351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KY, Chen YY, Smolke CD. A yeast-based rapid prototype platform for gene control elements in mammalian cells. Biotechnol Bioeng. 2013;110:1201–1210. doi: 10.1002/bit.24792. [DOI] [PubMed] [Google Scholar]
- Win MN, Liang JC, Smolke CD. Frameworks for programming biological function through RNA parts and devices. Chem Biol. 2009;16:298–310. doi: 10.1016/j.chembiol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci U S A. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U S A. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- Zhao H, Arnold FH. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Symons J, Hoppes R, Krueger C, Berens C, Hillen W, Berkhout B, Das AT. Improved single-chain transactivators of the Tet-On gene expression system. BMC Biotechnol. 2007;7:6. doi: 10.1186/1472-6750-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci U S A. 2014;111:E2310–2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv N, Brandt NJ, Gresham D. The use of chemostats in microbial systems biology. Journal of visualized experiments: JoVE. 2013 doi: 10.3791/50168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.