Abstract
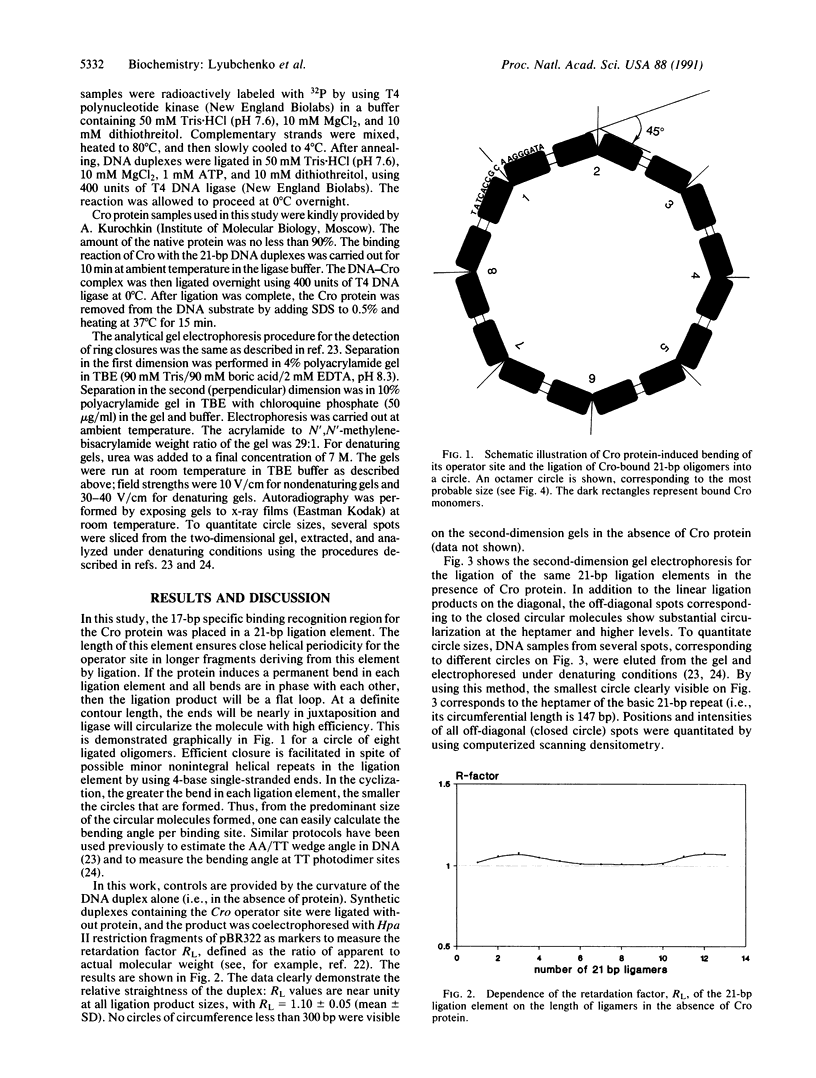
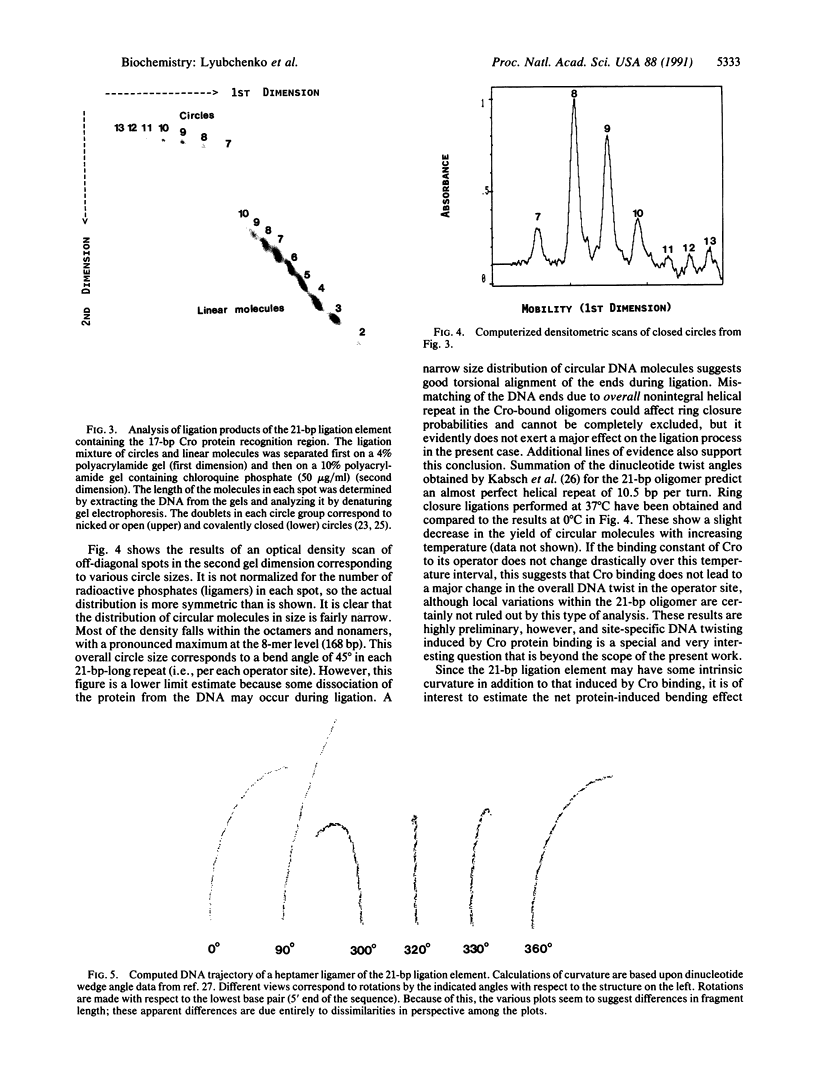
We report an approach for studying protein-induced DNA bends in solution that is based on measuring the sizes of circular DNA molecules by using two-dimensional gel electrophoresis. These circular fragments are obtained by ligating short synthetic oligonucleotides containing a protein-recognition region in the presence of protein. Oligonucleotides 21-base-pairs-long containing the OR3 recognition site were synthesized and ligated in both the presence and the absence of the Cro repressor from lambda phage. We show that in the presence of Cro protein, circular DNA molecules are formed with substantial frequency. No circular molecules are observed in the DNA samples ligated in the absence of Cro. These experiments clearly demonstrate that DNA bending is induced by Cro in this operator site. The sum of inherent plus Cro-induced bending is estimated as 45 degrees.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F. Proposed alpha-helical super-secondary structure associated with protein-dna recognition. J Mol Biol. 1982 Aug 25;159(4):745–751. doi: 10.1016/0022-2836(82)90111-5. [DOI] [PubMed] [Google Scholar]
- Barber A. M., Zhurkin V. B. CAP binding sites reveal pyrimidine-purine pattern characteristic of DNA bending. J Biomol Struct Dyn. 1990 Oct;8(2):213–232. doi: 10.1080/07391102.1990.10507803. [DOI] [PubMed] [Google Scholar]
- Bolshoy A., McNamara P., Harrington R. E., Trifonov E. N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Roderick S. L., Takeda Y., Matthews B. W. Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8165–8169. doi: 10.1073/pnas.87.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Dripps D., Wartell R. M. DNA bending induced by the catabolite activator protein allows ring formation of a 144 bp DNA. J Biomol Struct Dyn. 1987 Aug;5(1):1–13. doi: 10.1080/07391102.1987.10506370. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. CAP and RNA polymerase interactions with the lac promoter: binding stoichiometry and long range effects. Nucleic Acids Res. 1983 Jan 11;11(1):141–158. doi: 10.1093/nar/11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Husain I., Griffith J., Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C., Trifonov E. N. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982 Feb 11;10(3):1097–1104. doi: 10.1093/nar/10.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Kirpichnikov M. P., Hahn K. D., Buck F., Rüterjans H., Chernov B. K., Kurochkin A. V., Skryabin K. G., Bayev A. A. 1H NMR study of the interaction of bacteriophage lambda Cro protein with the OR3 operator. Evidence for a change of the conformation of the OR3 operator on binding. Nucleic Acids Res. 1984 Apr 25;12(8):3551–3561. doi: 10.1093/nar/12.8.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz D., Fritsch A., Buc H. Variations of intramolecular ligation rates allow the detection of protein-induced bends in DNA. EMBO J. 1986 Apr;5(4):799–803. doi: 10.1002/j.1460-2075.1986.tb04284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- McNamara P. T., Bolshoy A., Trifonov E. N., Harrington R. E. Sequence-dependent kinks induced in curved DNA. J Biomol Struct Dyn. 1990 Dec;8(3):529–538. doi: 10.1080/07391102.1990.10507827. [DOI] [PubMed] [Google Scholar]
- Metzler W. J., Arndt K., Tecza E., Wasilewski J., Lu P. Lambda phage cro repressor interaction with its operator DNA: 2'-deoxy-5-fluorouracil OR3 analogues. Biochemistry. 1985 Mar 12;24(6):1418–1424. doi: 10.1021/bi00327a020. [DOI] [PubMed] [Google Scholar]
- Metzler W. J., Lu P. Lambda cro repressor complex with OR3 operator DNA. 19F nuclear magnetic resonance observations. J Mol Biol. 1989 Jan 5;205(1):149–164. doi: 10.1016/0022-2836(89)90372-0. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Shliakhtenko L. S., Liubchenko Iu L., Chernov B. K., Zhurkin V. B. Vliianie temperatury i ionnoi sily na élektroforeticheskuiu podvizhnost' sinteticheskikh fragmentov DNK. Mol Biol (Mosk) 1990 Jan-Feb;24(1):79–95. [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Comparative gel electrophoresis measurement of the DNA bend angle induced by the catabolite activator protein. Biopolymers. 1990 Jan;29(1):29–38. doi: 10.1002/bip.360290106. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]