Significance
Gastric cancer (GC) is a leading and lethal malignancy in the world. Unfortunately, therapeutics targeting GC lag behind those of many other cancers, and the effective treatment options are limited. Here we report that increased expression of growth hormone-releasing hormone receptor (GHRH-R) in tumor specimens is significantly associated with tumorigenic progression and poor clinical outcome of GC patients. MIA-602, a highly potent GHRH-R antagonist, effectively inhibits GC growth in vitro and in vivo. The anti-neoplastic effects were mediated by blockage of the p21-activated kinase 1 (PAK1)–signal transducer and activator of transcription 3 (STAT3)/nuclear factor-κB (NF-κB) axis. Thus, GHRH-R presents a molecular marker and therapeutic target of GC, and MIA-602 demonstrates excellent therapeutic potential against human GC.
Keywords: GHRH receptor, GHRH-R antagonist, PAK1, stomach cancer, prognostic predictor
Abstract
Gastric cancer (GC) ranks as the fourth most frequent in incidence and second in mortality among all cancers worldwide. The development of effective treatment approaches is an urgent requirement. Growth hormone-releasing hormone (GHRH) and GHRH receptor (GHRH-R) have been found to be present in a variety of tumoral tissues and cell lines. Therefore the inhibition of GHRH-R was proposed as a promising approach for the treatment of these cancers. However, little is known about GHRH-R and the relevant therapy in human GC. By survival analyses of multiple cohorts of GC patients, we identified that increased GHRH-R in tumor specimens correlates with poor survival and is an independent predictor of patient prognosis. We next showed that MIA-602, a highly potent GHRH-R antagonist, effectively inhibited GC growth in cultured cells. Further, this inhibitory effect was verified in multiple models of human GC cell lines xenografted into nude mice. Mechanistically, GHRH-R antagonists target GHRH-R and down-regulate the p21-activated kinase 1 (PAK1)-mediated signal transducer and activator of transcription 3 (STAT3)/nuclear factor-κB (NF-κB) inflammatory pathway. Overall, our studies establish GHRH-R as a potential molecular target in human GC and suggest treatment with GHRH-R antagonist as a promising therapeutic intervention for this cancer.
Gastric cancer (GC) ranks as the fourth most common cancer in incidence and the second most frequent in mortality among all cancers worldwide (1). Surgery remains the only curative therapy for GC, and it must be accomplished in a timely manner. The 5-y relative survival rate of GC is less than 25% (2). Despite recent advances, the molecular mechanisms underlying gastric tumorigenesis remain largely unknown. Several molecular targets including human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), c-MET, VEGR2, HGF, and mTOR have been proposed in human GC (2–4). However, the response rates related to these therapeutic approaches vary considerably (4). Thus, the development of novel molecular targets and derived therapeutic strategies is an urgent need.
Increasing evidence suggests that GC is a type of inflammation-associated cancer caused by the complex interaction between host and environmental factors (5). Infection with the pathogen Helicobacter pylori, which triggers chronic gastritis, remains the strongest single risk factor for human GC (6). Numerous cellular and molecular pathways, which converge at the level of the signal transducer and activator of transcription 3 (STAT3) and nuclear factor-κB (NF-κB) (7–10), are involved in this inflammation-driven gastric tumorigenesis and progression.
Growth hormone-releasing hormone (GHRH) is a neuropeptide produced in the hypothalamus. In the anterior pituitary, GHRH regulates the synthesis and secretion of growth hormone (GH) (11, 12) upon binding to GHRH receptor (GHRH-R) and subsequently exerts mitogenic activity for pituitary cells (11, 12). GHRH and GHRH-R had been demonstrated to be expressed predominantly in the anterior pituitary gland but also found modestly in other somatic cells. However, accumulating evidence shows that both GHRH and GHRH-R are significantly present in various cancers including breast, prostate, ovarian, pancreatic, colon, gastric, and lung cancers, lymphoma, and glioblastoma (11–19). The GHRH/GHRH-R pathway is considered a growth factor-signaling pathway in these cancers and may modulate the activities of multiple intracellular pathways (11–13). Thus, targeting the GHRH/GHRH-R pathway has been proposed for the treatment of cancer (11, 12). Over the past three decades, various classes of GHRH-R antagonists have been developed that have shown strong growth-inhibitory effects in cancer both in vitro and in vivo (11–14, 20). MIA-602 (14) represents the latest in a series of GHRH-R antagonists and has been chosen for clinical development. We previously reported that GHRH-R is present in gastric mucosa (13), and GHRH-R mRNA is detectable in two GC cell lines (17). However, little is known about the effects of a GHRH-R antagonist, particularly on GC in vivo, and the mechanisms by which GHRH-R antagonist achieves anti-neoplastic effects in GC. Moreover, although GHRH-R expression has been discovered in multiple cancers, the clinical relevance of GHRH-R in tumorigenic progression and in clinical outcomes is largely elusive.
Among the multiple oncoproteins, a serine/threonine protein kinase designated “p21-activated kinase 1” (PAK1), which is stimulated by active Rac1 and Cdc42-GTPases, works as a node of cancer-signaling networks (21, 22). PAK1 overexpression has been observed frequently in a variety of cancers. Aberrant PAK1 plays a critical role in tumor cell proliferation and invasiveness, thereby modulating oncogenesis and tumorigenic progression (21, 22). Because PAK1 has been recognized as a potential pharmacological molecular target, the clinical pipeline of molecular therapy targeting PAK1 has been growing (21–23). Our previous studies and those of other investigators demonstrated that PAK1 is up-regulated in cancers of the gastrointestinal tract, including GC (23–28). Intriguingly, PAK1 also activates the inflammatory signaling induced by H. pylori infection in epithelial cells, further linking PAK1 to human GC pathogenesis (29). Indeed, PAK1 not only induces the oncogenic signaling but also promotes inflammatory pathways, such as STAT3 and NF-κB (29–32), which are associated with inflammation-associated malignancy (7–10), suggesting that PAK1 has a function in inflammation-related tumor progression.
In this study, we show that aberrant GHRH-R is clinically important in GC progression and patient prognosis. Targeting GHRH-R by using MIA-602 induces the inhibition of GC growth both in vitro and in vivo. The inhibitory effects of GHRH-R antagonist are mediated by targeting the inflammatory signaling pathway of PAK1–STAT3/NF-κB.
Results
Overexpression of GHRH-R Is Clinically Important for Tumorigenic Progression and Is Associated with Overall Survival in Human GC.
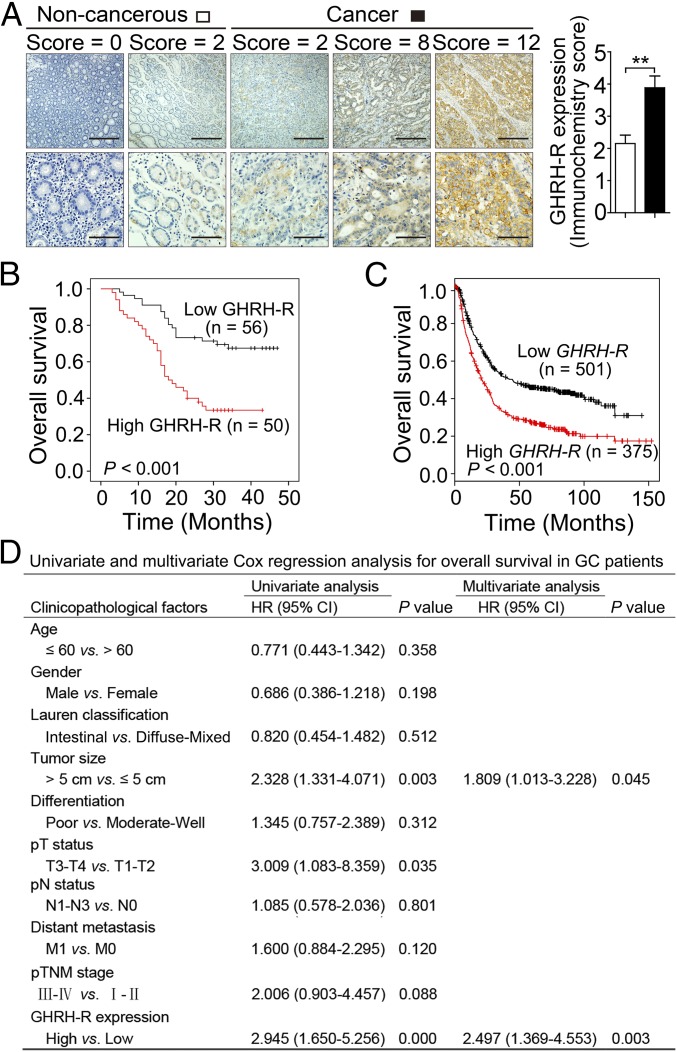
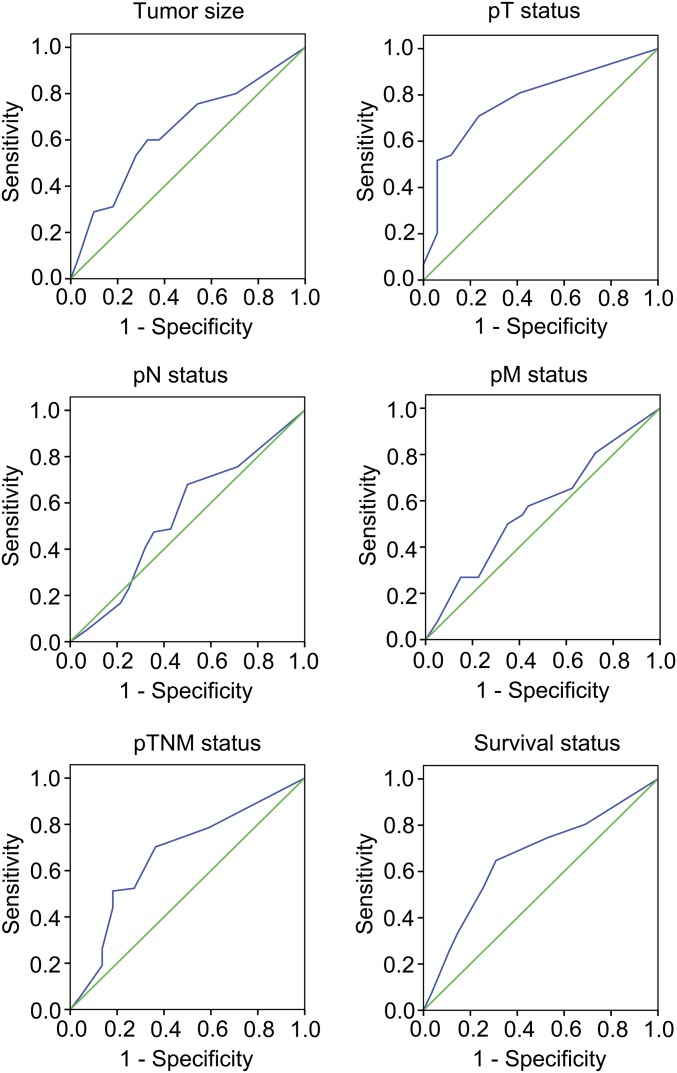
To evaluate the clinical significance of GHRH-R in human GC, we investigated GHRH-R expression in GC specimens from a 106-patient cohort. By analysis of specimens immunohistochemically stained for GHRH-R in primary GC specimens, along with paired adjacent normal tissues, we discovered that the GC tissues exhibited robust expression of GHRH-R compared with normal tissues (P < 0.01) (Fig. 1A). To validate a reasonable cutoff score for GHRH-R overexpression, a receiver operating characteristic (ROC) curve analysis was performed (Fig. S1). Accordingly, the cutoff score for GHRH-R overexpression was 2.5 (immunochemistry score), which was closest to the point with both maximum sensitivity and specificity. The cases with scores ≤2.5 were defined as having low expression of GHRH-R, and those with scores >2.5 were defined as having overexpression of GHRH-R. Overexpression of GHRH-R was detected in 50 of 106 human primary GCs (47.17%). We further examined the association of GHRH-R expression with clinicopathological features of malignant behavior. Correlation analysis demonstrated that the overexpression of GHRH-R was positively correlated with tumor size (P = 0.031) and pathological tumor (pT) status (P = 0.001) (Table S1). Thus, GHRH-R is highly enriched in human GC tissues and correlates closely with cancer progression.
Fig. 1.
GHRH-R overexpression in GC patients is associated with poor survival. (A, Left) Immunohistochemical staining of GHRH-R (brown) on GC sections (n = 106) and paired noncancerous tissues. Nuclei were counterstained with hematoxylin (blue). (Scale bars: Top, 200 μm; Bottom, 50 μm.) (Right) The immunohistochemistry score of GHRH-R in GC (filled bar) and paired noncancerous (open bar) tissues was plotted. Error bars indicate SEM. **P < 0.01 by paired t test. (B) Kaplan–Meier curves compared the overall survival in GC patients with high and low protein levels of GHRH-R. (C) The relationship between overall survival and mRNA levels of GHRH-R in GC by Kaplan–Meier survival analysis. (D) Univariate and multivariate Cox regression analysis of clinicopathological factors in GC patients (n = 106).
Fig. S1.
ROC curve analysis was used to determine the cutoff score for the overexpression of GHRH-R. The sensitivity and specificity for each clinical outcome were plotted. Immunohistochemistry-stained samples were split into groups with high (n = 50) and low (n = 56) GHRH-R expression by ROC analysis. The blue trace represents ROC curve and the green trace represents diagonal reference line.
Table S1.
Correlation between GHRH-R expression and clinicopathological factors in GC patients
| GHRH-R expression | ||||
| Clinicopathological factors | No. of patients | Low GHRH-R, n (%) | High GHRH-R, n (%) | P value |
| Total | 106 | 56 (52.8) | 50 (47.2) | |
| Age (y) | ||||
| ≤60 | 54 | 29 (53.7) | 25 (46.3) | 0.527 |
| >60 | 52 | 27 (51.9) | 25 (48.1) | |
| Sex | ||||
| Female | 31 | 18 (58.1) | 13 (41.9) | 1.000 |
| Male | 75 | 38 (50.7) | 37 (49.3) | |
| Lauren classification | ||||
| Intestinal | 37 | 20 (54.1) | 17 (45.9) | 0.731 |
| Diffuse | 53 | 29 (54.7) | 24 (45.3) | |
| Mixed | 16 | 7 (43.8) | 9 (56.2) | |
| Tumor size | ||||
| ≤5 cm | 61 | 38 (62.3) | 23 (37.7) | 0.031 |
| >5 cm | 45 | 18 (40.0) | 27 (60.0) | |
| Differentiation | ||||
| Moderate-well | 43 | 23 (53.5) | 20 (46.5) | 1.000 |
| Poor | 63 | 33 (52.4) | 30 (47.6) | |
| pT status | ||||
| T1–T2 | 17 | 15 (88.2) | 2 (11.8) | 0.001 |
| T3–T4 | 89 | 41 (46.1) | 48 (53.9) | |
| pN status | ||||
| N0 | 28 | 16 (53.3) | 12 (46.7) | 0.662 |
| N1–N3 | 78 | 40 (51.3) | 38 (48.7) | |
| Distant metastasis | ||||
| M0 | 80 | 45 (56.2) | 35 (43.8) | 0.261 |
| M1 | 26 | 11 (42.3) | 15 (57.7) | |
| pTNM stage | ||||
| I–II | 22 | 16 (72.7) | 6 (27.3) | 0.054 |
| III–IV | 84 | 40 (47.6) | 44 (52.4) | |
pT, pathological tumor; pN, pathological nodal; pTNM, pathological tumor-node-metastasis.
Further, Kaplan–Meier analysis demonstrated that GC patients with increasing expression of GHRH-R showed a poor overall survival (P < 0.001, log-rank test) (Fig. 1B). We further confirmed our findings in a larger cohort containing 876 GC patients from a published dataset (kmplot.com/analysis/index.php?p=service&cancer=gastric) (33), which was multi-institutional and represented various races and countries, including Germany, Australia, Brazil, Korea, Singapore, and China. In line with above observations, this dataset showed that higher GHRH-R expression is associated with a poorer overall survival of GC patients (P < 0.001, log-rank test) (Fig. 1C). Multivariate Cox regression analysis of the 106-patient cohort determined that GHRH-R expression is an independent predictor of prognosis for GC patients [hazard ratio (HR) = 2.497, 95% confidence interval (CI) = 1.369–4.553, P = 0.003] (Fig. 1D). The survival analyses from these multiple cohorts confirmed that GHRH-R overexpression is positively associated with poor overall survival of GC patients and is a potential predictor for GC patients.
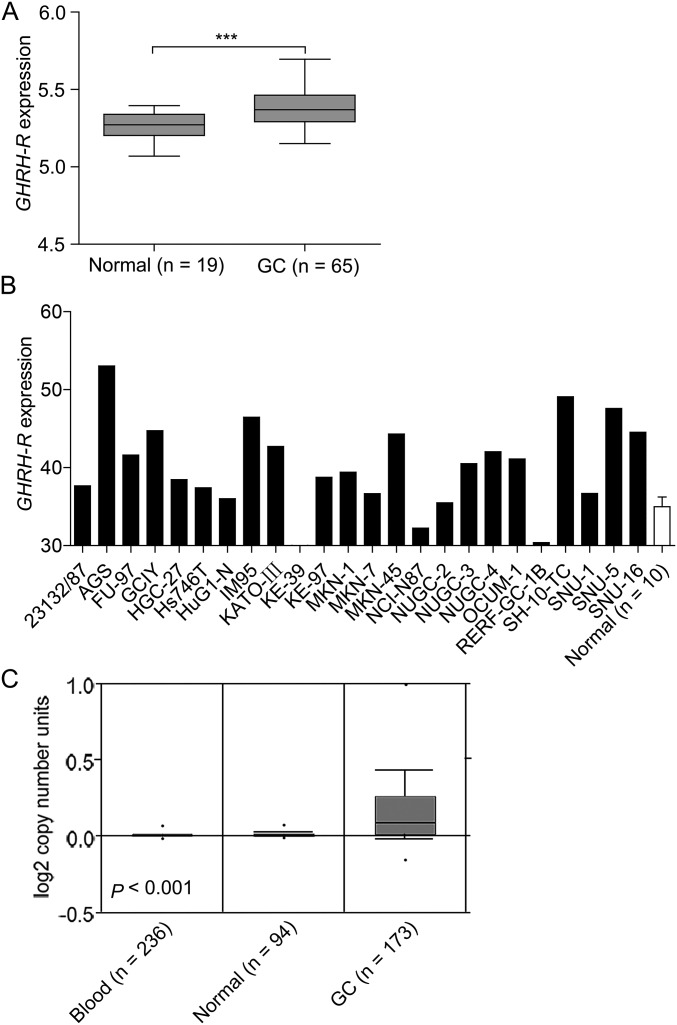
Because increased transcription and gene copy number gain or amplification are alternative mechanisms of oncoprotein overexpression, we examined the status of mRNA expression and gene copy number of GHRH-R in human GC. In a published dataset (GSE13861) including 65 GC specimens and 19 adjacent normal tissues (34), GHRH-R mRNA was found to be significantly elevated in GC specimens vs. normal controls (P < 0.001) (Fig. S2A). Supporting the above findings, 21 of 24 GC cell lines had higher expression of GHRH-R mRNA than did normal gastric tissues (n = 10, MERAV database, merav.wi.mit.edu/) (Fig. S2B) (35). Next, the amplification of the GHRH-R gene in human GC was observed by analyzing DNA copy number using the Oncomine database (Fig. S2C) (36). Thus, in addition to protein overexpression, GHRH-R mRNA is also overexpressed in human GC tissues, along with gene amplification. Together these findings show that increased GHRH-R is clinically important in tumorigenic progression in human GC and correlates closely with patient outcome.
Fig. S2.
mRNA expression and the DNA copy number of GHRH-R was increased in human GC cells and specimens. (A) mRNA levels of GHRH-R in GC specimens were analyzed in the GC dataset from GEO, GSE13861. (B) mRNA levels of GHRH-R in a panel of GC cell lines (filled bars) and normal gastric tissues (open bar) (n = 10) was analyzed using the MERAV database (merav.wi.mit.edu/). Error bar indicates SEM. (C) Gene copy number of GHRH-R in GC was obtained from ONCOMINE. ***P < 0.001 by Student’s t test.
The GHRH-R Antagonist MIA-602 Inhibits the Growth of Human GC Cells in Vitro.
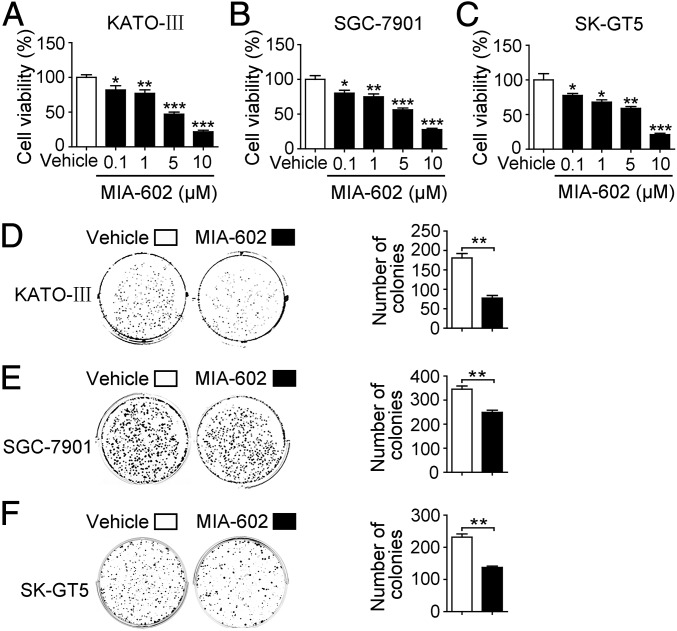
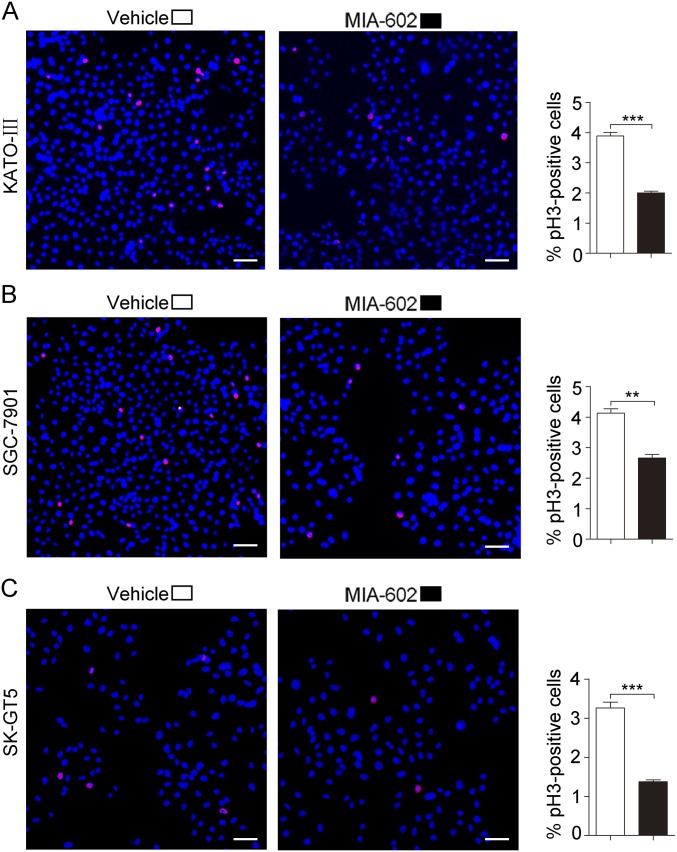
The clinical importance of GHRH-R involvement in tumor progression and patient outcomes underscores the great therapeutic promise of targeting GHRH-R in human GC. We therefore determined the inhibitory efficacy of MIA-602, one of the latest GHRH-R antagonists, on human GC cells. We treated three human GC cell lines, KATO-III, SGC-7901, and SK-GT5, with MIA-602 at 0.1-, 1-, 5-, and 10-μM concentrations for 48 h (15, 16, 37). MIA-602 treatment resulted in reduced cell viability in all cell lines in a dose-dependent manner, as compared with vehicle controls (P < 0.05 for all) (Fig. 2 A–C). Because a striking decrease, by ∼25% (IC25), was seen in cell viability at 1 μM following MIA-602 treatment (Fig. 2 A–C), we chose the dosage of 1 μM for subsequent studies. After exposure to 1 μM MIA-602 for 9 d, all treated cell lines formed fewer colonies than the same cell lines treated with vehicle (control) (P < 0.01 for all) (Fig. 2 D–F). Cellular proliferation represented by phospho-Histone H3 (pH3) immunofluorescence was decreased in all MIA-602–treated cells compared with controls (P < 0.01 for all) (Fig. S3). Therefore, the GHRH-R antagonist MIA-602 inhibits GC cell growth.
Fig. 2.
The inhibitory effect of MIA-602 on the viability of GC cells in vitro. (A–C) The viability of KATO-III (A), SGC-7901 (B), and SK-GT5 (C) cells treated with MIA-602 (0.1, 1, 5, or 10 μM) or vehicle solution for 48 h was measured by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. (D–F) Colony formation of KATO-III (D), SGC-7901 (E), and SK-GT5 (F) cells was decreased by treatment with MIA-602 (filled bar) compared with vehicle treatment (open bar). Quantitative analysis of colony numbers is given on the right. Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by a one-way ANOVA with post hoc intergroup comparisons (A–C) or Student’s t test (D–F); n = 3 in each group (A–F).
Fig. S3.
The expression of pH3 in GC cells was decreased by MIA-602 treatment. (Left) Representative images of immunofluorescence analysis of pH3 in KATO-III (A), SGC-7901 (B), and SK-GT5 (C) cells. (Right) The percentages of pH3+ cells in treated (filled bars) and untreated (open bars) cells were plotted. (Scale bars, 200 μm.) Error bars indicate SEM. **P < 0.01, ***P < 0.001 by Student’s t test; n = 3 in each group.
The GHRH-R Antagonist MIA-602 Inhibits the Growth of Human GC in Vivo.
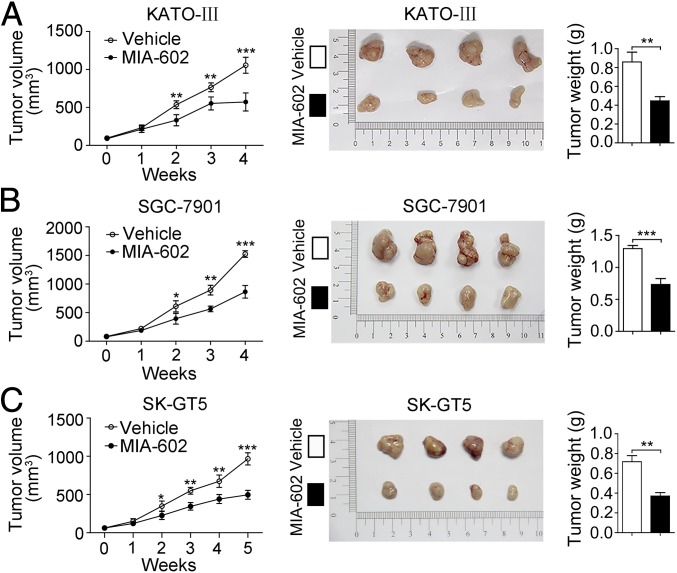
To evaluate the therapeutic potential of the GHRH-R antagonist MIA-602 in a more clinically relevant GC model system, three mouse xenograft models were generated by s.c. inoculation of KATO-III, SGC-7901, or SK-GT5 cells. When individual tumors reached a volume of 75.89 mm3, mice were treated with MIA-602 at a dose of 5 μg/d by s.c. injection (KATO-III- and SGC-7901-mice were treated for 4 wk; SK-GT5-mice was treated for 5 wk). MIA-602 exhibited remarkable inhibitory effects on tumor growth in vivo (P < 0.001 for all) (Fig. 3), as evidenced by the decreased tumor size and weight (Fig. 3). These data indicate that MIA-602 effectively inhibits GC growth in vivo, further indicating that GHRH-R is a potential therapeutic target in human GC.
Fig. 3.
Inhibition of gastric tumor growth in vivo by MIA-602. (A–C, Left) Tumor growth curves of KATO-III (A), SGC-7901 (B), and SK-GT5 (C) models (n = 12 in each group) treated with MIA-602 administration or vehicle. (Center) Tumors were harvested at the end of the experiments. (Right) The average weights of tumors derived from treatment with MIA-602 (filled bar) or control (open bar). Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test.
The GHRH-R Antagonist MIA-602 Inhibits the Growth and Progression of Human GC by Blocking the PAK1–STAT3/NF-κB Axis.
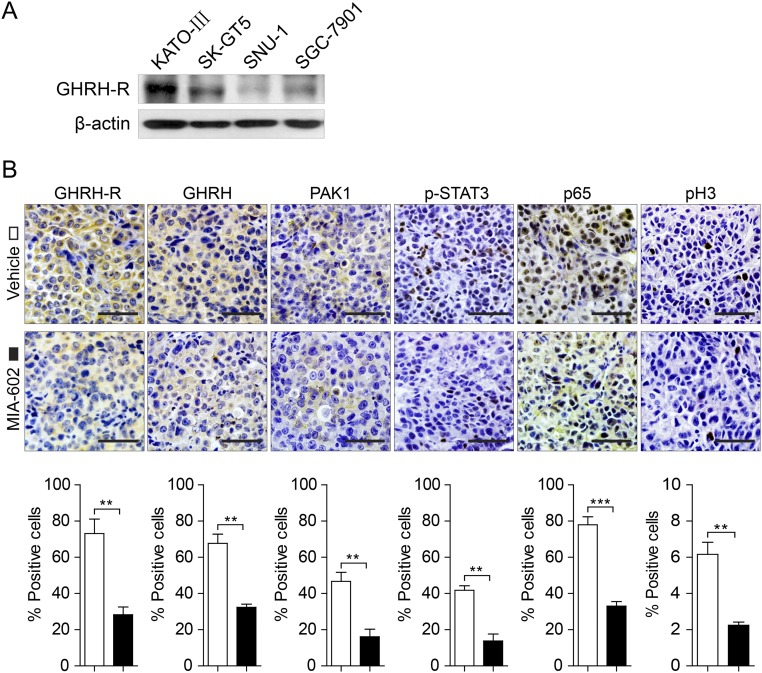
To generate a cellular model for studying the underlying mechanisms responsible for the effects of MIA-602, we examined the expression of GHRH-R in a panel of GC cell lines by Western blotting. Full-length GHRH-R glycosylated protein (55 kDa) was present in all examined cell lines. KATO-III cells had the highest level of GHRH-R; SGC-7901 and SK-GT5 cells expressed moderate levels of GHRH-R; SNU-1 cells expressed a low level of GHRH-R (Fig. S4A).
Fig. S4.
Expression of GHRH-R in GC cells and effects of MIA-602 on inflammatory signaling in vivo. (A) The status of GHRH-R in four GC cell lines was analyzed by Western blotting. β-Actin was used as an internal control. (B, Upper) Representative images of immunohistochemistry of GHRH-R, GHRH, PAK1, p-STAT3, p65, and pH3 in tumors derived from SK-GT5 cells treated with MIA-602. (Scale bars: 50 μm.) (Lower) The percentages of treated (filled bars) and untreated (open bars) cells expressing the indicated proteins were plotted. Error bars indicate SEM. **P < 0.01, ***P < 0.001 by Student’s t test; n = 12 in each group.
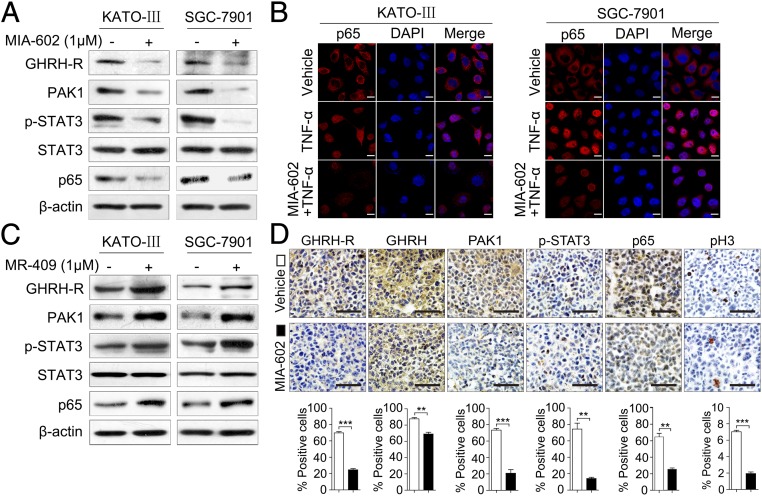
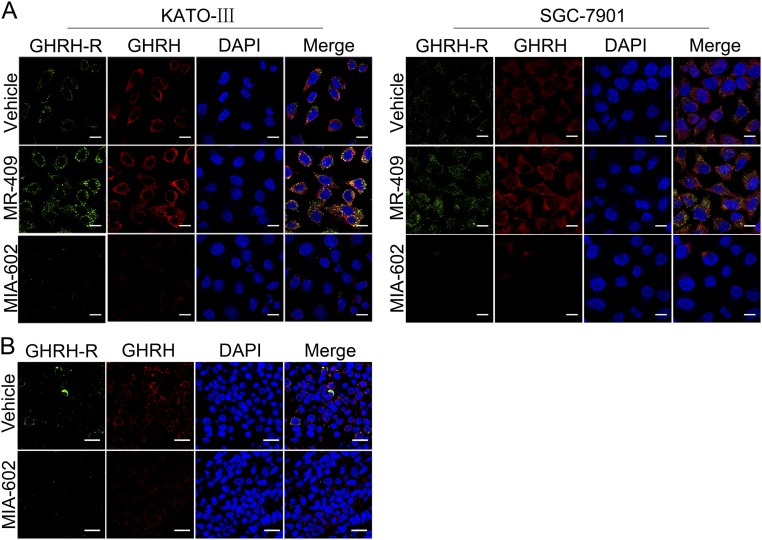
Given that GC is an inflammation-driven cancer, and that MIA-602 has been shown to inhibit inflammatory signaling (14, 38), we investigated whether MIA-602 could suppress inflammatory signaling, specifically in GC. STAT3 and NF-κB, two critical inflammation regulators, were suspected to be the molecular surrogates of downstream GHRH-R signaling. Of note, MIA-602 dramatically inhibited the levels of phosphorylated STAT3 (p-STAT3) and p65, a critical unit of NF-κB, in KATO-III and SGC-7901 cells (Fig. 4A). As expected, treatment with MIA-602 suppressed GHRH-R expression in both cell lines (Fig. 4A). Importantly, TNF-α stimulation, which activates the canonical NF-κB pathway, increased the translocation of p65 to the nucleus in both KATO-III and SGC-7901 cells, whereas MIA-602 strongly inhibited the nuclear translocation (Fig. 4B), indicating that the activation of NF-κB signaling was blocked by MIA-602. Contrarily, p-STAT3 and p65 were both activated by the GHRH-R agonist MR-409 (Fig. 4C), further indicating that targeting GHRH-R can nullify the inflammatory signals of STAT3/NF-κB. In addition, after MIA-602 administration, the expression of p-STAT3 and of p65 was significantly suppressed in SGC-7901 and SK-GT5 xenografted tumors (P < 0.01 for all) (Fig. 4D and Fig. S4B). Cellular proliferation, as evidenced by the number of pH3+ cells, was also decreased significantly by MIA-602 in both xenografted tumors (P < 0.01 for both) (Fig. 4D and Fig. S4B). Interestingly, the expression of both GHRH-R and GHRH was remarkably decreased by MIA-602 treatment in vitro and in vivo, as shown by immunohistochemistry and immunofluorescence confocal microscopy (Fig. 4D and Figs. S4B and S5), whereas treatment by the GHRH-R agonist MR-409 increased the expression of both GHRH-R and GHRH in GC cells (Fig. S5A). These results collectively strongly support the notion that MIA-602 is a suppressor of inflammatory signaling in human GC.
Fig. 4.
MIA-602 suppresses inflammatory signaling in GC. (A) KATO-III and SGC-7901 cells were treated with MIA-602 or vehicle and then were harvested, and equal amounts of lysates were immunoblotted for the labeled antigens. β-Actin was used as an internal control. (B) Subcellular localization of p65 (red) in the KATO-III and SGC-7901 cells, as analyzed by an immunofluorescence confocal assay. Nuclei were stained with DAPI (blue). (C) KATO-III and SGC-7901 cells were treated by MR-409 (1 μM) or vehicle and then were harvested. Equal amounts of lysates were immunoblotted for the labeled antigens. β-Actin was used as an internal control. (D, Upper) Representative images of immunohistochemistry of GHRH-R, GHRH, PAK1, p-STAT3, p65, and pH3 in tumors derived from SGC-7901 cells. (Lower) The percentages of treated (filled bars) and untreated (open bars) cells expressing the indicated proteins were plotted. Error bars indicate SEM. **P < 0.01, ***P < 0.001 by Student’s t test; n = 12 in each group. (Scale bars: B, 10 μm; D, 50 μm.)
Fig. S5.
Effects of MIA-602 or MR-409 on the expression of GHRH-R and GHRH. (A) Representative images of confocal microscopy analysis of GHRH-R (green) and GHRH (red) in KATO-III and SGC-7901 cells treated with MIA-602 or MR-409. Nuclei were stained with DAPI (blue). (B) Representative images of confocal microscopy analysis of GHRH-R (green) and GHRH (red) in tumors derived from SGC-7901 cells. Nuclei were stained with DAPI (blue). (Scale bars: A, 10 μm; B, 25 μm.)
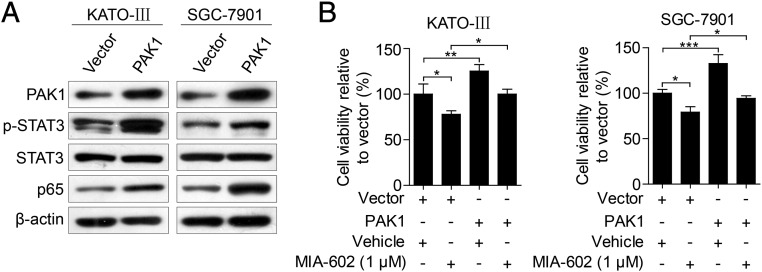
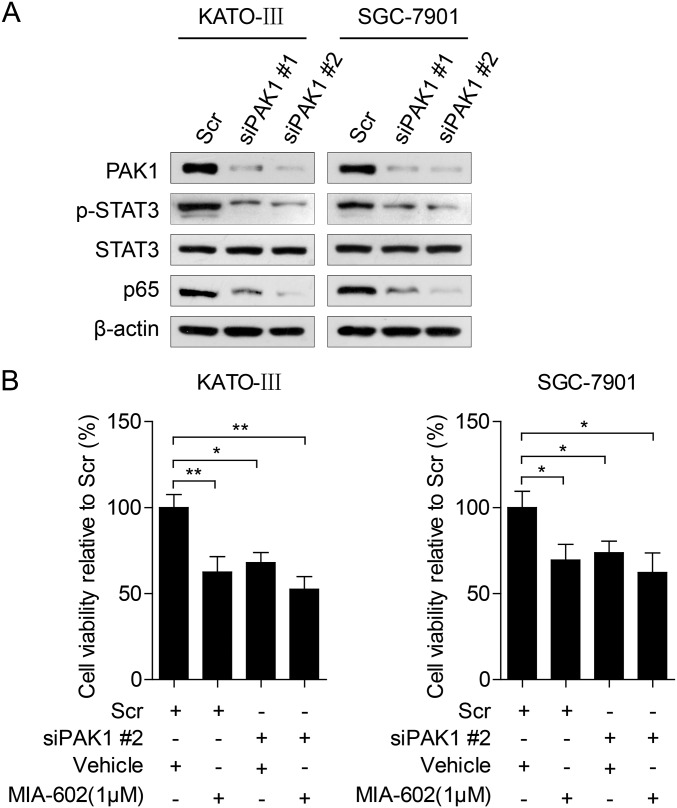
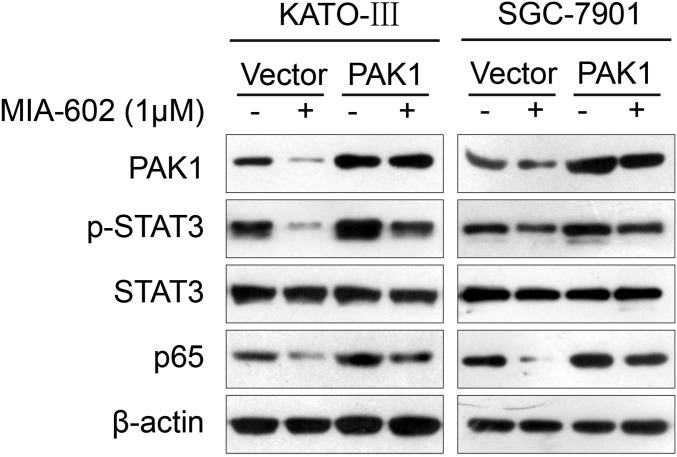
As a nodal regulator of cancer-inflammation signaling networks (21), PAK1 has been recognized as an activator of STAT3 and NF-κB (29–32). We next tested whether the anti-neoplastic effects of GHRH-R on GC depend on PAK1-mediated STAT3/NF-κB activity. MIA-602 administration suppressed PAK1 expression in cultured KATO-III and SGC-7901 cells (Fig. 4A) and also in xenograft tumors (P < 0.01 for both) (Fig. 4D and Fig. S4B). Contrarily, the expression of PAK1 was up-regulated by the GHRH-R agonist MR-409 in both GC cells lines (Fig. 4C). Thus, experimental results using both a GHRH-R antagonist and a GHRH-R agonist indicate that pharmacological targeting of GHRH-R is able to regulate oncogenic kinase PAK1 effectively, suggesting that PAK1 may be a downstream effector of GHRH-R in GC. Furthermore, ectopic expression of PAK1 obviously enhanced the expression levels of p-STAT3 and p65 in KATO-III and SGC-7901 cells (Fig. 5A), whereas the knockdown of PAK1 by two specific siRNAs remarkably down-regulated the expression of p-STAT3 and p65 in both cell lines (Fig. S6A). We further investigated whether overexpression of PAK1 can rescue the MIA-602–induced anti-neoplastic effects in GC cells in PAK1-overexpressing KATO-III and SGC-7901 cells treated with MIA-602. The results showed that overexpression of PAK1 virtually abrogated MIA-602–induced anti-neoplastic effects in both GC cells as well as the inhibition of p-STAT3 and p65 (Fig. 5B and Fig. S7). In contrast, knockdown of PAK1 did not dramatically potentiate MIA-602–induced inhibition of cell viability (Fig. S6B). Thus, these data further demonstrate that MIA-602 exerts the anti-neoplastic effects by blocking PAK1-mediated STAT3/NF-κB signaling.
Fig. 5.
PAK1 overexpression reverses the anti-neoplastic effects of MIA-602. (A) KATO-III and SGC-7901 cells were transiently transfected with PAK1-overexpressing plasmid or control vector and were subjected to Western blotting for the indicated antigens. β-Actin was used as an internal control. (B) The viability of PAK1-overexpressing cells that were treated with MIA-602 (1 μM) or vehicle for 48 h was assessed by MTT assay. Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with post hoc intergroup comparisons; n = 3 in each group.
Fig. S6.
Effects of MIA-602 in PAK1-silenced GC cells. (A) KATO-III (Left) and SGC-7901 (Right) cells were transiently transfected with PAK1 siRNAs or scramble (Scr) siRNA for 48 h and then were subjected to Western blotting for the indicated antigens. β-Actin was used as an internal control. (B) Cell viabilities of PAK1-silenced cells that were treated with MIA-602 (1 μM) or vehicle for 48 h were assessed by MTT assays. Error bars indicate SEM. *P < 0.05, **P < 0.01 by a one-way ANOVA with post hoc intergroup comparisons; n = 3 in each group.
Fig. S7.
PAK1 overexpression reverses the inhibitory effects of MIA-602 on inflammatory signatures. KATO-III (Left) and SGC-7901 (Right) cells transiently transfected with PAK1 overexpression plasmid or control vector were treated with MIA-602 (1 μM) or vehicle for 48 h and then subjected to Western blotting for the indicated antigens. β-Actin was used as an internal control.
Discussion
This study has provided experimental and clinical evidence demonstrating the clinical importance of the association of GHRH-R with human GC progression and with patient survival and has revealed the targeting effects of a GHRH-R antagonist, MIA-602, on GC in vitro and in vivo. We also revealed the previously unrecognized mechanisms by which MIA-602 blocks the PAK1–STAT3/NF-κB axis and thereby suppresses growth of GC.
Although the presence of GHRH-R has been previously identified in multiple types of human cancers (11–19), the clinical importance of GHRH-R in the tumorigenic progression and patient outcome was unappreciated prior to this study. In this study, by analyzing multiple cohorts of GC patients of different races, we defined the heretofore undocumented associations between GHRH-R and poor survival of human GC. Interestingly, by analysis of a local patient cohort that contains clear clinicopathological information, we identified the overexpression of GHRH-R as an independent predictive factor for patient prognosis, although this finding needs to be validated across the spectrum of multiple cohorts in the future. The observed amplification of the GHRH-R gene in GC tissues suggests that the gain in gene copy number may be a mechanism that contributes to GHRH-R overexpression in human GC. These data predict the contribution of GHRH-R to human cancer progression and demonstrate the rationale for using GHRH-R as a therapeutic target in GC.
MIA-602 is one of the latest in a series of highly potent GHRH-R antagonists and is based on an earlier antagonist, JMR-132 (11, 15, 16, 37). MIA-602 has been reported to possess greater potency and stability in antitumor activity than JMR-132 (15). Using MIA-602, we systemically examined its therapeutic effects by targeting GHRH-R in human GC cells in vitro and in vivo and found that MIA-602 exerts higher anti-neoplastic effects than previous antagonists, as we previously demonstrated in cell lines (11, 17). In the current study we validated the beneficial effects of MIA-602 in multiple cellular models and three independent human GC xenograft models, establishing MIA-602 as an effective anticancer agent in human GC.
Studies of different classes of GHRH-R antagonists in cancers demonstrate the GHRH-R antagonists’ ability to modulate multiple intracellular pathways involved in cellular proliferation, survival, metastasis, apoptosis, and inflammation (11–14). The identified mechanisms of GHRH-R antagonists include suppression of the canonical pituitary–hepatic GH–insulin-like growth factor I (IGF-I) axis (11), inhibition of autocrine and/or paracrine production of IGFs in tumors, and blocking the direct stimulatory action of local GHRH on tumors (11). Beyond these canonical pathways, accumulating evidence indicates that additional intracellular molecules and pathways can be modulated by GHRH-R antagonists. JMR-132 induced cell-growth inhibition and apoptosis through PKCδ-mediated activation of p53/p21 in endometrial cancer (39) and caused p21-mediated S-phase arrest in colon cancer (40). Another GHRH-R antagonist, MZ-J-7-138, up-regulated wild-type p53 but down-regulated mutant p53 and p21 in prostate cancer (41). Similarly, the GHRH-R antagonists JV-1-65 and JV-1-63 inhibited mutant p53 expression in small cell lung carcinoma (42). MZ-5-156 inhibited the release of matrix metalloproteinases, which led to the suppression of invasion in lung cancer (43). In lung cancer and prostate cancer, MZ-J-7-118 and RC-J-29-18 decreased the expression of the EGFR/HER family (44, 45). In ovarian cancer, JMR-132 inactivated the EGFR–Akt pathway (46). Of note, GHRH-R antagonists also modulate multiple inflammation-relevant molecules and pathways. JMR-132 exhibited antioxidant activity by decreasing NF-κB and COX-2 in prostate cancer (47) and inactivated the JAK2/STAT3 pathway in prostate hyperplasia (48). JMR-132, MIA-313, and MIA-459 suppressed the expression of IL-1β, NF-κB, and COX-2 in prostatic hyperplasia (49). MIA-602 inhibited NF-κB in glioblastoma, breast cancer, and ovarian cancer (37). Most recently, MIA-602 was shown to inactivate STAT3 in mouse bone marrow-derived mesenchymal stem cells (50) and to down-regulate the expression of inflammatory cytokines in experimental ocular inflammation (38) and triple-negative breast cancer (14).
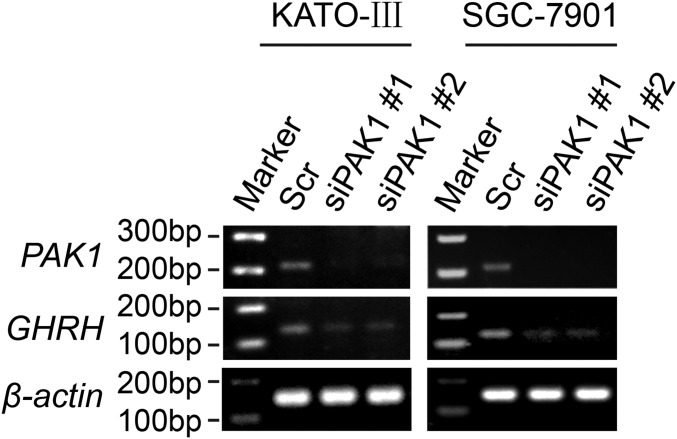
Although multiple inflammation-associated molecules and pathways have been reported to be modulated by GHRH-R antagonists, the key molecule that links GHRH-R to these downstream pathways remains to be discovered. In our study, the STAT3/NF-κB signaling pathway was markedly inhibited by MIA-602. Furthermore, the inhibited STAT3/NF-κB signaling in MIA-602–treated cells was attributed to the down-regulation of PAK1. These data provide mechanistic insights into the actions of GHRH-R antagonists on human cancers. Given that PAK1 has been reported to participate in the activation of inflammatory signaling in H. pylori-infected epithelial cells (29), it is reasonable to predict that MIA-602–induced down-regulation of PAK1 is a critical mechanism by which GHRH-R antagonists inhibit inflammatory signaling pathways in human GC. Because STAT3 may be activated by multiple growth factors and cytokines in GC cells (8, 51), the STAT3 inactivation induced by MIA-602 may involve a more complex regulation. Future studies are required to examine whether MIA-602 interferes with the identified signaling in other mechanistic manners. Moreover, the mechanism by which MIA-602 alters the GHRH–GHRH-R axis and PAK1-signaling pathways may be more complicated. The GHRH–GHRH-R axis and other growth factors may mediate the down-regulation of PAK1 induced by MIA-602 (52–54), whereas PAK1 may regulate GHRH in the feedback regulation (Fig. S8).
Fig. S8.
Knockdown of PAK1 reduces the expression of GHRH. KATO-III (Left) and SGC-7901 (Right) cells were transiently transfected with PAK1 siRNAs or scramble (Scr) siRNA for 48 h, and then RNA extracted from these cells was subjected to RT-PCR for PAK1 and GHRH. β-Actin was used as an internal control.
In summary, these studies have illuminated long-standing questions on how GHRH-R expression correlates with clinical outcomes in human cancers, establishing the role of GHRH-R as a molecular signature as well as a potential prognostic predictor of GC. MIA-602 remarkably inhibits the growth of human GC in vitro and in vivo through the suppression of PAK1–STAT3/NF-κB signaling. Our study strongly highlights the therapeutic potential of GHRH-R antagonists in the treatment of GC patients. Knowledge gained in our study will shed light on how to select the appropriate patients for personalized cancer therapy using GHRH-R antagonists.
Materials and Methods
Clinical Patients and Samples.
A total of 106 paraffin-embedded specimens of human primary GCs and paired noncancerous tissues were collected from the Sun Yat-sen University Cancer Center during 2011–2012. All samples were surgical cases. All samples were histopathologically and clinically confirmed as GC. No patients in the study had undergone preoperative radiotherapy or chemotherapy for GC. Clinical research protocol of this study was reviewed and approved by the Ethics Committee of Sun Yat-sen University Cancer Center. Written informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Peptides and Chemicals.
The GHRH-R antagonist MIA-602 and the GHRH-R agonist MR-409 were synthesized by R.C. and A.V.S at the Miami Veterans Administration hospital and the University of Miami. For in vitro experiments, MIA-602 and MR-409 were dissolved in 100% DMSO [American Chemical Society (ACS) grade; Sigma-Aldrich] and diluted with appropriate incubation medium. The concentration of DMSO never exceeded 0.1%. The details of MIA-602 dilution for in vivo experiments are given in SI Materials and Methods.
Tumor Xenografts.
A total of 2 × 106 KATO-III, SGC-7901, or SK-GT5 cells were resuspended in 150 μL PBS and were injected s.c. into the flanks of 5- to 6-wk-old nude mice (Vital River Laboratory Animal Technology Co. Ltd.). Details of experiments with the tumor xenografts are given in SI Materials and Methods. These animal experiments were reviewed and approved by the Ethics Committee of Shantou University Medical College.
Materials and methods are described further in SI Materials and Methods.
SI Materials and Methods
Immunohistochemistry.
Immunohistochemistry staining was performed as previously described (55). Briefly, 4-μm sections were cut either from clinical GC specimens or from the tumor xenograft tissues that had been fixed in 10% (vol/vol) neutral-buffered formalin. The sections then were processed, deparaffinized, rehydrated, and incubated with primary antibodies overnight at 4 °C, followed by incubation with the HRP-conjugated secondary antibodies at 37 °C for 1 h. Staining was visualized by incubation with 3, 3′-diaminobenzidine (DAB) substrate, which resulted in a brown-colored precipitate at the antigen site. Nuclei were counterstained with hematoxylin. Immunostaining results were graded semiquantitatively by counting the percentage of positive cells and the intensity of cells. The specimens were evaluated by two independent observers who were unaware of the clinical information.
Peptides and Chemicals.
The GHRH-R antagonist MIA-602 and the GHRH-R agonist MR-409 were synthesized by R.C. and A.V.S. at the Miami Veterans Administration hospital and the University of Miami. For in vitro experiments, MIA-602 and MR-409 were dissolved in 100% DMSO (ACS grade; Sigma-Aldrich) and diluted with appropriate incubation medium. The concentration of DMSO never exceeded 0.1% (vol/vol). For in vivo experiments, MIA-602 was dissolved in 100% DMSO and then was diluted 1:500 in 5.5% mannitol (Sigma-Aldrich) in sterile water (vehicle solution) for daily s.c. injection.
Tumor Xenografts.
A total of 2 × 106 KATO-III, SGC-7901, or SK-GT5 cells were resuspended in 150 μL PBS and were injected s.c. into the flanks of 5- to 6-wk-old nude mice (Vital River Laboratory Animal Technology Co. Ltd.). Once an individual tumor volume reached an average volume of 75.89 mm3, the mice were randomized into two groups (n = 12). Both groups were treated s.c. with 5 μg of the GHRH-R antagonist MIA-602 or with vehicle solution daily for 4 wk (KATO-III and SGC-7901) or 5 wk (SK-GT5). Tumor volumes were measured weekly, and tumor mass was calculated by the following formula: volume = 0.5236 × length × width2. Mice were killed after 4 or 5 wk of treatment, and tumors were excised for paraffin block preservation. These animal experiments were reviewed and approved by the Ethics Committee of Shantou University Medical College.
Cell Culture and Reagents.
The human GC cell lines KATO-III and SNU-1 were obtained from the American Type Culture Collection (ATCC). The human GC cell line SGC-7901 was obtained from the Cell Bank of the Chinese Academy of Sciences. The human GC cell line SK-GT5 was kindly provided by X. C. Xu, University of Texas MD Anderson Cancer Center, Houston, TX. All cell lines were cultured in RPMI 1640 (Gibco/Invitrogen) supplemented with 10% FBS (Gibco/Invitrogen).
Antibodies against PAK1 (catalog no. 2602), p-STAT3 (catalog no. 9145), STAT3 (catalog no. 9132), p65 (catalog no. 8242), and β-actin (catalog no. 4970) were purchased from Cell Signaling Technology. An antibody against GHRH-R (catalog no. sc-54201) was obtained from Santa Cruz Biotechnology. An antibody against GHRH (catalog no. ab187512) was purchased from Abcam. An antibody against pH3 (catalog no. 06-570) was obtained from Upstate Biotechnology. An Alexa Fluor 594 (Red)–conjugated donkey anti-rabbit secondary antibody (catalog no. A21207) and an Alexa Fluor 488 (Green)–conjugated donkey anti-goat secondary antibody (catalog no. A21206) were purchased from Invitrogen. TNF-α (catalog no. H8916) was obtained from Sigma-Aldrich.
Transfection Experiments.
Transfections were performed according to the manufacturer’s instructions using Lipofectamine 3000 (Life Technologies). The transfections were performed as described previously (10). Briefly, KATO-III and SGC-7901 cells were seeded onto a six-well plate and then were transfected with 2.5 μg pCMV6M-PAK1 plasmid DNA or 100 pM siRNA (siPAK1 #1 sense: 5′-GAUGCUUUGACCCGGAAUATT-3′ and antisense: 5′-UAUUCCGGGUCAAAGCAUCTT-3′; siPAK1 #2 sense: 5′-GGCGAUCCUAAGAAGAAAUTT-3′ and antisense: 5′-AUUUCUUCUUAGGAUCGCCTT-3′; scramble siRNA sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense: 5′-ACGUGACACGUUCGGAGAATT-3′) using Lipofectamine 3000 (Life Technologies). Forty-eight hours after transfection, cells were subjected to the indicated experiment.
Western Blotting.
Western blotting was performed as described previously (56). Briefly, the cells were lysed in RIPA buffer, and then equivalent amounts of the protein extracts were resolved using 10% SDS/PAGE and were transferred to a PVDF membrane. The membranes were blocked in 5% skim milk in TBS containing 0.1% Tween-20 (TBST) buffer and then were incubated with the primary antibodies at 4 °C overnight. After incubation with the primary antibodies, the secondary antibodies were added and incubated for 2 h at room temperature. The immunoreactive bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and were exposed to X-ray film (Eastman Kodak).
RT-PCR.
Total RNA was isolated from cells transfected with PAK1 siRNAs or scramble siRNA by the TRIzol reagent (Invitrogen) and was reverse transcribed using the M-MLV First Strand kit (Invitrogen) according to the manufacturer’s instructions. The cDNA was subjected to PCR with the following primers: PAK1 forward: 5′-CACCACTCCACCAGATGCTT-3′ and reverse: 5′-GCTTAATGGCCACCTCCTGT-3′; GHRH forward: 5′-GAGAGAGCAACCAAGAGCGA-3′ and reverse: 5′-GAGTTCCTGCTGTGCTTCTGC-3′; β-actin forward: 5′-GAACCCCAAGGCCAACCGCGAGA-3′ and reverse: 5′-TGACCCCGTCACCGGAGTCCATC-3′.
Immunofluorescence and Confocal Microscopy.
The cells or sections were processed before being subjected to immunofluorescence analysis as described previously (55) and then were incubated with specific primary antibodies overnight at 4 °C, followed by Alexa Fluor 594 (red)–conjugated donkey anti-rabbit secondary antibody or Alexa Fluor 488 (green)–conjugated donkey anti-goat secondary antibody for 1 h in darkness. Nuclei were stained with DAPI. Images were captured using Zeiss Imager A2a fluorescence microscope or an Olympus FV-1000 confocal fluorescence microscope.
Cell-Viability Assays.
KATO-III, SGC-7901, or SK-GT5 cells were seeded into 96-well plates at a density of 2,000 cells per well, cultured overnight, starved for 24 h in medium without FBS, and then treated for 48 h with different concentrations (0.1, 1, 5, or 10 μM) of the GHRH-R antagonist MIA-602 in a medium containing 0.5% FBS. Cell viability was measured by MTT assays as described previously (57).
Colony-Formation Assays.
Colony-formation assays were performed as described previously (57). Briefly, KATO-III, SGC-7901, or SK-GT5 cells were seeded into six-well plates in triplicate at a concentration of 500 cells per well. After 24 h incubation, the cells were washed with PBS, and then RPMI 1640 (Gibco/Invitrogen) supplemented with either 2% FBS (Gibco/Invitrogen) with 1 μM MIA-602 or vehicle solution was added. The medium was changed every 3 d. Cells were treated for a total of 9 d. The cell colonies then were fixed by methanol and dyed with 0.1% crystal violet; the colonies that contained more than 50 cells were counted.
TNF-α Treatment.
Cells were seeded on glass coverslips for 24 h and then were pretreated with 1 μM MIA-602 or vehicle solution for 1 h, followed by treatment with 10 ng/mL TNF-α for 30 min. After the treatment, the cells were subjected to immunofluorescence analysis and visualized with confocal microscopy (FV-1000; Olympus).
Bioinformatic Analysis.
Kaplan–Meier survival analysis of mRNA for GHRH-R was performed with an online tool (kmplot.com/analysis/index.php?p=service&cancer=gastric) (33). The database was established using gene-expression data and survival information for 876 GC patients originating from seven independent microarray datasets: GSE22377 (n = 43, Germany), GSE51105 (n = 93, Australia), GSE14210 (n = 119, Korea), GSE62254 (n = 283, Korea), GSE38749 (n = 15, Brazil), GSE15459 (n = 197, Singapore), and GSE29272 (n = 126, China). A normalized GC microarray dataset, GSE13861 (34), was downloaded from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (58) and was used to analyze the mRNA levels of GHRH-R in GC and adjacent normal tissues. The probe used to detect GHRH-R transcript expression was ILMN_1740186. The expression of mRNA for GHRH-R in GC cell lines was obtained from the MERAV database (merav.wi.mit.edu/) (35). A dataset (titled “TCGA Gastric”) from the Oncomine database (https://www.oncomine.org//) (36), was used to investigate the DNA copy number levels of GHRH-R in GC tissues. The reporter used to detect GHRH-R DNA copy number was 07-030977914.
Statistical Analysis.
All statistical analyses were performed using the SPSS 13.0 statistical software package (SPSS Inc.). An ROC curve analysis was performed to define the cutoff score for the overexpression of GHRH-R. The correlation between GHRH-R expression and clinicopathological features of GC patients was analyzed by the χ2 test. Overall survival was estimated using the Kaplan–Meier method, and the difference in survival was evaluated using the log-rank test. Univariate and multivariate survival analyses were done with the Cox proportional hazards regression model. Comparisons between two groups were performed with a Student’s (or paired) t test, and comparisons among more than two groups were performed with one-way ANOVA with post hoc intergroup comparisons. All bar graphs show the mean ± SEM of at least three independent experiments. A P value of less than 0.05 was considered statistically significant.
Acknowledgments
We thank the Li Ka Shing Foundation for generous support. This work was supported by a block grant from the University Grants Committee Hong Kong (to C.P.P.) and in part by National Natural Science Foundation of China Grants 81071736, 30973508, and 81572876 and the Clinical Research Enhancement Initiative of Shantou University Medical College Grants 201412 and 201421 (all to H.Z.), by funding for the Collaborative and Creative Center, Molecular Diagnosis and Personalized Medicine, Shantou University, Guangdong Province (H.Z.), and by funding from the Department of Education, Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases. The work at the Veterans Affairs Medical Center, Miami, was supported by the Medical Research Service of the Department of Veterans Affairs (A.V.S.), the South Florida Veterans Affairs Foundation for Research and Education (A.V.S.), and the Sylvester Comprehensive Cancer Center, Miller School of Medicine of the University of Miami (A.V.S.).
Footnotes
Conflict of interest statement: A.V.S. and R.C. are listed as co-inventors on patents on GHRH analogs which were assigned to the University of Miami and Veterans Affairs Department.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618582114/-/DCSupplemental.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016:S0140-6736(16)30354-3. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 3.Wadhwa R, et al. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10(11):643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lordick F, Janjigian YY. Clinical impact of tumour biology in the management of gastroesophageal cancer. Nat Rev Clin Oncol. 2016;13(6):348–360. doi: 10.1038/nrclinonc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elinav E, et al. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 6.Polk DB, Peek RM., Jr Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: Central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10(12):1314–1319. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 9.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, et al. Metformin promotes autophagy and apoptosis in esophageal squamous cell carcinoma by downregulating Stat3 signaling. Cell Death Dis. 2014;5:e1088. doi: 10.1038/cddis.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schally AV, Varga JL, Engel JB. Antagonists of growth-hormone-releasing hormone: An emerging new therapy for cancer. Nat Clin Pract Endocrinol Metab. 2008;4(1):33–43. doi: 10.1038/ncpendmet0677. [DOI] [PubMed] [Google Scholar]
- 12.Schally AV, Varga JL. Antagonistic analogs of growth hormone-releasing hormone: New potential antitumor agents. Trends Endocrinol Metab. 1999;10(10):383–391. doi: 10.1016/s1043-2760(99)00209-x. [DOI] [PubMed] [Google Scholar]
- 13.Barabutis N, Schally AV. Growth hormone-releasing hormone: Extrapituitary effects in physiology and pathology. Cell Cycle. 2010;9(20):4110–4116. doi: 10.4161/cc.9.20.13787. [DOI] [PubMed] [Google Scholar]
- 14.Perez R, et al. Antagonists of growth hormone-releasing hormone suppress in vivo tumor growth and gene expression in triple negative breast cancers. Oncotarget. 2012;3(9):988–997. doi: 10.18632/oncotarget.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozsgai E, et al. The effect of GHRH antagonists on human glioblastomas and their mechanism of action. Int J Cancer. 2010;127(10):2313–2322. doi: 10.1002/ijc.25259. [DOI] [PubMed] [Google Scholar]
- 16.Klukovits A, et al. Novel antagonists of growth hormone-releasing hormone inhibit growth and vascularization of human experimental ovarian cancers. Cancer. 2012;118(3):670–680. doi: 10.1002/cncr.26291. [DOI] [PubMed] [Google Scholar]
- 17.Busto R, et al. The expression of growth hormone-releasing hormone (GHRH) and splice variants of its receptor in human gastroenteropancreatic carcinomas. Proc Natl Acad Sci USA. 2002;99(18):11866–11871. doi: 10.1073/pnas.182433099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahrenholtz CD, et al. Preclinical efficacy of growth hormone-releasing hormone antagonists for androgen-dependent and castration-resistant human prostate cancer. Proc Natl Acad Sci USA. 2014;111(3):1084–1089. doi: 10.1073/pnas.1323102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havt A, et al. The expression of the pituitary growth hormone-releasing hormone receptor and its splice variants in normal and neoplastic human tissues. Proc Natl Acad Sci USA. 2005;102(48):17424–17429. doi: 10.1073/pnas.0506844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarandi M, et al. Synthesis and biological activities of highly potent antagonists of growth hormone-releasing hormone. Proc Natl Acad Sci USA. 1994;91(25):12298–12302. doi: 10.1073/pnas.91.25.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dammann K, Khare V, Gasche C. Tracing PAKs from GI inflammation to cancer. Gut. 2014;63(7):1173–1184. doi: 10.1136/gutjnl-2014-306768. [DOI] [PubMed] [Google Scholar]
- 22.Vadlamudi RK, et al. An essential role of Pak1 phosphorylation of SHARP in Notch signaling. Oncogene. 2005;24(28):4591–4596. doi: 10.1038/sj.onc.1208672. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, et al. Personalizing risk stratification by addition of PAK1 expression to TNM staging: Improving the accuracy of clinical decision for gastroesophageal junction adenocarcinoma. Int J Cancer. 2015;136(7):1636–1645. doi: 10.1002/ijc.29167. [DOI] [PubMed] [Google Scholar]
- 24.Ching YP, et al. P21-activated protein kinase is overexpressed in hepatocellular carcinoma and enhances cancer metastasis involving c-Jun NH2-terminal kinase activation and paxillin phosphorylation. Cancer Res. 2007;67(8):3601–3608. doi: 10.1158/0008-5472.CAN-06-3994. [DOI] [PubMed] [Google Scholar]
- 25.Carter JH, et al. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res. 2004;10(10):3448–3456. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, et al. PAK1-mediated MORC2 phosphorylation promotes gastric tumorigenesis. Oncotarget. 2015;6(12):9877–9886. doi: 10.18632/oncotarget.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, et al. Prognostic importance and therapeutic implications of PAK1, a drugable protein kinase, in gastroesophageal junction adenocarcinoma. PLoS One. 2013;8(11):e80665. doi: 10.1371/journal.pone.0080665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gan J, et al. Dysregulation of PAK1 is associated with DNA damage and is of prognostic importance in primary esophageal small cell carcinoma. Int J Mol Sci. 2015;16(6):12035–12050. doi: 10.3390/ijms160612035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foryst-Ludwig A, Naumann M. p21-activated kinase 1 activates the nuclear factor kappa B (NF-kappa B)-inducing kinase-Ikappa B kinases NF-kappa B pathway and proinflammatory cytokines in Helicobacter pylori infection. J Biol Chem. 2000;275(50):39779–39785. doi: 10.1074/jbc.M007617200. [DOI] [PubMed] [Google Scholar]
- 30.Shrestha Y, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene. 2012;31(29):3397–3408. doi: 10.1038/onc.2011.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, et al. PAK1 mediates pancreatic cancer cell migration and resistance to MET inhibition. J Pathol. 2014;234(4):502–513. doi: 10.1002/path.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dammann K, et al. PAK1 modulates a PPARγ/NF-κB cascade in intestinal inflammation. Biochim Biophys Acta. 2015;1853(10 Pt A):2349–2360. doi: 10.1016/j.bbamcr.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szász AM, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016 doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho JY, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaul YD, et al. MERAV: A tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res. 2016;44(D1):D560–D566. doi: 10.1093/nar/gkv1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhodes DR, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellyei S, et al. GHRH antagonists reduce the invasive and metastatic potential of human cancer cell lines in vitro. Cancer Lett. 2010;293(1):31–40. doi: 10.1016/j.canlet.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 38.Qin YJ, et al. Antagonist of GH-releasing hormone receptors alleviates experimental ocular inflammation. Proc Natl Acad Sci USA. 2014;111(51):18303–18308. doi: 10.1073/pnas.1421815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu HM, et al. Growth hormone-releasing hormone antagonist induces apoptosis of human endometrial cancer cells through PKCδ-mediated activation of p53/p21. Cancer Lett. 2010;298(1):16–25. doi: 10.1016/j.canlet.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Hohla F, et al. GHRH antagonist causes DNA damage leading to p21 mediated cell cycle arrest and apoptosis in human colon cancer cells. Cell Cycle. 2009;8(19):3149–3156. doi: 10.4161/cc.8.19.9698. [DOI] [PubMed] [Google Scholar]
- 41.Stangelberger A, et al. Inhibitory effects of antagonists of growth hormone releasing hormone on experimental prostate cancers are associated with upregulation of wild-type p53 and decrease in p21 and mutant p53 proteins. Prostate. 2012;72(5):555–565. doi: 10.1002/pros.21458. [DOI] [PubMed] [Google Scholar]
- 42.Kanashiro CA, et al. Inhibition of mutant p53 expression and growth of DMS-153 small cell lung carcinoma by antagonists of growth hormone-releasing hormone and bombesin. Proc Natl Acad Sci USA. 2003;100(26):15836–15841. doi: 10.1073/pnas.2536558100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siejka A, Barabutis N, Schally AV. GHRH antagonist inhibits focal adhesion kinase (FAK) and decreases expression of vascular endothelial growth factor (VEGF) in human lung cancer cells in vitro. Peptides. 2012;37(1):63–68. doi: 10.1016/j.peptides.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Stangelberger A, et al. Antagonists of growth hormone releasing hormone (GHRH) and of bombesin/gastrin releasing peptide (BN/GRP) suppress the expression of VEGF, bFGF, and receptors of the EGF/HER family in PC-3 and DU-145 human androgen-independent prostate cancers. Prostate. 2005;64(3):303–315. doi: 10.1002/pros.20262. [DOI] [PubMed] [Google Scholar]
- 45.Kanashiro CA, Schally AV, Zarandi M, Hammann BD, Varga JL. Alterations of EGFR/HER, angiogenesis and apoptosis pathways after therapy with antagonists of growth hormone releasing hormone and bombesin in non-small cell lung cancer. Int J Oncol. 2007;30(4):1019–1028. [PubMed] [Google Scholar]
- 46.Guo J, Schally AV, Zarandi M, Varga J, Leung PC. Antiproliferative effect of growth hormone-releasing hormone (GHRH) antagonist on ovarian cancer cells through the EGFR-Akt pathway. Reprod Biol Endocrinol. 2010;8:54. doi: 10.1186/1477-7827-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barabutis N, Schally AV. Antioxidant activity of growth hormone-releasing hormone antagonists in LNCaP human prostate cancer line. Proc Natl Acad Sci USA. 2008;105(51):20470–20475. doi: 10.1073/pnas.0811209106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siejka A, Schally AV, Block NL, Barabutis N. Antagonists of growth hormone-releasing hormone inhibit the proliferation of human benign prostatic hyperplasia cells. Prostate. 2010;70(10):1087–1093. doi: 10.1002/pros.21142. [DOI] [PubMed] [Google Scholar]
- 49.Rick FG, et al. Antagonists of growth hormone-releasing hormone (GHRH) reduce prostate size in experimental benign prostatic hyperplasia. Proc Natl Acad Sci USA. 2011;108(9):3755–3760. doi: 10.1073/pnas.1018086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Q, et al. Profound actions of an agonist of growth hormone-releasing hormone on angiogenic therapy by mesenchymal stem cells. Arterioscler Thromb Vasc Biol. 2016;36(4):663–672. doi: 10.1161/ATVBAHA.116.307126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giraud AS, Menheniott TR, Judd LM. Targeting STAT3 in gastric cancer. Expert Opin Ther Targets. 2012;16(9):889–901. doi: 10.1517/14728222.2012.709238. [DOI] [PubMed] [Google Scholar]
- 52.Rider L, Diakonova M. Adapter protein SH2B1beta binds filamin A to regulate prolactin-dependent cytoskeletal reorganization and cell motility. Mol Endocrinol. 2011;25(7):1231–1243. doi: 10.1210/me.2011-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiao M, Shapiro P, Kumar R, Passaniti A. Insulin-like growth factor-1 regulates endogenous RUNX2 activity in endothelial cells through a phosphatidylinositol 3-kinase/ERK-dependent and Akt-independent signaling pathway. J Biol Chem. 2004;279(41):42709–42718. doi: 10.1074/jbc.M404480200. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, et al. Activation of Rac1-PI3K/Akt is required for epidermal growth factor-induced PAK1 activation and cell migration in MDA-MB-231 breast cancer cells. J Biomed Res. 2011;25(4):237–245. doi: 10.1016/S1674-8301(11)60032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong H, et al. PTPRO represses ERBB2-driven breast oncogenesis by dephosphorylation and endosomal internalization of ERBB2. Oncogene. 2016 doi: 10.1038/onc.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, et al. Aberrant chimeric RNA GOLM1-MAK10 encoding a secreted fusion protein as a molecular signature for human esophageal squamous cell carcinoma. Oncotarget. 2013;4(11):2135–2143. doi: 10.18632/oncotarget.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Q, et al. Resveratrol-induced apoptosis is enhanced by inhibition of autophagy in esophageal squamous cell carcinoma. Cancer Lett. 2013;336(2):325–337. doi: 10.1016/j.canlet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 58.Barrett T, et al. NCBI GEO: Mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]