Significance
The RNA lariat debranching enzyme Dbr1 cleaves the 2′,5′-phosphodiester linkages in intron lariats generated during pre-mRNA splicing. The enzyme is central to RNA metabolism because its activity is required for intron turnover and for the production of small nucleolar RNAs and microRNAs encoded in intronic RNA. Here, the kinetics of Dbr1-mediated debranching of a synthetic RNA substrate are measured by using apoenzyme reconstituted with various divalent cations. The results suggest Fe and Zn are preferred cofactors. Structures of a binuclear catalytic mutant in complex with bona fide branched RNAs reveal a metal-bridging hydroxide positioned to attack the scissile phosphate. The results clarify structure/function relationships in Dbr1 enzymes and are guiding the search for inhibitors that hold promise as therapies for retroviral infections and neurodegenerative disease.
Keywords: RNA debranching, intron lariat, enzyme kinetics, X-ray crystallography, Dbr1
Abstract
Intron lariats are circular, branched RNAs (bRNAs) produced during pre-mRNA splicing. Their unusual chemical and topological properties arise from branch-point nucleotides harboring vicinal 2′,5′- and 3′,5′-phosphodiester linkages. The 2′,5′-bonds must be hydrolyzed by the RNA debranching enzyme Dbr1 before spliced introns can be degraded or processed into small nucleolar RNA and microRNA derived from intronic RNA. Here, we measure the activity of Dbr1 from Entamoeba histolytica by using a synthetic, dark-quenched bRNA substrate that fluoresces upon hydrolysis. Purified enzyme contains nearly stoichiometric equivalents of Fe and Zn per polypeptide and demonstrates turnover rates of ∼3 s−1. Similar rates are observed when apo-Dbr1 is reconstituted with Fe(II)+Zn(II) under aerobic conditions. Under anaerobic conditions, a rate of ∼4.0 s−1 is observed when apoenzyme is reconstituted with Fe(II). In contrast, apo-Dbr1 reconstituted with Mn(II) or Fe(II) under aerobic conditions is inactive. Diffraction data from crystals of purified enzyme using X-rays tuned to the Fe absorption edge show Fe partitions primarily to the β-pocket and Zn to the α-pocket. Structures of the catalytic mutant H91A in complex with 7-mer and 16-mer synthetic bRNAs reveal bona fide RNA branchpoints in the Dbr1 active site. A bridging hydroxide is in optimal position for nucleophilic attack of the scissile phosphate. The results clarify uncertainties regarding structure/function relationships in Dbr1 enzymes, and the fluorogenic probe permits high-throughput screening for inhibitors that may hold promise as treatments for retroviral infections and neurodegenerative disease.
The enzymatic processing of diverse RNA molecules requires selective recognition of their unique physicochemical properties. The sequential trans-esterification reactions catalyzed by the spliceosome yield mature messenger RNA (mRNA) and excised intron lariats (1, 2), the latter of which contain internal branchpoint adenosine nucleotides harboring vicinal 2′,5′- and 3′,5′-phosphodiester linkages (3). Mature mRNA transcripts are exported to the cytosol for protein synthesis, but lariat introns must be linearized before they can be turned over or processed into the subset of small nucleolar RNAs and microRNAs that are derived from intronic RNA (4, 5). The lariat forms when the 2′-hydroxyl group of an adenosine nucleotide near the 3′-end of the intron acts as the nucleophile to attack the 5′-splice site, producing 5′-exon-3′-OH and intron lariat/3′-exon intermediates. The 3′-hydroxyl group of the 5′-exon-3′-OH intermediate subsequently acts as the nucleophile to attack the 3′-splice site, resulting in intron excision and exon ligation (6, 7) (Fig. 1A). The resulting vicinal 2′,5′- and 3′,5′-phosphodiester linkages confer unique topological and chemical features to the branchpoint and flanking nucleotides, and these lariats persist in yeast cells lacking active Dbr1 (RNA lariat debranching enzyme) protein (8), the metallophosphoesterase known as the RNA (intron) debranching enzyme. Intron lariats accumulate to unnaturally high levels in ∆dbr1 strains of Saccharomyces cerevisiae, suggesting hydrolysis of the 2′,5′-linkages is rate-limiting in intron processing and turnover after splicing (8). A central role for Dbr1 activity in normal RNA metabolism is supported by findings that ∆dbr1 strains of Schizosaccharomyces pombe exhibit severe morphological and growth defects (9), whereas ablation of the DBR1 gene in higher eukaryotes is lethal (10, 11), presumably due to their need for a larger array of essential intron-derived small nucleolar RNAs and microRNAs.
Fig. 1.
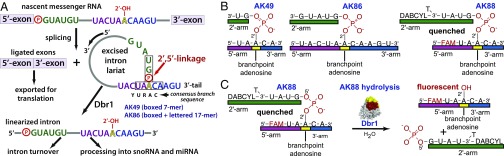
Dbr1 in pre-mRNA splicing and synthetic bRNAs used in this study. (A) The phosphate of the branching 2′,5′-phosphodiester bond is circled in red. The coloring of the branchpoint and flanking nucleotides is preserved throughout the manuscript. The sequence of the synthetic bRNAs AK86 is displayed, and the shorter AK49 sequence is boxed. (B) Cartoon representation of AK49, AK86, and AK88. (C) Dbr1-mediated hydrolysis of the fluorogenic bRNA AK88 permits the 2′- and 5′-arms to separate, relieving the quenching of the 5′-FAM fluorophore by the 2′-DABCYL moiety.
RNA debranching activity was first detected in HeLa cell extracts in 1985 (3). Hydrolysis was selective for 2′,5′-phosphodiester linkages, but RNA nucleotides linked solely by 2′,5′-phosphodiester bonds were not substrates, suggesting the vicinal 2′,5′- and 3′,5′-phosphodiester linkages act as specificity determinants in RNA lariat branchpoint recognition (3). The addition of 10 mM EDTA abolished activity, suggesting a requirement for divalent cations (12). The gene encoding the enzyme was subsequently identified in a yeast genetic screen designed to identify factors involved in Ty1 retrotransposon mobility. Ablation of the DBR1 gene reduced the frequency of transposition with the concomitant accumulation of intron lariats (8). More recently, knockdown of human Dbr1 (hDbr1) gene expression was reported to significantly diminish retroviral replication in cells infected with the HIV (13, 14). This observation is consistent with the evolutionarily conserved genetic structures and replication mechanisms of retrotransposons and retroviruses (15, 16). Together, these studies implicate the RNA lariat debranching enzyme as a participant in retroelement and retroviral replication, although the precise mechanism(s) through which it facilitates these processes remains unclear. Hypotheses regarding the molecular mechanism(s) of Dbr1 in these systems are discussed (17–21).
Yeast Dbr1 isolated from Escherichia coli was reported to possess biochemical properties indistinguishable from the enzyme isolated from yeast (22). Substrates included Y-like trans-splicing intermediates, group II intron lariats, and multicopy single-stranded DNAs (msDNAs) in which the RNA components are attached to the 5′-DNA portions via 2′,5′-phosphodiester linkages (22). Yeast Dbr1 (yDbr1) was active “as-purified,” although EDTA inhibited activity at concentrations of 25 mM or greater, reinforcing the suggestion that a bound metal ion is required for debranching activity. Debranching experiments using msDNAs and “Y”-shaped synthetic bRNAs harboring various branchpoint and flanking nucleotides revealed a strong preference for purines at the 2′-flanking nucleotide position and branchpoints flanked by single 2′, 3′, and 5′ nucleotides were sufficient as substrates for yDbr1, suggesting extensive lariat RNA/enzyme interactions are not a prerequisite for branchpoint recognition (22). In subsequent work using comparative sequence analyses, site-directed mutagenesis, and gel-based RNA debranching assays, Khalid et al. concluded that yDbr1 is a manganese-dependent member of the MPE superfamily of enzymes. The addition of exogenous Zn and other divalent cations was found to suppress debranching activity (23).
The first Dbr1 structures determined were from Entamoeba histolytica (EhDbr1) (24). EhDbr1 is shorter than Dbr1 from higher eukaryotes (354 vs. ∼550 residues) that thus far have been recalcitrant to crystallization. The structures revealed the first 261 residues adopt the canonical MPE fold with adjacent α and β metal-binding pockets. Features unique to Dbr1 include a 28-residue insertion loop immediately adjacent to the active site (residues 130–158) and an invariant cysteine residue (Cys14) in the position of the canonical Asp in the α pocket (25, 26). Because previous work suggested yDbr1 is Mn-dependent (23), 1 mM MnCl2 was added to the EDTA-treated enzyme before crystallization. The resulting wild type and inactive C14S EhDbr1 structures were unexpectedly mononuclear, with Mn(II) in the β-pocket and a water molecule coordinated to the Mn(II) ion occupying the position expected for the putative α-site metal (26). Alignment of the backbones of structures of EhDbr1 in complex with product analog 5′-GMP and with a synthetic RNA possessing a 2′-phosphate monoester at the branchpoint adenosine (AK65), superimposed their 5′- and 2′-phosphate moieties, respectively, suggesting a model of an intact lariat branchpoint with flanking nucleotides bound at the active site (24). The conserved 28-residue insertion loop interacts extensively with nucleotides in the putative 3′-arm of the branchpoint and is thus termed the “lariat recognition loop.” The data suggested a mononuclear mechanism of hydrolysis in which the invariant α-pocket Cys serves as a catalytic base to activate the metal-bound water nucleophile for attack on the scissile phosphorus atom, with H91 poised to protonate the 2′-O leaving group. However, because essentially all characterized MPEs harbor metal ions in both binding pockets (26) and because Cys residues are excellent ligands for certain metal ions, a binuclear mechanism could not be ruled out (25).
Since the discovery of RNA debranching activity more than 30 years ago, measurement of this activity depended upon gel-based autoradiographic assays in which enzyme and intron lariat substrates are preincubated followed by examination of the mobility of the RNA species postincubation (3, 9, 12, 22, 23, 25). Importantly, however, the metal contents of purified Dbr1 proteins have never been characterized and in the absence of endogenous metal binding information, existing gel-based debranching activity data in which exogenous metal ions were added to purified (and potentially metal-replete) Dbr1 proteins are difficult to interpret (12, 22, 23, 27). More recently, gel-based complementation assays in which Dbr1 and its variants are expressed in trans have been developed to probe structure/function relationships via the ability of these proteins to alleviate the intron lariat accumulation phenotype in ∆dbr1 yeast (23, 25, 28). Although both types of electrophoretic assays are useful as qualitative monitors of debranching activity and as probes of the participation of various amino acid residues in substrate binding and catalysis, they have shed little light upon the kinetic parameters of the enzyme and are complicated by the task of obtaining uniform (and stable) intron lariat substrates.
Here, we measure the kinetics of EhDbr1-mediated debranching of a fluorogenic bRNA substrate that permits real-time, continuous monitoring of the hydrolysis of 2′,5′-phosphodiester linkages. Inductively coupled plasma mass spectrometry (ICP-MS) analyses reveal that highly active, wild-type EhDbr1 contains stoichiometric amounts of Ca, Fe, and Zn, but undetectable levels of Mn, Mg, Ni, Co, and Cu. Wild-type EhDbr1, apo-EhDbr1 reconstituted with a mixture of Fe and Zn, and apoenzyme reconstituted with Fe(II) hydrolyze the fluorogenic probe at rates between 3–4 s−1. Apo-EhDbr1 reconstituted with Zn alone yields rates 25–33% slower, whereas apoenzyme reconstituted with Mn alone does not support debranching activity. Crystal structures of the inactive EhDbr1 variant H91A in complex with 7-mer and 16-mer bRNAs reveal extensive protein interactions with flanking 2′- and 3′-nucleotides but a paucity of interactions with the 5′-arm. All structures except the apoenzyme are binuclear with a water/hydroxide bridging the Fe and Zn ions in optimal position to attack the scissile phosphate. The results resolve previous uncertainties regarding Dbr1 active site structure and the newly developed dark-quenched bRNA probe is facilitating the screening of large small molecule libraries for Dbr1 inhibitors that may hold promise as therapeutic agents for neurodegenerative diseases such as ALS (29) and for retroviral infections such as HIV (13, 14, 17).
Results
Synthetic bRNAs for Kinetic and Structural Studies.
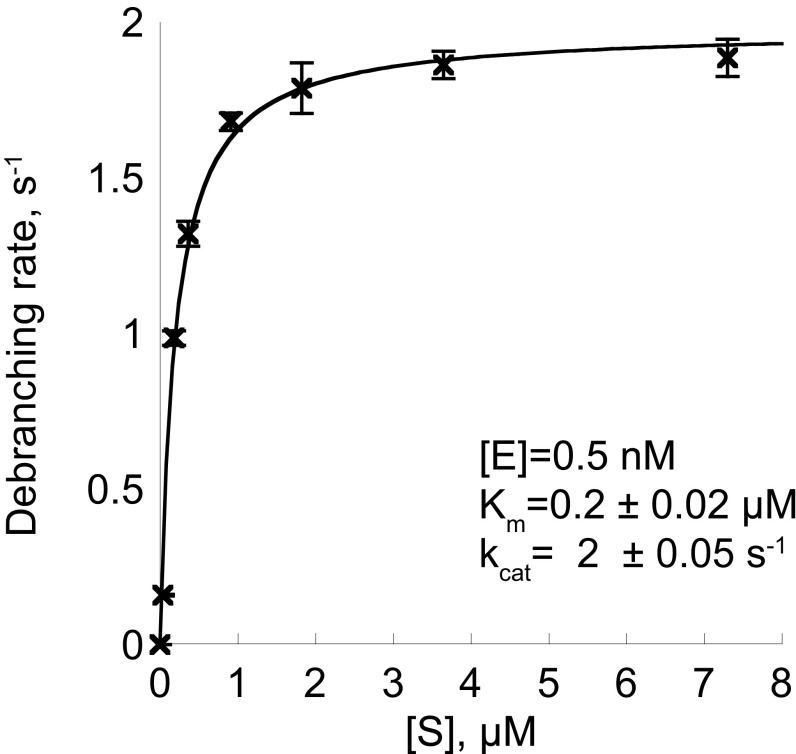
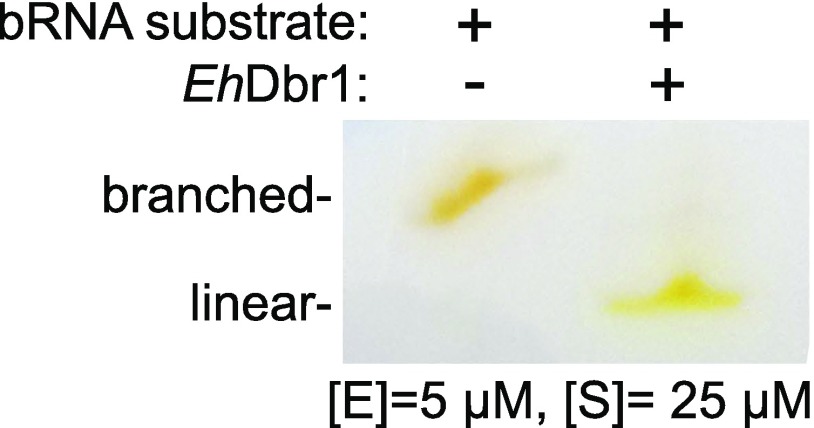
The 10-mer AK88, the 7-mer AK49, and the 16-mer AK86 bRNAs used here were synthesized via solid-phase methods that yield branched oligoribonucleotides of desired length, base composition, and regiochemistry at the branchpoint junction (25) (Fig. 1B). The sequence integrity of the synthetic products was cross-validated by using recombinant Dbr1-mediated debranching assays, HPLC, and electrospray ionization mass spectrometry (25, 30). AK88 is designed as a reporter of Dbr1 activity. It possesses a 6-carboxylfluorescein (6-FAM) fluorophore covalently linked to the 5′-arm and a 4-([4-(Dimethylamino)phenyl]azo)benzoic acid (DABCYL) quencher covalently linked to the 2′-arm. The proximity of the DABCYL to the 6-FAM suppresses the latter’s fluorescence, but this quenching is relieved upon Dbr1-mediated hydrolysis of the 2′,5′-phosphodiester bond (Fig. 1C). All three constructs possess the canonical adenosine branchpoint and flanking nucleotides generated when an adenosine nucleotide in the consensus yeast YURAC branch sequence (Y, pyrimidine; R, purine) positioned near the C terminus of the intron acts as the nucleophile to attack the 5′-phosphate of the consensus GU sequence in the donor site positioned at the 5′-end of the intron (Fig. 1A, boxed). AK88 was used as a substrate for EhDbr1 in a continuous fluorometric assay to obtain values of 0.2 ± 0.02 μM for Km, and 2 ± 0.05 s−1 for kcat (Fig. S1). This Km value agrees with a previous report of a Kd of 0.5 μM for bRNA binding to yDbr1 (31). Electrophoretic analysis of the substrate before and after exposure to Dbr1 indicated that the substrate was completely debranched by Dbr1 (Fig. S2).
Fig. S1.
Steady-state kinetics of EhDbr1 with the fluorogenic bRNA substrate AK88. The data were fit with the Michaelis–Menten equation, and error bars represent 1 SD for triplicate measurements.
Fig. S2.
Polyacrylamide gel electrophoresis of the fluorogenic bRNA substrate AK88 before and after exposure to EhDbr1. The mobility shift suggests that the substrate was completely debranched. The gel was visualized by using white light, taking advantage of the absorption properties of the covalently linked fluorophores.
Metal Content and Enzymatic Activity.
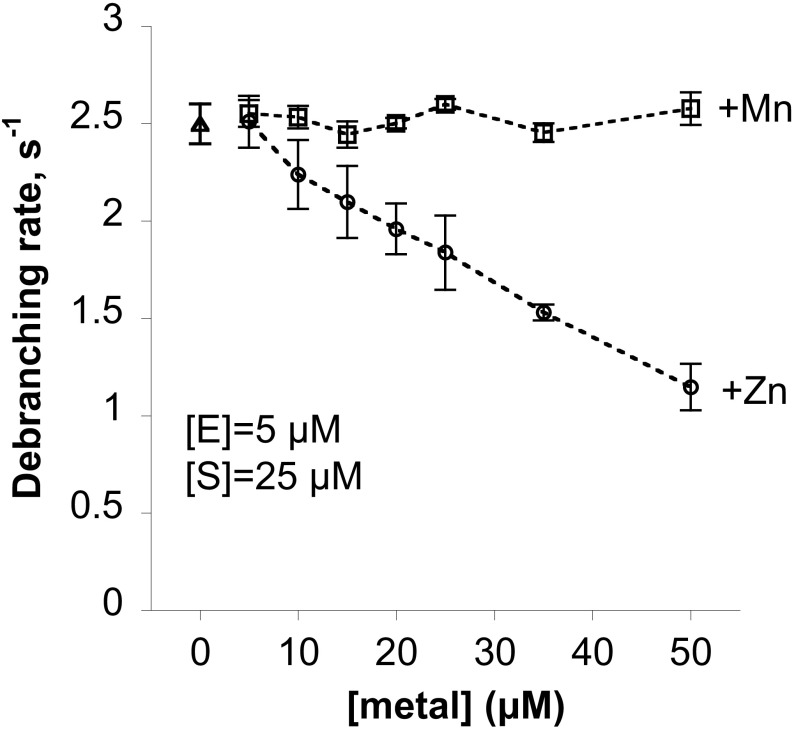
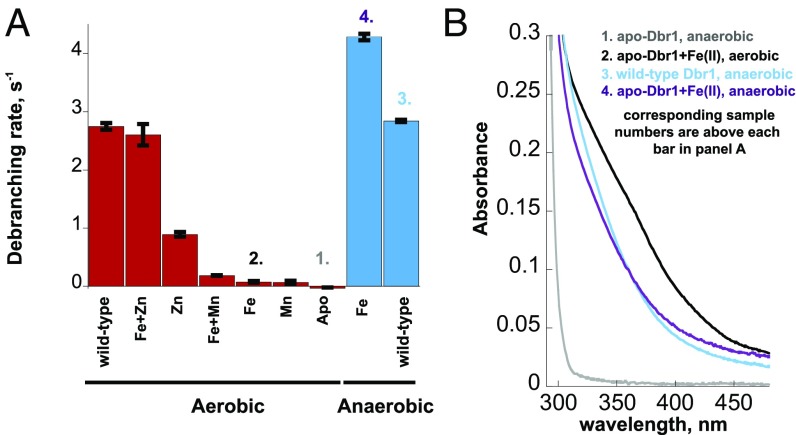
In independent purification runs, wild-type EhDbr1 expressed in E. coli contained, on average, 0.7 eq. Fe, 0.7 eq. Zn, and 1.0 eq. Ca per polypeptide as measured by ICP-MS. Mn, Mg, Ni, Co, Cu, and other divalent cations were undetectable. Addition of Mn, Fe, or Zn to the growth medium did not alter the metal content of the enzyme. Adding exogenous Mn(II) to standard assays did not alter the activity of the purified wild-type enzyme, but the activity of the enzyme decreased in the presence of Zn(II) (Fig. S3). Metal-free EhDbr1 was prepared by extensive dialysis of freshly purified enzyme against EDTA/NTA-containing buffer (Materials and Methods and SI Materials and Methods). Reconstitution of the apoenzyme with a mixture of Fe(II) and Zn(II) under aerobic conditions followed by gel filtration to remove excess metal yielded enzyme that catalyzed AK88 hydrolysis at the same rate as the freshly purified holoenzyme, ∼3 s−1 (Fig. 2A). In contrast, apo-Dbr1 reconstituted with Zn(II) alone had a significantly slower rate of less than 1 s−1, and apo-Dbr1 reconstituted aerobically with Mn(II) or Fe(II) alone did not have significant debranching activity (Fig. 2A). Of the metals tested (Fe, Zn, Mn), only Fe(II) is sensitive to oxidation by O2, so the Fe(II) reconstitution was also performed anaerobically. When the apoenzyme was mixed with Fe(II) under anaerobic conditions, the rate exceeded 4 s−1 (Fig. 2A). The results suggest that Fe(II) added to the apoenzyme under aerobic conditions was oxidized to Fe(III), and that the activity seen with Fe in the β-pocket of the wild-type enzyme (Fig. 3 A and C) is due to Fe(II).
Fig. S3.
Effects of addition of exogenous Zn and Mn to wild-type (Fe and Zn containing) EhDbr1 in aerobic assays. Increasing Mn(II) did not alter the debranching activity, but increasing Zn concentrations decreased the activity of the wild-type enzyme. Error bars represent 1 SD of triplicate measurements.
Fig. 2.
Effect of added metals on apo-EhDbr1. (A) The catalytic activity of EhDbr1 with AK88 as substrate was determined after adding the indicated metals to the apoenzyme as described in Materials and Methods. The experiments were done aerobically for the samples in red and anaerobically for those in blue. The column labeled wild-type refers to the enzyme before treatment with chelators. (B) UV/Vis spectra of selected samples from A. The numbering of each trace is shown above its corresponding bar in A.
Fig. 3.
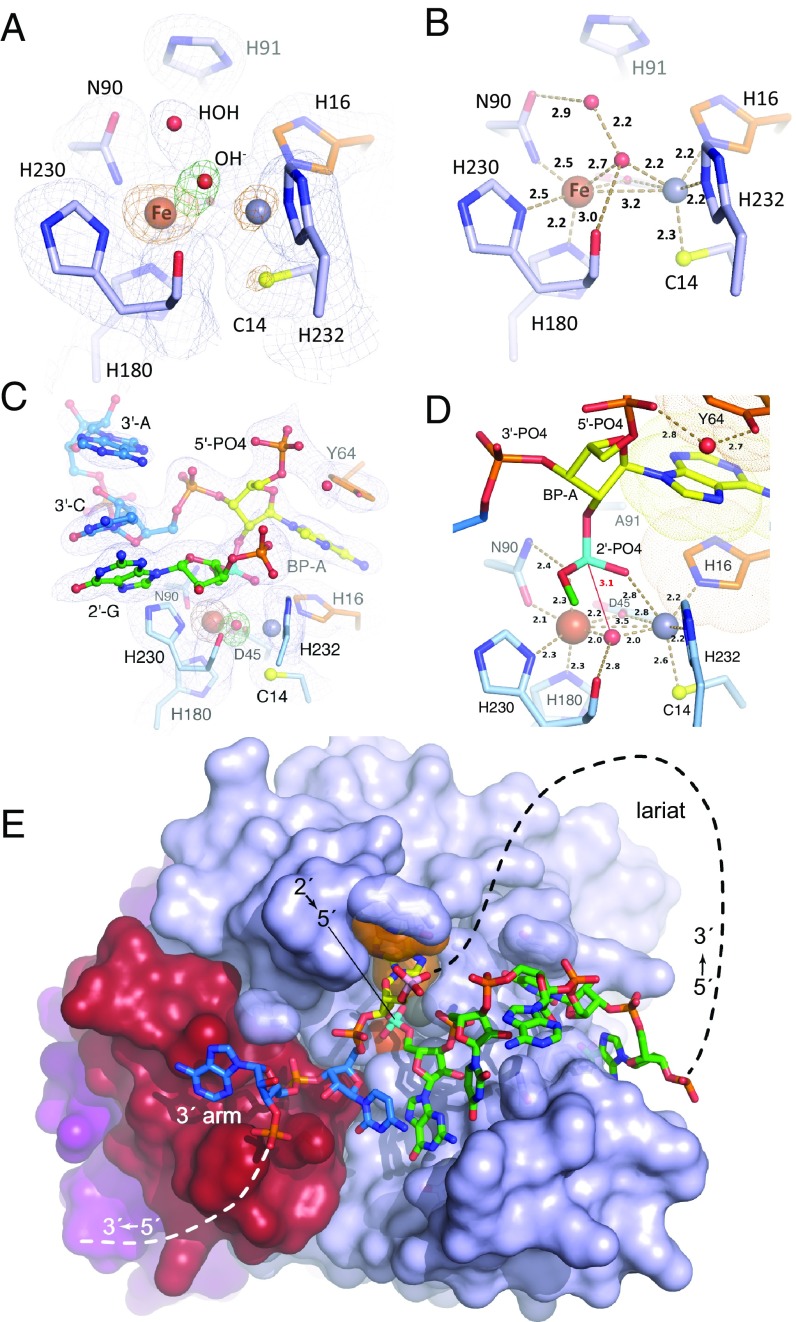
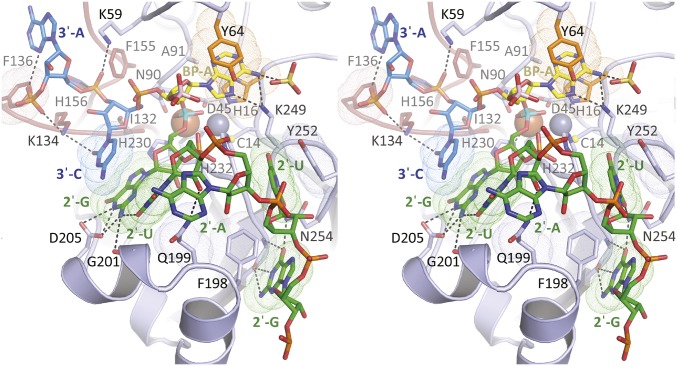
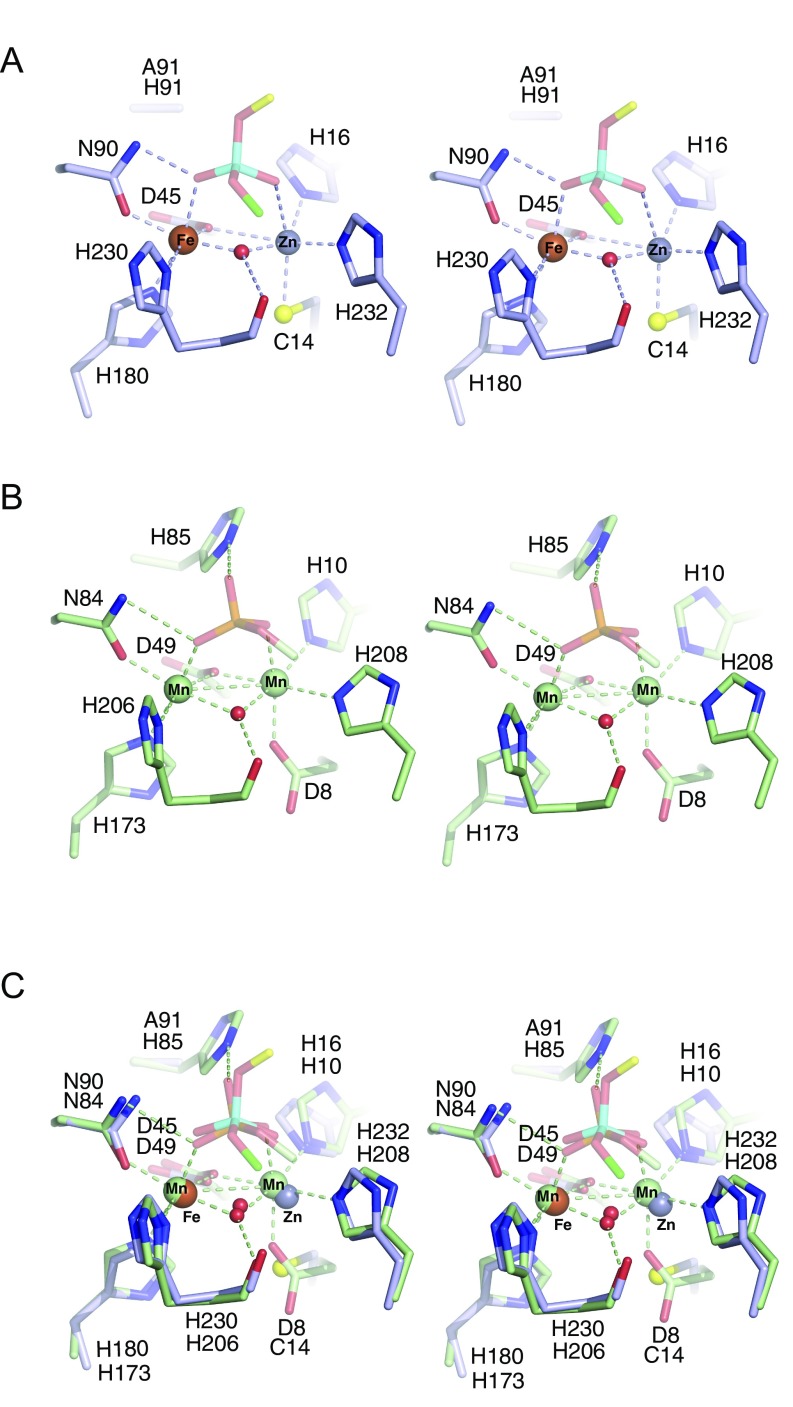
Structures of wild-type EhDbr1 alone and its H91A variant in complex with AK49 and AK86. The nucleotide color coding is as in Fig. 1. H16 and Y64 engage in aromatic stacking interactions with the branchpoint adenine and are orange, Fe and Zn ions are rust and gray spheres, respectively, the 2′,5′-linkage is cyan, and the 5′-phosphate of the branchpoint nucleotide is pink. (A) σA-weighted 2Fo-Fc (light blue mesh), Fo-Fc (green mesh), and anomalous difference (orange mesh) electron density maps superimposed on the refined structure of wild-type EhDbr1 contoured at 1.2, 5.0, and 5.0 σ, respectively. The density for the bridging hydroxide is obscured by the metal ion density in the 2Fo-Fc map. The Fo-Fc electron density revealing its position was calculated after removing the hydroxide from the refined model. The anomalous difference map was calculated by using X-rays tuned to the Fe absorption edge (see Results). (B) Coordination distances (Å) for the metal ions in wild-type EhDbr1. The view is the same as in A. (C) σA-weighted 2Fo-Fc, Fo-Fc, and anomalous difference electron density maps superimposed on the refined structure of H91A EhDbr1 in complex with the synthetic 7-mer bRNA AK49 at 1.0, 5.0, and 5.0σ, respectively. (D) Coordination distances for the metal ions shown in C. For clarity, only the 5′-carbon atom of the flanking 2′-G is shown. (E) The 16-mer bRNA AK86 in complex with H91A EhDbr1. The MPE domain is light blue, the lariat recognition loop is red, and the C-terminal domain is purple. The dashed lines indicate the connectivity in the full intron lariat.
Crystal Structures of EhDbr1.
The new structures described here include the following: (i) wild-type EhDbr1 containing Fe+Zn (Fig. 3 A and B); (ii) wild-type enzyme stripped of its metal ions (apo-EhDbr1, Fig. S4); (iii) H91A EhDbr1 in complex with the 7-mer bRNA AK49 (Fig. 3 C and D and Fig. S5A); and (iv) H91A EhDbr1 in complex with the 16-mer bRNA AK86 (Fig. 3E and Fig. S6). All EhDbr1 structures determined to date are isomorphous in space group P212121 with five protomers in the asymmetric unit. The 20 protomers coming from the four EhDbr1 structures reported here superimpose with an average rmsd of 0.21 Å for all backbone atoms. Diffraction data and protein structure refinement statistics are given in Table S1. The metal content of H91A EhDbr1 was similar to that of the wild-type enzyme. The identities of the metal ions in the α and β pockets in the wild-type and AK49/H91A EhDbr1 complex structures were visualized directly through inspection of anomalous difference Fourier electron density maps by using X-rays tuned to the Fe absorption edge (7.11 keV, 1.74 Å). At this wavelength, iron is illuminated but zinc (9.66 keV, 1.28 Å) is not. The magnitudes of the peaks in the anomalous difference Fourier map reveal that iron partitions predominantly to the β pocket in these structures (Fig. 3 A and C and Table S2), but Fe is present to varying degrees in the α pocket in the wild-type enzyme (Fig. 3A and Table S2). The structures of wild-type EhDbr1 and H91A EhDbr1 in complex with AK49 reveal a water/hydroxide bridging the two metal ions, although the positions of the bridging species differ in the RNA-bound versus free forms (compare Fig. 3 A and C). The Fe ions in the β-sites of both structures are coordinated in a geometry best described as distorted octahedral with D45, N90, H230, and the bridging OH− serving as equatorial ligands, whereas H180 and either a water molecule (wild-type) or phosphate oxygen atom (AK49 complex) act as axial ligands. In the wild-type enzyme, the α-pocket Zn ion is coordinated in a pseudotrigonal bipyramidal geometry with Cys14, H16, and the bridging OH− acting as equatorial ligands and D45 and H232 acting as axial ligands. In the AK49 complex, the Zn ion is bound in the α pocket in a distorted octahedral geometry, with H16, D45, H232, and the bridging OH− serving as equatorial ligands and Cys14 and an oxygen atom coming from the scissile phosphate serving as axial ligands. The distortion in octahedral geometry arises because the scissile phosphate oxygen, the Zn ion, and the Sγ atom of Cys14 are not colinear, with the position of the scissile phosphate oxygen atom displaced from colinearity by ∼23° (compare Fig. 3 B and D). Coordination distances for each of the five protomers in the asymmetric unit are given in Table S2.
Fig. S4.
Divergent stereoview of the structure of metal-free (apo) EhDbr1. The σA-weighted 2Fo-Fc (light blue mesh) is contoured at 1.2 sigma. In the absence of metals, a water binds in the β-pocket, and a sulfate from the crystallization solution hydrogen bonds to the catalytic acid H91. His16 adopts an “out” position, rotating away from the position where it could stack with a branchpoint adenosine and ligate the α-pocket metal. Hydrogen bonds are dashed lines.
Fig. S5.
The requirement for vicinal 2′,5′- and 3′,5′-phospodiester linkages for recognition by Dbr1 and the molecular basis for why 2′,5′-linked poly-A RNA is not a substrate. (A) The structure of the AK49/EhDbr1 complex. The two flanking nucleotides of the 3′-arm (blue sticks) participate in five hydrogen-bonding interactions (yellow dashes) with the enzyme, two aromatic stacking interactions (with F136 and the 2′-G), and numerous van der Waals interactions. (B) Model of 2′,5′-linked poly-A RNA bound as in A. 2′,5′-linked (cyan) poly-A RNA lacks a 3′-arm and therefore is devoid of the interactions contributed by the blue sticks in A. The 2′,5′-phosphodiester bond indicated at the lower left is predicted to clash with F202 (see Discussion). Adenosine (magenta) in place of the guanosine at the 2′-position is predicted to engage in two fewer hydrogen bonds with the enzyme and lacks the 5′-phosphate shown for the 2′-G in A).
Fig. S6.
Divergent stereoview of the 16-mer bRNA AK86 in complex with H91A EhDbr1. The nucleotide color coding is as in Fig. 1. H16 and Y64 that engage in aromatic stacking interactions with the branchpoint adenine are orange, Fe and Zn ions are rust and gray spheres, respectively, the 2′,5′-linkage is cyan, and the 5′-phosphate of the branchpoint nucleotide is pink. van der Waals surfaces of bases engaging in aromatic stacking interactions are shown as dots. The third nucleotide of the 2′ arm makes no interactions with the protein. The elements that form this protrusion include the fourth and fifth nucleotides in the 2′ arm, which make aromatic stacking interactions with Y252 and F198. Hydrogen bonds are shown as dashed lines.
Table S1.
X-ray diffraction and protein structure refinement statistics for the four Dbr1 structures determined in this work
| Wild-type Fe edge | AK49 Se edge | AK49 Fe edge | AK86 Zn edge | Apo Se edge | |
| PDB code | 5K73 | 5K77 | — | 5K78 | 5K71 |
| Ligand | None | AK49 | AK49 | AK86 | None |
| Data collection | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit Cell, Å | 73.2, 141.7, 213.8 | 73.2, 141.9, 212.5 | 73.3, 142.3, 212.7 | 72.8, 141.8, 214.3 | 73.1, 142.2, 214.3 |
| Wavelength, Å | 1.73161 | 0.97950 | 1.73160 | 1.28081 | 0.97918 |
| Resolution, Å | 2.08 (2.19–2.08)* | 2.17 (2.29–2.17) | 3.1 (3.27–3.10) | 2.64 (2.78–2.64) | 2.57 (2.63–2.57) |
| Rsym, % | 4.4 (55.9) | 11.8 (72.0) | 12.9 (87.3) | 15.1 (176.5) | 11.0 (92.2) |
| I/σI | 15.4 (1.5) | 10.5 (1.4) | 10.4 (1.8) | 10.3 (1.1) | 12.6 (1.6) |
| CC(1/2)† | 99.7 (61.6) | 0.984 (0.594) | 0.988 (0.670) | 0.988 (0.574) | 0.992 (0.543) |
| Completeness (%) | 90.9 (66.4) | 99.7 (99.8) | 98.7 (97.7) | 99.1 (99.8) | 99.4 (99.7) |
| Unique Refl. | 121,755 (12,734) | 117,264 (16,995) | 40,579 (5,760) | 66,902 (9,693) | 71,512 (4,575) |
| Redundancy | 3.5 (2.6) | 4.6 (3.8) | 4.7 (4.3) | 5.6 (5.8) | 4.9 (5.0) |
| Refinement | |||||
| Resolution, Å | 2.08 | 2.17 | — | 2.64 | 2.57 |
| Rwork/ Rfree, % | 17.8/21.3 | 18.2/22.3 | — | 19.5/24.6 | 19.4/24.2 |
| No. atoms | 15,014 | 15,528 | — | 14,679 | 14,760 |
| Protein + RNA | 14,350 | 14,870 | — | 14,579 | 14,305 |
| Ligand/ion | 85 | 140 | — | 75 | 80 |
| Water | 579 | 518 | — | 25 | 375 |
| B-factors, Å2 | 50.4 | 49.0 | — | 74.4 | 55.83 |
| Protein + RNA | 50.4 | 48.8 | — | 74.4 | 56.01 |
| Ligand/ion | 78.7 | 85.25 | — | 95.5 | 88.7 |
| Water | 46.7 | 44.61 | — | 37.73 | 42.00 |
| rms deviations | |||||
| Bond lengths, Å | 0.002 | 0.003 | — | 0.003 | 0.003 |
| Bond angles, ° | 0.50 | 0.49 | — | 0.55 | 0.64 |
Highest resolution shell is shown in parentheses.
CC(1/2) = correlation (I1, I2), where I1 and I2 are randomly assigned half-sets of reflections.
Table S2.
Anomalous difference Fourier peak heights calculated at the Fe edge, and bond lengths of coordinating ligands to the Fe and Zn atoms in the five protomers in each asymmetric unit
| Atom(s) | Chain A | Chain B | Chain C | Chain D | Chain E | Avg | STD |
| Wild-type | |||||||
| Intermetal | 3.3 | 3.4 | 3.3 | 3.4 | 3.3 | 3.3 | 0.1 |
| Bridging OH−–H2O | 2.2 | 2.2 | 2.2 | 2.3 | 2.3 | 2.3 | 0.0 |
| Bridging OH−–O230 | 3.0 | 3.0 | 3.2 | 3.6 | 3.0 | 3.2 | 0.2 |
| Bridging OH−–ZN | 2.2 | 2.2 | 2.1 | 2.1 | 2.1 | 2.1 | 0.1 |
| C14–Zn | 2.4 | 2.4 | 2.5 | 2.3 | 2.3 | 2.4 | 0.1 |
| H16–Zn | 2.3 | 2.3 | 2.3 | 2.4 | 2.3 | 2.3 | 0.1 |
| D45(Oδ2)–Zn | 2.6 | 2.6 | 2.7 | 2.7 | 2.7 | 2.7 | 0.0 |
| H232–Zn | 2.3 | 2.2 | 2.2 | 2.6 | 2.3 | 2.3 | 0.1 |
| Bridging OH−–Fe | 2.7 | 2.7 | 2.7 | 2.7 | 2.7 | 2.7 | 0.0 |
| H2O Fe | 3.6 | 3.3 | 3.3 | 3.3 | 3.4 | 3.4 | 0.1 |
| N90 (Oδ1)–Fe | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 0.0 |
| H230–Fe | 2.5 | 2.6 | 2.6 | 2.6 | 2.5 | 2.6 | 0.0 |
| H180 -Fe | 2.2 | 2.2 | 2.3 | 2.3 | 2.2 | 2.2 | 0.0 |
| D45 (Oδ2)–Fe | 2.4 | 2.3 | 2.2 | 2.5 | 2.2 | 2.3 | 0.1 |
| D45 (Oδ1)–Fe | 3.3 | 3.6 | 3.2 | 3.4 | 3.3 | 3.4 | 0.1 |
| Fe ano α (σ) | 4.0 | 7.4 | 5.7 | 4.9 | 9.1 | — | — |
| Fe ano β (σ) | 12.5 | 11.7 | 15.1 | 9.6 | 14.1 | — | — |
| AK49 | |||||||
| Intermetal Å | 3.6 | 3.5 | 3.6 | 3.6 | 3.5 | 3.6 | 0.1 |
| Bridging OH−–G510 OP1 | 2.7 | 2.7 | 2.8 | 2.6 | 2.7 | 2.7 | 0.1 |
| Bridging OH−–G510 OP2 | 2.9 | 3.0 | 2.9 | 2.9 | 2.8 | 2.9 | 0.1 |
| Bridging OH−–Carbonyl 230 | 2.8 | 2.9 | 3.1 | 2.8 | 2.9 | 2.9 | 0.1 |
| Bridging OH−–G510 P | 3.1 | 3.2 | 3.2 | 3.1 | 3.1 | 3.1 | 0.0 |
| Bridging OH−–ZN | 2.0 | 2.0 | 2.0 | 2.0 | 1.9 | 2.0 | 0.0 |
| C14–Zn | 2.6 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 0.0 |
| H16–Zn | 2.3 | 2.3 | 2.1 | 2.2 | 2.2 | 2.2 | 0.0 |
| D45(Oδ2)–Zn | 2.9 | 2.9 | 2.8 | 2.8 | 2.8 | 2.8 | 0.0 |
| H232–Zn | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 0.0 |
| G510 OP1–Zn | 2.8 | 2.6 | 2.8 | 2.8 | 2.7 | 2.8 | 0.1 |
| Bridging OH−–Fe | 2.0 | 1.9 | 1.9 | 2.0 | 2.0 | 2.0 | 0.0 |
| N90 (Oδ1)–Fe | 2.1 | 2.2 | 2.1 | 2.1 | 2.3 | 2.2 | 0.1 |
| H230–Fe | 2.3 | 2.3 | 2.2 | 2.2 | 2.3 | 2.3 | 0.0 |
| H180–Fe | 2.3 | 2.2 | 2.1 | 2.2 | 2.1 | 2.2 | 0.0 |
| D45 (Oδ2)–Fe | 2.2 | 2.2 | 2.3 | 2.3 | 2.2 | 2.2 | 0.0 |
| D45 (Oδ1)–Fe | 3.4 | 3.5 | 3.3 | 3.4 | 3.3 | 3.4 | 0.1 |
| G510 OP2–Fe | 2.3 | 2.3 | 2.3 | 2.4 | 2.3 | 2.3 | 0.0 |
| Fe anom α (>3 σ) | — | — | — | — | — | — | — |
| Fe anom β (>3 σ) | 8.7 | 5.1 | 8.1 | 6.2 | 6.3 | — | — |
| AK86 | |||||||
| Intermetal Å | 3.3 | 3.3 | 3.5 | 3.3 | 3.3 | 3.3 | 0.1 |
| C14–Zn | 2.6 | 2.5 | 2.6 | 2.6 | 2.6 | 2.6 | 0.0 |
| H16–Zn | 2.3 | 2.5 | 2.4 | 2.4 | 2.4 | 2.4 | 0.0 |
| D45(Oδ2)–Zn | 2.7 | 2.9 | 2.7 | 2.9 | 2.8 | 2.8 | 0.1 |
| D232–Zn | 2.2 | 2.3 | 2.2 | 2.4 | 2.1 | 2.3 | 0.1 |
| G510 OP1–Zn | — | — | 2.2 | 2.6 | — | 2.4 | 0.2 |
| N90 (Oδ1)–Fe | 2.3 | 2.3 | 2.2 | 2.1 | 2.3 | 2.2 | 0.1 |
| H230–Fe | 2.4 | 2.4 | 2.4 | 2.5 | 2.4 | 2.4 | 0.1 |
| H180–Fe | 2.3 | 2.2 | 2.3 | 2.1 | 2.1 | 2.2 | 0.1 |
| D45 (Oδ2)–Fe | 2.5 | 2.4 | 2.4 | 2.4 | 2.4 | 2.4 | 0.0 |
| D45 (Oδ1)–Fe | 3.2 | 3.3 | 3.1 | 3.1 | 3.2 | 3.2 | 0.1 |
| G510 OP2–Fe | — | — | 2.5 | 2.3 | — | 2.4 | 0.1 |
Measurements in angstroms. STD, standard deviation.
SI Materials and Methods
Cloning, Expression, and Purification.
The DBR1 gene from E. histolytica was codon optimized, subcloned into a modified pJ411 expression plasmid (DNA2.0, Genescript), and transformed into Escherichia coli strain BL21 (New England Biolabs). This construct encodes an 8×-histidine tag fused to the Dbr1 N terminus separated by a tobacco etch virus (TEV) protease cleavage site. Transformed cells were cultured in 1-L volumes in 3-L fernbach flasks shaken at 30 °C. When the OD600 reached ∼3, IPTG was added to achieve a final concentration of 1 mM and the cells were grown for another 6 h before harvesting by centrifugation. The pelleted cells were flash-frozen in liquid nitrogen and stored at −80 °C until protein purification. For each purification run, ∼250 g of thawed cell paste were resuspended in 1 L of Ni-A buffer (50 mM Tris pH 8, 500 mM NaCl, 1 mM TCEP, 20 mM imidazole). Resuspended cells were lysed by sonication using a 1-inch high-gain sonication horn (Qsonica), and the resulting lysate was clarified by centrifugation at 15,000 × g for 20 min. Clarified supernatant was loaded on a 25-mL Ni-Sepharose Fast Flow column (GE Healthcare) and washed with Ni-A buffer. Proteins were eluted by using a shallow linear gradient with Ni-B (Ni-B buffer is Ni-A buffer made 400 mM in imidazole). Eluted fractions containing Dbr1 were pooled, and TEV protease was added at a stoichiometric ratio of ∼10:1 over Dbr1. The sample was dialyzed overnight against heparin-A buffer (20 mM Hepes pH 7, 50 mM NaCl, and 1 mM TCEP), filtered using a 0.4-μm filter (Millipore), and loaded onto a 5-mL HiTrap-heparin column (GE Healthcare). After washing, Dbr1 was eluted by using a shallow linear gradient of heparin-B buffer (heparin A buffer made 1 M in NaCl). Two Dbr1-containing peaks were resolved. The first contained nearly metal-free protein, and the Dbr1 in the second contained ∼0.7 eq. Fe and ∼0.7 eq. Zn per monomer as determined by ICP-MS (see below). Peak fractions were concentrated and loaded onto a Superdex 200 sizing column (GE Healthcare) equilibrated with 50 mM Hepes pH 7, 150 mM NaCl, 1 mM TCEP. Eluted fractions were concentrated in a spin concentrator (Millipore). After concentration to ∼1 mL, the samples were diluted ∼15-fold into crystallization buffer (20 mM Hepes pH 7, 1 mM TCEP) and concentrated again. The concentrating procedure was terminated when the samples reached 10–20 mg/mL (ε280 = 69,200 M−1⋅cm−1). Small aliquots were then frozen at −80 °C until used in kinetic and structural analyses. The purification procedure was identical for wild-type and H91A Dbr1 except H91A possesses a C-terminal noncleavable 6× His tag. All metal analyses and reconstitutions used N-terminal fusion proteins with the His tag proteolytically removed. The yield per liter of E. coli culture of C-terminal Dbr1 fusion protein was ∼twofold higher than the yield of N-terminal fusion proteins with cleavable tags. We found no difference in structure or stability of N- vs. C-terminally tagged proteins.
Metal Content of Dbr1 Preparations by ICP-MS.
Protein samples were diluted to ∼5 μM in 20 mM Hepes pH 7. Fifteen-milliliter falcon tubes were washed with 5 mM EDTA and low salt buffer before sample addition. Dilution buffer was used as the “blank,” and all values reported were blank-subtracted. Concentrations of Mg, Mn, Ca, Fe, Co, Ni, Cu, and Zn were measured by using ICP-MS analyses performed by the Chemical Analysis Laboratory at the University of Georgia.
Metal-Free (apo) Dbr1.
Dbr1-containing fractions were dialyzed against stripping buffer (10 mM EDTA, 10 mM nitrilotriacetic acid, 20 mM Hepes pH 7, 50 mM NaCl, and 1 mM TCEP) at 4 °C for 72 h. The dialysis bag (50-mL sample) was transferred to a room-temperature beaker containing 1-L Chelex-treated buffer (50 mM Hepes pH 7, 100 mM NaCl, and 1 mM TCEP) to remove EDTA and nitrilotriacetic acid. The dialysate was replaced with fresh buffer every 2 h. After the fourth exchange, the dialysis setup was moved to 4 °C and equilibrated overnight. Before activity measurements, metal-free Dbr1 was dialyzed against Chelex-treated assay buffer (50 mM Hepes pH 7, 100 mM NaCl). The resulting protein samples were analyzed by ICP-MS and were found to contain ∼0.07 eq. Zn and transition metals such as Mn, Fe, Co, Ni, and Cu were present at fewer than 0.01 equivalents.
Aerobic Metal-Reconstitution and Kinetic Assays.
One molar stocks of Zn(II)SO4, Mn(II)SO4, and Fe(II)(NH4)2(SO4)2 in 2 mM HCL were prepared immediately before activity measurements. Fe, Zn, and Mn at 1 mM, 40 μM, and 40 μM, respectively, were mixed with apoenzyme. Wild-type Dbr1 samples were subjected to a mock reconstitution protocol as a control. After 30 min of incubation, the mixtures were passed over a PD-10 desalting column (GE Healthcare) equilibrated with assay buffer. The desalted samples at 5 µM were submitted for ICP-MS analyses. All kinetic assays were performed at room temperature. The AK88 substrate was made 50 μM in assay buffer and loaded into the injection syringe on the plate reader. Ten microliters of each Dbr1 sample was added to a low-volume 384-well plate in quadruplicate, and the reactions initiated upon injection of 10 μL of substrate into each well. The fluorescence intensity (excitation at 488 nm and emission at 520 nm) was measured every 20 ms for 10 s. Initial velocities were derived by fitting the slope of the linear portion (typically 0.5–1.5 s) of the reaction. Rates in relative fluorescence units/s were converted to μM/s by using a final relative fluorescence unit value of completed reactions corresponding to 25 μM product and the background fluorescence as 0 μM product. Turnover rates were obtained after dividing the rates by the final enzyme concentration. The final reaction conditions were 25 μM substrate and 5 μM enzyme. Total reaction times were 10 s, and most reactions were completed by 2 s. A relatively high enzyme concentration of 5 μM was selected to prevent trace metal ions (typically present at levels of 100–500 nM in our buffers) from interfering in our metal-dependence assays.
Anaerobic Metal-Reconstitution Kinetic Assays.
Fe(II) is rapidly oxidized to Fe(III) at neutral pH by molecular oxygen, and the resulting Fe(III) is insoluble at neutral pH. We therefore used an anaerobically conditioned stopped-flow instrument (Applied Photophysics) configured for kinetic analyses of iron enzymes (47, 48). Solutions containing apo-Dbr1 and Fe(II) were added to the main chamber and side-arms of the tonometer, respectively. The contents of the tonometer were made anaerobic by 10 cycles of vacuum/argon gas, after which the contents of the side arm (e.g., Fe solution or 2 mM HCl alone in the case of positive and negative control experiments) were mixed with apo-Dbr1 to effect the reconstitution. After an additional five cycles of vacuum/argon gas cycles, the tonometer was connected to the luer fitting of the stopped flow apparatus. A 50 μM solution containing the AK88 substrate in assay buffer was made anaerobic by bubbling with argon gas in a Hamilton gas-tight syringe for 10 min. The stopped-flow instrument was scrubbed of oxygen by incubating for 2 h with an anaerobic solution of glucose and glucose oxidase. The instrument was maintained at 25 °C with a circulating water bath, and the bath was continually sparged with nitrogen gas to prevent the diffusion of oxygen into the samples through the bath water. Reaction curves for each sample were measured in quadruplicate. The excitation wavelength was 480, and the emitted light was filtered with a 495-nm high-pass filter. The reactions were observed for 5 s and typically came to completion in ∼1.5 s. Rates were calculated as described in the previous section. Final reaction conditions were 25-μM substrate, 5-μM enzyme, and 50 μM Fe(II).
Crystallization, Data Collection, and Protein Structure Determination and Refinement.
apoenzyme, wild-type enzyme, and H91A Dbr1 variants were all crystallized at 10–20 mg/mL by using Nextal hanging drop plates (Qiagen). The mother liquor was 0.1 M Bis-Tris pH 5.5 and made 10–17% (wt/vol) PEG 3350, 0.4 M LiSO4, 10% (vol/vol) glycerol. Mother liquor (0.5 mL) was added to the reservoir and 4 drops were dispensed on the screw-top lid (typically 2 μL of protein and 2 μL of mother liquor per drop). The next day, drops were streak seeded from previously grown crystals at 20 °C. Needle-like crystals suitable for single crystal X-ray work appeared ∼3–5 d after seeding. For the AK49/H91A Dbr1 structure, drops consisted of 1.3 μL of H91A Dbr1, 1.3 μL of 2 mM AK49, and 1.3 μL of mother liquor. The AK49/H91A drops were not seeded. For the AK86 structure, crystals of H91A Dbr1 were soaked overnight in 1 mM AK86 in cryo solution [30% (wt/vol) PEG 3350, 0.4 M LiSO4, 0.1 M Bis-Tris pH 5.5, 20% (vol/vol) glycerol]. Suitable specimens were extracted from their drop by using nylon loops and then flash-frozen in liquid nitrogen for storage until use. All datasets were collected at NE-CAT beamline 24-IDC at the Advanced Photon Source, Argonne National Laboratory. Intensities were integrated and scaled by using the program XDS (43) and merged with the program POINTLESS (44). Structure determination and refinement were performed by using the PHENIX program suite (45) using the previously determined Dbr1 structure 4PEF (24) with metal ions removed as the starting model. All crystals were isomorphous in space group P212121 with five Dbr1 protomers in the asymmetric unit. Metal ions, solvent molecules, and RNA ligands were built into positive difference density in σ-A–weighted Fo-Fc maps. Restraints for the 2′,5′-phosphodiester linkage were defined manually. Model building was performed with the program COOT (46), and the RNA components were built by using the RCrane plugin (49). Iterative cycles of model building in COOT and refinement in PHENIX were continued until Rfree ceased to decrease.
Discussion
A Fluorogenic Assay for Debranching Activity and the Metal Preferences of Dbr1.
Until now, characterization of RNA debranching activity depended on gel-based autoradiographic assays in which enzyme and intron lariat substrates were preincubated with millimolar concentrations of MgCl2 (3, 9, 12, 22, 23, 25). Several groups have reported that Dbr1 was required at concentrations 10- to 100-fold over substrate to observe a mobility shift in a reasonable time frame (e.g., 30–60 min). One exception was yDbr1, which acted catalytically in the presence of Mn but not Mg (23). The requirement for a large excess of enzyme in most cases suggests that only a small subset of Dbr1 molecules were active in those assays. The availability of the bRNA substrate AK88 described here permits hydrolysis by purified and reconstituted Dbr1 proteins to be monitored in real time by using a flourometer, thereby providing a simple and convenient way to measure activity.
The data presented here suggest that EhDbr1 is a binuclear enzyme, with Fe(II) and Zn(II) conferring the highest rates of hydrolysis. First, EhDbr1 purified after expression in E. coli possesses nearly stoichiometric amounts of Fe and Zn, but undetectable levels of Mg, Mn, Co, Ni, Cu, or Cd. Second, the metal centers in the structures of wild-type EhDbr1 and H91A EhDbr1 in complex with AK49 reveal binuclear centers with a bridging water molecule/hydroxide ion (Fig. 3 A–D). Third, EhDbr1 containing homobinuclear centers Fe(II) or Zn(II) support debranching activities of 4 and 1 s−1, respectively, whereas enzyme containing a heterobinuclear Fe(II)Zn(II) center has a rate of 3 s−1 (Fig. 2A). Finally, diffraction data from crystals of enzyme using X-rays tuned to the Fe absorption edge reveal that Fe partitions primarily to the β-pocket (Fig. 3 A and C and Table S2). The observation that metal-free EhDbr1 reconstituted with a solution of freshly made Fe(II) under aerobic conditions is inactive can be attributed to oxidation to Fe(III), which does not support activity (Fig. 2 A and B). Taken together, the kinetic and structural data suggest the enzyme requires Fe(II) and/or Zn(II) in the α-pocket for activity.
Yeast Dbr1 was reported to be a manganese-dependent enzyme (23), in contrast to the results reported here with EhDbr1. Thus, available evidence indicates that yDbr1 and EhDbr1 have different metal cofactor requirements. We cannot rule out the possibility that EhDbr1 is active with Mn, but the affinity of that metal for the enzyme is much lower than the affinity for Fe or Zn (Fe was not tested with the yeast enzyme). We have found that EhDbr1 precipitates in the presence of mM Zn; this precipitation may explain the lack of observed activity of the yeast enzyme in the presence of 2.5 mM Zn. The lack of an effect of added Mn on wild-type EhDbr1 (Fig. S3) can be attributed to that enzyme already having a full complement of metal bound.
Mechanism of Dbr1-Mediated Intron Debranching.
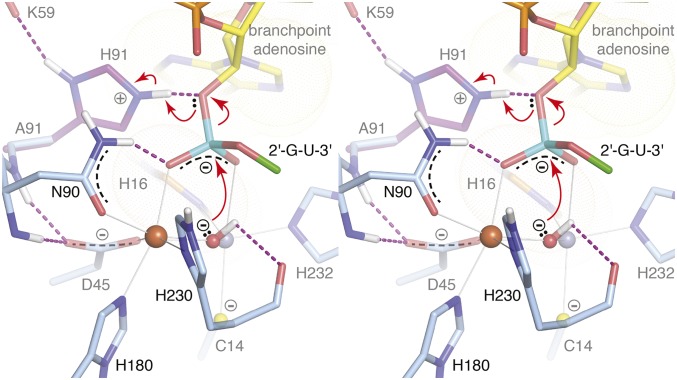
The structure of the Fe+Zn-containing wild-type enzyme represents the resting state (Fig. 3 A and B), whereas the structure of the EhDbr1 variant H91A in complex with AK49 represents a snapshot of the bRNA/enzyme complex immediately before hydrolysis. Our previous structural work on mononuclear EhDbr1 implicated H91 as the catalytic acid (24) because it appears poised to protonate the 2′-O leaving group during hydrolysis in the structure of the EhDbr1/AK65 complex. Indeed, H91A EhDbr1 is inactive, enabling the first direct visualizations of bona fide 2′,5′-phosphodiester linkages bound to the enzyme. The position of the bridging water/hydroxide in the resting state (Fig. 3 A and B) is approximately the same as the scissile phosphorous atom of the 2′,5′-phosphodiester linkage in the EhDbr1/AK49 complex (Fig. 3 C and D). Upon binding of AK49, the bridging aquo species is displaced to a new bridging position where it is stabilized by a hydrogen bond to the carbonyl oxygen of H230, a ligand to the β-pocket Fe atom (Fig. 3 C and D). The active site structure suggests a SN2 mechanism of hydrolysis in which the metal ions serve as Lewis acids to polarize the bridging oxygen-phosphorous bonds, increasing the electrophilic character of the scissile phosphorous (Fig. 4). The position of the bridging OH− species appears optimal for nucleophilic attack at a distance of 3.1 Å. The reaction is predicted to proceed through a trigonal bipyramidal pentacoordinate transition state with inversion of configuration and protonation of the 2'-O leaving group by H91. The proposed flow of electrons during catalysis is shown in Fig. 4.
Fig. 4.
Divergent stereoview of the proposed mechanism of EhDbr1-mediated hydrolysis of intron lariats. The bridging OH− is in position for nucleophilic attack on the 2′, 5′-phospodiester linkage. The reaction proceeds via a SN2 mechanism of hydrolysis through a trigonal bipyramidal pentacoordinate intermediate, followed by inversion of configuration and concludes when the 2′-O leaving group is protonated by H91. The red arrows indicate electron flow during the reaction.
Dbr1/bRNA Interactions.
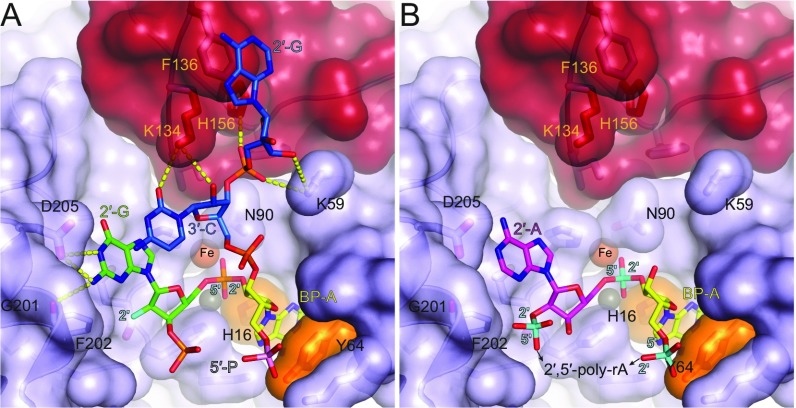
The structures of the 7-mer and 16-mer bRNAs AK49 and AK86 in complex with the EhDbr1 variant H91A provide insight into the molecular basis for bRNA recognition. AK49 possesses two nucleotides in its 2′-, 3′-, and 5′-arms, whereas AK86 possesses five nucleotides in each arm. The branchpoint and flanking nucleotides of both constructs mimic those found in the S. cerevisiae consensus splice site (Fig. 1 A and B). The 5′-arms are disordered in both structures, suggesting they do not play a significant role in branchpoint recognition. In contrast, extensive enzyme/bRNA interactions occur at the branchpoint adenosine, the flanking guanosine in the 2′-arm and the flanking cytidine and adenosine of the 3′-arm. Together, these four nucleotides participate in eight protein/RNA hydrogen bonds, three protein/RNA aromatic stacking interactions, and one RNA/RNA base stacking interaction (Fig. S5A). The flanking 2′-G forms hydrogen bonds with the side chains of G101 and D205 and base stacks with the cytidine of the flanking 3′-C that, in turn, donates two hydrogen bonds to the side chain of K134 of the lariat recognition loop (LRL). LRL residues I132, F155, and H156 make extensive contacts with the ribose ring and the 3′-,5′-phosphodiester bond linking the 3′-C and terminal 3′-A (Fig. S5A). The second flanking nucleotide in the 3′-arm (3′-A) base-stacks with the side chain of F136 of the LRL. Although two nucleotides are present in the 2′-arm of AK49, only the flanking guanosine is visible in the structure of the complex (Fig. S5A). The extent of these interactions is consistent with previous findings that a trinucleotide consisting of a branchpoint adenosine with single flanking 2′- and 3′-nucleotides is recognized and cleaved by Dbr1 (12).
In contrast, in the structure of the AK86/EhDbr1 complex, all five nucleotides of the 2′-arm are visible and these nucleotides engage the enzyme at the junction of a β-hairpin formed by residues 233–249 and a helix-turn-helix insertion element formed by residues 179–212 (Fig. S6). Together, these structural elements form a platform that projects from the metal center around which the nucleotides of the 2′-arm wrap. The third nucleotide of the 2′-arm (adenosine) is completely devoid of interactions with protein (Fig. 3E and Fig. S6). The fourth and fifth nucleotides in the 2′-arm make aromatic stacking interactions with Y252 and F198, respectively. The aromatic base-stacking between the flanking 2′-G, and 3′-C (Fig. 3E and Figs. S5 and S6) combined with extensive interactions with the enzyme cause the branchpoint adenine base to be exposed and to protrude in a direction opposite to the flanking stacking 2′- and 3′-bases, where it is sandwiched in aromatic stacking interactions between H16 and Y64. In their seminal 1985 paper, Ruskin and Green (3) reported poly-A RNA linked solely by 2′,5′-phosphodiester bonds is not a substrate for human Dbr1. The structure of the AK49/EhDbr1 complex permits rationalization of this observation. If 2′,5′-linked poly-A RNA binds analogously to the branchpoint adenosine in AK49 and AK86, the 2′-G is replaced by a 2′-A with the loss of two protein–RNA hydrogen-bonding interactions (Fig. S5B). Because 2′,5′-linked poly-A RNA does not possess the equivalent of a 3′-arm, it would be devoid of all of the previously stabilizing interactions. In addition, the 5′,3′-phosphodiester bond linking the 2′-G and 2′-U in AK86 and AK49 (Fig. 3E and Fig. S5A) would be replaced by a 2′,5′-phosphodiester linkage between adenosine nucleotides, a linkage predicted to clash with F202 and other nearby residues (Fig. S5B).
The Invariant Cys in the α-Pocket of Dbr1 Enzymes.
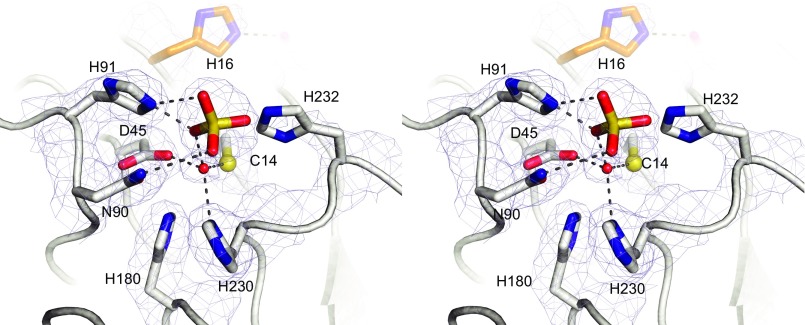
MPEs possess the conserved signature sequence Dα[X]Hα [X]n GDα+β [X]n GNβH[D/E] [X]n Hβ [X]n GHα[X]Hβ, where underlined/italicized letters correspond to metal ligands, α and β to the binding pockets in which the ligands reside, and X to any amino acid (26). Despite the conservation of metal-binding residues, the repertoire used by various MPEs is diverse and signature sequence-containing enzymes bind Mn(II), Fe(II), Fe(III), Zn(II), Co(II), Cd(II), and Ni(II) (reviewed in ref. 26). Normally, Fe(II) and Mn(II) tend to prefer similar coordination environments, making it difficult for some proteins to distinguish between them on the basis of active site structures alone (32). Indeed, the insertion of the “correct” metal ion into a metalloprotein often depends on the relative concentrations of various metal ions in the immediate environment during and after protein folding as well as the affinity and selectivity of the nascent (apo) metalloprotein for its metal cofactor (reviewed in ref. 32). Fe(II) and Zn(II) are abundant in biological systems, at levels ∼50-fold higher than Mn(II) [∼300 μM vs. 6 μM; ref. 33). Still, Mn(II) is commonly found in MPEs and was reported to support the highest phosphoesterase activity among divalent cations tested in Mre11 (34, 35), 5′-nucleotidase (36, 37), MPPED2 (38, 39), and yeast Dbr1 (23). Superposition of the Mre11 and Dbr1 binuclear centers reveals they are indeed similar, including the position of the bridging nucleophile, except for the substitution of the first aspartic acid in the signature sequence with Cys in Dbr1 (Fig. S7). Examples of Cys serving as a ligand to Mn(II) are rare, with the RNA ligase RtcB being a notable example (40). In the MESPEUS database of metal sites in proteins (mined from the PDB), there are 35 examples of Cys coordinating Mn ions, 6,023 examples of Cys coordinating Fe ions, and 8,069 examples of Cys coordinating Zn ions (41). The inability of the α-pocket of Dbr1 to bind Mn(II) was evident in previous work in which EDTA-treated EhDbr1 crystallized in the presence of 1 mM MnSO4 with Mn(II) only in the β-pocket (24). Even when the concentration of MnSO4 in the crystallization buffer was increased to 10 mM, there was no Mn in the α-pocket.
Fig. S7.
Alignment of EhDbr1 and MRE11. (A) Complex of the 7-mer bRNA AK46 with H91A EhDbr1. (B) Mre11 (PDB ID code 1II7) in complex with dAMP. (C) Alignment of Mre11 and EhDbr1. The active sites are similar, including the position of the bridging nucleophile, despite the substitution of the first aspartic acid in the signature MPE sequence for a cysteine.
We speculate that the presence of the α-pocket Cys ligand reflects the metal preferences of Dbr1. Although Fe(II) and Zn(II) provide the highest turnover rates in our kinetic assays with EhDbr1 (Fig. 2), it remains to be demonstrated that other Dbr1 orthologs (such as yeast Dbr1) also prefer Fe(II) and Zn(II). More experiments are required to explain why Dbr1 enzymes harbor a Cys in the α-pocket, instead of the Asp found in other MPEs. Whether modulation of Fe(II) and Zn(II) levels affects Dbr1 activity in vivo also remains to be demonstrated.
Materials and Methods
Detailed procedures are given in SI Materials and Methods. Briefly, the DBR1 gene from E. histolytica was codon optimized (DNA 2.0; Genescript), expressed as a His-tagged fusion protein in E. coli strain BL21 (New England Biolabs), and purified by using a Ni-Sepharose Fast Flow column (GE Healthcare). The metal content was determined by ICP-MS at the Chemical Analysis Laboratory at the University of Georgia. Apoenzyme was obtained by dialyzing Dbr1-containing fractions against stripping buffer [10 mM EDTA, 10 mM NTA, 20 mM Hepes pH 7, 50 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP)] at 4 °C for 72 h followed by dialysis against assay buffer (50 mM Hepes pH 7, 100 mM NaCl) that had been treated with 10 g of Chelex resin (Bio-Rad) per liter of buffer. Kinetic assays were performed by using 5 μM enzyme (final concentration) to eliminate effects of trace metal ions remaining in the assay buffer. A Pherastar plate reader (BMG Labtech) in rapid kinetic mode and a stopped-flow mixing spectrophotometer (Applied Photophysics) were used for aerobic and anaerobic kinetic assays, respectively. For measurement of steady-state kinetic parameters, the inner-filter effect was corrected for using the method described in ref. 42. Crystals of EhDbr1 grew in 0.1 M Bis-Tris pH 5.5 made 10–17% (wt/vol) PEG 3350, 0.4 M LiSO4, 10% (vol/vol) glycerol in hanging-drop plates. Diffraction data were taken at the Advanced Photon Source 24 ID-C beamline with a Pilatus-6MF Pixel Array Detector operated by Northeast Collaborative Access Team (NE-CAT). The programs XDS (43), AIMLESS (44), PHENIX (45), and COOT (46) were used for data integration, scaling, model refinement, and model building, respectively. Structure factors and model coordinates are deposited in the PDB with codes: 5K71, 5K73, 5K77, and 5K78 for apo-, wild-type, AK49/H91A, and AK86/H91A Dbr1 structures, respectively.
Acknowledgments
P.J.H. was supported in part by Merit Review Award I01 BX0025801 from the US Department of Veterans Affairs, Biomedical Laboratory Research and Development Service, S. Texas Veterans Health Care System, and in part by a Welch Foundation Grant AQ-1399 and by the Judith and Jean Pape Adams Charitable Foundation. M.J.D. was supported by a Discovery Grant from the National Sciences and Engineering Research Council of Canada. S.W.S. was supported by National Institutes of Health (NIH) Grant GM084246. N.E.C. and E.J.M. were supported by NIH Grant T-32 AG021890 through the Barshop Institute for Longevity and Aging Studies. E.J.M. was supported by Grant DBI-0905865 through the National Science Foundation. P.F.F. and K.M.R. were supported by Grant AQ-1245 from The Welch Foundation. Support for NE-CAT beamline 24-ID-E is provided by NIH Grant P41 GM103403 and US Department of Energy Grant DE–AC02–06CH11357. The X-Ray Core Laboratory at University of Texas Health Science Center, San Antonio (UTHSCSA) is supported in part by the Office of the Vice President for Research and by San Antonio Cancer Institute Grant P30 CA054174. Equipment and technical expertise were provided by the Center for Innovative Drug Discovery and High Throughput Screening Facility at UTHSCSA, which is supported by Grant UL1 TR001120 from the National Center for Advancing Translational Sciences, NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5K71, 5K73, 5K77, and 5K78).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612729114/-/DCSupplemental.
References
- 1.Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365(6444):364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- 2.Konarska MM, Grabowski PJ, Padgett RA, Sharp PA. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- 3.Ruskin B, Green MR. An RNA processing activity that debranches RNA lariats. Science. 1985;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- 4.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA. 1998;4(9):1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domdey H, et al. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- 7.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15(2):108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65(3):483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 9.Nam K, Lee G, Trambley J, Devine SE, Boeke JD. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol. 1997;17(2):809–818. doi: 10.1128/mcb.17.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Hill K, Perry SE. An Arabidopsis RNA lariat debranching enzyme is essential for embryogenesis. J Biol Chem. 2004;279(2):1468–1473. doi: 10.1074/jbc.M309106200. [DOI] [PubMed] [Google Scholar]
- 11.Zheng S, et al. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell. 2015;161(4):762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arenas J, Hurwitz J. Purification of a RNA debranching activity from HeLa cells. J Biol Chem. 1987;262(9):4274–4279. [PubMed] [Google Scholar]
- 13.Galvis AE, Fisher HE, Nitta T, Fan H, Camerini D. Impairment of HIV-1 cDNA synthesis by DBR1 knockdown. J Virol. 2014;88(12):7054–7069. doi: 10.1128/JVI.00704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Y, De Leon J, Yokoyama N, Naidu Y, Camerini D. DBR1 siRNA inhibition of HIV-1 replication. Retrovirology. 2005;2:63. doi: 10.1186/1742-4690-2-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12(10):1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Z, Menees TM. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science. 2004;303(5655):240–243. doi: 10.1126/science.1087023. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Z, Menees TM. RNA splicing and debranching viewed through analysis of RNA lariats. Mol Genet Genomics. 2011;286(5-6):395–410. doi: 10.1007/s00438-011-0635-y. [DOI] [PubMed] [Google Scholar]
- 19.Salem LA, Boucher CL, Menees TM. Relationship between RNA lariat debranching and Ty1 element retrotransposition. J Virol. 2003;77(23):12795–12806. doi: 10.1128/JVI.77.23.12795-12806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coombes CE, Boeke JD. An evaluation of detection methods for large lariat RNAs. RNA. 2005;11(3):323–331. doi: 10.1261/rna.7124405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi SL, et al. RNA lariat debranching enzyme. Methods Enzymol. 2001;342:233–248. doi: 10.1016/s0076-6879(01)42548-1. [DOI] [PubMed] [Google Scholar]
- 22.Nam K, et al. Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem. 1994;269(32):20613–20621. [PubMed] [Google Scholar]
- 23.Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res. 2005;33(19):6349–6360. doi: 10.1093/nar/gki934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montemayor EJ, et al. Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. Nucleic Acids Res. 2014;42(16):10845–10855. doi: 10.1093/nar/gku725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katolik A, et al. Regiospecific solid-phase synthesis of branched oligoribonucleotides that mimic intronic lariat RNA intermediates. J Org Chem. 2014;79(3):963–975. doi: 10.1021/jo4024182. [DOI] [PubMed] [Google Scholar]
- 26.Matange N, Podobnik M, Visweswariah SS. Metallophosphoesterases: Structural fidelity with functional promiscuity. Biochem J. 2015;467(2):201–216. doi: 10.1042/BJ20150028. [DOI] [PubMed] [Google Scholar]
- 27.Ruskin B, Green MR. RNA lariat debranching enzyme as tool for analyzing RNA structure. Methods Enzymol. 1990;181:180–188. doi: 10.1016/0076-6879(90)81120-j. [DOI] [PubMed] [Google Scholar]
- 28.Kim JW, et al. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res. 2000;28(18):3666–3673. doi: 10.1093/nar/28.18.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armakola M, Hart MP, Gitler AD. TDP-43 toxicity in yeast. Methods. 2011;53(3):238–245. doi: 10.1016/j.ymeth.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tago N, et al. Design, synthesis, and properties of Phosphoramidate 2′,5′-linked branched RNA: Toward the rational design of inhibitors of the RNA lariat debranching enzyme. J Org Chem. 2015;80(20):10108–10118. doi: 10.1021/acs.joc.5b01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrey SM, et al. A homolog of lariat-debranching enzyme modulates turnover of branched RNA. RNA. 2014;20(8):1337–1348. doi: 10.1261/rna.044602.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotruvo JA, Jr, Stubbe J. Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: The class I ribonucleotide reductases as a case study. Metallomics. 2012;4(10):1020–1036. doi: 10.1039/c2mt20142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunzhi H, et al. Serum and tissue levels of six trace elements and copper/zinc ratio in patients with cervical cancer and uterine myoma. Biol Trace Elem Res. 2003;94(2):113–122. doi: 10.1385/BTER:94:2:113. [DOI] [PubMed] [Google Scholar]
- 34.Hopfner KP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105(4):473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 35.Hopfner KP, et al. Mre11 and Rad50 from Pyrococcus furiosus: Cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182(21):6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knöfel T, Sträter N. Mechanism of hydrolysis of phosphate esters by the dimetal center of 5′-nucleotidase based on crystal structures. J Mol Biol. 2001;309(1):239–254. doi: 10.1006/jmbi.2001.4656. [DOI] [PubMed] [Google Scholar]
- 37.McMillen L, Beacham IR, Burns DM. Cobalt activation of Escherichia coli 5′-nucleotidase is due to zinc ion displacement at only one of two metal-ion-binding sites. Biochem J. 2003;372(Pt 2):625–630. doi: 10.1042/BJ20021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dermol U, Janardan V, Tyagi R, Visweswariah SS, Podobnik M. Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome. J Mol Biol. 2011;412(3):481–494. doi: 10.1016/j.jmb.2011.07.060. [DOI] [PubMed] [Google Scholar]
- 39.Tyagi R, Shenoy AR, Visweswariah SS. Characterization of an evolutionarily conserved metallophosphoesterase that is expressed in the fetal brain and associated with the WAGR syndrome. J Biol Chem. 2009;284(8):5217–5228. doi: 10.1074/jbc.M805996200. [DOI] [PubMed] [Google Scholar]
- 40.Desai KK, Bingman CA, Phillips GN, Jr, Raines RT. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013;52(15):2518–2525. doi: 10.1021/bi4002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsin K, Sheng Y, Harding MM, Taylor P, Walkinshaw MD. MESPEUS: A database of the geometry of metal sites in proteins. J Appl Cryst. 2008;41:963–968. [Google Scholar]
- 42.Liu Y, et al. Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Anal Biochem. 1999;267(2):331–335. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts KM, Pavon JA, Fitzpatrick PF. Kinetic mechanism of phenylalanine hydroxylase: Intrinsic binding and rate constants from single-turnover experiments. Biochemistry. 2013;52(6):1062–1073. doi: 10.1021/bi301675e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis HR, McCusker KP, Fitzpatrick PF. Use of a tyrosine hydroxylase mutant enzyme with reduced metal affinity allows detection of activity with cobalt in place of iron. Arch Biochem Biophys. 2002;408(2):305–307. doi: 10.1016/s0003-9861(02)00568-4. [DOI] [PubMed] [Google Scholar]
- 49.Keating KS, Pyle AM. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc Natl Acad Sci USA. 2010;107(18):8177–8182. doi: 10.1073/pnas.0911888107. [DOI] [PMC free article] [PubMed] [Google Scholar]