Can you imagine New York’s Central Park as a source of the next generation of antibiotics that can treat hundreds of millions of people? The work by Charlop-Powers et al. in PNAS (1) suggests this possibility. Antibiotic-resistant microbes pose one of the main health threats of the 21st century (2). According to the Centers for Disease Control and Prevention, more people die today of bacterial infections than HIV (3) and more people will die of bacterial infections than cancer by 2050 (4). Because antibiotic resistance is a global threat that affects hundreds of millions of people worldwide (5), it has become a key issue for the White House and the United Nations. In 2015, the White House developed an action plan to combat antibiotic resistance and the General Assembly of the United Nations had a high-level meeting addressing worldwide antibiotic resistance (6, 7).
Most (∼75%) of today’s antibiotics are derived from natural products (2, 8). Penicillin was isolated from a fungus, and azithromycin, commonly prescribed in the form of a Z-Pak, was isolated from a bacterium (8). The period from the 1930s through the 1980s represented the “Golden Age” for the discovery of antibiotics. Since 1975, very few antibiotics have emerged (rare exceptions include vancomycin and daptomycin), and some pathogens are resistant to all known antibiotics. To combat these “superbugs,” researchers are exploring new natural sources that were missed in previous screens (3). Exotic areas, usually in regions of the planet minimally impacted by people and with high biodiversity, such as rainforests or coral reefs, are most often explored for natural sources of next-generation antibiotics. Charlop-Powers et al. (1) extracted DNA from the environment, followed by high-throughput sequencing of its genetic instructions, to reveal that your local park is an untapped resource with enormous potential for discovery of new therapeutics. Additionally, it is a lot easier to get to than a rainforest or a reef. The results raise a critical question: What other urban settings harbor organisms that make new classes of antibiotics? Perhaps the chair you are sitting on, the floor you are walking on, the restaurant you eat at, or the subway railing you touch on your way to work contains bacteria or fungi whose genomes encode instructions to make the next antibiotic that could cure hundreds of millions of people.
In PNAS, Charlop-Powers et al. (1) build on the wealth of knowledge built up by many laboratories that identify DNA signatures encoding proteins involved in biosynthesis of natural products produced by microbes, as well as advances in DNA sequencing technology and extraction protocols that make it feasible to identify these sequences in environmental samples in high throughput. One can think of these DNA signatures as barcodes that can be identified in any DNA database. To find the signatures of molecules that have therapeutic potential in environmental DNA (eDNA), they created the environmental Surveyor of Natural Product Diversity (eSNaPD) (9), a searchable database of annotated DNA sequences, where new sequences can be matched to known biogenetic gene clusters for antimicrobial natural products. With this tool in hand, they sampled the soil of urban parks of the New York City (NYC) area and extracted total DNA from each sample (Fig. 1). To enrich sequences most likely to encode new antimicrobial natural products, they selectively amplified genes that encode polyketide (PKS) and nonribosomal peptide synthetases (NRPS), which are the enzymes that synthesize PKS and nonribosomal peptides, respectively, both important classes of natural antimicrobials, and then sequenced the resulting enriched libraries. Some of the better known products of PKS and NRPS include the antibiotics penicillin and daptomycin (nonribosomal peptides) and the cholesterol-lowering agent lovastatin (a PKS), but many other therapeutics are made by these biosynthetic pathways (10).
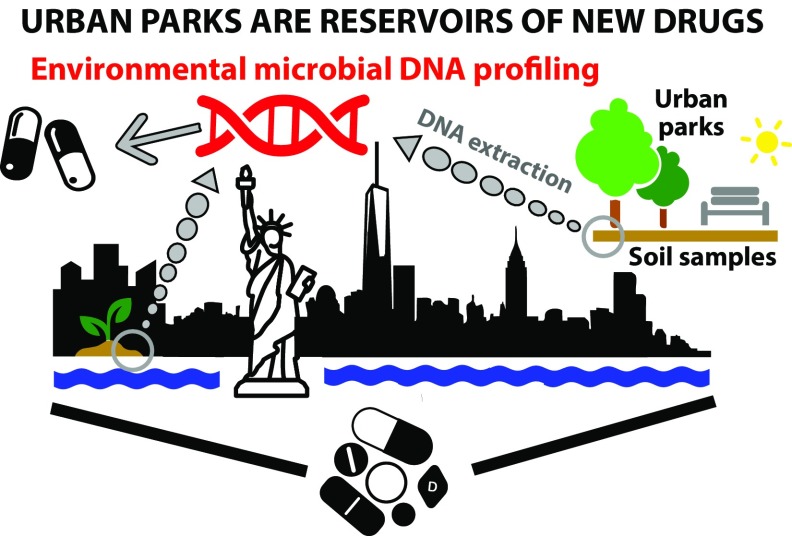
Fig. 1.
Soil urban parks of NYC, an urban area of 10 million inhabitants, were found to contain biosynthetic genes clusters involved in the biosynthesis of many widely used drugs, revealing an untapped reservoir of new therapeutics. Images courtesy of The Noun Project under public domain license or Creative Commons (CC 3.0); DNA courtesy of Warslab, Freedom Tower courtesy of Chris McDonnell, city plant courtesy of GRACE Communications Foundation and Mother Jones, New York city courtesy of Raf Verbraeken, Statue of Liberty courtesy of Shashank Singh, park bench courtesy of Roma, and pills courtesy of Tonielle Purdy.
The authors compared the urban NYC parks’ biosynthetic potential with soil samples from nonurban environments in upstate New York, the Midwest, and the western United States. Excitingly, eDNA from soil samples of the NYC urban parks contained many diverse genes related to the biosynthesis of clinically important antibiotics, antifungal agents, immunosuppressants, and anticancer agents. What was even more remarkable is that the diversity of potential therapeutic agents was just as great between soils within tens of miles as between NYC park soils and desert soils more than 2,500 miles away, suggesting that closer inspection of more samples closer to hand might be more rewarding, and certainly less expensive, than a quest for pristine and exotic locations to sample new antimicrobials. Even the NYC urban park soil samples harbored tremendous diversity in the types of molecules that could be discovered, yet they can still be grouped according to their ecotype (maritime forest, upland grass, etc.) rather than clustering as urban versus nonurban. Interestingly, samples belonging to the same ecotype were similar across multiple NYC boroughs, suggesting that the diversity of natural product synthesis is primarily driven by ecotype (rather than being random or driven by the urban-nonurban split). However, high levels of intra- and interpark heterogeneity were observed, suggesting that geography might be one of many factors driving microbial population differences among different urban park localities (11). Some parks also had idiosyncratic features: For example, although having the general features of other parks from the maritime forest ecotype, some of the park soil samples from Staten Island were biosynthetically distinct. This area is the most suburban and least populated of the urban park samples studied.
Among the NRPS and PKS sequence data obtained, about 1% were found to encode 11 classes of natural products used in the clinic, or relatives of these natural products, including those natural products related to the anticancer drug epothilone or to the antibiotics erythromycin and rapamycin. Perhaps even more important than finding the biosynthetic capacity for known molecules, most of the barcodes obtained belonged to unknown NRPS and PKS biogenetic gene clusters. This finding suggests that there is an enormous therapeutic discovery that is found in NYC parks, essentially the backyard of 10 million people. Furthermore, as annotations continue to be improved as more reference sequences are added to the eSNaPD database, this discovery potential will only continue to increase (9). These observations highlight an example in which therapeutics may be discovered in an urban environment, but only if there is a collective effort by the scientific community to organize this knowledge and develop software to harvest the results and make them accessible. Ultimately, we will need to access the molecules and their structures to evaluate their therapeutic potential, and doing so remains an important challenge that has not been accelerated by DNA sequencing technology.
There are two key approaches to get access to the molecules to screen them for therapeutic potential. One approach is to culture the microbes that produce them. In some cases, innovative culturing methods are required to grow organisms with specialized requirements to get them to produce these important molecules (12, 13). Culturing efforts can even be distributed through citizen science projects, such as the Small World Initiative (www.smallworldinitiative.org/) and the Natural Product Discovery Group (npdg.ou.edu/citizenscience) (14), and direct analysis of the metabolites’ content can now be done by anyone using open crowd-sourced analysis platforms, such as Global Natural Product Social Molecular Networking (gnps.ucsd.edu) (15). One promising strategy for the valorization of potential therapeutical discovery from eDNA is to
Charlop-Powers et al. extracted DNA from the environment, followed by high-throughput sequencing of its genetic instructions, to reveal that your local park is an untapped resource with enormous potential for discovery of new therapeutics.
clone or synthesize de novo the gene cluster(s) involved in biosynthesis of a specific product, and to express it in a host organism (16, 17). Although still not trivial, the improvement in the past decade in methods for assembling long genetic constructs in a different species host (17), yield optimization by pathway manipulation (18), and combinatorial biosynthesis (19) offer an exciting toolbox for accessing this molecular biodiversity.
Important though the implications of this work for antibiotic discovery are, it also offers a glimpse into future understanding of geographic patterns associated with antibiotic resistance. Microbes that produce an antibiotic molecule usually also contain the DNA sequences responsible for resistance to it, and these resistance genes have been found in soil samples (20). It would be of great interest to test for relationships between the gene clusters coding for antibiotic NRPS or PKS in eDNA and the occurrence rate of clinical antibiotic resistance in the area. If such relationships exist, then understanding patterns of the presence of antibiotic biosynthetic machineries, as shown by Charlop-Powers et al. (1), could perhaps one day be used to guide antibiotic use in the clinic based on geography, avoiding antibiotics for which resistance genes can be found in a nearby park (or garden or decorative plant’s soil, which is not studied here but is potentially important) and where they might be tracked in on shoes and moved to clinically relevant strains via horizontal gene transfer.
Although this study reveals the potential for discovering antibiotics in our backyards, many questions still remain. Are these molecules actually produced by the microbes in soil? If they are, when and where? If they are produced, what are their normal functions in soil (or in the other habitats where the same microbes live)? How do people and the built environment influence the natural product biosynthetic potential? What impact, if any, do the molecules have on a child who eats the dirt, especially in the context of our increasing understanding of long-term effects of early childhood antibiotic use and given the cross-cultural practice of geophagia for health reasons (21)?
It is clear from the work by Charlop-Powers et al. (1) that we have overlooked some obvious areas for therapeutic discovery and that biodiversity hotspots and exotic environments are not the only habitats that researchers should consider when looking for new microbial natural products. Indeed, although the postindustrial era has endangered many natural habitats worldwide, the features of metropolitan areas introduce unique microbial environments and perhaps even create new opportunities to fend off the looming worldwide threat of antibiotic resistance. Although human creativity and ingenuity will likely find ways to deal with this urgent problem, the solution may ultimately be found by taking a walk in the park.
Acknowledgments
Work in the laboratories of P.C.D. and R.K. is supported, in part, by the Alfred P. Sloan Foundation Program for the Microbiology of the Built Environment, by the National Institutes of Justice, and by the NIH.
Footnotes
The authors declare no conflict of interest.
See companion article on page 14811.
References
- 1.Charlop-Powers Z, et al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc Natl Acad Sci USA. 2016;113:14811–14816. doi: 10.1073/pnas.1615581113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventola CL. The antibiotic resistance crisis: Part 1: Causes and threats. P T. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline in 2013. J Antibiot (Tokyo) 2013;66(10):571–591. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]
- 4.Walsh F. 2014 Superbugs to kill more than cancer by 2050. BBC News. Available at www.bbc.com/news/health-30416844. Accessed November 11, 2016.
- 5.Arias CA, Murray BE. A new antibiotic and the evolution of resistance. N Engl J Med. 2015;372(12):1168–1170. doi: 10.1056/NEJMcibr1500292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.United Nations 2016 High-level meeting on antimicrobial resistance. General Assembly of the United Nations. Available at www.un.org/pga/71/event-latest/high-level-meeting-on-antimicrobial-resistance/. Accessed November 11, 2016.
- 7.White House Office of the Press Secretary 2015 FACT SHEET: Obama Administration Releases National Action Plan to Combat Antibiotic-Resistant Bacteria. Available at https://www.whitehouse.gov/the-press-office/2015/03/27/fact-sheet-obama-administration-releases-national-action-plan-combat-ant. Accessed November 11, 2016.
- 8.Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Curr Biol. 2009;19(11):R437–R441. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy BVB, Milshteyn A, Charlop-Powers Z, Brady SF. eSNaPD: A versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem Biol. 2014;21(8):1023–1033. doi: 10.1016/j.chembiol.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14(2):111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez KS, et al. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc Biol Sci. 2014;1795(281):20141988. doi: 10.1098/rspb.2014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13(8):509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 13.Nichols D, et al. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol. 2010;76(8):2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du L, et al. Crowdsourcing natural products discovery to access uncharted dimensions of fungal metabolite diversity. Angew Chem Int Ed Engl. 2014;53(3):804–809. doi: 10.1002/anie.201306549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber T, et al. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015;33(1):15–26. doi: 10.1016/j.tibtech.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, Enghiad B, Zhao H. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat Prod Rep. 2016;33(2):174–182. doi: 10.1039/c5np00085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones JA, Koffas MAG. Optimizing metabolic pathways for the improved production of natural products. In: O’Connor SE, editor. Methods in Enzymology, Synthetic Biology and Metabolic Engineering in Plants and Microbes. Part A: Metabolism in Microbes. Elsevier; Amsterdam: 2016. pp. 179–193. [DOI] [PubMed] [Google Scholar]
- 19.Kim E, Moore BS, Yoon YJ. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol. 2015;11(9):649–659. doi: 10.1038/nchembio.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311(5759):374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 21.Blaser MJ. Missing Microbes: How the Overuse of Antibiotics Is Fueling Our Modern Plagues. Henry Holt and Company; New York: 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]