Abstract
Broad-spectrum analysis of DNA and RNA samples is of increasing importance in the growing field of biotechnology. We show that nanopore measurements may be used to assess the purity, phosphorylation state, and chemical integrity of nucleic acid preparations. In contrast with gel electrophoresis and mass spectrometry, an unprecedented dynamic range of DNA sizes and concentrations can be evaluated in a single data acquisition process that spans minutes. Because the molecule information is quantized and digitally recorded with single-molecule resolution, the sensitivity of the system can be adjusted in real time to detect trace amounts of a particular DNA species.
The purity of a nucleic acid preparation and the chemical integrity of nucleic acid bases affect the efficiency of hybridization procedures, enzymatic reactions, and chemical modifications. These processes dictate the accuracy and reliability of routine biochemical and clinical investigations as well as the expanding field of array technologies. Although traditional techniques of electrophoresis, chromatography, and mass spectrometry can assess DNA or RNA sample purity and chemical integrity, the sensitivity of these methods is limited by the relative size and quantity of contaminating nucleic acids. More importantly, the resolution of these methods decreases with increasing DNA or RNA length. Sample evaluation is difficult for nucleic acids with >100 nt and is virtually impossible for those with >1,000 nt. In a process we call single-molecule electrophoresis, we show that a transmembrane nanopore can evaluate polynucleotides with <100 bases or >1,000 bases. The ensemble pattern produced by the individual interaction between the polymer and the nanopore reveals DNA sample properties in a manner unparalleled by other detection systems.
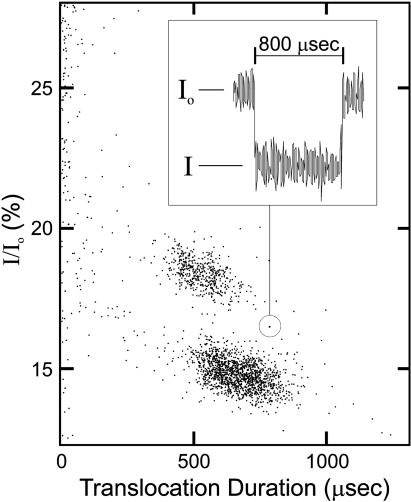
The nanopore was formed by self-assembly of α-hemolysin (1), a robust channel-forming protein that has been used and engineered for stochastic sensing, characterization of small molecules, and detection and discrimination of individual DNA strands (2–7). A single α-hemolysin channel was embedded in a lipid membrane that partitioned a conducting solution into two chambers. A voltage bias across the membrane produced an ion current through the α-hemolysin and also drove negatively charged single-stranded DNA from the cis to the trans side of the channel. Translocations occurred on the microsecond timescale and were signaled by the partial blockage of ion current to 10–25% of the open pore current value (Fig. 1). Blockage duration reflected both the length of the single-stranded DNA and its retardation characteristics by the pore. At 120 mV and 16–25°C, thousands of single-molecule translocations could be monitored in minutes. Custom data analysis software displayed the translocation data as scatter plots showing each translocation event's normalized average current (I/Io) as a function of that event's translocation duration (Fig. 1). Here, we illustrate the DNA sample properties revealed by the translocation scatter plots.
Fig. 1.
Scatter plot displaying individual translocation events at 17°C of poly dC500. The ion current trace for the encircled event is shown in Inset, where I is the ionic current during polymer translocation and Io is the open (unblocked) nanopore current. Each data point on the scatter plot represents a single DNA translocation through the α-hemolysin pore.
Materials and Methods
Nucleic Acid Preparations. Synthetic DNA oligonucleotides were purchased from different commercial suppliers. DNA prepared by PCR was amplified with synthetic primers from synthetic templates by using Ready-To-Go PCR Beads (Amersham Biosciences), and the synthetic segments were removed from the final products by restriction digests with Bsp1286I and SfcI (NEB). The resulting 70-mer (dS70) and 78-mer restriction products were separated by denaturing PAGE to generate single-stranded PCR products. dC500 and dA1300 were purchased from Amersham Biosciences. All DNA except for dA1300 and dC500 were purified by PAGE under denaturing conditions. PCR products and long homopolymers were generated with 5′ phosphorylation. Most synthetic oligonucleotides were 5′ phosphorylated with phosphoramidite during synthesis. Some samples were again phosphorylated with T4 polynucleotide kinase. Dephosphorylation was performed with calf intestine alkaline phosphatase. Phosphorylation at the 3′ end of polynucleotides was achieved during synthesis by using Glen Research (Sterling, VA) chemical phosphorylation reagent. Any unphosphorylated strands were removed with exonuclease I. Diethylpyrocarbonate (DEPC) reactions were performed at room temperature with 5–0.05% DEPC with 2 μM DNA for 0.5–5 h. All enzymatic reactions were performed by using standard protocols described by the commercial enzyme suppliers. All samples assayed with the nanopore were also evaluated with denaturing PAGE.
The sequence for dS70 was as follows: 5′-CCACAAACAAACAACCACACAAACACACAACCACAACACCAACACACAAACAAACCAACACACAAACTCC-3′.
Construction of Symmetric Molecules. Symmetric molecules with two 3′ termini were constructed by oxidation of identical oligonucleotides with deprotected 5′ thiomodifier phosphoramidites. The symmetric molecules with two 5′ termini were constructed by oxidation of oligonuclotides with deprotected 3′ thiomodifier phosphoramidite (Glen Research). Oxidation products were purified and characterized by denaturing PAGE. The symmetric molecules we tested were constructed from dA homopolymers or 48-mer with the following sequence: CAAACAAACCAACACACA A ACTCC-S-S-CCTCA A ACACACA ACCA A ACAAAC, where S-S indicates disulfide bonds. In each case, the respective control molecules with one 5′ terminus and one 3′ terminus had the same sequence but did not contain disulfide bonds. The dA homopolymers were constructed from either two 24-mer segments or 98-mer segments. Phosphorylations were performed with T4 polynucleotide kinase.
Nanopore Setup and Data Acquisition. Single-channel formation, instrument setup, and data acquisition are described in ref. 7. All experiments were performed in 1 M KCl/10 mM Tris·HCl, pH 8/1 mM EDTA at a 2-μs sampling rate. A 120-mV bias was applied across the channel at 17°C unless otherwise specified. The amplified signals were low-pass filtered at 100 KHz.
Data Analysis. The software data analysis, implemented in mat-lab r12 (Mathworks, Natick, MA), consisted of three stages: preprocessing, event extraction, and postprocessing. During the preprocessing stage, the experimental data were read from Axon Binary Files (Axon Instruments, Union City, CA) into a data array and then smoothed with a Daubechies Wavelet Filter (Mathworks). The characteristic pattern of the translocation events was then extracted with a heuristic threshold method by defining two threshold levels: an upper level and a lower level. A translocation event was defined by a current trace that (i) dropped below the upper level and remained below the upper level until crossing below the lower threshold level, then (ii) rose successively past the lower and upper threshold. Translocation duration time was calculated for each translocation event as the time between the crossings of the lower threshold at both the falling and rising edges. The average translocation current level was the arithmetic mean of all of the current value data points sampled at 2-μs intervals within this translocation duration. After all possible translocation events were extracted, the post-processing step was to tag and discard the flawed events. These events included those with clearly unreasonable durations or clearly unreasonable average translocation current values, with dramatic fluctuations during the translocation duration, and with or near voltage spikes or voltage reversal. Voltage reversals were sometimes manually introduced to dislodge molecules that appeared to be stuck in the channel. During software development, individual accepted and rejected events were continuously evaluated visually. These evaluations eliminated accepting unreasonable signals as translocation and minimized rejections, which would be identified as a translocation event by a person who has superior pattern recognition ability. The two threshold levels (as defined above) for the data presented here were 85% and 33% of the open-pore current.
Cluster scores were calculated by dividing the scatter plot into rectangular grids of 20 μs and 0.2% current units. The data point density for each rectangle was assigned as a percentage of the densest rectangle. The total number of data points in the rectangles with >50% density was then divided by the total number of data points in the ≤50% density rectangles. The quotient multiplied by 100 was the cluster score. The tighter the cluster, the higher the cluster score. This cluster scoring method was designed for comparing samples with scattered clusters relative to high-quality DNA. The cluster score fails if the very short events (presumably caused by backbone scission) form tighter and larger clusters than the expected clusters from intact DNA.
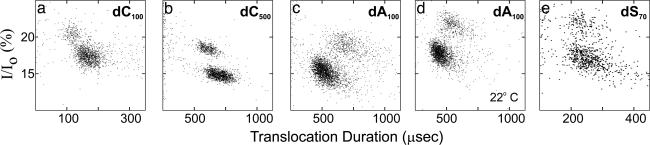
Molecular Profile Patterns Can Be Generated from Translocation Signals. Homopolymers with 40 to >1,000 nt and heteropolymers (dSn, where n ranged from 48 to 87 nt) were assayed at different temperatures. None of the sequences contained stable base-pairing structures. For most of the polynucleotide sequences the data points consistently separated themselves into two well defined clusters (Fig. 2). The two-cluster behavior was reproducible but not universal. Polymers that were expected to have more difficulty threading through the pore because of base stacking or transient base pairing (8, 9) translocated in one rather diffuse cluster. For example, RNA heteropolymers as well as DNA heteropolymers containing many guanines translocated as one cluster (data not shown). These samples also translocated more slowly (11–21 μs per nucleotide at 17°C) than did the two-cluster samples (1.2–4.6 μs per nucleotide at 17°C).
Fig. 2.
Cluster profiles reflect molecular properties. The translocation duration varied with DNA length (the number of nucleotides in the polymers that were tested is shown in the subscript of each graph label); the translocation rate and the relative positioning of the two clusters varied with base sequence. Increasing the polymer length (a vs. b) or temperature (c vs. d) affected only the mean translocation duration and current blockage, not the relative position of the two clusters. Unstructured heteropolymers also exhibited the two-cluster pattern (e). Data were obtained at 17°C, except for d (22°C). Values for the lower and upper clusters (with SD), respectively, were as follows: a, 170 + 20 vs. 117 + 13 μs and 17 + 1 vs. 20 + 1% I/Io; b, 666 + 76 vs. 530 + 62 μs and 15 + 1 vs. 18 + 1% I/Io; c, 455 + 89 vs. 760 + 144 μs and 15 + 1 vs. 19 + 1% I/Io; d, 312 + 62 vs. 463 + 85 μs and 18 + 1 vs. 22 + 1% I/Io; e, 276 + 62 vs. 246 + 47 μs and 17 + 1 vs. 22 + 1% I/Io.
The more densely populated cluster always blocked more ionic current than the less densely populated cluster, and, hence, this more densely populated cluster always appeared in the lower portion of the plot. On the other hand, the relative translocation durations of the two clusters depended on the DNA sequence. For example, molecules in the more densely populated clusters of 100-mer dC100 and dA100 both blocked more ionic current than those in their respective less densely populated clusters. Whereas dA100 molecules in the more densely populated cluster consistently translocated faster than the ones in the less densely populated cluster, the reverse was true for dC100 (Fig. 2 a and c). Although uncontrollable differences in pore behavior caused the open pore current as well as the mean translocation duration and current to vary in different assays (usually within ±5%, but sometimes by as much as 20%), the relative position of the two clusters to each other was always conserved in the translocation pattern. This behavior was independent of DNA length and assay temperature. Increasing the length of a given DNA sequence or lowering the assay temperature resulted in proportional increases of translocation duration and ion current for both clusters (Fig. 2). Thus, the conserved discrete clustering patterns in the translocation profiles can serve to identify DNA samples.
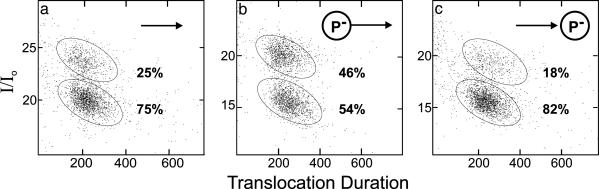
Nanopore Measurements Indicate the Phosphorylation State of the DNA Sample. The observation that translocation data from a homogeneous DNA sample forms two distinct clusters has been a topic of continued speculation since the first observed DNA translocations (6, 7). Our observations indicated that phosphorylation of single-stranded DNA changed the distribution of events in the two clusters. For example, the presence of phosphate on the 5′ end increased the fraction of events in the usually less densely populated upper cluster from ≈25% for DNA bearing no 5′ end phosphate to ≈50% for DNA bearing 5′ end phosphate (Fig. 3 a and b). This result suggested that the upper group represented translocation events initiated by the 5′ end because the additional negative charge on the phosphorylated 5′ end would likely increase the probability of this end being captured by the electrical bias across the pore. This hypothesis was strengthened by the converse observation: the fraction of events in the lower cluster increased from 75% for heteropolymer bearing no 3′ end phosphate to ≈82% for DNA bearing 3′ end phosphate (Fig. 3c). Heteropolymers with both 3′ and 5′ phosphorylation translocated as the 5′ phosphorylated molecules, with 47% of the events in the upper cluster.‡
Fig. 3.
Phosphorylation changed the distribution of events in the two clusters. The phosphorylation state of each dS70 sample is indicated by the cartoon in each panel (arrowhead denotes the 3′ end, P– denotes phosphorylation). Of all clustered events (≈2,300 in each plot), the percentage of events in each cluster is shown. The boundaries of our interpretation of each cluster are shown by the ellipses. Phosphorylation did not affect the mean translocation durations and current levels of the clusters.
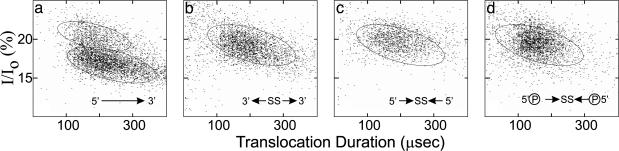
To confirm the hypothesis that phosphorylation influences capture probability and, hence, translocation direction, translocations of symmetric homopolymer and heteropolymer molecules were examined. Oligonucleotides with either two 3′ termini or two 5′ termini were constructed by linking two 3′ or 5′ sugar–phosphate backbones of palindromic sequences together with a disulfide bond. The symmetric homopolymers molecules contained either 48 or 198 nt, and the symmetric heteropolymers contained 48 nt. As expected, and exemplified by the results shown in Fig. 4, the translocation profiles of the symmetric molecules all exhibited a single cluster positioned at the current values corresponding to approximately the average values of the two clusters observed with equivalent 3′ to 5′ control sequences. Moreover, these cluster positions were not affected by phosphorylation. The single cluster produced by 5′ phosphorylated symmetric molecules had the same translocation current and duration as the nonphosphorylated symmetric molecules.
Fig. 4.
Symmetric molecules support the directionality hypothesis. Molecules with identical sequence but different backbone connectivities were compared. Backbone connectivity is shown symbolically, as in Fig. 3, in the lower right of each panel. (a) The control 48-mer heteropolymer containing palindromic nucleotide sequence translocated as two clusters. (b) The homologous but symmetric molecule with its backbone modified to form a molecule with two 3′ ends translocated as one cluster. (c) As in b, but with two 5′ ends. (d) As in c, but with both 5′ ends phosphorylated.
Final evidence that phosphorylation influences capture probability and translocation direction came from distinguishing between successful and unsuccessful translocation events. Unsuccessful translocation events were defined as those that exhibited only partial current blockages. These partial current blockages probably represent collisions between the polymer and the channel or brief polymer visits into only the channel “vestibule.” [The assembled α-hemolysin channel opening is a relatively large vestibule that narrows to the transmembrane “neck” region that traverses the membrane bilayer (1).] The ratio of successful to failed translocation events was compared for the symmetric 3′ and 5′ ended molecules. For the 3′ ended symmetric molecule, 30 ± 10% of translocation attempts failed, whereas for the symmetric 5′ ended molecules, 50 ± 4% failed. Phosphorylation of the 5′ ended molecules reduced the failure rate to 22 ± 4%. This result indicates that DNA entrance from the 5′ end often fails to translocate and that phosphorylation reduced this problem. This observation accounted for the cluster density bias and suggested that alterations of cluster densities could be used to reveal phosphorylation.
For both dA100 and dA1300, 5′ phosphorylation changed the fraction of translocation events in the usually less densely populated upper cluster from ≈15% to 25%. For dS70, dC100, and dC500, the fraction changed from ≈25% to 50%. The cluster density change was immediately evident by visual examination of the translocation profile. To explore the utility of these observations, heteropolymers containing 0%, 25%, 50%, 75%, and 100% 5′ phosphorylated strands were assayed. The fractions of events in the upper clusters were 21 ± 1%, 24 ± 2%, 27 ± 1%, 33 ± 1%, and 39 ± 1%, respectively. Thus, once the distribution ratio is determined for a given sequence, the nanopore can determine the phosphorylation state of that sequence. Fewer than 1,000 molecules need to be sampled and the measurement is rapid, because there is no need for enzymatic analysis or chemical modifications. The striking effect that terminal phosphorylation had on guiding the molecules through the nanopore may be related to end effects that have been postulated to enhance the electrophoretically effective charge at the end of a polyelectrolyte (10).
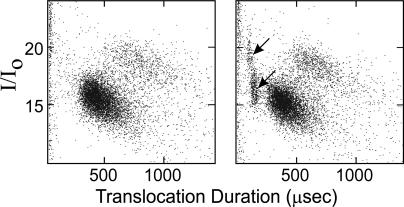
Traces of Nontarget Molecules Are Readily Detected. The clusters also reflected any heterogeneity of the DNA sample. Because translocation duration is sequence-sensitive and proportional to the length of DNA, data points outside of the target molecule clusters can reveal sample heterogeneity. Scattered minor species of different lengths that are usually invisible even on overloaded or isotope-labeled gel electrophoretograms are plainly displayed by the translocation profile. For example, when a sample of pure dA100 doped with trace amounts of dC100 (≈3 molecules of dC100 to 97 molecules of dA100 as determined by absorbance extinction coefficient calculations), the dC100 molecules were readily detected as distinct clusters by the nanopore (Fig. 5). Conversely, the dC100 in the same mixture was not visible in electrophoretograms of 32P-labeled mixtures where labeling can be significantly compromised by the sequence bias of the T4 polynucleotide kinase (11).
Fig. 5.
Minor components can be readily detected by cluster profiles. (a) The translocation profile of pure dA100.(b) The same sample as in a but spiked with 3% dC100. The presence of only 3% dC100 was clearly indicated by the two clusters of poly dC100 data points (arrows).
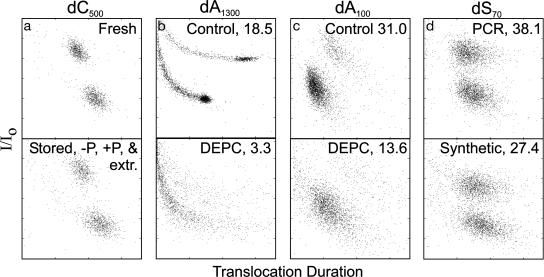
The nanopore did exhibit some sequence and size biases, which can be amplified at low temperatures (≤10°C), but the presence of nonhomogeneous species is qualitatively displayed. An example of this heterogeneity detection utility was in revealing the presence of degradation and backbone scission. The level of breakdown of a homogeneous sample was clearly measurable by comparing the translocation profile of freshly prepared dC500 and the same molecule after storage, dephosphorylation, phosphorylation, and phenol extractions (Fig. 6a). Another example of this utility was seen for the translocation profile of a commercially prepared adenine homopolymer generated by nonspecific terminal transferase elongation. As expected, this preparation (Fig. 6b, control) showed a predominant 1,300-nt product as well as data points generated by the range of expected shorter molecules.§ DNA molecules as small as 10 nt, whose ratio to the main species was <1:600, were potentially as visible as any other molecule in this mixture. Even though the capacity for absolute quantification remains to be optimized because of some observed sequence and size bias in translocation frequency, the sensitivity of the nanopore system to detecting low abundance products can be adjusted by simply sampling translocations for a few extra minutes to increase the number of sampled molecules from hundreds to tens of thousands.
Fig. 6.
DNA sample quality reflected by clustering. Pairs of samples (labeled at the top of each pair) were compared, with the higher quality of each pair in Upper. The axes for each vertical pair are identical, but for visual clarity, the absolute values that differed between pairs are not shown. (a) Serially storing, dephosphorylating, phosphorylating, and phenol extracting the sample caused minor degradations that are reflected in the percentage of translocation events that fell outside of the lower and upper clusters: 12% (Upper) to 20% (Lower). (b and c) DEPC treatment clearly disrupted clustering and was evident even in long polymers (b) with length heterogeneity. dA1300 was treated with 0.05% DEPC for 270 min, and dA100 was treated with 0.96% DEPC for 260 min. (d) Sensitivity to even the minor differences was demonstrated by comparing cluster scores of enzymatically prepared and chemically prepared DNA of identical sequence. Gels (data not shown) failed to demonstrate these differences. For b–d, the data were evaluated by cluster scores (shown in upper right corner) that are independent of translocation duration or current values.
Chemical Integrity of DNA Samples Is Revealed by Translocation Profiles. Because each data point on the translocation profile is generated by the unique interaction between one DNA molecule and one protein channel, minor changes in the chemical integrity of the DNA molecule can affect the electrical signals. This conclusion explained the disparity between the distinct data clustering behavior seen in our figures here and those from a previous report where the translocated dA100 molecules distributed themselves into one well defined group and one scattered group (7). The more distinct clustering presented here may partly be due to improved data analysis. But more significantly, studies of newly synthesized dA100 molecules demonstrated that the more distinct clustering behavior was the result of improved DNA sample integrity. Detection of chemical integrity is illustrated by the translocation profile of dA1300 and dA100 after DEPC modification (Fig. 6 b and c). DEPC reacts preferentially with N7 of adenine. This base modification results in the opening of the purine ring and destabilizes the nucleic acid backbone. The resulting sample was a mixture of randomly modified adenine bases and cleaved backbones. DEPC-treated DNA was more scattered and contained a greater number of short events than the untreated sample. The treated molecules generally behaved as though they had difficulty threading through the pore and exhibited a larger number of very short aborted events, more frequent prolonged blockages, and more variable current blockages than did untreated molecules. The same effect was observed in homopolymers and heteropolymers. In other experiments, we correlated the translocation profiles of polymers with their transcription efficiencies before and after DEPC treatment. Enzymatic reactions such as transcription are sensitive to the quality of the nucleic acids (12). Polymers that exhibited poorly clustered translocation profile were poor templates for transcription (data not shown).
To demonstrate the applicability of this quality evaluation, we applied a simple cluster scoring method to objectively evaluate quality differences between samples with identical sequence and length (Fig. 6 b–d). Only two of several synthetic DNA suppliers provided DNA that translocated through the channel to yield the tightly clustered data points characteristic of high-quality DNA (Fig. 6c, control). These samples produced clear, tight, and distinct bands when run on denaturing polyacrylamide gels. Translocation of DNA from the other suppliers produce less clustered scatter plots and appeared as less distinct and less sharp bands in denaturing gel analysis. Cluster scores of identical DNA from the different suppliers ranged from >32 to <20. The nanopore cluster assay for quality was not confined by specificity of chemical alteration or DNA size and sequence. Single-stranded DNA generated enzymatically in a PCR clustered more tightly than the equivalent chemically synthesized molecules from a high-quality supplier (Fig. 6d). It is well known that synthesis chemistry and postsynthesis processing can affect DNA base quality, especially for longer oligonucleotides, but making the quality distinctions with the nanopore required fewer sample manipulations to visualize the few variably degraded molecules in a DNA sample. Evaluating chemical quality by the DNA band morphology on denaturing gels or by chromatography is constrained by polynucleotide length, whereas the nanopore has no known length limitations.
Discussion
Our findings provide proof-of-principle applications showing that a nanopore can serve as a single-molecule electrophoretic device. Because the information is quantized at the single-molecule level, an unprecedented range of DNA lengths and quantities can simultaneously be characterized at equal resolution in a single data acquisition process. Thus, in the sample shown by Fig. 5b, the few molecules of poly dC were easily visualized as a distinct species in the poly dA sample. More significantly, the nanopore assay enables the detection of variable or randomly located chemical damage as well as other important attributes of a nucleic acid sample, such as phosphorylation or contamination, in one single assay without any chemical or enzymatic manipulation. Such assessments will be of increasing importance in many developing areas of biotechnology, such as the preparation of synthetic genes.
Although the empirical results presented here yield valuable information on the phosphorylation state, length diversity, and chemical integrity of the DNA sample, further investigations are needed to explain the physical basis for the different current blockage levels and translocation rates that characterize the observed event clusters. For now, many of our results remain as a series of reproducible empirical observations whose apparent inconsistencies await explanation even as they inform us about the probant molecules. For example, the difference in clustering behavior between phosphorylated heteropolymers and phosphorylated homopolymers‡ is perplexing yet informs us about the phosphorylation state of the molecules. Better molecular explanations and models for the observed electrical signals may yield new information about DNA's electrical properties (13) and structural dynamics. This knowledge will expand the potential for additional laboratory applications of single-molecule electrophoresis in nanopores.
Acknowledgments
We thank the Harvard Nanopore Group, in particular, Eric Brandin and Jeremy Katz; Drs. A. Kavcic, D. Stein, and A. Meller for insight; and Drs. J. Sampson, J. Myerson, D. Deamer, and M. Akeson for valuable feedback on the manuscript. This work was supported by Agilent Technologies, the U.S. Department of the Air Force, and National Institutes of Health Grant RO1 HG02338.
Abbreviation: DEPC, diethylpyrocarbonate.
Footnotes
Although 5′ phosphorylation of the dA or dC homopolymers increased upper cluster density, 3′ phosphorylation of these homopolymers produced only a single well formed cluster (data not shown). Although this single cluster may represent DNA that translocated 3′ end first, the position of the cluster on the current axis was unexpectedly shifted to fall midway between the two clusters usually observed with unphosphorylated homopolymers. This single-cluster behavior was hard to understand, particularly because homopolymers with both 3′ and 5′ ends phosphorylated had the same two-cluster translocation profile as samples in which neither polymer end was phosphorylated.
An upward curved pattern is observed on the scatter plot because molecules with shorter translocation durations also consistently exhibited proportionally less ion current blockage than molecules with longer translocation durations.
References
- 1.Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H. & Gouaux, J. E. (1996) Science 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
- 2.Howorka, S., Cheley, S. & Bayley, H. (2001) Nat. Biotechnol. 19, 636–639. [DOI] [PubMed] [Google Scholar]
- 3.Slater, G. W., Guillouzic, S., Gauthier, M. G., Mercier, J.-F., Kenward, M., McCormick, L. C. & Tessier, F. (2002) Electrophoresis 23, 3791–3816. [DOI] [PubMed] [Google Scholar]
- 4.Bayley, H. & Cremer, P. S. (2001) Nature 413, 226–230. [DOI] [PubMed] [Google Scholar]
- 5.Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. (1996) Proc. Natl. Acad. Sci. USA 93, 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akeson, M., Branton, D., Kasianowicz, J. J., Brandin, E. & Deamer, D. W. (1999) Biophys. J. 77, 3227–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meller, A., Nivon, L., Brandin, E., Golovchenko, J. & Branton, D. (2000) Proc. Natl. Acad. Sci. USA 97, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adler, A., Grossman, L. & Fasman, G. D. (1967) Proc. Nat. Acad. Sci. 57, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiner, S. J., Kollman, P. A., Case, D. A., Singh, U. C., Ghio, C., Alagona, G., Profeta, S. & Weiner, P. (1984) J. Am. Chem. Soc. 106, 765–784. [Google Scholar]
- 10.Olmsted, M. C., Anderson, C. F. & Record, M. T., Jr. (1989) Proc. Natl. Acad. Sci. USA 86, 7766–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Houten, V., Denkers, F., van Dijk, M., van den Brekel, M. & Brakenhoff, R. (1998) Anal. Biochem. 265, 386–389. [DOI] [PubMed] [Google Scholar]
- 12.Wang, H., Di Gate, R. J. & Seeman, N. C. (1998) in Structure, Motion, Interaction, and Expression of Biological Macromolecules, eds. Sarma, R. H. & Sarma, M. H. (Adenine, Schenectady, NY), pp. 103–116.
- 13.Porschke, D. (1997) Biophys. Chem. 66, 241–257. [DOI] [PubMed] [Google Scholar]