Abstract
Over the past three decades, massive bleaching events of zooxanthellate corals have been documented across the range of global distribution. Although the phenomenon is correlated with relatively small increases in sea-surface temperature and enhanced light intensity, the underlying physiological mechanism remains unknown. In this article we demonstrate that thylakoid membrane lipid composition is a key determinate of thermal-stress sensitivity in symbiotic algae of cnidarians. Analyses of thylakoid membranes reveal that the critical threshold temperature separating thermally tolerant from sensitive species of zooxanthellae is determined by the saturation of the lipids. The lipid composition is potentially diagnostic of the differential nature of thermally induced bleaching found in scleractinian corals. Measurements of variable chlorophyll fluorescence kinetic transients indicate that thermally damaged membranes are energetically uncoupled but remain capable of splitting water. Consequently, a fraction of the photosynthetically produced oxygen is reduced by photosystem I through the Mehler reaction to form reactive oxygen species, which rapidly accumulate at high irradiance levels and trigger death and expulsion of the endosymbiotic algae. Differential sensitivity to thermal stress among the various species of Symbiodinium seems to be distributed across all clades. A clocked molecular phylogenetic analysis suggests that the evolutionary history of symbiotic algae in cnidarians selected for a reduced tolerance to elevated temperatures in the latter portion of the Cenozoic.
Coral bleaching on a global scale is a growing concern because of both the reduction in essential ecological services provided by zooxanthellate corals within reef communities (1, 2) and the potentially devastating economic impacts accompanying the phenomenon (3). Small, positive deviations in temperature of <2°C can trigger massive losses of symbiotic algae, Symbiodinium spp., from their cnidarian host cells (4). However, not all corals within a reef are equally susceptible to elevated temperature stress (5, 6). Although elevated temperatures often lead to a reduction in the quantum yield of photochemistry, a concomitant increase in the rate of protein turnover in oxygen-generating reaction center, photosystem (PS)II (7–9), and an increase in the production of reactive oxygen species (ROS) (10–12), no mechanism has been elucidated. Here we show that thermal sensitivity in isolated clones of zooxanthellae and in symbiotic animal hosts is correlated with the degree of saturation of the lipids in the thylakoid membranes in the algal plastids. Our results provide a mechanistic basis for understanding and diagnosing coral bleaching patterns in nature.
Materials and Methods
Cultures and Corals. Cultures of Symbiodinium spp., obtained from culture collections or isolated from hosts, were grown in F/2 medium under a 10/14-h light/dark cycle and illuminated with 100 μmol quanta m–2·s–1. Corals were grown at 26°C in 800 liters of aquaria with running artificial seawater (Instant Ocean sea salt, Aquarium Systems, Mentor, OH) as described (13). For thermal-stress experiments, duplicate colonies were transferred to 300 liters of aquaria that were heated to 32°C and maintained at that temperature for 2 months or until the colonies died. Light, at 200 μmol quanta m–2·s–1 on a 12/12-h light/dark cycle was provided by 400-W metal halide bulbs (Iwasaki Electric, Tokyo). Nutrients (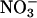 ,
, 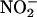 ,
, 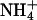 , and
, and 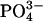 ) were kept at submicromolar concentrations by foam fractioning and biological filtration (e.g., live sand).
) were kept at submicromolar concentrations by foam fractioning and biological filtration (e.g., live sand).
Variable Fluorescence. Variable chlorophyll fluorescence kinetic transients were measured with a custom-built fast repetition-rate fluorometer using protocols described by Kolber et al. (14).
Lipid Analysis. Lipids were saponified, methylated, and extracted into hexane/methyl tertiary butyl ether as described (15). Fatty acid methyl esters were analyzed by GC/MS with an Agilent series 6890 GC system and 5973 mass selective detector, equipped with an HP5MS capillary column (i.d., 30 m × 0.25 mm; film thickness, 0.25 μm) with helium as the carrier gas.
Membrane Inlet MS. Light-dependent production and consumption of oxygen was measured by using a membrane inlet system attached to a Prisma QMS-200 (Pfeiffer, Nashua, NH) quadruple mass spectrometer with closed ion source recording at mass/charge (m/z) ratios of 32 (16O16O), 36 (18O18O), and 40 (Ar). The membrane inlet system was modified from a water-jacketed DW/2 oxygen electrode chamber (Hansatech Instruments, Pentney King's Lynn, U.K.) in which the electrode base plate was replaced by a stainless-steel base plate with a gas port drilled through the center. The standard Teflon membrane (thickness, 12.5 μm) supplied with the DW/2 oxygen electrode system was used. Illumination was provided by a high-pressure halogen arc source at 300 μmol quanta m–2·s–1. Temperature was maintained at 26°C. Oxygen signals were calibrated with O2-saturated water and zero (plus sodium dithionite) O2 water and normalized to Ar. Oxygen production and consumption rates were calculated by linear regression analysis.
ROS. Cultures were harvested by centrifugation and resuspended in culture medium that had been stripped of O2 by bubbling with N2 gas. Subsamples were incubated for 3 h at 150 μmol quanta m–2·s–1 in 96-well plates in the presence of 15 μM dihydrorhodamine 123, a dye that fluoresces green in the presence of ROS (10). Fluorescence (i.e., ROS production) was measured kinetically with a plate reader (Molecular Devices) at excitation λ = 488 nm and emission λ = 525 nm.
Transmission Electron Microscopy. Cells were harvested by centrifugation (15 min at 7,000 × g) and fixed in cacodylate buffer containing 4% glutaraldehyde and 8.6% sucrose. Pellets were washed in a series of cacodylate buffers with descending sucrose concentration and postfixed in OsO4 for 2 h. After dehydration in an ascending ethanol series (70–100%), samples were embedded in agar and Epon, sectioned (50-nm thickness) with a Reichert ultramicrotome, stained with uranyl acetate and lead citrate, and examined with a JEOL 100 CX transmission electron microscope.
Large Subunit rRNA-Encoding DNA (rDNA) Sequencing and Phylogenetic Analyses. Genomic DNA was extracted from zooxanthellae by using the DNeasy plant minikit (Qiagen, Valencia, CA). Standard PCR amplification of nuclear ribosomal DNA was performed by using two sets of primers: (i) S-DINO (cgctcctaccgattgagtga) and l-DIN-1 (aacgatttgcacgtcagtaccgc), which are Symbiodinium-specific and cover the ITS-1/5.8S/ITS-2/partial large subunit (LSU) rDNA, and (ii) D1R (acccgctgaatttaagcatat) and D2C (ccttggtccgtgttt), which are dinoflagellate-specific and target a 5′ fragment of the LSU rDNA. PCR products were purified by using shrimp alkaline phosphatase and exonuclease I and directly sequenced by using an Applied Biosystems 3100-Avant automatic sequencer.
The D1 and D2 sequences of the LSU rDNA were aligned manually to the 294 homologous gene fragments from Symbiodinium spp. available in GenBank. All redundant, identical sequences were removed from the alignment, which resulted in a final DNA matrix containing 84 sequences and 556 nucleotide sites (297 parsimony informative characters). Hierarchical likelihood ratio tests were applied to our data set to select the most appropriate DNA substitution model: a general time-reversible model considering the proportion of invariant sites as well as rate heterogeneity among sites (γ-shaped distribution, γ = 1.2581) (16). Phylogenetic trees were inferred by using Bayesian (1 million MCMC generations, substitution model parameters = GTR+G+I), maximum-likelihood (substitution model parameters = TIM+G+I), and neighbor-joining (substitution model parameters = Tamura and Nei+G) statistics with mrbayes, paup*, and lintree, respectively (17, 18). To give a time dimension to our tree, the 13 consensus, highly resolved clades (thick branches in the tree of Fig. 4) were tested for molecular clock deviation by using relative rate tests (20), with clade A used as an outgroup. None of the LSU rDNA Symbiodinium clades evolve significantly faster than others (threshold risk for 12 clades and 66 tests, P < 0.08%). Consequently, we used lintree to infer a clock-enforced, linearized tree (see Fig. 4), which was calibrated in time by a “dinoflagellate” rate of LSU rDNA substitution based on a previously published DNA–fossil comparative data set (19).
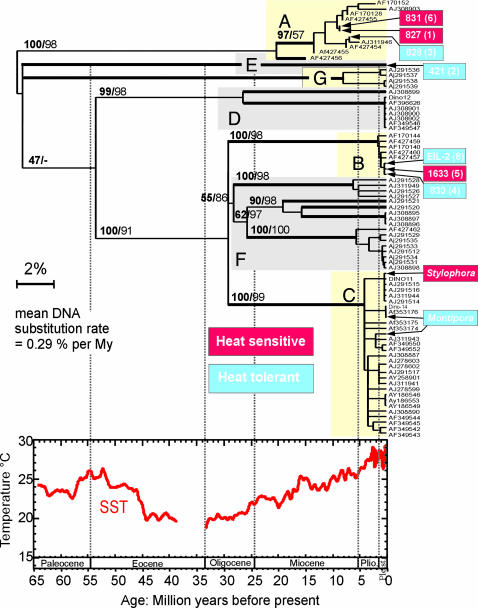
Fig. 4.
LSU rDNA-based evolution of the Symbiodinium species complex (SSC) and phylogenetic position of the zooxanthellae isolates analyzed in Figs. 1, 2, 3. Heat-sensitive and resilient phylotypes are shown in red and blue, respectively. Clades A–G are the seven recognized Symbiodinium phylogenetic groups (35), with A and B (shaded yellow) being typically considered as bleaching-resistant, shallow-water types, and C (shaded pink) as bleaching-sensitive, deeper-living types. Our analysis suggests that at least 13 clades can be recognized based on genetic distances (thick branches in the tree) and that thermal sensitivity is not clade-specific. The ultrametric, linearized tree shown here allowed us to apply a crude clock and calibrate the evolution of the SSC in time. The sea-surface temperature curve, based on tropical planktonic foraminifera δ18O, serves as an approximate time scale for SSC evolution. Note that two to three DNA substitutions in the LSU rDNA correspond to 1 million years of evolution; thus, speciation events in the last 500,000 years may not be detectable by using this genetic marker. Neighbor-joining (1,000 replicates) and Bayesian (1 million generations) statistical values are indicated on the main internal branches.
Results and Discussion
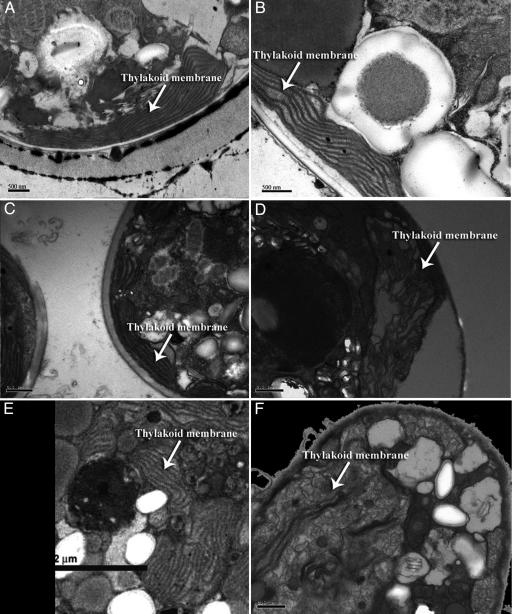
Representative transmission electron micrographs, selected from thousands of zooxanthellae cells, revealed that when thermally tolerant clones of Symbiodinium spp. grown at 26°C were transferred to 32°C (a thermal stress that induces bleaching), the stacking properties and ultrastructural integrity of thylakoid membranes remained unaffected (Fig. 1 A–C and E; Table 1, which is published as supporting information on the PNAS web site). In contrast, thylakoid membranes of thermally sensitive clones subjected to the higher temperature were significantly disrupted, and the organized stacking pattern, which is essential for efficient photochemical energy transduction, was compromised (Fig. 1 D and F). This process is not reversible and was further observed in zooxanthellae in hospite in heat-sensitive corals cultivated in the laboratory before bleaching.
Fig. 1.
Effects of elevated temperatures on the structure of thylakoid membranes in zooxanthellae. Transmission electron micrographs of thin sections of Symbiodinium spp. isolated from Tridacna spp. [Provasoli–Guillard National Center for Culture of Marine Phytoplankton (CCMP) (West Boothbay Harbor, ME) no. 828] (A and B), the sea anemone Aiptasia sp. (CCMP no. 831) (C and D), the coral M. samarensis (E), and the coral S. pistillata (F). Samples were incubated at 26°C(A and C) and 32°C(B and D–F). All cultures were grown in F/2 medium (36) under a 12/12-h light/dark cycle. The corals were grown in a closed system supported by a biological filtration system under a 10/14-h light/dark cycle. Note the degradation of the thylakoid membranes within the plastids of the heat sensitive strains.
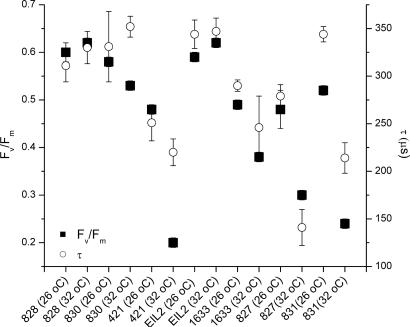
The effect of thermal stress on the photochemical energy-conversion efficiency was confirmed by fast repetition-rate fluorometer measurements (14) on a variety of isolated, cultured clones of zooxanthellae (Fig. 2). Thermally induced changes in membrane integrity were initially accompanied by both an increase in the rate of electron transport on the acceptor side of PSII and a simultaneous decrease in the maximum quantum yield of photochemistry within the reaction center (Table 2, which is published as supporting information on the PNAS web site). In energetically coupled thylakoids, the fastest component of fluorescence decay corresponds to a single electron transfer from the primary electron acceptor, QA, to the secondary quinone, QB or 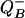 (21), and occurs with a time constant ranging from 300 to 500 μs (22). In temperature-sensitive clones of zooxanthellae, the measured time constant fell from an average of 304 ± 54 to 200 ± 46 μs, whereas in thermally tolerant clones the time constant remained statistically unchanged, averaging 318 ± 24 μsat26°C and 341 ± 9 μsat32°C. The marked change in electron-transfer times in thermally sensitive clones was accompanied by a 40% decrease in (but not loss of) photochemical energy-conversion efficiency in PSII reaction centers. These two phenomena are diagnostic of an energetically uncoupled system in which the transmembrane proton gradient, established by the photochemical reactions in the functional reaction centers, is dissipated without generating ATP (23). This fluorescence kinetic pattern, uniquely found in thermally sensitive zooxanthellae, qualitatively differs from photoinhibition (24–26), with which the time constant for electron transfer increases as the reaction centers become increasingly impaired (27). Moreover, in thermally sensitive clones of zooxanthellae, the pattern of change in photochemical energy conversion occurs over a very narrow thermal window of <2°C. These results not only demonstrate that high-resolution, kinetic measurements of variable chlorophyll fluorescence can be used to rapidly assess the sensitivity of zooxanthellae to thermal stress, but moreover suggest that thylakoid membrane integrity is potentially a critical determinant of thermal tolerance.
(21), and occurs with a time constant ranging from 300 to 500 μs (22). In temperature-sensitive clones of zooxanthellae, the measured time constant fell from an average of 304 ± 54 to 200 ± 46 μs, whereas in thermally tolerant clones the time constant remained statistically unchanged, averaging 318 ± 24 μsat26°C and 341 ± 9 μsat32°C. The marked change in electron-transfer times in thermally sensitive clones was accompanied by a 40% decrease in (but not loss of) photochemical energy-conversion efficiency in PSII reaction centers. These two phenomena are diagnostic of an energetically uncoupled system in which the transmembrane proton gradient, established by the photochemical reactions in the functional reaction centers, is dissipated without generating ATP (23). This fluorescence kinetic pattern, uniquely found in thermally sensitive zooxanthellae, qualitatively differs from photoinhibition (24–26), with which the time constant for electron transfer increases as the reaction centers become increasingly impaired (27). Moreover, in thermally sensitive clones of zooxanthellae, the pattern of change in photochemical energy conversion occurs over a very narrow thermal window of <2°C. These results not only demonstrate that high-resolution, kinetic measurements of variable chlorophyll fluorescence can be used to rapidly assess the sensitivity of zooxanthellae to thermal stress, but moreover suggest that thylakoid membrane integrity is potentially a critical determinant of thermal tolerance.
Fig. 2.
Maximum quantum yields of fluorescence (Fv/Fm, dimensionless) and electron-transfer rates (τ, μs) from the primary electron acceptor in PSII, QA, to the secondary quinone, QB, for all clones of zooxanthellae. Fluorescence parameters were derived from measurements with a custom-built fast repetition-rate fluorometer (14, 24). All cultures were grown in F/2 medium; cultures were incubated for up to 224 h (to verify resilience and nonreversibility of thermally damaged cultures) under a 10/14-h light/dark cycle at 26 and 32°C for each species tested. Maximum quantum yields of photochemistry (Fv/Fm) of the thermally tolerant clones averaged 0.57 ± 0.05 at 26°C and 0.55 ± 0.01 at 32°C; the corresponding electron-transfer rates (τ) were 318 ± 24 and 341 ± 9 μs. In heat-sensitive clones, the maximum quantum yields averaged 0.50 ± 0.07 at 26°C and 0.31 ± 0.03 at 32°C; the corresponding electron-transfer rates were 304 ± 54 and 200 ± 46 μs.
We further examined the patterns of thermal sensitivity and bleaching in colonies of the zooxanthellate corals Stylophora pistillata and Montipora samarensis and the symbiotic anemone Aiptasia sp. cultivated ex situ. S. pistillata and Aiptasia sp. both lost >50% of their symbiotic algae within 72 h after exposure to waters of 32°C. In contrast, M. samarensis retained zooxanthellae at the elevated temperature for >2 months. In the thermally sensitive species, not only was there a change in membrane integrity (e.g., Fig. 1F) and loss of photochemical competence, but production of ROS in isolated zooxanthellae also increased by >2-fold at high irradiance levels. The production of ROS corresponded to a light-dependent increase in O2 consumption as measured by membrane inlet MS using 10% 18O18O as a tracer (data not shown) (28). These results strongly suggest that the production of ROS is caused by the Mehler reaction, i.e., the photochemical reduction of O2 in photosystem I (29). Moreover, the dye-tracer measurements clearly indicate that ROS produced in the algae leaks out of the cells. If this phenomenon happens in hospite, ROS would be transferred directly to the animal host, inducing a physiological stress (12).
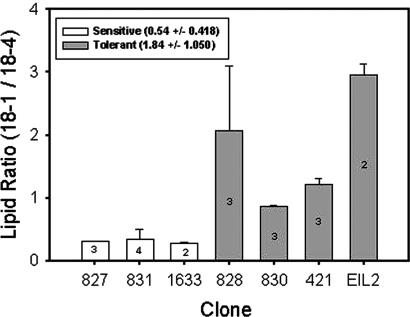
GC/MS analysis of seven zooxanthellae isolates revealed a striking contrast in the relative composition of lipids associated with thylakoid membranes between thermally sensitive and resilient clones (Table 3, which is published as supporting information on the PNAS web site). Specifically, thermally tolerant, cultured Symbiodinium clones and zooxanthellae freshly isolated from corals that did not bleach after experimental thermal stress (Table 1) have a markedly lower content of the major polyunsaturated fatty acid, Δ6,9,12,15-cis-octadecatetraenoic acid (18:4), in relation to Δ9-cis-octadecatetraenoic (18:1) acid, independent of the experimental temperature (Fig. 3). The differences in this lipid profile are statistically significant at the 0.001 level (ANOVA). The higher relative concentration of the saturated polyunsaturated fatty acid enhances thermal stability in eukaryotic thylakoid membranes (30) and simultaneously reduces the susceptibility of the membrane lipids to attack by ROS (31–33). These experimental results strongly suggest that the wide variety of Symbiodinium spp. we analyzed have a limited ability to acclimate physiologically to changes in temperature by significantly modifying their thylakoid lipid composition and hence, unlike most eukaryotic algae, are confined to relatively narrow thermal regimes. The absence of qualitative differences in thylakoid lipid composition between the heat-sensitive and tolerant species suggests that differential susceptibility to elevated temperature results from changes in lipid biosynthetic pathways not associated with lipid desaturases per se but rather with regulatory elements of the enzyme(s) that controls the relative amount of desaturation in specific pools of fatty acids.
Fig. 3.
Ratios of Δ9-cis-octadecatetraenoic (18:1) acid to Δ6,9,12,15-cis-octadecatetraenoic acid (18:4) for seven clones of Symbiodinium spp. ANOVA of the log-transformed data indicates a statistically significant difference between heat-sensitive and heat-tolerant clones.
Phylogenetic analyses of the zooxanthellae isolates used in this study clearly show that thermal tolerance is not associated with a single, monophyletic clade. Heat-sensitive Symbiodinium spp. are found in totally different subdivisions of the LSU rDNA-based tree (Fig. 4 A–C and E), in which thermally tolerant phylotypes systematically branch as closely related sister species. This evolutionary pattern suggests that the reduced physiological ability to acclimate to elevated temperatures by enhancing thylakoid lipid-saturation levels was either acquired in the common ancestor of all modern Symbiodinium clades and was subsequently lost independently in individual taxa within each clade or was selected multiple times in independent lineages belonging to different clades.
The application of a molecular clock to the Symbiodinium spp. phylogenetic tree suggests that the ancestor of the species complex appeared at the Cretaceous–Tertiary boundary, which corresponds to a major transition time from the extinct Mesozoic, rudist-based, reefs to the modern scleractinian-dominated reefs. Juxtaposition of the clocked Symbiodinium spp. phylogenetic tree with a sea-surface temperature curve derived from oxygen isotope analysis of tropical planktonic foraminifera for the last 65 million years (34) suggests that for the first several million years in the Cenozoic Era, zoox-anthellate-based symbioses evolved in warm tropical waters. We hypothesize that extensive cooling periods, starting in the Eocene, selected for cold-tolerant, heat-sensitive, Symbiodinium species, which may have been subject to negative selection (bleaching) later in the Pleistocene and even more strongly in the contemporary Anthropocene period.
Our combined physiological, biochemical, and molecular data confirm that the widely accepted but rather arbitrarily defined Symbiodinium taxonomic “clades” (35), often referred to as genetic or functional units, are in fact multimillion-year-old groups containing a broad diversity of modern species that are differentiated physiologically. Phylotypes belonging to different “clades” can present similar patterns of sensitivity to elevated temperatures but differ from their closely related sister phylotypes. This analysis clearly indicates that a priori rDNA genotyping is not diagnostic of thermal sensitivity in zooxanthellate symbiotic associations.
Our results suggest that the physiological basis of bleaching is initiated when thylakoid membrane integrity is compromised at elevated temperatures, leading to an uncoupling of photosynthetic energy transduction. The accompanying proton leak and loss of ATP restricts photosynthetic carbon assimilation; however, O2 generated by PSII can react with the photochemically generated electrons in PSI to form ROS, which in turn oxidizes membrane lipids. The oxidized lipids initiate a positive feedback of ROS production that is accelerated by high light. Ultimately the ROS kills the intracellular algal symbionts and damages the host cells. The symbiotic algae literally are bleached and/or expelled from their hosts. These results provide an experimental demonstration of a biochemical adaptation associated with thermal tolerance in zooxanthellae and suggest that lipid analysis could potentially provide a rapid, sensitive tool for diagnosing the susceptibility of corals to thermally induced bleaching.
Supplementary Material
Acknowledgments
We thank Sam Jones and the Osborn Laboratories at the New York Aquarium for coral cultivation; Jim Wright for the oxygen isotope data; Thomas Haines, Xavier Pochon, Uwe Johns, and Kevin Wyman for discussions; and Judith Grassle and two anonymous reviewers for comments. This research was supported by the Strategic Environmental Research and Development Program and the National Science Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PS, photosystem; ROS, reactive oxygen species; LSU, large subunit; rDNA, rRNA-encoding DNA.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY684261–AY684270).
References
- 1.Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., Grosberg, R., Hoegh-Guldberg, O., Jackson, J. B., Kleypas, J., et al. (2003) Science 301, 929–933. [DOI] [PubMed] [Google Scholar]
- 2.Ostrander, G. K., Armstrong, K. M., Knobbe, E. T., Gerace, D. & Scully, E. P. (2000) Proc. Natl. Acad. Sci. USA 97, 5297–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson, C. (2000) Status of Coral Reefs of the World: 2000 (Global Coral Reef Monitoring Network and Australian Institute of Marine Science, Townsville, Australia).
- 4.Podesta, G. P. & Glynn, P. W. (2001) Bull. Mar. Sci. 69, 43–59. [Google Scholar]
- 5.Baker, A. C. (2003) Annu. Rev. Ecol. Evol. Syst. 34, 661–689. [Google Scholar]
- 6.Rowan, R., Knowlton, N., Baker, A. & Jara, J. (1997) Nature 388, 265–269. [DOI] [PubMed] [Google Scholar]
- 7.Jones, R. J., Hoegh-Guldberg, O., Larkum, A. W. D. & Schreiber, U. (1998) Plant Cell Environ. 21, 1219–1230. [Google Scholar]
- 8.Warner, M. E., Fitt, W. K. & Schmidt, G. W. (1999) Proc. Natl. Acad. Sci. USA 96, 8007–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner, M. E., Chilcoat, G. C., McFarland, F. K. & Fitt, W. K. (2002) Mar. Biol. (Berlin) 141, 31–38. [Google Scholar]
- 10.Lesser, M. P. (1996) Limnol. Oceanogr. 41, 271–283. [Google Scholar]
- 11.Shick, J. M., Lesser, M. P., Dunlap, W. C., Stochaj, W. R., Chalker, B. E. & Won, J. W. (1995) Mar. Biol. (Berlin) 122, 41–51. [Google Scholar]
- 12.Downs, C. A., Fauth, J. E., Halas, J. C., Dustan, P., Bemiss, J. & Woodley, C. M. (2002) Free Radical Biol. Med. 33, 533–543. [DOI] [PubMed] [Google Scholar]
- 13.Rinkevich, B. & Shafir, S. (1998) Aquat. Sci. Conserv. 2, 237–250. [Google Scholar]
- 14.Kolber, Z. S., Prasil, O. & Falkowski, P. G. (1998) Biochim. Biophys. Acta 1367, 88–106. [DOI] [PubMed] [Google Scholar]
- 15.Ruess, L., Häggblom, M., Garcia-Zapara, E. & Dighton, J. (2002) Soil Biol. Biochem. 34, 745–756. [Google Scholar]
- 16.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 17.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 18.Takezaki, N., Rzhetsky, A. & Nei, M. (1995) Mol. Biol. Evol. 12, 823–833. [DOI] [PubMed] [Google Scholar]
- 19.John, U., Fensome, R. A. & Medlin, L. K. (2003) Mol. Biol. Evol. 20, 1015–1027. [DOI] [PubMed] [Google Scholar]
- 20.Robinson-Rechavi, M. & Huchon, D. (2000) Bioinformatics 16, 296–297. [DOI] [PubMed] [Google Scholar]
- 21.Crofts, A. R. & Wraight, C. A. (1983) Biochim. Biophys. Acta 726, 149–185. [Google Scholar]
- 22.Falkowski, P. G., Wyman, K., Ley, A. C. & Mauzerall, D. C. (1986) Biochim. Biophys. Acta 849, 183–192. [Google Scholar]
- 23.Finazzi, G., Ehrenheim, A. M. & Forti, G. (1993) Biochim. Biophys. Acta 1142, 123–128. [Google Scholar]
- 24.Gorbunov, M. Y., Kolber, Z. S., Lesser, M. P. & Falkowski, P. G. (2001) Limnol. Oceanogr. 46, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown, B. E., Dunne, R. P., Warner, M. E., Ambarsari, I., Fitt, W. K., Gibb, S. W. & Cummings, D. G. (2000) Mar. Ecol. Prog. Ser. 195, 117–124. [Google Scholar]
- 26.Hawkridge, J. M., Pipe, R. K. & Brown, B. E. (2000) Mar. Biol. 137, 1–9. [Google Scholar]
- 27.Long, S. P., Humphries, S. & Falkowski, P. G. (1994) Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 633–662. [Google Scholar]
- 28.Helman, Y., Tchernov, D., Reinhold, L., Shibata, M., Ogawa, T., Schwarz, R., Ohad, I. & Kaplan, A. (2003) Curr. Biol. 13, 230–235. [DOI] [PubMed] [Google Scholar]
- 29.Falkowski, P. G. & Raven, J. A. (1997) Aquatic Photosynthesis (Blackwell, Oxford).
- 30.Hazel, J. R. (1995) Annu. Rev. Physiol. 57, 19–42. [DOI] [PubMed] [Google Scholar]
- 31.Murakami, Y., Tsuyama, M., Kobayashi, Y., Kodama, H. & Iba, K. (2000) Science 287, 476–479. [DOI] [PubMed] [Google Scholar]
- 32.Gombos, Z., Wada, H., Hideg, E. & Murata, N. (1994) Plant Physiol. 104, 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato, N., Sonoike, K., Kawaguchi, A. & Tsuzuki, M. (1996) J. Photochem. Photobiol. 36, 333–337. [Google Scholar]
- 34.Wright, J. D. (2001) Nature 411, 142–143. [DOI] [PubMed] [Google Scholar]
- 35.Pochon X., Pawlowski, J., Zaninetti L. & Rowan, R. (2001) Mar. Biol. 139, 1069–1078. [Google Scholar]
- 36.Guillard, R. R. L. & Ryther, J. H. (1962) Can. J. Microbiol. 8, 437–445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.