Abstract
Catecholaminergic neurons control diverse cognitive, motor, and endocrine functions and are associated with multiple psychiatric and neurodegenerative disorders. We present global gene-expression profiles that define the four major classes of dopaminergic (DA) and noradrenergic neurons in the brain. Hypothalamic DA neurons and noradrenergic neurons in the locus coeruleus display distinct group-specific signatures of transporters, channels, transcription, plasticity, axon-guidance, and survival factors. In contrast, the transcriptomes of midbrain DA neurons of the substantia nigra and the ventral tegmental area are closely related with <1% of differentially expressed genes. Transcripts implicated in neural plasticity and survival are enriched in ventral tegmental area neurons, consistent with their role in schizophrenia and addiction and their decreased vulnerability in Parkinson's disease. The molecular profiles presented provide a basis for understanding the common and population-specific properties of catecholaminergic neurons and will facilitate the development of selective drugs.
Catecholaminergic (CA) neurons producing the neurotransmitters dopamine (DA) and noradrenalin (NA) are organized in anatomically discrete groups and constitute ≈1 of 107 cells in the vertebrate CNS (1, 2). The most prominent groups of DA neurons reside in the substantia nigra (SN; A9 cell group; ≈10,000 neurons in the rat) and the ventral tegmental area (VTA; A10; ≈25,000 neurons) of the midbrain. The SN neurons provide the nigrostriatal ascending inputs to the telencephalon and comprise a key component of the extrapyramidal motor system controlling postural reflexes and initiation of movement. DA neurons in the adjacent VTA give rise to mesocortical and mesolimbic pathways that are implicated in control of emotional balance, reward-associated behavior, attention, and memory. Additional groups of DA neurons reside in the medial zona incerta of the hypothalamus (A13; ≈900 neurons) and participate in the regulation of endocrine functions. The largest collection of NA neurons resides in the pontine locus coeruleus (LC; ≈1,500 neurons). These neurons form a modulatory projection system to most CNS areas and contribute to the regulation of emotional status, sensory perception, arousal, sleep–wake patterns, and most autonomic functions.
Consistent with their varied functions, the CA neurons are associated with multiple neurodegenerative, psychiatric, and endocrine disorders. Selective degeneration of DA neurons in the SN but not in the VTA or the hypothalamus leads to Parkinson's disease (PD) (3–7), whereas abnormal function of the VTA DA neurons has been linked to schizophrenia, attention deficit, addiction, and hyperactivity disorders (8–11). In addition, dysfunction of hypothalamic DA neurons can cause hyperprolactinemia, an endocrine disorder of the reproductive system (12), whereas changes in the activity of the NA system have been linked to depression as well as sleep disorders (13, 14).
The drugs currently available for therapy reflect the critical role of CA systems in disease states. Compounds substituting for the diminished levels of DA in SN neurons are palliative for PD (15–17), whereas drugs blocking the function of VTA neurons are the mainstay therapy for schizophrenia (18). DA drugs are also used to control hyperprolactinemia, whereas antidepressant drugs (13) and also drugs of abuse, such as amphetamines and cocaine, act on transporter systems of CA neurons (8, 9). Because these drugs do not distinguish between classes of CA neurons, the desired effects in one condition become the adverse effects in another disease. For example, PD-like extrapyramidal motor disturbances are common in schizophrenia drug therapy, whereas hallucinations and paranoia are common side effects of DA drug therapy for PD and hyperprolactinemia. Here, we report the genome-wide expression profiles of four major subpopulations of adult CA neurons. This study provides a foundation for mechanistic understanding of the development, stereotypic positions, control of innervation targets, function, and disease vulnerability of these neuronal classes and for the identification of selective drug targets.
Materials and Methods
Tissue Preparation and Immunohistochemistry. Adult (7- to 9-month-old) female Sprague–Dawley rats were anesthetized and killed by decapitation. The brains were rapidly dissected and immediately frozen on dry ice. We mounted 12-μm cryosections on polyethylene-naphthalene membrane slides pretreated with 0.1% poly-l-lysine for 5 min, followed by 30 min of UV irradiation. The sections were fixed immediately in 100% ethanol for 30 s, followed by acetone for 3 s, and air-dried. After rehydration in PBS (pH 7.0) for 5 s, the sections were stained for 2 min in PBS (pH 7.0) containing 100 μg/ml anti-tyrosine hydroxylase (TH) Ab (clone TH-16; Sigma) that had been labeled with the Alexa Fluor 488 mAb-labeling kit (Molecular Probes) according to the manufacturer's instructions. Rehydration and staining were performed in the presence of 1 unit/μl RNase inhibitor (Roche). The sections were washed twice in PBS for 5 s; dehydrated for 30 s in 75%, 95%, and 100% ethanol, respectively; and air-dried at room temperature.
Laser Microdissection, RNA Isolation, and Amplification. Single neurons were isolated from immunostained cryosections by using a PALM Robot–Microbeam system (PALM Microlaser Technology, Bernried, Germany). To facilitate detection of fluorescent neurons, a drop of 100% ethanol was applied to the section during cell selection. The sections were allowed to air dry, and neurons were dissected and catapulted into 30 μl of lysis buffer. Total RNA from 200 pooled neurons was isolated by using the PicoPure kit (Arcturus, Mountain View, CA), and contaminating genomic DNA was removed during the isolation by an on-column DNase digestion step. The common reference RNA was generated from three whole brains of age-matched female rats. RNA was isolated by using RNA-Bee (Tel-Test, Friendswood, TX), followed by DNase digestion with the DNAfree kit (Ambion, Austin, TX). The RNA from dissected neurons and a common reference was amplified by two rounds of T7-based linear amplification (19) using the RiboAmp kit (Arcturus) with the following modifications: to minimize generation of template-independent amplification product from the T7 primer, a 1:5 dilution of primer A was used for the first round of cDNA synthesis, and the reaction volume was scaled down by 50%. The yield and size distribution of the amplified RNA product were evaluated by microfluidic gel electrophoresis with a bioanalyzer (Agilent, Palo Alto, CA).
RNA Labeling, Microarray Hybridization, and Data Analysis. Detailed protocols for probe synthesis and DNA microarray hybridization are available at http://cmgm.stanford.edu/pbrown/protocols/index.html. In short, 2 μg of amplified RNA was random primed to generate single-stranded aminoallyl-dUTP cDNA targets, which were subsequently coupled with either Cy3 (whole-brain reference) or Cy5 (experimental sample). Images were analyzed by using agilent feature extraction software (version A.6.1.1). Processing included local background subtraction and a rank consistency-based probe selection for lowess normalization. The data were filtered with respect to signal significance. A two-tailed t test was used to determine significance of the signal versus background, and spots with a P value of >0.01 in the red or green channel were omitted. Data were log2-transformed and analyzed by using cluster and treeview (20). Statistical analysis was performed by using various functions of the significance analysis of microarrays algorithm sam (21), with a false-discovery rate set at <1%. Only genes for which information was available for >80% of arrays were included in the analysis. Four independent experiments were conducted for every cell type, and in each experiment, cells were isolated from different animals and RNA extraction, amplification, labeling, and hybridization were carried out separately. The mean correlation coefficient of the expression ratios (log2-isolated neurons/whole brain) between replicates was 0.86, and values ranged from 0.81 to 0.93.
In Situ Hybridization. Probes were amplified from rat brain RNA by nested RT-PCR with T3 promoter sequences attached to the 5′ end of the 3′ nested primers. The sequence-confirmed PCR products were used as templates for synthesis of digoxigenin-labeled RNA probes. Coronal cryosections (20 μm) of adult rat brains were air-dried for 30 min and fixed in 4% paraformaldehyde for 15 min. The sections were bleached in 6% H2O2 for 10 min, digested with 1 μg/ml proteinase K in PBS for 5 min, and refixed in 4% paraformaldehyde, followed by a 10-min acetylation step in 0.25% acetic anhydride/100 mM Tris, pH 7.5, and two washes in 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 5). The sections were prehybridized in hybridization buffer [5× SSC, pH 5/1% blocking reagent (Roche)/50% formamide/5 mM EDTA/0.1% Tween 20/10% dextrane sulfate/100 μg/ml salmon sperm DNA/100 μg/ml tRNA/100 μg/ml heparin] for 1 h at 65°C and hybridized for 20 h at 65°C in 100 μl of hybridization buffer containing 1 μg/ml digoxigenin-labeled probe. The slides were washed at 60°C two times for 10 min in 5× SSC/50% formamide, two times for 15 min in 1× SSC, and for 30 min in 0.2× SSC, followed by digestion with 10 μg/μl RNase for 15 min at 37°C. Digoxigenin epitopes were detected with alkaline phosphatase-coupled antidigoxigenin Fab fragments (Roche) and developed with BM purple (Roche) for 24–48 h.
Results
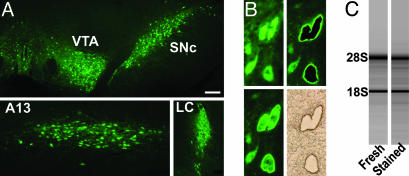
Profiling Individual Classes of CA Neurons. We have determined the gene-expression profiles of DA neurons in the SN, VTA, and dorsal hypothalamus, and of NA neurons in the LC of normal adult rats. The neurons were identified in cryosections of the respective brain regions by a rapid one-step immunostaining of TH, which is the rate-limiting enzyme for DA and NA biosynthesis (Fig. 1A). From each class, 200 neurons were isolated by laser microdissection (Fig. 1B), and the RNA integrity of the tissue section was confirmed by quantification of the 18S and 28S ribosomal RNAs (Fig. 1C). The RNA of microdissected neurons was subjected to two rounds of T7-based linear amplification and labeling (19) and was hybridized to 14,800 element rat cDNA microarrays. A common reference was generated by pooling and amplifying RNA from three whole brains of age-matched rats and used for competitive hybridization.
Fig. 1.
Isolation of intact RNA from CA neurons. (A) Identification of CA neurons in the SN, VTA, zona incerta, and LC in coronal brain sections of the rat by rapid TH immunostaining. (B) Laser microdissection of individual neurons in the SN. (C) Preserved integrity of RNA in immunostained brain sections. Scale bar indicates 250 μm(A Upper and Lower Right), 75 μm(A Lower Left), and 15 μm(B).
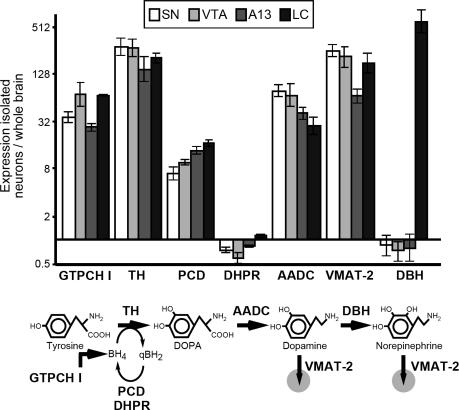
To assess the validity of our approach, we first focused on the expression patterns of known key enzymes involved in DA and NA biosynthesis and vesicular transport (Fig. 2). TH was the most highly enriched transcript in all three examined DA cell groups (>150-fold compared with the whole-brain reference) and the second most highly enriched in the NA neurons. In contrast, DA-β-hydroxylase (DBH), which catalyzes the conversion of DA to NA, was exclusively enriched in LC neurons (>500-fold over reference). Additional genes that were expressed at high levels in all CA populations included the CA-synthesis enzymes aromatic amino acid decarboxylase, GTP cyclohydrolase I, and pterin-4-α-carbinolamine dehydratase, and the vesicular monoamine transporter 2, which mediates the transport of monoamine neurotransmitters into synaptic vesicles. As expected, the ubiquitously expressed dihydropteridine reductase did not show significant enrichment in any of the cell populations.
Fig. 2.
Expression of CA biosynthetic enzymes and transporters in DA and NA cell groups. The ratios of expression in the respective cell group compared with the whole-brain reference are shown. Error bars indicate 95% confidence interval. AADC, aromatic amino acid decarboxylase; DHPR, dihydropteridine reductase; GTPCH I, GTP cyclohydrolase I; PCD, pterin-4-α-carbinolamine dehydratase; VMAT-2, vesicular monoamine transporter 2.
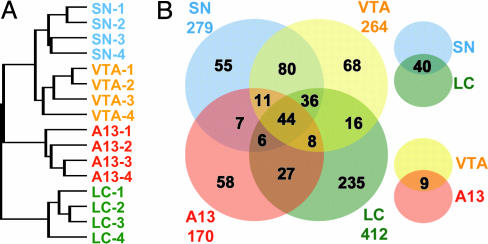
Lineage Relationships Between CA Neuron Subclasses. Having validated our approach with known genes, we next sought to establish the lineage relationships between the different classes of CA neurons as revealed by overlapping and distinct patterns of gene expression. To this end, we used unsupervised hierarchical clustering (20) to group the four CA cell classes based on all of the genes represented on the array. Independent biological replicates representing a given cell group clustered together, indicating the existence of specific transcriptomes in each subgroup of CA neurons. The phylogenetic relationship between the individual cell classes is shown in a dendrogram (Fig. 3A). The SN and VTA DA neurons displayed highly similar signatures of gene expression, suggesting that these anatomically adjacent cell groups are closely related at the molecular level. In contrast, the profile of incertohypothalamic DA neurons was related only distantly to those of the SN and VTA neurons, despite the fact that all three groups of neurons use the same transmitter. In fact, hypothalamic A13 DA neurons are not positioned markedly closer to midbrain DA neurons in the phylogenetic tree than the NA neurons are.
Fig. 3.
Relationship among classes of CA neurons. (A) Dendrogram showing the relationship of the gene-expression profiles of all experiments. The tree was generated by unsupervised hierarchical clustering based on all features that were significant (P < 0.01) and >2.6 SDs over background in >80% of all experiments (13,173 of 14,797 genes). (B) Venn diagram showing shared and distinct expression of genes enriched in CA neurons. Genes with >4-fold expression compared with the whole-brain reference were selected for each group of neurons, with a false-discovery rate of <1%.
To illustrate shared and cell type-specific gene expression in the CA system, we selected transcripts with at least 4-fold higher expression compared with whole brain in any one of the four cell groups and organized them in a Venn diagram (Fig. 3B). NA neurons in the LC expressed the highest number of enriched transcripts (412), followed by SN (279), VTA (264), and hypothalamic DA (170) neurons. As expected from the phylogenetic tree, there was a large overlap between SN and VTA (46%; 171/372). In contrast, SN and A13 neurons shared 18% (68/381), SN and LC shared 22% (126/565), and A13 and LC shared 17% (85/497) of their enriched transcripts. Only 44 transcripts were enriched in all four examined CA groups.
In an alternative approach to assess the molecular phylogeny, we determined the percentage of transcripts with differences between any two given cell groups by significance analysis (21). SN and VTA differed in only 122 (<1%) of their expressed genes. In contrast, there were 766 (>5%) differentially expressed transcripts between SN and A13, 1,079 (>7%) differentially expressed transcripts between SN and LC, and 1,453 (>10%) differentially expressed transcripts between LC and A13 neurons. Together, these findings demonstrate that each group of CA neurons displays a unique set of expressed genes, and they support the hypothesis that SN and VTA neurons are closely related by lineage and/or function.
Transcripts Enriched in All CA Neurons. To identify transcripts that define CA cell identity irrespective of subclass, we examined the genes that are enriched in all groups of CA neurons (see Table 1, which is published as supporting information on the PNAS web site). The most prominent class included transcripts that are known to be involved in neurotransmitter synthesis and transport, such as TH, aromatic amino acid decarboxylase, GTP cyclohydrolase I, pterin-4-α-carbinolamine dehydratase, and vesicular monoamine transporter 2. In addition, we identified genes coding for proteins that counteract stress-induced cell damage (glutathione peroxidase, ARC, and ORP150), inflammation-related genes (DAF and MHC class I heavy chain), genes involved in neural-cell adhesion (neural cell adhesion molecule and polysialyltransferase 1), and modulators of DA-receptor activity (calcyon and chloride intracellular channel 3). Of these pan-CA transcripts, 38% were ESTs or coding for hypothetical proteins, indicating that a large portion of the transcriptome of CA neurons is still uncharacterized.
All CA neurons were found to express aldehyde dehydrogenase enzymes; however, they expressed different members of this protein family (Fig. 4). ALDH1A3 was expressed in the hypothalamus, whereas ALDH1A1 was expressed in the SN/VTA. Both enzymes can convert retinaldehyde to retinoic acid. Signaling of retinoic acid was shown to be involved in many developmental processes, including the specification of motorneuron subclasses (22). LC neurons express ALDH3A1, which is not capable of synthesizing retinoic acid but may be involved in neurotransmitter metabolism and detoxification.
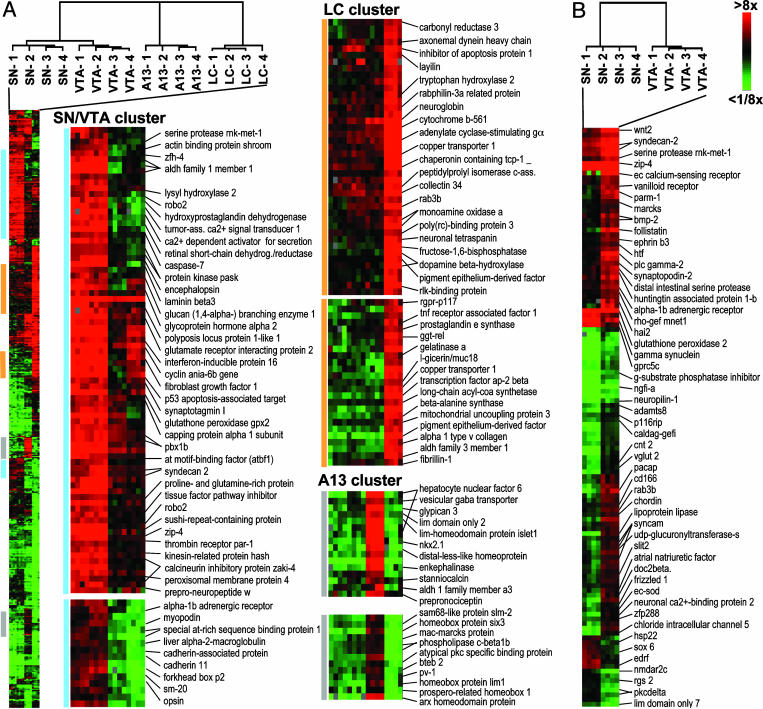
Fig. 4.
Supervised cluster analysis of genes with differential expression between CA cell groups. (A) Genes were filtered based on their expression level relative to the whole-brain reference (>4-fold higher or lower in at least 3 of 16 experiments), and transcripts with significant differences in expression between at least two cell groups were selected by multiclass sam, with a false-discovery rate of <1%. The resulting set of 534 genes and the experimental samples were grouped based on their similarities of gene expression by two-dimensional hierarchical clustering (Pearson correlation, average linkage). Selected gene clusters with preferential expression in SN/VTA, LC, or A13 cell groups are shown, and known annotated genes are listed. The complete list of genes is provided in Table 2, which is published as supporting information on the PNAS web site. (B) Differential gene expression between SN and VTA neurons. Genes with significant differences in expression were identified by sam (false-discovery rate of <1%) and sorted by hierarchical clustering. Selected genes are annotated. The complete list of genes is provided in Table 3, which is published as supporting information on the PNAS web site.
A Common Signature of Midbrain DA Neurons. Having identified a pan-CA signature, we further sought to identify genes that are differentially expressed in subsets of CA neurons. Gene filtering and multiclass significance analysis (21) revealed a set of 534 genes that differ in expression between at least two groups of CA neurons. Genes and experimental samples were grouped by supervised two-dimensional hierarchical clustering (Fig. 4A). This analysis again demonstrated the high degree of similarity between SN and VTA DA neurons, as well as the distant relationship of the SN/VTA with both A13 DA and LC neurons.
The cluster of genes with enriched expression in SN and VTA neurons contained a large number of transcriptional regulators (ZFH-4, ATBF1, PBX1, FOXP2, IFI-16, and SATB1) and regulators of synaptic signaling and/or plasticity [the calcineurin inhibitor ZAKI-4, synaptotagmin I, the kinesin-related protein Hash, the calcium-activated protein for secretion, and the glutamate receptor-interacting protein 2, which is involved in the synaptic targeting of (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, AMPA) receptors]. Three apoptosis-related transcripts, caspase 7, SM-20, and PERP (p53-effector related to pmp-22), were also highly enriched in the midbrain DA cluster. SM-20 is a mitochondrial protein that promotes caspase-dependent cell death in neurons, whereas PERP is a positive effector of p53-induced neuronal apoptosis. Aldehyde dehydrogenase 1, which is known to be highly and specifically expressed in the SN and VTA, served as a validating marker for this gene cluster.
A Complex NA Signature in the LC. As expected from the Venn diagram, the LC cluster contained the largest collection of cell group-specific transcripts (Fig. 4A). Marker genes for this cluster included DBH, monoamine oxidase A, and cytochrome b561, which is a major transmembrane protein of CA secretory vesicles that provides reducing equivalents for the DBH reaction. AP-2β, a member of the AP-2 family of retinoic acid-induced transcription factors was highly enriched in LC neurons. The closely related transcription factor AP-2α, which recognizes the same target sequence and shares a highly conserved DNA-binding and dimerization domain with AP-2β, has been shown to activate the expression of TH and DBH (23) and to be essential for the development of NA LC neurons in zebrafish (24). We also observed high specific expression of potential vulnerability factors, the Cu transporter 1, the γ-glutamyltranspeptidase-related enzyme, and prostaglandin E synthase. Cu is an essential cofactor for various enzymes including Cu/Zn superoxide dismutase, cytochrome oxidase, and DBH. However, excess of Cu combined with glutathione metabolites leads to free-radical damage and possible neuronal dysfunction (25). LC neurons also expressed factors known to protect from oxidative damage and/or apoptosis-like pigment epithelium-derived factor, neuroglobin as well as IAP and TRAF, which can both mediate antiapoptotic TNF signaling.
Transcripts Defining Hypothalamic A13 DA Neurons. The A13 DA neurons were characterized by high and specific expression of multiple transcriptional regulators (Hnf-6, Lmo2, Bteb2 Isl-1, Nkx2.1, Dlx, Six3, Lim1, and Prox1 Arx; Fig. 4A). Six3 has been shown to alter the regional responses to Fgf8 and Shh, which is required for development of the hypothalamus (26). The Arx, Dlx, Isl-1, Lim1, and Nkx2.1 are important regulators of proliferation, migration, and differentiation of neurons in the embryonic forebrain (27). In Dlx1/2 mutants, for example, the A13 DA neurons do not form (28). The functions of Hnf-6, Lmo2, Bteb2, and Prox1 in the A13 DA neurons are currently not known. The fact that expression of multiple developmental regulators is sustained suggests additional functions of these genes in the adult brain.
Differential Gene Expression in SN and VTA Neurons. Even though SN and VTA neurons are located in close proximity, develop in response to the same inductive signals, and have partially overlapping axonal projection domains, they mature to innervate distinct targets, control different brain functions, and display different vulnerabilities to neurodegeneration. We identified 122 genes with differential expression between SN and VTA neurons by two-class significance analysis (Fig. 4B). Among these factors were transcripts from various categories, including transcriptional regulators (Sox-6, Zfp 288, HTF, and NGFI-A), molecules involved in vesicle trafficking [DOC2B, rab3B, and MARCKS (myristoylated alanine-rich C kinase substrate)], axon guidance (neuropilin 1, slit-2, and ephrin B3), ion channels [CLIC5 (chloride intracellular channel 5), VR1 (vanilloid receptor 1), and NMDAR2C], transporters (VGLUT2 and CNT2), and G protein-coupled receptors (α-1B-adrenergic receptor and GPRC5C).
The two most prominent functional classes encoded factors involved in synaptic plasticity and in cell survival and protection. Strikingly, most of these transcripts were expressed at a higher level in the VTA neurons. The factors with a reported or anticipated function in synaptic plasticity included the synaptic-adhesion molecules synCAM and syndecan 2 (29) and the actin-associated synaptopodin 2, which is required for the formation of the dendritic-spine apparatus (30). The MARCKS and G substrate have been implicated in learning and long-term potentiation. Likewise, we found elevated expression of PLCγ2 (phospholipase-Cγ-2), an isoform of PLCγ1 (phospholipase-Cγ-1), which is thought to be involved in the maintenance of long-term potentiation (31) and NGFI-A, an immediate-early gene that is associated with learning and plasticity. Finally, we have identified the serine proteases RNK-Met 1 and DISP, as well as the serine protease inhibitor Hai2, which might contribute to synaptic plasticity by modulation of the extracellular environment.
VTA-enriched transcripts associated with neuroprotective functions included the neuropeptides PACAP (pituitary adenylate cyclase-activating peptide) and ANP (atrial natriuretic peptide), the growth factor BMP-2 (bone morphogenetic protein 2), and PARM-1 (prostatic androgen-repressed message 1). PACAP and BMP-2 are known survival factors for ventral mesencephalic DA neurons that can protect from 6-hydroxydopamine and MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) (32–34). ANP can counteract oxidative stress and excess NO (35, 36), whereas PARM-1 is implicated in suppression of apoptosis (37). The BMP-inducible antagonists follistatin and chordin were also selectively expressed in the VTA, which indicates active BMP-signaling controlled by autoregulatory feedback loops. Enriched expression in VTA over SN neurons was also observed for enzymes with detoxifying properties: extracellular superoxide dismutase, an antioxidant enzyme that attenuates brain and lung injury from oxidative stress (38); lipoprotein lipase, a key enzyme involved in the metabolism of lipoproteins, which protects from cell death induced by oxidized lipoproteins (39); and UDP-glucuronosyltransferase, which detoxifies compounds by conjugation to glucuronic acid. However, expression of PKC-δ, which has been shown to mediate DA cell apoptosis induced by MPTP or pesticides was significantly lower in VTA neurons compared with the SN (40, 41).
Two additional genes that could play an important role in the pathobiology of PD were γ-synuclein and the zinc transporter ZIP4. Interestingly, γ-synuclein transcripts are highly enriched in both SN and LC (28- and 29-fold, respectively), which are vulnerable to PD and is dramatically lower in the less vulnerable VTA (4-fold) and A13 (2-fold) neurons. ZIP4 is a member of the Zn2+ uptake transporter family that has not been described in brain tissues before. We detected highly specific expression of the ZIP4 messenger in the neurons of the VTA (>250-fold) and the SN (>100-fold).
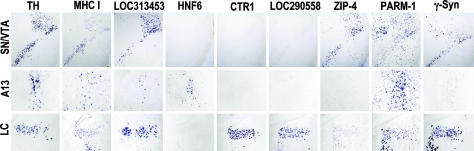
Validation of Cell Group-Specific Gene Expression. To independently validate the microarray-derived molecular signatures of the CA neurons we have analyzed the distribution of 14 transcripts by in situ hybridization (Fig. 5). We found a tight correlation between the expression patterns that were predicted by microarray and the expression patterns determined by in situ hybridization for nine transcripts, whereas in five cases, we were unable to detect signal in situ, possibly because of low mRNA abundance. Consistent with the microarray analysis, the in situ hybridization revealed expression of the MHC class I heavy chain and the hypothetical protein LOC313453 in all four CA populations. In contrast, expression of the transcription factor HNF-6 was confined to the hypothalamic A13 cell group, whereas the Cu transporter Ctr1 and the hypothetical protein LOC290558 were restricted to the LC. Likewise, the zinc transporter ZIP4 was highly expressed in the VTA and SN. The γ-synuclein transcripts were abundant in SN and LC and the PARM-1 message was enriched in the VTA and A13 cell groups. Together, these findings confirm the authenticity of the microarray analysis.
Fig. 5.
Validation of the gene-expression profiles by in situ hybridization. As predicted by microarray analysis, MHC class I heavy chain and the hypothetical protein LOC313453 were detected in all four cell groups, whereas expression of HNF-6 was restricted to the hypothalamus, and Ctr1 and the hypothetical protein LOC290558 were restricted to the LC. ZIP4 expression was strong in SN and VTA, whereas γ-synuclein transcripts were abundant in the SN and LC but low in the VTA and A13 cell groups. The PARM-1 message was abundant in the VTA and A13 cell groups, with lower levels in the SN and LC. Expression of PARM-1 was not confined to CA cell groups, as expected from the low ratios of expression measured by microarray.
Discussion
In this study, we have analyzed the molecular signatures that define the four major subpopulations of CA neurons. We have shown that individual neurons can be identified, isolated from brain tissue, and subjected to microarray analysis. Phylogenetic examination revealed that, despite considerable heterogeneity between the SN and VTA DA system with respect to cell morphology, target innervation, electrophysiological properties, and disease susceptibility, these two neuronal cell types differ by <1% of their transcripts. In contrast, 5% of the transcripts in the hypothalamic DA neurons differed from those of the SN or VTA neurons. DA neurons in the midbrain and hypothalamus each expressed specific sets of transcriptional regulators, suggesting that although both neuronal classes depend on Fgf8 and Shh for their early specification (42), their later phenotype is maintained, in part, by independent regulatory cascades. Such a cascade would include the DA midbrain- and forebrain-specific transcription factors PBX1, ZFH-4, IFI 116, Bteb2, Lmo2, Prox1, and Hnf-6 that have been identified here, as well as the known transcriptional regulators of DA differentiation and survival Nurr1, Lmx1b, and Pitx3.
The complexity of a given gene profile seems to be correlated with diversity of projections and complexity of biological functions. The LC NA system, which innervates much of the CNS and regulates emotional, cognitive, and sleep–wake functions, expressed the highest number of specific genes. In contrast, hypothalamic A13 neurons, which have simpler projections and control mainly endocrine functions, displayed the lowest transcript complexity.
Despite the high similarity of the transcriptomes in SN and VTA neurons, we were able to identify a number of subpopulation-specific genes. Among the gene transcripts enriched in the VTA, several encoded proteins that are implicated in synaptic plasticity. These factors may contribute to the long-term synaptic plasticity elicited by psychostimulants, leading to drug addiction (43). A critical role of PLCγ1 in the regulation of long-term adaptations to drugs has recently been demonstrated by overexpression experiments in the VTA (44). Likewise, the expression of the learning- and plasticity-associated immediate-early gene Zif268 is induced in VTA neurons upon drug-conditioned stimulation and decreases during prolonged withdrawal (45, 46).
VTA neurons were also enriched for several factors that are involved in axonal pathfinding and neuronal migration (neuropilin-1, slit-2, and ephrin B3). During development, SN neurons target mainly the dorsolateral striatum, whereas VTA neurons mainly innervate the ventromedial striatum. The molecular signals that regulate the development of these pathways have been characterized only partially (47). The differential expression of multiple members of the ephrin/Eph and slit/robo families identified here could have important functions in the maintenance and stability of neuronal networks, in adult plasticity and remodeling, and possibly in schizophrenia, a disease that is most likely linked to abnormal development of cortical areas innervated by VTA neurons (48).
One of our goals was to identify genes that may influence the selective vulnerability of CA neurons in PD. The SN is most susceptible to PD pathology, whereas the adjacent VTA DA neurons are less vulnerable and hypothalamic DA neurons are spared (3–6, 49). The same selective vulnerability of DA neuron subpopulations has been observed in rodent and primate models of PD (7, 50–52). The sparing of VTA neurons could be mediated by selective expression of neuroprotective factors, including neurotrophic factors (BMP-2, PACAP, and ANP), detoxifying enzymes (EC-SOD, lipoprotein lipase, and UDP-glucuronosyltransferase), the antiapoptotic factor PARM-1, and decreased levels of the proapoptotic PKC-δ. We also observed selective high expression of γ-synuclein in neurons of the SN and in LC NA that degenerate in PD, which may modify the toxic effects of the widely expressed α-synuclein protein. Likewise, selective expression of the Zn2+ transporter by the SN and VTA may play a role in the pathophysiology of PD. Low concentrations of Zn2+ can exert a cell-protective effect; however, excess of Zn2+ is neurotoxic and has been shown to promote degeneration of midbrain DA neurons (53).
In summary, the molecular signatures of the major classes of CA neurons can advance our understanding of the characteristic features and functions of these neurons and facilitate the discovery of subgroup-selective drug targets.
Supplementary Material
Abbreviations: CA, catecholamine/catecholaminergic; DA, dopamine/dopaminergic; LC, locus coeruleus; NA, noradrenalin/noradrenergic; PD, Parkinson's disease; SN, substantia nigra; VTA, ventral tegmental area; TH, tyrosine hydroxylase; DBH, DA-β-hydroxylase.
References
- 1.Dahlstroem, A. & Fuxe, K. (1964) Acta Physiol. Scand. Suppl. 232, 1–55. [PubMed] [Google Scholar]
- 2.Björklund, A. & Lindvall, O. (1984) in Handbook of Chemical Neuroanatomy, eds. Björklund, A. & Hökfelt, T. (Elsevier, New York), Vol. 2, Part 1, pp. 55–122. [Google Scholar]
- 3.Hirsch, E., Graybiel, A. M. & Agid, Y. A. (1988) Nature 334, 345–348. [DOI] [PubMed] [Google Scholar]
- 4.Uhl, G. R., Hedreen, J. C. & Price, D. L. (1985) Neurology 35, 1215–1218. [DOI] [PubMed] [Google Scholar]
- 5.Purba, J. S., Hofman, M. A. & Swaab, D. F. (1994) Neurology 44, 84–89. [DOI] [PubMed] [Google Scholar]
- 6.Matzuk, M. M. & Saper, C. B. (1985) Ann. Neurol. 18, 552–555. [DOI] [PubMed] [Google Scholar]
- 7.Varastet, M., Riche, D., Maziere, M. & Hantraye, P. (1994) Neuroscience 63, 47–56. [DOI] [PubMed] [Google Scholar]
- 8.Bonci, A., Bernardi, G., Grillner, P. & Mercuri, N. B. (2003) Trends Pharmacol. Sci. 24, 172–177. [DOI] [PubMed] [Google Scholar]
- 9.Viggiano, D., Vallone, D., Ruocco, L. A. & Sadile, A. G. (2003) Neurosci. Biobehav. Rev. 27, 683–689. [DOI] [PubMed] [Google Scholar]
- 10.Goto, Y. & O'Donnell, P. (2002) J. Neurosci. 22, 9070–9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer-Lindenberg, A., Miletich, R. S., Kohn, P. D., Esposito, G., Carson, R. E., Quarantelli, M., Weinberger, D. R. & Berman, K. F. (2002) Nat. Neurosci. 5, 267–271. [DOI] [PubMed] [Google Scholar]
- 12.Mah, P. M. & Webster, J. (2002) Semin. Reprod. Med. 20, 365–374. [DOI] [PubMed] [Google Scholar]
- 13.Nestler, E. J., Barrot, M., DiLeone, R. J., Eisch, A. J., Gold, S. J. & Monteggia, L. M. (2002) Neuron 34, 13–25. [DOI] [PubMed] [Google Scholar]
- 14.Pace-Schott, E. F. & Hobson, J. A. (2002) Nat. Rev. Neurosci. 3, 591–605. [DOI] [PubMed] [Google Scholar]
- 15.Dauer, W. & Przedborski, S. (2003) Neuron 39, 889–909. [DOI] [PubMed] [Google Scholar]
- 16.Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819–822. [DOI] [PubMed] [Google Scholar]
- 17.Fahn, S. (2003) Ann. N.Y. Acad. Sci. 991, 1–14. [DOI] [PubMed] [Google Scholar]
- 18.Bennett, M. R. (1998) J. Psychopharmacol. 12, 289–304. [DOI] [PubMed] [Google Scholar]
- 19.Van Gelder, R. N., von Zastrow, M. E., Yool, A., Dement, W. C., Barchas, J. D. & Eberwine, J. H. (1990) Proc. Natl. Acad. Sci. USA 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sockanathan, S. & Jessell, T. M. (1998) Cell 94, 503–514. [DOI] [PubMed] [Google Scholar]
- 23.Kim, H. S., Hong, S. J., LeDoux, M. S. & Kim, K. S. (2001) J. Neurochem. 76, 280–294. [DOI] [PubMed] [Google Scholar]
- 24.Holzschuh, J., Barrallo-Gimeno, A., Ettl, A. K., Durr, K., Knapik, E. W. & Driever, W. (2003) Development (Cambridge, U.K.) 130, 5741–5754. [DOI] [PubMed] [Google Scholar]
- 25.Enoiu, M., Aberkane, H., Salazar, J. F., Leroy, P., Groffen, J., Siest, G. & Wellman, M. (2000) Free Radical Biol. Med. 29, 825–833. [DOI] [PubMed] [Google Scholar]
- 26.Kimura, S., Hara, Y., Pineau, T., Fernandez-Salguero, P., Fox, C. H., Ward, J. M. & Gonzalez, F. J. (1996) Genes Dev. 10, 60–69. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura, K., Yanazawa, M., Sugiyama, N., Miura, H., Iizuka-Kogo, A., Kusaka, M., Omichi, K., Suzuki, R., Kato-Fukui, Y., Kamiirisa, K., et al. (2002) Nat. Genet. 32, 359–369. [DOI] [PubMed] [Google Scholar]
- 28.Andrews, G. L., Yun, K., Rubenstein, J. L. & Mastick, G. S. (2003) Mol. Cell. Neurosci. 23, 107–120. [DOI] [PubMed] [Google Scholar]
- 29.Yamagata, M., Sanes, J. R. & Weiner, J. A. (2003) Curr. Opin. Cell Biol. 15, 621–632. [DOI] [PubMed] [Google Scholar]
- 30.Deller, T., Korte, M., Chabanis, S., Drakew, A., Schwegler, H., Stefani, G. G., Zuniga, A., Schwarz, K., Bonhoeffer, T., Zeller, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernfors, P. & Bramham, C. R. (2003) Trends Neurosci. 26, 171–173. [DOI] [PubMed] [Google Scholar]
- 32.Espejo, M., Cutillas, B., Ventura, F. & Ambrosio, S. (1999) Neurosci. Lett. 275, 13–16. [DOI] [PubMed] [Google Scholar]
- 33.Reiriz, J., Espejo, M., Ventura, F., Ambrosio, S. & Alberch, J. (1999) J. Neurobiol. 38, 161–170. [PubMed] [Google Scholar]
- 34.Takei, N., Skoglosa, Y. & Lindholm, D. (1998) J. Neurosci. Res. 54, 698–706. [DOI] [PubMed] [Google Scholar]
- 35.Vaudry, D., Pamantung, T. F., Basille, M., Rousselle, C., Fournier, A., Vaudry, H., Beauvillain, J. C. & Gonzalez, B. J. (2002) Eur. J. Neurosci. 15, 1451–1460. [DOI] [PubMed] [Google Scholar]
- 36.Fiscus, R. R. (2002) Neurosignals 11, 175–190. [DOI] [PubMed] [Google Scholar]
- 37.Bruyninx, M., Hennuy, B., Cornet, A., Houssa, P., Daukandt, M., Reiter, E., Poncin, J., Closset, J. & Hennen, G. (1999) Endocrinology 140, 4789–4799. [DOI] [PubMed] [Google Scholar]
- 38.Sheng, H., Kudo, M., Mackensen, G. B., Pearlstein, R. D., Crapo, J. D. & Warner, D. S. (2000) Exp. Neurol. 163, 392–398. [DOI] [PubMed] [Google Scholar]
- 39.Paradis, E., Clement, S., Julien, P. & Ven Murthy, M. R. (2003) J. Biol. Chem. 278, 9698–9705. [DOI] [PubMed] [Google Scholar]
- 40.Kaul, S., Kanthasamy, A., Kitazawa, M., Anantharam, V. & Kanthasamy, A. G. (2003) Eur. J. Neurosci. 18, 1387–1401. [DOI] [PubMed] [Google Scholar]
- 41.Kitazawa, M., Anantharam, V. & Kanthasamy, A. G. (2003) Neuroscience 119, 945–964. [DOI] [PubMed] [Google Scholar]
- 42.Ye, W., Shimamura, K., Rubenstein, J. L., Hynes, M. A. & Rosenthal, A. (1998) Cell 93, 755–766. [DOI] [PubMed] [Google Scholar]
- 43.Gerdeman, G. L., Partridge, J. G., Lupica, C. R. & Lovinger, D. M. (2003) Trends Neurosci. 26, 184–192. [DOI] [PubMed] [Google Scholar]
- 44.Bolanos, C. A., Perrotti, L. I., Edwards, S., Eisch, A. J., Barrot, M., Olson, V. G., Russell, D. S., Neve, R. L. & Nestler, E. J. (2003) J. Neurosci. 23, 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, K. L., Arroyo, M. & Everitt, B. J. (2003) Eur. J. Neurosci. 17, 1964–1972. [DOI] [PubMed] [Google Scholar]
- 46.Mutschler, N. H., Miczek, K. A. & Hammer, R. P., Jr. (2000) Neuroscience 100, 531–538. [DOI] [PubMed] [Google Scholar]
- 47.Yue, Y., Widmer, D. A., Halladay, A. K., Cerretti, D. P., Wagner, G. C., Dreyer, J. L. & Zhou, R. (1999) J. Neurosci. 19, 2090–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis, D. A. & Levitt, P. (2002) Annu. Rev. Neurosci. 25, 409–432. [DOI] [PubMed] [Google Scholar]
- 49.Fearnley, J. M. & Lees, A. J. (1991) Brain 114, 2283–2301. [DOI] [PubMed] [Google Scholar]
- 50.Melamed, E., Rosenthal, J., Globus, M., Cohen, O., Frucht, Y. & Uzzan, A. (1985) Eur. J. Pharmacol. 114, 97–100. [DOI] [PubMed] [Google Scholar]
- 51.Mogi, M., Harada, M., Kojima, K., Kiuchi, K. & Nagatsu, T. (1988) J. Neurochem. 50, 1053–1056. [DOI] [PubMed] [Google Scholar]
- 52.Zuddas, A., Corsini, G. U., Schinelli, S., Johannessen, J. N., di Porzio, U. & Kopin, I. J. (1989) Brain Res. 501, 1–10. [DOI] [PubMed] [Google Scholar]
- 53.Lin, A. M., Fan, S. F., Yang, D. M., Hsu, L. L. & Yang, C. H. (2003) Free Radical Biol. Med. 34, 1416–1425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.