Abstract
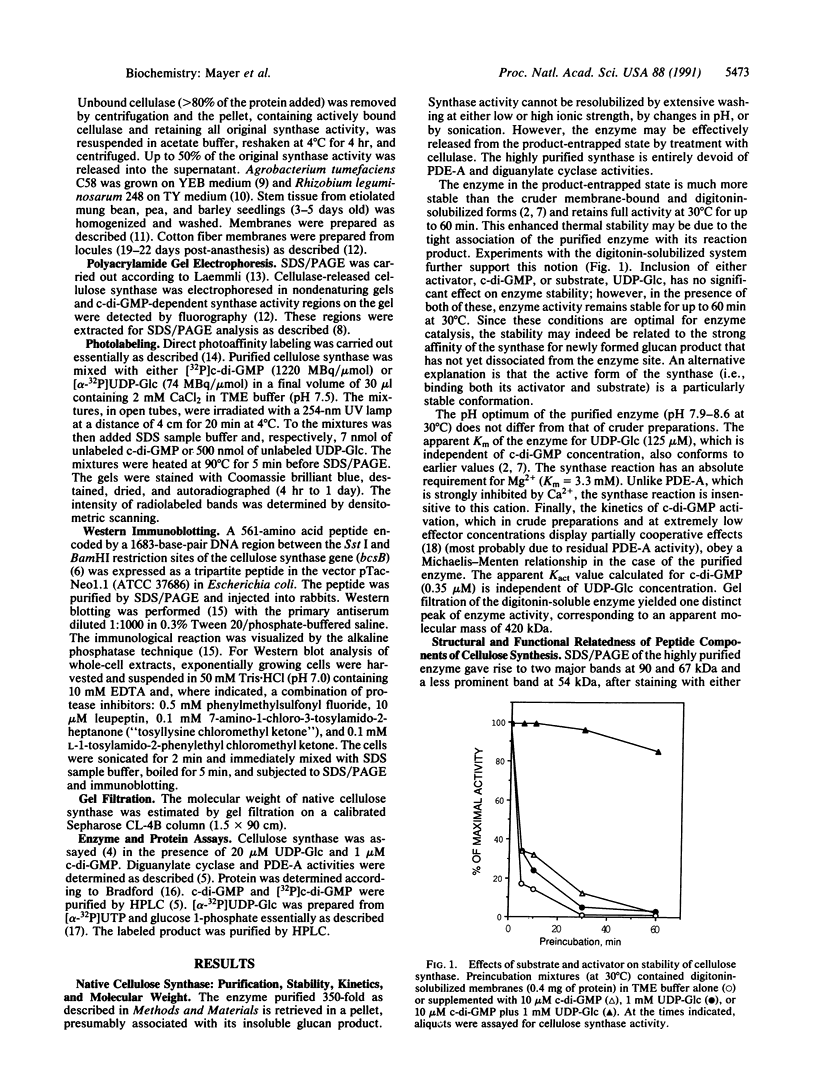
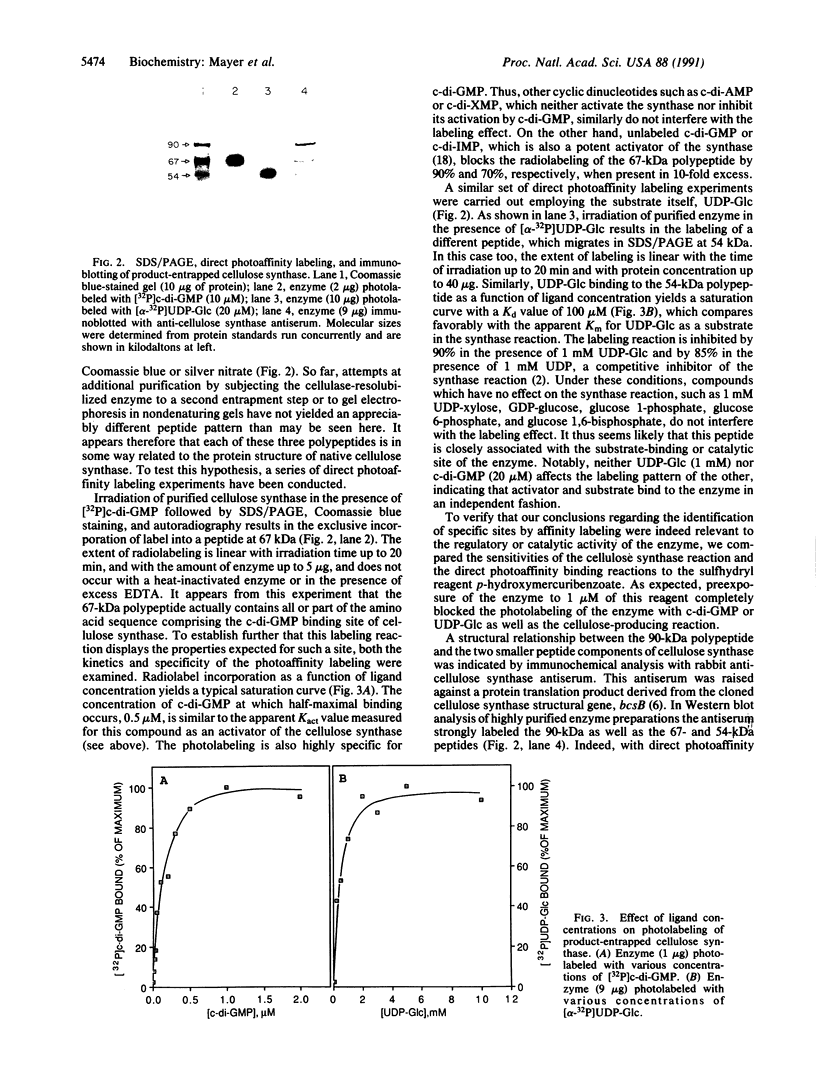
To comprehend the catalytic and regulatory mechanism of the cyclic diguanylic acid (c-di-GMP)-dependent cellulose synthase of Acetobacter xylinum and its relatedness to similar enzymes in other organisms, the structure of this enzyme was analyzed at the polypeptide level. The enzyme, purified 350-fold by enzyme-product entrapment, contains three major peptides (90, 67, and 54 kDa), which, based on direct photoaffinity and immunochemical labeling and amino acid sequence analysis, are constituents of the native cellulose synthase. Labeling of purified synthase with either [32P]c-di-GMP or [alpha-32P]UDP-glucose indicates that activator- and substrate-specific binding sites are most closely associated with the 67- and 54-kDa peptides, respectively, whereas marginal photolabeling is detected in the 90-kDa peptide. However, antibodies raised against a protein derived from the cellulose synthase structural gene (bcsB) specifically label all three peptides. Further, the N-terminal amino acid sequences determined for the 90- and 67-kDa peptides share a high degree of homology with the amino acid sequence deduced from the gene. We suggest that the structurally related 67- and 54-kDa peptides are fragments proteolytically derived from the 90-kDa peptide encoded by bcsB. The anti-cellulose synthase antibodies crossreact with a similar set of peptides derived from other cellulose-producing microorganisms and plants such as Agrobacterium tumefaciens, Rhizobium leguminosarum, mung bean, peas, barley, and cotton. The occurrence of such cellulose synthase-like structures in plant species suggests that a common enzymatic mechanism for cellulose biogenesis is employed throughout nature.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Cohen R., Benziman M., Delmer D. Solubilization of the UDP-glucose:1,4-beta-D-glucan 4-beta-D-glucosyltransferase (cellulose synthase) from Acetobacter xylinum. A comparison of regulatory properties with those of the membrane-bound form of the enzyme. J Biol Chem. 1983 Apr 10;258(7):4419–4423. [PubMed] [Google Scholar]
- Aloni Y., Delmer D. P., Benziman M. Achievement of high rates of in vitro synthesis of 1,4-beta-D-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6448–6452. doi: 10.1073/pnas.79.21.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D., Benziman M. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1989 Dec;171(12):6649–6655. doi: 10.1128/jb.171.12.6649-6655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J. S., Butler P. E. Intracellular proteases. Annu Rev Biochem. 1987;56:333–364. doi: 10.1146/annurev.bi.56.070187.002001. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bulawa C. E., Slater M., Cabib E., Au-Young J., Sburlati A., Adair W. L., Jr, Robbins P. W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986 Jul 18;46(2):213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Edelman A. M., Blumenthal D. K., Krebs E. G. Protein serine/threonine kinases. Annu Rev Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- Groppi V. E., Steinberg F., Kaslow H. R., Walker N., Bourne H. R. Identification by direct photoaffinity labeling of an altered phosphodiesterase in a mutant S49 lymphoma cell. J Biol Chem. 1983 Aug 25;258(16):9717–9723. [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Kang M. S., Elango N., Mattia E., Au-Young J., Robbins P. W., Cabib E. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J Biol Chem. 1984 Dec 10;259(23):14966–14972. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin F. C., Brown R. M., Jr, Drake R. R., Jr, Haley B. E. Identification of the uridine 5'-diphosphoglucose (UDP-Glc) binding subunit of cellulose synthase in Acetobacter xylinum using the photoaffinity probe 5-azido-UDP-Glc. J Biol Chem. 1990 Mar 25;265(9):4782–4784. [PubMed] [Google Scholar]
- Ross P., Mayer R., Weinhouse H., Amikam D., Huggirat Y., Benziman M., de Vroom E., Fidder A., de Paus P., Sliedregt L. A. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J Biol Chem. 1990 Nov 5;265(31):18933–18943. [PubMed] [Google Scholar]
- Smit G., Kijne J. W., Lugtenberg B. J. Involvement of both cellulose fibrils and a Ca2+-dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J Bacteriol. 1987 Sep;169(9):4294–4301. doi: 10.1128/jb.169.9.4294-4301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen M. P., Delmer D. P. Gel-Electrophoretic Separation, Detection, and Characterization of Plant and Bacterial UDP-Glucose Glucosyltransferases. Plant Physiol. 1986 Jul;81(3):913–918. doi: 10.1104/pp.81.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. A., Schlender K. K., Larner J. A rapid filter paper assay for UDPglucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968 Oct 24;25(1):486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- Wong H. C., Fear A. L., Calhoon R. D., Eichinger G. H., Mayer R., Amikam D., Benziman M., Gelfand D. H., Meade J. H., Emerick A. W. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]