Abstract
A salinity and dehydration stress-responsive calcium-dependent protein kinase (CDPK) was isolated from the common ice plant (Mesembryanthemum crystallinum; McCPK1). McCPK1 undergoes myristoylation, but not palmitoylation in vitro. Removal of the N-terminal myristate acceptor site partially reduced McCPK1 plasma membrane (PM) localization as determined by transient expression of green fluorescent protein fusions in microprojectile-bombarded cells. Removal of the N-terminal domain (amino acids 1–70) completely abolished PM localization, suggesting that myristoylation and possibly the N-terminal domain contribute to membrane association of the kinase. The recombinant, Escherichia coli-expressed, full-length McCPK1 protein was catalytically active in a calcium-dependent manner (K0.5 = 0.15 μm). Autophosphorylation of recombinant McCPK1 was observed in vitro on at least two different Ser residues, with the location of two sites being mapped to Ser-62 and Ser-420. An Ala substitution at the Ser-62 or Ser-420 autophosphorylation site resulted in a slight increase in kinase activity relative to wild-type McCPK1 against a histone H1 substrate. In contrast, Ala substitutions at both sites resulted in a dramatic decrease in kinase activity relative to wild-type McCPK1 using histone H1 as substrate. McCPK1 undergoes a reversible change in subcellular localization from the PM to the nucleus, endoplasmic reticulum, and actin microfilaments of the cytoskeleton in response to reductions in humidity, as determined by transient expression of McCPK1-green fluorescent protein fusions in microprojectile-bombarded cells and confirmed by subcellular fractionation and western-blot analysis of 6× His-tagged McCPK1.
Calcium is a ubiquitous and pivotal second messenger in the signal transduction networks that plants use to respond to a wide variety of physiological stimuli. Cytosolic Ca2+ fluctuations have been observed in response to a number of stimuli, including red light, abscisic acid (ABA), GA, drought, hyper- and hypo-osmotic stress, ionic stress, touch, cold, heat shock, oxidative stress, fungal elicitors, and nodulation factors (Sanders et al., 2002). Ca2+-dependent or calmodulin-like domain protein kinases (CDPKs), which are found only in plants, green algae, and certain protists, are an important group of sensor-responder proteins that function through intramolecular interactions to decode Ca2+ signals. In Arabidopsis, CDPKs form one of the largest Ca2+-binding protein families, with 34 unique CDPK genes plus numerous CDPK-related kinases (Harmon et al., 2000, 2001; Hrabak, 2000; Hrabak et al., 2003).
CDPKs are composed of a single polypeptide chain with a catalytic kinase domain at the N terminus, an intervening junction domain, and a calmodulin-like domain at the C terminus containing up to four functional Ca2+-binding EF hands. The junction domain acts as an autoinhibitor in a pseudosubstrate fashion (Harper et al., 1994; Vitart et al., 2000; Weljie et al., 2000). Binding of Ca2+ to the calmodulin-like domain results in a conformational change leading to the release of the autoinhibitor domain from the active site and kinase activation (Harmon et al., 1994; Harper et al., 1994). Many CDPKs also have additional N-terminal leader and C-terminal domains composed of highly variable amino acid sequences. AtCPK25 and some CDPK-related kinases possess degenerate calmodulin domains (Lindzen and Choi, 1995; Zhang and Lu, 2003) and apparently do not require Ca2+ activation (Furumoto et al., 1996).
CDPKs exhibit diverse subcellular localization patterns, suggesting that they participate in a wide variety of signaling pathways. CDPKs have been localized to the cytosol nucleus (Roberts and Harmon, 1992; Dammann et al., 2003), cytoskeleton (Putnam-Evans et al., 1989; Dammann et al., 2003), oil bodies of endosperm cells (Anil et al., 2003), endoplasmic reticulum (ER; Lu and Hrabak, 2002; Dammann et al., 2003), plasma membrane (PM; Schaller et al., 1992; Verhey et al., 1993; Baizabal-Aguirre and de la Vara, 1997; Iwata et al., 1998; Yoon et al., 1999; Dammann et al., 2003), and peroxisomes (Dammann et al., 2003). A majority of CDPKs from Arabidopsis contain the proper consensus sequence context for N-myristoylation at the second position Gly residue (MGNXX[ACGSTV][^DE] or MG[^DEFKRVWY]XX[ACGSTV][KR]; Boisson et al., 2003). To date, three CDPKs have been shown to be myristoylated at the N-terminal Gly residue: CpCDPK1 from zucchini (Cucurbita pepo; Ellard-Ivey et al., 1999), AtCPK2 from Arabidopsis (Lu and Hrabak, 2002), and LeCPK1 from tomato (Lycopersicon esculentum; Rutschmann et al., 2002). Such myristoylation events are critical for localization to the ER (Lu and Hrabak, 2002) or the PM (Rutschmann et al., 2002). Membrane localization correlates well with the presence of N-terminal myristoylation consensus motifs, suggesting that myristoylation is an important factor in membrane association (Yoon et al., 1999; Martín and Busconi, 2000; Dammann et al., 2003). CDPKs containing a myristoylation consensus motif also contain at least one potential palmitoylation site at several Cys residues (Cheng et al., 2002; Hrabak et al., 2003). Myristoylation of both the N-terminal Gly and palmitoylation at the C4 and C5 positions of OsCPK2 has been demonstrated to occur in vivo in maize (Zea mays) leaf protoplasts with both acyl modifications contributing to full membrane association (Martín and Busconi, 2000).
Recent studies have begun to clarify the specific roles of individual CDPKs in signal transduction cascades. Direct evidence for CDPK participation in biotic, abiotic, and ABA signaling pathways has been reported in only a few instances (Sheen, 1996; Saijo et al., 2000, 2001; Romeis et al., 2001). For example, Sheen (1996) showed that two out of eight Arabidopsis CDPK isoforms could activate expression from a cold, high-salt, dark, and ABA-regulated promoter (HVA-1) in a maize leaf protoplast transient expression system. More recently, Saijo et al. (2000, 2001) demonstrated that overexpression of a single CDPK conferred improved cold, salt, and drought tolerance in rice (Oryza sativa). Pharmacological studies have implicated CDPK participation in fungal pathogen defense (Romeis et al., 2000), wound signaling (Schaller and Oecking, 1999), and osmotic stress signaling (Pestenácz and Erdei, 1996; Taybi and Cushman, 1999). Furthermore, the expression patterns or activity changes of some CDPKs have suggested functional roles in low-temperature, ionic, osmotic, or water deficit stress signaling (Urao et al., 1994; Botella et al., 1996; Pestenácz and Erdei, 1996; Yoon et al., 1999; Komatsu et al., 2001; Chico et al., 2002; Llop-Tous et al., 2002), light responses (Pagnussat et al. 2002), jasmonate signaling (Ulloa et al., 2002), and elicitor-induced signaling (Yoon et al., 1999; Murillo et al., 2001; Romeis et al., 2001).
The wide variety of potential CDPK substrates described to date suggests diverse functional roles for this class of protein kinases (Harmon et al., 2000; Hrabak, 2000; Cheng et al., 2002). One approach to elucidate the functional roles of specific CDPKs is reverse genetics. To this end, Arabidopsis plants harboring T-DNA insertions in many different CDPK isogenes have now been isolated (Cheng et al., 2002). However, this approach may be hampered by functional redundancy of related CDPKs. Another approach to understanding CDPK function is to define the specific substrates or other proteins with which particular kinases interact. Although many potential CDPK substrates have been identified (e.g. soluble enzymes, ion and water channels, pumps, and transcription factors), there are few reports of identified substrates for specific CDPKs (Harmon et al., 2000; Hrabak, 2000; Cheng et al., 2002). A guard cell CDPK has been characterized that can phosphorylate the KAT1 potassium channel and thus is likely to participate in Ca2+ regulation of stomatal responses to environmental stimuli (Li et al., 1998). Phosphorylation of AtACA2, a calmodulin-stimulated Ca2+ pump, by CPK1 inhibits Ca2+ pumping, leading to alterations in Ca2+ signatures arising from ER stores (Hwang et al., 2000). Using a maize protoplast transient expression system, AtCPK1 can phosphorylate in vivo recombinant Phe ammonia lyase, a key enzyme in pathogen defense, stress response, and secondary metabolism (Cheng et al., 2001). The yeast (Saccharomyces cerevisiae) two-hybrid system has also been used to directly identify CDPK substrates. Patharkar and Cushman (2000) identified a pseudoresponse regulator (McCSP1) as a substrate of a salt and drought stress-inducible CDPK from ice plant (Mesembryanthemum crystallinum; McCPK1). More recently, a regulatory subunit of 26S proteasome (NtRpn3) was found to be a substrate for a tobacco (Nicotiana tabacum) CDPK (NtCPK1; Lee et al., 2003).
In this article, we show that the mRNA abundance of McCPK1 is transiently up-regulated by ionic and osmotic stress, implicating its involvement in abiotic stress signaling pathways. Biochemical analysis of McCPK1 demonstrates that this kinase is activated by Ca2+ and undergoes autophosphorylation on at least two different Ser residues, with the location of two such sites being mapped to Ser-62 and Ser-420. We used fluorescence imaging of McCPK1-synthetic green fluorescent protein (sGFP) fusions to demonstrate that McCPK1 localizes to the PM, nucleus, and cytoskeleton structures in living cells. Subcellular fractionation studies corroborated these observations. We also demonstrate that McCPK1 undergoes cotranslational N-myristoylation, which appears to be necessary for subcellular localization to the PM. Moreover, we show that exposure to low (40%) relative humidity causes McCPK1-sGFP to move from the PM to the nucleus. Under constant conditions, this subcellular localization is reset to the PM. These results demonstrate that McCPK1 undergoes dynamic changes in subcellular localization following abiotic stress.
RESULTS AND DISCUSSION
Isolation and Sequence Analysis of McCpk1
A λ Uni-ZapXR cDNA expression library was prepared from leaf tissue of ice plants exposed to 0.5 m NaCl stress for 30 h, as described previously (Forsthoefel et al., 1995), and probed with a partial cDNA product isolated by degenerate primer reverse transcription (RT)-PCR. Sequence analysis of the longest cDNA clone recovered by library screening revealed that it did not encode a full-length cDNA. Therefore, 5′ RACE was used to recover the full-length transcript. The full-length McCpk1 cDNA (2,364 bp) contained a single open reading frame (ORF) of 1,605 bp flanked by 5′ and 3′ untranslated sequences of 336 and 423 bp, respectively, including a poly(A+) tail of 48 adenine residues (accession no. AF090835). The ORF predicted a polypeptide of 534 amino acids with a calculated molecular mass of 60 kD. Analysis of the deduced amino acid sequence indicated that the protein contains the domain structure typical of CDPKs, including an N-variable domain (amino acids 1–84), a protein kinase domain (amino acids 85–346), a junction or autoinhibitory (pseudosubstrate) domain (amino acids 347–379), and four EF hands within the calmodulin-like domain (amino acids 386–534). Alignment and phylogenetic analysis using the entire length of the deduced McCPK1 protein sequence showed it fell within the branch 2 cluster of plant and algal CDPKs (see Fig. 2; Harmon et al., 2000) and, from among Arabidopsis CDPKs is most closely related to AtCPK9 (accession no. AAB03242; Hrabak et al., 2003).
Figure 2.
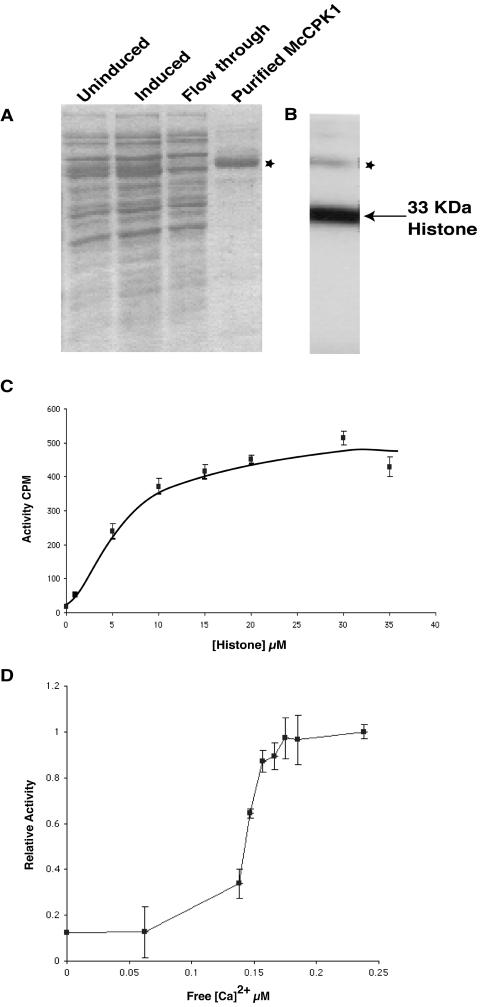
Purification of recombinant McCPK1 and calcium-dependent phosphorylation of histone H1. A, The full-length McCPK1 cDNA was expressed in E. coli as a C-terminal 6× His-tag fusion protein and the purification was monitored by SDS-PAGE followed by Coomassie Brilliant Blue staining. The uninduced (uninduced) lane provides a control for nonspecifically expressed E. coli proteins. The isopropyl thio-β-d-galactoside-induced (induced) crude extracts were purified by affinity chromatography on a His-Bind resin. The bound fusion protein, indicated with an asterisk following elution of the McCPK1-6× His fusion (purified McCPK1), was purified from the column (flow through). The uninduced, induced, and flow-through lanes contained 1.2 μg of protein, whereas the purified McCPK1 lane contained 0.8 μg of protein. B, Autoradiogram of SDS-polyacrylamide gel of purified, recombinant McCPK1-6×His fusion protein following kinase reaction in the presence of histone H1, Ca2+, and γ-32P-ATP. The asterisk indicates autophosphorylation of McCPK1 and arrow indicates the position of the 33-kD histone H1. C, Phosphorylation of histone H1 by McCPK1. Each data point was the average of triplicate kinase assays, and the bars indicate sd. The specific activity of the purified kinase was 5,275 ± 531 nm min−1 mg−1. D, Characterization of McCPK1 activation in the presence of various Ca2+ concentrations. The relative kinase activity of McCPK1 was determined at various concentrations of free Ca2+ concentrations with 1 mg mL−1 histone H1 as a substrate. The point plotted on the ordinate indicates activity in the absence of added Ca2+. Each point is the average of triplicate assays and the bars indicate the sd.
McCpk1 Is Transiently Induced by Salinity and Water Deficit Stress
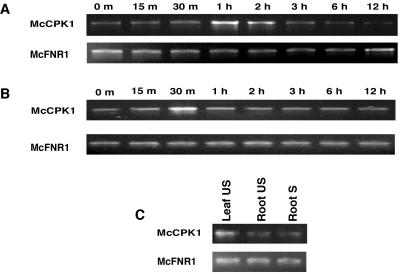
Semiquantitative RT-PCR analysis of McCpk1 mRNA abundance in detached leaves subjected to dehydration and salinity stress treatments showed that McCpk1 steady-state transcripts increased in abundance after 1 to 2.5 h following high-salinity (0.4 m NaCl) and dehydration stress, respectively (Fig. 1, A and B). Dehydration stress caused a more rapid induction of McCpk1 transcripts than salinity stress. Detached leaves of ice plant have been used successfully to monitor gene expression or enzyme activities related to Crassulacean acid metabolism induction by osmotic and ionic stress, suggesting that they can sense and respond to such stimuli in the absence of root signals (Taybi and Cushman, 1999). Comparison of McCpk1 expression in different ice plant organs revealed that steady-state McCpk1 transcripts were approximately 3-fold more abundant in leaves than in roots from well-watered or soil-grown plants subjected to salinity stress (500 mm NaCl) for 5 d (Fig. 1C). As a control, the transcript abundance of a gene coding for ferredoxin NADP+ reductase (Fnr1) was monitored using the same RNA samples and remained unchanged following stress consistent with earlier observations that Fnr1 expression was unaffected by dehydration and ionic stress (Michalowski et al., 1989; Taybi and Cushman, 1999). It should be noted that McCpk1 transcripts were more than 130-fold less abundant than Fnr1 transcripts.
Figure 1.
McCpk1 expression in detached leaves of salinity and water-deficit stressed ice plant. A, Ethidium bromide-stained gel of RT-PCR products from total RNA isolated from detached leaves exposed to salt stress (0.4 m NaCl) for times indicated. Gene-specific primers for McCpk1 and Fnr1 generated RT-PCR products of 850 and 600 bp, respectively. B, Time course of RT-PCR analyses of McCpk1 expression in detached leaves exposed to dehydration stress. C, Comparison of RT-PCR analysis of McCpk1 expression in unstressed (US) leaves and unstressed (US) and salinity-stressed (S) roots.
These results suggest that McCPK1 may be involved in decoding stress-induced Ca2+ signatures and linking them to adaptive responses. Stress-responsive mRNA expression correlates well with a role in stress-adaptive responses in several cases. For example, transcripts for the AtCPK10 and AtCPK11 genes increased in abundance following water deficit and salinity stress (Urao et al., 1994). Expression of a constitutively active form of AtCPK10 in maize protoplasts activated expression from the stress-inducible HVA1 promoter (Sheen 1996). The salt-, water deficit-, and cold-induced OsCPK7 gene from rice, when overexpressed in transgenic rice, conferred improved cold, salt, and drought tolerance (Saijo et al., 2000). Many other examples of multiple stimuli inducing the expression of individual CPKs are known. Increased steady-state transcripts, abundance, or activities of various CPKs in response to low-temperature, ionic, osmotic, water deficit, wounding, and elicitor treatments implicate their possible involvement in signaling response networks for these environmental stimuli (Botella et al., 1996; Pestenácz and Erdei, 1996; Yoon et al., 1999; Komatsu et al., 2001; Murillo et al., 2001; Romeis et al., 2001; Chico et al., 2002; Llop-Tous et al., 2002). The remaining challenge is to determine the specific contribution to signaling pathways made by each individual CDPK and the degree of overlap or redundancy among these different signaling cascades.
Biochemical Characterization of McCPK1
To assess the biochemical activity of McCPK1, the full-length cDNA of McCpk1 was expressed as a C-terminal 6× His-tag fusion protein and purified from Escherichia coli to near homogeneity using His-Bind resin tag affinity chromatography (Fig. 2A). The substrate specificity of the recombinant, purified kinase was determined using a variety of exogenous substrates known to be phosphorylated by CDPKs. Histone H1 was phosphorylated (Km = 5.5 μm) by McCPK1 (Fig. 2, B and C) and served as a better substrate than all synthetic peptide (e.g. syntide-2, autocamtide, autocamtide-2, and kemptide) substrates tested (data not shown). Therefore, histone H1 was used to determine how Ca2+ binding affects McCPK1 activity. Recombinant McCPK1 kinase activity was tested over a range of Ca2+ concentrations in the presence of histone H1 as a substrate (Fig. 2D). The Ca2+ stimulated McCPK1 activity by more than 6-fold. The [Ca2+] for half-maximal activity (K0.5) was 0.15 μm of free Ca2+ with histone H1 as the substrate. This value was lower than previously reported values for soybean (Glycine max) CDPK-β (K0.5 = 0.4 μm) and CDPK-γ (K0.5 = 1.0 μm), with only CDPK-α (K0.5 = 0.06 μm), measured in the presence of the peptide substrate syntide-2, having a lower value (Lee et al., 1998). This value was also higher than that obtained for sandalwood (Santalum album) CDPK (0.7 μm) with histone III-S as a substrate (Anil and Rao, 2001); however, it was lower than the K0.5 for histone III-S phosphorylation (about 4 μm) by soybean CDPK-α or for casein phosphorylation by Plasmodium falciparum CDPK1 (Zhao et al., 1994).
Various signals modulate intracellular Ca2+ levels (Rudd and Franklin-Tong, 2001; Sanders et al., 2002). However, Ca2+ does not move freely within the cell (Trewavas, 1999). To transduce these signals intracellularly into downstream effects, CDPKs are thought to be the primary sensors of these Ca2+ signatures. The resting concentration for cytosolic Ca2+ is estimated to be approximately 100 nm (Rudd and Franklin-Tong, 2001). Since McCPK1 has a Ca2+ half-maximal activity value at 150 nm, which is only 50 nm above the resting concentration, this suggests that McCPK1 may sense relatively small changes in cytosolic Ca2+ concentrations and link these changes to appropriate downstream adaptive responses. Vitart et al. (2000) proposed two models for the activation of CPKs by Ca2+. In both models, Ca2+ binding to the calmodulin-like domain is proposed to result in conformational changes that lead to kinase activation. However, it is difficult to predict the Ca2+-binding properties of a calmodulin-like domain from its amino acid sequence alone. Some CDPKs appear to have defective EF hands that alter their Ca2+-binding properties. For example, Zhao et al. (1994) showed that only the first EF hand of PfCPK1 needed to be functional in order to undergo Ca2+ activation. Rutschmann et al. (2002) classified two EF hands of LeCPK1 as high Ca2+-binding sites and the other two as low Ca2+-binding sites, with the first group of EF hands believed to be responsible for the Ca2+ activation of LeCPK1. Based on these observations, Ca2+ is thought to bind more readily to the EF hands proximal to the kinase domain. To verify this hypothesis, site-directed mutagenesis of the calmodulin-like domain of McCPK1 could be used to mutagenize each of the four EF hands alone and in various combinations, and then to test the activity of the mutant forms in response to Ca2+.
Mapping the Autophosphorylation Sites of McCPK1
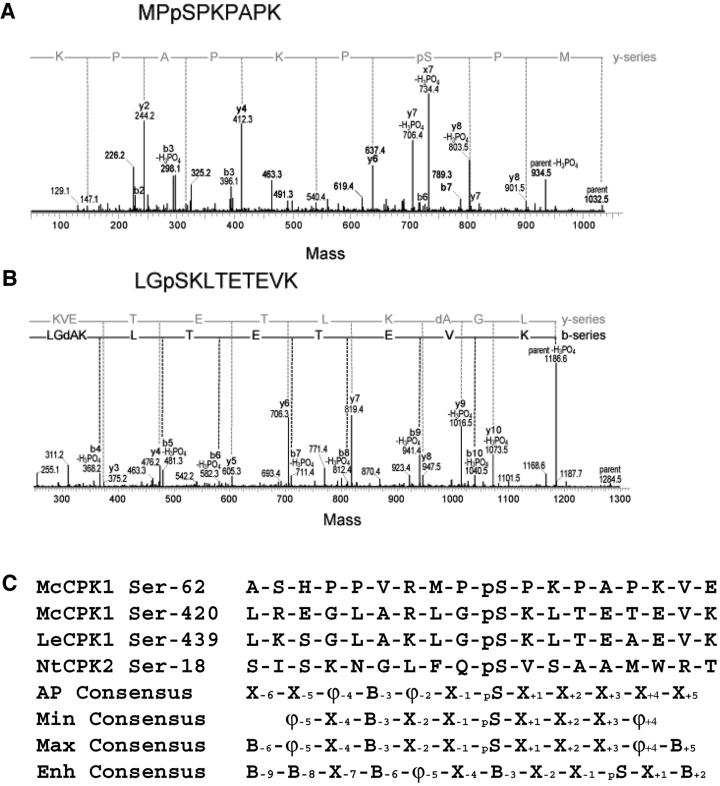
In addition to Ca2+, intramolecular autophosphorylation on Ser/Thr residues may play a role in influencing the activity of CDPK. As a first step toward understanding how autophosphorylation events may affect CDPK activity, we mapped two autophosphorylation sites on McCPK1. Recombinant McCPK1 was subjected to in vitro autophosphorylation without prior phosphatase treatment, after which the autophosphorylation sites were mapped by quadrupole-time-of-flight mass spectrometry. We identified Ser-62 and Ser-420 as two of the targets for autophosphorylation in McCPK1 (Fig. 3, A and B). The Ser-62 site mapped to the N-variable domain, similar to the Ser-18 autophosphorylation site mapped recently in NtCPK2 (Glinski et al., 2003). The Ser-420 is located within the calmodulin-like domain between the first and second EF hands nearest the kinase domain. This location is analogous to the autophosphorylation site Ser-439 mapped in LeCPK1 from tomato (Rutschmann et al., 2002). The amino acid sequences surrounding the phosphophorylation sites of McCPK1 were compared with those of NtCPK2 and LeCPK1 (Fig. 3C). These sites of phosphorylation conform largely to a consensus autophosphorylation motif (X−6-X−5-ϕ−4-Basic−3-ϕ−2-X−1-pS-X+1-X+2-X+3-X+4-X+5), where ϕ indicates hydrophobic residues and pS indicates the phosphorylated Ser residue. This consensus sequence most closely resembles the maximal phosphorylation consensus described by Huang and Huber (2001) except for the hydrophobic residues present at positions −2 and −4, the lack of consensus hydrophobic residues at position −5 and +4, and the lack of consensus basic residues at positions −6 and +5.
Figure 3.
Processed MS/MS spectra of phosphopeptides of autophosphorylated McCPK1. Mass spectrometry analysis identified two tryptic peptides of autophosphorylated McCPK1 as indicated in A and B. A, First autophosphorylated McCPK1 peptide (m/z = 516.74; double charged; mass = 1,032.5 D) whose y-ion series is labeled as such and the deduced amino acid sequence is indicated. The mass difference (803.5 − 637.4) indicates the presence of a phosphorylated Ser residue. B, Second autophosphorylated McCPK1 peptide is of m/z = 642.78; double charged; mass = 1,284.5. The y- and b-ion series are labeled as such and the deduced amino acid sequence is indicated. The mass difference (1,016.5 − 947.5) indicates the presence of dA (dehydroalanine, indicative of Ser) minus H3PO4. C, Comparison of autophosphorylation sequences from McCPK1 (Ser-62 and Ser-420) to those identified for LeCPK1 (Ser-439; Rutschmann et al., 2002) and NtCPK2 (Ser-18; Glinski et al., 2003), and resulting autophosphorylation (AP) consensus sequence where ϕ indicates hydrophobic residues, B indicates basic residues, and p indicates the phosphorylated Ser residue. Previously described minimal (Min; Bachmann et al., 1996), maximal (Max), and enhanced (Enh) consensus phosphorylation target sites of spinach leaf CDPK PKI and PKII (Huang and Huber, 2001; Huang et al., 2001).
Activity of McCPK1 Autophosphorylation Mutants
Recombinant and native CDPKs exhibit intramolecular autophosphorylation. However, the role of autophosphorylation in regulating CDPK activity remains unclear. Autophosphorylation of a CDPK purified from winged bean was shown to inhibit its activity (Saha and Singh, 1995). On the other hand, autophosphorylation of groundnut CDPK was suggested to be a prerequisite for its activity, but autophosphorylation occurs at low Ca2+ concentrations and might not have a regulatory role in vivo (Chaudhuri et al., 1999). Preautophosphorylation of sandalwood CDPK neither inhibited nor stimulated the kinase's activity, suggesting that autophosphorylation is inconsequential to its substrate activity (Anil and Rao, 2001). Activation in vitro of CPK2 from tobacco was accompanied by autophosphorylation, which was not inhibited by W-7, a calmodulin and CDPK antagonist, suggesting that direct phosphorylation by an upstream protein kinase and not autophosphorylation was responsible for activation (Romeis et al., 2000, 2001). Therefore, clear evidence for the dependence of activity on the autophosphorylation has not emerged to date.
With the identification of two residues that undergo autophosphorylation in McCPK1, we examined how autokinase and substrate kinase activities may be changed by mutating these autophosphorylation sites to Ala. Both the single mutants (S62A McCPK1 and S420A McCPK1) and the double mutant (S62A/S420A McCPK1) were examined. Autophosphorylation was analyzed by in vitro kinase reactions using affinity-purified, recombinant McCPK1 (Table I). All mutant forms retained calcium-dependent activity. However, quantitative analysis of autokinase activities revealed that, without substrate, although there were differences among genotypes in their ability to autophosphorylate both with (P = 0.0164) and without (P = 0.0349) calcium, such differences were only slightly significant. The double-mutant form of the kinase containing both S62A and S420A substitutions was still able to exhibit autophosphorylation (data not shown). However, attempts to map these alternative sites have proved unsuccessful thus far.
Table I.
Autophosphorylation activity for bacterially expressed and purified wild type and autophosphorylation site mutants of McCPK1-6× His fusion proteins assayed in the absence of substrate
| McCPK1 Variant | Specific Activitya
|
X-Fold Ca2+ Activation | Activityb versus Wild-Type McCPK1 | |
|---|---|---|---|---|
| −Ca2+ | +Ca2+ | |||
| nm min−1 mg−1 | % | |||
| Wild type | 0.26 ± 0.006c | 0.36 ± 0.006d | 1.38 | 100 |
| S62A | 0.27 ± 0.005* | 0.38 ± 0.008 | 1.40 | 105.3 |
| S420A | 0.29 ± 0.006* | 0.41 ± 0.007* | 1.41 | 111.2 |
| S62A/S420A | 0.30 ± 0.006* | 0.40 ± 0.005* | 1.33 | 110 |
Specific activity data are represented as the mean ± se and were derived from the average of three replicate assays with each of four replicate enzyme preparations. Kinase activity assays contain 50 ng kinase and were performed as described in “Materials and Methods.”
In the presence of Ca2+.
In the absence of calcium (R2 = 0.5), kinase mutants indicated with an asterisk displayed only slightly significantly different kinase activity from wild type (P < 0.0349).
In the presence of calcium (R2 = 0.53), kinase mutants indicated with an asterisk displayed significantly different kinase activity (P < 0.0164) from wild type.
Autophosphorylation site amino acid substitutions may result in minute conformational changes in kinase structure, leading to the observed changes in activity. Currently, such conformational changes would be difficult to predict, since the crystal structure of CDPKs is still unknown. However, one of the models proposed by Vitart et al. (2000) for the activation of CDPKs suggests that the inactive and active states of the kinase are closely related conformationally. Once Ca2+ binds to the EF hands and/or autophosphorylation takes place, subtle changes in the conformation of the kinase are thought to cause the removal of the pseudosubstrate and the activation of the enzyme. Nevertheless, such predicted conformational changes cannot be confirmed until the structure of wild-type CDPK and its mutant forms is resolved. It will also be interesting and informative to map the alternative autophosphorylation sites in these mutants. The phosphorylation of alternative sites also suggests that a particular autophosphorylation site may not be entirely essential for autophosphorylation activity per se.
Next, we examined the effect of mutating the autophosphorylation sites on the ability of the enzymes to phosphorylate histone H1 (Table II). There was a notable interaction between the influence of enzyme preparation and kinase genotype on kinase activity that was more pronounced in the presence of Ca2+ (P = 0.0004) than in its absence (P = 0.03). Lack of autophosphorylation at Ser-62 or Ser-420 increased activity of the kinase slightly relative to wild-type McCPK1. In contrast, the double substitution S62A/S420A McCPK1 dramatically decreased the activity relative to the wild-type kinase. This suggests that simultaneous autophosphorylation of both Ser-62 and Ser-420 may be important for the activity of the enzyme. Ser-62 is located in the N-terminal variable domain. Ser-420 is located in the calmodulin-like domain between EF hand 1 and EF hand 2. Lack of autophosphorylation at these sites may limit the ability of the mutant kinase to undergo subtle conformational changes or impair the efficiency of Ca2+ binding required for the removal of the pseudosubstrate and activation of the enzyme relative to the wild-type kinase. Alternatively, the introduced double S62A/S420A substitution may have resulted in structural alterations in the kinase itself that could account for the reduced activity of the kinase.
Table II.
Phosphorylation activity for bacterially expressed and purified wild type and autophosphorylation site mutants of McCPK1-6× His fusion proteins assayed in the presence of histone H1 substrate
| McCPK1 Variant | Specific Activitya
|
X-Fold Ca2+ Activation | Activitybversus Wild-Type McCPK1 | |
|---|---|---|---|---|
| −Ca2+ | +Ca2+ | |||
| nm min−1 mg−1 | % | |||
| Wild type | 0.54 ± 0.010c | 83.11 ± 0.696d | 153.90 | 100 |
| S62A | 0.54 ± 0.019 | 89.81 ± 2.37* | 166.31 | 107.5 |
| S420A | 0.62 ± 0.011* | 96.28 ± 1.255* | 155.29 | 101.1 |
| S62A/S420A | 0.45 ± 0.016* | 3.28 ± 0.028* | 7.28 | 4.7 |
Specific activity data are represented as the mean ± se and were derived from the average of three replicate assays with each of four replicate enzyme preparations. Kinase activity assays contain 50 ng kinase with 30 μm histone H1 substrate and were performed as described in “Materials and Methods.”
In the presence of Ca2+.
In the absence of calcium (R2 = 0.76), kinase mutants indicated with an asterisk displayed significantly different kinase activity from wild type (P < 0.0001).
In the presence of calcium (R2 = 0.99), kinase mutants indicated with an asterisk displayed significantly different kinase activity (P < 0.0001) from wild type.
It is highly unlikely that the autophosphorylation and kinase activities obtained here were influenced by a contaminating kinase copurifying with the recombinant, purified forms of McCPK1 used in these experiments for several reasons. The relative purity of the different forms of the recombinant kinases was estimated using NIH image software to be very high (S62A McCPK1-6× His 95%, S420A McCPK1-6× His 96%, S62A S420A McCPK1-6× His 96%, and the wild-type McCPK1-6× His 95%). The relative abundance of any contaminating kinase activity copurifying with the McCPK1-6× His tagged after nickel resin affinity chromatography would be expected to be very low. Second, all of the activities measured were Ca2+ dependent. An inspection of all of the kinases encoded on the E. coli genome at the Bioinformatics Center Institute for Chemical Research, Kyoto University Web site (www.genome.ad.jp) revealed that the E. coli genome does not encode predicted Ca2+- or calmodulin-dependent protein kinases.
McCPK1 Myristoylation In Vitro and Membrane Localization Requirements
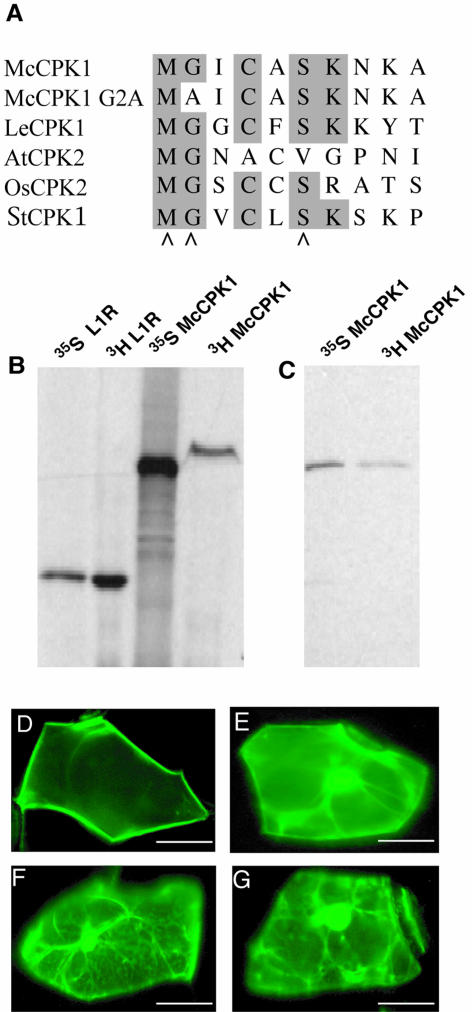
McCPK1 contains an N-terminal consensus sequence context for myristoylation at the N-terminal second position Gly residue (MGxxxS) similar to several other CDPKs that have been experimentally determined to undergo this cotranslational modification (Fig. 4A). We confirmed that McCPK1 undergoes N-terminal myristoylation in vitro by transcribing and translating the full-length cDNA containing a C-terminal 6× His-tag fusion using a coupled transcription-translation system from wheat germ extract, which contains N-myristoyltransferase activity (Heuckeroth et al., 1988; Ellard-Ivey et al., 1999), in the presence of [35S]Met for detecting protein synthesis or [3H]Myristic acid for detecting myristoylation (Fig. 4B). A 25-kD vaccinia viral protein (L1R) known to be myristoylated (Franke et al., 1990) was used as a positive control (Fig. 4B). The identity of the labeled products from the McCPK1 programmed reactions was confirmed by immunoprecipitation with anti-6× His-tag monoclonal antibodies. A protein of the expected size of the McCPK1-6× His-tag fusion protein (approximately 60 kD) was recovered from both the [35S]Met and [3H]Myristic acid-labeling reactions (Fig. 4C).
Figure 4.
McCPK1 undergoes myristoylation in vitro and PM localization of McCPK1-sGFP fusion depends upon myristoylation of McCPK1. A, Comparison of N-terminal amino acid sequences of McCPK1 (McCPK1) and its G2A (McCPK1-G2A) mutant form with other CPKs confirmed to undergo myristoylation or where myristoylation was found to be important for membrane localization, including LeCPK1 (Rutschmann et al., 2002), AtCPK2 (Lu and Hrabak, 2002), OsCPK2 (Martín and Busconi, 2000), and StCPK1 (Raíces et al., 2001). Identical amino acids are indicated with gray shading. Arrowheads indicate the consensus sequence (MGXXXS) for N-myristoylation. B, Wheat germ-coupled in vitro transcription-translation reactions programmed with L1R protein (L1R), a 25-kD vaccinia viral protein known to undergo myristoylation (positive control), and McCPK1-6× His-tag fusion (McCPK1) in the presence of either [35S]Met (lanes 35S L1R, 35S McCPK1) or [3H]myristate (lanes 3H L1R, 3H McCPK1). C, Immunoprecipitation of McCPK1 from in vitro transcription-translation reaction mixtures with the 6× His-tag antibody. Ice plant cells from plants grown in 80% relative humidity were transiently transfected by microprojectile bombardment with McCPK1-sGFP (D), G2A mutant of McCPK1-sGFP (E), N-terminal deletion mutant Δ1-70McCPK-sGFP (F), and sGFP (G). All cells were imaged 13 h after bombardment. Bars represent 10 μm.
To detect activity for N-myristoyltransferase, Gly is absolutely required at position 2 relative to the N-terminal Met (Yalovsky et al., 1999). Furthermore, N-myristoylation in Arabidopsis requires the consensus sequence (MGNXX[ACGSTV][^DE] or MG[^DEFKRVWY]XX[ACGSTV][KR]; Boisson et al., 2003). All of the CDPK shown to undergo N-terminal myristoylation conform to one of these two consensus sequences (Fig. 4A). The N-variable domain of most CDPKs plays an important role in the subcellular localization to the PM via N-terminal myristoylation or in the interaction with other proteins or substrates and may thereby confer specificity to discrete Ca2+-dependent signaling pathways. For example, Dammann et al. (2003) showed that two CDPKs from Arabidopsis (AtCPK3 and AtCPK4) without putative acylation sites localize to the soluble fraction of the cell extracts, whereas seven other CDPKs (AtCPK1, AtCPK7, AtCPK8, AtCPK9, AtCPK16, AtCPK21, and AtCPK28) harboring potential acylation sites (myristoylation and palmitoylation) were membrane associated.
CDPKs containing a myristoylation consensus motif also contain at least one potential palmitoylation site at several Cys residues (i.e. C4 or C5 in Fig. 4A; Cheng et al., 2002; Hrabak et al., 2003). Since McCPK1 has a Cys residue at position C4, we tested whether or not McCPK1 could undergo palmitoylation in the presence of [3H]Palmitic acid using the wheat germ transcription-translation system; however, we could detect no evidence of labeling (data not shown). Protein palmitoylation is achieved through esterification of Cys thiol groups by palmitate (Yalovsky et al., 1999). The primary sequence motif predictive of palmitoylation has not been identified; however, palmitoylation is often found associated with myristate or near-hydrophobic sequences. To our knowledge, N-palmitoyltransferase activity has not been reported in wheat germ extracts; therefore, it remains uncertain if McCPK1 failed to become palmitoylated because the activity is absent from the extracts or because McCPK1 is normally not palmitoylated in vivo. Future experiments using a protoplast system known to contain N-palmitoyltransferase activity, such as that employed by Martín and Busconi (2000), are needed.
Since myristoylation by itself normally is not sufficient for membrane localization (Shahinian and Silvius, 1995), we further investigated the membrane localization requirements for McCPK1 by fusing the wild-type kinase to sGFP (Sheen et al., 1995; Chiu et al., 1996) under the control of the cauliflower mosaic virus 35S promoter. Fusions were introduced into the cells of plants exposed to 80% relative humidity (unstressed conditions) via microprojectile bombardment and examined using fluorescence microscopy. The fusion localized preferentially to the PM (Fig. 4D). We then mutated the N-terminal Gly residue and replaced it with an unmyristoylatable Ala residue. Substitution of the N-terminal Gly residue results in the abolishment of both myristoylation and palmitoylation, suggesting that myristoylation may be a prerequisite for palmitolyation (Martín and Busconi, 2000). The G2A-McCPK1-sGFP fusion protein displayed a marked reduction in localization to the PM (Fig. 4E). This observation was consistent with the major role N-terminal myristoylation (and possibly palmitoylation) is known to play in the localization of CPKs to the PM. However, some PM localization was retained by the G2A-McCPK1-sGFP fusion protein, suggesting that myristoylation alone (and possibly palmitoylation) cannot account fully for the observed membrane localization. An additional feature for stable membrane anchoring for many fatty acylated proteins is the presence of a polybasic domain with a cluster of positively charged amino acids (Resh, 1999). The basic amino acids form electrostatic interactions with the head groups of acidic, negatively charged phospholipids of the membrane bilayer (Murray et al., 1997). The N-terminal domain (amino acids 1–70) of McCPK1 contains 8 basic amino acids (11.4%), which may aid in membrane binding. To test this possibility, we removed the first 70 amino acids of McCPK1-sGFP fusion protein and found that the remaining PM localization was lost (Fig. 4F), such that the N-terminal Δ1 to 70 deletion mutant of the McCPK1-sGFP fusion protein assumed a subcellular distribution pattern similar to the sGFP alone, which exhibited no PM localization (Fig. 4G). Since McCPK1 does not undergo palmitoylation in vitro, this suggests that stable anchoring of McCPK1 to the PM may require a second feature at the N terminus, such as N-terminal polybasic domains with clusters of positively charged amino acids to anchor it to the PM. The N-terminal domain (amino acids 1–70) of McCPK1 contains 8 positively charged basic residues. Such amino acids may interact with the head groups of acidic phospholipids that impart a negative charge to the cytoplasmic surface of the PM. Similarly, Rutschmann et al. (2002) observed a partial loss in the localization of LeCPK1 to the PM in suspension-cultured tomato cells transiently expressing a G2A-GFP fusion of LeCPK1 protein. The disruption of one acylation site (G2) in AtCPK2 was reported to result in only a partial loss (50%) of membrane association (Lu and Hrabak, 2002). sGFP is strongly expressed throughout the cytoplasm in both unstressed (Fig. 4G) and stressed (Fig. 5M) ice plant epidermal cells. SGFP exhibits no PM localization (Fig. 4G), suggesting that PM localization is not being driven by the sGFP portion of the McCPK1-sGFP fusion.
Figure 5.
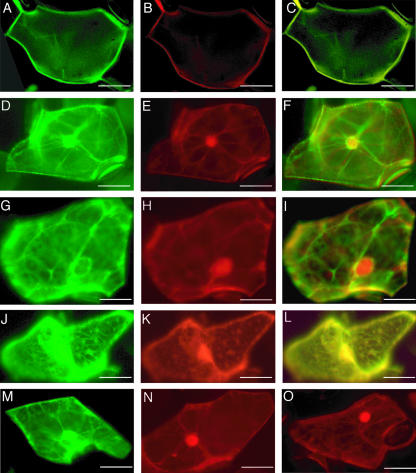
Identification of the subcellular structures to which McCPK1 localizes. Representative cell from ice plant leaves that were cobombarded and exhibited expression of AHA2-GFP (PM marker; A), McCPK1-DsRed (B), and superimposition of McCPK1-DsRed and AHA2-GFP expression (C). Plants were grown in 80% relative humidity (A–C). Representative expression of BiP-GFP (ER marker; D), McCPK1-DsRed (E), and superimposition of McCPK1-DsRed and BiP-GFP expression (F). Representative expression of tubulin-GFP (G), McCPK1-DsRed (H), and superimposition of McCPK1-DsRed and tubulin-GFP expression (I). Representative expression of talin-GFP (actin cytoskeleton marker; J), McCPK1-DsRed (K), and superimposition of McCPK1-DsRed and talin-GFP expression (L). Plants were grown in 40% relative humidity (D–L). sGFP expression in a representative cell from bombarded leaves of ice plants grown in 40% relative humidity (M). DsRed expression in cells from ice plants grown in 80% (N) and 40% relative humidity (O), respectively. Bars represent 10 μm.
Given the potential for CDPK myristoylation, Ca2+ binding may affect conformational changes in the modified protein that change the accessibility of the attached myristoyl group, leading to increased membrane affinity in a mechanism known as the Ca2+-myristoyl switch (Zozulya and Stryer, 1992). Real-time imaging of a GFP-tagged visinin-like protein 1 (VILIP-1) in living cells showed that this protein fusion undergoes a Ca2+-myristoyl switch enabling it to localize to distinct membrane compartments (Spilker et al., 2002). A similar observation was reported for recoverin, a myristoylated Ca2+ sensor in retinal rod cells (Ames et al., 1997). Recoverin has a compact three-dimensional structure consisting of two domains, each of which harbors one functional and one nonfunctional EF hand. Each EF hand becomes occupied by Ca2+ in a sequential process, wherein one functional EF hand is occupied first, triggering a rearrangement of the domain interface and subsequent binding of Ca2+ to the other functional EF hand. Occupation of both sites leads to the full exposition of the buried myristoyl group (Matsuda et al., 1998; Permyakov et al., 2000; Senin et al., 2002, 2003). McCPK1 may operate in a similar way; however, detailed structural analyses of myristoylated McCPK1 in the presence and absence of Ca2+ are needed to confirm this hypothesis.
McCPK1 Subcellular Localization under High and Low Relative Humidity Conditions
To gain a better understanding of the subcellular location of the McCPK1-sGFP, we conducted a series of colocalization studies with well-characterized marker protein-GFP fusions. Previously, Patharkar and Cushman (2000) demonstrated that McCPK1-sGFP undergoes a change in subcellular localization from the PM to the nucleus following 4 h of 0.5 m NaCl stress treatment to the plant. However, McCPK1-sGFP also localized to other unidentified subcellular structures besides the nucleus. To confirm the localization of McCPK1 to the PM in plants grown under 80% relative humidity (unstressed conditions), we coexpressed McCPK1-DsRed fusions with the AHA2 (Arabidopsis plasma membrane H+-ATPase)-GFP fusion, a PM marker (kindly supplied by Dr. Jeffery Harper, Scripps Research Institute, La Jolla, CA). McCPK1-DsRed colocalized with AHA2-GFP (Fig. 5, A–C). To identify the subcellular structures to which McCPK1 localized following exposure to plants under 40% relative humidity (low-humidity stress) conditions for 13 h (Fig. 5E), we cobombarded McCPK1-DsRed fusions with the ER marker BiP-GFP (kindly provided by Inhan Hwang, Gyeongsang National University, Korea; Kim et al., 2001). McCPK1-DsRed colocalized with the ER marker in large part consistent with subcellular fractionation studies (see below); however, there was also partial colocalization with filamentous structures (Fig. 5, D–F). The structures resembled the transcytoplasmic ER under the control of the actin cytoskeleton (Boevink et al., 1998), so we next tested if McCPK1-DsRed would colocalize with well-known cytoskeletal components. First, we coexpressed McCPK1-DsRed fusion with the microtubule marker tubulin-GFP (kindly provided by Seiichiro Hasezawa, University of Tokyo; Kumagai et al., 2001) and observed little colocalization (Fig. 5, G–I). Next, we coexpressed McCPK1-DsRed fusion with actin microfilament marker talin-GFP (kindly provided by Nam-Hai Chua, Rockefeller University, New York; Rees et al., 1990; McCann and Craig, 1997; Kost et al., 1998) and observed nearly complete colocalization (Fig. 5, J–L). Control panels show sGFP subcellular localization in an epidermal cell from an ice plant grown at 40% relative humidity (Fig. 5M), and DsRed subcellular localization in an epidermal cell from ice plants grown at 80% relative humidity (Fig. 5N) or 40% relative humidity (Fig. 5O). In summary, McCPK1-sGFP or McCPK1-DsRed fusions undergo a pronounced shift in subcellular location from the PM to the ER, the nucleus, and actin microfilaments in plants transferred from high to low relative humidity conditions, respectively. The diverse subcellular localization of CDPKs throughout the plant cell suggests that different CDPK isoforms regulate distinct signaling pathways. In the case of McCPK1, environmental changes, such as salinity stress (Patharkar and Cushman, 2000) and reductions in relative humidity result in a dynamic change in the subcellular location of the kinase from the PM to the nucleus, the transcytoplasmic ER, and the actin microfilaments (Fig. 5, B and E; O.R. Patharkar and J.C. Cushman, unpublished data).
Subcellular Fractionation
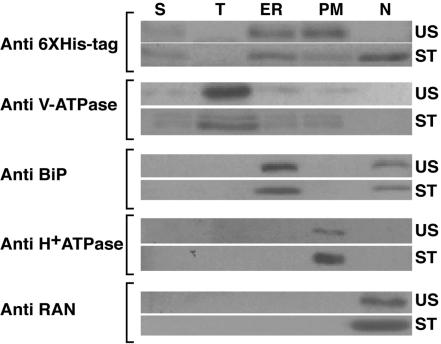
Subcellular localization studies using GFP fusion may not reflect a biologically relevant subcellular location due to the large size of the GFP, which may influence the final destination of the fusion protein within the cell. Therefore, we confirmed the subcellular localization patterns obtained using McCPK1-sGFP or McCPK1-DsRed fusions by conducting subcellular fractionation studies with McCPK1 tagged with a much smaller epitope tag. The ORF of McCPK1 was cloned in front of a 6× His tag under the control of cauliflower mosaic virus 35S promoter. Plasmid DNA containing McCPK1-6× His-tag fusion gene was introduced into ice plant epidermal cells by microprojectile bombardment, and then epidermal tissue was resolved into soluble, tonoplast, ER, and PM fractions by Suc step gradient centrifugation (Barkla et al., 1995; Vera-Estrella et al., 1999) or into a nuclear fraction by isolating intact nuclei (Cushman, 1995). Protein fractions were resolved by SDS-PAGE and analyzed by western blotting using a monoclonal antibody raised against the 6× His tag. This monoclonal antibody cross-reacted with a single 60-kD polypeptide that corresponds to the predicted mass of the McCPK1-6× His-tagged gene product. We observed that the McCPK1-6× His-tag fusion protein localized primarily to the PM and ER in leaves from plants subjected to 80% relative humidity (unstressed conditions; Fig. 6). However, in leaves exposed to 40% relative humidity (stressed conditions), the McCPK1-6× His-tag fusion protein localized to the PM, ER, and nucleus (Fig. 6). The isolation of the PM, ER, and tonoplast fractions was confirmed by probing them with anti-H+-ATPase antibodies (Pardo and Serrano, 1989), anti-BiP antibodies (Anderson et al., 1994a, 1994b), and anti-V-ATPase antibodies (Zhigang et al., 1996), respectively. The isolation of the nuclear fraction was confirmed by probing it with anti-Ras-related GTPase (RAN) antibodies (Melchior et al., 1993; Moore and Blobel, 1993; Melchior and Gerace, 1998). The observed change in the subcellular location of the McCPK1-6× His-tag fusion protein from the PM to the nucleus following the transition from high to low humidity was consistent with the results of sGFP-DsRed fusion localization studies (Fig. 5, B and E, respectively). These studies also validated the presence of McCPK1-6× His-tag fusion protein in the ER under both unstressed and stressed conditions, an observation not immediately evident in sGFP-DsRed fusion studies with unstressed leaves (Fig. 5, A–C), but evident in stressed leaves (Fig. 5, D–F). The localization of McCPK1 from the PM under unstressed conditions to the actin microfilaments, transcytoplasmic ER, and nucleus under stressed conditions suggests that McCPK1 is regulating a stress response pathway. The localization of McCPK1 to the nucleus is consistent with a previous observation by Patharkar and Cushman (2000) in which McCSP1, a substrate for McCPK1, is localized to the nucleus.
Figure 6.
McCPK1-6× His localization in subcellular fractions. Epidermal extracts of leaves from ice plants grown in either 80% (unstressed conditions, US) or 40% (stressed conditions, ST) relative humidity were fractionated into soluble (S), tonoplast (T), endoplasmic reticulum (ER), plasma membrane (PM), and nuclear (N) fractions. Protein extracts prepared from 30 epidermal peels for the lanes labeled S, T, ER, PM, and from 24 epidermal peels for the lane labeled N were separated by SDS-PAGE and detected using different marker-specific antibodies. Anti-6× His tag, anti-V-ATPase, anti-BiP, anti-H+ATPase, and anti-Ran antibodies were used to detect the location of McCPK1-6× His, or tonoplast, ER, PM, and nuclear envelope markers, respectively.
Resetting of McCPK1 Localization under Constant Conditions
The observed change in the subcellular localization of McCPK1-sGFP-DsRed-6× His fusion proteins from the PM to the nucleus and actin cytoskeleton following the imposition of either salinity or low-humidity stress suggests that McCPK1 may play a role as a Ca2+ sensor responder to these environmental stimuli. We hypothesized that, after McCPK1 moves to the nucleus and the cytoskeleton upon the application of stress, it must return to the PM at some later time to sense new environmental changes. This resetting phenomenon is analogous to the prerequisite of repetitive Ca2+ signaling wherein elevated cytosolic Ca2+ concentrations must return rapidly back to a resting state so that cell Ca2+ signaling can be repeated (Trewavas, 1999). Assuming that the default sensing of Ca2+ changes by McCPK1 occurs at the PM or other subcellular locations, such as the peroxisome (Dammann et al., 2003), McCPK1 must at some point reset to the PM so that it can repeat its sensor-responder role. To test this hypothesis, plasmid DNA containing the McCPK1-sGFP fusion gene was introduced into ice plant leaves by microprojectile bombardment and the plants were subjected to 40% relative humidity for 12 h, after which time the bombarded leaves were detached and subjected to constant water, dehydration, or high-salinity (500 mm NaCl) treatment conditions at 40% relative humidity for an additional 12 h. Directly after detachment, transiently transfected cells were scored for subcellular McCPK1-sGFP localization. As expected, in only 38% to 40% of cells did McCPK1-sGFP exhibit a PM subcellular localization pattern. By comparison, in plants held under unstressed conditions (70%–80% relative humidity), McCPK1-sGFP displayed a predominantly PM localization pattern in 88% to 96% of cells (O.R. Patharkar and J.C. Cushman, unpublished data). The same leaves were then resurveyed after 6 and 12 h in water at 40% humidity (Table III), water deficit stress (Table IV), or high-salinity (500 mm NaCl; Table V) stress conditions. After 6 h, PM localization increased to 49% to 62% of cells. After 12 h, the trend toward PM localization increased to 65% to 73% of cells displaying a predominantly PM subcellular localization pattern (Tables III–V). Cochran-Mantel-Haenzel (C-M-H) statistical analysis showed that there were significant differences in the mean frequency of localization of McCPK1-sGFP between the PM and the cytoskeleton/nucleus in the cell populations counted either at 0 or 12 h for each of the treatments. The C-M-H statistic for these treatments was greater than 7.88, the chi-squared distribution value for 1 degree of freedom at 95% confidence level (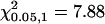 ). However, this observation was not true for the cell populations counted at 6 h of treatment because the C-M-H statistic at this time point was less than 7.88 for the different treatments. These results show that, in leaves exposed to constant conditions, whether held in water at 40% relative humidity, or exposed to dehydration or salinity stress, McCPK1-sGFP changed location from the nucleus, transcytoplasmic ER, and actin cytoskeleton to the PM, ready to repeat the sensor-transducer cycle should another differential stress condition be encountered. Because similar results were obtained under all three conditions, the type of stress treatment used was considered less important than keeping the conditions constant.
). However, this observation was not true for the cell populations counted at 6 h of treatment because the C-M-H statistic at this time point was less than 7.88 for the different treatments. These results show that, in leaves exposed to constant conditions, whether held in water at 40% relative humidity, or exposed to dehydration or salinity stress, McCPK1-sGFP changed location from the nucleus, transcytoplasmic ER, and actin cytoskeleton to the PM, ready to repeat the sensor-transducer cycle should another differential stress condition be encountered. Because similar results were obtained under all three conditions, the type of stress treatment used was considered less important than keeping the conditions constant.
Table III.
Resetting of McCPK1 localization in cells of bombarded ice plant leaves detached and subjected to water treatment at 40% relative humidity
| 0 ha
|
6 h
|
12 h
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma Membrane | Cyto/Nucleus | Total No. Cells | Plasma Membrane | Cyto/Nucleus | Total No. Cells | Percent Relative Change to PM | Plasma Membrane | Cyto/Nucleus | Total No. Cells | Percent Relative Change to PM | |
| Leaf 1 | 42 | 58 | 100 | 42 | 42 | 84 | 11b(±)3 | 72 | 32 | 104 | 29(±)2 |
| Leaf 2 | 37 | 65 | 102 | 67 | 72 | 139 | 58 | 33 | 91 | ||
| Leaf 3 | 42 | 78 | 120 | 43 | 44 | 87 | 63 | 31 | 94 | ||
| C-M-H χ2 = 12.04c | C-M-H χ2 = 4.44 | C-M-H χ2=14.79 | |||||||||
The post-treatment time is indicated (0, 6, and 12 h).
The relative shift in localization from the cytoskeleton/nucleus to the PM is calculated as the difference in the mean percentage of the PM-localized signal at 6 or 12 h and PM-localized signal at 0 h for each treatment with sd indicated.
The C-M-H statistic for significant differences in localization at different treatments and time points is shown.  .
.
Table IV.
Resetting of McCPK1 localization in cells of bombarded ice plant leaves detached and subjected to water deficit stress treatment at 40% relative humidity
| 0 ha
|
6 h
|
12 h
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma Membrane | Cyto/Nucleus | Total No. Cells | Plasma Membrane | Cyto/Nucleus | Total No. Cells | Percent Relative Change to PM | Plasma Membrane | Cyto/Nucleus | Total No. Cells | Percent Relative Change to PM | |
| Leaf 1 | 42 | 58 | 100 | 46 | 36 | 82 | 18(±)5b | 65 | 20 | 85 | 34(±)2 |
| Leaf 2 | 33 | 58 | 91 | 50 | 45 | 95 | 64 | 24 | 88 | ||
| Leaf 3 | 37 | 55 | 92 | 50 | 51 | 101 | 64 | 25 | 89 | ||
| C-M-H χ2 = 13.44c | C-M-H χ2 = 0.01 | C-M-H χ2 = 17.09 | |||||||||
The post-treatment time is indicated (0, 6, and 12 h).
The relative shift in localization from the cytoskeleton/nucleus to the PM is calculated as the difference in the mean percentage of the PM-localized signal at 6 or 12 h and PM-localized signal at 0 h for each treatment with sd indicated.
The C-M-H statistic for significant differences in localization at different treatments and time points is shown. 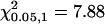 .
.
Table V.
Resetting of McCPK1 localization in cells of bombarded ice plant leaves detached and subjected to 500 mm NaCl stress treatment at 40% relative humidity
| 0 ha
|
6 h
|
12 h
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma Membrane | Cyto/Nucleus | Total No. Cells | Plasma Membrane | Cyto/Nucleus | Total No. Cells | Percent Relative Change to PM | Plasma Membrane | Cyto/Nucleus | Total No. Cells | Percent Relative Change to PM | |
| Leaf 1 | 43 | 61 | 104 | 49 | 32 | 81 | 22(±)3b | 43 | 21 | 64 | 24(±)2 |
| Leaf 2 | 40 | 67 | 107 | 58 | 33 | 91 | 58 | 32 | 90 | ||
| Leaf 3 | 32 | 46 | 78 | 55 | 35 | 90 | 55 | 31 | 86 | ||
| C-M-H χ2 = 10.08c | C-M-H χ2 = 0.01 | C-M-H χ2 = 10.89 | |||||||||
The post-treatment time is indicated (0, 6, and 12 h).
The relative shift in localization from the cytoskeleton/nucleus to the PM is calculated as the difference in the mean percentage of the PM-localized signal at 6 or 12 h and PM-localized signal at 0 h for each treatment with sd indicated.
The C-M-H statistic for significant differences in localization at different treatments and time points is shown. 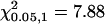 .
.
The resetting of McCPK1 back to the PM following application of constant conditions ensures that the cell can react again to the same or different stresses. Returning plants exposed to low-humidity conditions back to high-humidity conditions results in a reversal of McCPK1 localization from the nucleus, the transcytoplasmic ER, and the actin microfilaments to the PM within 2 to 4 h (O.R. Patharkar and J.C. Cushman, unpublished data). However, resetting can also occur to a more limited, yet significant, extent in leaves held under constant conditions of 40% humidity, high-salinity, or water deficit stress treatments (Tables III–V). The resetting of McCPK1 might be due to new protein synthesis of McCPK1 moving along the cytoskeletal structures to reach the PM, or due to movement of preexisting protein back to the PM via the cytoskeleton. Future experimentation will focus on discerning these possibilities.
MATERIALS AND METHODS
Plant Growth Conditions and Stress Treatments
Common ice plant (Mesembryanthemum crystallinum) seeds were germinated in Metromix 200 (Scotts Sierra Horticultural Products, Marysville, OH) in a growth chamber on a 12-h light (26°C)/12-h dark (18°C) cycle. Fluorescent and incandescent lighting provided a photon flux density of 450 to 500 μE m−2 s−1. Ten-day-old seedlings were transplanted individually to 1-L styrofoam pots containing Metromix 200 and irrigated once daily with 0.5× Hoagland solution. Leaves from 5-week-old plants were used for detached leaf assays according to Taybi and Cushman (1999). Placing the detached leaves in Magenta boxes without nutrient solution imposed drought treatment. Transferring the detached leaves to 400 mm NaCl imposed ionic stress. Plant material was harvested at various times after the start of various stress treatments (as indicated in figure legends), frozen in liquid nitrogen, and stored at −80°C until use.
cDNA Library Construction and Screening
A lambda Uni-ZapXR cDNA expression library (300,000 pfu) prepared from leaf tissue of plants exposed to 0.5 m NaCl stress for 30 h, as described previously (Forsthoefel et al., 1995), was screened with a 807-bp cDNA CDPK probe obtained by RT-RCR using degenerate primers corresponding to the subdomain VIb (RDLKPEN) of the protein Ser/Thr kinase catalytic domain (Hanks and Hunter, 1995; 5′-GNGATCTNAAGCTNGAAAA-3′), and a region between EF hand 1 and EF hand 2 of the calmodulin-like domain (5′-CN(G/C)(A/T)NCC(G/A)TC(T/C)TT(G/A)TC(G/A)AA-3′), respectively. Positively hybridizing plaques were excised and propagated as plasmids according to the manufacturer's instructions (Stratagene, La Jolla, CA). Plasmids having the largest inserts were identified by restriction analysis and sequenced.
Isolation and Sequence Analysis of McCpk1
One putative CDPK clone (designated CH 1) having the largest insert was completely sequenced and found to lack an in-frame initiation codon. To recover a full-length cDNA of McCpk1, the 5′ RACE system (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions, using gene-specific primer, GSP1 (5′-TGTAGTACACCTTCACATCCTC-3′) and anchoring primer (5′-(CUA)4GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGII-3′). Total RNA was isolated as previously described (Taybi and Cushman, 1999). The identity of 5′ RACE products was confirmed by DNA sequence analysis. The nucleotide sequence of both strands of the full-length cDNA (accession no. AF090835) was determined using the dideoxy chain-termination method on an Applied Biosystems 373A automated DNA sequencing system using the Prism Ready Reaction Dyedeoxy Terminator Cycle Sequencing kit (Applied Biosystems Division, Perkin-Elmer, Foster City, CA).
DNA sequence data were analyzed using the MacVector/AssemblyLIGN sequence analysis programs (Accelrys, San Diego). Database searches were conducted using the National Center for Biotechnology Information network version of BLAST 2.0 (Altschul et al., 1997). Multiple sequence alignments were conducted with the ClustalX multiple alignment program (Thompson et al., 1997).
Plasmid Constructs for Recombinant Proteins
The vector pET-30b(+) (Novagen, Madison, WI) was used to express the full-length McCPK1 fusion construct having a 6× His tag on its C terminus. The McCpk1 sequence was amplified by PCR using the following primers (5′-AATTCTAGACTTTAAGAAGGAGATATACATATGGGGATTTGTGCTAGC-3′ and 5′-ATACTCGAGAATGACTTTGGTAACCGGG-3′). The amplified DNA was digested with XhoI and XbaI and subcloned into the XhoI/XbaI site of the pET-30b(+) vector. The cloned fragment was confirmed to be error free by DNA sequencing.
The vector pET-30b(+) (Novagen) was used to express the myristoylation mutant of McCPK1 fusion construct having 6× His tag on its C terminus (McCPK1-6× His). The McCpk1 sequence was amplified by PCR using Taq DNA polymerase and the following primers: 5′-AATTCTAGACTTTAAGAAGGAGATATACATATGGCGATTTGTGCTAGC-3′ and 5′-ATACTCGAGAATGACTTTGGTAACCGGG-3′ and cloned as above.
The three different constructs expressing McCPK1 mutants, S62A, S420A, and S62A/S420A, were prepared using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Amplification of the plasmid-expressing S62A McCPK1-6× His was obtained using wild-type McCpk1-6× His DNA template and the following primers: 5′-CCATTAGGATGCCCGCCCCAAAACCAGCCCC-3′ and 5′-GGGGCTGGTTTTGGGGCGGGCATCCTAATGG-3′. Plasmids expressing S420A McCPK1-6× His and S62A/S420A McCPK1-6× His were obtained using the following primers: 5′-GATTGGCTCGACTTGGAGCAAAGCTTACTGAAACTG-3′ and 5′-CAGTTTCAGTAAGCTTTGCTCCAAGTCGAGCCAATC-3′. For the single-mutant S420A McCpk1-6× His, wild-type McCpk1-6× His DNA was used as a template, whereas for the double-mutant S62A/S420A McCpk1-6× His the mutant construct S62A McCpk1-6× His was used as the template. All site-directed mutation constructs were confirmed to be correct by sequence analysis before and after conducting kinase activity measurements.
RNA Isolation and Semiquantitative RT-PCR Assays
Semiquantitative RT-PCR assays were conducted as previously described (Taybi and Cushman, 1999) using 1.25 μg or 91 ng of DNase I-treated RNA for McCpk1 and Fnr1 products, respectively. A 850-bp amplicon was obtained using McCpk1-specific primers: 5′-AAACTACCACCAGCAGCC-3′ (forward) and 5′-TTGAGCAGCAGTAATCCG-3′ (reverse). A 600-bp amplicon was obtained using Fnr1-specific primers: 5′-ATTGCCAGCAGGCCCTTG-3′ (forward) and 5′-GAACCAGTCAATACCATCT-3′ (reverse). RT-PCR reaction was conducted in a 25-μL reaction containing 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 2.5 μm MgCl2, 10 mm dithiothreitol (DTT), 100 μm each dNTP, 400 nm forward primer, 400 nm reverse primer, 20 units of RNase Out (Invitrogen), 0.5 unit of Taq DNA polymerase (Fisher Scientific, Pittsburgh), and either of RNA for McCpk1 or Fnr1 amplification, respectively. Reactions were conducted using a PTC-200 thermal cycler (MJ Research, Watertown, MA) at 50°C for 30 min, at 94°C for 2 min to denature the RT, followed by either 21 PCR cycles for Fnr1 or 30 PCR cycles for McCpk1: 95°C for 1 min, 94°C for 15 s, either 50°C for Fnr1 or 57°C for McCpk1 for 30 s, 72°C for 45 s, and 5 min of extension step at 72°C. After amplification, the reaction products were resolved by electrophoresis on the same 1.2% (w/v) agarose gel and stained with ethidium bromide. Images were captured using a Gel-Doc 1000 DNA Gel Analysis and Documentation system (Bio-Rad Laboratories, Hercules, CA). All semiquantitative RT-PCR experiments were repeated twice and representative data were shown.
Expression and Purification of McCPK1
Transformed Escherichia coli cells (BL21) expressing a wild-type or mutated version of McCPK1 were grown overnight at 37°C in 2 mL of Luria-Bertani/kanamycin medium (50 μg/mL), transferred to 250 mL of Luria-Bertani/kanamycin medium, and cultured until OD600 was 0.7. Expression was induced by the addition of 1 mm isopropyl thio-β-d-galactoside for 1 h. Cells were harvested, resuspended in ice-cold B-PER lysis buffer (Pierce, Rockford, IL), and proteins were collected as specified by the manufacturer. The fusion protein was purified by affinity chromatography with nickel resin (Novagen) that had been prepared by prerunning it with charge buffer (50 mm NiSO4) and then with binding buffer (5 mm imidazole, 0.5 m NaCl, 20 mm Tris-HCl, pH 7.9). After loading the column with the recombinant protein, it was washed with wash buffer (60 mm imidazole, 0.5 mm NaCl, 20 mm Tris-HCl, pH 7.9) and eluted with elution buffer (1 m imidazole, 0.5 m NaCl, 20 mm Tris-HCl, pH 7.9). Protein concentrations were determined by Bradford assays. The purity of all recombinant proteins was judged to be within ±20% of the wild-type control.
Myristoylation Analysis of McCPK1
The cDNAs encoding either McCPK1 or L1R, a vaccinia viral protein known to be myristoylated (Franke et al., 1990), in pET30b or pBR322, respectively, were transcribed under the control of the bacteriophage T7 promoter and translated in a T7 TNT Coupled Transcription/Translation Wheat Germ Extract system, as specified by manufacturer (Promega, Madison, WI). Radiolabeling was performed with [35S]Met (50 μCi of 1194 Ci/mm; Amersham-Pharmacia Biotech, Piscataway, NJ), [3H]Myristic acid (48 μCi of 55 Ci/mm; Amersham-Pharmacia Biotech), or [3H]Palmitic acid (48 μCi of 55 Ci/mm; Amersham-Pharmacia Biotech). Labeled translation products were resolved by electrophoresis on a 12% SDS-polyacrylamide gel. The gels were fixed for 30 min in 10% (v/v) acetic acid and 30% (v/v) methanol, rinsed briefly in distilled water, then treated with Enlightning Autoradiography Enhancer (NEN Life Science Products, Boston), dried, and subjected to autofluorography at −70°C using Biomax MS x-ray Film (Eastman-Kodak, Rochester, NY). Anti-6× His-tag monoclonal antibodies (BabCO, Richmond, CA) were used at a 1:1,000 dilution in 1× Tris-buffered saline (TBS; 20 mm Tris-HCl, pH 7.6, 137 mm NaCl) to immunoprecipitate the [35S]- and [3H]-labeled McCPK1-6× His-tag fusion proteins. Antigen-antibody complexes were collected with protein A agarose beads (Santa Cruz Biotech, Santa Cruz, CA) and washed as specified by the manufacturer, resolved on a 12% SDS-polyacrylamide gel, and visualized by autofluorography.
Kinase Activity Assays
The protein kinase activity of the fusion protein (McCPK1-6× His) was assayed in a 50-μL reaction mixture containing 50 mm HEPES (pH 7.5), 100 mm KCl, 2.5 mm MgCl2, 0.25 mm EGTA, 0.275 mm CaCl2, 0.1 mg mL−1 bovine serum albumin, various concentrations of histone H1 (product no. 38205; Calbiochem, San Diego) as indicated in Figure 2, and 40 ng of recombinant McCPK1. Histone H1 was used at a concentration of 30 μm as indicated in Table II. Reactions were initiated by the addition of 10 μCi of [γ-32P] ATP (60 μm final concentration) and incubated for 15 min at 30°C. Reactions were stopped by spotting 10 μL aliquot on P81 paper (Whatman, Clifton, NJ) and washed three times with 150 mm H3PO4, once with 95% ethanol, and once with ethyl ether. To determine Ca2+ dependence, the above assay buffer was supplemented with 0.25 mm EGTA and the amount of free Ca2+ was calculated by simple equilibrium calculations from the amount of CaCl2 supplied. After incubation for 15 min at 30°C, adding warm SDS sample buffer and boiling for 5 min stopped the reactions. Then, the proteins were separated by 12% SDS-PAGE and stained with the biosafe GelCode Blue Stain Reagent (Pierce) as specified by the manufacturer. The gel was dried and exposed directly to Hyperfilm ECL X-Ray film (Amersham-Pharmacia Biotech) for 12 to 18 h.
For quantitative kinase assays, the reactions were stopped by spotting 10 μL aliquot on P81 paper (Whatman) and washed three times with 150 mm H3PO4, three times with 70% ethanol, and once with ethyl ether. Filters were dried and counted in EcoLume Scintillation Liquid (ICN, Irvine, CA) in a liquid scintillation counter (Liquid Scintillation Systems LS 3801, Beckman, Fullerton, CA). Specific activities reported are the average of three replicate assays with each of four replicate enzyme preparations. Data are represented as the mean ± se of specific activity. Kinase activity assays contained 50 ng recombinant McCPK1 kinase with or without (to assay autophosphorylation) 30 μm histone substrate. ANOVA was performed separately for assays in the presence or absence of calcium, followed by multiple comparison of means by the Tukey's honestly significant difference mean-separation test, using JMP software (SAS, Cary, NC).
Determination of McCPK1 Autophosphorylation Sites
Purified, recombinant McCPK1-6× His fusion protein (30 μg) was autophosphorylated in kinase buffer containing (50 mm Tris, pH 7.5, 10 mm MgCl2, 1 mm EGTA, 1.2 mm CaCl2) with 1 mm ATP at room temperature (approximately 22°C) for 24 h. The protein concentration was kept at 1 mg mL−1 to improve stability of reaction components during the long reaction time. The kinase (30 μg) was pelleted with acetone, dissolved in 8 m urea, then diluted to 1.5 m urea with 50 mm ammonium bicarbonate, pH 7.5, 1 mm DTT. Proteolysis was initiated by the addition of 0.5 μg of sequencing-grade trypsin (Promega) and incubated at room temperature for 14 h. The entire digest was subjected to purification using a C18 Zip Tip (Millipore, Bedford, MA) prior to being loaded onto a two-dimensional (SCX/C18) microcapillary column. Three C18 eluting gradients were performed following the loading of increasing concentrations of 250 mm and then 1 m ammonium acetate. The phosphopeptides in question were eluted using 250 mm ammonium acetate. Samples were analyzed on a Micromass Q-TOF2 mass spectrometer (Waters, Milford, MA) equipped with an in-house liquid chromatography-nanoelectrospray source.
Transient Transfection and Fluorescent Microscopy
The McCPK1-sGFP and DsRed constructs were prepared as described previously (Patharkar and Cushman, 2000). The G to A mutant at position 2 for McCPK1-sGFP was generated using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol, as described above using wild-type McCPK1-sGFP plasmid DNA as template and the following primers: Forward primer: 5′-ATAGTCGACAAGATGGCGATTTGTGCT-3′ and reverse primer: 5′-ATACTGCAGTTACTTGTACAGCTCGTCCATGC-3′. The deletion amino acids 1–70 construct was created by the PCR reaction using Taq DNA polymerase and forward primer: 5′-ATAGTCGACAAGATGGGTAAACCCTTTGAGG-3′ and reverse primer: 5′-ATAAGATCTTGCTCCTGCTCCTGCTCCTGCTCCTGCTCCAATGAGTTTGGTAACCGGG-3′. The amplification product was digested with SalI/BglII and inserted into the SalI/BamHI site of the 35S-sGFP-TYG-nos (pUC19) vector kindly provided by Jen Sheen (Massachusetts General Hospital, Boston), resulting in the creation of a McCPK1-sGFP fusion wherein the truncated McCPK1 and sGFP proteins were separated by a flexible (GA)5 repeat linker. The construct expressing AHA2-GFP (35S-AHA2(D>A)-GFP-TAP2) was kindly supplied by Dr. Jeffery Harper (Scripps Research Institute, La Jolla, CA). The construct expressing GFP-tubulin (pUC-35S-GFP-α-tubulin) was kindly supplied by Seiichiro Hasezawa (University of Tokyo, Kashiwa, Japan), the ER-targeted BiP-GFP (35S-BiP-GFP) construct was kindly supplied by Inhan Hwang (Gyeongsang National University, Kyungbuk, Korea), and the GFP-talin construct (PYSC14) was kindly supplied by Nam-Hai Chua (Rockefeller University, New York). Five-week-old ice plants were bombarded with a hand-held Helios Gene Gun (Bio-Rad Laboratories) according to the manufacturer's instructions, except that after DNA was precipitated onto gold particles, the gold was washed five times with absolute ethanol. Each shot fired contained 0.2 mg of 1.6-μm gold particles. A force of 1,034 kPa was used to deliver the gold particles to leaves on intact plants. A diffuser screen was used to minimize tissue damage. Cobombardment of McCPK1-DsRed and each of tubulin-GFP, talin-GFP, and BiP-GFP constructs was accomplished by coating the 0.2 mg of gold with 0.8 μg McCPK1-DsRed and 0.2 μg of the GFP constructs. Plants were grown under unstressed conditions in an environment that maintained 80% relative humidity. To stress the plants, they were removed from this high-humidity environment, bombarded, and then placed in a lower humidity environment (approximately 40% relative humidity) for 13 h. Whole leaves were excised from plants and individual cells visualized with a Nikon Eclipse E400 epifluorescence microscope (Nikon, Tokyo). GFP was visualized with a 450- to 490-nm excitation filter, a 495-nm dichroic mirror, and a 500- to 550-nm emission filter. DsRed was visualized with a 528- to 553-nm excitation filter, a 565-nm dichroic mirror, and a 600- to 660-nm emission filter.
Cell Fractionation
The construct expressing McCPK1-6× His in pET30b was excised and cloned between the XhoI and SacI sites of the vector p35MCS kindly provided by Nam-Hai Chua (Rockefeller University, New York), thus putting the construct under the control of 35S promoter. Five-week-old plants were bombarded with the construct expressing McCPK1-6× His. Each shot was composed of 0.2 mg of 1.6 μm gold particles and 1 μg of plasmid. The epidermal peels from 30 shots of the bombarded area were harvested and homogenized in 2 mL of ice-cold homogenization buffer. The homogenization buffer consisted of 30 mm Tris, 50% (w/v) bovine serum albumin, 10% (v/v) glycerol, 0.25 mm butylated hydroxytoluene, 1.0 mm phenylmethylsulfonyl fluoride, 5.0 mm EGTA, 5.0 mm MgSO4, 0.25 mm mannitol, 2.0 mm K+-metabisulfite, 0.25 mm dibucaine, 1.0 mm benzamidine, and 5% (w/v) soluble PVP (PVP-10) adjusted to pH 8.0 with H2SO4. All subsequent steps were carried out at 4°C. Membrane fractions were isolated and purified as described by Barkla et al. (1995). The microsomal portion was fractionated through a discontinuous Suc gradient, the tonoplast and the PM were collected at the 0 to 31% and the 31% to 38% (w/v) Suc interfaces, respectively. The ER fraction was collected at the 38% to 44% interface. After fractionation, membranes were frozen in liquid nitrogen and stored at −80°C in 30-μL aliquots.
For nuclei isolation, 24 bombarded epidermal peels were homogenized in 2 mL of ice-cold nuclei isolation buffer (NIB; 250 mm Suc, 10 mm [MES]-KOH, pH 5.4, 10 mm NaCl, 10 mm spermine, 0.5 mm spermidine, 10 mm 2-mercaptoethanol) until completely disrupted. All subsequent manipulations were conducted at 4°C. Homogenate was filtered through two layers of Miracloth and then through a series of nylon meshes of decreasing size (54 μm, then 25 μm). Ten percent (v/v) Triton X-100 was gradually added dropwise to the filtrate to a final concentration of 0.4% (v/v). The resulting solution was incubated on ice for 5 min to solubilize the chloroplast membranes. The lysate was overlaid onto a two-step gradient consisting of 25% to 75% Percoll prepared in NIB without Triton X-100, and was centrifuged at 2,000g for 20 min in a swinging bucket rotor (Sorvall HB-4; Sorvall Products, Newtown, CT). Using a serological pipette, the nuclei, which appeared as a white fluffy band at the 25% to 75% Percoll interface, were carefully removed, resuspended with three volumes of NIB, and centrifuged at 1,000g for 10 min in a swinging bucket rotor (Sorvall HB-4), then resuspended again in 2 mL of NIB and centrifuged at 1,000g for 10 min to remove residual Percoll. The final nuclear pellet was resuspended in 50 μL of nuclei storage buffer, aliquoted, frozen and stored at −80°C. Nuclear storage buffer consisted of 20% (v/v) glycerol, 20 mm (HEPES)-KOH, pH 7.2, 5 mm MgCl2, 1 mm DTT, and 10% Triton X-100 (Cushman, 1995).
Western-Blot Analysis
After the fractions were isolated from leaf tissue as previously described, they were run and separated by 12% SDS-PAGE and analyzed by western-blot analysis using the appropriate antibodies and a western-Light Plus kit (Applied Biosystems). Anti- H+-ATPase antibody was used to identify the PM fraction (1:5,000 dilution in TBS, kindly provided by Dr. Serrano, Camino de Vera, Valencia, Spain; Pardo and Serrano, 1989). Anti-V-ATPase subunit A antibody was used to identify the tonoplast fraction (1:500 dilution in TBS, kindly provided by Dr. Ratajczak, Technische Hochschule Darmstadt, Darmstadt, Germany; Zhigang et al., 1996). Anti-NPC antibody was used to target nuclei (1:200 dilution in TBS; BabCO, Richmond, CA). Anti-Hsc70 (BiP; StressGen, Victoria, Canada) was used to identify the ER fraction at a dilution of 1 μg of antibody in 1 mL of TBS. Anti-6× His antibody (BabCO) was used to detect McCPK1-6× His (1:1,000 dilution in TBS).
Availability of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial purposes.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AF090835, AAB03242, and AF090835.
Acknowledgments
The authors thank Dr. Jeff Harper for providing us with the AHA2-GFP construct, Dr. Inhan Hwang for providing the BiP-GFP construct, Dr. Seiichiro Hasezawa for providing the tubulin-GFP construct, Dr. Nam-Hai Chua for providing the p35MCS and talin-GFP constructs, and Dr. Jen Sheen for providing us with the 35S-sGFP-TYG-nos construct. The authors also thank Alice Harmon for helpful discussions and the Nevada Genomics Center for providing automated DNA sequencing services.
This work was supported in part by the U.S. Department of Agriculture, National Research Initiative, Competitive Grants Program (grant no. 98–35100–6035 to J.C.C), by the National Science Foundation (grant no. MCB–0114769), and by the Nevada Agricultural Experiment Station (publication no. 03042812 of the Nevada Agricultural Experiment Station).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.035238.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M (1997) Molecular mechanics of calcium-myristoyl switches. Nature 389: 198–202 [DOI] [PubMed] [Google Scholar]
- Anderson JV, Haskell DW, Guy CL (1994. a) Differential influence of ATP on native spinach 70-kilodalton heat-shock cognates. Plant Physiol 104: 1371–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JV, Haskell DW, Guy CL (1994. b) Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol 104: 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anil VS, Harmon AC, Rao KS (2003) Ca2+-dependent protein kinase in oil bodies: its biochemical characterization and significance. Plant Cell Physiol 44: 367–376 [DOI] [PubMed] [Google Scholar]
- Anil VS, Rao KS (2001) Purification and characterization of a Ca2+-dependent protein kinase from sandalwood (Santalum album L.): evidence for Ca2+-induced conformational changes. Phytochemistry 58: 203–212 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo B-C, Harmon AC, Huber SC (1996) Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell 8: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baizabal-Aguirre VM, de la Vara LEG (1997) Purification and characterization of a calcium-regulated protein kinase from beet root (Beta vulgaris) plasma membranes. Physiol Plant 99: 135–143 [Google Scholar]
- Barkla BJ, Zingarelli L, Blumwald E, Smith JAC (1995) Tonoplast Na+/H+ antiport activity and its energization by the vacuolar H+-ATPase in the halophytic plant M. crystallinum L. Plant Physiol 109: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Cruz SS, Martin B, Betterridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Boisson B, Giglione C, Meinnel T (2003) Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J Biol Chem 278: 43418–43429 [DOI] [PubMed] [Google Scholar]
- Botella JR, Arteca JM, Somodevilla M, Arteca RN (1996) Calcium-dependent protein kinase gene expression in response to physical and chemical stimuli in mungbean (Vigna radiata). Plant Mol Biol 30: 1129–1137 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Seal A, DasGupta M (1999) Autophosphorylation-dependent activation of a calcium-dependent protein kinase from groundnut. Plant Physiol 104: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-H, Sheen J, Gerrish C, Bolwell PG (2001) Molecular identification of phenylalanine ammonia-lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS Lett 503: 185–188 [DOI] [PubMed] [Google Scholar]
- Cheng S-H, Willman MR, Chen H-C, Sheen J (2002) Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Raices M, Tellez-Inon MT, Ulloa RM (2002) A calcium dependent protein kinase is systemically induced upon wounding in tomato plants. Plant Physiol 128: 256–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Cushman JC (1995) Isolation of nuclei suitable for in vitro transcriptional studies. In D Galbraith, D Bourque, H Bohnert, eds, Methods in Cell Biology: Plant Cell Biology, Part B, Vol 50. Academic Press, Inc., San Diego, CA, pp 113–128 [DOI] [PubMed]
- Dammann D, Ichida A, Hong B, Romanowsky S, Hrabak EM, Harmon AC, Pickard BG, Harper JF (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellard-Ivey M, Hopkins RB, White TJ, Lomax TL (1999) Cloning, expression and N-terminal myristoylation of CpCPK1, a calcium-dependent protein kinase from zucchini (Cucurbita pepo L.). Plant Mol Biol 39: 199–208 [DOI] [PubMed] [Google Scholar]
- Forsthoefel NR, Cushman MAF, Cushman JC (1995) Post-transcriptional and post-translational control of enolase expression in the facultative CAM plant, Mesembryanthemum crystallinum. Plant Physiol 108: 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke CA, Wilson EM, Hruby DE (1990) Use of a cell-free system to identify the vaccinia virus L1R gene product as the major late myristoylated virion protein M25. J Virol 64: 5988–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto T, Ogawa N, Hata S, Izui K (1996) Plant calcium-dependent protein kinase-related kinases (CRKs) do not require calcium for their activities. FEBS Lett 396: 147–151 [DOI] [PubMed] [Google Scholar]
- Glinski M, Romeis T, Witte CP, Wienkoop S, Weckwerth W (2003) Stable isotope labeling of phosphopeptides for multiparallel kinase target analysis and identification of phosphorylation sites. Rapid Commun Mass Spectrom 17: 1579–1584 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151: 175–183 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF (2000) CDPK's – A kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Yoo BC, McCaffery C (1994) Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7278–7287 [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang JF, Lloyd SJ (1994) Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7267–7277 [DOI] [PubMed] [Google Scholar]
- Heuckeroth RO, Towler DA, Adams SP, Glaser L, Gordon JI (1988) 11-(Ethylthio)undecanoic acid. A myristic acid analogue of altered hydrophobicity which is functional for peptide N-myristoylation with wheat germ and yeast acyltransferase. J Biol Chem 263: 2127–2133 [PubMed] [Google Scholar]
- Hrabak EM (2000) Calcium-dependent protein kinases and their relatives. Adv Bot Res 32: 185–223 [Google Scholar]
- Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford H, Kudla J, Luan S, Nimmo HG, Sussman MR, et al (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-Z, Hardin SC, Huber SC (2001) Identification of a novel phosphorylation motif for CDPKs: phosphorylation of synthetic peptides lacking basic residues at P-3/P-4. Arch Biochem Biophys 393: 61–66 [DOI] [PubMed] [Google Scholar]
- Huang J-Z, Huber SC (2001) Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase. Influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol 42: 1079–1087 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sze H, Harper JF (2000) A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc Natl Acad Sci USA 97: 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Kuriyama M, Nakakita M, Kojima H, Ohto M, Nakamura K (1998) Characterization of a calcium-dependent protein kinase of tobacco leaves that is associated with the plasma membrane and is inducible by sucrose. Plant Cell Physiol 39: 1176–1183 [DOI] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Li W, Konishi H, Yoshikawa M, Konishi T, Yang G (2001) Characterization of a calcium-dependent protein kinase from rice root: differential response to cold and regulation by abscisic acid. Biol Pharm Bull 24: 1316–1319 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kumagai F, Yoneda A, Tomida T, Sano T, Nagta T, Hasezawa S (2001) Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP-tubulin: time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant Cell Physiol 42: 723–732 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yoo BC, Harmon AC (1998) Kinetic and calcium-binding properties of three calcium dependent protein kinase isoenzymes from soybean. Biochemistry 37: 6801–6809 [DOI] [PubMed] [Google Scholar]
- Lee SS, Cho HS, Yoon GM, Ahn J-W, Kim H-H, Pai H-S (2003) Interaction of NtCDPK1 calcium-dependent protein kinase with NtRpn3 regulatory subunit of the 26S proteasome in Nicotiana tabacum. Plant J 33: 825–840 [DOI] [PubMed] [Google Scholar]
- Li J, Lee YR, Assmann SM (1998) Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol 116: 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindzen E, Choi JH (1995) A carrot cDNA encoding an atypical protein kinase homologous to plant calcium-dependent protein kinases. Plant Mol Biol 28: 785–797 [DOI] [PubMed] [Google Scholar]
- Llop-Tous I, Dominquez-Puigjaner E, Vendrell M (2002) Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during ripening and affected by low temperature. J Exp Bot 53: 2283–2285 [DOI] [PubMed] [Google Scholar]
- Lu SX, Hrabak E (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128: 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín ML, Busconi L (2000) Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J 24: 429–435 [DOI] [PubMed] [Google Scholar]
- Matsuda S, Hisatomi O, Ishino T, Kobayashi Y, Tokunaga F (1998) The role of calcium-binding sites in S-modulin function. J Biol Chem 273: 20223–20227 [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW (1997) The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA 94: 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Gerace L (1998) Two-way trafficking with Ran. Trends Cell Biol 8: 175–179 [DOI] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L (1993) Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol 123: 1649–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski CB, Schmitt JM, Bohnert HJ (1989) Expression during salt stress and nucleotide sequence of cDNA for ferredoxin-NADP+ reductase from Mesembryanthemum crystallinum. Plant Physiol 89: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G (1993) The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365: 661–663 [DOI] [PubMed] [Google Scholar]
- Murray D, Ben-Tal N, Honig B, McLaughlin S (1997) Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure 5: 985–989 [DOI] [PubMed] [Google Scholar]
- Murillo I, Jaeck E, Cordero MJ, San Segundo B (2001) Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitors and infection. Plant Mol Biol 45: 145–158 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Fiol DF, Salerno GL (2002) A CDPK type protein kinase is involved in rice SPS light modulation. Physiol Plant 115: 183–189 [DOI] [PubMed] [Google Scholar]
- Pardo J, Serrano R (1989) Structure of a plasma membrane H+ATPase gene from the plant Arabidopsis thaliana. J Biol Chem 264: 8557–8562 [PubMed] [Google Scholar]
- Patharkar OR, Cushman JC (2000) A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J 24: 679–692 [DOI] [PubMed] [Google Scholar]
- Permyakov SE, Cherskaya AM, Senin I, Zargarov AA, Shulga-Morskoy SV, Alekseev AM, Zinchenko DV, Lipkin VM, Philippov PP, Uversky VN, et al (2000) Effects of mutations in the calcium-binding sites of recoverin on its calcium affinity: evidence for successive filling of the calcium binding sites. Protein Eng 13: 783–790 [DOI] [PubMed] [Google Scholar]
- Pestenácz A, Erdei L (1996) Calcium-dependent protein kinase in maize and sorghum induced by polyethylene glycol. Physiol Plant 97: 360–364 [Google Scholar]
- Putnam-Evans C, Harmon AC, Palevitz BA, Fechheimer M, Cormier MJ (1989) Calcium-dependent protein kinase is localized with F-actin in plant cells. Cell Motil Cytoskeleton 12: 12–22 [Google Scholar]
- Raíces M, Chico JM, Tellez-Inon MT, Ulloa RM (2001) Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development. Plant Mol Biol 46: 591–601 [DOI] [PubMed] [Google Scholar]
- Rees DJ, Ades SE, Singer SJ, Hynes RO (1990) Sequence and domain structure of talin. Nature 347: 685–689 [DOI] [PubMed] [Google Scholar]
- Resh MD (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Roberts DM, Harmon AC (1992) Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol 43: 375–414 [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JDG (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JDG (2000) Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE (2001) Unraveling response-specificity in Ca2+ signaling pathways in plant cells. New Phytol 151: 7–33 [DOI] [PubMed] [Google Scholar]
- Rutschmann F, Stalder U, Piotrowski M, Oecking C, Schaller A (2002) LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol 129: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P, Singh M (1995) Characterization of a winged bean (Phosphoocarpus tetragonolobus) protein-kinase with calmodulin-like domain regulation by autophosphorylation. Biochem J 305: 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23: 319–327 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: 401–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Oecking C (1999) Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen responses in tomato plants. Plant Cell Physiol 39: 895–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller EG, Harmon AC, Sussman MR (1992) Characterization of a calcium and lipid-dependent protein kinase associated with the plasma membrane of oat. Biochemistry 31: 1721–1727 [DOI] [PubMed] [Google Scholar]
- Senin II, Fischer T, Komolov KE, Zinchenko DV, Philippov PP, Koch KW (2002) Ca2+-myristoyl switch in the neuronal calcium sensor recoverin requires different functions of Ca2+-binding sites. J Biol Chem 277: 50365–50372 [DOI] [PubMed] [Google Scholar]
- Senin II, Vaganova SA, Weiergraber OH, Ergorov N, Philippov PP, Koch KW (2003) Functional restoration of the Ca2+-myristoyl switch in a recoverin mutant. J Mol Biol 330: 409–418 [DOI] [PubMed] [Google Scholar]
- Shahinian S, Silvius JR (1995) Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry 34: 3813–3822 [DOI] [PubMed] [Google Scholar]
- Sheen J (1996) Ca2+-dependent protein kinases and signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J 8: 777–784 [DOI] [PubMed] [Google Scholar]
- Spilker C, Gundelfinger ED, Braunewell KH (2002) Evidence for different functional properties of the neuronal calcium sensor proteins VILIP-1 and VILIP-3: from subcellular localization to cellular function. Biochim Biophys Acta 1600: 118–127 [DOI] [PubMed] [Google Scholar]
- Taybi T, Cushman JC (1999) Signaling events leading to Crassulacean acid metabolism induction in the common ice plant. Plant Physiol 121: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas A (1999) How plants learn. Proc Natl Acad Sci USA 96: 4216–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa RM, Raíces M, MacIntosh GC, Maldonado S, Tellez-Inon MT (2002) Jasmonic acid affects plant morphology and calcium-dependent protein kinase expression and activity in Solanum tuberosum. Physiol Plant 115: 417–427 [DOI] [PubMed] [Google Scholar]
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K (1994) Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol Gen Genet 224: 331–340 [DOI] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Bohnert HJ, Pantoja O (1999) Salt stress in Mesembryanthemum crystallinum L. cell suspensions activates adaptive mechanisms similar to those observed in the whole plant. Planta 207: 426–435 [DOI] [PubMed] [Google Scholar]
- Verhey SD, Gaiser JC, Lomax TL (1993) Protein kinases in zucchini: characterization of calcium-requiring plasma membrane kinases. Plant Physiol 103: 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart V, Christodoulou J, Huang JF, Chazin WJ, Harper JF (2000) Intramolecular activation of a Ca2+-dependent protein kinase is disrupted by insertions in the tether that connects the calmodulin-like domain to the kinase. Biochemistry 39: 4004–4011 [DOI] [PubMed] [Google Scholar]
- Weljie AM, Clarke TE, Juffer AH, Harmon AC, Vogel HJ (2000) Comparative modeling studies of the calmodulin-like domain of calcium-dependent protein kinase from soybean. Proteins 39: 343–357 [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Rodriguez-Concepcion M, Gruissem W (1999) Lipid modifications of proteins - slipping in and out of membranes. Trends Plant Sci 4: 439–445 [DOI] [PubMed] [Google Scholar]
- Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HS (1999) Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39: 991–1001 [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu YT (2003) Calmodulin-binding protein kinases in plants. Trends Plant Sci 8: 123–127 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Pokutta S, Maurer P, Lindt M, Franklin RM, Kappes B (1994) Calcium-binding properties of a calcium-dependent protein kinase from Plasmodium falciparum and the significance of individual calcium-binding sites for kinase activation. Biochemistry 33: 3714–3721 [DOI] [PubMed] [Google Scholar]
- Zhigang A, Low R, Rausch T, Luttge U, Ratajczak R (1996) The 32 kDa tonoplast polypeptide Di associated with the V-type H+-ATPase of Mesembryanthemum crystallinum L. in the CAM state: a proteolytically processed subunit B? FEBS Lett 389: 314–318 [DOI] [PubMed] [Google Scholar]
- Zozulya S, Stryer L (1992) Calcium-myristoyl protein switch. Proc Natl Acad Sci USA 89: 11569–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]