Abstract
Although length of the telomeric DNA tract varies widely across evolution, a species-specific set point is established and maintained by unknown mechanisms. To investigate how telomere length is controlled in Arabidopsis thaliana, we analyzed bulk telomere length in 14 wild-type accessions. We found that telomere tracts in Arabidopsis are fairly uniformly distributed throughout a size range of 2 to 9 kb. Unexpectedly, telomeres in plants of the Wassilewskija ecotype displayed a bimodal size distribution, with some individuals harboring telomeres of 2 to 5 kb and others telomeres of 4 to 9 kb. F1 and F2 progeny of a cross between long and short telomere parents had intermediate telomeres, implying that telomere length in Arabidopsis is not controlled by a single genetic factor. We provide evidence that although global telomere length is strictly regulated within an ecotype-specific range, telomere tracts on individual chromosome ends do not occupy a predetermined length territory. We also demonstrate that individual telomere tracts on homologous chromosomes are coordinately regulated throughout development and that telomerase acts preferentially on the shortest telomeres. We propose that an optimal size for telomere tracts is established and maintained for each Arabidopsis ecotype.
INTRODUCTION
Telomeres are nucleoprotein complexes that distinguish the natural ends of chromosomes from damage-induced double-strand DNA breaks. Maintenance of the telomere not only is essential for genome stability but also is required to promote the long-term proliferation capacity associated with immortalized and undifferentiated cell populations. Telomere architecture is well conserved across evolution and consists of tandem arrays of simple G-rich repeats, with the 3′ terminus of the G-rich strand forming a single-strand overhang (McEachern et al., 2000). Most plant telomeres are comprised of TTTAGGG repeats (Fuchs et al., 1995). Telomere length varies considerably among different plant species. For example, telomeres in the Columbia ecotype of Arabidopsis thaliana span 2 to 5 kb (Richards and Ausubel, 1988), whereas in tobacco (Nicotiana tabacum), telomeres are much longer and reach 150 kb (Fajkus et al., 1995). In all cases, telomere tracts are strictly maintained at a species-specific set point; hence, length homeostasis is achieved.
Although mechanisms governing telomere size are poorly understood, dynamic forces can both shorten and lengthen the repeat array (McEachern et al., 2000). Shortening occurs largely as a result of incomplete replication by conventional DNA replication machinery (Watson, 1972; Olovnikov, 1973). Each time a cell divides a few nucleotides are lost from the 5′ end of the daughter strand synthesized by the lagging strand machinery. Telomeres can also be shortened through exonucleolytic degradation (Maringele and Lydall, 2002; Hackett and Greider, 2003). In the absence of a compensating mechanism, telomere loss proceeds unabated until the chromosome terminus elicits a DNA damage checkpoint response leading to cell-cycle arrest. Cells that escape the arrest undergo illegitimate repair and face the consequences of genome instability (Hackett et al., 2001). Fortunately, the damaging effects of telomere shortening can be circumvented by the action of telomerase, a ribonucleoprotein reverse transcriptase that catalyzes the addition of telomere repeats onto the 3′ terminus of the chromosome. Telomerase not only rebuilds shortened telomere tracts but can also extend an existing array for a net increase in telomere length (McEachern et al., 2000).
Telomere length differs not only between evolutionarily distant species but also within species of the same genera. For instance, telomeres in wild-derived mouse strains are similar in length to telomeres in humans (10 to 15 kb) (Hemann and Greider, 2000), whereas established inbred mouse strains have much longer telomere lengths of ∼40 kb (Zijlmans et al., 1997). How is telomere homeostasis achieved? Current models propose that telomere length is modulated by telomere-specific proteins, which regulate telomerase access to the terminus (Evans and Lundblad, 1999; Taggart et al., 2002). Telomeres that reach an optimal length carry a full complement of telomere binding proteins and exist in a closed conformation largely inaccessible to telomerase. In the absence of telomerase action, the tract gradually shortens, resulting in the loss of telomere protein binding sites and as a consequence shifts to a more open accessible conformation for telomerase (Shore, 2001; de Lange, 2002; Teixeira et al., 2004).
Although this model provides a useful framework for investigating aspects of telomere homeostasis, many questions remain unanswered. For instance, how is the optimal telomere length established for different organisms? What is the fate of individual telomere tracts through successive generations? Do individual telomeres tend to reside in a preset size territory or is the length of each tract dynamic? Finally, is the length of the telomere tract on homologous chromosome arms coordinately regulated? To address some of these issues, we examined telomere length regulation in wild-type Arabidopsis. Arabidopsis has emerged as a useful model for telomere biology (Riha and Shippen, 2003). Particularly relevant is the fact that subtelomeric DNA sequences are unique on 7 of the 10 chromosome arms, making Arabidopsis very well suited for analysis of individual telomere tracts. In addition, previous studies indicate that telomere lengths differ in the Columbia and Wassilewskija (Ws) ecotypes of Arabidopsis (Richards et al., 1992; Riha et al., 2002), providing an opportunity to evaluate telomere length dynamics in different genetic settings.
Here, we examine telomere length in 14 different Arabidopsis ecotypes. We find significant size differences among these accessions and a striking bimodal size distribution of telomeres in individual plants of the Ws ecotype. In this study, we also employ unique subtelomeric sequences as probes to follow the fate of individual chromosome ends in both Columbia and Ws ecotypes through successive plant generations. Our data illustrate the dynamic nature of telomere maintenance and suggest that whereas the global telomere length is strictly regulated within an ecotype-specific range, individual telomere tracts are not limited to a set size within this range. Our results also indicate that telomere length homeostasis in Arabidopsis occurs through intermittent telomerase action on shorter telomeres to achieve an optimal, ecotype-specific size.
RESULTS
Ecotype-Specific Telomere Lengths
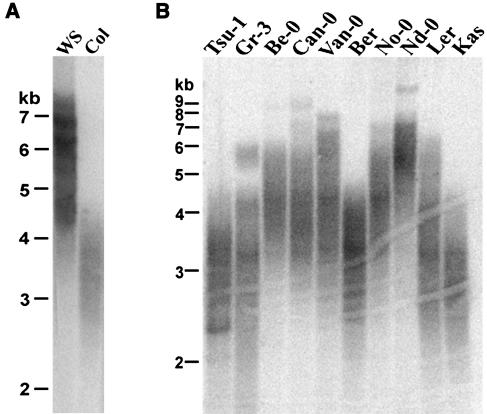
To determine the extent of telomere length variation among wild-type Arabidopsis, we performed terminal restriction fragment (TRF) analysis on 14 different Arabidopsis accessions. DNA samples were digested with Tru1I, which cleaves immediately adjacent to the telomeric DNA tract, and then hybridized with a (TTTAGGG)4 probe. As previously noted, telomeres in plants of the Columbia and Ws ecotypes were not the same size (Riha et al., 2002). Telomeres in Columbia plants ranged from 2 to 5 kb in length, whereas telomeres in Ws plants were longer, spanning from 3.5 to 8 kb (Figure 1A). Among all the ecotypes we studied, telomere tracts were distributed fairly uniformly throughout a range of 2 to 9 kb. For example, telomeres in Tsu-1, Ber, and Kas were similar to those of Columbia and spanned 2 to 4.5 kb. Telomeres were somewhat longer in Be-0 (3 to 6 kb) and Van-0 (2.7 to 8 kb) and longer still in Nd-0 (3.5 to 9 kb) (Figure 1B, Table 1). We conclude that telomeres in wild-type Arabidopsis can span a 2- to 9-kb range, with no obvious bias toward the short or long end of this distribution.
Figure 1.
Arabidopsis Ecotypes Display Different Telomere Size Distributions.
DNA isolated from 10 to 12 individual plants of the ecotypes indicated was pooled and subjected to TRF analysis. Telomere tracts for the different ecotypes displayed a relatively uniform size distribution, spanning 2 to 9 kb.
(A) TRF analysis of ecotypes often used to generate mutants by insertional mutagenesis.
(B) TRF analysis for 10 randomly selected ecotypes.
Table 1.
Telomere Length in Wild-Type Arabidopsis Accessions
| Ecotype | Telomere Length (kb) |
|---|---|
| Tsu-1 | 2 to 4.5 |
| Gr-3 | 2 to 6 |
| Be-0 | 3 to 6 |
| Can-0 | 2.8 to 9 |
| Van-0 | 2.7 to 8 |
| Ber | 2 to 4.5 |
| No-0 | 2.8 to 7 |
| Nd-0 | 3.5 to 9 |
| Ler | 2 to 6 |
| Kas | 2 to 4.5 |
| La-0 | 3 to 7 |
| Cvi-0 | 3.5 to 9 |
| Wsa | 2 to 8 |
| Columbia | 2 to 5 |
The majority of the Ws plants harbors telomeres in the 3.5- to 8-kb range.
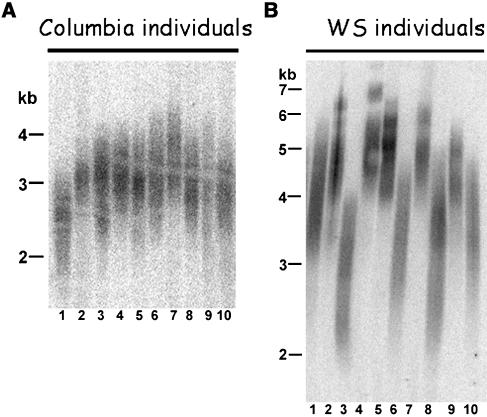
These initial experiments were conducted on DNA from pooled populations of Arabidopsis plants. To study telomere length regulation in more detail, we examined telomeres in individual plants. As expected, individual Columbia plants displayed a homogeneous profile of telomere length, with the majority of plants bearing telomeres of 2 to 4 kb (Figure 2A). Only occasionally were individuals with slightly longer or shorter telomere arrays observed (Figure 2A, compare lanes 1 and 7). A similar result was obtained for Landsberg erecta (Ler) individuals (Richards et al., 1992). In this case, telomere tracts were even more regular and spanned 2 to 6 kb (data not shown).
Figure 2.
Telomere Size Distribution in Individual Plants of the Columbia and Ws Ecotypes.
TRF analysis was performed on genomic DNA isolated from 10 individual plants of the Columbia (A) or Ws (B) ecotype.
A strikingly different result was observed with Ws individuals. Whereas the majority of plants we examined harbored telomeres in the 3.5- to 8-kb size range, as previously reported (Gallego and White, 2001; Bundock and Hooykaas, 2002; Riha et al., 2002; Gallego et al., 2003), a significant proportion harbored shorter telomeres of 2 to 5 kb (Figure 2B). All of the Ws plants used for this analysis displayed the distinct Ws morphology, ruling out cross-contamination with Columbia seeds. This bimodal telomere length distribution was also observed in Ws plants from Wisconsin's T-DNA insertion collection (data not shown). To determine whether the bimodal size distribution was characteristic of other ecotypes with longer telomeres, TRF analysis was also performed on individual plants of Nd-0 and Cvi-0 accessions. Interestingly, telomere size distribution in Nd-0 and Cvi-0 plants was uniform among different individuals (data not shown), implying that Ws is unusual among Arabidopsis accessions in harboring a bimodal distribution of telomere lengths.
Genetic Analysis of Telomere Length Variation in the Ws Ecotype
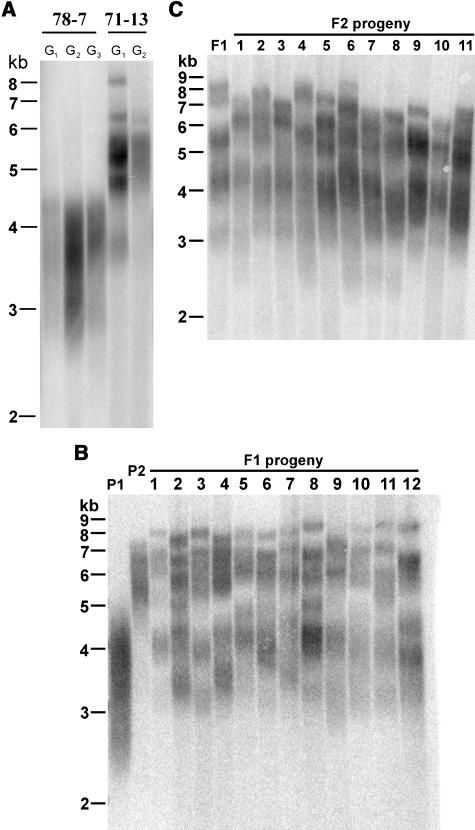
Given the striking variation in telomere length among Ws individuals, we next asked whether their telomere size was stably inherited. TRF analysis was performed on Ws parents bearing either short (line 78-7) or long (line 71-13) telomeres and their progeny obtained by self-pollination. For both lines, the telomere tract of the parent was maintained at the same size for at least three successive generations (Figure 3A; data not shown). Similar results were obtained for Columbia plants; bulk telomere length in the progeny differed from the parents by no more than 0.5 kb (data not shown). Thus, telomere size distribution in wild-type Ws and Columbia accessions is tightly regulated and heritable through successive generations.
Figure 3.
Telomere Length Is Not Established by a Single Dominant Factor.
(A) TRF analysis of successive generations of Ws individuals derived from the 78-7 short telomere line and 71-13 long telomere line.
(B) TRF analysis of F1 progeny from a cross between lines 78-7 (P1) and 71-13 (P2).
(C) TRF analysis of F2 individuals. Telomeres do not segregate into long and short size distributions.
To further explore the molecular basis for the bimodal size distribution of telomeres in Ws plants, we crossed individuals from lines 78-7 (P1) and 71-13 (P2) to obtain plants heterozygous with respect to parental telomere length (Figure 3B). Telomeres in F1 progeny displayed intermediate length with respect to telomeres in their parents and ranged from 2.7 to 8.5 kb. Although the overall size distribution was broadened relative to either parent, the shortest F1 telomeres were ∼0.6 kb longer than the shortest telomeres in P1, and the longest F1 telomeres were slightly longer than the longest telomeres in P2 (Figure 3B). Moreover, instead of the more homogenous smear associated with parental DNA, telomeres in the progeny exhibited a more discrete banding pattern, which may reflect the contribution of one short and one long telomere from each parent (see below).
If a single dominant factor was responsible for establishing telomere length in Ws plants, we would expect segregation of telomere lengths in an F2 population with some plants bearing telomere length similar to the original parents. To test this prediction, F1 plants were self-pollinated to generate F2 progeny. Telomeres in the F2 plants displayed roughly the same size distribution as their F1 parent (Figure 3C). In 9 of 11 F2 individuals, the longest telomeres were slightly shorter than in F1, and in approximately half of the F2 plants, the shortest telomeres were slightly shorter than in F1. In our experience, this subtle difference is not significant because some telomere length variation is commonly observed even among siblings. We conclude that the overall range of telomere tracts was remarkably similar in F1 and F2 populations, arguing that like the situation in maize (Zea mays) (Burr et al., 1992), the length of the telomere tract in Arabidopsis is not controlled by a single genetic factor. Instead, we speculate that epigenetic factors may be responsible for the establishment of different telomere lengths in Ws individuals (see below). If this is the case, the intermediate telomere length observed in the F1 and F2 progeny of the cross can be explained by the contribution of epigenetic factors present in both parents.
Telomere Length Dynamics at Individual Chromosome Ends
Although it is clear that the overall size of telomere tracts is maintained within preset boundaries, relatively little is known about how length homeostasis is achieved for individual telomere tracts. To investigate this question, we followed the fate of individual telomeres in parents and their progeny using chromosome end–specific probes. For analysis of individual chromosome ends, TRF analysis was performed on DNA isolated from whole plants that was digested with PvuII and SpeI, enzymes that cleave in the subtelomeric regions to release the telomere tract along with the unique sequences adjacent to it. We used probes specific for the South or right arm of chromosome two (2R) and the South (right) and North (left) arms of chromosome 5 (5R and 5L, respectively). The actual size of the telomere tract can be determined from the TRF signal by subtracting the distance from the subtelomere to the telomere. These values are 1.3 kb for 2R, 1.5 kb for 5L, and 2.6 kb for 5R.
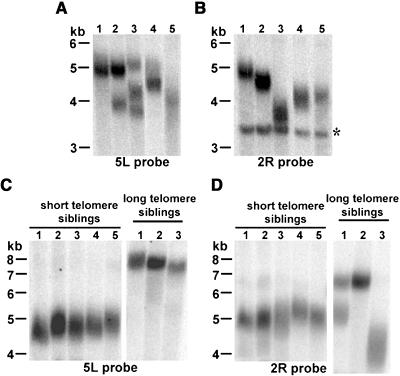
As shown in Figure 4, a single discrete band was detected in most Columbia plants, indicating that the length of individual telomere tracts on homologous chromosomes is coordinately controlled throughout development. In a few plants, two (Figure 4A, lane 2) or even three bands (Figure 4A, lane 3) were observed. Whereas two bands would be consistent with differentially sized telomeres on homologous chromosomes, the presence of three 5L bands in one plant implies the existence of distinct cell populations bearing 5L telomeres of different lengths. Although the size of individual telomere tracts is tightly regulated in a single plant, we noted dramatic variation in telomere length among unrelated individuals. As shown in Figures 4A and 4B, the 2R telomere in Columbia plant 3 was quite short (2.4 kb), but in plant 1 it was 1.3 kb longer. Similarly, the 5L telomeres of plants 4 and 5 were much shorter than in plant 1. For both Columbia and Ws individuals, telomere lengths were more similar among siblings than unrelated individuals (Figure 4; data not shown).
Figure 4.
Individual Telomeres in Arabidopsis Vary in Length.
(A) and (B) TRF analysis of individual chromosome arms from unrelated Columbia plants was conducted using probe-specific subtelomeric sequences at 5L (A) and 2R (B). Asterisk indicates interstitial band cross-hybridizing to 2R probe.
(C) and (D) TRF analysis of DNA from Ws siblings from short and long telomere lines hybridized with 5L (C) or 2R (D) probes is shown.
We also compared telomere size in Ws siblings derived from parents bearing either long or short telomeres. Telomeres 5L and 2R in the siblings of the short telomere line (78-7) were remarkably uniform in size, with most tracts measuring between 3.3 and 3.7 kb (Figures 4C and 4D). The same result was obtained for the 5L telomeres in siblings from the long telomere line (71-13) (Figure 4C). By contrast, the 2R telomeres from this same line (71-13) varied dramatically in size among long telomere siblings. The 2R telomere in plant 3 was 2.5 kb shorter than in plant 2, whereas for plant 1, two discrete 2R telomere populations were observed, one corresponding to 2R in plant 2 and the other intermediate in size between 2R for plants 2 and 3 (Figure 4D). Although much shorter than in other siblings, the 2R telomere in sibling 3 is still within the acceptable lower size limit for this line and likely to be extended in the next generation (see below). We conclude from these data that although global telomere length is strictly regulated within an ecotype-specific range, individual telomere tracts are not limited to a set size within this range and are subject to lengthening and shortening events.
Telomeres Lengthen and Shorten in Progeny to Achieve an Optimal Size
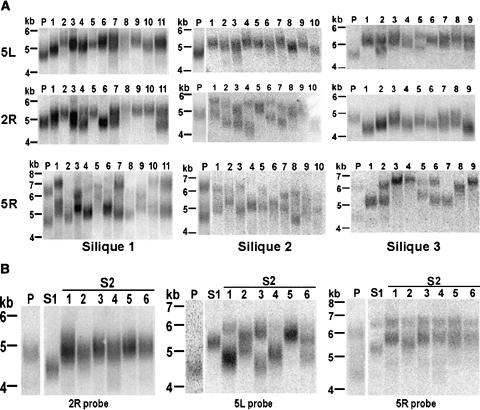
The data presented thus far demonstrate the dynamic nature of individual telomere tracts. However, the variations appeared to be stochastic and provided no insight into how the upper and lower size limits are established and maintained. To address this question, we compared telomere length in parents and their progeny to follow the fate of individual telomere tracts through successive generations of Columbia and Ws plants. Figure 5 shows the results for Columbia plants derived from different siliques of the same parent. The parental 5L telomere was 3.2 kb, but in essentially all of the progeny (from three different siliques), telomeres were lengthened for a net increase of 0.3 to 0.8 kb (Figure 5A). In the majority of the progeny, only a single prominent population of 5L telomeres was detected. However, in plants 2 and 5 from silique 3, two populations of 5L telomeres were observed, implying that the 5L telomere was subjected to differential processing in these plants.
Figure 5.
Parent-Progeny Analysis of Plants from the Columbia Ecotype.
(A) One Columbia plant (parent) was self-pollinated, and the progeny from three random siliques were chosen for TRF analysis using the probes indicated.
(B) TRF analysis was performed on DNA from three successive generations of Columbia plants to measure 2R, 5L, and 5R telomere lengths. The S1 lane shows results for DNA obtained from the progeny of the parent (P). S1 was self-pollinated to produce seeds for S2 plants.
Analysis of the 2R telomere yielded a strikingly different result. In this case, the parental telomere was 3.5 kb, ∼300 bp longer than the 5L telomere. For silique 1, 2R telomeres in a subset of the progeny were elongated (up to 0.6 kb), but this was not the case in all plants. In several individuals, 2R telomeres were approximately the same size as in the parent, and in 6 of 10 progeny from silique 2, 2R telomere split into two populations, with one telomere signal being shorter than the parent and one longer. Remarkably, for silique 3 all of the 2R telomeres were slightly shorter than the parent and ranged in size from 2.9 to 3.3 kb for a net decrease of 0.2 to 0.6 kb. This decrease corresponds to the amount of telomeric DNA lost per generation in a telomerase-deficient mutant (Riha et al., 2001), arguing that the 2R telomere was not an efficient substrate for telomerase action in this generation.
The fate of the 5R telomere was particularly interesting. The parent displayed two distinct 5R telomere populations, one at 3.7 kb and a second at only 1.9 kb, which represents the lower size limit of telomeres in all of the wild-type accessions we examined. Although it is not possible to unambiguously connect the lower band in the progeny to the lower band in the parent, it is striking that in most of the progeny, 5R telomeres were extended relative to this short parental 5R telomere. The only exception was individual 8 from silique 2, whose shorter 5R telomere was slightly shorter than that of the parent, indicating that this particular telomere was not extended in this individual. Remarkably, in silique 3 the 5R telomere converged in six of nine progeny to compose only a single size distribution that was up to 2 kb longer than the short parental telomere. These findings indicate that telomerase is acting preferentially on the shortest telomere in the population, allowing it to enter a more favorable size range.
The dynamic nature of this telomere-measuring mechanism was even more evident when we followed the fate of telomeres through three consecutive generations (Figure 5B). The 3.5-kb 2R telomere dropped in size in the second generation (S1) to 3 kb but then in the third generation (S2) was restored to approximately the same size as in the first generation. By contrast, the 3.2-kb 5L telomere was extended to 3.7 kb in generation 2, but then in generation 3 telomeres in most of the progeny split into two size classes, one longer and one shorter than the generation 2 parent. For 5R, two populations of telomeres were maintained throughout the three subsequent generations. However, the shorter telomere, which was only 1.9 kb in the first generation, was extended to 2.6 kb in the second generation and then to 3.1 kb in the third generation. Conversely, the longer 5R telomere was extended from 3.7 to 4 kb in the second generation but remained almost the same length in the third generation.
All together these data indicate that the length of telomere tracts in Arabidopsis is actively monitored and reset in each generation to maintain an optimal size. For the Columbia ecotype, the optimal size appears to be ∼3.5 kb. Telomeres shorter than this are likely to be acted on by telomerase in the next generation, whereas longer telomeres are less inclined to be telomerase substrates.
A New Optimal Telomere Length in Ws Plants
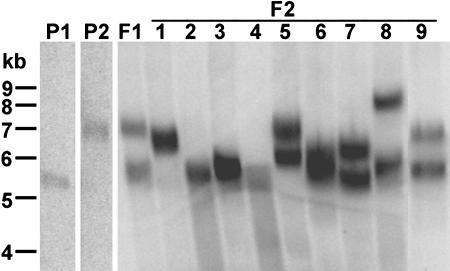
We found that telomeres in Ws accession were subjected to the same type of measuring mechanism, but different length optima were observed depending on whether the plants were derived from a short (average ∼3.5 kb) or a long telomere parental line (average ∼6.0 kb) (data not shown). To explore how an ideal telomere length is established, we examined telomere dynamics in Ws plants derived from the cross between parents with short and long telomeres (Figure 6). As expected, the 2R telomere in each parent fell within predicted size range, 4.0 kb for the short telomere parent and 5.6 kb for the long (Figure 6, parents P1 and P2). In F1, two populations of 2R telomeres were detected, one population of the same size as the long telomere parent and a second slightly longer than in the short telomere parent (Figure 6, lane F1). Thus, the broad telomere size distribution in F1 plants (Figure 3B) reflects the contribution of one short and one long telomere in the cross.
Figure 6.
A Broader Telomere Length Range Is Established in the Progeny of a Cross between Ws Plants Bearing Long and Short Telomeres.
TRF analysis was used to measure 2R telomere lengths in three successive generations of Ws plants. The F1 plant was derived from the cross between short (78-7, P1) and long (71-13, P2) telomere parents. F1 was self-pollinated to produce seeds for F2 plants. Variations in hybridization intensities reflect subtle differences in the amount of genomic DNA loaded in each lane and not a difference in target sequence between samples.
A different profile was observed in F2. In most plants, the two homologous 2R telomeres became shorter than the original long telomere parent, with many approaching the size of the short telomere parent. In only one F2 plant (Figure 6, plant 8) was there evidence for significant telomere elongation. However, in this case, this telomere was 6.9 kb, still within the acceptable size range for the original long telomere parent (Figure 3A). For the most part, 2R telomeres in F2 ranged in size from 4.2 to 5.5 kb. This observation suggests that a new broader set point had been established that is intermediate in size relative to the original long and short Ws telomere parents.
DISCUSSION
Natural Telomere Length Variation in Arabidopsis Accessions
Eukaryotes use telomeres as a general mechanism for chromosome end protection. Although the overall length of the telomeric tract varies from species to species, each organism maintains its telomeres within a defined, species-specific limit. Recent studies have established that perturbations in the telomere length maintenance machinery profoundly affect cell survival (Riha and Shippen, 2003). Thus, it is important to gain an understanding of how telomere length is established and maintained.
Arabidopsis thaliana is an excellent model for investigating natural variation. Arabidopsis accessions, collected from various natural habitats, display a wide variety of evolutionary traits (reviewed in Alonso-Blanco and Koornneef, 2000); here, we show that these variations include differences in telomere length set points. We found that telomeres in Arabidopsis ecotypes display a relatively uniform size distribution that ranges from 2 to 9 kb, with some ecotypes representing the shorter end of the spectrum (2 to 5 kb as in the Columbia ecotype) and other ecotypes, such as Nd-0 and Cvi-0, representing the longer end (3.5 to 9 kb). Thus, natural variation of telomere lengths in Arabidopsis appears to be widespread and unlinked to morphological traits that distinguish ecotypes. In addition, seeds for most ecotypes used in this study come from bulk seed propagation, which allows for genetic diversity. However, we observed no evidence for significant plant-to-plant telomere length variation between representatives of the same ecotype, with the remarkable exception of Ws plants.
Because most of the Arabidopsis insertional mutagenesis facilities employ Ws and Columbia ecotypes, it was of interest from a practical standpoint to more thoroughly investigate telomere length regulation in these accessions. Telomeres in Columbia plants are homogeneous in their overall length, but this is not the situation for Ws, where striking differences in telomere length can be observed among individual plants. In our experience, Ws plants bearing shorter telomeres are less common in the population than plants bearing longer telomeres. This may explain why, in all of the published studies that examine the role of telomere-related genes, Ws plants have had longer telomeres (Gallego and White, 2001; Bundock and Hooykaas, 2002; Riha et al., 2002; Gallego et al., 2003). Nonetheless, the presence of a bimodal size distribution of telomeres in Ws individuals could significantly confound the interpretation of phenotypes in mutants with perturbations in telomere-related genes. We recommend that TRF analysis performed on mutant progeny derived from a heterozygous Ws parent be compared with wild-type siblings and not with unrelated wild-type plants.
Telomere Dynamics on Individual Chromosome Ends
In contrast with the situation in yeast and mammals, popular models for telomere biology, most of the chromosome arms in Arabidopsis harbor unique subtelomere sequences that can be used to assess behavior of individual telomeres. We exploited this feature of Arabidopsis to examine the fate of individual telomeres through successive generations. Our data underscore the dynamic nature of telomere maintenance and demonstrate that individual telomeres do not occupy a predetermined length territory. Instead, each telomere is free to move within the ecotype-specific size boundary. We also discovered that the length of telomere tracts on homologous chromosomes is coordinately regulated because in most cases TRF analysis detected only a single band corresponding to the telomere on homologous chromosome arms.
The regulated nature of telomere tracts in Arabidopsis is quite remarkable, considering that DNA for our analysis was isolated from entire plants containing cells with different proliferation histories. The length of an individual telomere is expected to vary significantly depending on how often telomerase engaged the telomere and how much telomeric DNA was added in each elongation event. This is true for a clonal population of human cells, which display a dramatic difference of up to 6 kb for a particular chromosome end (Baird et al., 2003). By contrast, we observed very limited length variation for a given telomere end, and in most cases only one telomere-specific band was observed. The limited variation of telomere length may reflect a low number of cell divisions required for generation of an Arabidopsis plant relative to cell divisions required to maintain human cells in culture. In addition, the action of telomerase may be strongly influenced by the size of the telomere tract in the parent. In support of this idea, we found that the lengths of individual telomere tracts in siblings are much more similar than in unrelated individuals. The uniformity of telomere tracts may also be a consequence of telomerase action at a defined stage in the life cycle, perhaps in premeiotic or postmeiotic cells. If this is the case, then the dramatic extension of some telomere tracts (up to 2 kb within one generation) is striking. Whether telomere addition happens as a consequence of a single elongation event early in plant development or through multiple interactions later on remains to be determined.
In Arabidopsis, as in yeast and humans, telomerase appears to be preferentially recruited to the shortest telomeres (Marcand et al., 1999; Hemann et al., 2001; Samper et al., 2001; Hathcock et al., 2002; Liu et al., 2002; Teixeira et al., 2004). Because the maintenance of critically shortened telomeres is crucial for cell viability and chromosome stability (Hemann et al., 2001), this observation indicates that organisms across eukaryotic evolution have evolved a common solution to these problems. In wild-type Arabidopsis, most long telomeres shorten at a rate of ∼200 to 500 bp per generation. This attrition occurs at the same rate as in a telomerase mutant (Riha et al., 2001). Hence, it appears that some chromosome ends with long telomeres may be inaccessible to telomerase throughout the entire lifespan of the plant. This observation is consistent with a recent report showing that yeast telomerase does not act on every telomere in each cell cycle (Teixeira et al., 2004). The slow rate of telomere shortening further suggests that telomeres in wild-type Arabidopsis, in contrast with those in humans (Hackett and Greider, 2003), are not subjects to nuclease attack and hence may exist in a more sheltered configuration during much of the plant life cycle.
Although telomere shortening by the end-replication problem appears to be the major mechanism of telomere attrition in most organisms, including plants, an alternative, recombination-based pathway might also be involved in resetting the length of long Arabidopsis telomeres. In our parent-progeny analysis we often saw the loss of >1 kb from long telomeres in a single plant generation, which is far greater than the rate of telomere shortening simply caused by the end-replication problem (Fitzgerald et al., 1999; Riha et al., 2001). Whether this active but yet unknown mechanism of telomere shortening is similar to the telomere rapid deletion phenomenon observed in certain yeast and mammalian genetic backgrounds (reviewed in Lustig, 2003) remains to be determined.
Establishing and Maintaining an Optimal Telomere Length in Arabidopsis
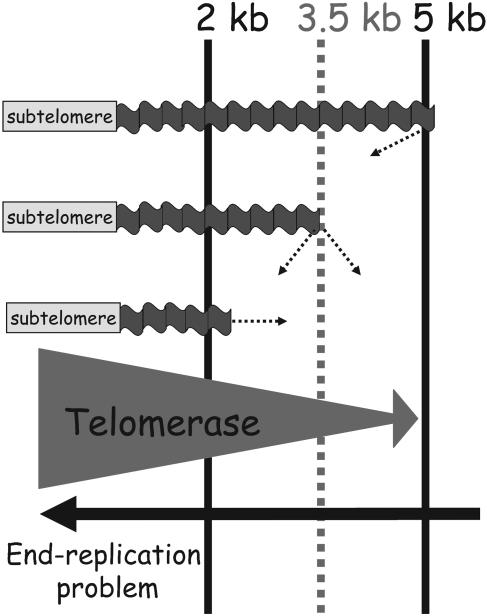
An optimal size for telomere tracts is established and maintained in each Arabidopsis ecotype. For Columbia telomeres, this size is 3.5 kb. Our data provide strong evidence that telomeres are acted upon by two opposing forces driving them to this size (Figure 7). Telomeres shorter than 3.5 kb are preferentially extended by telomerase, whereas longer telomeres tend to be poor substrates for telomerase and passively drift down to the optimal length as a consequence of the end replication problem. At the optimal size, an equilibrium is reached between shortening and lengthening activities, giving the telomere an equal chance of being elongated or shortened. In support of this idea, we observed telomere splitting in which a uniform population of telomeres in the optimal size range is divided into two subpopulations of shorter and longer sizes in the next generation (Figure 7). Convergence of two chromosome-specific bands into one also occurred, an outcome expected when a shorter and longer telomere are simultaneously brought closer in size to reach the optimal length.
Figure 7.
Model for Telomere Length Homeostasis in the Columbia Ecotype of Arabidopsis.
Telomeres are brought to an optimal, ecotype-specific size by the opposing consequences of telomerase action and the end-replication problem. When telomere length reaches the lower threshold (2 kb), telomerase is more likely to act (bottom telomere). As telomeres approach the optimum threshold (3.5 kb), there is an equal chance of telomerase action or inaction, resulting in telomere splitting (middle telomere). At the maximum size (5 kb, top telomere), telomerase is less likely to act, and telomeres shorten because of the end-replication problem.
The bimodal distribution of telomeres within the Ws ecotype raises several interesting questions about the mechanisms that establish a species-specific telomere size. Both populations of Ws plants maintain their telomere lengths from generation to generation. Because the majority of wild-type Ws plants have telomeres in the longer range (3.5 to 8 kb), it seems likely that plants with shorter telomeres have somehow reset their telomere length ranges to the lower Columbia-type limit (2 to 5 kb). The inability to segregate short and long telomere lengths in the F2 progeny of a cross between short and long telomere Ws parents argues that the establishment and maintenance of ecotype-specific telomere length are distinct processes and are not regulated by a single genetic factor. Rather, a new intermediate set point of telomere length appears to be established in F1 plants that is maintained in F2 progeny. Telomeres inherited from the parental lines are no longer restricted by the parental telomere set points but are able to move freely within the broader size range of 2 to 8 kb. Further analysis will be necessary to elucidate possible genetic factors involved in telomere length regulation in Arabidopsis.
It is conceivable that epigenetic factors contribute to telomere length regulation. In this scenario, we would predict that genetic factors, such as putative homologs of mammalian double-strand telomere binding proteins (Broccoli et al., 1997), would be involved in the maintenance of telomere length but not in its establishment. In light of the recent discoveries that Schizosaccharomyces pombe has mechanistically distinguishable processes leading to the establishment and maintenance of the functional telomere complex (Sadaie et al., 2003) and that mammalian histone methyltransferases are involved in epigenetic regulation of telomere length (Garcia-Cao et al., 2004), the potential contribution of epigenetic control in telomere length regulation in higher eukaryotes remains an intriguing possibility.
METHODS
Plant Material
Wild-type Arabidopsis thaliana seeds (ecotype Ws) were purchased from Lehle Seeds (Round Rock, TX), catalog number WT-8A. Parental Ws lines 71-13 and 78-7 were randomly selected from this population based on the differences in the overall length of their telomeres. Arabidopsis plants of ecotypes Col-6, Ler, La-0, Cvi-0, Tsu-1, Nd-0, No-0, Can-0, Be-0, Ber, Van-0, Gr-3, and Kas-1 were obtained from the ABRC (catalog numbers CS8155, CS8581, CS1299, CS1096, CS6926, CS1390, CS1394, CS1064, CS965, CS8068, CS1584, CS3179, and CS1264, respectively).
DNA Isolation and TRF Analysis
DNA from individual 6-week-old whole plants was extracted as described (Cocciolone and Cone, 1993). TRF analysis was performed with DNA digested with Tru1I (Fermentas, Hanover, MD) restriction enzyme. 32P 5′ end–labeled (T3AG3)4 oligonucleotide was used as a probe (Fitzgerald et al., 1999). Single telomere analysis was performed as follows: 1 μg of genomic DNA was digested with PvuII and SpeI, and DNA was separated by electrophoresis in a 0.8% agarose gel and blotted onto a nylon membrane. Telomere-adjacent DNA sequences were amplified with primers PAT51-5, 5′-CAACATGGCCCATTTAAGATTGAACG-3′, and PAT51-3, 5′-CACATATATGTTTGTTGAGTGTCGC-3′, for the 2R probe; TAS5R-F1, 5′-TACGGTTTAGAGTTTAGGGT-3′, and TAS5R-R1, 5′-CGCTCTCATTGCGAGTGGTA-3′, for the 5R probe; and TAS5L-F2, 5′-TGAGTTTGCATAAAGCGTCACG-3′, and TAS5L-R2, 5′-CGACAACGACGACGAATGACAC-3′, for the 5L probe and were used for hybridization. Radioactive signals were scanned by a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA), and the data were analyzed by IMAGEQUANT software (Molecular Dynamics).
Acknowledgments
We thank Tom McKnight and members of the Shippen lab for many helpful discussions. We are also grateful to Tom McKnight, Laurent Vespa, and Matt Watson for critically reading the manuscript. This work was supported by National Institutes of Health Grant GM65383 to D.E.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dorothy E. Shippen (dshippen@tamu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023093.
References
- Alonso-Blanco, C., and Koornneef, M. (2000). Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 5, 22–29. [DOI] [PubMed] [Google Scholar]
- Baird, D.M., Rowson, J., Wynford-Thomas, D., and Kipling, D. (2003). Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 33, 203–207. [DOI] [PubMed] [Google Scholar]
- Broccoli, D., Smogorzewska, A., Chong, L., and de Lange, T. (1997). Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Bundock, P., and Hooykaas, P. (2002). Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14, 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, B., Burr, F.A., Matz, E.C., and Romero-Severson, J. (1992). Pinning down loose ends: Mapping telomeres and factors affecting their length. Plant Cell 4, 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone, S.M., and Cone, K.C. (1993). Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange, T. (2002). Protection of mammalian telomeres. Oncogene 21, 532–540. [DOI] [PubMed] [Google Scholar]
- Evans, S.K., and Lundblad, V. (1999). Est1 and Cdc13 as comediators of telomerase access. Science 286, 117–120. [DOI] [PubMed] [Google Scholar]
- Fajkus, J., Kovarik, A., Kralovics, R., and Bezdek, M. (1995). Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 247, 633–638. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M.S., Riha, K., Gao, F., Ren, S., McKnight, T.D., and Shippen, D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, J., Brandes, A., and Schubert, I. (1995). Telomere sequence localization and karyotype evolution in higher plants. Plant Syst. Evol. 196, 227–241. [Google Scholar]
- Gallego, M.E., Jalut, N., and White, C.I. (2003). Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell 15, 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., and White, C.I. (2001). RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao, M., O'Sullivan, R., Peters, A.H., Jenuwein, T., and Blasco, M.A. (2004). Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36, 94–99. [DOI] [PubMed] [Google Scholar]
- Hackett, J.A., Feldser, D.M., and Greider, C.W. (2001). Telomere dysfunction increases mutation rate and genomic instability. Cell 106, 275–286. [DOI] [PubMed] [Google Scholar]
- Hackett, J.A., and Greider, C.W. (2003). End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 23, 8450–8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathcock, K.S., Hemann, M.T., Opperman, K.K., Strong, M.A., Greider, C.W., and Hodes, R.J. (2002). Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl. Acad. Sci. USA 99, 3591–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann, M.T., and Greider, C.W. (2000). Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 28, 4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann, M.T., Strong, M.A., Hao, L.Y., and Greider, C.W. (2001). The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67–77. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Kha, H., Ungrin, M., Robinson, M.O., and Harrington, L. (2002). Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. USA 99, 3597–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig, A.J. (2003). Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4, 916–923. [DOI] [PubMed] [Google Scholar]
- Marcand, S., Brevet, V., and Gilson, E. (1999). Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele, L., and Lydall, D. (2002). EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16, 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M.J., Krauskopf, A., and Blackburn, E.H. (2000). Telomeres and their control. Annu. Rev. Genet. 34, 331–358. [DOI] [PubMed] [Google Scholar]
- Olovnikov, A.M. (1973). A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41, 181–190. [DOI] [PubMed] [Google Scholar]
- Richards, E.J., and Ausubel, F.M. (1988). Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53, 127–136. [DOI] [PubMed] [Google Scholar]
- Richards, E.J., Chao, S., Vongs, A., and Yang, J. (1992). Characterization of Arabidopsis thaliana telomeres isolated in yeast. Nucleic Acids Res. 20, 4039–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha, K., and Shippen, D.E. (2003). Telomere structure, function and maintenance in Arabidopsis. Chromosome Res. 11, 263–275. [DOI] [PubMed] [Google Scholar]
- Riha, K., Watson, J.M., Parkey, J., and Shippen, D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie, M., Naito, T., and Ishikawa, F. (2003). Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev. 17, 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper, E., Flores, J.M., and Blasco, M.A. (2001). Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc−/− mice with short telomeres. EMBO Rep. 2, 800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, D. (2001). Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11, 189–198. [DOI] [PubMed] [Google Scholar]
- Taggart, A.K., Teng, S.C., and Zakian, V.A. (2002). Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297, 1023–1026. [DOI] [PubMed] [Google Scholar]
- Teixeira, M.T., Arneric, M., Sperisen, P., and Lingner, J. (2004). Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117, 323–335. [DOI] [PubMed] [Google Scholar]
- Watson, J.D. (1972). Origin of concatemeric T7 DNA. Nat. New Biol. 239, 197–201. [DOI] [PubMed] [Google Scholar]
- Zijlmans, J.M., Martens, U.M., Poon, S.S., Raap, A.K., Tanke, H.J., Ward, R.K., and Lansdorp, P.M. (1997). Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]