Abstract
Alcohol (ethanol) dependence is a chronic relapsing brain disorder partially influenced by genetics and characterized by an inability to regulate harmful levels of drinking. Emerging evidence has linked genes that encode KV7, KIR, and KCa2 K+ channels with variation in alcohol-related behaviors in rodents and humans. This led us to experimentally test relations between K+ channel genes and escalation of drinking in a chronic intermittent ethanol (CIE) exposure model of dependence in BXD recombinant inbred strains of mice. Transcript levels for K+ channel genes in the prefrontal cortex (PFC) and nucleus accumbens (NAc) covary with voluntary ethanol drinking in a non-dependent cohort. Transcripts that encode KV7 channels covary negatively with drinking in non-dependent BXD strains. Using a pharmacological approach to validate the genetic findings, C57BL/6J mice were allowed intermittent access to ethanol to establish baseline consumption before they were treated with retigabine, an FDA-approved KV7 channel positive modulator. Systemic administration significantly reduced drinking, and consistent with previous evidence, retigabine was more effective at reducing voluntary consumption in high-drinking than low-drinking subjects. We evaluated the specific K+ channel genes that were most sensitive to CIE exposure and identified a gene subset in the NAc and PFC dysregulated in the alcohol-dependent BXD cohort. CIE-induced modulation of nine genes in the NAc and six genes in the PFC covaried well with the changes in drinking induced by ethanol dependence. Here we identified novel candidate genes in the NAc and PFC that are regulated by ethanol dependence and correlate with voluntary drinking in non-dependent and dependent BXD mice. The findings that Kcnq expression correlate with drinking and that retigabine reduces consumption suggest that KV7 channels could be pharmacogenetic targets to treat individuals with alcohol addiction.
Introduction
Alcohol use disorder (AUD) is a chronic, relapsing brain disease with comparatively high heritability of 40–60% that imposes an enormous social and economic burden, and a host of costly and serious health consequences (NIAAA, 2015). Despite the deleterious effects on individual health and society, there are currently only three FDA approved pharmacological treatments for AUD, all of which have limited success at reducing the high rates of relapse (Clapp, 2012; Williams, 2005). This clearly identified gap in treatment options highlights our limited understanding of the underling vulnerabilities for developing an AUD and demonstrates the need for more research to define neurobiological mechanisms of alcohol dependence in an effort to identify novel pharmacotherapeutics.
Recent evidence has shown that a number of well-tolerated anticonvulsants may prove useful in the prevention of relapse and reducing alcohol consumption (Book & Myrick, 2005; Clapp, 2012; De Sousa, 2010; Malcolm, Myrick, Brady, & Ballenger, 2001; Padula et al., 2013). Originally implicated for their role in epilepsy (Maljevic & Lerche, 2014; Smets et al., 2015), studies show that calcium-activated (KCa), voltage-dependent (KV), and G protein-coupled inwardly-rectifying (Kir) K+ channels are targets for both acute and chronic effects of ethanol (Hopf et al., 2011; Mayfield, Blednov, & Harris, 2015; Mulholland, 2012; Mulholland, Hopf, et al., 2009). Additionally, preclinical studies have demonstrated that chronic ethanol reduces the function and trafficking of KCa2 (Kcnn), KV4.2 (Kcnd2), and KV7.2 (Kcnq2) channels in the nucleus accumbens (NAc) and hippocampus (Hopf et al., 2010; McGuier et al., 2015; Mulholland, Spencer, Hu, Kroener, & Chandler, 2015; Padula et al., 2015; Spencer, Mulholland, & Chandler, In Press), and pharmacologically enhancing KCa2 and KV7 channel function attenuated voluntary drinking in rodents (Hopf et al., 2011; Knapp, O’Malley, Datta, & Ciraulo, 2014; McGuier et al., 2015; Padula et al., 2013). Ethanol actions on voltage- and calcium-dependent KCa1.1 (Kcnma1) channels are involved in acute ethanol tolerance, dependence, and heavy ethanol consumption (Bukiya et al., 2014; Ghezzi, Pohl, Wang, & Atkinson, 2010; Kreifeldt, Le, Treistman, Koob, & Contet, 2013; Treistman & Martin, 2009), and Kcnma1 and Kcnq5 were identified as major hub genes for the acute actions of ethanol (Wolen et al., 2012). Deletion of the gene that encodes Kir3.3 channels (Kcnj9) enhanced ethanol conditioned place preference (Tipps, Raybuck, Kozell, Lattal, & Buck, 2016) and blunted ethanol-induced excitation of dopamine neurons and increased binge-like ethanol consumption in mice in both a limited-access 2-bottle choice (LA-2BC) model (15% v/v ethanol vs. water) and with limited-access to a single bottle of 20% v/v ethanol (Herman et al., 2015). In addition to ethanol actions on these K+ channels, recent studies reveal that variation in genes that encode KCa2, Kir3 and KV7 channels may modulate risk for susceptibility of developing an AUD (Clarke et al., 2011; Edenberg & Foroud, 2013; Kozell, Walter, Milner, Wickman, & Buck, 2009; McGuier et al., 2015; Padula et al., 2015). Thus, drugs that target K+ channels may prove useful in the treatment of AUD.
To experimentally test associations between K+ channel genes and ethanol drinking, we generated and analyzed data as part of an extensive collaborative behavioral and expression genetics study on a well-characterized model of dependence-induced escalation of voluntary LA-2BC drinking in a large cohort of genetically diverse BXD strains of mice. The BXDs are an effective genetic reference population to study both causes and correlates of complex behaviors, such as ethanol consumption in dependent and non-dependent mice (Philip et al., 2010). A full description of the chronic intermittent ethanol (CIE) exposure model in BXD RI mice and the analysis of variation in drinking and gene expression are reported elsewhere in this issue (Lopez, Miles, Williams, & Becker, This issue; van der Vaart et al., This issue). We utilized the GeneNetwork database (www.genenetwork.org) to generate and analyze correlations in K+ channel transcript levels in two key regions of the addiction circuitry, the prefrontal cortex (PFC) and NAc, with voluntary drinking in non-dependent and dependent BXD mice. We then used a computational modeling approach to determine how CIE-induced adaptations in K+ channel gene expression impacts function of deep-layer PFC projection neurons. Finally, we validated one identified target using a pharmacological approach in C57BL/6J mice.
Methods
Animals
As described in Lopez et al., (this issue), adult male and female C57BL/6J (B6) mice (10 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME), and BXD RI strains (12–16 weeks old) and DBA/2(UT) mice were obtained from the University of Tennessee Health Science Center or Jackson Laboratory. The sex, treatment groups, strains and the numbers of mice/strain are shown in Supplemental Table 1 (75 total mice from 28 unique strains; 54 males, 21 females; 36 control and 39 CIE-exposed mice). While littermates were used for some of the BXD strains, males and females from the same strain were not used in the present experiment. After 1–3 weeks acclimation in group housing conditions on a 12 hr light/dark cycle, mice were individually housed in temperature and humidity controlled environments for 72 hr prior to the start of alcohol access in their home cages. Food and water were available ad libitum during all procedures. The Medical University of South Carolina Institutional Animal Care and Use Committee approved all procedures in accordance with NIH guidelines for the humane care and use of laboratory animals.
CIE exposure and limited-access 2-bottle choice drinking
To establish baseline drinking, mice consumed ethanol in their home cage using a standard limited-access (2 hr) 2-bottle choice (15% ethanol (v/v) vs. water) protocol (LA-2BC) for 6 weeks following routine procedures (Becker & Lopez, 2004; Lopez et al., This issue) (Figure 1A). The reader is referred to Lopez et al., (this issue) for a full description of the drinking data in these BXD strains. At 30 min before the beginning of the dark cycle, water bottles were removed from the home cage and replaced with 15 ml drinking bottles that contained water or a 15% ethanol solution. After a 2-hr access period, the drinking bottles were removed from the home cage and replaced with water bottles. Mice then underwent 4 cycles of chronic intermittent ethanol (CIE or dependent) exposure in vapor inhalation chambers. CIE treated mice underwent sixteen hours of vapor exposure followed by eight hours of withdrawal, and weekly CIE exposure in vapor inhalation chambers was alternated weekly with home cage ethanol (15% v/v) or water drinking sessions (Becker & Lopez, 2004; Lopez et al., This issue). Chamber ethanol concentrations were monitored daily and air flow was adjusted to maintain ethanol concentrations within a range that yields stable blood ethanol levels (~175 mg/dl) throughout exposure for the C57BL/6J strain (see Lopez et al. (this issue) for explanation and potential confounds of this experimental design). Prior to entry into the ethanol chambers, EtOH mice were administered ethanol (1.6 g/kg; 8% w/v) and the alcohol dehydrogenase inhibitor pyrazole (1 mmol/kg; ip, dose volume 20 ml/kg). Control mice were handled similarly, but received injections of saline and pyrazole and were not exposed to CIE (Air or non-dependent) and, thus, had a total of 10 weeks of LA-2BC drinking. Mice were put through one final cycle of CIE exposure after a week of drinking, and tissue was collected for microarray analysis after a 72-hr withdrawal period. As described in Lopez et al. (this issue), all mice were exposed to the same experimental conditions in ethanol vapor inhalation chambers, and BXD RI strains of mice display considerable amounts of variability in their blood ethanol concentrations during the CIE exposure cycles. As a consequence of the study design, 24 mice died during exposure in the inhalation chambers or during withdrawal.
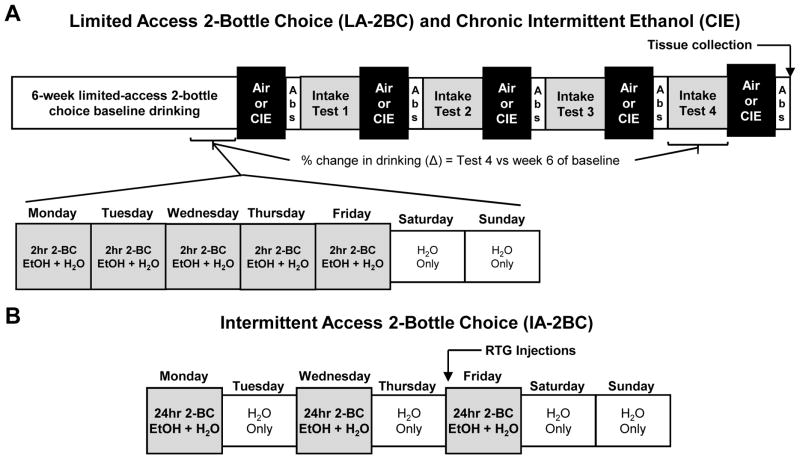
Figure 1. Schematic of the drinking models used in BXD RI strains of mice and the C57BL/6J parental strain.
(A) The two-bottle choice limited-access voluntary drinking and chronic intermittent ethanol (CIE) exposure paradigm used in BXD RI strains of mice. Mice were given access to ethanol (15% v/v) for 2 hr using a limited-access 2-bottle choice (LA-2BC) procedure for 6 weeks to establish baseline ethanol intake. Mice then received either CIE vapor in inhalation chambers or air in control chambers. After chamber exposure, mice entered a 72 hr abstinence (Abs) period and were then tested for ethanol drinking for 5 days. This pattern of chamber exposure, abstinence, and voluntary drinking was repeated for 4 cycles. Brain tissue was collected 72 hr after the final chamber exposure. The percent change in drinking (Δ) was calculated as the percent change between Intake Test 4 and the 6th week of baseline drinking. (B) Schematic of the intermittent access 2-bottle choice (IA-2BC) paradigm used in C57BL/6J mice. In this model, mice had access to 20% ethanol (v/v) for 24 hr three days a week with a 24 hr forced abstinence period in between drinking sessions. Mice were then treated with retigabine after 7 weeks of baseline drinking.
Microarray data generation and analysis
The reader is referred to van der Vaart et al., (this issue) for a full description of the tissue collection and microarray data generation and analysis. Briefly, tissue punch dissections, RNA isolation, and hybridization/scanning procedures with Affymetrix Mouse Genome 430 2.0 arrays (Affymetrix, Cat #900497) were performed as described previously (van der Vaart et al., This issue; Wolen et al., 2012). To minimize the risk of technical variation confounding the detection of an alcohol response, an Air- and CIE-exposed mouse from each strain were processed simultaneously. Annotation data for Mouse Genome 430 2.0 probe-sets were obtained from the GeneNetwork Data Sharing Zone (genenetwork.org/share/annotations). Expression data from the Air and CIE treatment groups were background corrected, quantile normalized and summarized using the robust multi-array average (RMA) expression measure. To directly assess CIE-responsive expression changes across BXD strains, the S-score algorithm (Kennedy, Kerns, Kong, Archer, & Miles, 2006; Kerns, Zhang, & Miles, 2003; Zhang, Wang, Ravindranathan, & Miles, 2002) was used for probe-level comparison of CIE vs. Air samples. S-scores represent a significance score in a similar manner to Z-scores, with a mean of 0 and a standard deviation of 1 across an entire array. Positive S-scores denote an increase in expression with CIE. S-scores were generated using the S-score R package from each BXD RI strain, C57BL/6J, DBA/2(UT), and C57BL/6J+DBA/2J F1 mice separately. Because only 1 CIE and 1 control sample was available for most BXD RI strains, significance of gene expression response across the BXD cohort was determined using Fisher’s Combined Probability Test with S-scores as previously described (Wolen et al., 2012). All datasets generated for this paper are accessible on GeneNetwork (www.genenetwork.org). All data are MIAME compliant.
Bioinformatics analyses
We performed three sets of analyses to determine the relationship between K+ channel gene expression, voluntary drinking, and ethanol dependence in the LA-2BC paradigm (Figure 1A): 1) correlation of ethanol consumption in non-dependent (Air exposed) controls (Supplemental Figure 1) and K+ channel gene expression in the PFC and NAc; 2) dependence-induced changes in K+ channel gene expression (difference in gene expression between CIE and Air, i.e., S-scores); and 3) correlation of dependence-induced change in ethanol consumption (calculated across all strains as a percent change between intake during Test 4 and intake during week 6 of baseline drinking) and S-scores. Specifically, using standard linear regression analysis, we conducted a phenotype correlation analysis of K+ channel RMA levels (196 probe sets for 79 K+ channel genes) in the PFC (GeneNetwork data set: VCU BXD PFC CIE Air M430 2.0 (Jan11) RMA) and NAc (GeneNetwork data set: VCU BXD NAc CIE Air M430 2.0 (Jan13) RMA) and ethanol intake values after 10 weeks of 2-bottle choice drinking (GeneNetwork record ID 12961: Ethanol intake (5 day average in g/kg/2 hr) after cycle 4 of Air exposure) obtained from non-dependent BXD RI mice. Next, to determine which K+ channel genes were responsive (i.e., differentially regulated) to CIE exposure in drinking mice, we measured the impact of CIE exposure on K+ channel transcript abundance using the Significance-score (S-score) algorithm, as described previously (Wolen et al., 2012; Zhang et al., 2002). This analysis utilized the S-score algorithm to compare K+ channel expression levels across BXD strains and between the two treatment groups to generate an Air vs CIE S-score for each K+ channel probe set in the PFC (GeneNetwork data set: VCU BXD PFC EtOH vs CIE Air M430 2.0 (Jan11) Sscore) and NAc (GeneNetwork data set: VCU BXD NAc EtOH vs CIE Air M430 2.0 (Jan13) Sscore). A positive S-score indicates up-regulation by CIE exposure and vice versa. Statistical significance of the K+ channel changes following CIE exposure across strains were assessed using Fisher’s combined probability test. To correct for multiple testing, q values were generated from the empirical p values, and probe-sets with q values <.05 were considered significantly CIE-responsive. As a final step, we determine how K+ channel S-scores in the PFC and NAc correlated with CIE-induced changes in alcohol consumption (GeneNetwork Record ID: 12741). CIE-induced changes in drinking were calculated across all strains as a percent change between intake at Test 4 and intake at week 6 of baseline drinking (Figure 1A), and the percent change highly correlated with the difference (in g/kg) in intake between Test 4 and baseline week 6 (F1,27 = 169.2, R2 = 0.862, p < 0.0001, Supplemental Figure 2). Standard linear regression analysis was used, and p values <.05 for the slope were considered significant. Data from male and female subjects were collapsed for all analyses. The hypergeometric functional enrichment analysis to determine if K+ channel genes that correlated with drinking were over-represented was performed using the ‘phyper’ function in R software (https://www.R-project.org).
Computational modeling
The NEURON 7.1 simulation environment (Carnevale & Hines, 2006) was used to generate a computational model containing 10 compartments (soma with proximal and apical dendrites) to determine how coordinated changes in CIE-responsive K+ channel genes in the PFC affect firing of pyramidal neurons. Computer simulations were performed on a modified deep-layer PFC pyramidal neuron model taken from ModelDb accession number 82849 (Durstewitz, Seamans, & Sejnowski, 2000). This model incorporated passive leak conductance, incorporated Hodgkin-Huxley fast sodium channel currents and delayed-rectifier K+ channel currents. Calcium-activated K+ channel currents were modeled according to the parameters used in ModelDb accession number 123453 (Akemann, Lundby, Mutoh, & Knopfel, 2009), delayed rectifier K+ channel currents were derived using the criteria in ModelDb accession number 74298 (Gillies & Willshaw, 2006), and inwardly-rectifying K+ channel kinetics were based on ModelDb accession number 147514 (van der Velden, van Hooft, & Chameau, 2012). The reversal potential of the passive leak conductance was −70 mV. Passive parameters included Cm (ranging from 0.9 to 1.2 μF*cm−2), Rm (ranging from 36.4 to 44.9 kΩ*cm2), Ri (ranging from 70 to 150 Ω*cm), and temperature (32–36°C). Our past studies t hat have measured functional changes in K+ channels in neurons revealed that chronic ethanol exposure (both in vitro and in vivo) reduced currents by 56.4 ± 5.3% (Mulholland, Becker, Woodward, & Chandler, 2011; Mulholland et al., 2015; Nimitvilai, Lopez, Mulholland, & Woodward, 2016; Padula et al., 2015). To model the coordinated gene expression changes in the PFC of ethanol dependent BXD mice, currents mediated by KV3.1, KV7, KCa1.1, and KV2.1 channels were either simultaneously decreased by 0.5-fold or were enhanced 1.5-fold in the pyramidal neuron model to mimic published functional changes in K+ channel currents. Current clamp simulations examined evoked firing in response to current steps of increasing amplitude (100 pA steps; 1 sec duration) injected at the soma starting with −1.0 nA and ending with +1.0 nA. Resting membrane potential was held at −65 mV, and active and passive membrane properties were analyzed. One-way ANOVA was used to analyze afterhyperpolarization (AHP) duration and interspike interval (ISI) with a Newman-Keuls post-hoc test and a significance level of p < 0.05.
Intermittent 2-bottle choice drinking
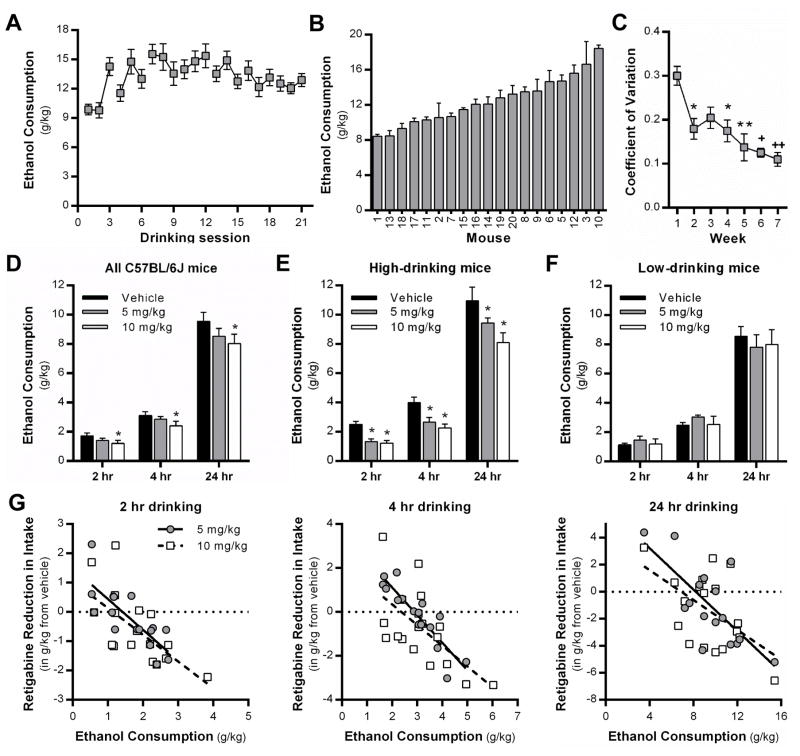
To pharmacologically validate and demonstrate the potential utility of using this database to identify novel pharmacotherapeutic targets for reducing ethanol consumption, we chose to utilize the intermittent-access 2-bottle choice (IA-2BC) model of voluntary ethanol drinking (Figure 1B). In a separate cohort of male C57BL/6J mice (n = 19), approximately 8 weeks old at the start of the study, were given chronic, intermittent access to 20% ethanol (v/v) and water in a 2-bottle choice design on Mondays, Wednesdays and Fridays for 24 hr, with two bottles of water on alternate days and weekends. Fluid access began 3 h into the dark cycle. Mice were allowed to drink for 7 weeks to establish stable baseline ethanol consumption prior to testing. On test days, mice were injected with retigabine (RTG, dihydrochloride, Alomone Labs; 0, 5, 10 mg/kg – doses reflect the base, IP, 10 ml/kg in sterile saline) approximately 30 min prior to bottle access in a repeated measures fashion (i.e., each animal received each dose across a 3 week testing period in a counterbalanced order). Ethanol consumption was measured at 2, 4 and 24 hr after bottles were placed on the cages. Prior data indicate that RTG is most effective in rats with a high-drinking phenotype (McGuier et al., 2015), and despite the fact they are inbred and nearly genetically identical, C57BL/6 mice exhibit dramatic intra-strain variability in drinking behavior (Wolstenholme et al., 2011), thus mice were divided into “high-drinking” and “low-drinking” groups. To identify “high-drinking” mice, we initially split animals around the median (12.1 g/kg) of average consumption during week 7 of baseline drinking. But, because C57BL/6J mice tend to front-load in most drinking paradigms (Griffin, Lopez, Yanke, Middaugh, & Becker, 2009; Wilcox et al., 2014) and generally drink in excess of 1.5 g/kg in the first 2 hr of two-bottle choice access (Linsenbardt & Boehm, 2014), we set the additional criteria that mice also had to drink a minimum of 1.5 g/kg in the first 2 hr after vehicle injections. A mixed-model analysis of variance accounting for within-subjects effects was used in the statistical software language SAS (SAS Institute, Cary, NC) to analyze all drinking data. On occasions where intake data were not obtainable, these individual data points were removed from analyses entirely, as opposed to mean substitution or listwise deletion (a total of 12 individual data points were removed from analysis, 9 in the 5 mg/kg dose and 3 in the 10 mg/kg dose). To follow up significant treatment effects, Tukey’s post-hoc tests were used. To determine the variability in drinking across the 7 weeks of baseline drinking, the coefficient of variation (CV = SD/mean) was calculated each week for the individual mice, and the weekly averages were analyzed by repeated measures one-way ANOVA with a Tukey’s post-hoc test (GraphPad Prism, version 6.04). All data are reported as mean ± SEM and statistical significance was established with p < .05, and effect sizes were calculated according to corrected Hedges’s g. Additionally, the dependence of RTG effects on baseline drinking was assessed in a 3-level hierarchical linear model (HLM 7.0) with subject at level 3, dose at level 2, and vehicle drinking and time and their interaction at level 1. The response to KV7 activation by RTG was indicated by the difference between ethanol consumption after vehicle injection and consumption after RTG injections.
Results
K+ channel genes correlate with voluntary drinking
We have previously reported that differences in Kcnn3 and Kcnq2/3 gene expression correlates with voluntary alcohol drinking and that these genes are positional candidates for ethanol consumption in the BXD family of strains (McGuier et al., 2015; Padula et al., 2015). To identify novel K+ channel genes that correlate with drinking in non-dependent and dependent mice, we conducted an extensive analysis of K+ channel genes in the NAc and PFC from C57BL/6J and DBA/2 mice and BXD strains that experienced the LA-2BC and CIE (or Air) paradigm, which results in dependence-induced escalation in 2BC drinking. We first determined significant correlations between K+ channel transcript expression levels and voluntary drinking in non-dependent (Air controls) BXD cases. Thirty-five probe sets corresponding to 16 unique genes in the NAc (Table 1) and 8 probe sets and 7 unique genes in the PFC (Table 2) correlate with voluntary ethanol (15%, v/v) consumption in the non-dependent mice that had 10 weeks of LA-2BC home cage drinking. Although we identified multiple K+ channel genes that correlated with drinking in non-dependent BXD strains, functional enrichment analysis revealed that K+ channel genes were not over-represented in the total gene set that correlated with LA-2BC drinking (NAc: p = 0.196; PFC: p = 0.141; Supplemental Table 2). Tables 1–6 lists the functional class and predominant subcellular localization in brain for each K+ channel gene (Goldstein et al., 2005; Gutman et al., 2005; Kubo et al., 2005; Sun, Zaydman, & Cui, 2012; Trimmer, 2015; Wei et al., 2005). As shown in Tables 1 and 2, the majority of genes that covary with voluntary LA-2BC drinking in non-dependent BXDs encode voltage- and calcium-activated and delayed rectifier K+ channels that localize to dendrites, axons, or presynaptic terminals.
Table 1.
Genes for potassium channels in the nucleus accumbens that significantly correlated with voluntary consumption in non-dependent BXD strains of mice (n = 24 strains).
| Gene | Probe set | Protein | r | R2 | P value | Functional Class | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Kcna1 | 1455785_at | KV1.1 | 0.514 | 0.264 | 0.009 | Delayed rectifier voltage-gated | Axon/Terminals |
| Kcnab3 | 1436019_a_at | KVβ3 | 0.461 | 0.213 | 0.022 | β subunit, KV1.5 modifier | Axon/Terminals |
| Kcnb1 | 1440838_at | KV2.1 | −0.404 | 0.163 | 0.050 | Delayed rectifier voltage-gated | Somatodendritic |
| Kcnd2 | 1433015_at | KV4.2 | −0.684 | 0.468 | 1.25−04 | A-type voltage-gated | Dendrites |
| 1446065_at | −0.665 | 0.442 | 2.42−04 | ||||
| 1458167_at | −0.599 | 0.359 | 0.002 | ||||
| 1459288_at | −0.580 | 0.337 | 0.002 | ||||
| Kcne1l | 1416640_at | minK | −0.610 | 0.372 | 0.001 | β subunit modulator of KV7.1 and KV12.2 | Axon/Terminals |
| Kcnj15 | 1435354_at | Kir4.2 | 0.417 | 0.174 | 0.042 | Inwardly rectifying | Unknown |
| Kcnj3 | 1444025_at | Kir3.1 | 0.438 | 0.192 | 0.031 | G protein-gated, inwardly rectifying | Dendrites |
| 1455374_at | 0.430 | 0.185 | 0.035 | ||||
| Kcnk4 | 1421419_at | K2P4.1 | 0.453 | 0.205 | 0.025 | Two-pore domain open rectifier | Intracellular |
| Kcnma1 | 1440728_at | KCa1.1 | −0.502 | 0.252 | 0.011 | Large conductance, calcium- and voltage-activated | Axon/Terminals |
| 1444973_at | −0.466 | 0.217 | 0.021 | ||||
| 1457018_at | −0.426 | 0.182 | 0.037 | ||||
| 1445774_at | −0.415 | 0.173 | 0.043 | ||||
| 1430276_at | 0.522 | 0.272 | 0.008 | ||||
| Kcnn1 | 1419617_at | KCa2.1 | 0.469 | 0.220 | 0.020 | Small-conductance, calcium-activated | Dendrites |
| Kcnn3 | 1459308_at | KCa2.3 | −0.551 | 0.304 | 0.005 | Small-conductance, calcium-activated | Dendrites |
| Kcnq1 | 1458161_at | KV7.1 | −0.734 | 0.538 | 1.78−05 | M-current delayed rectifier | Axon/Terminals |
| 1457781_at | −0.644 | 0.415 | 4.52−04 | ||||
| 1441588_at | −0.545 | 0.297 | 0.005 | ||||
| 1442029_at | −0.476 | 0.226 | 0.018 | ||||
| 1446255_at | −0.406 | 0.165 | 0.048 | ||||
| Kcnq2 | 1440258_at | KV7.2 | −0.573 | 0.328 | 0.003 | M-current delayed rectifier | Axon/Terminals |
| 1451595_a_at | −0.537 | 0.289 | 0.006 | ||||
| 1438260_at | −0.515 | 0.265 | 0.009 | ||||
| Kcnq5 | 1446537_at | KV7.5 | −0.581 | 0.338 | 0.002 | M-current delayed rectifier | Axon/Terminals |
| 1445837_at | −0.572 | 0.328 | 0.003 | ||||
| 1457898_at | −0.561 | 0.315 | 0.004 | ||||
| 1433110_at | −0.523 | 0.274 | 0.008 | ||||
| Kcnt1 | 1458428_at | KCa4.1 | −0.477 | 0.227 | 0.017 | Sodium-activated, voltage-gated | Somatodendritic |
| 1439486_at | −0.470 | 0.220 | 0.020 | ||||
| Kcnv1 | 1453273_at | KV8.1 | 0.446 | 0.199 | 0.028 | Modifier/silencer of KV2 currents | Unknown |
| 1439333_at | 0.421 | 0.177 | 0.040 |
Table 2.
Potassium channel genes in the prefrontal cortex that significantly correlated with voluntary consumption in non-dependent BXD strains of mice (n = 24 strains).
| Gene | Probe set | Protein | r | R2 | P value | Functional Class | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Kcna5 | 1417680_at | KV1.5 | 0.559 | 0.312 | 0.005 | Delayed rectifier voltage-gated | Axon/Terminals |
| Kcnc3 | 1421980_at | KV3.3 | −0.448 | 0.200 | 0.031 | Delayed rectifier voltage-gated | Axon/Terminals |
| Kcnd2 | 1447764_at | KV4.2 | 0.630 | 0.397 | 0.001 | A-type voltage-gated | Dendrites |
| Kcnj6 | 1425044_at | Kir3.2 | −0.448 | 0.201 | 0.031 | G-protein gated, inwardly rectifying | Dendrites |
| Kcnma1 | 1445774_at | KCa1.1 | −0.456 | 0.208 | 0.028 | Large conductance, calcium- and voltage- activated | Axon/Terminals |
| 1425987_a_at | −0.442 | 0.196 | 0.033 | ||||
| Kcnq5 | 1445837_at | KV7.5 | −0.469 | 0.220 | 0.023 | M-current delayed rectifier | Axon/Terminals |
| Kcns1 | 1421518_at | KV9.1 | 0.560 | 0.314 | 0.005 | Modifier/silencer | Unknown |
Table 6.
Potassium channel gene S-scores in the prefrontal cortex of BXD mice that significantly correlated with the change in voluntary drinking induced by ethanol dependence.
| Gene | Probe set | Protein | r | R2 | P value | Functional Class | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Kcna7 | 1450490_at | Kv1.7 | −0.558 | 0.311 | 0.006 | Delayed rectifier voltage-gated | Axon/Terminals |
| Kcnc1 | 1456965_at | Kv3.1 | −0.459 | 0.211 | 0.028 | Delayed rectifier voltage-gated | Axon/Terminals |
| Kcnd2 | 1447764_at | Kv4.2 | −0.446 | 0.199 | 0.033 | A-type voltage-gated | Dendrites |
| Kcne1 | 1420672_at | minK | −0.460 | 0.211 | 0.027 | β subunit modulator of Kv7.1 and Kv12.2 | Axon/Terminals |
| Kcnh3 | 1459107_at | Kv12.2 | 0.482 | 0.233 | 0.020 | Voltage-gated | Unknown |
| 1445805_x_at | 0.477 | 0.228 | 0.021 | ||||
| Kcnj9 | 1450712_at | Kir3.3 | 0.449 | 0.202 | 0.032 | G-protein gated, inwardly rectifying | Dendrites |
| Kcnk3 | 1425341_at | K2P3.1 | 0.640 | 0.409 | 0.001 | Two-pore domain open rectifier | Intracellular |
| Kcnmb2 | 1431844_at | BK-β2 subunit | −0.420 | 0.176 | 0.046 | Auxillary subunit of KCa1 channels | Axon/Terminals |
Complementary to our previous findings (McGuier et al., 2015; Padula et al., 2015), Kcnn and Kcnq transcript levels in the NAc and PFC significantly correlate with voluntary LA-2BC drinking in non-dependent BXDs. In the current study, all of the 13 Kcnq probe sets showed moderate to strong negative covariation with voluntary consumption (r = −0.55 ± 0.02). Low Kcnq transcript levels are associated with high levels of drinking. This is consistent with pharmacological evidence showing that positive modulation of KV7 channel function reduces drinking in rats (Knapp et al., 2014; McGuier et al., 2015). To confirm the previous rat studies, we examined the ability of retigabine, an FDA-approved KV7 channel positive modulator, to reduce drinking in C57BL/6J mice that were allowed intermittent access (IA) to ethanol (20%, v/v). Although not considered a traditional model to induce ethanol dependence, mice and rats consume large amounts of ethanol in the IA-2BC model and show mild-to-moderate signs of physical dependence during acute withdrawal (Hwa et al., 2011; Li, Bian, Dave, & Ye, 2011; Steensland et al., 2012). Thus, we chose to use the IA-2BC model (Figure 1B) in a separate cohort of C57BL/6J mice as an initial screen to validate Kcnq as a target for reducing ethanol consumption in high-drinking mice. After 7 weeks of drinking in the IA-2BC model (Figure 2A), C57BL/6J mice displayed a large degree of intra-strain variability in ethanol consumption on week 7, ranging from 8.4 ± 0.21 to 18.4 ± 0.37 g/kg/24 hr (Figure 2B). However, analysis of the intra-individual coefficient of variation of drinking showed significant decreases from 0.30 ± 0.02 in week 1 to 0.11 ± 0.02 in week 7 (F4.216, 75.88 = 8.797, p < 0.0001, n = 19 mice; Hedges’s g = 2.34; Figure 2C), demonstrating that drinking in each mouse became more consistent across time. After reaching this stable baseline of intake, mice were injected with RTG (5 or 10 mg/kg, IP) or vehicle approximately 30 min before the start of the 24 hr drinking session, and consumption was measured at 2, 4 and 24 hr time points. There was a significant main effect of RTG treatment on ethanol drinking (F2,32 = 4.76, p = 0.0154, n = 19; Figure 2D), and post-hoc analysis revealed that 10 mg/kg RTG significantly reduced voluntary consumption (Hedges’s g = 0.57). Our previous studies demonstrated that RTG was more effective at reducing intake in Wistar rats with a high-drinking phenotype (McGuier et al., 2015). Consistent with this, both doses of RTG significantly reduced ethanol consumption in high-drinking C57BL/6J mice (F2,13 = 16.74, p = 0.0003; n = 8 mice; 5 mg/kg: Hedges’s g = 0.82, 10 mg/kg: Hedges’s g = 1.08; Figure 2E), whereas neither dose affected intake in low-drinking mice (F2,17 = 0.06, p = 0.9451; n = 11 mice; Figure 2F). To further examine responsivity of RTG, we assessed the relationship between baseline drinking after vehicle injection and RTG-induced changes in ethanol consumption using HLM. There was a strong negative linear relationship between vehicle drinking and RTG-induced alteration of intake across time (β = −0.99, SE = 0.26, t47 = 2.32, p = 0.025), that is, the greater the vehicle drinking, the more KV7 channel activation reduced ethanol consumption (Figure 2G). The two doses of RTG did not differ (β = 0.432, SE = 0.56, t47 = 0.444, p > 0.05) and similar effects were seen at each time point (all Dose X Time interactions, p > 0.5). Finally, treatment with RTG did not alter water consumption overall (F2,32 = 0.38, p = 0.6843; n = 19) or across time (F2,64 = 0.64, p = 0.6358; n = 19) (Supplemental Figure 3), and this remained true when mice were divided into high versus low drinkers (p = 0.2302 and 0.5548, respectively).
Figure 2. The Kv7 channel positive modulator, retigabine, reduced ethanol consumption in high-drinking, but not low-drinking male C57BL/6J mice.
(A) Average weekly ethanol consumption and (B) intra-strain variability of 24 hr ethanol consumption in C57BL/6J mice during week 7 of the intermittent access model. (C) Despite the large between-subjects variation, the coefficient of variance during 7 weeks of access to ethanol in the intermittent model indicates increased within-subject stability (n = 19 mice, * p < .05 vs week 1, ** p < .05 vs week 1, + p < .0001 vs week 1, ++ p < 0.001 vs week 3). (D) Overall, retigabine (10 mg/kg, IP) reduced drinking when measured at 2, 4 and 24 hr in C57BL/6J mice (*p < .005 vs vehicle, main effect of treatment). (E, F) Retigabine (5 and 10 mg/kg) significantly reduced drinking in high-drinking, but not low-drinking C57BL/6J mice (*p < .01 vs vehicle, main effect of treatment). (G) HLM analyses revealed a strong negative linear relationship (p = 0.025) between vehicle drinking and RTG-induced alteration of ethanol intake across time, in that the greater the vehicle drinking, the more KV7 channel activation reduced ethanol consumption.
Chronic intermittent ethanol-responsive K+ channel genes
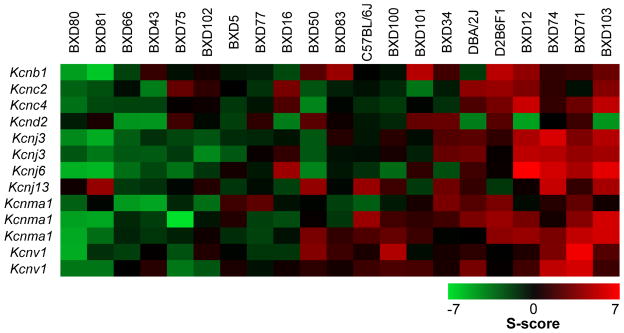
We previously reported that chronic ethanol exposure reduces expression and function of KCa2 (Kcnn) and KV4.2 (Kcnd2) channels in the NAc and hippocampus (Mulholland et al., 2015; Padula et al., 2015; Spencer et al., In Press). To identify novel K+ channel genes in the NAc and PFC that are responsive to ethanol dependence, we utilized the Significance-score (S-score) algorithm (Wolen et al., 2012; Zhang et al., 2002) for probe-level analysis of each strain’s K+ channel transcriptional response to CIE exposure. This analysis was followed by Fisher’s combined probability test to identify K+ channel genes that consistently respond to CIE exposure across BXD strains of mice, regardless of direction. In the NAc, CIE exposure significantly altered 13 probe sets and 9 unique K+ channel genes, including genes that encode KCa1.1 (Kcnma1) and KV4.2 channels (Figure 3 and Table 3). Our previous study demonstrated that high Kcnn3 expression levels in the NAc of BXD mice were protective against escalation of drinking in ethanol dependent BXD mice (Padula et al., 2015). Interestingly, BXD strains that showed a general increase in S-scores for these 13 probe sets (BXD12, BXD71, BXD74, and BXD103) did not escalate their LA-2BC drinking after CIE exposure (Δ = −5.9 ± 10.28%; Δ calculated as percent change between Intake Test 4 and Week 6 of baseline drinking). However, there wasn’t a clear relationship with LA-2BC drinking (Δ = 15.1 ± 41.81%) in BXD strains that showed a general down-regulation of these K+ channel genes.
Figure 3. K+ channel gene expression changes in the nucleus accumbens induced by chronic intermittent ethanol exposure of BXD strains of mice.
Strain heat maps of significantly different gene expression responses based on S-score analysis in the nucleus accumbens (n = 21 strains). Positive S-scores (shown in red) indicate up-regulation by CIE exposure and negative S-scores (shown in green) indicate down-regulation by CIE exposure.
Table 3.
Potassium channel genes in the nucleus accumbens that are significantly altered by chronic intermittent ethanol exposure of BXD strains of mice (n = 21 strains).
| Gene | Probe set | Protein | Fisher q-value | Empiric q-value | Functional Class | Subcellular Localization |
|---|---|---|---|---|---|---|
| Kcnb1 | 1423180_at | Kv2.1 | 2.93−19 | 0.0067 | Delayed rectifier voltage-gated | Somatodendritic |
| Kcnc2 | 1455258_at | Kv3.2 | 1.52−14 | 0.0242 | Delayed rectifier voltage-gated | Axons/terminals |
| Kcnc4 | 1425090_s_at | Kv3.4 | 2.65−14 | 0.0257 | Delayed rectifier voltage-gated | Axons/terminals |
| Kcnd2 | 1459288_at | Kv4.2 | 5.99−16 | 0.0171 | A-type voltage-gated | Dendrites |
| Kcnj3 | 1444025_at | Kir3.1 | 1.93−23 | 0.0015 | G protein-gated, inwardly rectifying | Dendrites |
| 1455374_at | 7.79−22 | 0.0026 | ||||
| Kcnj6 | 1441983_at | Kir3.2 | 6.71−40 | 1.30−5 | G-protein gated, inwardly rectifying | Dendrites |
| Kcnj13 | 1456418_at | Kir7.1 | 6.03−13 | 0.0346 | Inwardly rectifying | Unknown |
| Kcnma1 | 1424848_at | KCa1.1 | 1.71−27 | 4.62−4 | Large conductance, calcium- and voltage-activated | Axons/terminals |
| 1425987_a_at | 6.57−24 | 0.0013 | ||||
| 1457018_at | 1.83−11 | 0.0492 | ||||
| Kcnv1 | 1429741_at | Kv8.1 | 2.87−15 | 0.0203 | Modifier/silencer of Kv2 currents | Unknown |
| 1453273_at | 2.00−13 | 0.0312 |
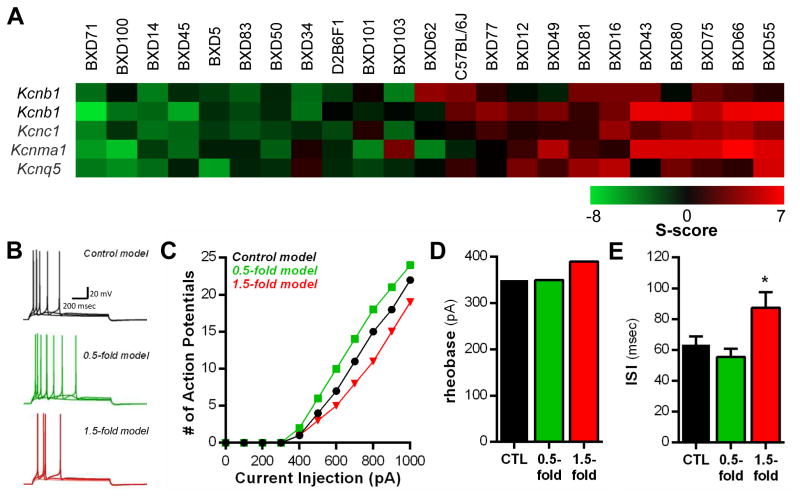
Analysis of array datasets in the PFC identified five probe sets and four unique K+ channel genes that encode KV2.1 (Kcnb1), KV3.1 (Kcnc1), KCa1.1 (Kcnma1), and KV7.5 (Kcnq5) channels that were responsive to CIE exposure (Figure 4A and Table 4). Interestingly, BXD strains (BXD14, BXD45, BXD71, and BXD100) that generally showed negative S-scores across these 5 probe sets were resistant to CIE-induced escalations in LA-2BC drinking. The average CIE-induced percent change from baseline drinking values in these strains was −13.4 ± 11.6%. In contrast, strains that generally showed positive S-scores across these 5 probe sets (BXD55, BXD66, BXD75, and BXD80) elevated their LA-2BC drinking after CIE exposure (Δ = 59.9 ± 31.74%). This suggests that coordinated adaptations in this set of four K+ channel genes may influence output neurons of the PFC that control drinking behavior in dependent BXDs.
Figure 4. Intrinsic excitability changes revealed by computational modeling of K+ channel gene expression adaptations in the prefrontal cortex of ethanol dependent BXD mice.
(A) Strain heat maps of significantly different gene expression responses based on S-score analysis in the prefrontal cortex (n = 23 strains). Positive S-scores (shown in red) indicate up-regulation by CIE exposure and negative S-scores (shown in green) indicate down-regulation by CIE exposure. (B) Traces showing simulated evoked action potentials in response to depolarizing current steps (300, 400, 500 pA) under control conditions (black traces), 0.5-fold down-regulation of channel activity (green traces), and 1.5-fold increase in channel conductance (red traces). (C) Input-output relationship for each simulation. The number of action potentials produced by each current step is compared across the simulations. The 1.5-fold model revealed a reduction in the number of action potentials elicited by current injection at the soma. (D, E) The rheobase current and ISI are increased in the 1.5-fold model simulations, resulting in fewer action potentials evoked by current injection (*p < 0.05 vs. CTL stimulations).
Table 4.
Potassium channel genes in the prefrontal cortex that are responsive to chronic intermittent ethanol exposure of BXD strains of mice (n = 23 strains).
| Gene | Probe set | Protein | Fisher q-value | Empiric q-value | Functional Class | Subcellular Localization |
|---|---|---|---|---|---|---|
| Kcnb1 | 1423179_at | Kv2.1 | 1.07−20 | .0382 | Delayed rectifier voltage-gated | Somatodendritic |
| 1423180_at | 8.77−62 | 5.33−05 | ||||
| Kcnc1 | 1423559_at | Kv3.1 | 1.94−23 | 0.0274 | Delayed rectifier voltage-gated | Axons/terminals |
| Kcnma1 | 1424848_at | KCa1.1 | 1.75−57 | 1.31−04 | Large conductance, calcium- and voltage-activated | Axons/terminals |
| Kcnq5 | 1433110_at | Kv7.5 | 1.18−27 | 0.0181 | M-current delayed rectifier | Axons/terminals |
To begin to understand how coordinated changes in KV3.1, KV7.5, KCa1.1, and KV2.1 channels affect PFC physiology, we modeled the passive and active membrane properties of a deep-layer PFC pyramidal neuron. The conductances of these four K+ channels of interest were then simultaneously decreased by 0.5-fold to model gene changes in ethanol-dependent BXD14, BXD45, BXD71, and BXD100 strains or were increased by 1.5-fold to model ethanol-dependent BXD55, BXD66, BXD75, and BXD80 strains (Figure 4B). The biophysical stimulations suggest minimal changes in the resting membrane potential (in mV; CTL: −63.9; 0.5-fold: −63.6; 1.5-fold: −62.9), action potential (AP) threshold (in mV: CTL: 48; 0.5-fold: 48; 1.5-fold: 49), AP amplitude (in mV: CTL: 91; 0.5-fold: 89; 1.5-fold: 90), or AHP duration measured at 500–1000 pA steps (in ms: CTL: 59.7 ± 4.6; 0.5-fold: 50.3 ± 6.7; 1.5-fold: 66.7 ± 5.3; one-way ANOVA, F2,15 = 0.55, p = 0.22) by coordinated up- or down-regulation of these four K+ channels. However, other measures of intrinsic excitability that could directly affect output of the PFC were affected. The number of evoked APs in response to depolarizing current steps delivered to the soma were reduced by 23.0 ± 2.5% in the 1.5-fold model simulation compared the 0.5-fold simulation, which resulted in 27.6 ± 6.5% increase in evoked firing (500–1000 pA steps; Figure 4B and C). Consistent with this, the rheobase current (lowest amount of current needed to produce an AP) was increased in the 1.5-fold simulation by 40 pA (Figure 4D), suggesting that the neuron is less likely to fire when depolarized. The ISI (Figure 4E; one-way ANOVA, F2,15 = 1.82, p = 0.019, post-hoc test, *p < 0.05) was markedly increased in the simulations with a 1.5-fold increase in channel conductance (500–1000 pA steps), suggesting that when the neurons reach threshold, fewer action potentials are produced due to changes in mechanisms that control AP generation. These modeling data demonstrate that coordinated regulation of KV3.1, KV7.5, KCa1.1, and KV2.1 channels by CIE exposure may affect intrinsic excitability of deep layer PFC pyramidal neurons.
CIE-responsive K+ channel genes that correlate with drinking in dependent mice
As a final analysis, we correlated S-scores (CIE-induced change in transcript levels) in the PFC and NAc with the percent change from baseline drinking (CIE-induced change in LA-2BC ethanol consumption) to further explore the relationship between dependence-induced regulation of K+ channel transcripts and drinking in CIE exposed BXD mice. As shown in Supplemental Figure 4 and Table 5, in the NAc S-scores for 13 probe sets and 12 unique K+ channel genes significantly correlated the change in drinking induced by chronic exposure to ethanol vapor in inhalation chambers, with S-scores for 7 of the probe sets demonstrating moderate-to-strong positive relationships. In the PFC, S-scores for 9 probe sets and 8 unique K+ channel genes significantly correlated the CIE-induced change in LA-2BC drinking (Supplemental Figure 5 and Table 6). Moderate-to-strong negative relationships were observed for 5 of the 9 S-scores for K+ channel genes, with the remaining S-scores for 4 genes showing moderate-to-strong positive relationships. Functional enrichment analysis showed that K+ channel genes in the NAc, but not PFC, were over-represented when compared with all of the genes that significantly correlated with the CIE-induced change in LA-2BC drinking in dependent BXD mice (NAc: p = 0.00063; PFC: p = 0.597; Supplemental Table 3).
Discussion
The purpose of this study was to use existing BXD RI mRNA data in GeneNetwork as an experimental resource to evaluate potential critical K+ channels genes that correlate with drinking in non-dependent and dependent subjects. We identified a number of novel K+ channel genes (e.g., Kcnd2) in the PFC and NAc that covary well with voluntary ethanol drinking in non-dependent BXD cases. We also identified a smaller subset of K+ channels genes in the PFC (Kcnc1) and NAc (Kcnv family) that are dysregulated by CIE exposure and that correlate with the CIE-induced changes in drinking. While this analysis fulfilled the goal of identifying new relations between K+ channels genes and dependence-induced drinking, this study also validated existing genetic findings on K+ channels genes—KCNN, KCNQ, and KCNJ—previously linked to AUD. As discussed below, many of these genes have been reported in human genetic studies of AUD risk, further validating the use of BXD cohort for experimentally testing the genetic underpinnings of complex behaviors, such as voluntary drinking associated with ethanol dependence. Finally, the strong overlap between the present findings and previous evidence validates this experimental genetic approach to investigate the link between dependence-induced drinking and other classes of ion channels, receptors, or signaling cascades.
Voltage-dependent K+ channels and risk for alcohol dependence
In non-dependent BXD RI mice, genes encoding voltage-dependent KV7 channels in the PFC (Kcnq5) and NAc (Kcnq1, Kcnq2, and Kcnq5) significantly correlate with voluntary LA-2BC drinking. This confirmed our previous findings linking Kcnq2 and Kcnq3 genes and alcohol-related behaviors, including ethanol preference and consumption in an IA-2BC model (McGuier et al., 2015). Using a pharmacological approach to validate a role for Kcnq in regulating drinking, systemic administration of the KV7 channel positive modulator retigabine reduced drinking in C57BL/6J mice given access to ethanol in the IA-2BC model. Consistent with existing findings in rats (McGuier et al., 2015), retigabine was most effective at reducing drinking in mice with a high-drinking, but not low-drinking phenotype. This functional genomic analysis also showed that Kcnq5 was differentially expressed in the PFC of ethanol-dependent BXD RI mice, and the dependence-induced regulation of Kcnq1 and Kcnq5 transcript levels in the NAc correlated with the change in LA-2BC drinking induced by CIE exposure. Interestingly, single nucleotide polymorphisms (SNPs) in KCNQ1 (rs12574151) and KCNQ5 (rs3799285) are associated with early onset and symptoms associated with alcohol dependence (Edenberg et al., 2010; Kendler et al., 2011). Together with our previous study, these data provide additional evidence that KCNQ genes are candidates for genetic diversity in alcohol-related behaviors, and suggest that differential expression or altered function of KCNQ produced by dependence in these populations may underlie the phenotypic differences that are risk factors for developing an AUD.
It is important to note that, while overlap in the genes that correlated with non-dependent and dependence-induced ethanol consumption was minimal, these differences could reflect compensatory neuroadaptations in different circuits as a mechanism to counteract changes in neural activity during repeated bouts of ethanol exposure and withdrawal. It is possible that correlations seen prior to dependence represent genes that are markers of a predisposition to heavy ethanol consumption, whereas changes seen after dependence are more reflective of the consequences of heavy alcohol exposure. For example, other voltage-dependent K+ channels (i.e., Kcnd2) correlated with LA-2BC drinking in non-dependent and dependent BXD RI strains of mice and were also differentially regulated following induction of ethanol dependence. Of particular interest are voltage-dependent K+ channels composed of KV4.2 α subunits that are encoded by Kcnd2. KV4.2 channels underlie the transient A-type K+ current in cortical and hippocampal dendrites where they shape postsynaptic responses, constrain coincidence detection, and control the amplitude of bAPs (Chen et al., 2006; Hoffman, Magee, Colbert, & Johnston, 1997), and recent evidence has implicated an important role of KV4.2 channels in both synaptic plasticity and cognition (Jung, Kim, & Hoffman, 2008; Lockridge & Yuan, 2011; Lugo, Brewster, Spencer, & Anderson, 2012; Truchet et al., 2012). Consistent with emerging data on chronic ethanol and KV4.2 channels, Kcnd2 transcript levels were significantly regulated by CIE exposure in the NAc. Prolonged withdrawal from intermittent ethanol exposure enhanced A-type K+ currents in medium spiny neurons in the NAc (Marty & Spigelman, 2012). Our previous studies showed that chronic ethanol reduced A-type K+ currents and KV4.2 channel surface expression in hippocampus, (Mulholland et al., 2015; Spencer et al., In Press), demonstrating differential effects of chronic ethanol on KV4.2 channels in across brain structures. Interestingly, two SNPs in KCND2 (rs728115 and rs17142876) were recently identified as a risk factor for developing alcohol and nicotine co-dependence (Buhler et al., 2015; Zuo et al., 2012), and decreased expression of KCND2 was reported in the frontal cortex and amygdala (central and basolateral nuclei) of alcoholics (Ponomarev, Wang, Zhang, Harris, & Mayfield, 2012). However, our evidence that Kcnd2 correlates with drinking in BXD RI mice is the first study to suggest that Kv4.2 channels in the PFC and NAc regulate voluntary drinking in non-dependent and dependent mice. Thus, future studies are necessary to determine if ethanol dependence alters KV4.2 channel expression in the PFC or NAc and if KV4.2 channels are a novel therapeutic target for reducing escalated drinking in dependence models.
In the PFC and NAc, Kncb1 (KV2.1) and the genes that encode KV2 channels auxiliary subunits (Kcns1/2 and Kcnv1/2) that act as KV2 channel modifier/silencers correlate with voluntary ethanol consumption and dependence-induced regulation of drinking in BXD mice. In addition, Kcnb1 and Kcnv1 were differentially regulated by CIE exposure in the PFC or NAc. KV2.1 channels are delayed-rectifiers that localize to somatodendritic domains and the axon initial segment of neocortical and hippocampal pyramidal neurons where they regulate intrinsic excitability during periods of repetitive, high-frequency activity. KV2.1 channel phosphorylation and surface clustering are regulated by NMDA receptor and glutamate transporter activity (Mulholland et al., 2008), and we previously demonstrated that acute and chronic ethanol exposure or ethanol withdrawal do not affect KV2.1 channel expression or phosphorylation in organotypic hippocampal cultures (Mulholland et al., 2008; Mulholland, Carpenter-Hyland, Woodward, & Chandler, 2009). It is unknown how KV2.1 channels and its modifiers influence excitability of accumbens neurons or if ethanol dependence affects their function or expression in the PFC or NAc. However, intriguing evidence from a meta-analysis using samples from the Collaborative Study on the Genetics of Alcoholism (COGA) and the Study of Addiction: Genetics and Environment (SAGE) revealed that two SNPs in KCNB1 (rs2128158 and rs2929567) showed significant associations with maximum number of alcoholic drinks consumed in a 24 hr period (Pan et al., 2013). Given these converging lines of evidence, future studies should determine if ethanol-induced dysregulation of or genetic polymorphisms in KV2.1 channels and their modifiers/silencers impact risk for developing an alcohol use disorder.
Calcium-activated K+ channels and ethanol dependence
Similar to KCNB1, recent studies identified two SNPs in KCNMA1 (rs717207 and rs12219105) as a genetic risk variant that associated with symptoms of alcohol dependence and early onset alcohol dependence in European- and African-Americans (Edenberg et al., 2010; Kendler et al., 2011). In our study, transcript levels in the PFC and NAc for Kcnma1 (large-conductance, voltage and calcium-activated KCa1.1 channels) significantly correlated with voluntary drinking in non-dependent BXD RI strains of mice. Kcnma1 in both regions was also dysregulated by ethanol dependence. In addition, dependence-induced regulation of the Kcnmb2 β2 auxiliary subunit of KCa1.1 channels correlated with the change in LA-2BC drinking in ethanol dependent BXD RI mice. Auxiliary β subunits confer resistance to acute ethanol-induced potentiation of KCa1.1 channel-mediated currents (Feinberg-Zadek & Treistman, 2007; Martin et al., 2004), and a recent report demonstrated that KCa1 β1 or β4 subunits did not influence voluntary consumption of ethanol in either a continuous access 2BC or the LA-2BC model in non-dependent C57BL/6J mice (Kreifeldt et al., 2013). However, genetic deletion of the KCa1 β4 subunit attenuated, whereas deletion of the KCa1 β1 subunit enhanced the escalation of ethanol drinking ethanol-dependent C57BL/6J mice in the LA-2BC model (Kreifeldt et al., 2013). In addition to KCa1 channels, Kcnn1 and Kcnn3 (small-conductance, calcium-activated KCa2.1/3 channels) transcript levels in the NAc significantly correlated with voluntary drinking in BXD RI strains of mice. Multiple studies in rats and mice provide strong evidence that KCa2 channel positive modulators are promising drugs for reducing drinking (Hopf et al., 2010; Hopf et al., 2011; Mulholland, 2012; Padula et al., 2013), and our previous study demonstrated that elevated NAc Kcnn3 expression levels protected against dependence-induced escalation of drinking in BXD RI mice (Padula et al., 2015). Thus, future studies are necessary to determine if calcium-activated KCa1 and KCa2 channels are effective pharmacogenetic targets for treating alcohol use disorder.
Computational modeling of PFC neurons
In the PFC, coordinated decreases in transcript expression levels of Kcnb1, Kcnc1, Kcnma1, and Kcnq5 protected against dependence-induced escalation of drinking, whereas enhanced expression of these same four genes was associated with general increases in consumption induced by dependence. Over-expression of these four K+ channels in a computational model of a deep-layer PFC pyramidal neuron reduced intrinsic excitability largely driven by an increase in rheobase and ISI duration. Recent studies have demonstrated that CIE exposure of C57BL/6J mice produces plasticity changes in passive and active membrane properties of layer V PFC pyramidal neurons (Hu, Morris, Carrasco, & Kroener, 2015; Nimitvilai, Lopez, Mulholland, & Woodward, 2015; Pleil et al., 2015). KV7 and KV2.1 channels are known regulators of intrinsic excitability and ISI in cortical neurons (Guan, Armstrong, & Foehring, 2013; Santini & Porter, 2010). Our bioinformatics and computational modeling analyses identified possible molecular substrates that may drive aberrant plasticity of intrinsic excitability of PFC projection neurons. We hypothesize that ethanol dependence produces functional adaptations in expression of these K+ channels in the PFC neurons that project to subcortical structures that regulate consummatory behaviors. Future studies are necessary to determine if CIE-induced regulation of K+ channels in the PFC or NAc work independently or in conjunction to protect against or promote escalation in drinking in dependent mice.
Summary and future directions
The major finding of this multifaceted approach was the identification of novel ethanol-sensitive K+ channel candidate genes that regulate long-term and escalated drinking in the PFC and NAc of BXD RI strains of mice. In addition, this targeted bioinformatics approach also confirmed previous studies identifying relationships between select K+ channel genes and alcoholism, providing further evidence that BXD strains of mice are a powerful model for studying complex phenotypes. We also demonstrated that these bioinformatics approaches are useful for identifying novel pharmacological targets by validating one of the implicated K+ channel genes using an in vivo pharmacological approach. And lastly, we established that these approaches can reveal coordinated changes in gene expression that, when modeled together, can reveal important cellular signaling patterns that explicate their role in behavior.
This study shows that select K+ channel genes in the PFC and NAc covary with voluntary drinking only in non-dependent BXD mice, while a smaller subset of K+ channel genes were sensitive to chronic ethanol exposure and correlated with drinking in ethanol dependent mice. Some of these K+ channel genes (e.g., KCND2, KCNK3, KCNMA1, KCNQ2/3) were also differentially expressed in postmortem human frontal cortex, amygdala, or NAc from alcohol dependent individuals (Flatscher-Bader, Harrison, Matsumoto, & Wilce, 2010; Liu et al., 2006; Ponomarev et al., 2012). Recent studies show that activity-dependent processes regulate K+ channel gene expression, including Kcnmb2, Kcnn1, Kcnq3, and Kcnv1 (Lee et al., 2015), and NMDA receptor activity can modulate surface trafficking and function of KCa2, KV2.1, and KV4.2 K+ channels (Kim, Jung, Clemens, Petralia, & Hoffman, 2007; Mulholland et al., 2008; Ngo-Anh et al., 2005). Because chronic ethanol exposure results in imbalances in NMDA receptor expression and synaptic transmission, we speculate that CIE-induced adaptations in some K+ channel genes are compensatory mechanisms to counterbalance chronic ethanol inhibition of NMDA receptor activity, whereas adaptations in other K+ channel genes may be protective against or facilitate escalation of drinking in dependent BXD mice.
Additionally, in an effort to pharmacologically validate one of these genetic targets, Kcnq, which has been implicated in a number of studies to influence ethanol consumption, we utilized a mouse model of ethanol consumption, the IA-2BC, which represents an intermediate stage of the transition to dependence. Using RTG, a nonselective Kcnq channel opener, we demonstrated that activation of this family of voltage-gated K+ channels can reduce heavy ethanol consumption in “high-drinking” mice. While future studies should test the remaining hypotheses generated from this bioinformatics approach, it is also necessary to determine which subregion in the PFC (e.g., prelimbic or infralimbic medial, orbitofrontal, or anterior cingulate cortex) and NAc (e.g., core or shell) these adaptations are occurring with an additional emphasis on identifying the specific subcellular compartment (e.g., axon vs dendrite) or cell type (e.g., interneuron, projection neuron). Finally, functional studies should measure how CIE exposure affects K+ channel influence on intrinsic excitability, action potential characteristics, and synaptic communication within the corticostriatal circuitry.
In recent years, personalized medicine approaches have proven useful for treating alcohol use disorders (Litten et al., 2015). For example, individuals with SNPs in OPRM1, HTR3A/B, or GRIK1 genes moderate the reduction in heavy drinking by naltrexone, ondansetron, and topiramate treatment, respectively (Anton et al., 2008; Johnson, Seneviratne, Wang, Ait-Daoud, & Li, 2013; Kranzler, Armeli, et al., 2014; Kranzler, Covault, et al., 2014; Oslin et al., 2003). There is evidence that SNPs in KCNB1, KCND2, KCNJ6, KCNMA1, and KCNQ1/5 are associated with alcohol dependence or increased risk for developing an alcohol use disorder (Buhler et al., 2015; Clarke et al., 2011; Edenberg et al., 2010; Kendler et al., 2011; Namkung, Kim, & Park, 2005; Pan et al., 2013; Zuo et al., 2012). This emerging evidence that K+ channel transcript levels correlate with drinking and polymorphisms in K+ channels are related to alcohol dependence implicate select K+ channels that may cause or maintain heavy alcohol intake and may represent a new relevant set of biomarkers for alcohol dependence. Thus, our preclinical data suggest that clinical laboratory studies are necessary to determine if these K+ channels are relevant pharmacogenetic targets that may advance personalized medicine approaches for treating alcohol use disorders.
Supplementary Material
Table 5.
Potassium channel gene S-scores in the nucleus accumbens that significantly correlated with the change in voluntary drinking after chronic intermittent ethanol exposure of BXD mice.
| Gene | Probe set | Protein | r | R2 | P value | Functional Class | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Kcna4 | 1422255_at | Kv1.4 | 0.477 | 0.227 | 0.029 | A-type voltage-gated | Axon/Terminals |
| Kcnh2 | 1449544_a_at | Kv11.1 | 0.527 | 0.277 | 0.014 | Inwardly rectifying voltage-gated | Unknown |
| Kcnh7 | 1427634_at | Kv11.3 | 0.467 | 0.218 | 0.033 | Voltage-gated | Unknown |
| Kcnj1 | 1418613_at | Kir1.1 | −0.538 | 0.289 | 0.012 | Inwardly rectifying | Unknown |
| Kcnj8 | 1418142_at | Kir6.1 | −0.451 | 0.203 | 0.04 | ATP-sensitive | Dendrites |
| Kcnj11 | 1455417_at | Kir6.2 | 0.484 | 0.235 | 0.026 | ATP-sensitive | Dendrites |
| Kcnj16 | 1435094_at | Kir5.1 | −0.455 | 0.207 | 0.038 | Inwardly rectifying | Unknown |
| Kcnq1 | 1459443_at | Kv7.1 | −0.507 | 0.257 | 0.019 | M-current delayed rectifier | Axon/Terminals |
| 1449464_at | −0.451 | 0.204 | 0.04 | ||||
| Kcnq5 | 1457587_at | Kv7.5 | 0.625 | 0.391 | 0.002 | M-current delayed rectifier | Axon/Terminals |
| Kcns1 | 1421518_at | Kv9.1 | −0.545 | 0.297 | 0.011 | Modifier/silencer of Kv2 currents | Unknown |
| Kcns2 | 1457325_at | Kv9.2 | 0.472 | 0.223 | 0.031 | Modifier/silencer of Kv2 currents | Unknown |
| Kcnv2 | 1440537_at | Kv8.2 | 0.538 | 0.29 | 0.012 | Modifier/silencer of Kv2 currents | Unknown |
Highlights.
Bioinformatic analyses of genetic regulation by ethanol dependence identified novel candidate K+ channel genes.
Analyses also confirmed the known genetic relationship between Kcnq, Kcnj, and Kcnn genes and alcohol drinking.
Pharmacological validation of Kcnq as a pharmacogenetic target with retigabine-induced reduction in alcohol consumption in “high-drinking” mice demonstrates the potential utility of the personalized medicine approach.
Acknowledgments
These studies were supported by NIH grants AA020930 (PJM), AA023288 (PJM), AA020929 (MFL), AA014095 (HCB), AA010761 (HCB), AA016667 (MFM), AA016662 (RWW), AA022701 (LJC), and AA022475 (HTD).
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akemann W, Lundby A, Mutoh H, Knopfel T. Effect of voltage sensitive fluorescent proteins on neuronal excitability. Biophys J. 2009;96(10):3959–3976. doi: 10.1016/j.bpj.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton RF, Oroszi G, O’Malley S, Couper D, Swift R, Pettinati H, Goldman D. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65(2):135–144. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 2004;28(12):1829–1838. doi: 10.1097/01.alc.0000149977.95306.3a. [DOI] [PubMed] [Google Scholar]
- Book SW, Myrick H. Novel anticonvulsants in the treatment of alcoholism. Expert Opin Investig Drugs. 2005;14(4):371–376. doi: 10.1517/13543784.14.4.371. [DOI] [PubMed] [Google Scholar]
- Buhler KM, Gine E, Echeverry-Alzate V, Calleja-Conde J, de Fonseca FR, Lopez-Moreno JA. Common single nucleotide variants underlying drug addiction: more than a decade of research. Addict Biol. 2015;20(5):845–871. doi: 10.1111/adb.12204. [DOI] [PubMed] [Google Scholar]
- Bukiya AN, Kuntamallappanavar G, Edwards J, Singh AK, Shivakumar B, Dopico AM. An alcohol-sensing site in the calcium- and voltage-gated, large conductance potassium (BK) channel. Proc Natl Acad Sci U S A. 2014;111(25):9313–9318. doi: 10.1073/pnas.1317363111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale NT, Hines ML. The NEURON Book. Cambridge, UK: Cambridge University Press; 2006. [Google Scholar]
- Chen X, Yuan LL, Zhao C, Birnbaum SG, Frick A, Jung WE, … Johnston D. Deletion of Kv4.2 gene eliminates dendritic A-type K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26(47):12143–12151. doi: 10.1523/JNEUROSCI.2667-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp P. Current progress in pharmacologic treatment strategies for alcohol dependence. Expert Rev Clin Pharmacol. 2012;5(4):427–435. doi: 10.1586/ecp.12.31. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Laucht M, Ridinger M, Wodarz N, Rietschel M, Maier W, … Schumann G. KCNJ6 is associated with adult alcohol dependence and involved in gene x early life stress interactions in adolescent alcohol drinking. Neuropsychopharmacology. 2011;36(6):1142–1148. doi: 10.1038/npp.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa A. The role of topiramate and other anticonvulsants in the treatment of alcohol dependence: a clinical review. CNS Neurol Disord Drug Targets. 2010;9(1):45–49. doi: 10.2174/187152710790966696. [DOI] [PubMed] [Google Scholar]
- Durstewitz D, Seamans JK, Sejnowski TJ. Dopamine-mediated stabilization of delay-period activity in a network model of prefrontal cortex. J Neurophysiol. 2000;83(3):1733–1750. doi: 10.1152/jn.2000.83.3.1733. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10(8):487–494. doi: 10.1038/nrgastro.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, … Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34(5):840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg-Zadek PL, Treistman SN. Beta-subunits are important modulators of the acute response to alcohol in human BK channels. Alcohol Clin Exp Res. 2007;31(5):737–744. doi: 10.1111/j.1530-0277.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcohol Clin Exp Res. 2010;34(7):1291–1302. doi: 10.1111/j.1530-0277.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Ghezzi A, Pohl JB, Wang Y, Atkinson NS. BK channels play a counter-adaptive role in drug tolerance and dependence. Proc Natl Acad Sci U S A. 2010;107(37):16360–16365. doi: 10.1073/pnas.1005439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies A, Willshaw D. Membrane channel interactions underlying rat subthalamic projection neuron rhythmic and bursting activity. J Neurophysiol. 2006;95(4):2352–2365. doi: 10.1152/jn.00525.2005. [DOI] [PubMed] [Google Scholar]
- Goldstein SA, Bayliss DA, Kim D, Lesage F, Plant LD, Rajan S. International Union of Pharmacology. LV. Nomenclature and molecular relationships of two-P potassium channels. Pharmacol Rev. 2005;57(4):527–540. doi: 10.1124/pr.57.4.12. [DOI] [PubMed] [Google Scholar]
- Griffin WC, 3rd, Lopez MF, Yanke AB, Middaugh LD, Becker HC. Repeated cycles of chronic intermittent ethanol exposure in mice increases voluntary ethanol drinking and ethanol concentrations in the nucleus accumbens. Psychopharmacology (Berl) 2009;201(4):569–580. doi: 10.1007/s00213-008-1324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Armstrong WE, Foehring RC. Kv2 channels regulate firing rate in pyramidal neurons from rat sensorimotor cortex. J Physiol. 2013;591(19):4807–4825. doi: 10.1113/jphysiol.2013.257253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman GA, Chandy KG, Grissmer S, Lazdunski M, McKinnon D, Pardo LA, … Wang X. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol Rev. 2005;57(4):473–508. doi: 10.1124/pr.57.4.10. [DOI] [PubMed] [Google Scholar]
- Herman MA, Sidhu H, Stouffer DG, Kreifeldt M, Le D, Cates-Gatto C, … Contet C. GIRK3 gates activation of the mesolimbic dopaminergic pathway by ethanol. Proc Natl Acad Sci U S A. 2015;112(22):7091–7096. doi: 10.1073/pnas.1416146112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387(6636):869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- Hopf FW, Bowers MS, Chang SJ, Chen BT, Martin M, Seif T, … Bonci A. Reduced nucleus accumbens SK channel activity enhances alcohol seeking during abstinence. Neuron. 2010;65(5):682–694. doi: 10.1016/j.neuron.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf FW, Simms JA, Chang SJ, Seif T, Bartlett SE, Bonci A. Chlorzoxazone, an SK-type potassium channel activator used in humans, reduces excessive alcohol intake in rats. Biol Psychiatry. 2011;69(7):618–624. doi: 10.1016/j.biopsych.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Morris B, Carrasco A, Kroener S. Effects of acamprosate on attentional set-shifting and cellular function in the prefrontal cortex of chronic alcohol-exposed mice. Alcohol Clin Exp Res. 2015;39(6):953–961. doi: 10.1111/acer.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa LS, Chu A, Levinson SA, Kayyali TM, DeBold JF, Miczek KA. Persistent escalation of alcohol drinking in C57BL/6J mice with intermittent access to 20% ethanol. Alcohol Clin Exp Res. 2011;35(11):1938–1947. doi: 10.1111/j.1530-0277.2011.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Seneviratne C, Wang XQ, Ait-Daoud N, Li MD. Determination of genotype combinations that can predict the outcome of the treatment of alcohol dependence using the 5-HT(3) antagonist ondansetron. Am J Psychiatry. 2013;170(9):1020–1031. doi: 10.1176/appi.ajp.2013.12091163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SC, Kim J, Hoffman DA. Rapid, bidirectional remodeling of synaptic NMDA receptor subunit composition by A-type K+ channel activity in hippocampal CA1 pyramidal neurons. Neuron. 2008;60(4):657–671. doi: 10.1016/j.neuron.2008.08.029. S0896-6273(08)00772-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, … Gejman PV. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res. 2011;35(5):963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RE, Kerns RT, Kong X, Archer KJ, Miles MF. SScore: an R package for detecting differential gene expression without gene expression summaries. Bioinformatics. 2006;22(10):1272–1274. doi: 10.1093/bioinformatics/btl108. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Zhang L, Miles MF. Application of the S-score algorithm for analysis of oligonucleotide microarrays. Methods. 2003;31(4):274–281. doi: 10.1016/s1046-2023(03)00156-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Jung SC, Clemens AM, Petralia RS, Hoffman DA. Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron. 2007;54(6):933–947. doi: 10.1016/j.neuron.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp CM, O’Malley M, Datta S, Ciraulo DA. The Kv7 potassium channel activator retigabine decreases alcohol consumption in rats. Am J Drug Alcohol Abuse. 2014;40(3):244–250. doi: 10.3109/00952990.2014.892951. [DOI] [PubMed] [Google Scholar]
- Kozell LB, Walter NA, Milner LC, Wickman K, Buck KJ. Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J Neurosci. 2009;29(37):11662–11673. doi: 10.1523/JNEUROSCI.1413-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Armeli S, Wetherill R, Feinn R, Tennen H, Gelernter J, … Pond T. Self-efficacy mediates the effects of topiramate and GRIK1 genotype on drinking. Addict Biol. 2014 doi: 10.1111/adb.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Covault J, Feinn R, Armeli S, Tennen H, Arias AJ, … Kampman KM. Topiramate treatment for heavy drinkers: moderation by a GRIK1 polymorphism. Am J Psychiatry. 2014;171(4):445–452. doi: 10.1176/appi.ajp.2013.13081014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreifeldt M, Le D, Treistman SN, Koob GF, Contet C. BK channel beta1 and beta4 auxiliary subunits exert opposite influences on escalated ethanol drinking in dependent mice. Front Integr Neurosci. 2013;7:105. doi: 10.3389/fnint.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, … Vandenberg CA. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev. 2005;57(4):509–526. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- Lee KY, Royston SE, Vest MO, Ley DJ, Lee S, Bolton EC, Chung HJ. N-methyl-D-aspartate receptors mediate activity-dependent down-regulation of potassium channel genes during the expression of homeostatic intrinsic plasticity. Mol Brain. 2015;8:4. doi: 10.1186/s13041-015-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bian W, Dave V, Ye JH. Blockade of GABA(A) receptors in the paraventricular nucleus of the hypothalamus attenuates voluntary ethanol intake and activates the hypothalamic-pituitary-adrenocortical axis. Addict Biol. 2011;16(4):600–614. doi: 10.1111/j.1369-1600.2011.00344.x. [DOI] [PubMed] [Google Scholar]
- Linsenbardt DN, Boehm SL., 2nd Alterations in the rate of binge ethanol consumption: implications for preclinical studies in mice. Addict Biol. 2014;19(5):812–825. doi: 10.1111/adb.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litten RZ, Ryan ML, Falk DE, Reilly M, Fertig JB, Koob GF. Heterogeneity of alcohol use disorder: understanding mechanisms to advance personalized treatment. Alcohol Clin Exp Res. 2015;39(4):579–584. doi: 10.1111/acer.12669. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Lockridge A, Yuan LL. Spatial learning deficits in mice lacking A-type K(+) channel subunits. Hippocampus. 2011;21(11):1152–1156. doi: 10.1002/hipo.20877. [DOI] [PubMed] [Google Scholar]
- Lopez MF, Miles MF, Williams RW, Becker HC. Effect of Chronic Intermittent Ethanol Exposure on Ethanol Drinking in BXD RI Strains. Alcohol. doi: 10.1016/j.alcohol.2016.09.003. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo JN, Brewster AL, Spencer CM, Anderson AE. Kv4.2 knockout mice have hippocampal-dependent learning and memory deficits. Learning & memory. 2012;19(5):182–189. doi: 10.1101/lm.023614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm R, Myrick H, Brady KT, Ballenger JC. Update on anticonvulsants for the treatment of alcohol withdrawal. Am J Addict. 2001;10(Suppl):16–23. doi: 10.1080/10550490150504100. [DOI] [PubMed] [Google Scholar]
- Maljevic S, Lerche H. Potassium channel genes and benign familial neonatal epilepsy. Prog Brain Res. 2014;213:17–53. doi: 10.1016/B978-0-444-63326-2.00002-8. [DOI] [PubMed] [Google Scholar]
- Martin G, Puig S, Pietrzykowski A, Zadek P, Emery P, Treistman S. Somatic localization of a specific large-conductance calcium-activated potassium channel subtype controls compartmentalized ethanol sensitivity in the nucleus accumbens. J Neurosci. 2004;24(29):6563–6572. doi: 10.1523/JNEUROSCI.0684-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty VN, Spigelman I. Effects of alcohol on the membrane excitability and synaptic transmission of medium spiny neurons in the nucleus accumbens. Alcohol. 2012;46(4):317–327. doi: 10.1016/j.alcohol.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J, Blednov YA, Harris RA. Behavioral and Genetic Evidence for GIRK Channels in the CNS: Role in Physiology, Pathophysiology, and Drug Addiction. Int Rev Neurobiol. 2015;123:279–313. doi: 10.1016/bs.irn.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuier NS, Griffin WC, 3rd, Gass JT, Padula AE, Chesler EJ, Mulholland PJ. Kv7 channels in the nucleus accumbens are altered by chronic drinking and are targets for reducing alcohol consumption. Addict Biol. 2015 doi: 10.1111/adb.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ. K(Ca)2 channels: novel therapeutic targets for treating alcohol withdrawal and escalation of alcohol consumption. Alcohol. 2012;46(4):309–315. doi: 10.1016/j.alcohol.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Becker HC, Woodward JJ, Chandler LJ. Small conductance calcium-activated potassium type 2 channels regulate alcohol-associated plasticity of glutamatergic synapses. Biol Psychiatry. 2011;69(7):625–632. doi: 10.1016/j.biopsych.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Carpenter-Hyland EP, Hearing MC, Becker HC, Woodward JJ, Chandler LJ. Glutamate transporters regulate extrasynaptic NMDA receptor modulation of Kv2.1 potassium channels. J Neurosci. 2008;28(35):8801–8809. doi: 10.1523/JNEUROSCI.2405-08.2008. 28/35/8801 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Carpenter-Hyland EP, Woodward JJ, Chandler LJ. Ethanol disrupts NMDA receptor and astroglial EAAT2 modulation of Kv2.1 potassium channels in hippocampus. Alcohol. 2009;43(1):45–50. doi: 10.1016/j.alcohol.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Hopf FW, Bukiya AN, Martin GE, Liu J, Dopico AM, … Chandler LJ. Sizing up ethanol-induced plasticity: the role of small and large conductance calcium-activated potassium channels. Alcohol Clin Exp Res. 2009;33(7):1125–1135. doi: 10.1111/j.1530-0277.2009.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Spencer KB, Hu W, Kroener S, Chandler LJ. Neuroplasticity of A-type potassium channel complexes induced by chronic alcohol exposure enhances dendritic calcium transients in hippocampus. Psychopharmacology (Berl) 2015;232(11):1995–2006. doi: 10.1007/s00213-014-3835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung J, Kim Y, Park T. Whole-genome association studies of alcoholism with loci linked to schizophrenia susceptibility. BMC Genet. 2005;6(Suppl 1):S9. doi: 10.1186/1471-2156-6-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo-Anh TJ, Bloodgood BL, Lin M, Sabatini BL, Maylie J, Adelman JP. SK channels and NMDA receptors form a Ca2+-mediated feedback loop in dendritic spines. Nat Neurosci. 2005;8(5):642–649. doi: 10.1038/nn1449. [DOI] [PubMed] [Google Scholar]
- NIAAA; N. I. o. Health, editor Alcohol Facts and Statistics. 2015. [Google Scholar]
- Nimitvilai S, Lopez MF, Mulholland PJ, Woodward JJ. Chronic Intermittent Ethanol Exposure Enhances the Excitability and Synaptic Plasticity of Lateral Orbitofrontal Cortex Neurons and Induces a Tolerance to the Acute Inhibitory Actions of Ethanol. Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimitvilai S, Lopez MF, Mulholland PJ, Woodward JJ. Chronic Intermittent Ethanol Exposure Enhances the Excitability and Synaptic Plasticity of Lateral Orbitofrontal Cortex Neurons and Induces a Tolerance to the Acute Inhibitory Actions of Ethanol. Neuropsychopharmacology. 2016;41(4):1112–1127. doi: 10.1038/npp.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, O’Brien CP. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28(8):1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- Padula AE, Griffin WC, 3rd, Lopez MF, Nimitvilai S, Cannady R, McGuier NS, … Mulholland PJ. KCNN Genes that Encode Small-Conductance Ca2+-Activated K+ Channels Influence Alcohol and Drug Addiction. Neuropsychopharmacology. 2015;40(8):1928–1939. doi: 10.1038/npp.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padula AE, McGuier N, Griffin W, Lopez M, Becker H, Mulholland P. Novel anticonvulsants for reducing alcohol consumption: A review of evidence from preclinical rodent drinking models. OA Alcohol. 2013;1(1):2. doi: 10.13172/2053-0285-1-1-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Luo X, Liu X, Wu LY, Zhang Q, Wang L, … Wang KS. Genome-wide association studies of maximum number of drinks. J Psychiatr Res. 2013;47(11):1717–1724. doi: 10.1016/j.jpsychires.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, … Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9(2):129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleil KE, Lowery-Gionta EG, Crowley NA, Li C, Marcinkiewcz CA, Rose JH, … Kash TL. Effects of chronic ethanol exposure on neuronal function in the prefrontal cortex and extended amygdala. Neuropharmacology. 2015;99:735–749. doi: 10.1016/j.neuropharm.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Porter JT. M-type potassium channels modulate the intrinsic excitability of infralimbic neurons and regulate fear expression and extinction. J Neurosci. 2010;30(37):12379–12386. doi: 10.1523/JNEUROSCI.1295-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets K, Duarri A, Deconinck T, Ceulemans B, van de Warrenburg BP, Zuchner S, … Baets J. First de novo KCND3 mutation causes severe Kv4.3 channel dysfunction leading to early onset cerebellar ataxia, intellectual disability, oral apraxia and epilepsy. BMC Med Genet. 2015;16:51. doi: 10.1186/s12881-015-0200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KB, Mulholland PJ, Chandler LJ. FMRP mediates chronic ethanol induced changes in NMDA, Kv4.2, and KChIP3 expression in hippocampus. Alcoholism: Clinical and Experimental Research. doi: 10.1111/acer.13060. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensland P, Fredriksson I, Holst S, Feltmann K, Franck J, Schilstrom B, Carlsson A. The monoamine stabilizer (−)-OSU6162 attenuates voluntary ethanol intake and ethanol-induced dopamine output in nucleus accumbens. Biol Psychiatry. 2012;72(10):823–831. doi: 10.1016/j.biopsych.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Sun X, Zaydman MA, Cui J. Regulation of Voltage-Activated K(+) Channel Gating by Transmembrane beta Subunits. Front Pharmacol. 2012;3:63. doi: 10.3389/fphar.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipps ME, Raybuck JD, Kozell LB, Lattal KM, Buck KJ. G Protein-Gated Inwardly Rectifying Potassium Channel Subunit 3 Knock-Out Mice Show Enhanced Ethanol Reward. Alcohol Clin Exp Res. 2016;40(4):857–864. doi: 10.1111/acer.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treistman SN, Martin GE. BK Channels: mediators and models for alcohol tolerance. Trends Neurosci. 2009;32(12):629–637. doi: 10.1016/j.tins.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS. Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron. 2015;85(2):238–256. doi: 10.1016/j.neuron.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet B, Manrique C, Sreng L, Chaillan FA, Roman FS, Mourre C. Kv4 potassium channels modulate hippocampal EPSP-spike potentiation and spatial memory in rats. Learning & memory. 2012;19(7):282–293. doi: 10.1101/lm.025411.111. [DOI] [PubMed] [Google Scholar]
- van der Vaart AD, Wolenholme MS, Smith M, Harris G, Lopez MF, Becker HC, Miles MF. Identification of expression patterns and eQTLs relating to progressive ethanol consumption in the BXD recombinant inbred panel. Alcohol This issue. [Google Scholar]
- van der Velden L, van Hooft JA, Chameau P. Altered dendritic complexity affects firing properties of cortical layer 2/3 pyramidal neurons in mice lacking the 5-HT3A receptor. J Neurophysiol. 2012;108(5):1521–1528. doi: 10.1152/jn.00829.2011. [DOI] [PubMed] [Google Scholar]
- Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, Wulff H. International Union of Pharmacology. LII. Nomenclature and molecular relationships of calcium-activated potassium channels. Pharmacol Rev. 2005;57(4):463–472. doi: 10.1124/pr.57.4.9. [DOI] [PubMed] [Google Scholar]
- Wilcox MV, Cuzon Carlson VC, Sherazee N, Sprow GM, Bock R, Thiele TE, … Alvarez VA. Repeated binge-like ethanol drinking alters ethanol drinking patterns and depresses striatal GABAergic transmission. Neuropsychopharmacology. 2014;39(3):579–594. doi: 10.1038/npp.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SH. Medications for treating alcohol dependence. Am Fam Physician. 2005;72(9):1775–1780. [PubMed] [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, … Miles MF. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PLoS One. 2012;7(4):e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6(6):e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang L, Ravindranathan A, Miles MF. A new algorithm for analysis of oligonucleotide arrays: application to expression profiling in mouse brain regions. J Mol Biol. 2002;317(2):225–235. doi: 10.1006/jmbi.2001.5350. [DOI] [PubMed] [Google Scholar]
- Zuo L, Zhang F, Zhang H, Zhang XY, Wang F, Li CS, … Luo X. Genome-wide search for replicable risk gene regions in alcohol and nicotine co-dependence. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(4):437–444. doi: 10.1002/ajmg.b.32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.