Abstract
The role of Notch signaling in prostate cancer has not been defined definitively. Several large scale tissue microarray studies revealed that the expression of some Notch signaling components including the Jagged1 ligand are upregulated in advanced human prostate cancer specimens. Jagged1 expressed by tumor cells may activate Notch signaling in both adjacent tumor cells and cells in tumor microenvironment. However, it remains undetermined whether increased Jagged1 expression reflects a cause for or a consequence of tumor progression in vivo. To address this question, we generated a novel R26-LSL-JAG1 mouse model that enables spatiotemporal Jagged1 expression. Prostate specific upregulation of Jagged1 neither interferes with prostate epithelial homeostasis nor significantly accelerates tumor initiation or progression in the prostate-specific Pten deletion mouse model for prostate cancer. However, Jagged1 upregulation results in increased inflammatory foci in tumors and incidence of intracystic adenocarcinoma. In addition, Jagged1 overexpression upregulates Tgfβ signaling in prostate stromal cells and promotes progression of a reactive stromal microenvironment in the Pten null prostate cancer model. Collectively, Jagged1 overexpression does not significantly accelerate prostate cancer initiation and progression in the context of loss-of-function of Pten, but alters tumor histopathology and microenvironment. Our study also highlights an understudied role of Notch signaling in regulating prostatic stromal homeostasis.
Keywords: Notch, Jagged1, Pten, prostate cancer, reactive stroma
Introduction
Notch is an evolutionarily conserved signaling pathway that plays a critical role in regulating tissue homeostasis and carcinogenesis (1–3). For example, Notch has been clearly defined as an oncogene in T-cell acute lymphoblastic leukemia (4) and a tumor suppressor in skin cancer (5). Notch signaling has been suggested to promote prostate cancer progression. Several large scale tissue microarray studies revealed that the expression of Notch receptors are upregulated in advanced human prostate cancer specimens (6–8). Notch activity is enriched in the side population (putative cancer stem cells) and docetaxel resistant populations in prostate cancer cell lines (9, 10). In addition, several cellular biological studies showed that inhibiting Notch signaling attenuates prostate cancer cell proliferation and migration (11–13). Finally, studies from our laboratory showed that increased Notch activity in mouse models promotes luminal cell proliferation, confers anoikis resistance, and drives tumor metastasis (14, 15) (Kwon et al 2016 in press). Nevertheless, there are also cellular biological evidence supporting an inhibitory role of Notch in prostate cancer cell growth (16).
Jagged1 is a major Notch ligand expressed in normal and cancerous prostate epithelial cells of rodent and human (17, 18). Increased Jagged1 expression is also correlated with prostate cancer metastasis and recurrence in two independent large-scale tissue microarray analyses (6, 7). This observation is very interesting because Jagged1 expressed by tumor cells is capable of activating Notch signaling in both adjacent tumor cells as well as cells in the tumor microenvironment. For example, in the breast cancer model, Jagged1-mediated Notch activation has been shown to promote tumor cell motility by inducing epithelial-mesenchymal transition (EMT) (19–21); On the other hand, Jagged1 expressed by breast cancer cells were also shown to activate Notch signaling in cells within their microenvironment such as osteoblasts, which secrete growth factors that in turn promotes survival, proliferation, and migration of tumor cells (22, 23). In contrast, the functional significance of Jagged1 upregulation in prostate cancer cells is still inadequately interrogated (12, 24, 25). It remains an open question whether Jagged1 upregulation plays a causative role in tumor progression in vivo or is only a consequence of tumor progression, as Jagged1 itself is a Notch downstream target. To address this question, we generated a novel mouse model that enables conditional Jagged1 upregulation. We bred this model with a prostate specific Pten deletion mouse model for prostate cancer and examined the biological consequence.
Results
Generation of a JAG1 conditional expression mouse model
We sought to generate a mouse model that is capable of enabling overexpression of JAG1 in a spatiotemporal manner. Fig. 1A shows the basic design of the targeting vector. Upon the targeting vector-mediated homologous recombination, JAG1 cDNA should be inserted into the ROSA26 locus. A floxed transcriptional stop sequence was put in front of the JAG1 cDNA to enable Cre-loxp mediated conditional activation. An IRES-Luciferase cassette was cloned downstream of JAG1 to facilitate detection of transgene activation via bioluminescent imaging. The targeting vector was electroporated into C57BL/6 ES cells and Southern blot analysis confirmed successful homologous recombination (Fig. 1B) in over 70% of the selected clones. Three recombined ES clones were injected into recipient blastocysts and seven founders with germ lines fully derived from injected ES cells were identified. This model was named the R26-LSL-JAG1 line and the data presented hereafter were obtained using one of the founders.
Figure 1. Generation of the R26-LSL-JAG1 model.
(A) Schematic illustration of the targeting vector. (B) Southern blot analysis identifies ES clone with correct homologous recombination. Wild type and recombined ES clones show bands of 10 and 4 kb, respectively.
JAG1 upregulation does not alter prostate epithelial homeostasis
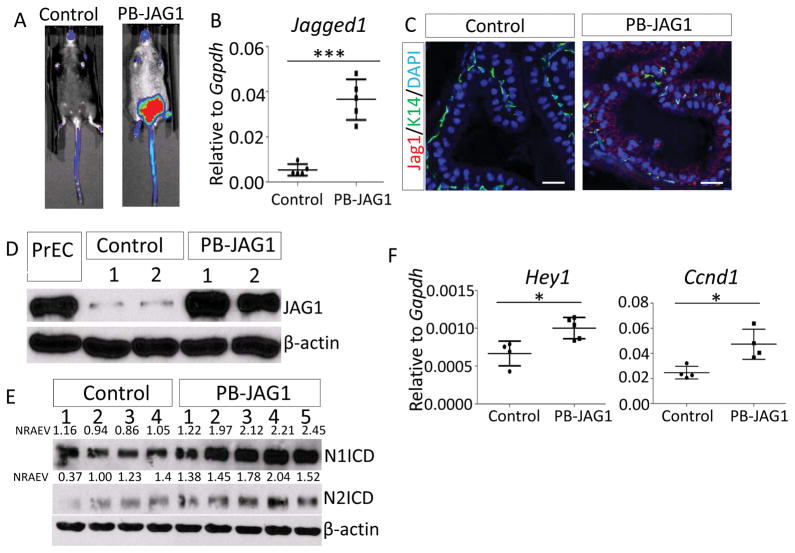
To enable spatiotemporal overexpression of JAG1 in the prostate, we bred R26-LSL-JAG1 mice with an ARR2PB-Cre (PB-Cre) line (26) and generated a cohort of PB-Cre;R26-LSL-JAG1Tg/Wt and control R26-LSL-JAG1 mice. Bioluminescent imaging confirmed specific expression of luciferase in lower abdomen, where major urogenital organs, including the prostate, are located (Fig. 2A). QRT-PCR analysis using a pair of primers recognizing both human and mouse Jagged1 revealed a 6-fold increase in expression in the PB-Cre;R26-LSL-Jag1 mice (Fig. 2B). Immunostaining confirmed prominent expression of membrane-bound JAG1 in the prostate tissues of 8-week-old PB-Cre;R26-LSL-JAG1 male mice as compared to those of age-matched littermate controls (Fig. 2C). Western blot analysis showed that the expression level of JAG1 in this model is comparable to that in the primary human prostate basal cells (PrEC) that express a high level of JAG1 (Fig. 2D). This suggests that JAG1 is expressed in this model at a physiologically relevant level. Western blot analysis confirms the increased production of both Notch 1 intracellular domain (1.99 fold, p<0.005) and Notch 2 intracellular domain (1.63 fold, p<0.05) in the prostates of PB-Cre;R26-LSL-JAG1 mice (Fig. 2E). In addition, qRT-PCR analysis showed that the typical Notch downstream target genes Hey1 and Ccnd1 were upregulated in the prostates of PB-Cre;R26-LSL-JAG1 mice (Fig. 2F), but the expression of other Notch target genes such as Hes1 and Jag1 were not altered (Supplementary Fig. 1, this set of Jag1 primers only recognize mouse Jag1). Collectively, these results demonstrate that overexpressed JAG1 is capable of activating Notch signaling.
Figure 2. Successful prostate specific JAG1 expression in the PB-JAG1 model.
(A) Bioluminescence imaging of a PB-JAG1 and a control mouse. (B) qRT-PCR analysis of Jagged1 in 52-wk-old PB-JAG1 and control mice. (C) Coimmunostaining of JAG1 and K14 in prostate tissues of PB-JAG1 and control mice. Bars=25μm. (D–E) Western blot analyses of JAG1 (D) and Notch1 and Notch2 intracellular domains (N1ICD and N2ICD) (E) in prostate tissues of PB-JAG1 and control mice. Individual lanes represent results from different mice. PrEC: primary human prostate basal epithelial cells. NRAEV: normalized relative arbitrary expression value. (F) qRT-PCR analysis of Hey1 and Ccnd1 in PB-JAG1 and control mice. *: p<0.05, ***: p<0.001.
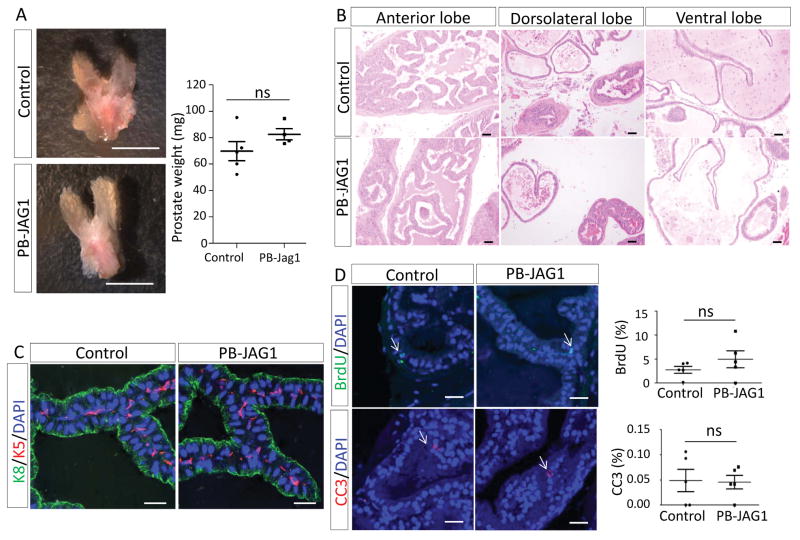
Fig. 3A shows that the prostatic weights of the PB-Cre;R26-LSL-JAG1 and the control mice are comparable at 1 year of age. H&E staining shows that all four prostatic lobes of PB-Cre;R26-LSL-JAG1 mice are histologically the same as those of the control mice (Fig. 3B). They all contained a layer of epithelial cells surrounding the lumen filled with eosinophilic secretions. Immunostaining further confirmed that prostate epithelia in both PB-Cre;R26-LSL-JAG1 and control mice were composed of a single layer of Keratin8-expressing luminal cells encapsulated by a discontinuous layer of basal cells that express Keratin5 (Fig. 3C). Finally, immunostaining of BrdU and the cleaved caspase 3 showed that the proliferative and apoptotic indices of the prostate tissues in the two groups of mice were similar (Fig. 3D). Collectively, these results indicate that JAG1 overexpression per se does not affect prostate epithelial homeostasis. We showed previously that prostate specific expression of the Notch1 intracellular domain promotes prostate epithelial proliferation and induces the formation of papillary prostatic intraepithelial neoplasia (14, 15). The different observations in the two models probably reflect the distinct levels of Notch activation.
Figure 3. JAG1 upregulation does not alter prostate homeostasis.
(A) Representative images of prostate tissues from 1-yr old control and PB-JAG1 mice. Dot plot shows quantification of prostate weights. Bars=5mm. (B) H&E staining of different prostate lobes of 1-yr old control and PB-JAG1 mice. Bars=50μm. (C) Immunostaining of K5 and K8 in prostate tissues of 1-yr old control and PB-JAG1 mice. Bars=25μm. (D) Immunostaining of BrdU and cleaved caspase 3 (CC3) in prostate tissues of 1-yr old control and PB-JAG1 mice. Arrows point to cells with positive staining. Bars=25μm. Dot plot shows means ± s.d.
JAG1 overexpression does not affect disease progression in a prostate specific Pten null mouse model for prostate cancer
JAG1 is expressed at a higher level in metastatic human prostate cancer specimens (6, 7). In addition, JAG1 expressed in breast cancer cells has been shown to activate signaling in osteoblast cells which in turn express IL-6 to promote tumor cell proliferation (22). Therefore, despite our finding that JAG1-induced Notch activation in prostate epithelial cells is insufficient to affect cell proliferation, we sought to investigate whether JAG1 expressed by prostate epithelial cells may interact with Notch receptors expressed by cells in tumor microenvironment or distal niches and help tumor cells migrate, adapt, survive and repopulate.
To test this hypothesis, we bred R26-LSL-JAG1 mice with a prostate specific Pten deletion model for prostate cancer and generated a cohort of mice including PB-Cre;Ptenfl/fl and PB-Cre;Ptenfl/fl;R26-LSL-JAG1Mut/Wt mice (hereafter referred to as PB-Pten and PB-Pten-JAG1 mice, respectively). The prostate tissues of the mice in the two groups had similar weights at the 32 (Supplementary Fig. 2A) and 52 weeks of age (Fig. 4A). JAG1 overexpression in the prostate tissues of PB-Pten-JAG1 mice was also confirmed by Western blot analysis (Supplementary Fig. 2B). All mice in both groups developed at least high grade prostatic intraepithelial neoplasia (PIN3/4) at 32 weeks (Supplementary Fig. 3 and Table 1). At one year of age, all mice developed PIN3/4 with focal adenocarcinoma (Fig. 4B and Table 1). Interestingly, areas of cystic structures lined by dysplastic epithelium were noted in the prostate tissues in both groups (Fig. 4C, Supplementary Fig. 4). But this phenotype is much more prominent in the PB-Pten-JAG1 mice. 15 out of 16 examined mice have large papillary structures with complex glandular architectures (Table 1, p<0.005, Fisher’s exact test). In addition, the PB-Pten-JAG1 mice developed higher grader reactive stroma with lymphocytic infiltration (Fig. 4D, Supplementary Fig. 4, and Table 1). These phenotypes are consistent with the intracystic carcinoma described previously (27). In addition, in 10 out of the 16 52-wk-old PB-Pten-JAG1 mice, we noticed that a mass of poorly differentiated adenocarcinoma extended into the lumen of seminal vesicles and replaced normal lining of the epithelium (Fig. 4E). In contrast, this is only seen in 20% of PB-Pten mice at the same age. This observation is in agreement with our recent study showing that Notch activation transformed seminal vesicle epithelial cells in the Pten null mouse model (Kwon et al 2016 in press).
Figure 4. JAG1 upregulation does not affect tumor progression in the Pten null model.
(A) Representative images of urogenital organs and prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Dot plot shows quantification of prostate weight. Bars=1cm. (B–D) H&E staining of prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice shows formation of high grade PIN lesions (B) and intracystic adenocarcinoma (arrow, C) with reactive stroma (D). Bars=50μm. (E) H&E staining of seminal vesicles of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Bars=300μm. (F–G) Immunostaining of Pten, pAKT, K5, K8, P63 and AR in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Black bars=50μm, white bars=25μm.
Table 1.
Summary of histopathology in prostates of PB-Pten and PB-Pten-JAG mice
| 32 weeks | |||
|---|---|---|---|
| PB-Pten (N=5) | PB-Pten-JAG1 (N=5) | ||
| PIN3/4 | 4 | 3 | |
| Cystic glands | 1 | 3 | |
| Intracystic adenocarcinoma | 1 | 2 | |
| Reactive stroma | Grade 1 | 4 | 2 |
| Grade 2 | 1 | 3 | |
| 52 weeks | |||
| PB-Pten (N=8) | PB-Pten-JAG1 (N=16) | ||
| PIN3/4 with focal adenocarcinoma | 8 | 16 | |
| Cystic glands | 2 | 15 | |
| Intracystic adenocarcinoma | 1 | 10 | |
| Reactive stroma | Grade 1 | 6 | 2 |
| Grade 2 | 1 | 2 | |
| Grade 3 | 1 | 12 | |
Note: grade 1, grade 2, and grade 3 reactive stroma are defined as focal stroma confined around ductal structures, more extensive stroma filling areas between ducts, and extensive stroma proliferation involving most of the prostates, respectively.
Immunostaining of BrdU and cleaved caspase 3 showed that there are no significant differences in proliferative and apoptotic indices in the prostate tissues between the two groups (Supplementary Figure 5). IHC analysis confirmed that Pten was not expressed but phosphorylated AKT was significantly expressed in the prostate tissues in both groups (Fig. 4F). Fig. 4G shows that the expression patterns of the lineage markers including K5, P63, K8 and the androgen receptor (AR) were all similar in prostate tissues of the two groups of mice. No distal metastasis was observed in PB-Pten-JAG1 and PB-Pten mice by bioluminescent imaging or H&E staining of major internal organs including lung and liver etc. (data not shown). Collectively, these results indicate that JAG1 upregulation does not promote progression and metastasis of prostate cancer in the Pten null model.
JAG1 overexpression promotes formation of reactive stroma
H&E staining revealed more inflammatory foci in the prostate tissues of PB-Pten-JAG1 mice (Fig. 5A). This is confirmed by flow cytometric analysis showing more CD45+ leukocytes in the prostates of PB-Pten-JAG1 mice (19.9±1.42% in PB-Pten-JAG1 mice versus 30.6±2.96% in PB-Pten mice, p<0.05, Fig. 5B). In addition, the blood vessel density in the prostates of PB-Pten-JAG1 mice is also 1.8- fold more than that in the control PB-Pten mice as shown by immunostaining of CD31 (Fig. 5C). Myeloid cells play a critical role in tumor progression (28). Notch signaling has been shown to promote maturation of tumor associated macrophages and expansion of the Gr-1+CD11b+ myeloid derived suppressor cells (29, 30). We examined whether JAG1 expressed by tumor cells may affect the composition of myeloid derived suppressor cells in blood circulation and primary tumors but find no significant difference (Fig. 5D).
Figure 5. JAG1 upregulation augments inflammatory foci and blood vessel density.
(A) H&E staining of prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Black arrows denote inflammatory foci. Bars=50μm. (B) FACS plots of CD45 staining in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Dot plot shows means ± s.d. of percentage of CD45+ cells. (C) Immunostaining of CD31 in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Red arrows point to CD31-expressing vessels. Bars=50μm. Dot plot shows means ± s.d. of blood vessel number per field. (D) FACS plots of myeloid derived suppressor cells (MDSCs) in blood and tumor tissues in 1-year-old PB-Pten-JAG1 and PB-Pten mice. Bar graphs show quantification. *: p<0.05, **: p<0.01.
Increased inflammatory foci and blood vessel density in the PB-Pten-JAG1 model prompted us to examine additional changes in the prostate stromal microenvironment. During cancer progression, the stromal microenvironment also undergoes substantial remodeling and becomes reactive stroma that is fundamentally different from normal tissue stroma. Reactive stroma in prostate cancer displays a significant decrease in differentiated smooth muscle cell content. It expresses both vimentin and smooth muscle α-actin, suggestive of a myofibroblast phenotype. Reactive stromal cells also exhibited increased production of specific ECM components such as Type I collagen and tenascin C etc (31). Trichrome staining revealed more collagen deposition at the prostatic peri-glandular spaces of PB-Pten-JAG1 mice (Fig. 6A). This is corroborated by stronger peri-glandular staining of Procollagen I in the PB-Pten-JAG1 model (Fig. 6B) Fig. 6C shows that the prostate epithelial glands of PB-Pten mice were immediately surrounded by a layer of stromal cells expressing smooth muscle actin. Very few stromal cells in the peri-glandular spaces expressed vimentin. In contrast, the smooth muscle actin-expressing stromal cell layer in PB-Pten-JAG1 mice was often disrupted and much thinner, whereas vimentin staining was frequently observed in peri-glandular stromal cells. Finally, Immunostaining also revealed stronger expression of Tenascin C in stromal cells of the PB-Pten-JAG1 mice (Fig. 6D). TGFβ signaling has been shown to play an inductive role in the formation reactive stroma (32, 33). We found that pSmad3 was expressed in a higher percentage of stromal cells in the PB-Pten-JAG1 model (Fig. 6E). qRT-PCR analysis showed a slight but consistent increase in expression of Tgfβ ligands in the prostate tissues of PB-Pten-JAG1 mice, although the increase did not reach statistical significance (Fig. 6F). Collectively, these results suggest a more advanced reactive stromal microenvironment in the PB-Pten-JAG1 model.
Figure 6. Jagged1 upregulation promotes formation of reactive stroma.
(A) Trichrome staining of prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Dot plot shows means ± s.d. of percentage of blue collagen staining per field. Bars=50μm. (B) Immunostaining of Procollagen1 in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Bars=50μm. (C) Coimmunostaining of Smooth muscle actin (SMA) and Vimentin (Vim) in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Bars=100μm. (D) Immunostaining of Tenascin C (TNC) in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Asterisks denote background staining in epithelial cells while arrows point to stronger staining in stromal cells. Bars=50μm. (E) Immunostaining of pSmad3 in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice. Dot plot shows means ± s.d. of percentage of pSmad3+ stromal cells. Bars=50μm. (F) Dot graphs show means ± s.d. of expression of Tgfβ1 and Tgfβ3 in prostate tissues of 1-yr old PB-Pten and PB-Pten-JAG1 mice by qRT-PCR. *: p<0.05.
RT-PCR analysis reveals that Notch receptors and downstream target genes are widely expressed in adult murine prostate stromal cells, among which Notch1 and Hes1 are abundantly expressed (Fig. 7A). This result suggests that Notch signaling may actively regulate stromal cell biology. We showed previously that Notch activation is capable of augmenting TGFβ activity in prostate basal epithelial cells (15). Therefore, we hypothesized that the formation of the reactive stroma could be due to the interaction of the prostate stromal cells with JAG1-expressing tumor cells. To test this hypothesis, we cocultured the immortalized human prostate stromal cells WPMY-1 with DU145 cells that overexpress JAG1 or control DU145 cells (Fig. 7B). The cocultured cells were differentially marked with fluorescent proteins. WPMY-1 cells were FACS separated from cocultured DU145 cells and RNA was extracted from WPMY-1 cells. Fig. 7C shows that the Notch downstream target genes HES1 and HEY1 were upregulated in WPMY-1 cells interacting with JAG1-expressing DU145 cells, corroborating the activation of the Notch signal. TGFβ ligands (TGFβ1 and TGFβ3) and several TGFβ downstream target genes (COL1A1, TIMP3, P21 and P15) were also upregulated in WPMY-1 cells cocultured with JAG1-expressing DU145 cells (Fig. 7C). Collectively, these results further support that Notch activation in prostate stromal cells promotes TGFβ signaling and formation of reactive stroma.
Figure 7. Notch signaling regulates prostate stromal cell biology.
(A) qRT-PCR analysis of expression of Notch receptors and target genes in FACS-sorted prostate stromal cells of 10-wk-old wild type mice. Bar graphs show means ± s.d. from 3 independent experiments. (B) Schematic illustration of coculture experimental approach. (C) qRT-PCR analysis of gene expression. Bar graphs show means ± s.d. of 1 out of 2 independent representative experiments. **: p<0.01, ***: p<0.05.
The probasin promoter has been shown to be active in some prostate stromal cells (34). Therefore, Notch signaling may also be activated in stromal cells through interactions between stromal cells with leaky JAG1 expression. To investigate whether JAG1 is expressed in prostate stromal cells, we prepared genomic DNA from FACS sorted prostate stromal cells of PB-JAG1 mice (Supplementary Fig. 6A) and performed PCR analysis to determine whether the Cre-mediated homologous recombination occurred at the R26-LSL-JAG1 locus. As shown in Supplementary Fig. 6B, strong bands of 540bp that represents the recombined allele were amplified from DNAs of FACS-sorted luminal cells. In contrast, the bands amplified from the stromal samples were faint. We cannot conclude definitively whether the amplification from the stromal samples reflects rare unspecific recombination occurred in stromal cells or a contamination of luminal cells in the FACS-sorted stromal cells. Because PB-JAG1 model did not develop extensive reactive stroma observed in the PB-Pten-JAG1 model, we reason that even if JAG1 is expressed by stromal cells it did not contribute substantially to the reactive stromal phenotype. Finally, regardless of the mechanisms, our study reveals a previously understudied role of Notch signaling in regulating prostate stromal homeostasis (34, 35).
Discussion
We have generated an R26-LSL-JAG1 model to enable spatiotemporal upregulation of JAG1. Increased expression of Notch ligands, has been shown to suppress, rather than activates, Notch activity either through cis-inhibition (36) or by competing for the proteolytic machineries essential for Notch receptor activation (37). We showed clearly that JAG1 upregulation increased the production of N1ICD and N2ICD as well as the expression of some Notch downstream target genes such as Hey1, which demonstrates that JAG1 at this expression level is capable of activating Notch.
We showed that although epithelial cell autonomous JAG1 upregulation altered tumor histopathology and microenvironment, it did not accelerate disease progression, especially metastasis, in the prostate specific Pten null mouse model. This result is distinct from previous reports showing that JAG1 overexpression in prostate cancer cell lines promotes proliferation and migration (25). Our unpublished study (Su Q and Xin L) also showed that JAG1 did not affect proliferation of DU145 cells but promoted lung colonization when they were inoculated into immuno-deficient mice via tail vein. In these studies tumor cells bypassed intravasation and entered blood circulation directly. In contrast, in the PB-Pten-JAG1 model, cells must invade through basement membrane and intravasate in order to develop distal metastasis. This implies that JAG1 is unlikely to play a role in promoting metastasis through basement membrane or intravasation. Future study will determine whether JAG1 accelerates distal metastasis in mouse models for prostate cancer that develop spontaneous local or distal metastatic disease more robustly, such as the mouse model with prostate-specific Pten deletion and K-Ras activation (38) etc.
Other potential explanations for the distinct conclusions between studies using human cell lines and our mouse model could be the intrinsic differences between the Pten null murine prostate epithelial cells and human prostate cancer cells lines. Additionally, the expression level of JAG1 in different studies may also affect the results. Distinct levels of JAG1 expression could lead to Notch activation at different degrees. Excessive JAG1 may lead to the activation of JAG1-mediated Notch-independent signaling (37, 39), although the physiological relevance of those mechanisms remains to be determined.
It is interesting that the most prominent phenotype in our study was noted in prostate stroma although JAG1 was expressed in epithelial cells. We proposed that focal interaction of Jagged1-expressing epithelial cells with stromal cells could turn on stromal Notch signaling, which in turn augments TGFβ signaling (15), activates downstream signaling cascades that accelerates the formation of the reactive stromal microenvironment (32, 33). Although we were not able to conclude definitively whether JAG1 was also expressed by prostate stromal cells, we reason that the contribution of potential leaky JAG1 expression in stromal cells to the reactive phenotype is minimal as PB-JAG1 model did not develop the phenotype. Nevertheless, regardless of the mechanisms, our study revealed a previously understudied role of Notch signaling in regulating prostate stromal homeostasis (34, 35).
JAG1 is not only upregulated in tumor cells but also in the cells in tumor microenvironment. For example, JAG1 is expressed at a higher level in endothelial cells associated with invasive ovarian carcinoma (40). Functional studies have demonstrated that targeting Jag1 in endothelial cells is capable of suppressing the growth of tumor cells in orthotopic ovarian cancer xenograft models (41) and the TRAMP mouse model for prostate cancer (42). These studies highlight that Jag1 expressed in tumor microenvironment may play an even more important role in tumor progression. Prostate cancer often progresses and metastasizes to the bone. Shiozawa Y et al showed that prostate cancer cells metastasized to the bone by engaging the hematopoietic stem cell niches (23). Jagged1-expressing endothelial cells serve as one of the niches for the hematopoietic stem cells in the bone (43, 44). It is tempting to hypothesize that Jagged1-expressing endothelial cells in the bone facilitate anchorage, survival and proliferation of circulating prostate cancer cells, resulting in a bone tropism. Future studies using the R26-LSL-JAG1 model should test this hypothesis.
Finally, our study showed that JAG1 overexpression promotes leukocyte infiltration, angiogenesis and formation of reactive stroma. Men with castration resistant prostate cancer did not respond to immune checkpoint therapy as a monotherapy because of several potential mechanisms such as low expression level of PD-L1 in tumor cells or inefficient infiltration of CD8+ T cells (45). Inflammatory cytokines such as IFNγ has been shown to upregulate PD-L1 (46). Therefore, the increased angiogenesis and reactive stroma in the PB-Pten-JAG1 model raise the possibility that these tumors may be more susceptible to immune checkpoint therapy. Future studies using this mouse model will provide additional preclinical evidence whether these hypotheses are valid.
Materials and Methods
Mouse and genotyping
All animals used in this study received humane care that adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NIH Publication, 1996 edition, and the protocol was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. The C57BL/6 mice were purchased from Charles River (Wilmington, MA). The Ptenfl/fl mice were from Dr. Hong Wu at the University of California Los Angeles (47). The R26-LSL-JAG1 mice were generated at the transgenic mouse core at the Baylor College of Medicine. The ARR2PB-Cre transgenic mice were from Dr. Fen Wang at the Institute of Bioscience and Technology, Texas A&M Health Science Center (26). The Ptenfl/fl mice were at a mixed background while the rest were all of C57Bl/6 background. Mice were randomly allocated to experimental groups and investigators were blind to the group allocation during the experiments. Mice were genotyped by polymerase chain reaction using mouse genomic DNA from tail biopsy specimens. The sequences of genotyping primers are listed in Supplementary Table 1. PCR products were separated electophoretically on 1% agarose gels and visualized via ethidium bromide under UV light.
Generation and verification of the R26-LSL-JAG1 model
The pBIGT and pROSA26pAm1 vectors were obtained from Addgene (Cambridge, MA). A synthesized oligonucleotide linker was used to convert the NheI-ScaI cloning site in the pBIGT vector into a NotI-NheI-SacI site. An IRES-Luciferase cassette was cloned into the modified pBIGT vector using NotI and NheI to generate pBIGT-IRES-Luc. Subsequently, the human Jagged1 cDNA, obtained from Dr. Spyros Artavanis-Tsakonas, was PCR amplified, sequenced, and cloned into pBIGT-IRES-Luc using NotI. Finally, the LSL-JAG1 cassette was released from pBIGT vector using PacI and AscI and cloned into pROSA26pAm1 vector to generate the targeting vector. The targeting vector was linearized with SalI and electroporated into C57Bl/6 ES cells. Neomycin-resistant clones were selected with G418.
Genotypes of the embryonic stem cell clones and mice were verified by Southern blot. Briefly, Genomic DNA were digested by EcoRV for 8 h, After overnight electrophoresis at 35 V, the digested DNA in the gel was depurinated, denaturated, neutralized, and transferred to a Hybond-N+ nylon membrane (GE Healthcare, Piscataway, NJ). Probes were made using a Prime-it II Random Labeling Kit (Stratagene, La Jolla, CA) with a 600-bp DNA genomic sequence adjacent to the left arm as a template, and hybridization was carried out overnight at 42°C in Ultrahyb Ultrasensitive Hybridization Buffer (Thermal Scientific, Rockford, IL). Signals were visualized by exposure to X-ray films.
Bioluminescence imaging
Animals were imaged weekly using an IVIS Lumina II (Advanced Molecular Vision) following the manufacturer’s recommended procedures and settings. D-Luciferin (Gold Biotechnology Inc., Olivette, MO) was injected intraperitoneally (2 mg per mouse). 6–10 minutes after injection, mice were imaged with an exposure time of 5–30 seconds.
Cell culture
DU145 was from American Type Culture Collection (Manassas, VA). WPMY-1 was a kind gift from Dr. Chawnshang Chang at University of Rochester. Cells were cultured in RPMI 1640 with 10% FBS with penicillin and streptomycin (Invitrogen, Carlsbad, CA) at 37 °C in a humid atmosphere containing 5% CO2. The primary PrEC cells were cultured in the PrEGM media and both were purchased from Lonza (Walkersville, MD).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from cells using the RNeasy Plus mini kit (Qiagen, Valencia, CA). Reverse transcription was performed using the iScript cDNA synthesis kit (BioRad, Hercules, CA). Quantitative RT-PCR was performed using the SYBR Premix Ex Taq (Perfect Real Time) (Takara Bio Inc., Otsu, Shiga, Japan) on a StepOne plus Real-Time PCR system (Applied Biosystems, Foster City, CA). Primer sequences for qPCR are listed in Supplementary Table 2. Analysis of Notch receptors and target genes in murine prostate stromal cells was performed on RT2 Profiler PCR Array Mouse Notch Signaling Pathway plates following the manufacturer’s instruction (SABiosciences, Frederick, MD).
Western blot
Prostate tissues were lysed in RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4) with protease inhibitors and phosphatase inhibitors (Roche Applied Science, Indianapolis, IN) using Tissuelyser LT (Qiagen, Valencia, CA). Protein concentrations were determined by a Bradford Assay kit (BioRad, Hercules, CA). Protein was separated by 10% SDS/PAGE and transferred onto a PVDF membrane (Amersham Biosciences, Arlington Heights, IL). The membrane was blocked in 5% skim milk, and subsequently incubated with primary antibodies listed in Supplementary Table 3 at 4°C overnight followed by incubation with peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Jackson ImmunoResearch, Inc., West Grove, PA), and developed with Pierce ECL reagent (Thermal Scientific, Rockford, IL).
FACS
For separating prostate stromal cells, dissociated murine prostate cells were prepared as described previously (15) and stained with Pacific blue-anti CD31, CD45 and Ter119 antibodies (eBioscience, San Diego, CA), PE-anti Sca-1 antibody (eBioscience, San Diego, CA), Alexa 647-anti CD49f antibody (Biolegend, San Diego, CA). Prostate stromal cells are CD31−CD45−Ter119−Sca-1+CD49f− (48). For detecting MDSC cells, dissociated murine prostate cells were stained with Pacific blue-anti CD11b (eBioscience, San Diego, CA), and PE-Cy7-anti Gr-1 antibodies (Biolegend, San Diego, CA). FACS analyses and sorting were performed by using the BD LSR Fortessa and Aria II, respectively (BD Biosciences, San Jose, CA).
Histology and immunostaining
H&E staining and immunofluorescence staining were performed using standard protocols on 5-μm paraffin sections. Trichrome staining was performed using Masson’s trichrome (Sigma-Aldrich, St. Louis, MO). For immunostaining, slides were incubated with 5% normal goat serum (Vector Labs, Burlingame, CA) and with primary antibodies diluted in 2.5% normal goat serum overnight at 4°C. Primary antibodies used in this study were listed in Supplementary Table 3. For chromogenic detection, secondary antibodies were ImmPRESS anti-mouse peroxidase (Vector MP-7402). Peroxidase activity was developed with ImmPACT DAB (Vector SK-4015) according to the manufacturer’s instructions. Sections were counterstained with hematoxylin and mounted with Permount. For fluorescent staining, slides were incubated with secondary antibodies (diluted 1:500 in PBST) labeled with Alexa Fluor 488 or 594 (Invitrogen/Molecular Probes, Eugene, OR). Sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO). Immunofluorescence staining was imaged using an Olympus BX60 fluorescence microscope (Olympus Optical Co Ltd, Tokyo, Japan) or a Leica EL6000 confocal microscope (Leica Microsystems, Wetzlar, Germany). Images of immunostaining were analyzed by Image-Pro Plus version 6.3 by Media Cybernetics. For quantification of immunostaining, cell number was determined by using the count feature in the software which asks for the user to indicate the color that would be used to indicate a positive cell (For example: blue would be indicated to count nuclei and thus indicate total numbers of cells). Borders were created such that only epithelial cells or stromal cells would be analyzed.
Statistics
Sample sizes were determined to ensure adequate power to detect a pre-specified effect size wherever applicable. Experiments were performed using 4–16 mice. All animals were included in the analysis. Data are presented as mean ± SD. Student’s t test was used to determine significance between groups. For all statistical tests, the 0.05 level of confidence was accepted for statistical significance.
Supplementary Material
Acknowledgments
Source of support: NCI
We thank Dr. Spyros Artavanis-Tsakonas for providing the human Jagged1 cDNA, the technical support by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom. This work is supported by NIH R01 CA190378 (L.X.), CA196570 (L.X.), NIH P30 CA125123 (the Cancer Center Shared Resources Grant), and the Mouse Embryonic Stem Cell Core at Baylor College of Medicine.
Footnotes
Author contribution:
Qingtai Su, Boyu Zhang, Li Zhang, and Truong Dong: collection and assembly of data, data analysis and interpretation
David Rowley and Michael Ittmann, data analysis and interpretation
Li Xin: Conception and design, manuscript writing, collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
Disclosure of potential conflicts of interest
The authors indicate no potential conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. Epub 2009/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nature reviews Cancer. 2003;3(10):756–67. doi: 10.1038/nrc1186. Epub 2003/10/23. [DOI] [PubMed] [Google Scholar]
- 3.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature reviews Cancer. 2011;11(5):338–51. doi: 10.1038/nrc3035. Epub 2011/04/22. [DOI] [PubMed] [Google Scholar]
- 4.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71. doi: 10.1126/science.1102160. Epub 2004/10/09. [DOI] [PubMed] [Google Scholar]
- 5.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16(1):55–66. doi: 10.1016/j.ccr.2009.05.016. Epub 2009/07/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H, Zhou X, Redfield S, Lewin J, Miele L. Elevated Jagged-1 and Notch-1 expression in high grade and metastatic prostate cancers. American journal of translational research. 2013;5(3):368–78. Epub 2013/05/02. [PMC free article] [PubMed] [Google Scholar]
- 7.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64(19):6854–7. doi: 10.1158/0008-5472.CAN-04-2500. Epub 2004/10/07. [DOI] [PubMed] [Google Scholar]
- 8.Danza G, Serio CD, Ambrosio MR, Sturli N, Lonetto G, Rosati F, et al. Notch3 is activated by chronic hypoxia and contributes to the progression of human prostate cancer. International journal of cancer Journal international du cancer. 2013 doi: 10.1002/ijc.28293. Epub 2013/06/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65(14):6207–19. doi: 10.1158/0008-5472.CAN-05-0592. Epub 2005/07/19. [DOI] [PubMed] [Google Scholar]
- 10.Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, et al. Suppression of Acquired Docetaxel Resistance in Prostate Cancer through Depletion of Notch- and Hedgehog-Dependent Tumor-Initiating Cells. Cancer Cell. 2012;22(3):373–88. doi: 10.1016/j.ccr.2012.07.016. Epub 2012/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, et al. Down-regulation of Notch-1 is associated with Akt and FoxM1 in inducing cell growth inhibition and apoptosis in prostate cancer cells. Journal of cellular biochemistry. 2011;112(1):78–88. doi: 10.1002/jcb.22770. Epub 2010/07/27. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Wang Z, Li Y, Banerjee S, Kong D, Ahmad A, Nogueira V, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. Journal of cellular biochemistry. 2010;109(4):726–36. doi: 10.1002/jcb.22451. Epub 2010/01/07. [DOI] [PubMed] [Google Scholar]
- 13.Yong T, Sun A, Henry MD, Meyers S, Davis JN. Down regulation of CSL activity inhibits cell proliferation in prostate and breast cancer cells. Journal of cellular biochemistry. 2011;112(9):2340–51. doi: 10.1002/jcb.23157. Epub 2011/04/27. [DOI] [PubMed] [Google Scholar]
- 14.Kwon OJ, Valdez JM, Zhang L, Zhang B, Wei X, Su Q, et al. Increased Notch signalling inhibits anoikis and stimulates proliferation of prostate luminal epithelial cells. Nature communications. 2014;5:4416. doi: 10.1038/ncomms5416. Epub 2014/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdez JM, Zhang L, Su Q, Dakhova O, Zhang Y, Shahi P, et al. Notch and TGFbeta form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell stem cell. 2012;11(5):676–88. doi: 10.1016/j.stem.2012.07.003. Epub 2012/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61(19):7291–7. Epub 2001/10/05. [PubMed] [Google Scholar]
- 17.Dalrymple S, Antony L, Xu Y, Uzgare AR, Arnold JT, Savaugeot J, et al. Role of notch-1 and E-cadherin in the differential response to calcium in culturing normal versus malignant prostate cells. Cancer Res. 2005;65(20):9269–79. doi: 10.1158/0008-5472.CAN-04-3989. Epub 2005/10/19. [DOI] [PubMed] [Google Scholar]
- 18.Velasco AM, Gillis KA, Li Y, Brown EL, Sadler TM, Achilleos M, et al. Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinology. 2004;145(8):3913–24. doi: 10.1210/en.2004-0311. Epub 2004/05/08. [DOI] [PubMed] [Google Scholar]
- 19.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. The Journal of experimental medicine. 2007;204(12):2935–48. doi: 10.1084/jem.20071082. Epub 2007/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. The EMBO journal. 2011;30(4):756–69. doi: 10.1038/emboj.2010.358. Epub 2011/01/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bin Hafeez B, Adhami VM, Asim M, Siddiqui IA, Bhat KM, Zhong W, et al. Targeted knockdown of Notch1 inhibits invasion of human prostate cancer cells concomitant with inhibition of matrix metalloproteinase-9 and urokinase plasminogen activator. Clin Cancer Res. 2009;15(2):452–9. doi: 10.1158/1078-0432.CCR-08-1631. Epub 2009/01/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19(2):192–205. doi: 10.1016/j.ccr.2010.12.022. Epub 2011/02/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiozawa Y, Pedersen EA, Havens AM, Jung Y, Mishra A, Joseph J, et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. The Journal of clinical investigation. 2011;121(4):1298–312. doi: 10.1172/JCI43414. Epub 2011/03/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terada N, Shiraishi T, Zeng Y, Aw-Yong KM, Mooney SM, Liu Z, et al. Correlation of Sprouty1 and Jagged1 with aggressive prostate cancer cells with different sensitivities to androgen deprivation. Journal of cellular biochemistry. 2014;115(9):1505–15. doi: 10.1002/jcb.24805. Epub 2014/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, Zhang Y, Guan W, Huang T, Kang J, Sheng X, et al. Androgen receptor promotes the oncogenic function of overexpressed Jagged1 in prostate cancer by enhancing cyclin B1 expression via Akt phosphorylation. Molecular cancer research: MCR. 2014;12(6):830–42. doi: 10.1158/1541-7786.MCR-13-0545. Epub 2014/02/28. [DOI] [PubMed] [Google Scholar]
- 26.Jin C, McKeehan K, Wang F. Transgenic mouse with high Cre recombinase activity in all prostate lobes, seminal vesicle, and ductus deferens. Prostate. 2003;57(2):160–4. doi: 10.1002/pros.10283. Epub 2003/09/02. [DOI] [PubMed] [Google Scholar]
- 27.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73(9):2718–36. doi: 10.1158/0008-5472.CAN-12-4213. Epub 2013/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nature reviews Cancer. 2013;13(10):739–52. doi: 10.1038/nrc3581. Epub 2013/09/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibb DR, Saleem SJ, Kang DJ, Subler MA, Conrad DH. ADAM10 overexpression shifts lympho- and myelopoiesis by dysregulating site 2/site 3 cleavage products of Notch. J Immunol. 2011;186(7):4244–52. doi: 10.4049/jimmunol.1003318. Epub 2011/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–5. doi: 10.1126/science.1252510. Epub 2014/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. The Journal of urology. 2001;166(6):2472–83. [PubMed] [Google Scholar]
- 32.Barron DA, Rowley DR. The reactive stroma microenvironment and prostate cancer progression. Endocrine-related cancer. 2012;19(6):R187–204. doi: 10.1530/ERC-12-0085. Epub 2012/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barron DA, Strand DW, Ressler SJ, Dang TD, Hayward SW, Yang F, et al. TGF-beta1 induces an age-dependent inflammation of nerve ganglia and fibroplasia in the prostate gland stroma of a novel transgenic mouse. PLoS One. 2010;5(10):e13751. doi: 10.1371/journal.pone.0013751. Epub 2010/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Xu K, Zhang L, Deng Y, Lee P, Shapiro E, et al. Differentiation of the ductal epithelium and smooth muscle in the prostate gland are regulated by the Notch/PTEN-dependent mechanism. Developmental biology. 2011;356(2):337–49. doi: 10.1016/j.ydbio.2011.05.659. Epub 2011/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr B, Grace OC, Vanpoucke G, Ashley GR, Thomson AA. A role for notch signaling in stromal survival and differentiation during prostate development. Endocrinology. 2009;150(1):463–72. doi: 10.1210/en.2008-0383. Epub 2008/09/20. [DOI] [PubMed] [Google Scholar]
- 36.del Alamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Current biology: CB. 2011;21(1):R40–7. doi: 10.1016/j.cub.2010.10.034. Epub 2011/01/11. [DOI] [PubMed] [Google Scholar]
- 37.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. The Journal of biological chemistry. 2003;278(36):34427–37. doi: 10.1074/jbc.M302659200. Epub 2003/06/27. [DOI] [PubMed] [Google Scholar]
- 38.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–89. doi: 10.1158/0008-5472.CAN-11-3132. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ascano JM, Beverly LJ, Capobianco AJ. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. The Journal of biological chemistry. 2003;278(10):8771–9. doi: 10.1074/jbc.M211427200. Epub 2002/12/24. [DOI] [PubMed] [Google Scholar]
- 40.Lu C, Bonome T, Li Y, Kamat AA, Han LY, Schmandt R, et al. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer Res. 2007;67(4):1757–68. doi: 10.1158/0008-5472.CAN-06-3700. Epub 2007/02/20. [DOI] [PubMed] [Google Scholar]
- 41.Steg AD, Katre AA, Goodman B, Han HD, Nick AM, Stone RL, et al. Targeting the notch ligand JAGGED1 in both tumor cells and stroma in ovarian cancer. Clin Cancer Res. 2011;17(17):5674–85. doi: 10.1158/1078-0432.CCR-11-0432. Epub 2011/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedrosa AR, Trindade A, Carvalho C, Graca J, Carvalho S, Peleteiro MC, et al. Endothelial Jagged1 promotes solid tumor growth through both pro-angiogenic and angiocrine functions. Oncotarget. 2015;6(27):24404–23. doi: 10.18632/oncotarget.4380. Epub 2015/07/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–34. doi: 10.1038/nature12984. Epub 2014/01/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulos MG, Guo P, Kofler NM, Pinho S, Gutkin MC, Tikhonova A, et al. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell reports. 2013;4(5):1022–34. doi: 10.1016/j.celrep.2013.07.048. Epub 2013/09/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alme AK, Karir BS, Faltas BM, Drake CG. Blocking immune checkpoints in prostate, kidney, and urothelial cancer: An overview. Urologic oncology. 2016;34(4):171–81. doi: 10.1016/j.urolonc.2016.01.006. Epub 2016/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. Epub 2002/07/02. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 48.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci U S A. 2007;104(1):181–6. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.