Abstract
DNA barcoding is a global initiative for species identification through sequencing of short DNA sequence markers. Sequences of two loci, ITS and LSU, were generated as barcode data for all (ca. 9k) yeast strains included in the CBS collection, originally assigned to ca. 2 000 species. Taxonomic sequence validation turned out to be the most severe bottleneck due to the large volume of generated trace files and lack of reference sequences. We have analysed and validated CBS strains and barcode sequences automatically. Our analysis shows that there were 6 and 9.5 % of CBS yeast species that could not be distinguished by ITS and LSU, respectively. Among them, ∼3 % were indistinguishable by both loci. Except for those species, both loci were successfully resolving yeast species as the grouping of yeast DNA barcodes with the predicted taxonomic thresholds was more than 90 % similar to the grouping with respect to the expected taxon names. The taxonomic thresholds predicted to discriminate yeast species were 98.41 % for ITS and 99.51 % for LSU. To discriminate current yeast genera, thresholds were 96.31 % for ITS and 97.11 % for LSU. Using ITS and LSU barcodes, we were also able to show that the recent reclassifications of basidiomycetous yeasts in 2015 have made a significant improvement for the generic taxonomy of those organisms. The barcodes of 4 730 (51 %) CBS yeast strains of 1 351 (80 %) accepted yeast species that were manually validated have been released to GenBank and the CBS-KNAW website as reference sequences for yeast identification.
Key words: Automated curation, D1/D2, Fungi, ITS, LSU, Taxonomy, Yeast
Introduction
DNA barcoding is a global initiative that aims at streamlining species identification through analysis of short DNA sequence markers (Hebert et al. 2003). The leading principles of the approach are (i) a general agreement on one or a few marker regions; (ii) the usage of vouchered material; (iii) the availability of trace files; and (iv) the assembly of sequence data and the specimen metadata in public databases (Stoeckle & Hebert 2008). The vision is that DNA barcoding will make species identification accessible to non-experts, and therefore will promote biological and medical progress at large. Although there is no generally applicable species concept (Wheeler & Meier 2000), especially for character-poor, not obligatory sexually reproducing organisms, such as most Fungi, species identification is a key step in many fields of biology, biotechnology, agriculture, ecology, medicine and commerce to describe biological interactions in the context of e.g. pathology, bioremediation, or biodiversity assessment (de Queiroz 2007).
The CBS-KNAW microbial biological resource centre is a large public collection that hosts more than 80 000 strains, of which 51 000 are publicly available on the CBS-KNAW website (www.cbs.knaw.nl). The collection is extensively used in basic and applied research. For example, many phylogenetic studies have used CBS strains, ex-type as well as non ex-type strains, to generate sequences for phylogenetic analyses. The CBS yeast collection currently has ca. 9 000 strains representing more than 1 700 currently recognized species, of which 7 581 strains are publicly available. Among them, ca. 2 200 strains are ex-type strains of ca. 1 600 currently recognized species. Each identified strain is assigned a taxon name linked to MycoBank (www.mycobank.org), an online registration system for fungal species and higher level taxon names. Strain identification was normally achieved with the knowledge and methods available at the time of accession with the oldest strains deposited in 1908. While taxon names of strains other than ex-type strains have been updated using current taxonomic concepts, it is often not possible to re-evaluate the original identification, because not all characters necessary for identification are expressed in culture. Thus, the CBS DNA barcoding project is aimed at generating barcode data for all strains included in the collection, 1) to validate these barcodes and (re-)assess strain identity, 2) to enhance the value of the collection and 3) to make these data publicly available. The lack of validated data is generally perceived as a setback for fungal and yeast research and its applications (Bidartondo 2008). With the CBS DNA barcoding project, we ultimately aim to publish a sizable ITS and LSU sequence reference dataset that is taxonomically validated.
For this study, all the yeast strains in the CBS-KNAW collection were sequenced for the Internal Transcribed Spacers (ITS) and D1/D2 domain of Large Subunit (LSU) ribosomal nuclear DNA to create a complete dataset of yeast barcodes. This study allows us to reduce the number of incorrect species assignments. When using ITS as a barcode, it has been estimated as more than 10 % of the public sequence databases such as GenBank (Nilsson et al. 2006). Although the ITS has been proposed and accepted as a universal DNA barcode for Fungi (Schoch et al. 2012), LSU was also sequenced to act as a secondary barcode. It has been widely used as a marker for yeast identification long before the concept of DNA barcoding was formulated and promoted (Peterson and Kurtzman, 1991, Kurtzman and Robnett, 1998). Also, other barcoding regions have recently been proposed (Stielow et al. 2015) to complement the data presented in this study. To manage and analyse the huge amount of data created in this study, we developed a dedicated laboratory information management system (LIMS; Vu et al. 2012) to keep track of the thousands of sequences generated, aligned, edited and stored on a daily basis.
Over the past seven years, the taxonomic assignment of 56 % of CBS yeast strains has been manually validated based on ITS or LSU barcodes. However, several sequence validation issues were experienced. In a routine sequence identification scheme, one searches for the best match of the sequence in the local database or other public databases such as GenBank (www.ncbi.nlm.nih.gov), whereby good taxonomic knowledge is required to interpret BLAST results (Altschul et al. 1997). This manual validation process becomes increasingly difficult, and sometimes impossible, stemming from the large number of generated sequences, the lack of reference sequences and limited human resources. Additionally, as the continuous development in taxonomy results in an on-going stream of reclassifications and introduction of new names, sequence validation will have to be regularly re-applied in order to update strain and sequence identification.
The task of assigning species names to newly generated sequences has inspired us to create a number of algorithms and tools for identification and classification (Vu et al. 2014). Our objectives were to assign a taxon name to unidentified strains and sequences, to suggest another taxon name for incorrectly labelled sets of strains and sequences, and to highlight the hidden yeast species diversity in the collection. Our expectation was that even among the identified strains, there would be a considerable portion of unrecognized and potentially undescribed taxa. The problems of satisfactory taxonomic assignment of newly generated ITS sequences have been addressed before. Kõljalg et al. (2013) proposed a useful paradigm for sequence-based identification of Fungi in which they introduced the term “species hypothesis” (SH) to generate a concise set of reference sequences for taxon discovery. Sequences were clustered into SHs with six given different thresholds from 97 % to 99.5 %. For each SH, a representative sequence was chosen automatically or manually. Unlike that approach, our representative sequences were chosen as representative sequences of yeast species. In particular, a representative sequence was chosen as the reference sequence of the ex-type strain of a species if this ex-type strain existed. Otherwise, it was chosen as the reference sequence of the central representative strain of the species. We automatically predicted a taxonomic threshold for clustering that produces the best match to the current taxonomic classification using yeast barcodes. Based on the predicted threshold, sequences were clustered and validated manually or automatically. Our approach in predicting a threshold to discriminate yeast species is also different from previous studies (Peterson and Kurtzman, 1991, Kurtzman and Robnett, 1998, Fell et al., 2000, Scorzetti et al., 2002, Sugita et al., 2002, Kurtzman, 2014) in yeast identification where the taxonomic threshold was based on the similarity of the strains within pre-defined yeast species.
Using the world's largest yeast barcode dataset (obtained from the CBS yeast collection), extensive analyses were performed to study the identification of yeast species and associated strains as the following metrics were assessed: (1) the similarity of strains based on DNA barcodes within and between yeast species; (2) the positioning of the ex-type strains within yeast species to assess how representative the ex-type strain is for the species; (3) the verification of synonymy and indistinguishable yeast species by ITS and/or LSU; (4) the correlation between the two similarity values based on ITS and LSU barcodes; as well as (5) the taxonomic thresholds to discriminate yeast species and genera using ITS and LSU barcodes. Based on the predicted taxonomic thresholds, yeast strains and sequences were automatically validated. All problematic sequences, strains, and species were highlighted to be further manually curated. The barcodes of 4 730 (51 %) CBS yeast strains of 1 351 (80 %) yeast species that were manually validated have been released to GenBank and the CBS-KNAW website as reference sequences for yeast identification.
Material and methods
Barcode sequences of ITS and LSU were generated for all CBS yeast strains and imported into the database. For the analysis presented in this paper, all the barcodes of yeast strains that were added to the database until June 2015, were included. The species names of the strains were also updated with the new names obtained from recent large-scale reclassifications in Basidiomycota (Liu et al., 2015, Wang et al., 2015a, Wang et al., 2015b, Wang et al., 2015c).
Generating and managing of barcode sequences
The protocol to generate DNA barcode sequences of the two loci ITS and LSU for CBS strains is given in Stielow et al. (2015). To be able to manage a large amount of sequence data and to keep track of the whole experimental procedure, a laboratory information management system (LIMS, Vu et al. 2012) was developed for the CBS-DNA barcoding workflow as a module of BioloMICS (Robert et al. 2011), a software package to manage, analyse and publish biological data. To have the dataset as complete as possible for the analysis, all ITS and LSU yeast sequences from GenBank (GB) (https://www.ncbi.nlm.nih.gov/nuccore/) were also downloaded with the queries txid4751[porgn] AND 5.8S [TITLE] AND “yeast”[ALL fields] and txid4751[porgn] AND (26s [TITLE] or 25s [TITLE] or 28s [TITLE] or Lsu[TITLE]) AND “yeast”[ALL fields] NOT 5.8[TITLE], and included in the database in June 2015.
Computation of DNA similarity between sequences
The similarity value of two DNA sequences was computed using our own implementation (Robert et al. 2011) of the BLAST algorithm (Altschul et al. 1997) as the percentage identity of the most similar region or overlap between the two sequences. To avoid problems associated with short sequences being similar to many reference sequences, the similarity value s between two sequences a and b, was recomputed when their overlap o was less than 150 bp as s = s × o/150.
Selecting reference sequences for strains and species
Theoretically, one strain should have only one sequence per locus. But in practice, many CBS strains had more than one sequence per locus, since they have been sequenced several times for different reasons and purposes. Besides, sequences of CBS strains from other collections and public databases were imported in the CBS collection as well. For each strain and locus, a reference sequence was selected manually based on its quality and length. If no sequence had been manually selected, the reference sequence will be selected automatically. We first employed the concept of a central representative sequence (Antonielli et al. 2011). The central representative sequence of a group was the sequence maximizing the similarity value to the other sequences of the same group. If a group had only two sequences, then the first sequence entered into the collection was selected. The reference sequence of a strain was the sequence maximizing the similarity value to the central representative sequence of the species. The representative sequence of a species was the reference sequence of the ex-type strain if it was available. Otherwise, it was the central representative sequence of all the reference sequences of the strains associated with the species. The strain associated with the (central) representative sequence was the (central) representative strain of the species. The similarity value of two strains was the similarity value of their reference sequences.
Probability of correct identification (PCI)
Given a locus, the identification of a species is correct if for every strain of the species, there is no other strain from another species such that the similarity value between them is greater or equal to the minimum similarity value between the strains of the species. The barcode gap PCI is the fraction of species correctly identified. To evaluate the resolving power of multiple loci for species discrimination, the similarity value of two strains using multiple loci was computed as the average similarity value of all similarity values computed for each locus (Hollingsworth et al., 2009, Schoch et al., 2012).
Clustering of strains or DNA sequences
To cluster strains or DNA sequences, we used an algorithm developed to find connected components (Hopcroft & Tarjan 1973) as it has shown to be highly accurate in comparison with other clustering algorithms (Vu et al. 2014). Given a similarity value or threshold, two strains or sequences of a dataset will be connected if there is a path of strains or sequences between them in which the similarity value of a strain or sequence to the next one is equal to or greater than the given threshold. The clustering algorithm places all connected strains or sequences of the dataset in the same group.
Quality of automatic clustering
Different thresholds lead to different groupings of the strains or sequences of a dataset. The strategy is to place the members of the dataset in the right taxonomic groups and automatically assign them to a taxon name. The F-measure function proposed by Paccanaro et al. (2006) was used to evaluate how similar an automatic clustering result is when compared to the manual taxonomic assignments. Let C = (C1,…,Cl) be the partition of a given set of strains or sequences obtained by taxonomic classification, and K = (K1,…,Km) be the partition obtained by clustering the dataset with a given threshold. The quality of clustering is computed by the F-measure function F (K, C), defined as follows
where n is the size of the dataset, ni is the size of Ki, nj is the size of Cj, and is the size of for 1 ≤ i ≤ m and 1 ≤ j ≤ l.
The F-measure ranges from 0 to 1. The higher the value of the F-measure, the more similar the clustering result is to the taxonomic classification. It is equal to 1 when the clustering result matches the taxonomic classification perfectly.
Predicting a taxonomic threshold for species identification
The taxonomic threshold to cluster a dataset of strains or sequences was calculated as the optimal threshold that produces the best quality for clustering in comparison with the taxonomic classification, in other words, the one having the highest F-measure.
Automatic species assignments of strains
To assign species names to the strains of a dataset automatically all the sequences of the dataset were clustered with the predicted taxonomic threshold. Strains or sequences of the same cluster will have a predicted taxon name, which is the most common or frequently used species name amongst the members of the cluster (Vu et al. 2012). If the given taxon name of a member is different from the predicted taxon name, the member is considered wrongly assigned if its similarity value to the associated representative is less than a given lower bound threshold. Otherwise, it is considered as a strain or sequence of a synonym or a closely related species. To reduce the problem of wrong species assignment because a dataset might contain a majority of similar sequences with the same wrong species assignment, for each strain, only the reference sequence was considered.
Results and discussion
DNA barcoding data
Ex-type strains at CBS
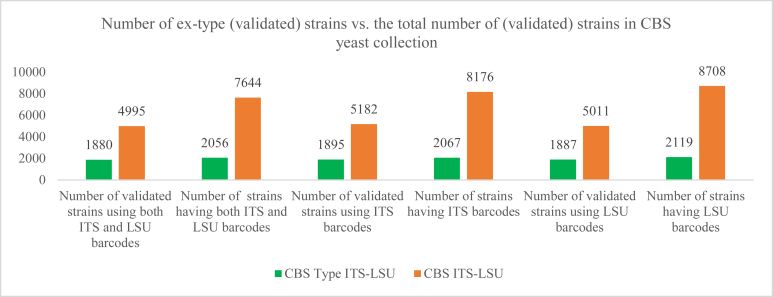
Firstly, barcode sequences of two loci ITS and LSU were generated for the total of 2 130 yeast ex-type strains. This included 1 571 ex-type strains of currently recognized species. There were 2 067 and 2 119 strains having at least one ITS and LSU sequences, respectively in which 2 056 strains had both barcodes available (see Fig. 1). The numbers of sequences, strains, and species of each CBS barcode type datasets are given in datasets T1 and T2 in Table 1.
Fig. 1.
Number of ex-type (validated) strains using ITS/LSU barcodes versus the total number of (validated) strains in the CBS yeast collection.
Table 1.
Numbers of species, strains, and sequences of different barcode datasets.
| Dataset | Abbr. | Number species | Number of strains identified at species level | Number of strains | Number of sequences |
|---|---|---|---|---|---|
| CBS type ITS | T1 | 1 436 | 2 067 | 2 067 | 6 108 |
| CBS ITS | C1 | 1 595 | 6 768 | 8 176 | 14 601 |
| Manually validated CBS ITS | M1 | 1 387 | 5 182 | 5 182 | 10 454 |
| GB ITS | N1 | 856 | 5 564 | 6 958 | 6 985 |
| CBS+GB ITS | CN1 | 1 782 | 11 768 | 17 994 | 21 134 |
| CBS type LSU | T2 | 1 463 | 2 119 | 2 119 | 8 795 |
| CBS LSU | C2 | 1 617 | 7 269 | 8 708 | 19 ,498 |
| Manually validated CBS LSU | M2 | 1 380 | 5 011 | 5 011 | 13 211 |
| GB LSU | N2 | 1 042 | 9 304 | 9 393 | 13 938 |
| CBS+GB LSU | CN2 | 1 804 | 15 679 | 21 678 | 32 546 |
| Manually validated dataset having both CBS ITS and CBS LSU sequences | M3 | 1 375 | 4 995 | 4 995 | 10 225 ITS sequences 13 188 LSU sequences |
The “CBS type datasets”, abbreviated as T1 for ITS and T2 for LSU, contained all the strains that were designated as ex-type strains for a currently accepted species or of a synonym species name. The “CBS datasets”, abbreviated as C1 for ITS and C2 for LSU, contained all the strains from the CBS collection including the ex-type strains T1 and T2. The “Manually validated datasets”, abbreviated as M1 for ITS, M2 for LSU and M3 for both ITS and LSU, contained all the CBS strains present in the C1 and C2 datasets that were manually checked by the curators to confirm their species assignments using ITS and/or LSU sequences. The “GB datasets”, abbreviated as N1 for ITS and N2 for LSU, contained all yeast sequences available from the GB database until June 2015. The “CBS+GB datasets”, abbreviated as CN1 for ITS and CN2 for LSU, contained all data from datasets C and N in which strains and sequences were accounted once.
All yeast strains at CBS
There were DNA sequences of both ITS and/or LSU loci for 9 240 CBS strains including ex-type strains. Of the 9 240 strains, 8 176 and 8 708 strains had ITS and LSU barcodes, respectively in which 7 644 strains had both barcodes available (see Fig. 1). The number of sequences, strains, and species of each CBS barcode dataset is given in datasets C1 and C2 in Table 1.
Manually validated yeast strains at CBS
Of 2 130 ex-type strains, 1 895 and 1 887 strains were manually validated using ITS and LSU barcodes, respectively in which 1 880 strains were manually validated using both barcodes. Of 9 240 strains, 5 182 and 5 011 strains were manually validated using ITS and LSU barcodes, respectively in which 4 995 strains were manually validated by both loci (see Fig. 1 and datasets M1, M2, and M3 in Table 1).
Yeast barcode sequences at GenBank (GB)
For ITS, there were 6 985 GB yeast sequences of which 668 sequences were cited with a CBS number; 5 740 sequences cited with some collection or personal numbers; 90 sequences labelled as uncultured yeast; 487 sequences labelled with some taxon name. In addition, 5 589 sequences were given a species name and identified to 856 species; 15 ITS sequences annotated as type sequences; and 27 CBS numbers cited with more than one GB sequences. For LSU, there were 13 938 GB sequences of which 1 151 sequences were cited with a CBS number; 8 023 sequences cited with some collection or personal numbers; 2 998 sequences labelled as uncultured yeast; and 1 766 sequences labelled with a taxon name. There were 9 393 sequences with a given species name identified to 1 042 species; 10 LSU sequences annotated as type sequences and 93 CBS numbers cited with more than one GB sequences. The number of sequences, strains and species of each GB and CBS together with GB barcode dataset are given in Table 1 (see datasets N1, N2, CN1 and CN2).
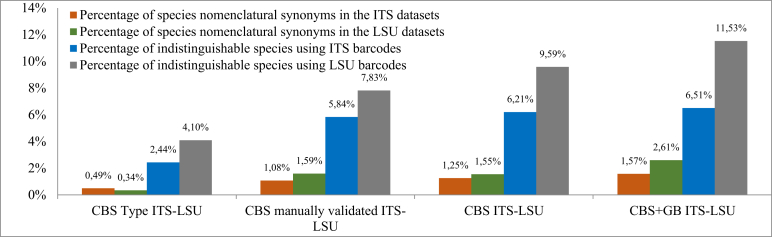
It must be noted that as many strains of the CBS collection have been sequenced many times for different projects and purposes, each CBS strain may include more than one sequence per locus. Furthermore, species were assumed to represent species complexes, i.e. all the expressions such as aff., cf., f. or var. were considered to belong to the same species. At the time of the analysis, we considered 1 684 taxa as accepted yeast species and 1 656 taxa as accepted species complexes of culturable yeasts in the CBS database. A further 116 yeast strains of the CBS yeast collection still await DNA barcodes due to various technical problems. Despite the effort to keep the species names up to date, a small fraction of species names in the CBS yeast database (∼1.5 %) represented species synonyms (see Fig. 7).
Fig. 7.
Percentages of yeast species synonyms and indistinguishable species by using ITS and LSU barcodes with the threshold of 100 %, respectively.
Analysis of DNA barcoding data
In our analysis, for each strain only the reference sequence was considered. Thus, for each dataset there was one sequence for each strain.
Similarity value within yeast species
To decide if a strain belongs to a given species, it is important to know the similarity of the strains within that species. The similarity of the strains within yeast species has been studied previously in many studies among which one can note of few outstanding studies like the analysis of 500 yeast species by Kurtzman & Robnett (1998), 337 strains of 230 yeast species in Fell et al. (2000), and 450 strains of 242 yeast species by Scorzetti et al. (2002). It was demonstrated in these studies that strains of yeast species showed less than 1 % dissimilarity in either ITS or LSU regions.
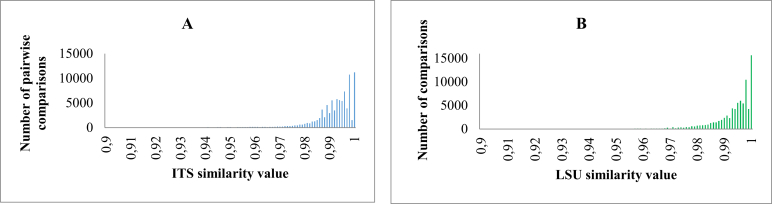
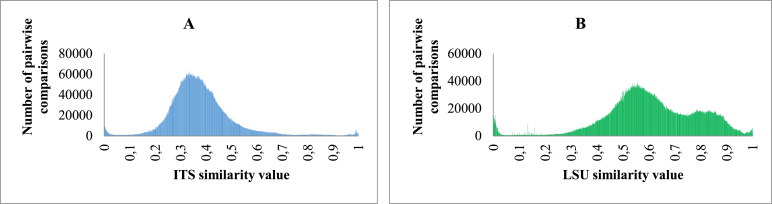
In our study, we first looked at the CBS manually validated datasets in which the data were evaluated by the curators. Fig. 2A shows the number of pairwise comparisons within yeast species of M1 when the associated ITS similarity value increased from 0.9 to 1 while Fig. 2B shows the number of pairwise comparisons within yeast species of M2 when the associated LSU similarity value increased from 0.9 to 1. It can be seen from Fig. 2A that the similarity values within yeast species varied mainly from 0.95 to 1 with ITS barcodes. For the latter, 97 % pairwise comparisons of strains of the same species in M1 had a similarity value of at least 95 %. Fig. 2B shows that the similarity values within yeast species varied mainly from 0.97 to 1 and 94 % pairwise comparisons of strains of the same species in M2 had a similarity value of at least 97 %.
Fig. 2.
The number of pairwise comparisons of manually validated strains of the same yeast species having ITS (A) and LSU (B) similarity values zoomed in the range from 0.9 to 1.
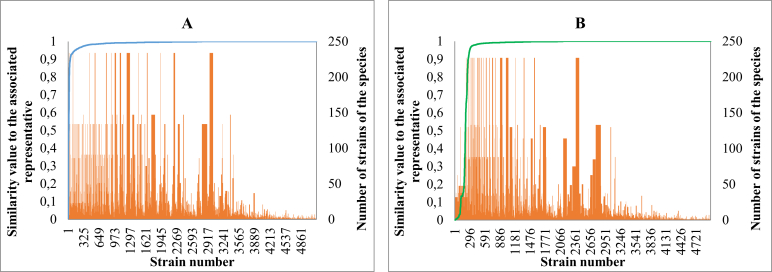
Fig. 3 shows the similarity value of each strain of M1 and M2 when compared to the associated species representative (ex-type if existed otherwise central) strain. Three and 6 % of the strains of M1 and M2 were less than 95 % and 97 % similar to their representative strain, respectively. Compared to the results of the previous studies, it is clear that some yeast species concepts have likely broadened and/or a number of CBS yeast strains and sequences were wrongly assigned as they were too distant to their representative strain.
Fig. 3.
The similarity value of each manually validated strain to the associated representative strain (in blue and green lines) and the number of strains within species (in red line, secondary axis) by using ITS (A) and LSU (B) barcodes, respectively.
When including all the CBS yeast strains (datasets C1 and C2), 88 % and 87 % of pairwise strains comparisons within yeast species showed at least 95 % and 97 % similarity using ITS and LSU barcodes, respectively (see Supplementary Figs S1A and S2A). The number of CBS strains that were less than 95 % and 97 % similar to their representative using ITS and LSU barcodes increased to 8 % and 9.5 % (see Supplementary Figs S1B and S2B). The problem was more obvious when all the sequences downloaded from GenBank were also taken into account (datasets CN1 and CN2). As seen in Supplementary Figs S3B and S4B, 19 % and 30 % of the sequences in CN1 and CN2 were less than 95 % and 97 % similar to their representative sequence, respectively. The peaks at the similarity value of 50 % in Supplementary Figs S3A and S4A indicate that a number of the sequences and strains from GenBank were not reliably identified as they were less than 50 % similar to other strains of the same species. This problem was also addressed for ITS sequences of fungal species in public databases such as UNITE and GenBank in Nilsson et al. (2006) and Kõljalg et al. (2013).
The positioning of ex-type strains within yeast species
As the ex-type strains are the reference points for species naming and identification (McNeil et al. 2012), it is essential to know if the ex-type strain of a species is the central representative strain of the species. If the ex-type strain is eccentrically positioned according to the barcode marker compared, the identification procedure based on sequence comparison with the ex-type strain related data is likely to assign strains that are not highly similar to the species as a whole, and it may exclude others. Using the CBS barcode datasets (C1 and C2), we addressed this question.
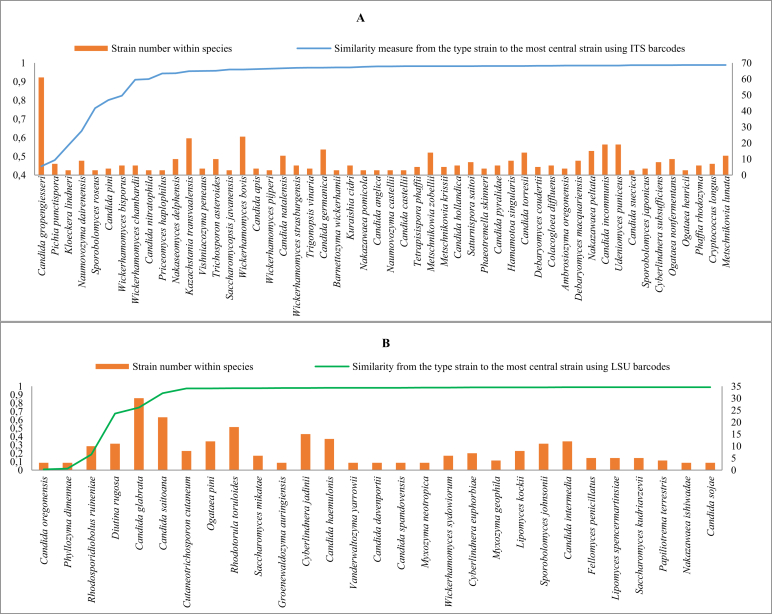
For the ITS dataset, there were 5 875 strains belonging to 1 397 species whose ex-type strain was available. For 969 species, only one or two strains were present. These species were excluded from the analysis. Of the 428 remaining species, 265 had an ex-type strain that was also the central representative strain of the species and 163 species (∼12 %) had an eccentric ex-type strain. Fig. 4A shows the species with the ITS similarity value lower than 99 % from the ex-type strain to the central representative strain, together with the number of the strains within the species. The were only 11 (∼0.8 %) species in which the ex-type strain was less than 95% similar to the central representative strain i.e. Papiliotrema laurentii (61 strains studied), Diutina rugosa (7), Ogataea trehaloabstinens (3), Pichia occidentalis (9), Cystobasidium pallidum (3), Rhodosporidium azoricum (4), Fellomyces penicillatus (6), Malassezia globosa (6), Myxozyma neotropica (3), Schizosaccharomyces octosporus (3), and Sporidiobolus johnsonii (10).
Fig. 4.
The yeast species with ITS (A) and LSU (B) similarity value less than 99 % from the ex-type to central representative strains. The number of the strains of the species is displayed in the secondary axis.
For the LSU dataset, there were 6 254 strains belonging to 1 427 species whose ex-type strain was available. For 978 species, only one or two strains were present. These species were excluded from the analysis. Of the 449 remaining species, 321 had an ex-type strain that was also the central one and 128 species (∼9 %) had an eccentric ex-type strain. Fig. 4B shows the species with the LSU similarity value lower than 99 % from the ex-type strain to the central representative strain, together with the number of the strains within the species. There were only six species (∼0.4 %) in which the ex-type strain was less than 97 % similar to the central representative strain i.e. Candida oregonensis (3 strains studied), Phyllozyma dimennae (3), Rhodosporidiobolus ruineniae (10), Diutina rugosa (11), Candida glabrata (30), and Candida saitoana (22).
The small number (0.8 % for ITS and 0.4 % for LSU) of species with a low similarity value between the ex-type and central representative strain shows that the identification procedure of most yeast species based on sequence comparison with ex-type strains will assign strains to highly similar species. The low similarity values between the ex-type and central representative strains of the listed species in both ITS and LSU datasets indicate that there is a need to re-evaluate the taxonomic assignment of the strains of these species. There can be three reasons for this problem: a) the strains or sequences were wrongly labelled; b) the characters formerly/traditionally used for species identification were not species specific and/or; c) species concepts have shifted emphasis away from the traditional species criteria with the accumulation of cryptically similar strains over time, without accounting for the distance to the ex-type strain. For example, Diutina rugosa has an ex-type strain that is 48.5 % and 67.5 % similar to the central representative strain using ITS and LSU respectively. The phylogenetic trees of the manually validated strains Diutina rugosa with the ex-type strain (CBS 1010) of Candida pararugosa using ITS and LSU barcodes are shown in Fig. 5. The strains of Diutina rugosa are split into two subgroups: one having the ex-type strain of the species (CBS 613), and the other one having strain CBS 2275 that now belongs to C. pararugosa or to a closely related species (Kurtzman et al. 2011).
Fig. 5.
Phylogenetic trees of Diutina rugosa complex strains with the ex-type strain of Candida pararugosa using ITS (A) and LSU (B) barcodes, respectively.
Similarity value between yeast species
The similarity between yeast species is just as important as the similarity values within yeast species for species identification. It is used to decide if a strain does not belong to a given species. Fig. 6 shows the numbers of pairwise comparisons of different yeast species of the CBS manually validated datasets M1 and M2 having an ITS and LSU similarity value increasing from 0 to 1, respectively. The similarity value between yeast species ranged from 0 % to 100 % by both loci. However, most of the similarity values ranged from 15 % to 70 % and from 30 % to 97 % using ITS and LSU barcodes, respectively. In particular, 96 % and 95 % pairwise comparisons of strains from different species had a similarity value between 15 % and 70 % using ITS and between 30 % and 97 % using LSU barcodes. The peaks around the 100 % similarity value in both figures show that a number of species were synonyms, or ITS and LSU loci were not discriminative enough in some yeast species.
Fig. 6.
The number of pairwise comparisons of manually validated strains between yeast species when the associated ITS (A) and LSU (B) similarity values increased from 0 to 1.
To examine the fraction of species that cannot be distinguished by ITS or LSU loci, sequences of different barcode datasets were clustered with the similarity value of 100 %. The species, whose sequences were grouped in the same cluster, were considered indistinguishable by the respective locus. To avoid two species being grouped on the basis of sequences that are distant from the representative (type and/or central representative sequence) of their own group, all the ITS and LSU sequences of the CBS and CBS+GB datasets having a similarity value lower than 95 % and 97 % to the associated representative were, respectively, removed. Supplementary Table 1 shows the results of clustering on different datasets after removing those sequences. The lists of species being grouped with the threshold 100 % in different datasets were checked by the specialists. There were a small number of species names occurring in each dataset (from 0.3 % to 1.5 %) that were grouped wrongly with the threshold of 100 % due to the problem of wrongly labelled strains or sequences. Except for these species, the obtained groups contained species names of species nomenclatural synonyms or of indistinguishable species (taxonomic synonyms or closely related species). Fig. 7 shows the percentage of species nomenclatural synonyms and indistinguishable species in the different datasets using ITS and LSU barcodes. There were a number of yeast species that could not be discriminated by ITS or LSU even in the type datasets. For the CBS+GB datasets, ∼11.5 % of the yeast species were indistinguishable using LSU barcodes, while ITS was shown to be more efficient with ∼6.5 %. It should be noted that to decide precisely which yeast species are taxonomic synonyms or closely related species, further studies are required.
Probability of correct identification of yeast species
To evaluate the resolving power of ITS and LSU for yeast species, the barcode gap PCI was computed as 88.4 % for ITS and 84.6 % for LSU using the manually validated datasets M1 and M2. When combining both loci, the barcode gap PCI was 85.83 % using the manually validated dataset M3. These values were much higher than the highest corresponding values computed for fungi (Schoch et al. 2012) which were 77 % for ITS, 75 % for LSU and 78 % for both, although our number of yeast species were six times larger (∼1 380 vs. 226). This shows that currently, ITS and LSU work better in species discrimination for yeasts than for filamentous fungi. With the high value of 88.4 %, ITS outperformed LSU (84.6 %) in yeast species discrimination. Despite the expectation that the barcode gap PCI of multiple loci will be higher than the barcode gap of a single locus as seen in Schoch et al. 2012, the barcode gap PCI of the two loci ITS and LSU was lower than the barcode gap PCI of ITS alone. This is because ITS was more variable than LSU as seen in the previous study of Nilsson et al. (2006) and later in Section Correlation of ITS and LSU of the current paper. When combining ITS and LSU, the average similarity value of the two loci was, in general, greater than the ITS similarity value alone. Therefore, although LSU could resolve a number of species that cannot be discriminated by ITS, there was a larger number of species that were resolved by ITS, being merged to another species using this combination.
Taxonomic thresholds for yeast species identification
In the previous sections, the similarity values within and between yeast species have been studied. However, the question of what should be the taxonomic similarity value (or threshold) to identify yeast species using ITS and LSU barcodes, remains. This question has been addressed before for yeasts and other microorganisms like bacteria and archaea. It was demonstrated (Kurtzman and Robnett, 1998, Fell et al., 2000, Scorzetti et al., 2002) that strains of yeast species showed less than 1 % dissimilarity in either ITS or LSU regions. To classify bacterial and archaeal species, a threshold of around 98.7 % was predicted using 16S rRNA gene sequences in (Stackebrandt & Ebers 2006). With the almost complete dataset of yeast barcodes with respect to recognized species, this question can be studied extensively. All the barcode datasets were clustered with different similarity values to find an optimal threshold that produced the best quality (F-measure) for clustering (Paccanaro et al. 2006). Here, we predicted the taxonomic threshold as the boundary similarity between the species, while the approaches of Kurtzman and others calculated the taxonomic threshold based on the documented similarity of the strains within species only. It must be noted that while the barcode gap PCI indicates the number of species being correctly identified, the F-measure evaluates the number of strains being correctly identified.
To optimize the taxonomic threshold, sequences of the CBS and CBS+GB datasets (C1, C2, CN1, and CN2), that were less than 95 % (for ITS) and 97 % (for LSU) similar to the associated representative sequence, were removed. Furthermore, as seen in the analyses above, there were yeast species sharing the same ITS and LSU sequences. In a set of analyses, sequences of those species were removed as well.
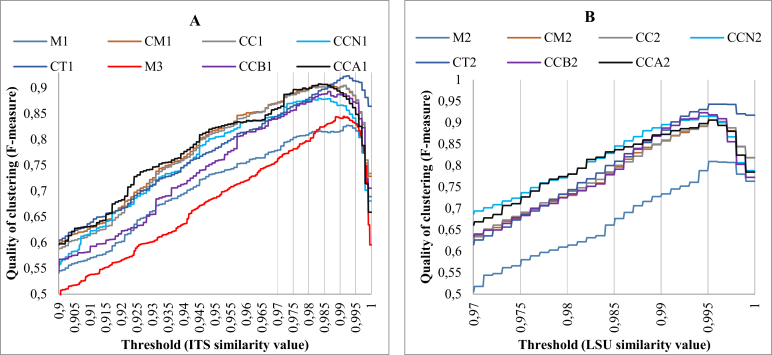
Fig. 8 shows the F-measures obtained by clustering different datasets with thresholds ranging from 0.9 to 1 for ITS and from 0.97 to 1 for LSU, respectively. The red line M3 in Fig. 8A shows the F-measures obtained when clustering the strains of the manually validated dataset M3 using the barcodes of both two loci ITS and LSU. The black (CCA1 and CCA2) and purple (CCB1 and CCB2) lines show the F-measures obtained when clustering ascomycetous and basidiomycetous yeasts using ITS and LSU barcodes, respectively. The vertical lines in the figures represent the thresholds proposed by UNITE for the species hypotheses (Kõljalg et al. 2013). The optimal thresholds and best F-measures computed for each dataset are displayed in Table 2.
Fig. 8.
Clustering qualities obtained when clustering different ITS (A) and LSU (B) barcode datasets with thresholds ranging from 0.9 and 0.97 to 1 using an incremental step of 0.0001. The red line M3 represents the qualities obtained where the similarity value was computed as the average similarity values of the two loci ITS and LSU.
Table 2.
Optimal thresholds and best F-measures obtained by clustering different clean barcode datasets.
| Dataset | Abbreviation | Optimal threshold | Best F-measure | Number of species | Number of strains |
|---|---|---|---|---|---|
| Manually validated ITS | M1 | 99.21 % | 82.73 % | 1 387 | 5 182 |
| Manually validated LSU | M2 | 99.51 % | 80.94 % | 1 380 | 5 011 |
| Manually validated having both ITS and LSU | M3 | 99.11 % | 84.47 % | 1 375 | 4 995 |
| Clean CBS type ITS | CT1 | 99.21 % | 92.33 % | 1 410 | 1 958 |
| Clean, manually validated ITS | CM1 | 98.11 % | 90.9 % | 1 318 | 4 022 |
| Clean CBS ITS | CC1 | 98.41 % | 90.67 % | 1 510 | 4 968 |
| Clean CBS Ascomycetous ITS | CCA1 | 98.31 % | 90.76 % | 981 | 3 029 |
| Clean CBS Basidiomycetous ITS | CCB1 | 98.61 % | 89.31 % | 5 34 | 2 097 |
| Clean CBS+GB ITS | CCN1 | 98.31 % | 88 % | 1 678 | 6 892 |
| Clean CBS type LSU | CT2 | 99.61 % | 94.31 % | 1 415 | 1 963 |
| Clean, manually validated LSU | CM2 | 99.51 % | 90.58 % | 1 285 | 3 835 |
| Clean CBS LSU | CC2 | 99.51 % | 91.48 % | 1 492 | 5 040 |
| Clean CBS Ascomycetous LSU | CCA2 | 99.51 % | 90.59 % | 959 | 3 101 |
| Clean CBS Basidiomycetous LSU | CCB2 | 99.41 % | 92.32 % | 515 | 1 929 |
| Clean CBS+GB LSU | CCN2 | 99.41 % | 91.49 % | 1 627 | 7 483 |
In the case of manually validated strains, the best clustering quality values (F-measures) obtained reduced significantly when distant sequences and sequences of indistinguishable species were not removed (see the results of clustering on datasets CM1, CM2, M1 and M2). For ITS, the optimal threshold produced for M1 was 99.21 %. When the dataset was clean (CM1), a lower threshold of 98.11 % was observed. For LSU, the optimal threshold of 99.51 % was produced for both dataset M2 and CM2.
When combining the two loci, the best clustering quality of M3 was better than the best clustering quality of each dataset M1 and M2 (84.47 % vs. 82.73 % and 80.94 %). This is because the number of the strains of species being merged using this combination was lower than the number of the strains of species being discriminated. Thus, although the barcode gap PCI of the combination of the two loci was lower than the barcode gap PCI of ITS alone as seen in the previous section, the two loci can be used together with the advantage of an additional discriminatory capability.
Although the similarity values within yeast species were variable as seen in the previous section and previous study (Nilsson et al. 2008), the high values of the best F-measures obtained by clustering different clean datasets indicate that except for indistinguishable species, ITS and LSU work well in differentiating yeast species. The CBS type datasets (CT1 and CT2) had the highest best F-measures of 92.33 % for ITS and 94.31 % for LSU, showing that the ex-type strains of current yeast species were clearly separated based on ITS and LSU barcodes.
When comparing the clustering results of all CBS yeast strains (CC1 and CC2) with the ones obtained from the manually validated datasets (CM1 and CM2), the best quality values of clustering of these two datasets were about the same (0.23 % changes for ITS and 0.9 % for LSU) although the numbers of strains were significantly higher (∼23.52 % for ITS and ∼31.42 % for LSU).
The best quality value obtained by clustering ascomycetous yeasts alone (90.76 %, CCA1) was higher than the best quality value obtained by clustering basidiomycetous yeasts alone (89.31 %, CCB1) using CBS ITS barcodes. While using CBS LSU barcodes, the other way around was observed (90.59 %, CCA2 vs. 92.32 %, CCB2). This shows that ITS worked better than LSU in species discrimination in Ascomycota, while in Basidiomycota, LSU outperformed ITS. The taxonomic threshold to discriminate yeast species in Ascomycota was lower than in Basidiomycota (98.31 % vs. 98.61 %) using ITS barcodes. When using LSU barcodes, they were slightly different (99.41 % vs. 99.51 %).
When including sequences from GenBank, the size of the datasets increased by ∼36.45 % for ITS and ∼36.7 % for LSU. The number of species raised by ∼11.19 % for ITS and ∼9 % for LSU. The best clustering quality values obtained were 88 % for ITS and 91.49 % for LSU.
For LSU, the predicted taxonomic thresholds to discriminate yeast species were consistent in different barcode datasets, which were 99.61 % for the type dataset, 99.51 % for the CBS datasets, and 99.41 % for CBS+GB dataset. These values are in agreement with previous studies (Kurtzman and Robnett, 1998, Fell et al., 2000, Scorzetti et al., 2002) stating that strains of yeast species showed less than 1 % dissimilarity in either ITS or LSU regions. For ITS, the type dataset produced a highest taxonomic threshold of 99.21 % which is also in the same line with the previous studies of Kurtzman and others. When including more strains and species, lower taxonomic thresholds were observed. The predicted taxonomic thresholds were 98.11 % for the manually validated dataset, 98.41 % for the CBS dataset, and 99.31 % for CBS+GB dataset. However, with the threshold of 99.11 %, the qualities of clustering produced were also high. They were 90.4 % for the manually validated dataset, 90.47 % for the CBS dataset and 86.23 % for the CBS+GB dataset.
Which UNITE cut-off thresholds should be used for yeast identification?
As mentioned earlier, unlike our approach, the reference sequences of UNITE were chosen automatically or manually from the species hypotheses (SHs), obtained by clustering the database with different thresholds 97 %, 97.5 %, 98 %, 98.5 %, 99 % and 99.5 %. It is up to the researcher using the UNITE database to decide which cut-off values are used for identification in ecological studies (Kõljalg et al. 2013). This section studies the best UNITE cut-off value to be used for yeast identification. We clustered the different ITS datasets with the given thresholds. The obtained F-measures are given in Table 3. It can be seen from Fig. 8A and Table 3 that these F-measures varied up to 7 % for the CBS type dataset (CT1) and 4 % for the other datasets. The best F-measures obtained from the CBS clean datasets (CT1, CM1, CC1) were more than 2 % higher than the one obtained when including GenBank sequences (CCN1). Among the given thresholds, the threshold of 98.5 % gave the best qualities for the clustering of the three largest datasets CM1, CC1 and CCN1. The obtained F-measures were not different from the F-measures obtained when clustering the datasets with the optimal threshold of 98.41 % predicted for the CBS ITS dataset (CC1).
Table 3.
The F-measures obtained by clustering different ITS datasets with the thresholds from 97 % to 100 % of the UNITE species hypotheses. The F-measure obtained by clustering the datasets with the optimal threshold of 98.41 % predicted for the CBS dataset are also given.
| Threshold | F-measure of M1 | F-measure of CT1 | F-measure of CM1 | F-measure of CC1 | F-measure of CCN1 |
|---|---|---|---|---|---|
| 97.0 % | 77.83 % | 84.44 % | 87.04 % | 86.92 % | 84.43 % |
| 97.5 % | 79.96 % | 86.34 % | 88.92 % | 88.65 % | 87.02 % |
| 98.0 % | 80.87 % | 87.75 % | 89.69 % | 89.48 % | 87.58 % |
| 98.5 % | 81.50 % | 89.74 % | 90.67 % | 90.21 % | 87.79 % |
| 99.0 % | 81.48 % | 91.47 % | 90.06 % | 89.80 % | 86.60 % |
| 99.5 % | 82.22 % | 91.40 % | 88.49 % | 88.60 % | 84.15 % |
| 98.41 % | 81.50 % | 89.74 % | 90.67 % | 90.21 % | 87.84 % |
Taxonomic thresholds for yeast genera identification
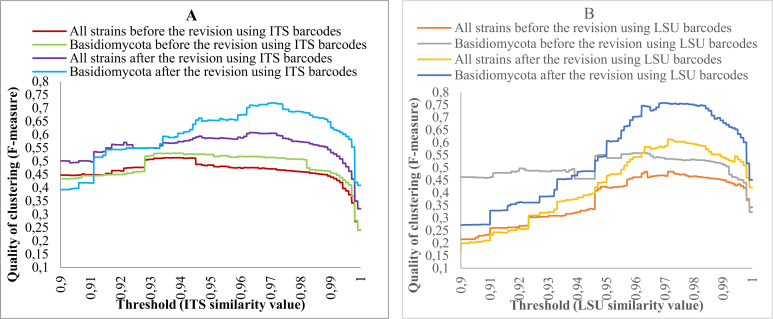
The same method to predict a taxonomic threshold for yeast identification was applied to study current classification of yeast genera based on ITS and LSU barcodes. Recent studies (Liu et al., 2015, Wang et al., 2015a, Wang et al., 2015b, Wang et al., 2015c) have revised the generic taxonomy of basidiomycetous yeasts that until recently was not concordant with the molecular phylogeny particularly as it relates to genus circumscriptions. To evaluate the revision of Basidiomycota and the current classification of yeasts at the genus level, reference sequences of CBS strains (datasets C1 and C2) were clustered to predict taxonomic thresholds that give the best match to the generic taxonomy of yeasts before and after the revision. The clustering results are given in Fig. 9.
Fig. 9.
Clustering qualities compared with the taxonomic classification at the genus level, obtained by clustering different ITS (A) and LSU (B) barcode datasets with thresholds ranging from 0.9 to 1 using an incremental step of 0.0001.
The taxonomic thresholds predicted to discriminate yeast genera in Basidiomycota before the revision in 2015 were 93.51 % with a quality of 53 % for ITS and 96.21 % with a quality of 56 % for LSU. After the revision, they were 97.01 % with a quality of 72 % for ITS and 96.91 % with a quality of 76 % for LSU. The significant increases of the best qualities of clustering strains in Basidiomycota, that rose from 53 % to 72 % for ITS and from 56 % to 76 % for LSU, show a significant improvement in the generic taxonomy of Basidiomycota, after the recent revisions. It must still be noted that in several cases the generic boundaries still may be too broadly defined. Future studies need to demonstrate this and the quality of the clustering could further increase.
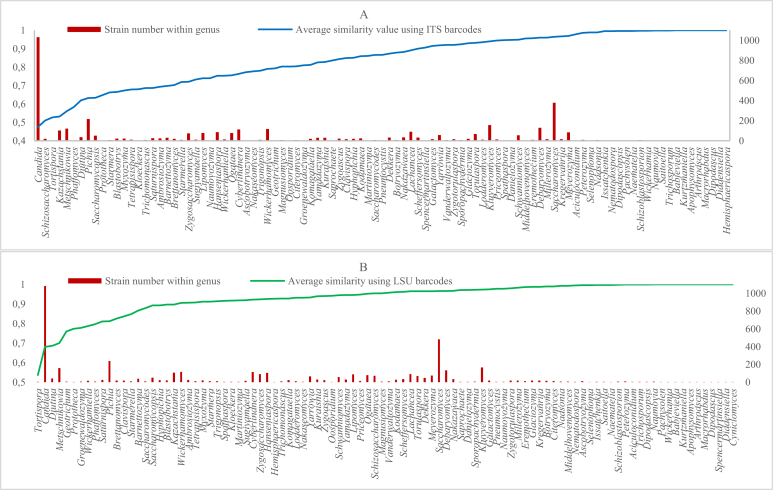
When including the strains of Ascomycota to the analysis, the taxonomic threshold to discriminate yeast genera before the revision were 93.81 % with a quality of 51 % for ITS and 97.21 % with a quality of 48 % for LSU. After the revision of Basidiomycota, they were 96.31 % with a quality of 61 % for ITS and 97.11 % with a quality of 63 % for LSU. These low clustering qualities indicate that there is a strong necessity to revise the generic taxonomy of ascomycetous yeasts as well. It will be challenging because of the complexity in reclassification of the largest yeast genus Candida (Daniel et al. 2014). Fig. 10 shows the ascomycetous yeasts with the associated average similarity values within genera ordered increasingly using ITS and LSU barcodes, respectively. The genera with a low average similarity value and a large number are likely to profit most from revision.
Fig. 10.
Ascomycetous genera with the associated average similarity values increasingly ordered by using ITS (A) and LSU (B) barcodes, respectively. The number of the strains within genera displayed in the secondary axis.
Detected versus described taxa
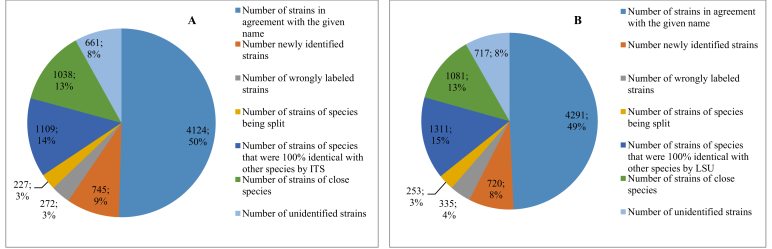
Based on the taxonomic thresholds predicted to identify yeast species, all CBS yeast strains were automatically validated using ITS and/or LSU barcodes (datasets C1 and C2). Reference sequences of the strains were clustered with the taxonomic thresholds 98.41 % for ITS (CM1) and 99.51 % for LSU (CM2). As seen above, 97 % and 94 % pairs of manually validated strains of the same species were more than 95 % and 97 % similar using ITS and LSU barcodes, respectively. To decide if a strain or sequence was wrongly labelled or not, we used the lower bound threshold of 95 % for ITS and of 97 % for LSU. Fig. 11 shows the statistics obtained after automatic clustering of all CBS yeast strains using the ITS and LSU barcode sequences.
Fig. 11.
Statistics obtained by clustering CBS ITS (A) and LSU (B) yeast barcodes with the threshold of 98.41 % and 99.51 %, respectively.
Based on the grouping of the sequences, 4 124 (50 %) strains of 1 366 species and 4 291 (49 %) strains of 1 434 species were in agreement with the current species name using ITS and LSU barcodes, respectively; 272 (3 %) strains by ITS and 335 (4 %) strains by LSU were found as wrongly labelled and have been suggested to another existing species name; 227 (3 %) strains by ITS and 253 (3 %) strains by LSU were of the species that must be split in the future as demonstrated by both loci, indicating that even among the identified strains, there was a considerable portion of potentially undescribed taxa; 1 038 (13 %) strains by ITS and 1 311 (15 %) strains by LSU were belonging to species synonyms and indistinguishable species; Species names have been suggested for 745 (9 %) strains by ITS and 720 (8 %) strains by LSU that had no species name given before the clustering; 661 (8 %) strains by ITS and 717 (8 %) strains by LSU remained unidentifiable and are potentially new species. It should be noted that one could increase the lower bound thresholds for yeast species identification. In this case, the number of wrongly labelled strains would increase and the number of closely related species would decrease.
ITS versus LSU
The ITS and LSU loci were not always in agreement regarding species assignment. Of the 99 and 155 species that were indistinguishable on the basis of ITS and LSU respectively, 46 species (∼3 %) were indistinguishable by both loci. Of the 120 and 104 species that were being split by ITS and LSU respectively, 48 species were in common. Of 272 and 335 strains that were wrongly labelled by ITS and LSU respectively, 102 strains were wrongly labelled by both loci. Among them, 71 strains were reassigned with the same name, and the other 31 strains were reassigned to a closely related species name. Finally, of 745 and 720 strains that have been newly identified by ITS and LSU respectively, 551 strains were identified by both whereas 345 strains were given the same species name. The remaining 206 strains were given a closely related species name.
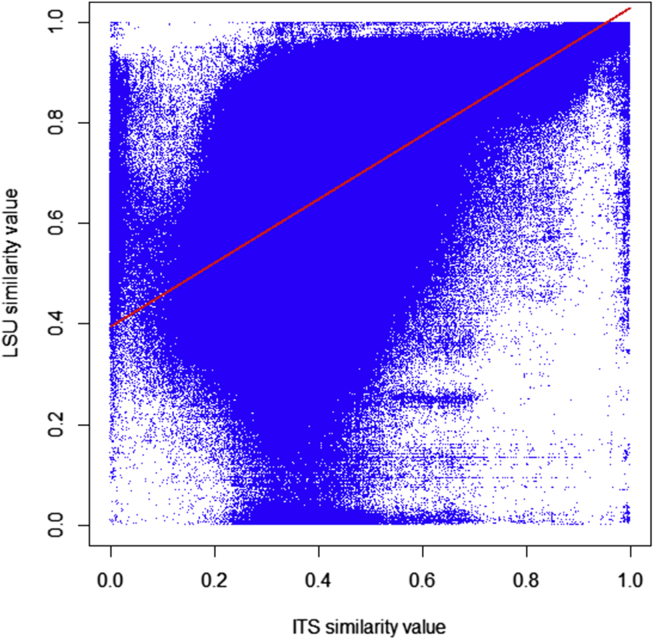
Correlation between ITS and LSU
To evaluate the level of correlation between the groupings obtained by both loci, all manually validated CBS strains (M3 dataset) belonging to 1 375 yeast species having both ITS and LSU sequences were aligned in a pairwise manner to produce two similarity matrices, one for each locus. Fig. 12 shows the scatter plot of all pairwise comparisons based on ITS and LSU similarity values. A Mantel test (i.e. a Pearson Correlation moment between two distance matrixes calculated on the basis of 999 permutations) between the two matrices was calculated and a correlation of 0.47 between ITS and LSU was observed. The red line in Fig. 12 is the best-fit linear regression between the two similarity values. The goodness of fit r2 of the linear regression model was measured as 0.3. Although this value was low indicating the independence of the two loci overall, a strong correlation of 0.84 between them was observed when ITS similarity value was greater than 60 % and LSU similarity value greater was than 89 % which happened only in 5 % of all cases.
Fig. 12.
2D scatter plots of ITS similarity values versus LSU similarity values. The goodness of fit of the linear regression model r2 is 0.3.
There was a small percentage of 0.006 % of pairwise compared strains of which the absolute difference of the associated ITS and LSU similarity values were more than 90 %. This can be caused by the following: a) some sequences were wrongly declared as ITS or LSU; b) some sequences were wrongly associated with the strain, and/or; c) since ITS and LSU have multiple copies of different lengths and variable sequences, the amplified/stored copy may not be the dominant one or there could be several versions that are very dissimilar from each other (Simon and Weiss, 2008, Kiss, 2012). The protocols that were implemented to generate all the barcoding data, together with the high level of curation (since M3 dataset was used to generate Fig. 12) have reduced the likelihood of a) and b), but they cannot be excluded. A number of randomly ran controls have shown that the cause c) was present. Fig. 12 also shows that ITS was more variable than LSU which has already been observed in previous sections of the current paper and by others before us (Nilsson et al. 2006).
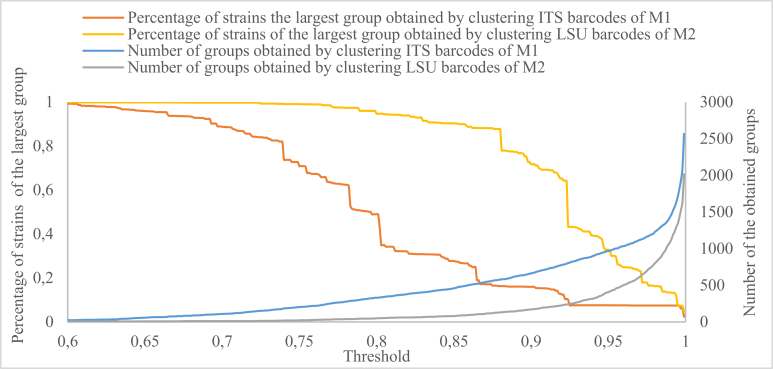
Distribution of yeast strains using ITS and LSU barcodes
To study the distribution of yeast strains based on ITS and LSU barcodes, we plotted the percentage of strains of the largest group and the number of groups obtained by clustering M1 and M2 with increasing thresholds in Fig. 13. The largest group contains more than ∼50 % of the strains even when the similarity threshold is equal to ∼79 % for ITS and ∼92 % for LSU, showing that yeast strains tended to be grouped together as one population at low ITS and LSU similarity values. The numbers of the groups obtained by clustering follow a smooth exponential curve, indicating a rather sudden increased of the number of taxa at higher ITS and LSU similarity values. Fig. 13 also shows that ITS was more variable than LSU as the number of the groups produced by ITS was always higher than the number of the groups produced by LSU when the threshold was increasing.
Fig. 13.
The number of the obtained groups (displayed in the secondary axis) and the percentage of strains of the largest group obtained by clustering M1 and M2 with thresholds increased from 0.6 to 1.
Data submission and release
The similarity values of each CBS strain to the associated ex-type and central representative strain together with the predicted taxon names using ITS and LSU barcodes were imported to highlight all the problematic strains and sequences in the CBS collection. Sequences of the manually validated datasets M1 and M2 that were less than 95 % for ITS and 97 % for LSU similar to the associated representative strain were removed. The short sequences that were not qualified for the submission portal of GenBank, were also removed. The remaining 4 456 ITS and 4 213 LSU sequences of 4 730 (51 %) CBS yeast strains of 1 351 (80 %) accepted yeast species have been released to the CBS website (www.cbs.knaw.nl/collections/) and GenBank (www.ncbi.nlm.nih.gov) as reference sequences for yeast identification. The GB accession numbers of these sequences are given in the files Supplementary_ITS_GBAccessionNumbers.txt and Supplementary_LSU_GBAccessionNumbers.txt.
Conclusions
With the almost complete dataset of yeast barcodes consisting of culturable yeasts, the taxonomic threshold predicted to discriminate yeast species was 99.51 % in the LSU region. This result was in agreement with the study of Kurtzman and others (Kurtzman and Robnett, 1998, Fell et al., 2000, Scorzetti et al., 2002) demonstrating that strains of yeast species showed less than 1 % dissimilarity in either ITS or LSU regions. For ITS, the taxonomic threshold predicted to discriminate yeast species was 99.21 % using ex-type strains only. When including all CBS yeast strains, a lower threshold of 98.41 % was observed. The taxonomic threshold to discriminate yeast species in Ascomycota was lower than in Basidiomycota (98.31 % vs. 98.61 %) using ITS barcodes. When using LSU barcodes, they were slightly different (99.41 % vs. 99.51 %). Except for 6 % and 9.5 % of yeast species sharing the same ITS and LSU sequences respectively, the high quality of clustering produced in different datasets shows that current yeast species can be clearly separated using ITS and LSU barcodes. Our analyses also showed that recent studies (Liu et al., 2015, Wang et al., 2015a, Wang et al., 2015b, Wang et al., 2015c) have made a significant improvement to the generic taxonomy of basidiomycetous yeasts. The taxonomic thresholds predicted to distinguish current yeast genera were 96.31 % for ITS and 97.11 % for LSU. However, there is a strong need for a reclassification of Ascomycota as the best quality of clustering at genus level was low using ITS and LSU barcodes. The predicted taxonomic thresholds can be used to flag potentially new yeast species and genera, and help the researchers of UNITE to decide which cut-off values are used for yeast species identification. They can also be used to estimate the diversity of yeast taxa from environmental samples that was demonstrated to be far outsizing the diversity of yeast taxa brought into culture to date using metagenomics approaches (Hawksworth, 2001, Handelsman, 2004, Hibbett, 2016).
All the strains of the CBS yeast collection have been updated with a predicted taxon name and the similarity values to the associated ex-type and central representative strains. The information helps to highlight the problematic strains and sequences for the curators, and therefore, speed up the validation process and improve the overall quality of the culture collection. The barcodes of 4 730 publicly available and manually validated CBS strains of 1 351 yeast species are released to GenBank (www.ncbi.nlm.nih.gov) to improve yeast barcodes and taxonomy at public databases. They are also publicly available from the CBS website (www.cbs.knaw.nl). Our next challenge is to apply the current method to validate the barcode data of the entire CBS fungal collection, where the amount of data is ten times bigger. Together with the metadata associated with the CBS strains ranging from the origin, media and growth condition, morphology, sexuality, physiology, and bibliography, this would accomplish our ultimate goal, that is to publish a reference dataset of ITS and LSU for fungal and yeast identification and classification.
As the CBS yeast collection contains almost all currently recognized species, it is a valuable dataset for the mycological community at large. The barcode data have already been used to study the taxonomic distribution of thermotolerant strains and species related to climate change and potential emerging pathogens (Robert et al. 2015). Beyond the expected taxonomic and identification applications, barcoding data combined with other data such as antibiotic resistance, ability to produce (for example) a number of metabolites or products of biotechnological or industrial interest, constitute a useful resource for researchers working with yeasts.
Acknowledgements
This study was financially supported by the “Fonds Economische Structuurversterking (FES)”, Dutch Ministry of Education, Culture and Science grant BEK/BPR-2009/137964-U, “Making the Tree of Life Work”.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.simyco.2016.11.007.
Contributor Information
D. Vu, Email: d.vu@cbs.knaw.nl.
V. Robert, Email: v.robert@cbs.knaw.nl.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Supplementary Fig. S1. The intraspecific similarity within CBS yeast species; (A) the similarity value of each CBS yeast strain to the associated representative strain; (B) and the interspecific similarity between CBS yeast species (C) using ITS barcodes.
Supplementary Fig. S2. The intraspecific similarity within CBS yeast species; (A); the similarity value of each CBS yeast strain to the associated representative strain; (B) and the interspecific similarity between CBS yeast species (C) using LSU barcodes.
Supplementary Fig. S3. The intraspecific similarity within CBS+GB yeast species; (A) the similarity value of each CBS+GB sequence to the associated representative sequence; (B) and the interspecific similarity between CBS+GB yeast species (C) using ITS barcodes.
Supplementary Fig. S4. The intraspecific similarity within CBS+GB yeast species; (A) the similarity value of each CBS+GB sequence to the associated representative sequence; (B) and the interspecific similarity between CBS+GB yeast species (C) using LSU barcodes.
References
- Altschul S.F., Madden T.L., Schäffer A.A. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonielli L., Robert V., Corte L. Centrality of objects in a multidimensional space and its effects on distance-based biological classifications. The Open Applied Informatics Journal. 2011;5:11–19. [Google Scholar]
- Bidartondo M.I. Preserving accuracy in Genbank. Science. 2008;319:1616. doi: 10.1126/science.319.5870.1616a. [DOI] [PubMed] [Google Scholar]
- Daniel H.M., Lachance M.A., Kurtzman C.P. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek. 2014;106:67–84. doi: 10.1007/s10482-014-0170-z. [DOI] [PubMed] [Google Scholar]
- de Queiroz K. Species concepts and species delimitation. Systematic Biology. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Boekhout T., Fonseca A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Mycological Research. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D.L. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycological Research. 2001;105:1422–1432. [Google Scholar]
- Hebert P.H., Cywinska A., Ball D.L. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett D. The invisible dimension of fungal diversity. Science. 2016;351(6278):1150–1151. doi: 10.1126/science.aae0380. [DOI] [PubMed] [Google Scholar]
- Hollingsworth P.M., Forresta L.L., Spouge J.L. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopcroft J., Tarjan R. Efficient algorithms for graph manipulation. Communications of the ACM. 1973;16:372–378. [Google Scholar]
- Kiss L. Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1811. doi: 10.1073/pnas.1207143109. author reply E1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U., Nilsson R.H., Abarenkov K. Towards a unified paradigm for sequence-based identification for fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P. Use of gene sequence analyses and genome comparisons for yeast systematics. International Journal of Systematic and Evolutionary Microbiology. 2014;64:325–332. doi: 10.1099/ijs.0.054197-0. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Fell J.W., Boekhout T. 5th edn. Elsevier Science; 2011. The yeasts: a taxonomic study. [Google Scholar]
- Kurtzman C.P., Robnett C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- Liu X.-Z., Wang Q.-M., Göker M. Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology. 2015;81:85–147. doi: 10.1016/j.simyco.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J, Barrrie FR, Buck WR, et al. (eds) (2012). International Code of Nomenclature for algae, fungi and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. [Regnum Vegetabile No. 154.]. Koeltz Scientific Books, Königstein.
- Nilsson R.H., Kristiansson E., Ryberg M. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One. 2006;1:59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R.H., Kristiansson E., Ryberg M. Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evolutionary Bioinformatics. 2008;4:193–201. doi: 10.4137/ebo.s653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccanaro P., Casbon J.A., Saqi M.A. Spectral clustering of proteins sequences. Nucleic Acids Research. 2006;34:1571. doi: 10.1093/nar/gkj515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Kurtzman C.P. Ribosomal RNA sequences divergence among sibling species of yeasts. Systematic and Applied Microbiology. 1991;14:124–129. [Google Scholar]
- Robert V., Cardinali G., Casadevall A. Distribution and impact of yeast thermal tolerance permissive for mammalian infection. BMC Biology. 2015;13:1–14. doi: 10.1186/s12915-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V., Szöke S., Jabas J. Biolomics software: biological data management, identification, classification and statistic. The Open Applied Informatics Journal. 2011;5:87–98. [Google Scholar]
- Schoch C., Seifert K.A., Huhndorf S. The nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzetti G., Fell J.W., Fonseca A., Statzell-Tallman A. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Simon U.K., Weiss M. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Molecular Biology and Evolution. 2008;25:2251–2254. doi: 10.1093/molbev/msn188. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today. 2006;8:6–9. [Google Scholar]
- Stielow J.B., Lévesque C.A., Seifert K.A. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 2015;35:242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle M.Y., Hebert P.D. Barcode of life. Scientific American. 2008;299:82–86. [PubMed] [Google Scholar]
- Sugita T., Nakajima M., Ikeda R. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. Journal of Clinical Microbiology. 2002;40:1826–1830. doi: 10.1128/JCM.40.5.1826-1830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Eberhardt U., Szöke S. A laboratory information management system for DNA barcoding workflows. Integrative Biology. 2012;4:744–755. doi: 10.1039/c2ib00146b. [DOI] [PubMed] [Google Scholar]
- Vu D., Szoke S., Wiwie C. Massive fungal biodiversity data re-annotation with multi-level clustering. Scientific Reports. 2014;4:6837. doi: 10.1038/srep06837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.-M., Begerow D., Groenewald M. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology. 2015;81:55–83. doi: 10.1016/j.simyco.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.-M., Groenewald M., Takashima M. Phylogeny of yeasts and related filamentous fungi within Pucciniomycotina determined from multigene sequence analyses. Studies in Mycology. 2015;81:27–53. doi: 10.1016/j.simyco.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.-M., Yurkov A.M., Göker M. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Studies in Mycology. 2015;81:149–189. doi: 10.1016/j.simyco.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.Q., Meier R. Columbia University Press; 2000. Species concepts and phylogenetic theory: a debate. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.