Abstract
Splicing factor 1 (SF1) recognizes 3′ splice sites of the major class of introns as a ternary complex with U2AF65 and U2AF35 splicing factors. A conserved SPSP motif in a coiled-coil domain of SF1 is highly phosphorylated in proliferating human cells and is required for cell proliferation. The UHM kinase 1 (UHMK1), also called KIS, double-phosphorylates both serines of this SF1 motif. Here, we use isothermal titration calorimetry to demonstrate that UHMK1 phosphorylation of the SF1 SPSP motif slightly enhances specific binding of phospho-SF1 to its cognate U2AF65 protein partner. Conversely, quantitative fluorescence anisotropy RNA binding assays and isothermal titration calorimetry experiments establish that double-SPSP phosphorylation reduces phospho-SF1 and phospho-SF1–U2AF65 binding affinities for either optimal or suboptimal splice-site RNAs. Domain-substitution and mutagenesis experiments further demonstrate that arginines surrounding the phosphorylated SF1 loop are required for cooperative 3′ splice site recognition by the SF1-U2AF65 complex (where cooperativity is defined as a nonadditive increase in RNA binding by the protein complex relative to the individual proteins). In the context of local, intracellular concentrations, the subtle effects of SF1 phosphorylation on its associations with U2AF65 and splice-site RNAs are likely to influence pre-mRNA splicing. However, considering roles for SF1 in pre-mRNA retention and transcriptional repression, as well as in splicing, future comprehensive investigations are needed to fully explain the requirement for SF1 SPSP phosphorylation in proliferating human cells.
Introduction
Expression of splicing factor 1 (SF1, also called ZFM1) is required for viability in yeast, flies, nematodes, and human cell lines (1, 2, 3, 4), and the SF1 gene locus is tightly linked to multiple endocrine neoplasia type I (5). SF1 associates with U2 small nuclear ribonucleoprotein (snRNP) auxiliary factor subunits (U2AF65 and U2AF35, also called U2AF2 and U2AF1) (1, 6, 7). Like SF1, the U2AF65 and U2AF35 splicing factors are essential for viability in multicellular eukaryotes (e.g., (8, 9).), and the U2AF35 subunit is frequently mutated in myelodysplastic syndromes (10, 11, 12). The ternary complex comprising the SF1 and U2AF subunits recognizes consensus pre-mRNA signals in the early stages of pre-mRNA splicing (reviewed in (13)) (Fig. 1 A). Specifically, a K-homology-quaking motif (KH-QUA2) of SF1, RNA recognition motifs (RRMs) of U2AF65, and zinc knuckles of U2AF35 recognize the branch-point sequence (BPS), polypyrimidine (Py) tract, and intron-exon junction of the major class of 3′ splice sites (14, 15, 16, 17, 18) (Fig. 1 B). The yeast homolog of SF1 also has been implicated in cross talk with the U1 snRNP (1, 19, 20, 21). Subsequently, ATP-dependent association of the U2 snRNP with the pre-mRNA displaces the SF1 subunit; instead, an SF3b155 subunit (also called SF3B1) of the U2 snRNP interacts with the U2AF65 subunit of the U2AF heterodimer (22, 23, 24, 25).
Figure 1.
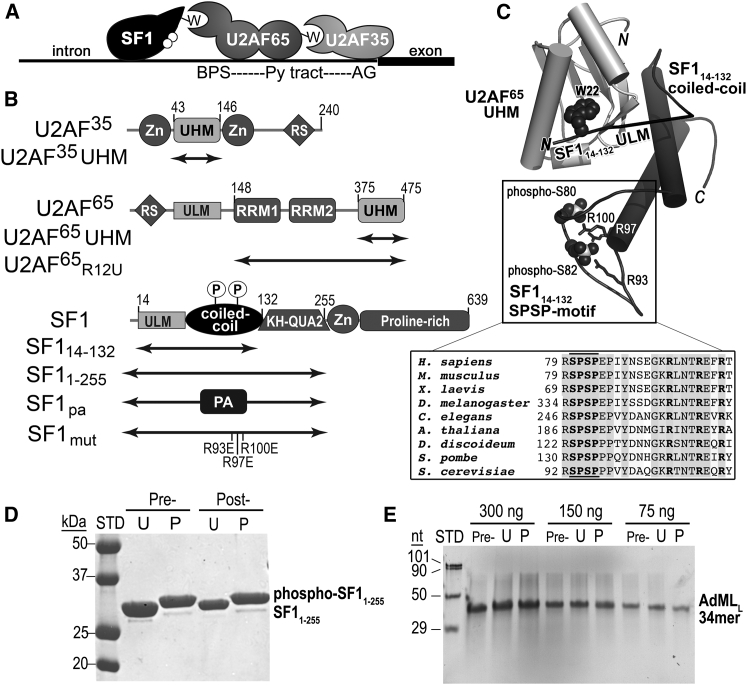
SF1 structure and function in pre-mRNA splicing. (A) Schematic diagram of the early-stage SF1-U2AF65-U2AF35 splicing-factor complex recognizing the 3′ splice site. ULMs are represented by “W.” White circles represent UHMK1-phosphorylation sites. BPS, branch point sequence; Py, polypyrimidine. (B) Domains of SF1, U2AF65, and U2AF35 splicing factors. The boundaries of constructs used in this study are indicated by double-headed arrows. Zn, CCCH zinc knuckle; UHM, U2AF homology motif; ULM, U2AF ligand motif; RRM, RNA recognition motif, RS, arginine-serine repeat; KH-QUA2, K homology with quaking-2 motif; PA, protein A; circled P, UHMK1-phosphorylation sites. (C) Structure of the phosphorylated SF114–132-U2AF65 UHM complex (PDB: 4FXW). A line representing residues that are absent from the electron density connects the C-terminus of the ULM to the coiled-coil motifs. The tryptophan of the ULM (W22) and phosphorylated serines are shown as spheres and arginine residues that are mutated in this study are shown in stick representation. The inset shows a sequence alignment of the UHMK1-phosphorylated SPSP motif (bold (between lines)). (D and E) Samples of (D) phospho-SF11–255 and (E) AdMLL RNA under experimental conditions. Samples either before (Pre-) or after (Post-) incubating at 23°C for 2 h in the presence of a 10-fold excess of SF1 (U) or phospho-SF1 (P) were analyzed by denaturing gel electrophoresis and stained using (D) Coomassie blue or (E) SYBR Gold, respectively. The phosphorylated form of SF11–255 runs higher by SDS-PAGE. STD, size markers; nt; nucleotides. Conditions were identical to those of fluorescence assays with the exception that the RNase inhibitor Superase-In (Ambion, Life Technologies) was omitted here to fully rule out RNase contamination.
The importance of the U2AF subunits for spliceosome assembly in mammals has been firmly established by biochemical reconstitution experiments (e.g., (15, 16), among others). In contrast, SF1 selectively regulates splicing of a subset of introns (26, 27, 28, 29). One possible explanation is a transient role and rapid recycling of SF1 (30, 31), such that trace amounts are sufficient to support splicing of traditional pre-mRNA substrates. Yet a direct relationship between the selective role for SF1 in pre-mRNA splicing and its requirement for cell viability is uncertain. Indeed, SF1 has been implicated in other major steps of gene expression, including nuclear pre-mRNA retention (2) and transcriptional repression (32, 33). The association of SF1 and U2AF in extra-spliceosomal complexes (6) and the involvement of U2AF65 in the nuclear pre-mRNA retention activity of SF1 (34, 35, 36) suggest that the SF1-U2AF65 complex contributes to these multiple SF1 functions.
The protein-protein interaction domains mediating the SF1-U2AF65-U2AF35 complex are called “U2AF homology motifs” (UHMs) based on their original discovery in the U2AF65 and U2AF35 prototypes (37, 38). The UHM shares the core topology of the RRM fold, yet the ribonucleoprotein consensus motifs (RNPs) diverge from typical RRM residues and have lost the ability to bind RNA (reviewed in (39)). Instead, an acidic α-helix and hydrophobic pocket of the UHM recognize basic residues and a tryptophan residue of a peptide ligand, called a “U2AF ligand motif” (ULM). Sequence searches extrapolating from the U2AF65 and U2AF35 UHMs identified putative UHMs in several nuclear proteins, many of which have now been confirmed to bind ULMs (e.g., (40, 41)). The ULMs share similar short, linear epitopes of positively charged and tryptophan amino acids, yet only a handful of ULM-containing proteins have been confirmed to date, and their interaction partners are remarkably specific. For example, the SF1 ULM targets the U2AF65 UHM, which also sequentially associates with an SF3b155 ULM, whereas the U2AF65 ULM mediates the heterodimer with the U2AF35 UHM. As such, the means for limiting off-target cross talk among UHMs and ULMs presents a molecular conundrum.
Extensions of the core ULM sequence offer one means to increase the interface, and hence specificity, of UHM-ULM complexes. For the U2AF35 UHM-U2AF65 ULM prototype, a proline-rich region at the C-terminus of the U2AF65 ULM sandwiches an otherwise exposed tryptophan side chain of the U2AF35 UHM (38). For SF1, a coiled-coil domain at the C-terminus of the SF1 ULM extends its interface with the U2AF65 UHM (42, 43) (Fig. 1 C). Despite extensive hydrophobic and electrostatic contacts, the SF1 coiled coil makes subtle contributions to the affinity of the SF1-U2AF65 complex (43). However, whether this SF1 coiled coil could discriminate against the SF1 ULM binding off-target UHMs remains unknown.
Phosphorylation within or adjacent to ULMs is a second mechanism for regulating the specificity of UHM interactions. In perhaps the earliest example of phosphorylation-sensitive ULM-UHM interactions, phosphorylation of a serine in the SF1 ULM by cGMP-dependent protein kinase I was shown to reduce SF1 association with U2AF65 (44). In a second, more recent example, phosphorylation near a ULM of ataxin-1 (ATX1) was found to promote its binding to the 14-3-3 protein, which in turn out-competes ATX1 association with the U2AF65 UHM (45).
SF1 also is phosphorylated on a conserved SPSP motif within its coiled-coil domain (46) (Fig. 1 C, inset). Both serines in the SPSP motif (S80 and S82) of endogenous SF1 isolated from proliferating cells are highly phosphorylated. Mutation of both serines in the SF1 SPSP motif to phosphorylation-resistant alanines significantly reduces NIH 3T3 cell proliferation (42) and leads to massive splicing changes in fission yeast strains (47). The SF1 SPSP motif is likely to be a substrate for several kinases. A UHM-containing kinase (UHMK1, also called KIS) highly phosphorylates both S80 and S82 of this SF1 motif (46). Yet, the SPSP-containing region of SF1 remains phosphorylated in more than half of the detected SF1 proteins in UHMK1-knockout mice (48), which suggests that other kinases regulate this SF1 motif. For example, SRPK2 detectably phosphorylates the second serine (S82) of the SF1 SPSP motif in vitro, and could act to phosphorylate SF1 in vivo (47). Accordingly, UHMK1 is differentially expressed in the brain (48) and may serve specific roles in the nervous system.
The functions of SF1 SPSP motif phosphorylation remain largely mysterious. Structurally, phosphorylation of the SF1 SPSP motif induces local folding of an arginine “claw” (R93, R97, and R100) surrounding the phosphorylated serines (42, 43). One hypothesis is that this structural change augments the SF1 ULM by promoting formation of the coiled-coil interface of SF1 with the U2AF65 UHM. Accordingly, the UHMK1-phosphorylated SPSP motif subtly promotes SF1 binding to U2AF65 in pull-down assays (46, 49). Although little phosphorylation-dependent increase in SF1 binding U2AF65 had been observed by isothermal titration calorimetry (ITC) (43), we were concerned that use of a phosphate-containing buffer could have reduced a potential response. Moreover, despite these studies of phosphorylated SF1 interactions with U2AF65, whether SPSP phosphorylation of SF1 could discriminate against noncognate binding to other UHMs was unknown. For association with 3′ splice sites, electrophoretic mobility shift assays (EMSAs) of SF1 in the presence of SRPK2 suggested that S82 phosphorylation slightly increased SF1 binding to RNAs containing suboptimal BPSs (47). However, SRPK2 preferentially phosphorylates S82 rather than both serines of the SPSP motif, and the EMSA is not an equilibrium method due to separation of the ligand and macromolecule during electrophoresis.
Here, we use ITC and fluorescence anisotropy RNA binding assays to measure the influence of SPSP phosphorylation on SF1 interactions with UHM-containing proteins and 3′-splice-site RNA. Altogether, SPSP phosphorylation subtly enhances the specificity of SF1 for binding the cognate U2AF65 UHM and discriminates against off-target UHMs. By either ITC or fluorescence-based RNA binding assays, phosphorylation of the SPSP motif slightly decreases the affinity of SF1 for splice-site RNAs. Domain substitution and mutagenesis further implicate arginines surrounding the phosphorylated motif as sites of SF1-RNA interactions, which could explain the discontinuous spacing of the BPS and Py tract consensus signals at the 3′ splice site. The reproducible effects of SPSP phosphorylation on SF1 binding to UHM-containing proteins and splice-site RNAs are likely to contribute to SF1 function. However, alternative functions may contribute to the significantly reduced proliferation of human cells in the presence of the SF1 APAP mutation (42).
Materials and Methods
Protein expression and purification
All proteins are the human homologs expressed as glutathione-S-transferase (GST) fusions in Escherichia coli using either pGEX-6p or an in-house pGEX variant encoding a TEV protease site. The nomenclature and boundaries of the SF1 (NCBI RefSeq: NP_004621) and U2AF65 (NCBI RefSeq: NP_009210) protein expression constructs used in this study are given in Fig. 1 B. Other recombinant proteins include the UHM domains of U2AF35 (residues 43–146 of NCBI RefSeq: NP_006749), CAPERα (residues 411–524 of NCBI RefSeq: NP_004893), and SPF45 (residues 301–401 of NCBI RefSeq: NP_116294). After an initial glutathione affinity step, we cleaved the GST-tags during overnight dialysis using PreScission Protease for SF1 and U2AF65 proteins and TEV protease for the U2AF35 UHM, CAPERα UHM, and SPF45 UHM. The splicing-factor proteins were further purified by subtractive GST-affinity chromatography followed by anion- or cation-exchange chromatography. Purified SF1 proteins were phosphorylated by UHMK1 under conditions similar to those described in (42, 50) and then separated from UHMK1 by size-exclusion chromatography. The phosphorylation of the SF1 protein appears complete as judged by a characteristic shift by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (42, 46) (Fig. 1 D). The complexes of SF1 variants with U2AF65 were prepared by mixing 1:1 molar ratios of the separately purified subunits followed by size-exclusion chromatography. All proteins used for RNA binding experiments, as well as phosphorylated SF1 or phospho/unphospho-SF1-U2AF65 complexes, were finally purified by size-exclusion chromatography using a HiLoad 16/600 Superdex 75 pg (GE Healthcare, Little Chalfont, United Kingdom) in 100 mM NaCl, 25 mM HEPES buffer (pH 7.4), and 0.2 mM TCEP buffer. Phospho-SF11–255-U2AF65R12U, SF1pa-U2AF65R12U, and SF1mut-U2AF65R12U showed no detectable difference in complex formation from unmodified SF11–255-U2AF65R12U by size-exclusion chromatography at ∼5 μM concentrations.
ITC methods
Protein samples for ITC were dialyzed against a >200-fold greater volume of 25 mM HEPES (pH 7.4), 0.2 mM TCEP, and either the indicated concentration of NaCl for U2AF65 titration into SF1 (Table S1 in the Supporting Material), 50 mM NaCl for off-target UHM titrations (Table 1), or 100 mM NaCl for RNA titrations (Tables 2 and 3). A control titration of B3P3SUBOPT RNA into SF1mut-U2AF65R12U in 100 mM NaCl and 25 mM sodium phosphate (pH 7.4) was compared to the HEPES buffer (Table 3; Fig. S6). The synthetic B3P3SUBOPT RNA of sequence 5′-UGGUAAC UUUCUCUUUCUCUCUCCC (branch site underlined) was deprotected and desalted according to manufacturer instructions (Dharmacon, GE Healthcare). The RNA was resuspended in water and diluted >50-fold in dialysis buffer. Proteins or RNA at the concentrations indicated in Figs. 2, 4, 6, 7, 8, S1, S2, and S4–S6 were titrated at 30°C with 4 min of relaxation time between injections using a VP-ITC (GE Healthcare). Control titrations of SPF45 UHM, U2AF35 UHM, SF11–255-U2AF65R12U complex, or B3P3SUBOPT RNA into buffer (Fig. S1) were linear and showed negligible heats of dilution. The CAPERα UHM titration into buffer is shown in (40). The average heats of the U2AF35 or CAPERα UHM dilution (Fig. S1 B and (40)) were used to correct the relatively low-affinity isotherms of these UHMs titrated into SF114-132 or phospho-SF114-132 (Fig. 2, E–G). All other isotherms were corrected for heats of dilution using an average of four to five data points at the saturated plateau of the titration. The isotherms were fit using Origin v7.0 (MicroCal, Northampton, MA). The average values and standard deviations of the tables and figures result from at least two independent titration experiments. The phosphorylated state of SF1 was stable throughout the ITC experiments (Fig. 1 D) and RNA oligonucleotide lacked detectable ribonuclease activity (Fig. 1 E). The c-values of the isotherms (where c = KA × [M], M is the macromolecule in the sample cell, and the stoichiometry is assumed to be 1) are given in the figure legends. A c-value between ∼5 and 500 is considered reliable for KD and n (apparent stoichiometry) measurement. Enthalpy changes may still be determined from isotherms with higher c-values (51).
Table 1.
Thermodynamics of U2AF Homology Motifs binding SF1 or Phospho-SF1
| Interactiona | KD (nM) | ΔGb (kcal mol−1) | ΔH (kcal mol−1) | −TΔSc (kcal mol−1) | nd |
|---|---|---|---|---|---|
| U2AF65 UHM Titrated intoe | |||||
| SF114–132 | 43 ± 1 | −10.2 ± 0.1 | −13.5 ± 0.1 | 3.3 ± 0.1 | 1.0 ± 0.1 |
| Phospho-SF114−132 | 24 ± 1 | −10.6 ± 0.1 | −20.6 ± 0.1 | 10.0 ± 0.1 | 0.9 ± 0.1 |
| SPF45 UHM Titrated intof | |||||
| SF114−132 | 99 ± 2 | −9.7 ± 0.1 | −15.2 ± 0.2 | 5.5 ± 0.2 | 1.0 ± 0.1 |
| Phospho-SF114−132 | 170 ± 8 | −9.4 ± 0.1 | −14.5 ± 0.1 | 5.1 ± 0.1 | 1.1 ± 0.1 |
| U2AF35 UHM Titrated intof | |||||
| SF114–132 | 2430 ± 350 | −7.8 ± 0.1 | −30.9 ± 1.2 | 23.1 ± 1.3 | 1.0 ± 0.1 |
| Phospho-SF114−132 | 3540 ± 1140 | −7.6 ± 0.2 | −38.2 ± 2.3 | 30.6 ± 2.1 | 0.9 ± 0.1 |
| CAPERα UHM Titrated intof,g | |||||
| SF114−132h | 11,000 ± 700 | −6.9 ± 0.1 | −11.5 ± 0.2 | 4.6 ± 0.2 | 1.0 ± 0.1 |
| Phospho-SF114–132 | 57,000 ± 42,000 | −6.0 ± 0.5 | −18.3 ± 11.4 | 12.3 ± 11.8 | 1.0 ± 0.3 |
Values are expressed as the mean ± SD of at least two independent titrations.
ΔG values were calculated using the equation ΔG = −RT ln (KD−1) at T = 303 K.
−TΔS values were calculated using the equation −TΔS = ΔG − ΔH.
Apparent stoichiometry of UHM/SF1 proteins.
Proteins in 100 mM NaCl, 25 mM HEPES (pH 7.4), and 0.2 mM TCEP.
Proteins in 50 mM NaCl, 25 mM HEPES (pH 7.4), and 0.2 mM TCEP.
Values for phospho-SF1 binding CAPERα UHM are estimates due to the very low c-value of 1.
Values for SF114-132 binding CAPERα UHM are from Loerch et al. (40).
Table 2.
Thermodynamics of Splicing Factor Subunits Binding B3P3-Splice Site RNA
| B3P3 RNA Titrated intoa | KD (nM) | ΔGb (kcal mol−1) | ΔH (kcal mol−1) | -TΔSc (kcal mol−1) | nd |
|---|---|---|---|---|---|
| U2AF65R12U | 509 ± 4 | −8.8 ± 0.1 | −52.1 ± 0.1 | 43.3 ± 0.1 | 0.8 ± 0.1 |
| SF11–255 | 40 ± 1 | −10.3 ± 0.1 | −23.3 ± 0.1 | 13.0 ± 0.1 | 0.9 ± 0.1 |
| Phospho-SF11–255 | 78 ± 9 | −9.8 ± 0.1 | −20.9 ± 0.1 | 11.1 ± 0.1 | 1.0 ± 0.1 |
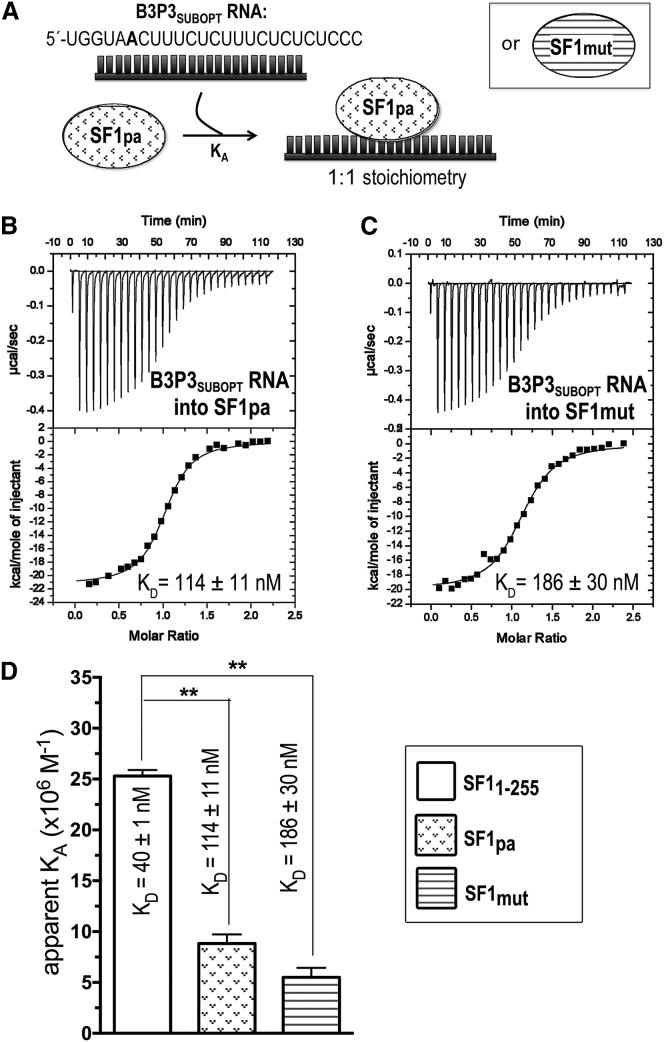
| SF1PA | 114 ± 11 | −9.7 ± 0.1 | −20.9 ± 0.1 | 11.2 ± 0.1 | 1.0 ± 0.1 |
| SF1mut | 186 ± 30 | −9.3 ± 0.1 | −20.0 ± 0.1 | 10.7 ± 0.1 | 1.0 ± 0.1 |
Values are expressed as the mean ± SD of at least two independent titrations.
ΔG values were calculated using the equation ΔG = −RT ln (KA), where T = 303 K.
−TΔS values were calculated using the equation −TΔS = ΔG − ΔH.
Fits are “ligand-in-cell” such that “n” is the apparent stoichiometry of protein/RNA.
Table 3.
Thermodynamics of Splicing Factor Complexes Binding B3P3-Splice Site RNA
| Interaction:a | KD (nM) | ΔGb (kcal mol−1) | ΔH (kcal mol−1) | −TΔSc (kcal mol−1) | nd |
|---|---|---|---|---|---|
| B3P3 RNA Titrated into Splicing-Factor Complexes | |||||
| SF11–255-U2AF65R12U, site 1 | 2 ± 1 | −12.1 ± 0.1 | −60.1 ± 0.1 | 47.9 ± 0.1 | 0.8 ± 0.1 |
| SF11–255-U2AF65R12U, site 2 | 500 ± 10 | −8.7 ± 0.1 | −65.3 ± 0.1 | 56.6 ± 0.1 | 0.3 ± 0.1 |
| Phospho-SF11–255-U2AF65R12U, site 1 | 7 ± 5 | −11.5 ± 0.1 | −60.4 ± 0.1 | 48.9 ± 0.1 | 0.8 ± 0.1 |
| Phospho-SF11–255-U2AF65R12U, site 2 | 600 ± 80 | −8.7 ± 0.1 | −65.3 ± 0.1 | 57.6 ± 0.1 | 0.4 ± 0.1 |
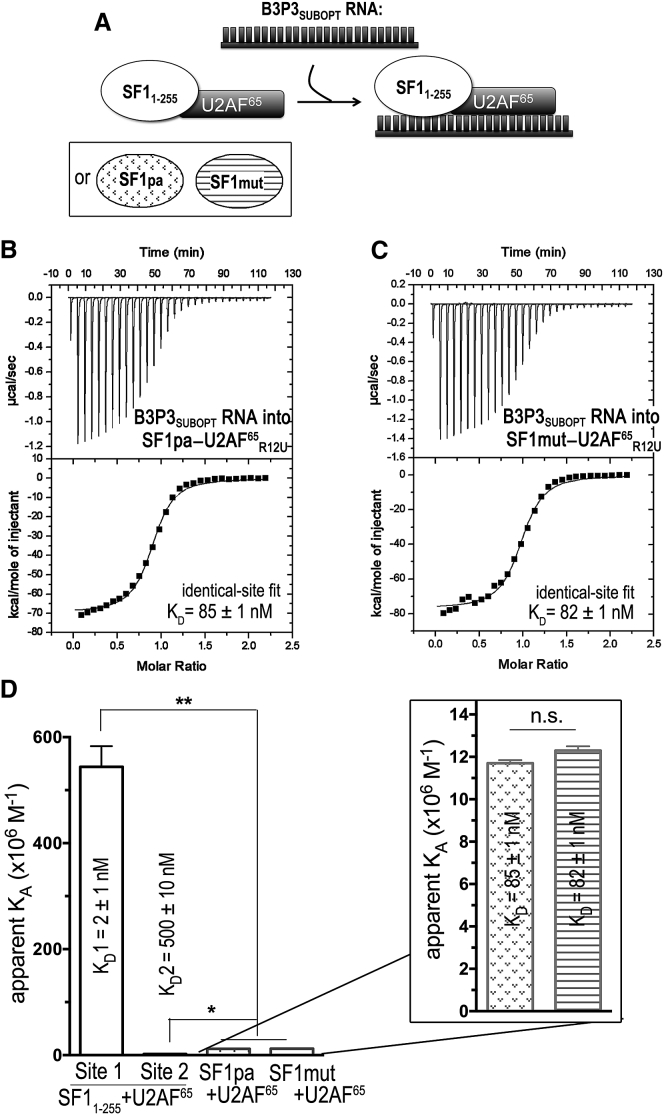
| SF1pa-U2AF65R12U | 85 ± 1 | −9.8 ± 0.1 | −67.5 ± 0.1 | 57.7 ± 0.1 | 0.9 ± 0.1 |
| SF1mut-U2AF65R12U | 82 ± 1 | −9.8 ± 0.1 | −76.7 ± 0.1 | 66.9 ± 0.1 | 0.9 ± 0.1 |
| SF1mut-U2AF65R12Ue | 104 ± 7 | −9.7 ± 0.2 | −68.4 ± 0.2 | 58.6 ± 0.2 | 1.0 ± 0.1 |
| Splicing-Factor Complexes Titrated into B3P3 RNAf | |||||
| SF11–255-U2AF65R12U, site 1 (reverse) | 160 ± 80 | −9.5 ± 0.1 | −22.3 ± 0.1 | 12.8 ± 0.1 | 0.6 ± 0.1 |
| SF11–255-U2AF65R12U, site 2 (reverse) | 7 ± 1 | −11.3 ± 0.1 | −53.2 ± 0.1 | 42.0 ± 0.1 | 0.9 ± 0.2 |
| Phospho-SF11–255-U2AF65R12U, site 2 (reverse) | 115 ± 5 | −9.8 ± 0.1 | −81.1 ± 4.9 | 71.4 ± 4.9 | 0.5 ± 0.1 |
| Phospho-SF11–255-U2AF65R12U, site 1 (reverse) | 18 ± 3 | −10.8 ± 0.1 | −36.2 ± 4.5 | 25.5 ± 4.7 | 0.7 ± 0.2 |
Values are expressed as the mean ± SD of at least two independent titrations.
ΔG values were calculated using the equation ΔG = −RT ln (KA) where T=303 K
−TΔS values were calculated using the equation −TΔS = ΔG − ΔH.
Fits are “ligand-in-cell” such that “n” is the apparent stoichiometry of protein/RNA.
Dialyzed into 100 mM NaCl and 25 mM sodium phosphate buffer (pH 7.4). Macromolecules for other experiments in this table were in 100 mM NaCl, 25 mM HEPES (pH 7.4).
Apparent stoichiometry of protein/RNA (n).
Figure 2.
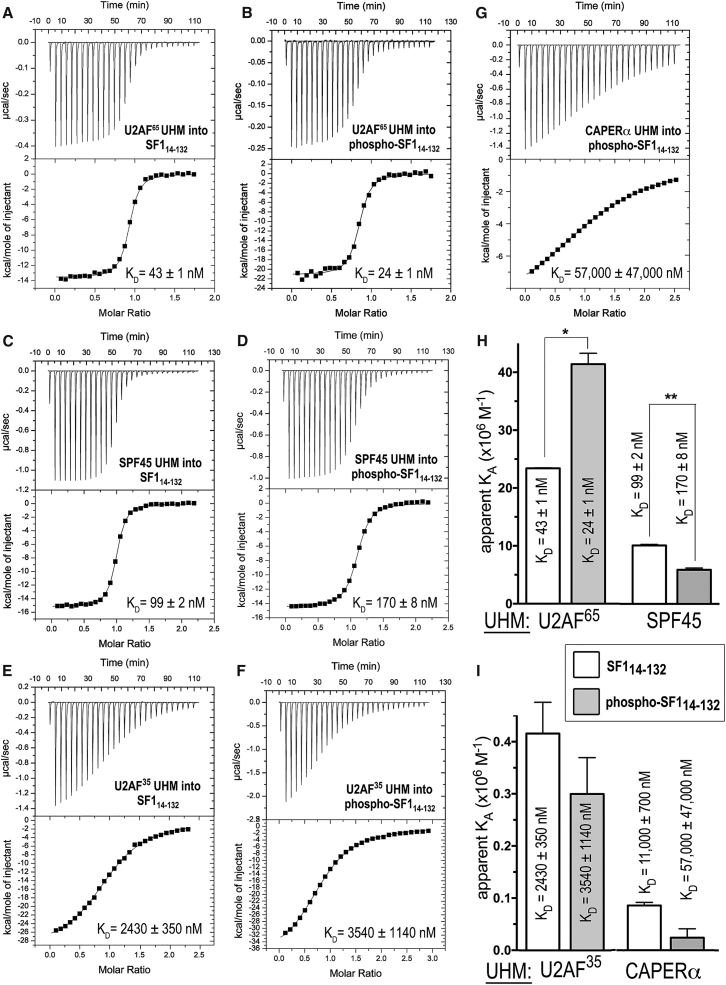
ITC results for SF114–132 or phospho-SF114–132 titrated with UHM proteins. (A–G) Representative isotherms fit with identical sites models. Titrations were of (A) 90 μM U2AF65 UHM into 10 μM SF114-132, c = 230; (B) 32 μM U2AF65 UHM into 4 μM phospho-SF114–132, c = 170; (C) 200 μM SPF45 UHM into 20 μM SF114–132, c = 200; (D) 200 μM SPF45 UHM into 20 μM phospho-SF114–132, c = 120; (E) 200 μM U2AF35 UHM into 20 μM SF114-132, c = 8; (F) 260 μM U2AF35 UHM into 20 μM phospho-SF114–132, c = 6; and (G) 600 μM CAPERα UHM into 50 μM phospho-SF114–132, c = 1. (H and I) Bar graphs showing the apparent equilibrium affinities (KA) of SF114–132 (white bars) and phospho-SF114–132 (gray bars) for the indicated UHM proteins. The apparent equilibrium dissociation constants (KD = KA−1) and the standard deviations of at least two replicates are given. Values from (G) are estimates due to the low affinity binding, which is at the limit for reliable fits of ITC data.
Figure 4.
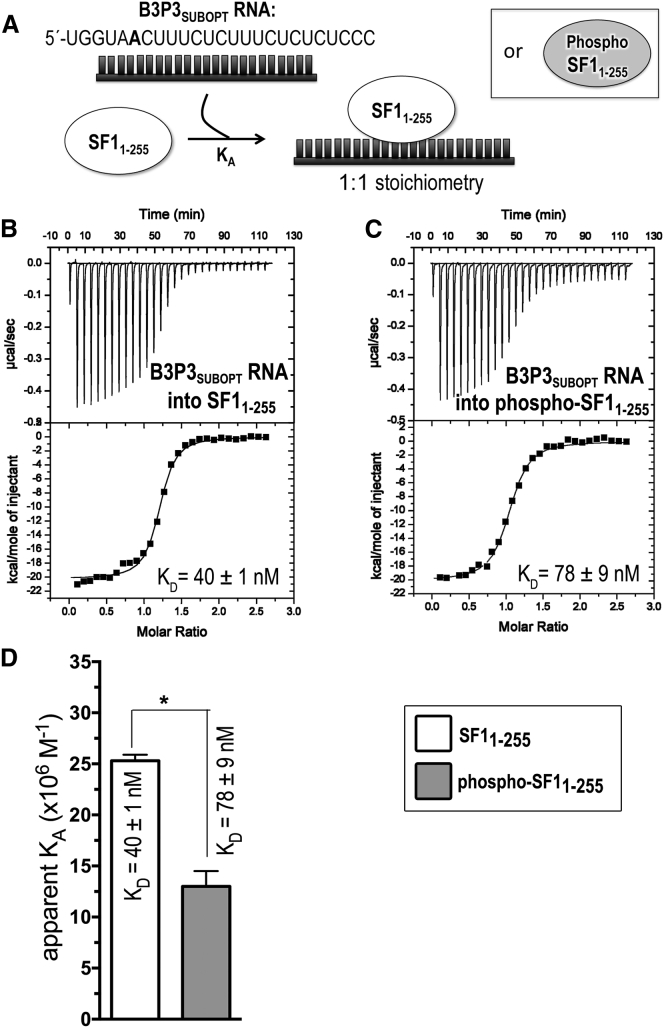
ITC results for SF11–255 (white) or phospho-SF11-255 (gray) titrated with B3P3 RNA. (A) Schematic diagram of the ITC experiment. (B and C) Representative isotherms fit with identical-site binding models. Titrations were of (B) 60 μM B3P3SUBOPT RNA into 5 μM SF11-255, c = 125; and (C) 60 μM B3P3SUBOPT RNA into 5 μM phospho-SF11–255, c = 64. Nonidentical-site models were discarded based on 1) little reduction in χ2 for nonidentical sites compared to identical sites; 2) lower data/parameter ratio; and 3) fitting errors greater than the resulting values. (D) Bar graph of apparent affinities (KA) of the interactions, colored as in (A). The apparent equilibrium dissociation constants (KD = KA−1) and the standard deviations between at least two replicates are given.
Figure 6.
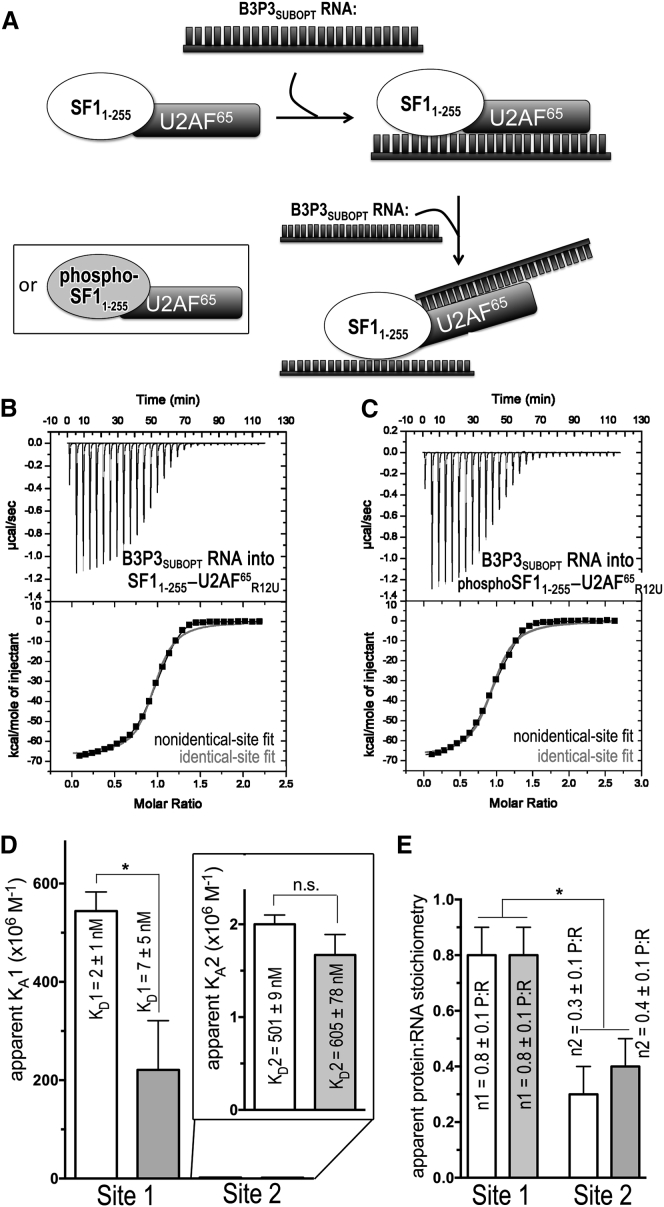
ITC results for SF11–255-U2AF65R12U (white) or phospho-SF11–255-U2AF65R12U (gray) protein complexes titrated with B3P3SUBOPT RNA. (A) Schematic diagram of the ITC experiment. (B and C) Representative isotherms fit with binding models for either nonidentical sites (black lines) or identical sites (red lines). Titrations were of (B) 35 μM B3P3 RNA into 5 μM SF11–255-U2AF65R12U complex (nonidentical sites, χ2 = 0.20E6 ± 0.16E6; identical sites, χ2 = 2.70E6 ± 0.29E6), c = 5000 for site 1 and c = 10 for site 2; and (C) 35 μM B3P3 RNA into 5 μM phospho-SF11–255-U2AF65R12U complex (nonidentical sites, χ2 = 0.23E6 ± 0.19E6; identical sites, χ2 = 3.13E6 ± 0.95E6), c = 700 for site 1 and c = 8 for site 2. The identical site models were discarded based on the large χ2 increase, poor fit, and nonidentical sites of reverse titrations (Fig. S5). (D) Bar graph of apparent affinities (KA) of the interactions, colored as for (A). The apparent equilibrium dissociation constants (KD = KA-1) and the standard deviations of at least two replicates are given. An expanded view of the lower affinity sites is inset. (E) Bar graph of apparent protein:RNA stoichiometries (n).
Figure 7.
ITC results for SF1mut (striped) or SF1pa (stippled) titrated with B3P3 RNA. (A) Schematic diagram of the ITC experiment. (B and C) Representative isotherms fit with identical-site binding models for (B) 50 μM B3P3SUBOPT RNA into 5 μM SF1pa, c = 44, and (C) 50 μM B3P3SUBOPT RNA into 5 μM SF1mut, c = 27. (D) Bar graph of apparent affinities (KA) of the interactions, colored as in (A). The KA of unmodified SF11–255 (white) is plotted for comparison. The inset shows apparent equilibrium dissociation constants (KD = KA−1) and standard deviations of at least two replicates.
Figure 8.
ITC results for SF1mut-U2AF65R12U (striped) or SF1pa-U2AF65R12U (stippled) complexes titrated with B3P3SUBOPT RNA. (A) Schematic diagram of the ITC experiment. (B and C) Representative isotherms fit with identical site models for (B) 50 μM B3P3 RNA into 5 μM SF1pa-U2AF65R12U complex, c = 59, and (C) 50 μM B3P3 RNA into 5 μM SF1mut-U2AF65R12U complex, c = 61. Nonidentical-site models were discarded based on 1) fitting errors greater than the resulting thermodynamic values; 2) stoichiometry values of nearly zero for one class of nonidentical sites; and 3) thermodynamic values for the remaining class of sites that matched the results of the identical-site fit. (D) Bar graph of apparent affinities (KA) of the interactions, colored as in (A). The apparent equilibrium dissociation constants (KD = KA−1) and standard deviations of at least two replicates are given. For comparison, the nonidentical-site affinities of the parent SF11–255-U2AF65R12U complex titrated with B3P3 RNA (details in Fig. 6) are plotted at left. The inset shows an expanded view of the B3P3 RNA affinities for the SF1mut- or SF1pa-U2AF65R12U complexes. A control titration of B3P3SUBOPT RNA into buffer is shown in Fig. S1 C and a titration of B3P3SUBOPT RNA into SF1mut-U2AF65R12U in phosphate buffer (as opposed to HEPES) is shown in Fig. S6.
Fluorescence anisotropy RNA binding assay
Synthetic RNAs with 5′-fluorescein (Fl) labels (Dharmacon, GE Healthcare) were deprotected and desalted according to manufacturer instructions and used without further purification. RNA sequences used for fluorescence measurements included AdMLS 5′-UACUUAUCCUGUCCCUUUUUUUUC, AdMLL 5′-UUCGUGCUGACCCUGUCCCUUUUUUUUCCACAGC, or BPSSUBOPT 5′-GAGUCAGACUAACUCGUUGUUUAG (branch site underlined). Fluorescence anisotropy changes during titration of 25 nM RNA with the indicated final concentrations of protein (Figs. 3 and 5) in a water-thermostatted cuvette at 23°C using a FluoroMax-3 spectrophotometer (HORIBA Jobin Yvon, Kyoto, Japan) were measured and analyzed in a manner similar to that described in (52, 53). Both macromolecules were diluted >50-fold in 150 mM NaCl, 25 mM HEPES (pH 7.4), 0.2 mM TCEP, and 0.1 U μL−1 Superase-In (Ambion, Life Technologies, Carlsbad, CA). Samples were excited at 490 nm and emission intensities were recorded at 520 nM with a slit width of 5 nm. Data were fit by non-linear regression assuming single-site binding to obtain the apparent equilibrium dissociation constant (KD) using the equation below, where X is the total protein concentration, [RNA] is the total RNA concentration, r is the observed anisotropy at the ith titration, rB is the anisotropy at zero protein concentration, and rF is the anisotropy at saturating protein concentration (floated in fit):
The KA was calculated using the relationship KD−1.
Figure 3.
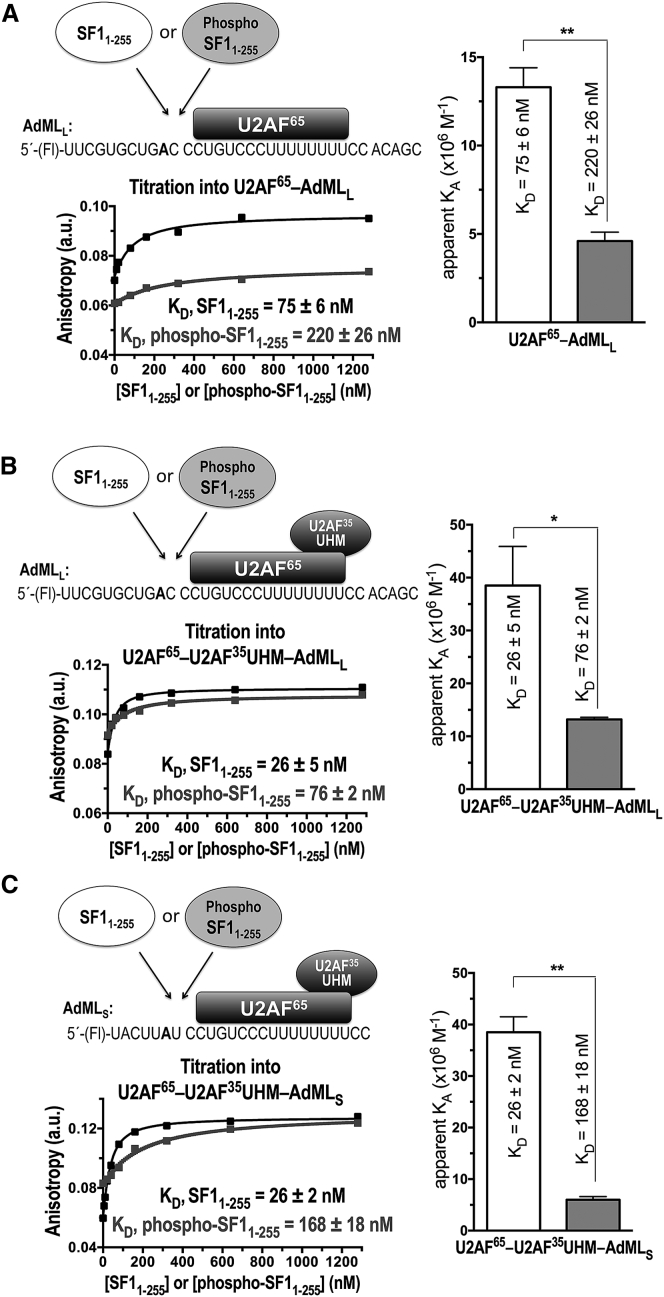
Fluorescence anisotropy RNA binding assay of SF11–255 (white bars, black binding curve) or phospho-SF11-255 (gray shading and binding curve) proteins titrated into fluorescein (Fl)-labeled AdML RNA oligonucleotides, which were pre-bound to full-length U2AF65 (either with or without the U2AF35 UHM subunit). The SF11–255 or phospho-SF11–255 proteins were titrated into protein-RNA complexes (A) Fl-AdMLL-U2AF65, (B) Fl-AdMLL-U2AF65-U2AF35 UHM, (C) Fl-AdMLS-U2AF65-U2AF35 UHM. Schematic diagrams of the experiments, shown above the plots, provide the RNA sequences (branchpoint in bold). The plots show representative fits of the fluorescence anisotropy changes during titration. Bar graphs of the apparent equilibrium affinity (KA) values are plotted on the right. The apparent equilibrium dissociation constants (KD = KA−1) and the standard deviations of three technical replicates are given.
Figure 5.
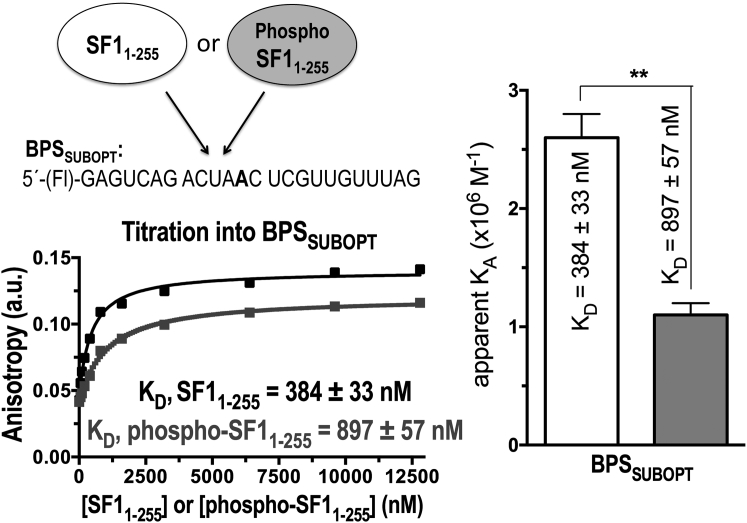
Fluorescence anisotropy RNA binding assays of SF11–255 or phospho-SF11–255 proteins titrated into BPSSUBOPT RNA, represented as for Fig. 3.
Statistical significance of results
We observe small differences between binding constants of the phosphorylated and those of the unmodified SF1 for UHM-containing proteins or RNA. These differences are reproducible in at least two ITC experiments with distinct protein preparations and at least triplicate technical replicates of fluorescence anisotropy RNA binding assays, which are customary sample sizes with the power to detect significant differences, if present. In Figs. 2, H and I, 3, 4 D, 5, 6, D and E, 7 D, and 8 D, we plot the KA values in a bar graph and provide the results of unpaired two-tailed t-tests with Welch’s correction as ∗p < 0.05, ∗∗p < 0.005. The t-test is robust and can withstand violations of model assumptions (e.g., (54)). However, the t-test assumes a normally distributed population that differs from the small number of replicates here and should be considered here only as a starting point for critical assessment.
Results
UHMK1 phosphorylation slightly enhances SF1 affinity for U2AF65
Originally, pull-down experiments showed that phosphorylation of the SF1 SPSP motif by UHMK1 increased retention of full-length U2AF65 by ∼2-fold (46). However, the affinities of SF1 (residues 1–260) and UHMK1-phosphorylated (phospho-) SF1 for a U2AF65 fragment comprising two central RRMs and the C-terminal UHM (U2AF65R12U, residues 148–475) appeared similar when measured by ITC in a phosphate-containing buffer (20 mM sodium phosphate (pH 6.5) and 50 mM NaCl) (43). To rule out competitive binding or possible allosteric effects of phosphate ions via the SF1 phosphorylation sites, we used ITC to measure the affinities of unmodified and phospho-SF11–255 (residues 1–255) for U2AF65R12U in 25 mM HEPES buffer (pH 7.4), 100 mM NaCl (Fig. S2, A and B). We used UHMK1 to phosphorylate the SF11–255 protein in a method identical to that used for our prior crystal structure (42), and the similar shift by SDS-PAGE suggests that both serines of the motif are phosphorylated as seen in our crystal structure. This phosphorylated state was stable under the conditions of the binding experiments (Fig. 1 D).
Similar to the ITC in phosphate buffer described in a previous study (43), UHMK1 phosphorylation increased the binding affinity of SF11–255 for U2AF65R12U by <2-fold (Table S1). We compared the affinity of a minimal U2AF65 UHM for an SF1 construct that included the ULM and coiled coil but lacked the RNA binding domain (residues 14–132, SF114-132) (Fig. 2, A and B). We observed a subtle enhancement after UHMK1 phosphorylation of phospho-SF114–132 binding to the U2AF65 UHM similar to that observed for the longer constructs (Table S1). Comparison of the SF11-255-U2AF65R12U and SF114-132-U2AF65 UHM affinities indicated that the SF1 and U2AF65 RNA binding domains make little contribution to, and even slightly interfere with (by twofold), formation of the SF1-U2AF65 complex.
Since these SF1-U2AF65 binding affinities were relatively high for reliable measurement by ITC (c-values of 100–230), we repeated the unmodified and phospho-SF114–132-U2AF65 UHM ITC at a higher salt concentration (25 mM HEPES (pH 7.4) and 250 mM NaCl) to reduce the slope (and hence improve the fits) of the isotherms (c-values of 16 and 24 for unmodified and phospho-SF114–132 in Fig. S2, C and D, respectively). As expected, the higher ionic strength conditions reduced the affinities of the SF114–132 or phospho-SF114–132 complexes with the U2AF65 UHM (Table S1), consistent with the previously noted importance of electrostatic interactions between charged residues of the ULM and UHM (37). The phosphorylation-induced increase in SF1-U2AF65 UHM affinity remained <2-fold, regardless of the higher salt concentration. Altogether, these data demonstrate that the free-energy change for SF1 binding to U2AF65 after UHMK1 phosphorylation relative to unmodified SF1 (ΔΔG) is less than expected for the gain of a single hydrogen-bond interaction. Nevertheless, the phosphorylation-induced enhancement of phospho-SF1-U2AF65 association is reproducible and consistent with an ∼2-fold greater retention of phosphorylated compared with unmodified SF1 by U2AF65 in prior pull-down experiments (46).
UHMK1 phosphorylation slightly reduces SF1 affinities for off-target UHM proteins
Based on the steric prominence of the SF1 coiled-coil domain (42, 43), we next hypothesized that phosphorylation-induced stabilization of this SF1 structure could reduce nonspecific binding to UHMs of other splicing factors. We focused on the UHMs of three splicing factors for which the cognate ULM partners are distinct from SF1: SPF45 and CAPERα, which are thought to recognize SF3b155 ULMs (40, 41), and U2AF35, which heterodimerizes with the U2AF65 ULM (38). We found that phosphorylation by UHMK1 subtly penalized SF1 binding to these off-target UHMs (Fig. 2). The net specificity of SF1 for binding the U2AF65 UHM improved from two- to sevenfold for the SPF45 UHM, from nearly 60- to 150-fold for the U2AF35 UHM, and from 250-fold to >2300-fold for the CAPERα UHM (Fig. 2, H and I; Table 1). A fivefold reduced affinity of phospho-SF114-132 compared to the unmodified SF1 for binding the CAPERα UHM was the greatest change, whereas UHMK1-phosphorylation reduced SF1 binding to the UHMs of SPF45 or U2AF35 by less than twofold. With the exception of the SPF45 UHM, SF1 phosphorylation consistently increased the magnitudes of both the enthalpy and entropy changes of UHM binding (Table 1). Considering a diminished effect in higher salt, the increase in the entropy-enthalpy compensation after phosphorylation of the SF114–132-U2AF65 UHM complex may result from enhanced folding and/or coupled ion or proton exchange with the SPSP-containing loop, for which phosphate-arginine salt bridges mediate an ordered structure (42, 43). Altogether, UHMK1-phosphorylation increased the net preference of phospho-SF1 slightly for binding to the U2AF65 UHM relative to the SPF45 or U2AF35 UHMs and showed a more substantial, sixfold net change in U2AF65 versus CAPERα UHM specificity after SF1 phosphorylation.
UHMK1 phosphorylation slightly reduces SF1 affinity for AdML RNA in the presence of U2AF65
Previously, others and we had shown that UHMK1 phosphorylation slightly enhances SF1 association with U2AF65 by EMSA in the presence of sub-stoichiometric amounts of a radiolabeled RNA oligonucleotide (43, 46). To further explore the influence of SPSP phosphorylation on the SF1 affinity for RNA in the presence of U2AF65 in solution, we completed a variation of this experiment, in which we titrated increasing amounts of SF11–255 or phospho-SF11–255 into an equimolar mixture of U2AF65 with 5′-fluorescein (Fl)-labeled RNA (Fig. 3).
We first tested the same protein constructs and RNA sequence as used in our prior EMSA, including phospho- or unmodified SF11–255, full-length U2AF65, and a 34mer RNA corresponding to the prototypical 3′ splice site of adenovirus major late promoter transcript (AdMLL) (Fig. 3 A). To our surprise, phosphorylation by UHMK1 slightly decreased (by threefold) rather than enhanced SF1 binding to the AdMLL RNA in the presence of U2AF65. To rule out whether the apparent phosphorylation-induced decrease in affinity was affected by aggregation of full-length U2AF65, we repeated the experiment after co-purification of U2AF65 with the U2AF35 UHM (Fig. 3 B), which improves the solubility and monodispersity of the U2AF protein preparation. Phosphorylation again decreased rather than enhanced SF1 binding to the AdMLL RNA in the presence of the U2AF65-U2AF35 UHM complex by the same amount (threefold) as for AdMLL RNA in the presence of U2AF65. To avoid possible alternative binding sites due to inclusion of sequences flanking the splice site, which none of the SF1, U2AF65, or U2AF35 UHM subunits are expected to recognize, we compared titrations of SF1 or phospho-SF1 into a stoichiometric mixture of U2AF65-U2AF35 UHM incubated with a 25mer 5′-Fl-RNA comprising the core BPS and Py tract of the AdML 3′ splice site (AdMLS) (Fig. 3 C). The affinities of unmodified SF1 for AdMLL and AdMLS in the presence of U2AF65-U2AF35-UHM were indistinguishable. The phosphorylation-induced decrease in protein-RNA affinity was slightly greater for the AdMLS RNA, possibly due to limited binding registers on the minimal sequence. The nearly sevenfold decrease in binding affinity of phosphorylated compared to unmodified SF1 for AdMLS in the presence of U2AF65-U2AF35 UHM corresponds to a ΔΔG that is similar in magnitude to the loss of a strong hydrogen bond.
A posteriori, we realized that in prior EMSA experiments, association of SF1 with U2AF65 dominated the apparent binding constants due to a large excess of U2AF65 compared to the radiolabeled RNA (>50-fold), coupled with the significantly higher RNA affinity of the SF1-U2AF65 complex compared to either subunit alone (55). As such, the increase in shifted RNA in the EMSA recapitulated the phosphorylation-induced increase in the affinity of SF1 for binding U2AF65, described above. In the titrations of SF1 into stoichiometric mixtures of fluorescently labeled RNA and U2AF65 here, approximately half of the U2AF65 and RNA molecules are dissociated in the sample cell before titration with SF1 (estimated from the ∼30 nM KD of the full-length U2AF65-U2AF35 UHM complex binding the AdML Py tract (56) and a 25 nM concentration of U2AF65-U2AF35 UHM protein and RNA in the sample cell). Considering the many competing equilibria (Fig. S3), only apparent binding constants for the experiments are given in Fig. 3. Regardless, we observed a consistent reduction in the apparent RNA affinity of phosphorylated compared to unmodified SF1 in all three U2AF65-containing RNA mixtures (U2AF65-AdMLL, U2AF65-U2AF35 UHM-AdMLL, U2AF65-U2AF35 UHM-AdMLS), which we then proceeded to test in better-defined systems.
UHMK1-phosphorylation slightly reduces SF1 affinity for splice-site RNAs
The decreased affinity of phospho-SF1 relative to unmodified SF1 for U2AF65-containing–RNA complexes, yet slightly enhanced phospho-SF1 binding to apo-U2AF65, implied that RNA binding by SF1 or SF1-U2AF65 was inhibited by UHMK1 phosphorylation. To test this possibility in a defined system, we measured the affinities of phosphorylated versus unmodified SF1 binding an RNA splice site by ITC (Fig. 4; Table 2), a quantitative method that fits the enthalpy changes of binding to obtain the apparent equilibrium dissociation constant (KD). In our prior EMSAs (42), phosphorylated versus unmodified SF1 bound with similar apparent affinities to the AdMLL RNA, which is a “strong,” near-consensus splice site. Yet in other hands, EMSAs suggested that SRPK2 phosphorylation of S82 slightly enhances SF1 binding to “weak” splice sites that poorly match the BPS consensus (47). With these considerations, we tested SF1 binding to a variant of a well-characterized “B3P3” splice site from α-tropomyosin (B3P3SUBOPT) (57), for which a -3/-4GG sequence preceding the branchpoint is expected to decrease SF1 affinity by approximately threefold (58). Phosphorylation of SF11–255 by UHMK1 decreased affinity for the B3P3SUBOPT RNA by twofold. This free-energy change (ΔΔG) of a ∼0.5 kcal mol−1 resulted from a 2.5 kcal mol−1 penalty for the enthalpy change (ΔΔH) and −2 kcal mol−1 favorable contribution to the entropy change [Δ(−TΔS)]. These thermodynamic differences agree with ordering of the SPSP-loop conformation after phosphorylation, and they further allude to the possibility of enthalpically favorable contacts between this region in the unphosphorylated state and the bound RNA site.
Although we weakened the BPS of our B3P3SUBOPT splice site, it remained possible that phosphorylation-enhanced RNA binding affinity would manifest only in the context of extreme suboptimal BPS such as that tested in EMSAs after SRPK2-phosphorylation of SF1 (47). To evaluate this possibility, we compared the affinities of UHMK1-phosphorylated and unmodified SF1 for binding an identical RNA oligonucleotide (BPSSUBOPT) as the prior, SRPK2-focused EMSA study. We used the fluorescence anisotropy RNA binding assay to determine the KD, since the affinities of RNA binding were relatively low for reliable measurement by ITC. The affinity of SF1 for the BPSSUBOPT oligonucleotide decreased more than twofold after UHMK1 phosphorylation, similar to the change we observed by ITC with the B3P3SUBOPT splice site (Fig. 5). The subtle, yet consistent, decrease in the affinity of phospho-SF1 relative to unmodified SF1 for splice-site RNA explained the reduced phospho-SF1 affinity for the pre-formed U2AF65-RNA complex that we had observed by fluorescence anisotropy assays.
UHMK1 phosphorylation slightly reduces SF1-U2AF65 affinity for a splice-site RNA
We next asked whether UHMK1 phosphorylation would affect RNA binding by the relevant, SF1-U2AF65 complex. To address this question, we used ITC to compare UHMK1-phosphorylated to unmodified SF11–255-U2AF65R12U complexes binding the B3P3SUBOPT RNA (Fig. 6; Table 3). First, we titrated the B3P3SUBOPT RNA into the unmodified or phospho-SF11–255-U2AF65R12U protein complex, which had been prepared by size-exclusion chromatography from a stoichiometric mixture of the pre-purified subunits. By placing the proteins in the sample cell ([SF11–255-U2AF65R12U] = 5000 nM), we avoided potential heats of dilution due to complex dissociation, which nevertheless were unlikely considering the high affinity of the SF1-U2AF65 complex (60 nM; Table 1). The isotherms were best fit by nonidentical-site models considering both visual inspection and reduction in χ2 (Fig. 6, B and C). The apparent stoichiometries of the two classes of sites (Fig. 6 E) suggested that SF1 (or phospho-SF1) and U2AF65 each bound separate RNA oligonucleotides in the presence of excess RNA (Fig. 6 A). Indeed, the B3P3SUBOPT RNA affinity of the second site agrees with the affinity of the isolated U2AF65 subunit (KD = 500 nM and 509 nM; Fig. S4), suggesting that a second RNA molecule replaces U2AF65 contacts with the intramolecular Py tract of B3P3SUBOPT bound to the SF11–255-U2AF65R12U complex (Fig. 6 A). Regardless of the model chosen for the fit (identical versus nonidentical sites), a similar subtle penalty for RNA binding was observed after SPSP-phosphorylation of SF1.
To confirm the nonidentical-binding-site model, we next reversed the ITC experiment and titrated the SF11–255-U2AF65R12U complex into B3P3SUBOPT RNA (Fig. S5). A control titration of SF11–255-U2AF65R12U into buffer was linear and showed negligible heats of dilution (Fig. S1 D). We expected that the isotherms of the reverse titrations would best fit nonidentical-binding-site models and that a low-affinity binding site with an apparent stoichiometry of two RNA molecules per protein complex would be evident during the early (as opposed to late) steps of the titration, for which the RNA is in molar excess over protein molecules. Accordingly, the isotherms were best fit by nonidentical-site binding models by visual inspection and reduction in χ2 (Fig. S5, B and C). The apparent stoichiometries of the two classes of binding sites (Fig. S5 E) agreed with initial binding of two RNA oligonucleotides per one SF11–255-U2AF65R12U (or phospho-SF11–255-U2AF65R12U) complex, presumably one RNA per SF1 or U2AF65 subunit (Fig. S5 A), when RNA is in large excess over protein during the early stage of this reverse titration. The higher-affinity class of binding sites with 1:1 stoichiometry of SF11–255-U2AF65R12U (or phospho-SF11–255-U2AF65R12U) per RNA oligonucleotide, becomes apparent in the later stages of titration as protein concentrations rise relative to RNA. The higher-affinity binding site of the reverse titration includes an unfavorable contribution from the dissociation of one RNA molecule from the 2:1 complex before rebinding to form the SF11–255-U2AF65R12U-RNA (or phospho-SF11–255-U2AF65R12U-RNA) complex with 1:1 stoichiometry. As such, we focus on interpreting the high-affinity binding site of the B3P3SUBOPT RNA titration into SF11–255-U2AF65R12U (or phospho-SF11–255-U2AF65R12U) complexes. Phosphorylation by UHMK1 consistently reduced the apparent B3P3SUBOPT RNA affinity of the SF1-U2AF65 complex by ∼3-fold for the high-affinity sites of either the forward or reverse titrations (Table 3), which is similar to the penalty observed for the isolated SF1 subunit.
SF1 coiled-coil domain contributes to SF1 or SF1-U2AF65 binding a splice-site RNA
The slight energetic, yet more significant enthalpic, penalties for RNA binding after phosphorylation of SF1 (ΔΔG = 0.5 kcal mol−1 and ΔΔH = 3 kcal mol−1) left open the possibility that the SF1 coiled coil might contact RNA. To explore this possibility, we first swapped the SF1 coiled coil for an unrelated three-helix bundle from Staphylococcus aureus protein A to generate a chimeric “SF1pa” protein (Fig. 1 B). We flanked the protein-A domain with 25-amino-acid glycine-rich linkers with the intention of structurally uncoupling the protein A region from the surrounding UHM-bound SF1 ULM and KH-QUA2 RNA binding domain. The total number of amino acids remained unchanged between the SF11–155 parent and SF1pa chimera. Likewise, the isoelectric points (pI), and hence net charges, of the phosphorylated SF1 coiled coil and the artificial protein-A domain were similar (pI = 5.1 and 5.5 compared with 6.8 for the unmodified coiled-coil domain). The substituted domain reduced the apparent affinity of the SF1pa protein for the B3P3SUBOPT RNA by ∼3-fold compared to unmodified SF11–255 (Fig. 7). This penalty for RNA binding, as well as the underlying enthalpy and entropy changes, were nearly indistinguishable between SF1pa and phospho-SF11–255 (Table 3), which suggested that the protein-A chimera had an effect on SF1-RNA binding similar to that observed for SPSP phosphorylation, at least in the context of the isolated SF1 subunit.
We next extended our investigation of the SF1 coiled coil to the consequences of its protein A substitution for RNA binding in the context of the SF1-U2AF65 complex. We monitored, by ITC, SF1pa-U2AF65R12U binding the B3P3SUBOPT RNA (Fig. 8). In contrast to the isotherms of wild-type SF1-U2AF65 complexes binding RNA, an identical-site binding model with an apparent 1:1 stoichiometry adequately fit the SF1pa-U2AF65R12U isotherms (Fig. 8, B and C). The apparent loss of the nonidentical RNA binding site suggested that steric hindrance or other effects prevented binding of two separate RNAs to the SF1pa-U2AF65R12U heterodimer at the concentrations used in our ITC experiment. Notably, the SF1pa-U2AF65R12U affinity for the B3P3SUBOPT RNA decreased by ∼40-fold relative to the high-affinity SF1-U2AF65R12U-B3P3SUBOPT RNA site (Fig. 8 D). This large penalty of the SF1 domain substitution for RNA binding by the SF1pa-U2AF65R12U complex significantly exceeded its threefold impact on RNA binding by the SF1 subunit, and also exceeded the threefold penalty incurred by UHMK1 phosphorylation of the protein complex. We conclude that the SF1 coiled-coil domain is required for high-affinity association of the SF1-U2AF65 complex with a representative B3P3SUBOPT splice site.
Arginines at the phosphorylation site contribute to SF1 or SF1-U2AF65 binding a splice-site RNA
We next tested a potential role in RNA binding for key arginine residues in the SPSP-containing SF1 loop (R93, R97, and R100), which otherwise coordinate the phosphorylated serines in a structured conformation for the phosphorylated state (42, 43) (Fig. 1 C). We previously had found that SF1 carrying glutamate mutations of these three arginines (R93E/R97E/R100E) was unable to restore cell proliferation after SF1 knockdown (42). We first used ITC to determine the thermodynamic consequences of the R93E/R97E/R100E mutations in the SF11–255 construct (SF1mut; Fig. 1 B) for binding the representative B3P3SUBOPT splice-site RNA. The arginine mutations penalized the RNA binding affinity of SF1mut by nearly fivefold relative to wild-type SF11–255 (Fig. 7). The enthalpy and entropy changes underlying B3P3SUBOPT RNA binding by the isolated SF1mut subunit resembled those of phospho-SF11–255 and SF1pa more than the unmodified SF1 (Table 2). An enthalpic penalty, ΔΔH, after introduction of the glutamates (3.3 kcal mol−1) overcame a lesser Δ(−TΔS) (−2.3 kcal mol−1) for the mutant compared to wild-type SF11–255 subunits. The slight ΔΔG of 1 kcal mol−1 between SF1mut and SF11–255 would agree with disruption of a single hydrogen bond rather than three ion pairs. On the other hand, the compensation of a large unfavorable entropy change by a larger enthalpy change for SF1mut-RNA binding would agree with structuring of the unphosphorylated, arginine-containing loop for RNA interactions.
We next tested the influence of the SF1 arginine mutations on B3P3SUBOPT RNA recognition in the context of the SF1mut-U2AF65R12U complex. The B3P3SUBOPT RNA affinity of SF1mut-U2AF65R12U was indistinguishable from that of SF1pa-U2AF65R12U (Table 3). In both cases, identical-binding-site models adequately fit the isotherms (Fig. 8, B and C). Also, as for SF1pa-U2AF65R12U, the B3P3SUBOPT RNA binding affinities of SF1mut-U2AF65R12U decreased by 40-fold relative to the high-affinity, stoichiometric site of unmodified SF1-U2AF65R12U (Fig. 8 D).
The enthalpy and entropy changes of all variant SF1-U2AF65R12U complexes binding the B3P3SUBOPT RNA (including those of the unmodified SF11–255, phospho-SF1, SF1pa, and SF1mut complexes with U2AF65) are remarkably large in magnitude (Table 3). In general, these thermodynamic changes are likely to be dominated by RNA association with the U2AF65 RRMs. In our previous ITC experiments that contained BES buffer, we had shown large enthalpy and entropy changes underpinning cognate RNA binding by tandem RRMs, including those of U2AF65R12U, Sex-Lethal, and poly-A binding protein (59). For comparison with other ITC experiments herein, we here recapitulated this result for B3P3SUBOPT RNA binding by U2AF65R12U in a HEPES-containing buffer (Fig. S4; Table 2).
Even so, the magnitudes of the enthalpy and entropy changes for B3P3SUBOPT RNA binding were notably greater for the SF1mut-U2AF65R12U compared to the unmodified SF11–255-U2AF65R12U complexes (ΔΔH of −16.6 kcal mol−1 and Δ(−TΔS) of 19 kcal mol−1). In comparison, these thermodynamic changes of RNA binding were similar between the phospho-SF11–255-U2AF65R12U and the unmodified complex (ΔΔH and Δ(−TΔS) were −0.3 kcal mol−1 and 1 kcal mol−1). Although the enthalpy and entropy changes of SF1pa-U2AF65R12U binding the B3P3SUBOPT RNA were slightly greater in magnitude than those of the SF11–255-U2AF65R12U complex (ΔΔH and Δ(−TΔS) were −7.4 kcal mol−1 and 9.8 kcal mol−1), they also were significantly less than those of SF1mut-U2AF65R12U.
One possible explanation for the larger magnitudes of the enthalpy and entropy changes for the SF1mut-U2AF65R12U complex binding RNA is that mutations of the ionizable arginine residues alter proton exchange between the RNA-bound complex and its surroundings. In an effort to avoid potential interactions between a phosphate buffer and allosteric or RNA binding sites of the proteins, we had used a HEPES buffer, which has a relatively large enthalpy of ionization (5 kcal mol−1 (60)). We repeated the experiment with the SF1mut-U2AF65R12U complex titrated with B3P3SUBOPT RNA in sodium phosphate buffer (Fig. S6), which has a relatively low ionization enthalpy (1 kcal mol−1 (60)). The magnitudes of the enthalpy and entropy changes for the mutant complex binding RNA in the phosphate buffer decreased to nearly match the values for the protein-A domain-substituted complex in HEPES buffer (ΔΔH of −0.9 kcal mol−1 and Δ(−TΔS) of 0.9 kcal mol−1 between RNA titrations of SF1pa-U2AF65R12U or SF1mut-U2AF65R12U complexes in respective phosphate or HEPES buffers) (Table 3). The buffer-dependent alteration in the enthalpy change was consistent with loss of approximately three additional protons from the mutant compared to the wild-type protein (site 1) on formation of the RNA complex, consistent with changed acid dissociation constants of three ionizable residues by the three arginine-to-glutamate mutations.
Altogether, these ITC experiments demonstrate that both the coiled-coil region of SF1 and the arginine residues surrounding the phosphorylation site are important for recognition of a representative splice site by the SF1-U2AF65 complex.
Discussion
Phosphorylation is an emerging means for regulating UHM-ULM interactions. Here, we show that phosphorylation of tandem serines in a conserved SPSP motif of SF1 enhances specific association with its U2AF65 partner and inhibits binding to representative splice-site RNAs. The effect of phosphorylation on the UHM-binding specificity of SF1 is subtle. Considering both the enhanced binding of phosphorylated SF1 to U2AF65 and its diminished association with other UHMs, the net increase in the binding specificity of U2AF65 for phosphorylated relative to unphosphorylated SF1 is typically three- to fourfold. The binding affinities of SF1 for the U2AF35 or CAPERα UHMs are significantly lower than for U2AF65, whereas the SF1 affinity for SPF45 approaches its affinity for U2AF65 (by ∼2-fold). As such, SPSP phosphorylation appears most relevant for avoiding unintended SF1 complexes with SPF45, although other UHMs also could compete for binding the SF1 ULM depending on their local relative abundance.
In a prior EMSA study, phosphorylation of SF1 by SRPK2, which selectively phosphorylates S82 of the SPSP motif as opposed to both S80 and S82, appeared to slightly enhance rather than inhibit SF1 binding to splice-site RNAs containing suboptimal BPSs (47). In contrast, our quantitative fluorescence anisotropy RNA binding assays here demonstrate that phosphorylation of both S80 and S82 by UHMK1 reduces SF1 affinity for a BPSSUBOPT RNA oligonucleotide that shares the same sequence as the major oligonucleotide of the prior study as well as a distinct B3P3-splice-site RNA. Several key differences distinguish our experiments from the prior study. A major distinction is that SRPK2 predominately phosphorylates S82 of human SF1 (47), whereas UHMK1 phosphorylates both serines of the SPSP motif (42, 46). The SPSP motif of SF1 is predominately found in the phosphorylated state in proliferating cells (46), and the majority of SF1 remains phosphorylated after knockout of UHMK1 in mice (48). As such, the SF1 SPSP motif is likely to be regulated by a multitude of different kinases and phosphatases, including, but not limited to, UHMK1 (46), SPRK2 (47), and possibly hPrp4 (61), for a potentially broad variety of distinct functions. Several technical differences also distinguish experiments in the prior study (47) from our experiments here, including the presence of SRPK2 and ATP in the prior binding experiment (rather than purified phospho-SF1 used in our procedures here), use of the isolated SF1 subunit rather than the SF1-U2AF65 complex, different migration of phosphorylated versus unmodified proteins (62), and the non-equilibrium nature of EMSAs, which may be affected by separation of the RNA ligand from the protein partner during the course of electrophoresis.
Thermodynamic changes are challenging to interpret in terms of protein structure, due to the multiple processes linked to the binding event that are simultaneously detected in an ITC experiment. Nonetheless, potential insights may be gleaned by examining the apparent enthalpy and entropy changes of the binding experiments, which are strengthened by comparisons among the variant complexes. The remarkably large magnitudes of the enthalpy and entropy changes for B3P3SUBOPT RNA binding by the SF1-U2AF65 complex (unmodified or phosphorylated) are likely to result from RNA binding by the U2AF65 RRMs, which have characteristically large enthalpy and entropy changes (59). Perhaps coincidentally, the enthalpy and entropy changes for the chimeric SF1pa-U2AF65 complex binding the B3P3SUBOPT RNA resemble those of the second RNA binding site of the unmodified SF1-U2AF65 complex, which appears to represent the different subunits of the complex each binding separate RNAs at high RNA/SF1-U2AF65 molar ratios. This thermodynamic similarity could reflect analogous uncoupling of the SF1 and U2AF65 RNA binding domains by the artificial protein-A replacement of the natural coiled coil.
Consistent with the protein-A domain replacement uncoupling the SF1 and U2AF65 RNA binding domains, cooperative RNA binding by the SF1-U2AF65 complex (as defined by increased RNA binding by the protein complex relative to the individual proteins) is lost in the SF1pa-U2AF65 complex. Specifically, the SF1-U2AF65 affinity for the B3P3SUBOPT RNA is nearly 20-fold greater than the summed affinities of the individual subunits (50E7 M−1 compared to 2.7E7 M−1), whereas the SF1pa-U2AF65 affinity for binding the B3P3SUBOPT RNA is indistinguishable from the summed RNA affinities of the separate subunits (1.2E7 M−1 compared to 1.1E7 M−1). Remarkably, mutation of the arginines at the phosphorylation site to glutamates results in a similar loss of cooperative RNA binding by the SF1mut-U2AF65 complex (1.2E7 M−1 compared to 0.74E7 M−1). Although the substituted domain from protein A is a three-helix bundle rather than a two-helix coiled coil, relatively long (25-residue) glycine-rich linkers were included to permit flexible orientations among the connected domains. Accordingly, the RNA/SF1pa-U2AF65 stoichiometry (as well as the RNA/SF1mut-U2AF65 complex) remains 1:1, supporting the capacity of both subunits to bind the same RNA in these mutated complexes. Although SPSP phosphorylation slightly decreases inter-subunit cooperativity (B3P3SUBOPT RNA affinity of phospho-SF1-U2AF65 is 14.3E7 M−1, compared to 1.5E7 M−1 from the summed RNA affinities of phospho-SF1 and U2AF65), the effect remains relatively subtle (a twofold decrease compared to the unphosphorylated complex). Altogether, these observations suggest that the SF1 coiled-coil domain and arginine-containing SPSP loop actively couple the SF1 and U2AF65 subunits for synergistic recognition of the 3′-splice-site RNA.
The reduced binding affinity of the phospho-SF1-U2AF65 complex for the negatively charged RNA is chemically intuitive, considering that two negatively charged phosphates are introduced in the UHMK1-phosphorylated SPSP motif. The large reduction in the apparent B3P3SUBOPT RNA affinity of SF1mut-U2AF65 relative to the wild-type SF1-U2AF65 complexes (50E7 M−1 compared to 1.2E7 M−1) further suggests that the mutated arginines, which otherwise coordinate the phosphorylated SPSP motif, could interact with RNA in the unmodified state. Given the many possible phosphorylation-dependent processes linked to the RNA binding event, such as global domain orientation and local conformation of the SPSP-containing loop, whether phosphorylation directly interferes with RNA contacts remains an open question. Direct RNA contacts by the arginine-rich SF1 loop in which the SPSP motif resides would reconcile a disparity between natural BPS-Py tract spacings and piecewise SF1-BPS and U2AF65-Py tract structures. The branched intron lariat preferentially forms over a range of ∼18–40 nucleotides from the 3′-splice-site junction (63, 64), and the branchpoint adenosine typically is separated from the start of the Py tract by eight poorly conserved nucleotides (64). Yet the known RNA interactions of the structurally characterized SF1 and U2AF65 domains encompass only three nucleotides after the branchpoint adenosine (65, 66) and nine pyrimidine nucleotides (56, 67), leaving approximately five intervening nucleotides unaccounted for. Based on available structures, the SF1 coiled coil could act as an inert ruler of the BPS-Py tract spacing. In light of the data presented here, we further propose that this SF1 region plays an active role in synergistic positioning of the SF1 and U2AF65 domains for RNA binding. It is possible that phosphorylation of the SF1 SPSP motif by UHMK1 reduces SF1-U2AF65-RNA recognition by diverting these key arginine residues for interactions with the phosphorylated serines as opposed to RNA.
Future experiments are needed to fully resolve the mechanisms underpinning the importance of SPSP phosphorylation for cell proliferation (42). At face value, the observations that SF1 phosphorylation promotes specific binding to its U2AF65 partner, yet reduces binding of the SF1-U2AF65 complex to 3′ splice sites, seem incongruous. One possible explanation is that the phosphorylation of the SF1 SPSP motif before pre-mRNA splicing might both facilitate specific assembly of the phospho-SF1-U2AF65 complex and also inhibit premature association of this complex with RNAs. Co-transcriptional dephosphorylation could then promote loading of SF1-U2AF65 on the emerging pre-mRNA during the early stages of splicing. Accordingly, SF1 associates with U2AF65 in extra-spliceosomal complexes (6), and phosphorylation and dephosphorylation events such as this one are required for pre-mRNA splicing (e.g., (68, 69, 70)). Indeed, the influences of SPSP phosphorylation on any SF1 functions other than UHM and RNA interactions have yet to be examined. The lack of SF1-dependent in vitro splicing substrates rules out biochemical reconstitution assays of pre-mRNA splicing activity. Whether SPSP phosphorylation could regulate SF1 roles in nuclear pre-mRNA retention (2) and transcriptional repression (32, 33) also remains unknown. It further remains possible that SPSP phosphorylation may promote relocalization of SF1 in the cell; the SRPK2 kinase that can phosphorylate S82 in the SPSP motif of human SF1 is known to redistribute the nuclear localization of other splicing factors (71). A precedent for phosphorylation-dependent ULM-protein interactions has been set for the case of ATX1 (45, 72). Phosphorylation of S776 adjacent to the ATX1 ULM slightly reduces its affinity for the U2AF65 or SPF45 UHM (45), comparable to the reduced binding of phospho-SF1 to the SPF45, U2AF35, or CAPERα UHM noted here. Conversely, ATX1 phosphorylation promotes its association with the 14-3-3 signaling protein. By analogy with the ATX1 precedent, a subtle influence of SPSP phosphorylation on SF1-UHM interactions could accompany a more drastic impact on SF1 binding to a hitherto unknown factor(s) in the cellular milieu.
Conclusions
Here, we investigated the consequences of UHMK1 phosphorylation of a conserved SPSP motif of human SF1 for two major classes of interactions: protein-protein interactions with UHM-containing splicing factors and protein-RNA interactions for recognition of the 3′ splice site. First, we confirmed that phosphorylation slightly enhances the binding affinity of SF1 for its cognate UHM partner, U2AF65. We further showed that UHMK1 phosphorylation slightly reduces binding of phospho-SF1 to other UHMs that are unlikely to participate in functional heterodimers, including SPF45, U2AF35, and CAPERα. Altogether, the net effect of phosphorylation is to subtly promote SF1 binding to U2AF65 and disfavor off-target complexes with other UHMs. Next, we quantified the influence of the UHMK1-phosphorylated SPSP motif on SF1 binding to splice-site RNAs. Regardless of the experimental context, including ITC or fluorescence anisotropy methods and the sequence of the splice-site RNA (AdML or suboptimal BPS variants of U6 or B3P3 RNAs), we found that phosphorylation by UHMK1 consistently but subtly reduced the affinities of SF1 or the SF1-U2AF65 complex for binding splice-site RNAs. Replacement of the SF1 coiled-coil domain for an unrelated domain from Staphylococcal protein A, or mutations of arginine residues surrounding the phosphorylated serines, reduced the affinities of SF1 or the SF1-U2AF65 complex for binding a representative, B3P3SUBOPT RNA, which suggests that this SF1 region functions in 3′-splice-site recognition. Moreover, we show that the SF1 coiled coil and arginine residues of the SPSP-containing loop contribute to cooperative RNA recognition by the SF1-U2AF65 complex, and that SPSP phosphorylation reduces this cooperativity. The relatively subtle effects of SPSP-motif phosphorylation on SF1 binding affinities for UHM-containing splicing factors or RNA is likely to be significant at intracellular levels of SF1. Nonetheless, considering that the SF1 SPSP motif is predominately phosphorylated in proliferating cells and is essential for cell proliferation, comprehensive investigations of the potential functions of SF1 SPSP phosphorylation should be pursued in the near future.
Author Contributions
R.C. accomplished the fluorescence anisotropy assays and ITC experiments. W.W., A.G., and S.L. accomplished additional ITC experiments. C.L.K. designed the research and wrote the paper with input from A.M., R.C., W.W., and A.G.
Acknowledgments
We thank Dr. B. Miller, A. Grossfield, J. Jenkins, S. Messing and her colleagues in the Biostatistics department for constructive discussions of our data and analysis.
This work was supported by a National Institutes of Health grant (no. R01 GM070503).
Editor: James Cole.
Footnotes
Wenhua Wang’s present address is Bristol-Myers Squibb, New Brunswick, New Jersey.
Ankit Gupta’s present address is NYU Langone Medical Center, New York, New York.
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)31034-7.
Supporting Material
References
- 1.Abovich N., Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 2.Rutz B., Séraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazroui R., Puoti A., Krämer A. Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA. 1999;5:1615–1631. doi: 10.1017/s1355838299991872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanackovic G., Krämer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toda T., Iida A., Imai T. Isolation and characterization of a novel gene encoding nuclear protein at a locus (D11S636) tightly linked to multiple endocrine neoplasia type 1 (MEN1) Hum. Mol. Genet. 1994;3:465–470. doi: 10.1093/hmg/3.3.465. [DOI] [PubMed] [Google Scholar]
- 6.Rino J., Desterro J.M., Carmo-Fonseca M. Splicing factors SF1 and U2AF associate in extraspliceosomal complexes. Mol. Cell. Biol. 2008;28:3045–3057. doi: 10.1128/MCB.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berglund J.A., Abovich N., Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudner D.Z., Kanaar R., Rio D.C. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol. Cell. Biol. 1998;18:1765–1773. doi: 10.1128/mcb.18.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golling G., Amsterdam A., Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K., Sanada M., Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 11.Graubert T.A., Shen D., Walter M.J. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makishima H., Visconte V., Maciejewski J.P. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berglund J.A., Chua K., Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 15.Zamore P.D., Patton J.G., Green M.R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 16.Wu S., Romfo C.M., Green M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 17.Zorio D.A., Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 18.Merendino L., Guth S., Valcárcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 19.Becerra S., Montes M., Suñé C. Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo. RNA. 2015;21:438–457. doi: 10.1261/rna.047258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwer B., Chang J., Shuman S. Structure-function analysis of the 5′ end of yeast U1 snRNA highlights genetic interactions with the Msl5∗Mud2 branchpoint-binding complex and other spliceosome assembly factors. Nucleic Acids Res. 2013;41:7485–7500. doi: 10.1093/nar/gkt490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang J., Schwer B., Shuman S. Structure-function analysis and genetic interactions of the yeast branchpoint binding protein Msl5. Nucleic Acids Res. 2012;40:4539–4552. doi: 10.1093/nar/gks049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gozani O., Potashkin J., Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thickman K.R., Swenson M.C., Kielkopf C.L. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 2006;356:664–683. doi: 10.1016/j.jmb.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cass D.M., Berglund J.A. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry. 2006;45:10092–10101. doi: 10.1021/bi060429o. [DOI] [PubMed] [Google Scholar]
- 25.Spadaccini R., Reidt U., Sattler M. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haraguchi N., Andoh T., Tani T. Mutations in the SF1-U2AF59-U2AF23 complex cause exon skipping in Schizosaccharomyces pombe. J. Biol. Chem. 2007;282:2221–2228. doi: 10.1074/jbc.M609430200. [DOI] [PubMed] [Google Scholar]
- 27.Shitashige M., Naishiro Y., Yamada T. Involvement of splicing factor-1 in β-catenin/T-cell factor-4-mediated gene transactivation and pre-mRNA splicing. Gastroenterology. 2007;132:1039–1054. doi: 10.1053/j.gastro.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Corioni M., Antih N., Krämer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2011;39:1868–1879. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang Y.H., Park H.Y., Kim J.K. A homolog of splicing factor SF1 is essential for development and is involved in the alternative splicing of pre-mRNA in Arabidopsis thaliana. Plant J. 2014;78:591–603. doi: 10.1111/tpj.12491. [DOI] [PubMed] [Google Scholar]
- 30.Rutz B., Séraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guth S., Valcárcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang D., Paley A.J., Childs G. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J. Biol. Chem. 1998;273:18086–18091. doi: 10.1074/jbc.273.29.18086. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D., Childs G. Human ZFM1 protein is a transcriptional repressor that interacts with the transcription activation domain of stage-specific activator protein. J. Biol. Chem. 1998;273:6868–6877. doi: 10.1074/jbc.273.12.6868. [DOI] [PubMed] [Google Scholar]
- 34.Rain J.C., Legrain P. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q., Zhang L., Rymond B.C. A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast. Nucleic Acids Res. 2008;36:2787–2798. doi: 10.1093/nar/gkn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takemura R., Takeiwa T., Ohno M. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells. 2011;16:1035–1049. doi: 10.1111/j.1365-2443.2011.01548.x. [DOI] [PubMed] [Google Scholar]
- 37.Selenko P., Gregorovic G., Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 38.Kielkopf C.L., Rodionova N.A., Burley S.K. A novel peptide recognition mode revealed by the x-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 39.Kielkopf C.L., Lücke S., Green M.R. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loerch S., Maucuer A., Kielkopf C.L. Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem. 2014;289:17325–17337. doi: 10.1074/jbc.M114.558825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corsini L., Bonnal S., Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 42.Wang W., Maucuer A., Kielkopf C.L. Structure of phosphorylated SF1 bound to U2AF65 in an essential splicing factor complex. Structure. 2013;21:197–208. doi: 10.1016/j.str.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Madl T., Sattler M. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 2013;41:1343–1354. doi: 10.1093/nar/gks1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Bruderer S., Robinson P.J. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 1999;18:4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Chiara C., Menon R.P., Pastore A. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 2009;4:e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manceau V., Swenson M., Maucuer A. Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. FEBS J. 2006;273:577–587. doi: 10.1111/j.1742-4658.2005.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipp J.J., Marvin M.C., Guthrie C. SR protein kinases promote splicing of nonconsensus introns. Nat. Struct. Mol. Biol. 2015;22:611–617. doi: 10.1038/nsmb.3057. [DOI] [PubMed] [Google Scholar]
- 48.Manceau V., Kremmer E., Maucuer A. The protein kinase KIS impacts gene expression during development and fear conditioning in adult mice. PLoS One. 2012;7:e43946. doi: 10.1371/journal.pone.0043946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manceau V., Kielkopf C.L., Maucuer A. Different requirements of the kinase and UHM domains of KIS for its nuclear localization and binding to splicing factors. J. Mol. Biol. 2008;381:748–762. doi: 10.1016/j.jmb.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maucuer A., Le Caer J.P., Sobel A. Specific Ser-Pro phosphorylation by the RNA-recognition motif containing kinase KIS. Eur. J. Biochem. 2000;267:4456–4464. doi: 10.1046/j.1432-1327.2000.01493.x. [DOI] [PubMed] [Google Scholar]
- 51.Wiseman T., Williston S., Lin L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 52.Jenkins J.L., Shen H., Kielkopf C.L. Solution conformation and thermodynamic characteristics of RNA binding by the splicing factor U2AF65. J. Biol. Chem. 2008;283:33641–33649. doi: 10.1074/jbc.M806297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okeyo-Owuor T., White B.S., Graubert T.A. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29:909–917. doi: 10.1038/leu.2014.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Winter, J. C. F. 2013. Using the Student’s t-test with extremely small sample sizes. Pract. Assess. Res. Eval. 18. Available online: http://pareonline.net/getvn.asp?v=18&n=10.
- 55.Gupta A., Jenkins J.L., Kielkopf C.L. RNA induces conformational changes in the SF1/U2AF65 splicing factor complex. J. Mol. Biol. 2011;405:1128–1138. doi: 10.1016/j.jmb.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agrawal A.A., Salsi E., Kielkopf C.L. An extended U2AF65-RNA-binding domain recognizes the 3′ splice site signal. Nat. Commun. 2016;7:10950. doi: 10.1038/ncomms10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullen M.P., Smith C.W., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991;5:642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- 58.Peled-Zehavi H., Berglund J.A., Frankel A.D. Recognition of RNA branch point sequences by the KH domain of splicing factor 1 (mammalian branch point binding protein) in a splicing factor complex. Mol. Cell. Biol. 2001;21:5232–5241. doi: 10.1128/MCB.21.15.5232-5241.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLaughlin K.J., Jenkins J.L., Kielkopf C.L. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–1431. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fukada H., Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 61.Gao Q., Mechin I., Huang S.M. Evaluation of cancer dependence and druggability of PRP4 kinase using cellular, biochemical, and structural approaches. J. Biol. Chem. 2013;288:30125–30138. doi: 10.1074/jbc.M113.473348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taggart A.J., DeSimone A.M., Fairbrother W.G. Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat. Struct. Mol. Biol. 2012;19:719–721. doi: 10.1038/nsmb.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mercer T.R., Clark M.B., Mattick J.S. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015;25:290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Z., Luyten I., Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 66.Jacewicz A., Chico L., Shuman S. Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs. RNA. 2015;21:401–414. doi: 10.1261/rna.048942.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackereth C.D., Madl T., Sattler M. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 68.Wang C., Chua K., Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanuma N., Kim S.E., Shima H. Nuclear inhibitor of protein phosphatase-1 (NIPP1) directs protein phosphatase-1 (PP1) to dephosphorylate the U2 small nuclear ribonucleoprotein particle (snRNP) component, spliceosome-associated protein 155 (Sap155) J. Biol. Chem. 2008;283:35805–35814. doi: 10.1074/jbc.M805468200. [DOI] [PubMed] [Google Scholar]
- 70.Zhou Z., Fu X.D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H.Y., Lin W., Fu X.D. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H.K., Fernandez-Funez P., Zoghbi H.Y. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.