Abstract
Small nucleolar and Cajal body ribonucleoprotein particles (RNPs) are required for the maturation of ribosomes and spliceosomes. They consist of small nucleolar RNA or Cajal body RNA combined with partner proteins and represent the most complex RNA modification enzymes. Recent advances in structure and function studies have revealed detailed information regarding ribonucleoprotein assembly and substrate binding. These enzymes form intertwined RNA–protein assemblies that facilitate reversible binding of the large ribosomal RNA or small nuclear RNA. These revelations explain the specificity among the components in enzyme assembly and substrate modification. The multiple conformations of individual components and those of complete RNPs suggest a dynamic assembly process and justify the requirement of many assembly factors in vivo.
1. Introduction
1.1 RNA-guided modification
The large majority of transcribed mammalian RNA is not translated into proteins, but rather becomes terminal gene expression products known as non-coding RNAs (ncRNAs) (Birney et al. 2007; Hannon et al. 2006; Matera et al. 2007). These ncRNAs participate in a wide range of gene expression and regulation activities that are crucial to cell growth and differentiation. An overwhelming number of ncRNAs act as guide RNAs in processing, chemical modification, degradation or functional interference of other nucleic acids. This is made possible because the guide RNAs form ribonucleoprotein particles (RNPs) with partner proteins. The RNP enzymes use RNA for recognition of the substrate and proteins for catalysis. From a biochemical perspective, RNA-guided enzymes comprise the most complex and fascinating class of modern enzymes.
Among the intensely studied RNA-guided enzymes are small nucleolar RNPs (snoRNPs) and small Cajal body RNPs (scaRNPs). The guide RNA component of these enzymes, snoRNA and scaRNA found in the nucleolus and Cajal bodies, respectively, serve as guides for protein enzymes to target site-specific modification of ribosomal RNA (rRNA) and small nuclear RNA (snRNA). In selected cases, these enzymes also facilitate the cleavage of rRNA (Kiss, 2002; Kiss et al. 2006; Matera et al. 2007; Maxwell & Fournier, 1995; Weinstein & Steitz, 1999).
Sno/scaRNAs are divided into two major types – box C/D and box H/ACA – according to their conserved secondary structural features and their associated modification reactions. The box C/D sno/scaRNAs serve as guides for site-specific 2′-O-methylation, whereas the box H/ACA sno/scaRNAs as guides for pseudouridylation of RNAs (Balakin et al. 1996; Kiss-Laszlo et al. 1996) (Fig. 1).
Fig. 1.
The two major modification reactions catalyzed by RNP modification enzymes: (a) 2′-O-methylation (box C/D RNPs) and (b) isomerization of uridine (pseudouridylation) (box H/ACA RNPs).
The reaction mechanisms of 2′-O-methylation and pseudouridylation have been studied extensively and much has been learned (Foster et al. 2000; Gu et al. 1999; Hamma & Ferre-D’Amare, 2006; Huang et al. 1998; Phannachet et al. 2005; Santi, 2000; Spedaliere et al. 2004). In the methyltransfer reaction, the thio-methyl group of the methyl donor, S-adenosyl-L-methionine (SAM), is transferred to the 2′-OH group with the conversion of SAM to S-adenosyl-L-homocysteine (SAH) (Fig. 1a). Pseudouridine synthases catalyze isomerization of uridine in which the N-glycosidic bond of the targeted uridine is first cleaved and the base is rotated and then reattached through its C-glycosidic bond (Fig. 1b). The resulting isomer, pseudouridine (Ψ), occurs most frequently than any other modified nucleotides and is known as the ‘fifth nucleotide ’ following the four primary nucleotides, A, G, C and U. Together, the box C/D RNPs and box H/ACA RNPs facilitate the most abundant modification reactions in functional RNAs. In archaea, homologs of sno/scaRNAs have been identified and are referred to as small RNAs (sRNAs) (Dennis & Omer, 2005; Omer et al. 2003). In this discussion, the terms ‘box C/D’ or ‘box H/ACA RNA(s)/RNP(s)’, ‘guide RNA(s)’ and ‘ substrate RNA(s)’ are collectively used for simplicity, unless specified otherwise.
In addition to the primary function of modification, several eukaryotic members of the box C/D and box H/ACA RNPs are known to facilitate site-specific cleavage processing of rRNA (Atzorn et al. 2004; Dragon et al. 2002; Eliceiri, 2006; Kass et al. 1990; Liang & Fournier, 1995; Peculis & Steitz, 1993), and one box H/ACA scaRNA is the mammalian telomerase RNA that guides telomere synthesis (Collins, 2006; Mitchell et al. 1999a). These processing RNPs share core components with the modification RNPs; however, they diverge substantially from modification RNPs in assembly and enzyme mechanisms. The focus of this review will be on the structure and function of the modification sno/scaRNPs responsible for methylating and pseudouridylating rRNA and snRNA.
1.2 Biological significance of RNA modifications
Site-specific 2′-O-methylation and isomerization of uridine are only two of more than 100 different types of chemical modifications that occur in various functional RNAs. However, eukaryotic rRNA and snRNAs contain a large number of 2′-O-methylation and pseudouridylation sites, supporting an evolutionary advantage of these two types of modifications. Lack of these modifications in eukaryotic rRNA impairs translation and delays pre-rRNA processing (Baudin-Baillieu et al. 2009; Liang et al. 2007b, 2009b; Piekna-Przybylska et al. 2008). Mis-targeted modification in rRNA negatively impacted ribosome synthasis and activity (Liu et al. 2008). Similar effects on snRNAs have also been observed (Yu et al. 1998; Zhao & Yu, 2004). Modified RNA has greater chemical and folding stability than canonical RNA and extends the chemical reservoir of RNA functional groups. For example, substitution of hydrogen for a methyl group at the 2′ ribose position is protective against cleavage (via alkaline hydrolysis or ribo-nucleolytic attack) and stabilizes the RNA fold by favoring the 3′ endo sugar pucker conformation (Kawai et al. 1992). 2′-O-methylation of adenosine also prevents A-to-I editing, suggesting a regulatory role of 2′-O-methylation (Beal et al. 2007). Similarly, pseudouridylation is believed to improve the stability of RNA by introducing extra hydrogen bond donor at the N1 position of uridine (Durant & Davis, 1999; Newby & Greenbaum, 2002; Yang et al. 2005a).
1.3 Advantages and limitations of RNA-guided modification
The levels of RNA modification in archaea and eukarya are significantly greater than RNA modifications seen in bacteria. Correspondingly, archaea and eukaryotes employ RNP enzymes, while bacteria have protein-only (or stand-alone) enzymes. Stand-alone enzymes are very simple and efficient. In contrast, the RNP enzymes have multiple subunits and require an accurate method of assembly. Binding and release of substrate RNA to RNP enzymes may involve topological barriers commonly found in nucleic acid interactions. It is difficult for a long substrate RNA to thread through the internal loops in order to base pair with the guide RNA. After modification reactions, a significant amount of activation energy is likely required in order to separate the substrate-guide RNA pairing. Thus, RNA-mediated modification faces unique challenges in enzyme assembly, substrate binding and substrate release. On the other hand, inclusion of the guide RNA as a cofactor in RNP modification enzymes allows easy expansion of the target sites without reinventing the protein catalytic subunit. In addition, it is believed that the use of the guide RNA to the nascent rRNA transcripts may prevent misfolding of the ribosome (Weinstein & Steitz, 1999).
2. Sno/scaRNP compositions
2.1 Core components of the box C/D RNP
Box C/D RNAs were first detected by nuclear fractionation as a class of RNAs specifically localized to the nucleolus (Reddy et al. 1981) and immunoprecipitated by the nucleolar protein fibrillarin (Tyc & Steitz, 1989). Box C/D RNAs contain two sets of conserved sequence motifs, box C or C′ (RUGAUGA, where R is any purine) and D or D′ (CUGA). In addition, a conserved stem is formed between the 5′ and 3′ ends of the box C/D RNA that forms a hairpin-like structure (Fig. 2). The internal loop flanked by the box C/D and box C′/D′ motifs is complementary to the sequence of substrate RNA and thus serves as the substrate specificity element. Remarkably, methylation takes place via a ruler mechanism in which the substrate nucleotide that is paired with the fifth nucleotide upstream of box D or D′ is modified. In archaea, an optimal length of the internal loop is found to be 10–12 nucleotides (Tran et al. 2005).
Fig. 2.
Conserved features of RNP modification enzymes. (a) Box C/D MTases comprise box C/D RNA and four core proteins. (b) Box H/ACA pseudouridylases comprise box H/ACA RNA and four core proteins. (c) hTR is a special member of the box H/ACA RNPs without known modification activities. The table lists names of the core proteins associated with each type of RNPs in both eukarya and archaea.
Following the identification of fibrillarin as the box C/D RNA-associated protein (Tyc & Steitz, 1989), genetic synthetic lethal screens with a fibrillarin mutant revealed two additional proteins, Nop56p and Nop58p, that are associated with box C/D RNA (Gautier et al. 1997; Lafontaine & Tollervey, 1999, 2000; Wu et al. 1998). Finally, tag-purification of U3-snoRNP followed by mass spectrometry identification revealed the fourth box C/D RNP protein, Snu13p (15·5 K in mammals and L7Ae in archaea) (Watkins et al. 2000). Fibrillarin, which contains an SAM binding domain, is the subunit responsible for the methyltransfer reaction. Nop56p and Nop58p are homologous to each other and to a single Nop56/58 protein in archaea (Lafontaine & Tollervey, 2000; Omer et al. 2000). These four proteins are found in all members of the box C/D RNPs including those required for rRNA cleavage. Notably, yeast U3 snoRNPs in isolated small ribosome processing complexes contain more than 20 additional proteins that are most likely required for U3-specific functions (Dragon et al. 2002; Grandi et al. 2002; Krogan et al. 2004; Perez-Fernandez et al. 2007).
2.2 Core components of the box H/ACA RNPs
Box H/ACA RNAs were also discovered through the isolation of nucleolus-specific RNAs (Kiss & Filipowicz, 1993), but were immunoprecipitated by another nucleolus protein, Gar1 (Girard et al. 1992). The box H/ACA sno/scaRNAs contains a central unit of the hairpin–hinge–hairpin–tail secondary structure as shown in Fig. 2. In archaea, box H/ACA RNAs can contain one, two, or three hairpin structures, each with an internal loop – the pseudouridine (Ψ) pocket – complementary to a substrate RNA (Rozhdestvensky et al. 2003; Tang et al. 2002). Box H refers to the conserved sequence (ANANNA, N means any nucleotide) in the hinge connecting two hairpin units and box ACA is the conserved trinucleotide sequence at the 3′ tail of the RNA. The uridine targeted for modification is positioned at the center of the Ψ pocket and is typically 14–16 nucleotides away from box H or box ACA.
The first H/ACA RNP protein identified is yeast Gar1p, from a screen for proteins containing glycine-arginine-rich domains similar to that of fibrillarin (Girard et al. 1992). Purification of Gar1p-bound RNPs revealed H/ACA RNAs and three additional proteins associated with H/ACA RNAs (Henras et al. 1998; Lafontaine et al. 1998; Watkins et al. 1998). In yeast, these correspond to Cbf5p (Dyskerin in human, NAP57 in mice, Cbf5 in archaea), Nhp2p (NHP2 in mammals, L7Ae in archaea) and Nop10p (Fig. 2). Cbf5p contains two of the three hallmark sequence motifs of pseudouridine synthases (motifs I and II) including the strictly conserved asparate residue in motif II (Koonin, 1996) and is thus the putative catalytic subunit. Nhp2p shares sequence similarity with the box C/D core protein, 15·5 kDa/Snu13p/L7Ae, but lacks K-turn binding specificity. This homology suggests that the two classes of sno/scaRNPs share a common origin. All four proteins are also components of the human telomerase (hTR) and are required for the biogenesis of hTR (Collins, 2006).
Mutations of the protein components of sno/scaRNPs have been implicated in multiple human disorders. However, it has not been firmly established whether these mutations cause defects in the modification function itself or other functions requiring the sno/scaRNP proteins. Fibrillarin, the enzymatic component of box C/D RNPs, is essential for development and its depletion is embryonic lethal (Newton et al. 2003). Mutations in the human dyskerin gene DKC1 (Cbf5) (Heiss et al. 1998; Marrone & Mason, 2003; Meier, 2005), NHP2 (Vulliamy et al. 2008) and NOP10 gene (Vulliamy et al. 2008; Walne & Dokal, 2008) are associated with the X-linked genetic disorder, dyskeratosis congenita (DC). This genetic disease is characterized by cutaneous pigmentation, bone marrow failure, nail dystrophy, white potentially malignant patches on the oral mucosa, continuous tear production and often abnormally low blood platelet counts, anemia and predispositions to epithelial cancers (Marrone & Mason, 2003). Importantly, the box H/ACA core proteins are also essential to hTR biogenesis (Collins, 2006; Mitchell et al. 1999b). They are associated with the 3′-end of the telomerase RNA and are required for the accumulation, 3′-end processing and nuclear localization of hTR in Cajal bodies. In DC patients, malfunction of rRNA and shortening of telomeres have been observed (Marrone et al. 2005; Mochizuki et al. 2004).
3. Substrates of sno/scaRNPs
In mammals, substrates for sno/scaRNPs include both snRNAs and rRNAs (but not tRNAs) and the modification reactions take place in the nucleolus and Cajal body, respectively. In archaea, which has no snRNA, substrates for sno/scaRNP homologs include tRNA and rRNA.
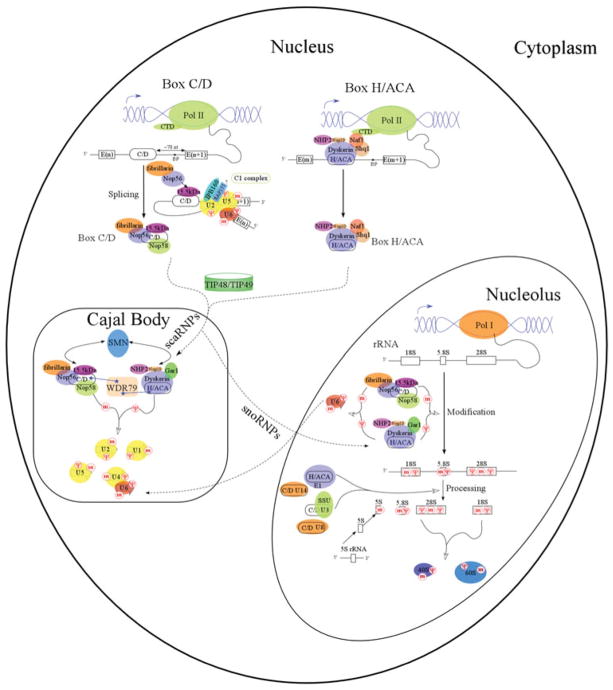
Chemical modification of snRNA by sno/scaRNPs is one of the many steps required for spliceosome maturation (Fatica & Tollervey, 2002; Hage & Tollervey, 2004; Kiss, 2006; Matera et al. 2007). Following their transcription, snRNAs are sequentially processed, assembled with essential spliceosome proteins, recruited to the nucleolus or Cajal body and finally modified by sno/scaRNPs. U1, U2, U4 and U5 snRNAs are transcribed by Pol II, assembled with the Sm ring, 3′-end processed, and 5′-end cap hypermethylated in the cytoplasm before being recruited to Cajal bodies for modification by box C/D and box H/ACA RNPs (Fig. 3). U6 snRNA is uniquely transcribed by RNA polymerase III (Pol III) in the nucleus, end processed and assembled with Lsm protein(s) in the cytoplasm and modified by box C/D and box H/ACA snoRNPs in the nucleolus. The mature U6 snRNP is then translocated into Cajal bodies where it forms the U4/U6 di-snRNP with U4 snRNP. The mature snRNPs perform intron splicing at perichromatin fibrils (PFs) or are stored in interchromatin granule clusters (IGCs) (Matera et al. 2007).
Fig. 3.
Expression and biogenesis of eukaryotic box C/D and box H/ACA RNPs. Key expression and biogenesis pathways and components are indicated. Mammalian box C/D and H/ACA RNAs are largely encoded as introns of functional genes. ‘E’ denotes ‘exon’ and ‘BP’ denotes the splicing branch point. Cajal body is the site of spliceosomal RNA modifications and nucleolus is the site of ribosomal RNA modifications. ‘m’ indicates 2′-O-methylation and ‘Ψ’ indicates pseudouridylation.
Ribosome maturation also has many steps, beginning with pre-rRNA transcription. The pre-rRNA, including 18S, 5·8S and 25/28S rRNAs, is transcribed by RNA polymerase I (Pol I) as a single transcript that is modified and cleaved by snoRNPs in the nucleolus. 5S rRNA is transcribed by RNA Pol III and also modified/cleaved by box C/D snoRNPs in nucleolus (Fig. 3). Chemically modified Pol I transcripts then undergo several site-specific cleavage steps in vertebrates that are facilitated by the processing snoRNPs, U3 and U8, along with U14 and a box H/ACA snoRNP, E1/U17 (snR30 in yeast) (Atzorn et al. 2004; Eliceiri, 2006; Gerbi, 1995; Maxwell & Fournier, 1995; Tollervey & Kiss, 1997). These cleavage steps are followed by further trimming by exonucleases, leading to the 5·8S, 18S and 25S/28S rRNAs (Tollervey & Kiss, 1997). The processed 18S rRNA begins to assemble with ribosomal proteins in the nucleolus to form the small pre-40S ribosomal subunit, whereas 5·8S, 28S rRNAs, together with 5S rRNA and ribosomal proteins form the large pre-60S ribosomal subunit. The pre-40S and pre-60S subunits are then transported separately through the nuclear membrane to the cytoplasm, where final maturation and protein translation take place.
In archaea, tRNAs can be methylated by box C/D RNPs (Clouet d’Orval et al. 2001; Joardar et al. 2008; Singh et al. 2008). In another example, Cbf5 alone or with Nop10 and Gar1 can function as a tRNA Ψ 55 synthase in a guide RNA-independent manner (Gurha et al. 2007).
4. Biogenesis of the sno/scaRNPs
4.1 Maturation of the sno/scaRNAs
The C/D and H/ACA RNAs are maturated through elaborative pathways that generally include transcription, 5′ and 3′ processing and localization. This process requires a number of biogenesis factors and depends on other RNA functional processes (Kiss et al. 2006; Matera et al. 2007).
The majority of mammalian box C/D and box H/ACA sno/scaRNAs responsible for guiding modifications are transcribed by RNA polymerase II (Pol II) as pre-mRNA introns (Fig. 3). Therefore, mRNA splicing also influences the generation of linear sno/scaRNAs precursors. Most box C/D RNAs are located 65–85 nucleotides upstream of the 3′ splice site and ~50 nucleotides upstream of the branch site (Hirose & Steitz, 2001), whereas H/ACA RNAs have no preferential intronic location relative to the 5′ or 3′ splice sites of the host intron (Richard et al. 2006; Schattner et al. 2006). Correspondingly, the processing of box C/D RNAs but not box H/ACA RNAs depends on general splicing factors (Hirose et al. 2006). The intronic sno/scaRNAs undergo debranching and exonucleolytic trimming of both 5′- and 3′-ends (Allmang et al. 1999). In lower eukaryotes, such as yeast and plants, sno/scaRNAs are processed from both non-intronic and splicing-independent intronic transcripts (Chanfreau et al. 1998; Leader et al. 1999; Qu et al. 1999).
Processed box C/D and box H/ACA RNAs are associated with either all or most RNP core proteins and are translocated into Cajal bodies before the mature snoRNPs relocate to nucleolus (Fig. 3). Factors or processes that facilitate the orderly localization events are yet to be completely characterized. Transport of human U3 snoRNA was shown to be facilitated by the snRNA export adaptor, PHAX, and the export receptor, CRM1 (Boulon et al. 2004). On the other hand, a conserved sequence box (CAB box) is responsible for the retention of scaRNAs in Cajal bodies through its specific interaction with a WD40 protein, WDR79. In general, the WD40 family of proteins has a circularized beta-propeller structure and forms platforms for the assembly of protein complexes associated with a wide range of biological pathways. WDR79 has been found to interact directly with the CAB box of both box C/D and box H/ACA scaRNAs, as well as telomerase RNA in Cajal bodies (Jady et al. 2004; Tycowski et al. 2009; Venteicher et al. 2009).
4.2 Assembly of sno/scaRNPs
The processed sno/scaRNAs are assembled with core proteins at the stage of pre-mRNA splicing or co-transcriptionally in order to avoid the degradation pathway that is known to target non-functional introns. A number of trans-acting factors have been identified that affect the assembly of mammalian sno/scaRNPs (Boulon et al. 2008; King et al. 2001; Newman et al. 2000; Venteicher et al. 2008), suggesting the existence of conformational barriers to achieve assembly. This property may explain why in vitro reconstituted eukaryotic sno/scaRNPs have no enzymatic activities (Maxwell, E. S. and Meier, U. T., personal communications) and the archaeal particles require high temperature to activate assembly and enzyme activities (Baker et al. 2005; Charpentier et al. 2005; Gagnon et al. 2006; Omer et al. 2002).
Box C/D sno/scaRNPs assembly begins at the C1 splicing complex stage at which the first transesterification reaction takes place with the recruitment of the 15·5 K protein (Hirose et al. 2003). The 15·5 K/NHPX protein interacts with box C/D RNA in a splicing-dependent manner (Hirose et al. 2003). In addition, in vitro studies with its archaeal homolog, L7Ae, identified this protein as the initiator of box C/D RNP assembly (Rashid et al. 2003; Tran et al. 2003). Thus, box C/D RNP assembly likely begins, while the spliceosome is still assembled around the pre-mRNA. In vivo chemical cross-linking experiments place 15·5 K/NHPX, fibrillarin and Nop58 close to the terminal box C/D motif and fibrillarin and Nop56 to the box C′/D′ motif (Cahill et al. 2002).
In vivo assembly of box H/ACA sno/scaRNPs is found to begin during transcription elongation (Richard et al. 2006) and depends on several conserved trans-acting factors (Grozdanov et al. 2009; Kittur et al. 2006; Yang et al. 2002, 2005b). The most extensively characterized factors include Naf1 and Shq1. Naf1, a protein specifically bound to the C-terminal domain (CTD) of Pol II, is associated with newly transcribed box H/ACA RNAs and dyskerin/Cbf5. Naf1 contains a domain structurally homologous to Gar1 and is believed to compete with Gar1 for binding to dyskerin/Cbf5 (Darzacq et al. 2006; Leulliot et al. 2007). Shq1 is another factor that is required for the accumulation of box H/ACA RNAs and forms a complex with Naf1 (Yang et al. 2002). The CTD of Shq1 is also shown to bind Cbf5 and have stand-alone chaperone activity in vitro (Godin et al. 2009). In addition, general chaperones and nucleic acid-associated ATPases are found to facilitate box H/ACA RNP assembly (Kiss et al. 2010).
5. RNA-guided versus stand-alone modification enzymes
5.1 Methyltransferases and fibrillarin
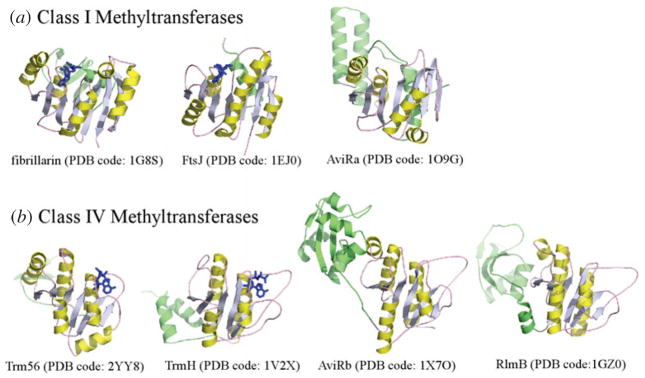
Methyltransferases (MTases) are found as proteins that function independently (stand-alone) or as ribonucleoprotein particles that rely upon RNA. The RNA-guided MTases have thus far been found to strictly target RNA ribose modification, while stand-alone enzymes target a much wider range of substrates and functional groups including proteins, DNA, RNA (ribose and base constituents), lipids and small molecules for methylation. In general, MTases can be SAM-dependent and SAM-independent. From primary structural features, and regardless of need for an RNA guide, the SAM-dependent MTases are divided into five classes (Class I–V MTases) (Schubert et al. 2003), with most of the 2′-O-MTases belonging to Class I and Class IV. The Class I MTases share little sequence identity, but have a common structural domain comprising a central seven β-sheet mixed with α-helices (Fig. 4a) (Cheng & Roberts, 2001; Martin & McMillan, 2002). The Class IV MTases, also known as SPOUT (SpoU-TrmD) superfamily MTases, exhibit a Rossmann α/β-fold with a deep trefoil knot (Anantharaman et al. 2002; Nureki et al. 2002) (Fig. 4b). The difference in the central domain fold reflects the different substrate specificity. Fibrillarin, the MTase in sno/scaRNPs, was found to have structural homologies to Class I bacterial SAM-dependent MTases. These include FtsJ (PDB code: 1EJ0) and AviRa (PDB code: 1O9G) (Bugl et al. 2000; Mosbacher et al. 2003) (Fig. 4a).
Fig. 4.
Fibrillarin resembles class I MTases. (a) Representative Class I MTase and fibrillarin structures. (b) Representative class IV MTase structures. The methylation donor, SAM, is colored in blue and the MTases are colored in yellow and green.
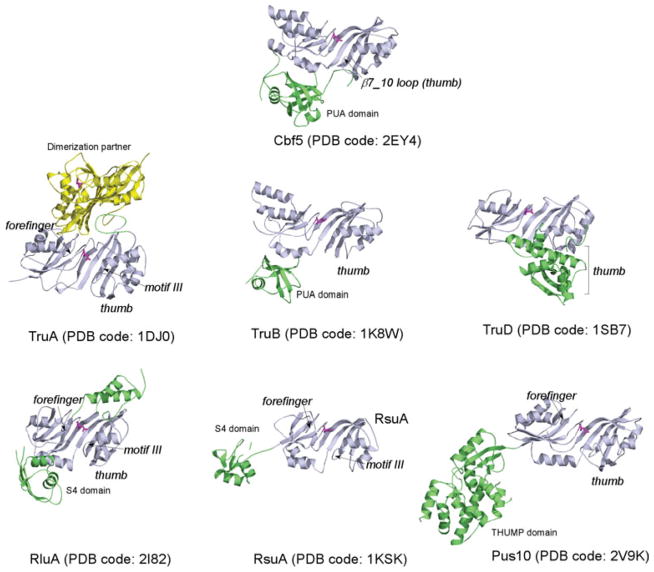
5.2 Pseudouridine synthases and Cbf5
In contrast to the widespread occurrence of methylation, pseudouridylation is limited to RNA molecules only. However, at least six families of pseudouridine synthases have been discovered that target one or few specific uridine(s) in each given tRNA or rRNA. Accordingly, they are named TruA, TruB, TruD, RsuA, RluA and Pus10, respectively. The Pus10 family is found in archaea and eukaryotes, but not in bacteria (McCleverty et al. 2007). Structures of all six families of pseudouridine synthases are available and clearly indicate a conserved catalytic core among them (Hamma & Ferre-D’Amare, 2006) (Fig. 5). Crystal structures of Cbf5 have been solved either as ternary (Cbf5-Nop10-Gar1) or binary (Cbf5-Nop10) complexes (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006) and these show that Cbf5 closely resembles TruB. In both Cbf5 and TruB, the catalytic domain comprises a nine strand β-sheet surrounded by α-helices and encompassing the active site which includes the strictly conserved aspartate. The C-terminal PseudoUridine synthase and Archaeosine transglycosylase (PUA) domain forms a mixed α/β-folding unit (Fig. 5). The PUA domain can interact with different RNA (Perez-Arellano et al. 2007), and in the case of Cbf5, it specifically interacts with the strictly conserved ACA trinucleotides of the guide RNA.
Fig. 5.
Cbf5 resembles the TruB-family of pseuoduridine synthases. Other known pseudouridylase structures are included for comparison. Key RNA-binding features are labeled where they are present. The catalytic domain is colored in light blue with a magenta catalytic aspartate and accessory domains are in green or yellow.
Stand-alone pseudouridine synthases have evolved a variety of elements to interact with the substrate RNA. These include peripheral domains for anchoring the main body of the substrate, a thumb loop, a forefinger and a motif III loop for gripping the target uridine and adjacent nucleotides (Hamma & Ferre-D’Amare, 2006). The target uridine is flipped out and tucked deep into the catalytic pocket so the catalytic residues can reach the pyrimidine base. In all enzymes, the adjacent nucleotides are remodeled in ways that depend on the RNA interaction elements in each enzyme. Dyskerin, the human Cbf5 homolog, contains extra N-terminal and C-terminal regions, but there is no structure available yet. Cbf5 lacks the forefinger and has a very short thumb loop (Fig. 5). The reduction in RNA-binding elements reflects the fact that Cbf5 functions with three other proteins and the guide RNA as discussed below.
6. Structures of box C/D RNPs
6.1 Overview
Structural studies of box C/D RNPs have been carried out with the simpler archaeal homologs. Two crystal structures of box C/D RNPs containing all core proteins and a halfmer box C/D RNA (a single C/D module) have been obtained and one of these also contains a bound substrate RNA (Xue et al. 2010; Ye et al. 2009). Due to limitations in crystallization, these RNP complexes are halfmer RNPs that are not active in methylation under normal in vitro experimental conditions. However, these structures have provided valuable information on protein–protein and protein–RNA interactions in an intact and functional box C/D RNP. Most significantly, when the high-resolution halfmer structural data are combined with the large body of biochemical data and an electron microscopy (EM) structure of the RNP containing full-length box C/D RNA (Bleichert et al. 2009), an appealing working model of box C/D function has emerged.
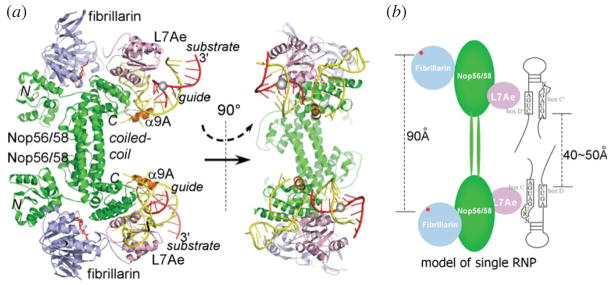
Each independent C/D RNP unit is organized around Nop56/58. Nop56/58 comprises an N-terminal domain, a coiled-coil domain and a CTD. Fibrillarin is bound to its N-terminal domain and this interaction is independent of RNA and L7Ae. The box C/D RNA–L7Ae complex, which forms independently of Nop56/58, is docked to the CTD of Nop56/58. Finally, the coiled-coil domain of Nop56/58 mediates self-dimerization of Nop56/58, leading to a homodimer of Nop56/58-fibrillarin-L7Ae-box C/D RNA complexes (Fig. 6a). The dyad symmetry of Nop56/58 was the basis for the initial proposal that the intact box C/D RNP has a bipartite architecture with the hairpin box C/D RNA placed along the Nop56/58 dimer (Fig. 6b) (Aittaleb et al. 2003; Oruganti et al. 2007).
Fig. 6.
Overview of the box C/D RNP MTases containing a halfmer box C/D RNA (yellow) and a bound substrate RNA (red) in ribbon (a) and cartoon (b) representations. (PDB code: 3NMU) (a) Nop56/58 is colored in green, L7Ae is in pink and fibrillarin in light blue. SAM is colored in red. Three domains of Nop56/58 are labeled as N (N-terminal), C (C-terminal) and coiled-coli domains, respectively. The Nop56/58 helix responsible for separating the two guide strands is colored in orange. The target nucleotide is marked by a gray sphere. (b) The classic bipartite assembly model based on the halfmer RNP structure indicates that the catalytic subunits will be too far apart (90 Å) to match the potential locations of the target nucleotides (40–50 Å).
The bipartite box C/D RNP model is met with several challenges. First, the length of an intact box C/D RNA is estimated to be shorter than the length of the protein platform in the complex, which is established by the long coiled-coil connector domains of Nop56/58. Second, it is expected to be topologically challenging for a large substrate RNA to form more than 10 bp with the central internal loop whose size is restricted by flanking proteins (Fig. 6b). The recent EM structure of a box C/D RNP containing an intact guide RNA revealed a surprising dual RNP model that changed the view on the traditional box C/D RNP assembly (Bleichert et al. 2009) and provided the basis for construction of a substrate-bound atomic model of diRNP (Xue et al. 2010) (Fig. 7). In the diRNP model, the box C/D RNA is poised to bind across the two RNPs instead of along a single RNP (Fig. 7). This model alleviates the constraint on the box C/D RNA in the traditional single RNP model shown in Fig. 6b without disrupting the known protein–protein and protein–RNA interactions. The diRNP model, though, has not yet been confirmed in vivo.
Fig. 7.
The substrate-bound diRNP model of box C/D MTase (same color scheme as in Fig. 6). (a) The atomic diRNP model built from the crystal structure of the halfmer RNP is consistent with the EM density of the fully assembled diRNP (Bleichert et al. 2009) (EMD-1643). Fibrillarin and the N-terminal domain of Nop56/58 could not be fitted, reflecting their observed variable positioning. Single letters refer to protein subunits. ‘N’ denotes Nop56/58, ‘F’ denotes fibrillarin and ‘L’ denotes L7Ae. (b) The substrate-bound diRNP model in surface representation. (c) Cartoon representation of the substrate-bound diRNP model.
6.2 Substrate recognition
The hallmark of the box C/D RNP MTases is the stringent specificity for the target nucleotide paired with the fifth nucleotide upstream of the guide box D or D′ (n+5 rule), suggesting an importance of the first five base pairs between the guide and substrate RNAs. The only available substrate-bound RNP structure is that of a halfmer box C/D RNP bound to a 13mer substrate RNA (Xue et al. 2010). In the substrate-bound structure, the guide-substrate duplex is sandwiched between a conserved Nop56/58 helix, α9A and L7Ae (Fig. 6a). The α9A helix is wedged between the guiding and non-guiding strands, creating alternating layers of protein and RNA elements. The unpaired 5′ nucleotides of the substrate RNA rest on the C-terminal end of the α9A, which further helps to direct the substrate RNA (Fig. 6a).
Surprisingly, this structure reveals that the substrate RNA is bound far from the nearest catalytic subunit fibrillarin (Fig. 6a, b) and can thus not be modified in the halfmer RNP. One solution to this problem is formation of the diRNP similar to that observed by EM(Bleichert et al. 2009) in which the substrate bound to one halfmer RNP may be easily placed into the active site of the opposing halfmer RNP, thereby suggesting a cross-RNP catalysis strategy (Fig. 7c). Therefore, the diRNP model provides topological solutions not only to the discrepancy in length between proteins and the guide RNA, but also to the problem of placing the substrate RNA close to the catalytic subunit.
6.3 Induced fit in box C/D RNP assembly
6.3.1 Induced fit formation of RNA guiding fork
Box C/D RNP structures reveal extraordinary structural changes during assembly and substrate binding. These changes imply the occurrence of dynamic processes in box C/D RNP function. The α9A helix of Nop56/58 important for substrate placement is the most conserved element in the entire region of Nop56/58. This region contains the strictly conserved motif comprising Gly-Ala-Glu-Lys tripeptide (GAEK motif). Interestingly, the structure of this ~20 amino acid region had not previously been observed in either RNA-free or RNA-bound structures, because it was disordered (Aittaleb et al. 2003; Oruganti et al. 2007; Ye et al. 2009) until a proper guide RNA was included (Xue et al. 2010). The mode of α9A folding explains the importance of the guide RNA in its stabilization. α9A and its C-terminal extension are wedged between the guide and the non-guide strands with nucleotides wrapping around the protein (RNA fork) (Fig. 6a). Conversely, the formation of α9A also leads to the separation of canonical RNA base pairs (Fig. 6a).
In addition to its role in substrate placement, the induced fit formation of the RNA fork has important implications in specific assembly of box C/D RNPs. Interesting comparisons have been drawn between the eukaryotic box C/D RNP and the spliceosomal 15·5 kDa–U4 snRNA complex. Mammalian U4 snRNA stem II loop binds 15·5 K protein similarly to box C/D RNA binding to 15·5 K (Watkins et al. 2000). However, U4 snRNA–15·5 K complex recruits the splicing protein Prp31, while box C/D RNA-15·5 K recruits Nop56 or Nop58 protein. Prp31 shares the same RNA binding domain, called NOP domain, with Nop56/Nop58 but lacks the GAEK motif. Furthermore, U4 snRNA stem II has a pentaloop in place of the open end in box C/D RNA and is, therefore, unable to form the RNA fork. The observed differences in the RNA secondary structure and in the GAEK motif explain the homologous but distinct assemblies of box C/D RNP and U4 snRNP.
6.3.2 RNA kink-turn specifies the order of assembly
Another observed structural change takes place at the box C/D motif and this change is required for sequential binding of box C/D proteins to the RNA. L7Ae or its eukaryotic homolog 15·5 K binds to the box C/D motif and initiates box C/D RNP assembly (Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003; Watkins et al. 2000). L7Ae-box C/D RNA complex structures revealed a sharp kink in the RNA (K-turn) as a result of a tandem GA pairs and three bulged nucleotides (Fig. 8a). Biophysical studies clearly demonstrated L7Ae’s ability to actively remodel the RNA (Goody et al. 2004; Suryadi et al. 2005). In contrast to RNA, L7Ae itself does not undergo conformational change and rigidly docks its conserved αβα fold onto the K-turn (Oruganti et al. 2005) (Fig. 8a).
Fig. 8.
Conformational changes in box C/D RNP assembly inferred from structural studies. (a) L7Ae induces formation of kink-turn by the conserved box C/D sequences. Similar kink-turns are also observed in H/ACA RNA and U4 snRNA. (b) Multiple conformations of fibrillarin are captured in crystal structures of the RNA-free Nop56/58-fibrillarin complexes indicate a hinge motion (indicated by arrows) in Nop56/58 that can place fibrillarin.
L7Ae–K-turn interaction is not unique to box C/D RNP assembly. L7Ae is itself a ribosomal protein and has a significant sequence and structural homology to eukaryotic 15·5 K/Snu13p (box C/D and spliceosomal protein), Nhp2p (box H/ACA protein), L30 (ribosomal protein), and SBP2 that are found to interact with similar RNA motifs (Allmang et al. 2002). Comparison with the available structures of L7Ae and its homologs bound with their respective substrates did not reveal obvious structural differences that explain their association with different complexes (Chao & Williamson, 2004; Hamma & Ferre-D’Amare, 2004; Moore et al. 2004; Vidovic et al. 2000) (Fig. 8a). It is possible that differences in the dynamics of binding contribute to their physiological specificity. In higher-order complexes, as illustrated in the box C/D and box H/ACA RNP assemblies, specificity is further defined by subsequently interacting proteins.
6.3.3 Multiple protein conformations imply a hinge motion that repositions fibrillarin
A wide range of fibrillarin positions are observed in both RNA-free and RNA-bound Nop56/58-fibrillarin complexes from three archaeal species (Aittaleb et al. 2003; Oruganti et al. 2007; Ye et al. 2009) (Fig. 8b). The difference in location is not a result of using different species because the position of fibrillarin of the same species was also demonstrated to vary (Ye et al. 2009). Fibrillarin interacts exclusively with the N-terminal domain of Nop56/58 that is tethered to its coiled-coil domain by a peptide linker. All observed fibrillarin positions are a result of simple hinge motions of the peptide linker (Fig. 8b). Computational normal mode analysis of one Nop56/58–fibrillarin complex yielded low-frequency motions of the hinge consistent with its experimentally observed conformations (Oruganti et al. 2007). The flexibility of the peptide linker in placing the catalytic subunit provides a means for fibrillarin to gain access to the target nucleotide. Strikingly, the position occupied by the Archaeoglobus fulgidus fibrillarin (Fig. 8b) is nearly perfect for methylation in the diRNP model (Xue et al. 2010). It is thus believed that in order for substrates to bind and to be modified, fibrillarin, and possibly L7Ae, need to be repositioned (Xue et al. 2010).
7. Structures of box H/ACA RNPs
7.1 Overview
Studies on H/ACA RNP structures have also been carried out with archaeal homologs reconstituted from recombinant and synthetic components. These studies not only provide molecular details for understanding the biochemical mechanisms of H/ACA RNPs, but also shed light on the assembly architecture of hTR holoenzyme.
In vitro biochemical studies of box H/ACA RNPs reveal that three of the four proteins are essential to pseudouridylation, while the fourth protein, Gar1, significantly improves the enzyme activity (Baker et al. 2005; Charpentier et al. 2005). Both Cbf5 and L7Ae interact with box H/ACA guide RNAs directly, while Nop10 and Gar1 interact with the guide RNAs indirectly through their interactions with Cbf5 (Baker et al. 2005; Charpentier et al. 2005). Structures of the box H/ACA RNP, especially those containing substrate RNA, provide the structural basis for these biochemical observations.
The structure of a substrate-free archaeal box H/ACA RNP containing all four proteins, Cbf5, Nop10, Gar1 and L7Ae and a single-hairpin H/ACA RNA revealed the first complete view of box H/ACA RNP architecture (Fig. 9) (Li & Ye, 2006). Cbf5 is the master organizer that interacts extensively with the guide RNA, Nop10 and Gar1. Nop10 is a small protein, containing an N-terminal zinc binding domain of four packed β-sheets, a C-terminal single helix domain and a linker loop. Nop10 is anchored against the catalytic domain of Cbf5. This interaction is required for its stabilization because Nop10 is significantly unfolded when in isolation (Hamma et al. 2005). Gar1 adopts a six β-strand barrel fold that is similar to that found in some RNA-binding proteins such as the translation elongation factor EF-Tu (Rashid et al. 2006). One study has found that Gar1 interacts with snoRNA directly in vitro (Bagni & Lapeyre, 1998) and a cross-link study placed mammalian Gar1 close to the substrate RNA (Wang & Meier, 2004). In the crystal structure of the box H/ACA RNP, it, however, binds to one side of Cbf5 that is away from the RNA. The guide RNA lies linearly along the protein surface comprised of Cbf5, Nop10 and L7Ae without contacting Gar1. The two ends of the box H/ACA RNA are secured by two specific RNA–protein interactions. The conserved ACA sequence of the guide RNA at the 3′-end are recognized specifically by the PUA domain of Cbf5 (Fig. 9) and the apical K-loop is recognized in the same manner as the box C/D motif by L7Ae (Fig. 8a). L7Ae also interacts with Cbf5-bound Nop10, which brings the upper stem of the guide RNA in close contact with Cbf5 and Nop10. Comparison of crystal structures of either binary or ternary protein complexes reveals no gross conformational changes in protein upon association with the guide RNA (Rashid et al. 2006), suggesting that proteins are rigidly docked to the guide RNA.
Fig. 9.
Overall view of the substrate-bound archaeal box H/ACA RNP (PDB code: 2HVY). Each subunit is labeled. The specific interaction between the trinucleotide ACA and the PUA domain of Cbf5 is highlighted.
7.2 Substrate recognition
Significant details are revealed by substrate-bound RNP structures containing different H/ACA proteins (Duan et al. 2009; Liang et al. 2007a, 2009a). Comparing these with the stand-alone pseudouridine synthase–RNA complexes (Alian et al. 2009; Hoang et al. 2006; Hoang & Ferre-D’Amare, 2001; Hur & Stroud, 2007; Pan et al. 2003) provides even greater insight. In all these studies, the target uridine is substituted by 5-fluorouridine, which the enzyme converts to 5-fluoro, 6-hydroxypseudouridine (5FhΨ). It is believed that extraction of the fluorine atom (or proton in uridine) and the C6 hydroxyl from 5FhΨ would lead to the final isomerized product. The gross features of enzyme–substrate interactions observed in this reaction intermediate complex are expected to remain throughout the reaction. However, detailed interactions between the substrate and the active site residues are expected to vary at different stages of the reaction.
Box H/ACA RNPs recruit substrates through base pair interactions between the guide and the substrate RNA. By pairing with both strands of the internal loop, the substrate RNA forms a three-way junction (Ω-like motif) with the guide RNA (Fig. 10a). This Ω-like motif is unconstrained in the absence of proteins (Jin et al. 2007; Wu & Feigon, 2007) but significantly remodeled when bound to proteins such that the target uridine and two adjacent nucleotides are sharply kinked (Duan et al. 2009; Liang et al. 2007a; Liang et al. 2009a) (Fig. 10a). The 3′ guide–substrate duplex mimics the TΨC stem of tRNA recognized by TruB. Like TruB, the thumb loop (β7_10 loop) interacts with the major groove of the stem and the motif III loop catches the approaching substrate kink (Fig. 10a). The substituted target nucleotide is converted to 5FhΨ and secured deep inside the active site pocket by a network of interactions with the catalytic residues (Fig. 10a) (Duan et al. 2009; Liang et al. 2009a). In both TruB and box H/ACA RNP, the thumb loop is open in the absence of substrate RNA but is closed in its presence (Duan et al. 2009; Hoang et al. 2005; Liang et al. 2009a). The short thumb loop of Cbf5 interacts only with one nucleobase, that of the +1 nucleotide, and two phosphate groups, those of the +1 and −4 nucleotides (the target nucleotide is assumed to be position 0) (Fig. 10a).
Fig. 10.
Substrate binding to box H/ACA RNP. (a) Overview of the substrate-bound box H/ACA RNP (PDB code: 3HJW or 3HAX). The asterisk indicates that Gar1 is modeled for the substrate-bound complex according its position in substrate-free RNP complex (PDB code: 2HVY) or that of the low-resolution structure of the fully assembly complex (PDB code: 3HAY). Interactions between the target nucleotide (5FhΨ) and the two flanking nucleotides are highlighted in the right upper panel. Protein residues are labeled according to Pyrococcus furiosus Cbf5 numbering. The right lower panel indicates the open and closed conformations of the β7_10 loop (thumb loop) of Cbf5 that accompany docking of the substrate RNA. (b) Comparison of guide-target RNA duplexes in protein-free (PDB code: 2PCW and 2P89), all protein-(+L7Ae) (PDB code: 3HJW), or those without L7Ae (−L7Ae)-bound (PDB code: 2RFK) states.
Owing to its dependence on the guide RNA, the RNA substrate of a box H/ACA RNP is therefore also influenced by proteins beyond the catalytic subunit. This is best illustrated by comparing the substrate-bound RNP structures and substrate fluorescence intensities in the presence and absence of L7Ae or Gar1 (Duan et al. 2009; Liang et al. 2009a). L7Ae interacts with the upper stem of the guide RNA adjacent to Nop10. In the absence of L7Ae, the upper stem is released from its anchored position, which leads to the departure of the substrate RNA from the active site (Liang et al. 2007a) (Fig. 10b).
The structural effect of Gar1 on substrate binding is not only more subtle, but also complex. The absence of Gar1 only slightly shifts the upper stem of the guide RNA (Liang et al. 2009a). Fluorescence studies on substrate docking showed that addition of Gar1 to the otherwise fully assembled RNP caused an initial undocking followed by redocking of the substrate RNA (Liang et al. 2008). These observations are consistent with the non-essential but accelerating role of Gar1 in pseudouridylation reactions (Baker et al. 2005; Charpentier et al. 2005; Muller et al. 2007). The fact that Gar1 does not directly contact either the guide or the substrate RNA indicates that Gar1 functions through its interaction with Cbf5, especially the Cbf5 thumb loop.
The flexibility of the guide RNA in directing substrate binding and modification has also been investigated. Comparing secondary structures of the available H/ACA guide RNAs indicates that the distance between the target uridine and the ACA trinucleotide may vary as a result of different numbers of paired bases in the 3′ substrate-guide helix (SH1) or in the guide RNA lower stem (P1) (Figs 1 and 10a). Comparing structures having two different distances (14 and 15 bp) in the P1 and SH1 revealed virtually no difference around the target uridine (Duan et al. 2009; Liang et al. 2009a). This indicates that the guide RNA is able to adjust its two junctions (J1 and J2, Fig. 10a) in order to correctly place the substrate RNA. Furthermore, mutational studies with altered guide RNA in its pseudouridine pocket, thus different SH1, and its lower stem P1 suggests an even wider range of guide flexibility in these regions (Liang et al. 2009a).
7.3 Pre-reactive, intermediate and post-reactive structures
In searching for the active site geometries at other stages of the isomerization reaction, crystal structures of the box H/ACA RNPs bound with substrate RNA containing various uridine analogs were obtained (Zhou et al. 2010a, b). These include two pre-reactive complexes containing 5-bromouridine (5BrU) and 3-methyluridine (3MU) and two product complexes containing 2-deoxyuridine (2dU) and 4-thiouridine (4SU) in place of the target uridine. The two product nucleotides, 2dU and 4SU, are not found in the active site, while the flanking nucleotides are, suggesting their low affinities for the enzyme. The two pre-reactive complex structures show that the substituted uridine residues are well ordered in the active site and that they bind less deep within the active site than 5FhΨ (Fig. 11, 5BrU, 3MU and 5FhΨ). Interestingly, in these complexes of different reaction status, the catalytic aspartate is oriented in a similar fashion with respect to the anomeric carbon C1′ but not to ring carbon C6. This result supports the possible role of C6 in catalysis as previously proposed (Gu et al. 1999).
Fig. 11.
Toward understanding of the catalytic steps of pseudouridylation. Substitution of C5 hydrogen by bromide (5BrU) or N3 hydrogen by methyl group (3MU) inhibited uridine isomerization while substitution of C5 hydrogen by fluoride did not inhibit the ring cleavage step, leading to 5,6-hydroxy-pseudouridine (5FhΨ) intermediate. Superimposed crystal structures of the Cbf5 active site with bound 5FhΨ-, 5BrU- and 3MU-substituted substrates (PDB codes: 3HJW, 3LWO and 3LWQ, respectively) indicate that the catalytic aspartate is positioned similarly to the anomeric carbon C1′ but dissimilarly to ring carbon C6, suggesting an important role of C6 in catalysis. Protein residues are labeled according to Pyrococcus furiosus Cbf5 numbering and colored in grey, blue and red. The target nucleotide is colored in red.
The structures of different reaction stages also reveal remodeling of the thumb loop of Cbf5. The thumb loop is disordered in the pre-reactive and product complexes but stabilized in the 5FhΨ complex, suggesting its distinct role in binding the reaction intermediate. The thumb loop of TruB is already known to take different positions in structures containing either catalytically deficient TruB complex or containing 5FhΨ (Hoang et al. 2005).
7.4 Structures of box H/ACA sno/scaRNP assembly factors
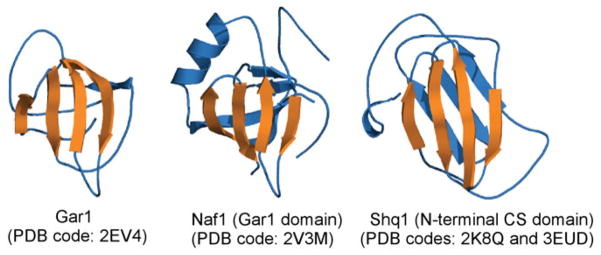
Shq1 and Naf1 are two conserved trans-acting factors involved in assembly of box H/ACA sca/snoRNPs. They bind early to Cbf5 during co-transcriptional H/ACA RNP assembly and are released from the mature RNPs. Structures of the N-terminal domain of Shq1 (PDB codes: 3EUD and 2K8Q) and the GAR1 domain of Naf1 (PDB code: 2V3M) have been solved (Godin et al. 2009; Leulliot et al. 2007; Singh et al. 2009). Superimposing Shq1 and Naf1 to Gar1 reveals a strikingly similar four β-strand sheet among these three proteins (Fig. 12). Gar1 uses this motif to interact with Cbf5. This structural comparison supports the proposed model of competition between Gar1 and Shq1/Naf1 for binding to Cbf5 in the late stages of box H/ACA RNP assembly (Darzacq et al. 2006; Godin et al. 2009; Grozdanov et al. 2009).
Fig. 12.
Biogenesis factors of box H/ACA RNPs share structural homologies with Gar1.
8. Concluding remarks
Since the identification of RNA-guided modification enzymes nearly a decade ago, we have seen significant advancement in their structure and function studies. Two different types of modification RNPs share some striking similarities in their organization and the method of substrate binding. Composite protein–protein and protein–RNA surfaces are used by RNPs to facilitate orderly assembly. Non-catalytic subunits play essential roles in placement of the substrate RNA through their conformational effects on the guide RNA or the catalytic subunit itself. On the other hand, the two RNPs differ dramatically in their specificities for assembly and the level of complexity. The key specificity determinant of the box H/ACA RNP assembly involves recognition of the ACA trinucleotide by the PUA domain of Cbf5, while that of the box C/D RNP assembly is the recognition of the box C/D motif (K-turn) by the L7Ae/15·5 kDa protein. The box H/ACA RNP is fully functional as a single RNP unit, while the box C/D RNP likely requires dual RNP units to function.
Sno/scaRNPs represent the most abundant and complex RNA modification enzymes. Their complexity is reflected in multiple processes that include their biogenesis, assembly and catalysis. Such complexity is paid off by their ability to locate and modify a large number of target sites efficiently. The enzymes make full use of proteins ’ ability to catalyze chemical reactions and to form structural support. As well, they fully exploit RNAs’ ability to both base pair and offer plasticity. Questions regarding how the core components assemble are now nearly answered. However, many other questions regarding how these enzymes are regulated, how they are localized and how they catalyze these remarkable reactions have just begun to be addressed.
References
- Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H. Structure and function of archaeal box C/D sRNP core proteins. Nature Structure Biology. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- Alian A, Degiovanni A, Griner SL, Finer-Moore JS, Stroud RM. Crystal structure of an RluF–RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome. Journal of Molecular Biology. 2009;388:785–800. doi: 10.1016/j.jmb.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Carbon P, Krol A. The SBP2 and 15·5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO Journal. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. Journal of Molecular Microbiology and Biotechnology. 2002;4:71–75. [PubMed] [Google Scholar]
- Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Molecular and Cellular Biology. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. Journal of Biological Chemistry. 1998;273:10868–10873. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes and Development. 2005;19:1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Research. 2009;37:7665–7677. doi: 10.1093/nar/gkp816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal PA, Maydanovych O, Pokharel S. The chemistry and biology of RNA editing by adenosine deaminases. Nucleic Acids Symposium Series (Oxford) 2007;51:83–84. doi: 10.1093/nass/nrm042. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, De Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, Mcdowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, De Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Gagnon KT, Brown BA, II, Maxwell ES, Leschziner AE, Unger VM, Baserga SJ. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science. 2009;325:1384–1387. doi: 10.1126/science.1176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, Bardoni B, Krol A, Branlant C, Allmang C, Bertrand E, Charpentier B. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. Journal of Cell Biology. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, Bertrand E. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Molecular Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Molecular Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Cahill NM, Friend K, Speckmann W, Li ZH, Terns RM, Terns MP, Steitz JA. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO Journal. 2002;21:3816–3828. doi: 10.1093/emboj/cdf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO Journal. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Williamson JR. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure. 2004;12:1165–1176. doi: 10.1016/j.str.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Research. 2005;33:3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Research. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d’Orval B, Bortolin ML, Gaspin C, Bachellerie JP. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Research. 2001;29:4518–4529. doi: 10.1093/nar/29.22.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nature Reviews Molecular Cell Biology. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. Journal of Cell Biology. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Omer A. Small non-coding RNAs in archaea. Current Opinion in Microbiology. 2005;8:685–694. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Molecular Cell. 2009;34:427–439. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. Journal of Molecular Biology. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- Eliceiri GL. The vertebrate E1/U17 small nucleolar ribonucleoprotein particle. Journal of Cellular Biochemistry. 2006;98:486–495. doi: 10.1002/jcb.20821. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Current Opinion in Cell Biology. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Foster PG, Huang L, Santi DV, Stroud RM. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nature Structural Biology. 2000;7:23–27. doi: 10.1038/71219. [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Zhang X, Agris PF, Maxwell ES. Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling. Journal of Molecular Biology. 2006;362:1025–1042. doi: 10.1016/j.jmb.2006.07.091. [DOI] [PubMed] [Google Scholar]
- Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Molecular Cell Biology. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA. Small nucleolar RNA. Biochemistry and Cell Biology. 1995;73:845–858. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- Girard JP, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO Journal. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin KS, Walbott H, Leulliot N, van Tilbeurgh H, Varani G. The Box H/ACA snoRNP assembly factor Shq1p is a chaperone protein homologous to Hsp90 cochaperones that binds to the Cbf5p enzyme. Journal of Molecular Biology. 2009;390:231–244. doi: 10.1016/j.jmb.2009.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody TA, Melcher SE, Norman DG, Lilley DM. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Molecular Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Grozdanov PN, Roy S, Kittur N, Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA. 2009;15:1188–1197. doi: 10.1261/rna.1532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Joardar A, Chaurasia P, Gupta R. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biology. 2007;4:101–109. doi: 10.4161/rna.4.2.5177. [DOI] [PubMed] [Google Scholar]
- Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria ? RNA Biology. 2004;1:10–15. [PubMed] [Google Scholar]
- Hamma T, Ferre-D’amare AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure (Cambridge) 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferre-D’amare AR. Pseudouridine synthases. Chemistry and Biology. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Hamma T, Reichow SL, Varani G, Ferre-D’amare AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nature Structural and Molecular Biology. 2005;12:1101–1107. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA. The expanding universe of noncoding RNAs. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:551–564. doi: 10.1101/sqb.2006.71.064. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genetics. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO Journal. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Molecular Cell. 2006;23:673–684. doi: 10.1016/j.molcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Molecular Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- Hirose T, Steitz JA. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12914–12919. doi: 10.1073/pnas.231490998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D’amare AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Molecular Cell. 2006;24:535–545. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Hoang C, Ferre-D’amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- Hoang C, Hamilton CS, Mueller EG, Ferre-D’amare AR. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Science. 2005;14:2201–2206. doi: 10.1110/ps.051493605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Molecular Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. Journal of Cell Biology. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Loria JP, Moore PB. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a Box H/ACA snoRNA. Molecular Cell. 2007;26:205–215. doi: 10.1016/j.molcel.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Joardar A, Gurha P, Skariah G, Gupta R. Box C/D RNA-guided 2′-O-methylations and the intron of tRNATrp are not essential for the viability of Haloferax volcanii. Journal of Bacteriol. 2008;190:7308–7313. doi: 10.1128/JB.00820-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Molecular and Cellular Biology. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kiss T. SnoRNP biogenesis meets Pre-mRNA splicing. Molecular Cell. 2006;23:775–776. doi: 10.1016/j.molcel.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Molecular Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO Journal. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittur N, Darzacq X, Roy S, Singer RH, Meier UT. Dynamic association and localization of human H/ACA RNP proteins. RNA. 2006;12:2057–2062. doi: 10.1261/rna.249306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Research. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt JF. High-definition macromolecular composition of yeast RNA-processing complexes. Molecular Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Bousquet-Antonelli A, Henry Y, Michèle Caizergues F, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes and Development. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Molecular and Cellular Biology. 2000;20:2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader DJ, Clark GP, Watters J, Beven AF, Shaw PJ, Brown JW. Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Molecular Biology. 1999;39:1091–1100. doi: 10.1023/a:1006157022319. [DOI] [PubMed] [Google Scholar]
- Leulliot N, Godin KS, Hoareau-Aveilla C, Quevillon-Cheruel S, Varani G, Henry Y, Van Tilbeurgh H. The Box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. Journal of Molecular Biology. 2007;371:1338–1353. doi: 10.1016/j.jmb.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- Liang B, Kahen EJ, Calvin K, Zhou J, Blanco M, Li H. Long-distance placement of substrate RNA by H/ACA proteins. RNA. 2008;14:2086–2094. doi: 10.1261/rna.1109808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Xue S, Terns RM, Terns MP, Li H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nature Structural and Molecular Biology. 2007a;14:1189–1195. doi: 10.1038/nsmb1336. [DOI] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nature Structural and Molecular Biology. 2009a;16:740–746. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WQ, Fournier MJ. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes and Development. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Molecular Cell. 2007b;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009b;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liang XH, Piekna-Przybylska D, Liu Q, Fournier MJ. Mis-targeted methylation in rRNA can severely impair ribosome synthesis and activity. RNA Biology. 2008;5:249–254. doi: 10.4161/rna.6916. [DOI] [PubMed] [Google Scholar]
- Manival X, Charron C, Fourmann JB, Godard F, Charpentier B, Branlant C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5–aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Research. 2006;34:826–839. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A, Mason PJ. Dyskeratosis congenita. Cellular and Molecular Life Sciences. 2003;60:507–517. doi: 10.1007/s000180300042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A, Walne A, Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Current Opinion in Genetics and Development. 2005;15:249–257. doi: 10.1016/j.gde.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Martin JL, Mcmillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Current Opinion in Structural Biology. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Noncoding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nature Reviews. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annual Review of Biochemistry. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. Journal of Molecular Biology. 2007;373:1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Molecular and Cellular Biology. 1999a;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999b;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA–protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure (Cambridge) 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Mosbacher TG, Bechthold A, Schulz GE. Crystal structure of the avilamycin resistance-conferring methyltransferase AviRa from Streptomyces viridochromogenes. Journal of Molecular Biology. 2003;329:147–157. doi: 10.1016/s0022-2836(03)00407-8. [DOI] [PubMed] [Google Scholar]
- Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:{Psi}55-synthase and RNA-guided RNA:{Psi}-synthase activities. Nucleic Acids Research. 2007;35:5610–5624. doi: 10.1093/nar/gkm606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12697–12702. doi: 10.1073/pnas.202477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DR, Kuhn JF, Shanab GM, Maxwell ES. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Petfalski E, Tollervey D, Caceres JF. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Molecular and Cellular Biology. 2003;23:8519–8527. doi: 10.1128/MCB.23.23.8519-8527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki O, Shirouzu M, Hashimoto K, Ishitani R, Terada T, Tamakoshi M, Oshima T, Chijimatsu M, Takio K, Vassylyev DG, Shibata T, Inoue Y, Kuramitsu S, Yokoyama S. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallographica D: Biological Crystallography. 2002;58:1129–1137. doi: 10.1107/s0907444902006601. [DOI] [PubMed] [Google Scholar]
- Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Decatur WA, Fournier MJ, Dennis PP. RNA-modifying machines in archaea. Molecular Microbiology. 2003;48:617–629. doi: 10.1046/j.1365-2958.2003.03483.x. [DOI] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruganti S, Zhang Y, Li H. Structural comparison of yeast snoRNP and spliceosomal protein Snu13p with its homologs. Biochemical and Biophysical Research Communications. 2005;333:550–554. doi: 10.1016/j.bbrc.2005.05.141. [DOI] [PubMed] [Google Scholar]
- Oruganti S, Zhang Y, Li H, Robinson H, Terns MP, Terns RM, Yang W, Li H. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. Journal of Molecular Biology. 2007;371:1141–1150. doi: 10.1016/j.jmb.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proceedings of the National Academy Sciences of the United States of America. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Perez-Arellano I, Gallego J, Cervera J. The PUA domain – a structural and functional overview. FEBS Journal. 2007;274:4972–4984. doi: 10.1111/j.1742-4658.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- Perez-Fernandez J, Roman A, de Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Molecular and Cellular Biology. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phannachet K, Elias Y, Huang RH. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry. 2005;44:15488–15494. doi: 10.1021/bi050961w. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. Journal of Biological Chemistry. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LH, Henras A, Lu YJ, Zhou H, Zhou WX, Zhu YQ, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie JP. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Molecular and Cellular Biology. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Aittaleb M, Chen Q, Spiegel K, Demeler B, Li H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. Journal of Molecular Biology. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Molecular Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Reddy R, Li WY, Henning D, Choi YC, Nohga K, Busch H. Characterization and subcellular localization of 7–8 S RNAs of Novikoff hepatoma. Journal of Biological Chemistry. 1981;256:8452–8457. [PubMed] [Google Scholar]
- Richard P, Kiss AM, Darzacq X, Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Molecular and Cellular Biology. 2006;26:2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in archaea. Nucleic Acids Research. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi DV. Mechanistic studies of RNA modifying enzymes. RNA pseudouridine synthase and m5cytosine methyl transferase. Nucleic Acids Symposium Series. 2000;44:147–148. doi: 10.1093/nass/44.1.147. [DOI] [PubMed] [Google Scholar]
- Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. Rna. 2006;12:15–25. doi: 10.1261/rna.2210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends in Biochemical Sciences. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Gonzales FA, Cascio D, Heckmann N, Chanfreau G, Feigon J. Structure and functional studies of the CS domain of the essential H/ACA ribonucleoparticle assembly protein SHQ1. Journal of Biological Chemistry. 2009;284:1906–1916. doi: 10.1074/jbc.M807337200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Gurha P, Gupta R. Dynamic guide–target interactions contribute to sequential 2′-O-methylation by a unique archaeal dual guide box C/D sRNP. RNA. 2008;14:1411–1423. doi: 10.1261/rna.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The pseudouridine synthases: revisiting a mechanism that seemed settled. Journal of the American Chemical Society. 2004;126:12758–12759. doi: 10.1021/ja046375s. [DOI] [PubMed] [Google Scholar]
- Suryadi J, Tran EJ, Maxwell ES, Brown II, BA The Crystal Structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the Box C/D RNAs(,) Biochemistry. 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Current Opinion in Cell Biology. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Tran E, Zhang X, Lackey L, Maxwell ES. Conserved spacing between the box C/D and ‘C′/’D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11:285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhang X, Maxwell ES. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and ‘C′/’′ RNPs. EMBO Journal. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO Journal. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Molecular Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, Mccann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15·5 kD protein bound to a U4 snRNA fragment. Molecular Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, Dokal I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Dokal I. Dyskeratosis congenita: a historical perspective. Mechanisms of Ageing and Development. 2008;129:48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO Journal. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Current Opinion in Cell Biology. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Wu H, Feigon J. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6655–6660. doi: 10.1073/pnas.0701534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Brockenbrough JS, Metcalfe AC, Chen S, Aris JP. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Journal of Biological Chemistry. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Wang R, Yang F, Terns RM, Terns MP, Zhang X, Maxwell ES, Li H. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Molecular Cell. 2010;39:939–949. doi: 10.1016/j.molcel.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Mcpheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2005a;280:6655–6662. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Molecular and Cellular Biology. 2005b;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. Journal of Biological Chemistry. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]
- Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L. Structural organization of box C/D RNA-guided RNA methyltransferase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13808–13813. doi: 10.1073/pnas.0905128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO Journal. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–690. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liang B, Li H. Functional and structural impact of target uridine substitutions on H/ACA ribonucleoprotein particle pseudouridine synthase. Biochemistry. 2010a;49:6276–6281. doi: 10.1021/bi1006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chao LV, Liang B, Chen M, Yang W, Li H. Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study. Journal of Molecular Biology. 2010b;401:690–695. doi: 10.1016/j.jmb.2010.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]