Abstract
Gibberellins (GAs) are crucial phytohormones involved in many aspects of plant growth and development, including plant-microbe interactions, which has led to GA production by plant-associated fungi and bacteria as well. While the GA biosynthetic pathways in plants and fungi have been elucidated and found to have independently arisen through convergent evolution, little has been uncovered about GA biosynthesis in bacteria. Some nitrogen-fixing/symbiotic, legume-associated rhizobia, including Bradyrhizobium japonicum, the symbiont of soybean, and Sinorhizobium fredii, a broad-host-nodulating species, contain a putative GA biosynthetic operon/gene cluster. Through functional characterization of five unknown genes, we demonstrate that this operon encodes the enzymes necessary to produce GA9, thereby elucidating bacterial GA biosynthesis. The distinct nature of these enzymes indicates that bacteria have independently evolved a third biosynthetic pathway for GA production. Furthermore, our results also reveal a central biochemical logic that is followed in all three convergently evolved GA biosynthetic pathways.
INTRODUCTION
Gibberellins (GAs) are complex diterpenoid phytohormones characterized by their 6-5-6-5 tetracyclic ent-gibberellane backbone (See Supplementary Results, Supplementary Fig. 1 for structure annotation). The common bioactive GAs (GA1, GA3, GA4, and GA7) are crucial to plant growth and development1,2, as well as plant-microbe interactions3,4, and other GAs have been implicated in species-specific bioactivity (e.g. GA5 and GA6 in Lolium sp. flower-induction2). Notably, the semi-dwarf crop varieties at the center of the “Green Revolution”, which led to significant increases in worldwide crop yields, were found to result from altered GA signaling5,6. Although endogenous to plants, GAs were initially discovered in the fungal plant pathogen Gibberella fujikuroi7 (also known as Fusarium fujikuroi), and subsequently, many plant-associated fungi and bacteria have been found to produce GAs8. Biosynthesis of GAs is elucidated in plants and fungi1,9 (see Supplementary Fig. 2a for a summary), and differences between the pathways and relevant enzymes indicate the independent evolution of GA production in each of these two biological kingdoms7.
As diterpenoids, GAs are derived from the 20-carbon isoprenyl precursor (E,E,E)-geranylgeranyl diphosphate (GGPP). In both plants and fungi, GA biosynthesis is initiated by two cyclization reactions, first through production of ent-copalyl diphosphate (CPP) from GGPP by ent-CPP synthase (CPS), then via formation of ent-kaurene by ent-kaurene synthase (KS)1. While these cyclizations are performed by two separate enzymes in plants2 and a bifunctional enzyme in fungi9, it is possible that these diterpene cyclases share a common evolutionary origin7,10. However, following ent-kaurene formation, GA biosynthesis between plants and fungi clearly diverges. In plants, ent-kaurene is oxygenated/oxidized by both cytochrome P450 monooxygenases (CYPs) and 2-oxoglutarate-dependent dioxygenases (2ODDs)2,11, while fungi make use of several CYPs and a desaturase9,12. Notably, the CYPs utilized by plants versus fungi do not share significant amino acid sequence identity and belong to separate CYP families13. Furthermore, the chronology of the peripheral hydroxylation reactions at C-3 and C-13 differs between plants and fungi. In particular, 3β-hydroxylation, which is essential for GA bioactivity2, occurs earlier in fungal biosynthesis, while occurring late in the plant pathway7. Therefore, the intervening biosynthetic intermediates are distinct in each pathway. Because of these differences it is clear that plants and fungi have convergently evolved the ability to produce bioactive GAs7.
GA biosynthesis in bacteria has remained uncharacterized, though it has been accepted for decades that bacteria can produce GAs8,14. The soybean symbiont, Bradyrhizobium japonicum USDA110 (now classified as Bradyrhizobium diazoefficiens USDA11015), contains an operon/gene cluster comprised of an isoprenoid synthase (IDS), two putative diterpene cyclases, three CYPs (CYP112, CYP114, and CYP117), a ferredoxin (FdGA), and a short-chain dehydrogenase/reductase (SDRGA)16,17 (see Supplementary Fig. 2b). The presence of the IDS and putative diterpene cyclases in conjunction with oxidative enzymes, particularly CYPs, led to speculation that this operon is involved in GA biosynthesis17,18, although it was only recently that B. japonicum was convincingly shown to produce GAs19. This operon was subsequently found to be distributed in many other symbiotic rhizobia such as Sinorhizobium fredii NGR234, Mesorhizobium loti MAFF303099, and Rhizobium etli CFN4218,20. Although there are additional genes present in the operons found in some of these other species, these appear to encode accessory enzymes that would not be directly involved in GA biosynthesis per se (e.g., an oft-found isopentenyl diphosphate δ-isomerase presumably balances the upstream supply of isoprenoid precursors). Accordingly, the collection of the aforementioned genes in B. japonicum appears to represent a “core operon”, as they are found in nearly all cases of this operon within rhizobia.
Previously, we have shown that the IDS produces GGPP, and is thus a GGPP synthase (GGPS), and that the two putative diterpene cyclases further act as a CPS and KS, whose sequential activity produces ent-kaurene10,20. Moreover, the function of these enzymes is conserved across four major genera of rhizobia10,20. In addition, recent isotope-labeled feeding studies with B. japonicum isolated from soybean nodules (termed “bacteroids”) demonstrated GA oxidase activity, and found that the major metabolites are ent-kaurenoic acid, GA12, GA15, GA24, and ultimately GA9, suggesting a pathway similar to that found in plants19. However, these bacteroids do not appear to be capable of producing 3β- or 13-hydroxylated GAs, despite 3β-hydroxylation being essential for classical bioactivity2. Here, through a combination of heterologous expression and knockout bacteroid incubations, we were able to unambiguously assign function to the remaining uncharacterized genes of the putative GA operon, elucidating their roles in bacterial biosynthesis of GA9.
RESULTS
General approach
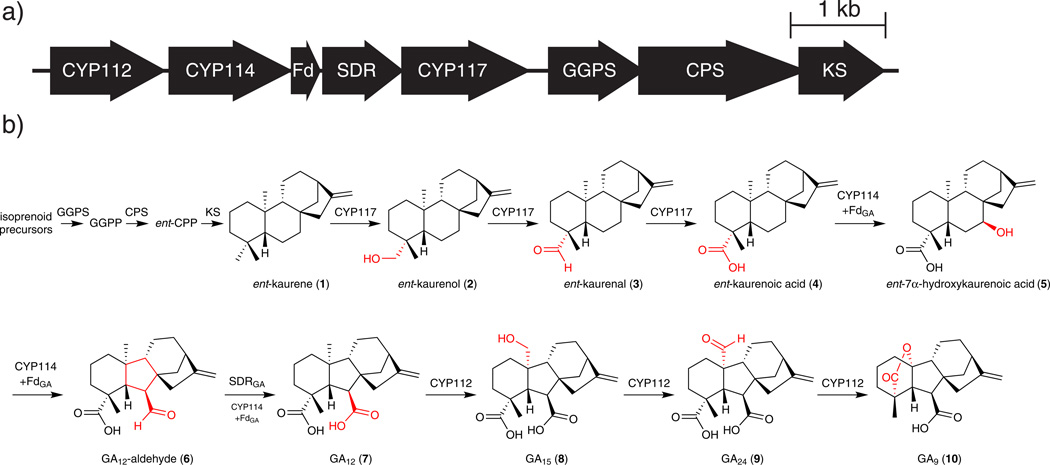
To determine the function of the five remaining proteins potentially involved in rhizobial GA biosynthesis (CYP112, CYP114, CYP117, SDRGA, and FdGA) we expressed the corresponding genes from the putative GA core operon of S. fredii NGR234 in the relatively closely related S. meliloti 1021 (Sm1021), which does not contain the operon21. In order to evaluate if FdGA is required to supply the electrons needed to activate one or more of the CYPs22, each CYP was expressed alone and in coexpression with FdGA. We incubated putative GA precursors/intermediates19 in these heterologous cultures and analyzed the extracts with gas chromatography-mass spectrometry (GC-MS), which allowed us to designate a function for each gene in GA biosynthesis (Fig. 1).
Figure 1. GA biosynthesis in rhizobia.
a) GA biosynthetic operon found in B. japonicum and S. fredii (1 kb=1 kilobase). b) Elucidated GA biosynthetic pathway in core operon-containing rhizobia.
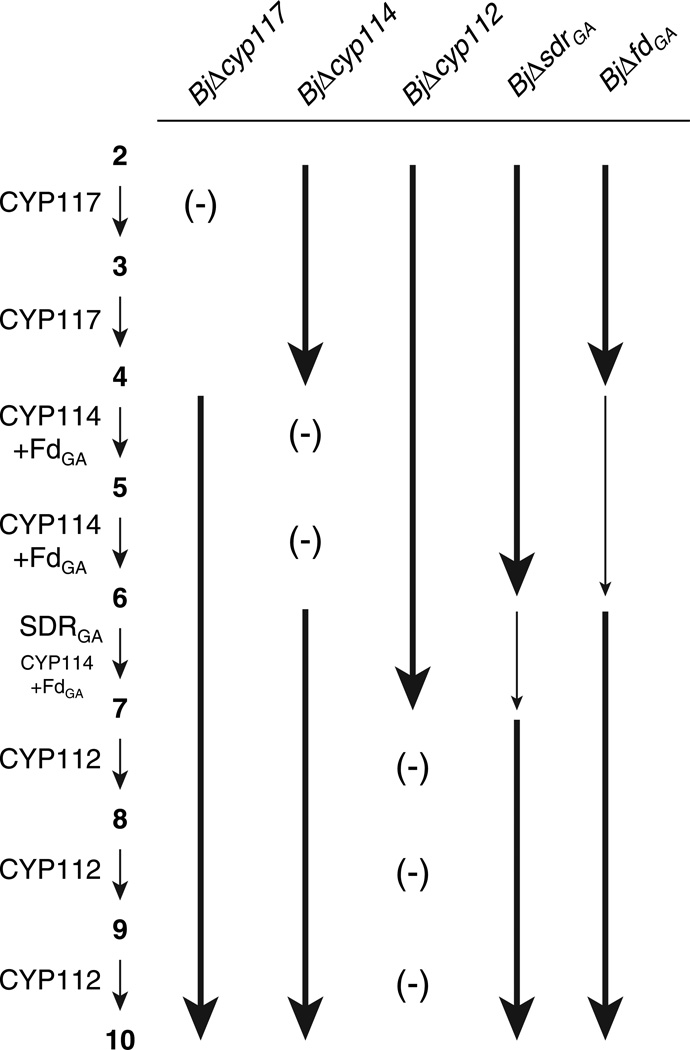
Additionally, we validated our results in vivo via a genetic (knock-out) approach23,24, in which we deleted each uncharacterized gene (in-frame) within the operons of both B. japonicum and S. fredii to create the following strains in each species: Δcyp112, Δcyp114, Δcyp117, ΔfdGA, and ΔsdrGA. These deletion strains were used to nodulate compatible legume hosts, B. japonicum with soybean (Glycine max) and S. fredii with cowpea (Vigna unguiculata), as functional expression of this operon and GA oxidase activity has only been detected in bacteroids19 (i.e. bacteria isolated from nodules). Isolated bacteroids were incubated with isotope-labeled GA intermediates, after which metabolites were extracted and analyzed using high performance liquid chromatography (HPLC) and GC-MS to determine any obstruction in GA metabolism corresponding to the deleted gene (summarized for B. japonicum in Fig. 2; see Supplementary Fig. 3 for full summary of both B. japonicum and S. fredii bacteroid incubation studies).
Figure 2. Summary of GA biosynthetic pathway reactions detected in incubations of B. japonicum gene-deletion bacteroids with GA precursors.
Arrows indicate observed reactions, with arrow thickness representing relative efficiency of the detected reactions. (−): reaction not detected. The underlying data is available in Supplementary Tables 1 – 4.
CYP117 is an ent-kaurene oxidase (KO)
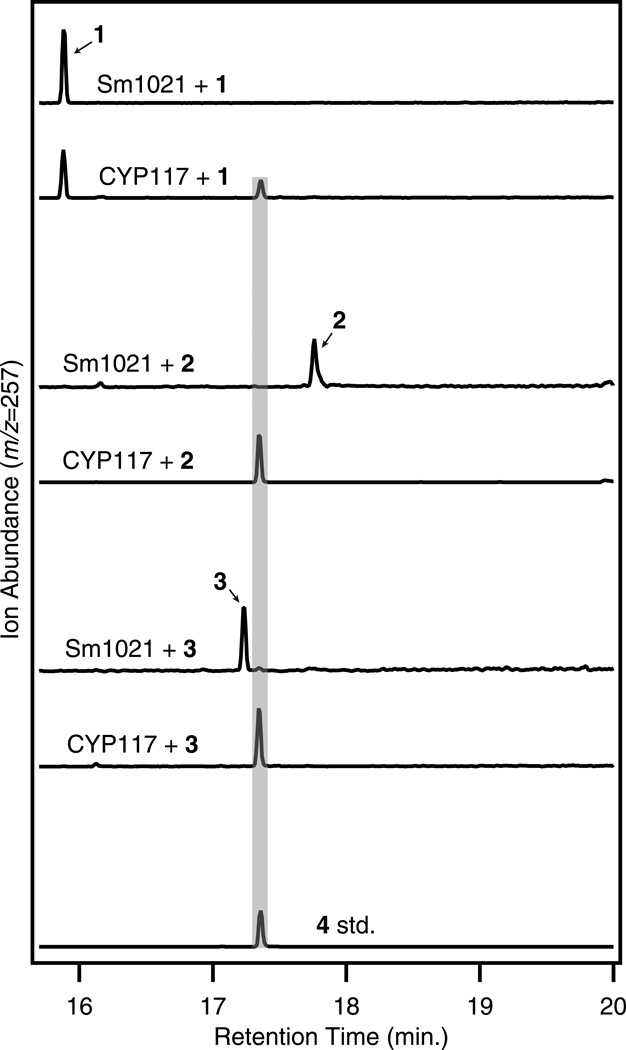
The olefin precursor ent-kaurene (1) is converted to ent-kaurenoic acid (4) by cultures expressing CYP117 (Fig. 3 & Supplementary Fig. 4), with trace amounts of the intermediates ent-kaurenol (2) and ent-kaurenal (3) also found (Supplementary Figs. 4 & 5), demonstrating that CYP117 acts as an ent-kaurene oxidase (KO). Furthermore, cells expressing CYP117 could efficiently convert either 2 or 3 to 4 (Fig. 3), confirming that CYP117 sequentially oxidizes 1 to the C-19 alcohol (2), aldehyde (3) and carboxylic acid (4). This activity did not depend on co-expression with FdGA, indicating that an endogenous redox system in the S. meliloti host could supply electrons to CYP117. Wild-type/untransformed S. meliloti 1021 did not metabolize any of these substrates, nor any of the subsequently tested GA intermediates, confirming that the chemical transformations were due to the expressed genes. In accordance with the heterologous expression, Δcyp117 bacteroids were unable to metabolize 2 but could react with 4 and other downstream intermediates (Fig. 2; see Supplementary Tables 1 & 2 for raw data).
Figure 3. CYP117 is an ent-kaurene oxidase (KO).
CYP117 converts ent-kaurene (1) into ent-kaurenoic acid (4), as shown here by GC-MS chromatograms with comparison to an authentic standard (std.) of 4 (the relevant mass spectra can be found in Supplementary Fig. 4). Additionally, individual incubations with ent-kaurenol (2) and ent-kaurenal (3) result in efficient conversion to 4, demonstrating the sequential oxidation of C-19 to the alcohol (2), aldehyde (3), and acid (4) by CYP117.
CYP114 (+FdGA) is an ent-kaurenoic acid oxidase (KAO)
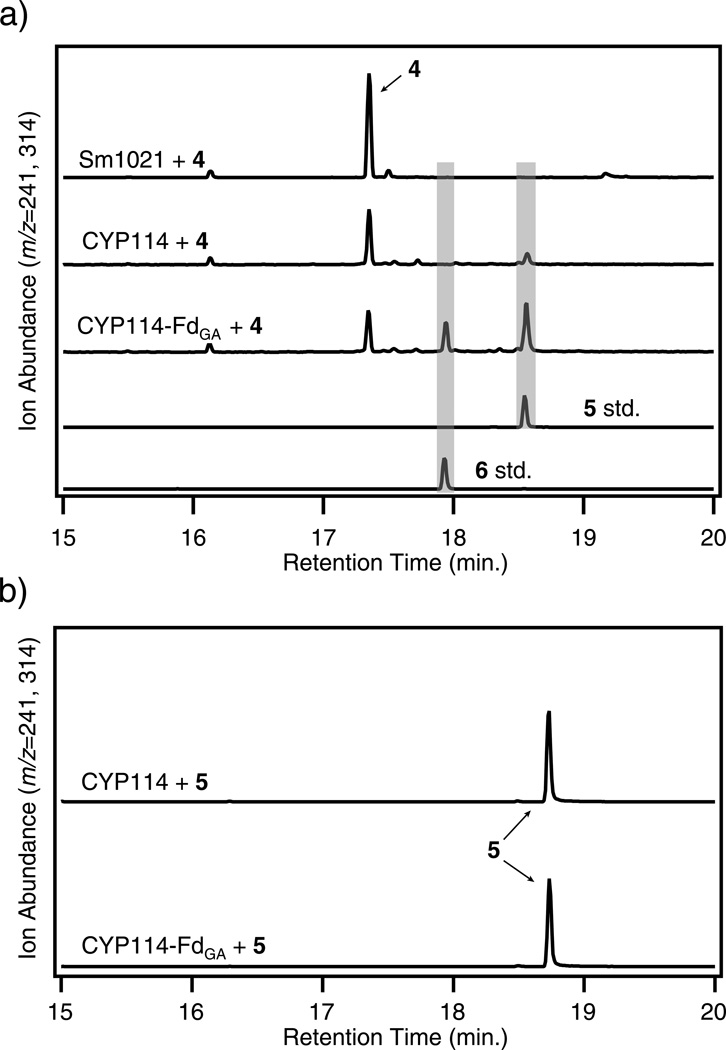
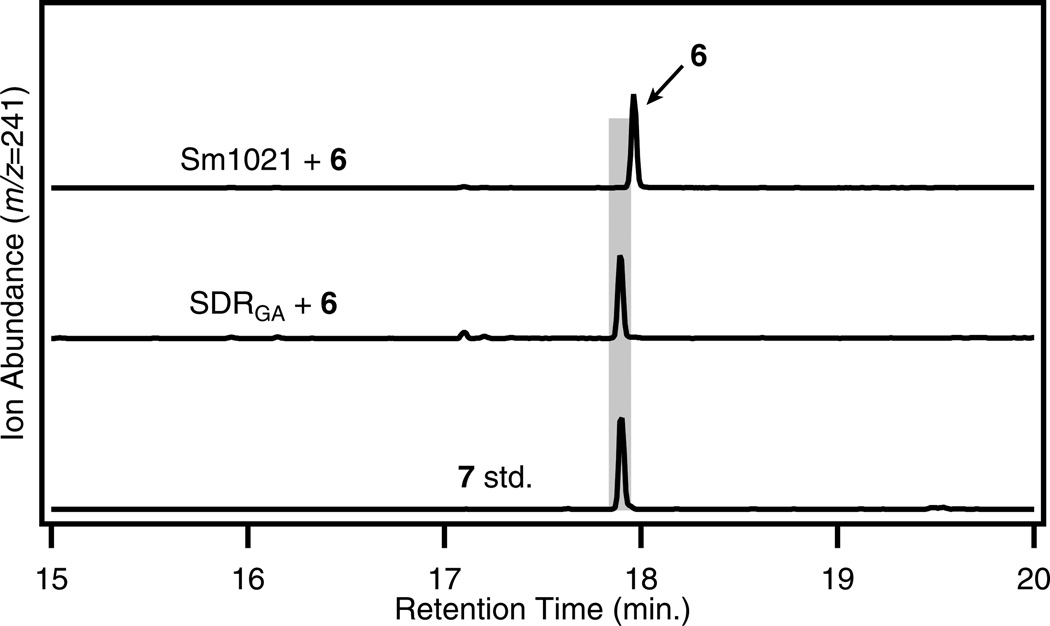
Incubation of 4 in cells expressing CYP114 resulted in production of ent-7α-hydroxykaurenoic acid (5; also referred to as 7β-hydroxy-ent-kaurenoic acid or ent-7α-OH-kaurenoic acid) (Fig. 4a & Supplementary Fig. 6). Interestingly, further chemical transformation was observed when CYP114 was co-expressed with FdGA (CYP114-FdGA), with these cultures converting 4 to both 5 and GA12-aldehyde (6), implicating FdGA as necessary for full CYP114 functionality (Fig. 4a & Supplementary Fig. 6). Trace amounts of GA12 (7) could be detected from these incubations (Supplementary Fig. 7), indicating that CYP114 is capable of full oxidation of C-7, albeit with low efficiency. Incubation of 6 in cells expressing CYP114-FdGA resulted in a detectable, but very low level of turnover to 7, demonstrating that while CYP114-FdGA can catalyze this reaction, it is quite inefficient. Δcyp114 bacteroids were blocked at the corresponding step as there was no transformation of 4 or 5 to 6 or other downstream compounds (Fig. 2 & Supplementary Tables 1 & 2). ΔfdGA bacteroids showed decreased activity, with upstream substrate feeding leading to accumulation of 5, but with no obstruction of downstream metabolism (Fig. 2 & Supplementary Tables 3 & 4), confirming the necessity of FdGA for full CYP114 functionality.
Figure 4. CYP114 is an ent-kaurenoic acid oxidase (KAO) and requires FdGA for full functionality.
a) CYP114 converts ent-kaurenoic acid (4) into ent-7α-hydroxykaurenoic acid (5) and ultimately GA12-aldehyde (6) as the major product, as shown here by GC-MS chromatograms with comparison to an authentic standard (std.) of 5 and 6 (the relevant mass spectra can be found in Supplementary Fig. 6), but production of 6 is only observed when FdGA is coexpressed. b) Incubation with 5 does not result in detectable conversion to 6.
To our surprise, feeding 5 to cells expressing either CYP114 or CYP114-FdGA did not result in any significant transformation to 6 (Fig. 4b). This did not appear to be due to lack of transportation of 5 into our heterologous cells, as this compound could be found in both the cell pellet and in the aqueous supernatant in substantial quantities (Supplementary Fig. 8). However, this substrate was metabolized by bacteroids into 6 and further downstream metabolites (Fig. 2 & Supplementary Tables 3 & 4), suggesting that 5 may indeed be an intermediate in the bacterial GA biosynthesis pathway, just as in plants and fungi7. If 5 is a true chemical intermediate between 4 and 6, it is possible that suboptimal conditions in our heterologous expression system may prevent turnover to a sufficient level for detection, as the natively-expressed CYP114 in B. japonicum and S. fredii appear capable of catalyzing this reaction (Fig. 2 & Supplementary Tables 3 & 4).
Molecular modeling indicates that the C-19 carboxylate of 4 is in close proximity to C-6 (Supplementary Figure 9), which we suggest may enable formation of a C-6 carbocation intermediate, thus allowing for the ring contraction to occur via a semipinacol rearrangement mechanism25–27. In support of this, neither 3 (which in the diol form sterically resembles 4) nor the methyl ester of 4, both of which lack a free C-19 carboxylate, were chemically transformed by cells expressing CYP114-FdGA (Supplementary Table 5), suggesting a need for the free carboxylate in this reaction.
Regardless of such speculation, it is clear that CYP114 acts as an ent-kaurenoic acid oxidase (KAO) and produces 6 as the major product from ent-kaurenoic acid. While it appears that CYP114 is capable of full oxidation of C-7 to form 7, the conversion of 6 to 7 does not seem to be efficiently catalyzed by CYP114. This is distinct from the KAOs of plants or fungi, which convert 4 to 7 or GA14, respectively, (both of which contain the fully oxidized C-7 carboxylate) without much production of the corresponding C-7 aldehyde28,29. In addition, CYP114 requires FdGA in order to catalyze the ring contraction reaction necessary for formation of 6 and 7 from 4, though it appears that endogenous Fd(s) from S. meliloti can supply electrons to CYP114 and elicit partial activity (i.e. oxidation of 4 to 5).
SDRGA is a GA 7-oxidase
When incubated with cultures expressing SDRGA, 6 is efficiently converted to 7 via C-7 oxidation (Fig. 5 & Supplementary Fig. 10), and thus SDRGA functions as an aldehyde dehydrogenase, and can be designated as a GA 7-oxidase. This was demonstrated by heterologous expression of SDRGA from both S. fredii and B. japonicum, with the latter expressed in E. coli (Supplementary Figs. 10 & 11). Consistent with these results, 6 accumulated in incubations of BjΔsdrGA bacteroids with 2 or 5 (Fig. 2 & Supplementary Tables 3 & 4). Transformation from 6 to 7 in BjΔsdrGA and SfΔsdrGA bacteroids was not entirely blocked (Fig. 2 & Supplementary Tables 3 & 4), and this is likely due to the low level conversion of 4 to 7 catalyzed by CYP114. Thus it seems that while CYP114 is catalytically capable of the full transformation of 4 to 7, SDRGA is necessary for the efficient oxidation of 6 to 7 to occur. The use of a dehydrogenase for this type of chemical transformation is not uncommon, as it has been shown in biosynthetic pathways for other metabolites that the relevant CYPs do not efficiently catalyze full oxidation to the carboxylate, leading to a requirement for alcohol and/or aldehyde dehydrogenases for complete conversion30–32. However, neither plant nor fungal GA biosynthesis rely on a dedicated dehydrogenase1, and therefore this enzyme is unique to the bacterial pathway.
Figure 5. SDRGA oxidizes GA12-aldehyde (6) to produce GA12 (7).
SDRGA is able to efficiently oxidize the extruded C-7 aldehyde of 6 to an acid, thus producing 7, as shown in this GC-MS chromatogram with comparison to an authentic standard (std.) of 7 (the relevant mass spectra can be found in Supplementary Fig. 10). SDRGA can therefore be referred to as a GA 7-oxidase.
CYP112 is a GA 20-oxidase
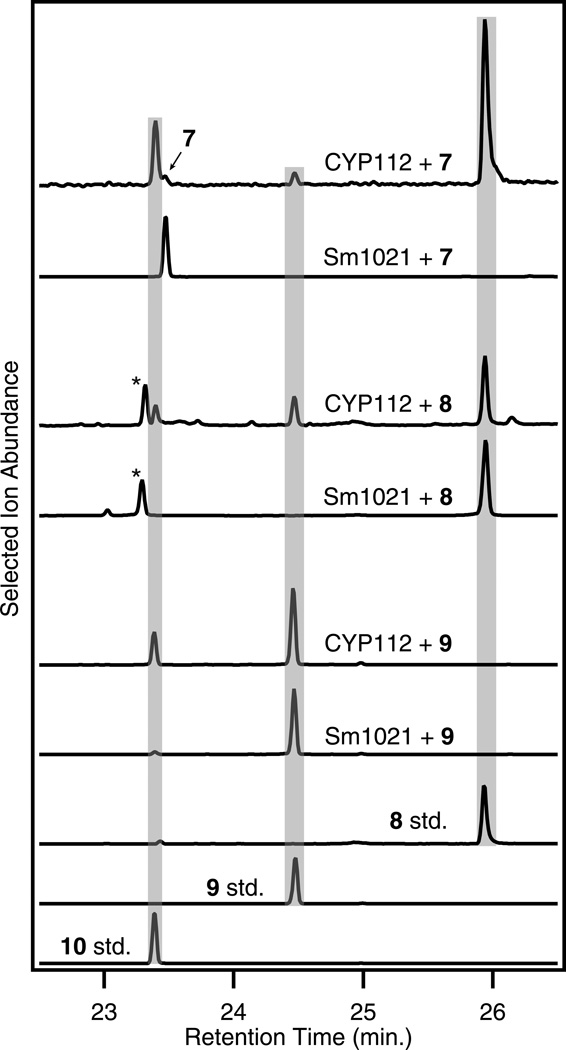
Cultures expressing CYP112 convert 7 to GA15 (8), GA24 (9) and, ultimately, GA9 (10), and this does not require FdGA coexpression (Fig. 6 & Supplementary Fig. 12). Separate incubations with 8 and 9 demonstrated that CYP112-expressing cells can convert both of these intermediates to the final product, 10 (Fig. 6). This demonstrates that 7 is oxidized by CYP112 first to the C-20 alcohol (8), then to the aldehyde (9) before loss of C-20 and formation of the lactone ring in 10. In correspondence with our heterologous expression results, Δcyp112 bacteroids were blocked in the conversion of 7 to 10, but could transform all upstream intermediates (Fig. 2 & Supplementary Tables 1 & 2). These results show that CYP112 acts as a GA 20-oxidase (GA20ox), catalyzing sequential oxygenation/oxidation of the C-20 methyl, culminating in demethylation and formation of the C-19,10-γ-lactone ring. Additionally, these results are consistent with a previous report indicating that 10 is the final product in these rhizobia19.
Figure 6. CYP112 is a GA 20-oxidase.
CYP112 converts GA12 (7) into GA9 (10) via GA15 (8) and GA24 (9) as shown here by GC-MS chromatograms with comparison to authentic standards (stds.) of 8, 9, and 10 (the relevant mass spectra can be found in Supplementary Fig. 12). 8 and 9 also both act as substrates for CYP112, showing sequential oxidation of C-20 to the alcohol (8) and aldehyde (9) before being lost to form 10. The selected ions for each chromatogram are as follows: +7 m/z = 226, 300; +8 m/z = 270, 284; +9 m/z = 226. *Contaminating substance found in 8 standard used for incubations.
The bacterial GA biosynthetic pathway is linear
Our initial results indicated that rhizobial GA biosynthesis likely proceeds via a direct pathway. However, GA metabolism in fungi and, especially, plants forms a more complex network2. To investigate this possibility within bacterial GA biosynthesis, all identified intermediates were incubated with each heterologously expressed enzyme to evaluate the possibility of promiscuous activity (e.g., could CYP112 form the C-19,10-γ-lactone ring with 4 or 6). Detectable activity was only observed with each previously described substrate, strongly suggesting linearity of GA biosynthesis in rhizobia (Supplementary Table 5). Thus, we were able to unequivocally assign the biosynthetic function of each gene found in the core GA operon along with demonstrating that these specific functions are relevant in vivo/planta, as shown through the knockout bacteroid experiments. Moreover, our results validate the functional equivalence of this operon in two distinct rhizobia from separate genera, and therefore conservation of the assigned catalytic activity of these enzymes in GA biosynthesis.
DISCUSSION
Elucidation of this pathway indicates that bacteria have independently evolved a third route for GA production that is distinct from either plant or fungal biosynthesis (Supplementary Fig. 2). Although the intermediates in bacterial GA biosynthesis are identical to those in the non-13-hydroxylation pathway found in plants, bacteria utilize a distinct set of enzymes to catalyze the relevant transformations. Other than the two diterpene cyclases (CPS and KS), which share some (albeit distant) sequence and structural homology with the plant and fungal cyclases10,33,34, the enzymes involved in rhizobial GA biosynthesis share little or no homology with their functionally analogous counterparts in plants and fungi.
CYP112, CYP114, and CYP117 seem to clearly be of bacterial ancestry35, and the CYPs utilized by each kingdom are more similar to each other than to their functional analogs from the other kingdoms (Supplementary Table 6). Specifically, CYP117, the bacterial KO, is distinct from that of plants and fungi, as they belong to the CYP701 and CYP503 families, respectively2,9, and CYP114, the bacterial KAO, is also distinct, as the KAO from plants falls within the CYP88 family, and that of fungi in the CYP68A sub-family2,9. Furthermore, bacteria use an SDR to efficiently convert 6 to 7. This is unique to bacterial GA biosynthesis, as the full oxidation of C-7 appears to be accomplished by the KAO of plants and fungi without the assistance of a separate dehydrogenase. However, in some plants (e.g., within the Cucurbitaceae family), a 2ODD has been shown to act as a GA 7-oxidase2,36, converting 6 to 7 and perhaps suggesting that oxidation of C-7 to the carboxylate by KAOs is also inefficient in those species. For GA20ox activity, rhizobia nominally resemble fungi in that they use a CYP, but again, CYP112 is distinct from the fungal enzyme, which is from the CYP68B sub-family2,9, and this is completely dissimilar from plants, which use a 2ODD to carry out this reaction2. This appears to provide an extreme example for convergent evolution of such complex metabolism in three kingdoms, as over ten chemical transformations are necessary to reach 10 from the general diterpene precursor GGPP (Fig. 1b). Furthermore, there appear to be few examples of organisms having independently converged on identical biosynthetic pathways37, as is the case with the bacterial and plant pathways, which share common intermediates but not enzymes.
With three examples of independently evolved GA biosynthetic pathways now known, it can be appreciated that these all follow a common order of central chemical transformations, perhaps reflecting biochemical constraints that may have shaped GA production. In particular, within all three pathways, 4 is formed prior to the neighboring B-ring contraction. We suggest that the C-19 carboxylate of this molecule may provide anchimeric assistance via stabilization of a non-classical C-6 carbocation intermediate that has been invoked in these CYP (KO) catalyzed ring contraction reactions38. Specifically, it has been proposed that B-ring contraction may proceed via a pinacol-type rearrangement (as has been utilized in chemical synthesis of GAs)25–27, and the presence of the nearby C-19 carboxylate could presumably promote selective formation of the carbocation at C-639, although a radical mechanism is also plausible27. Notably, the inability of CYP114 to catalyze this ring contraction with 3 or the methyl ester of 4, both of which lack a free C-19 carboxylate, is consistent with this hypothesis (Supplementary Table 5).
Furthermore, this ring contraction always precedes incorporation of the C-19 carboxylate into the γ-lactone ring. 3D structural modeling shows that B-ring contraction brings the C-19 carboxylate closer to C-20 (Supplementary Fig. 9), perhaps assisting the coupled demethylation (i.e., loss of C-20) and C-19,10-γ-lactone ring formation catalyzed by the GA20ox. However, such incorporation of the carboxylate into the lactone also eliminates the negative charge that may guide the ring contraction reaction. Thus, these transformations potentially revolve around the early formation and availability of the C-19 carboxylate. We propose that γ-lactone formation cannot precede ring contraction because this results in loss of the free C-19 carboxylate. Indeed, the ancient origin of the C-19 carboxylate in pathway evolution is supported by evidence that 4 was an ancient plant metabolite, as indicated by its use as a precursor to non-GA signaling molecules in early diverging land plants40. Accordingly, the early presence of the C-19 carboxylate may have shaped the complex metabolic evolutionary process leading to contemporary GA biosynthesis.
While the role for GA production in symbiotic rhizobia is still unclear, as operon knockouts of B. japonicum have been shown to be unhindered in nodule formation and symbiosis16, it is known that phytopathogens use GA as a virulence factor, likely through suppression of the jasmonic acid (JA) induced defense responses41,42. This has been shown for two rice pathogens, specifically the fungus G. fujikuroi43 and the bacterial species Xanthomonas oryzae pv. oryzicola (which possesses a homologous operon to that studied here)44. It is tempting to speculate that production of ancient GA precursor hormones by plant-associated microbes, presumably to manipulate the plant, may have driven the elaboration of plant GA biosynthesis to the extant bioactive molecules in an “arms race”-like scenario45. However, we cannot rule out the possibility that bacteria and/or fungi only developed their GA biosynthetic pathways following full evolution of GA biosynthesis and signaling by plants, with the observed independent evolution simply a reflection of the value that production of GA offers these microbes in interaction with their plant hosts.
ONLINE METHODS
Culture and growth conditions
All chemicals were purchased from Thermo Fisher Scientific and Sigma-Aldrich. Where indicated and unless noted otherwise, antibiotics were used at the following concentrations: kanamycin (Km) 50 µg/mL, chloramphenicol (Cm) 25 µg/mL, streptomycin (Sm) 500 µg/mL, spectinomycin (Sp) 50 µg/mL, tetracycline (Tc) 20 µg/mL, rifampicin (Rf) 25 µg/mL. E. coli was grown at 37° C in NZY media (5 g/L yeast extract, 10 g/L casein hydrolysate, 5 g/L NaCl, and 1 g/L MgSO4·7H2O). S. meliloti 102146 was grown at 30° C in LB/MC media (Luria-Bertani media supplemented with 2.5 mM MgSO4·7H2O and 2.5 mM CaCl2·2H2O) using Sm as the selecting antibiotic. S. fredii NGR23447 was grown at 30°C in TY media (5 g/L tryptone, 5 g/L yeast extract, and 2.5 mM CaCl2·2H2O) using Rf as the selecting antibiotic. B. japonicum USDA110 was grown in AG media (1 g/L arabinose, 1 g/L gluconate, and 1 g/L yeast extract) with Cm (34 ug/mL) as the selecting antibiotic. See Supplementary Table 7 for all strains used in this study.
GA operon in B. japonicum USDA110 and S. fredii NGR234
The core GA operon is found in both B. japonicum USDA110 and S. fredii NGR234. Each operon contains CYP112, CYP114, FdGA, SDRGA, CYP117, GGPS, CPS, and KS. The locus tags for the genes characterized in this manuscript in B. japonicum are as follows: cyp112, blr2144; cyp114, blr2145, fdGA (not annotated in NCBI); sdrGA, blr2146; cyp117, blr2147. The locus tags for these genes in S. fredii are as follows: cyp112, cpxP/NGR_a02700; cyp114, cpxR/NGR_a02710; fdGA, NGR_a02720; sdrGA, NGR_a02730; cyp117, cpxU/NGR_a02740.
CYP117 falls into class II/class E (subgroup IV) within the CYP superfamily, which archetypically have a two-component electron transfer system in which an NADPH-cytochrome P450 reductase (CPR) directly supplies electrons to the CYP22, although in several cases it has been demonstrated that such CYPs can be reduced by the more typical bacterial 3-component system48,49. CYP114 and CYP112 belong to class B of the CYP superfamily. CYPs in this group typically utilize a 3-component electron transfer system consisting of a Fd reductase (FdR), ferredoxins (Fd), and a CYP22. Even given co-expression of the FdGA, this requires reduction by an endogenous FdR (e.g., from the S. meliloti host in the exogenous expression system used here), and it appears that an endogenous Fd or flavodoxin also can reduce these CYPs (i.e., even CYP114 exhibits some, albeit not full, activity in the absence of FdGA).
Cloning of operon genes
Unless noted otherwise, all molecular biology reagents were purchased from Thermo Fisher Scientific. CYP114 (cpxR; NGR_a02710) and FdGA (NGR_a02720) were amplified directly from S. fredii NGR234 gDNA as a single construct using primers that anneal to the 5’ end of CYP114 and the 3’ end of FdGA (See Supplementary Table 8 for all primers used here). Synthetic clones of CYP112 (cpxP; NGR_a02700) and CYP117 (cpxU; NGR_a02740) were created with GeneArt™ gene synthesis via Thermo Fisher Scientific, in which each CYP was followed directly by the non-coding region following CYP114, along with FdGA. These synthetic constructs were made to contain silent point mutations to remove EcoRI and BamHI from the coding sequences to allow for use of these restriction sites for downstream cloning (G606A and A738G in CYP112; G202A in CY117). S. fredii NGR234 SDRGA (NGR_a02730) was cloned directly from gDNA. Constructs were initially amplified through PCR with AccuPrime Pfx and cloned into pCR™-Blunt II-TOPO® (KmR) for subsequent propagation. For S. meliloti expression, constructs were amplified with forward primers that annealed to the 5’ end of the gene and also contained a “linker” sequence containing stop codons and a ribosome binding site along with a 5’ BamHI restriction site, and a reverse primer that added a 3’ EcoRI site to the end of the gene (Supplementary Table 8). For individual CYP expression, the reverse primer was specific to the 3’ end of the CYP, whereas for CYP coexpression with FdGA, the reverse primer was specific to the 3’ end of FdGA. The S. fredii NGR234 CYP constructs with and without FdGA were digested and subsequently ligated into the pstb-LAFR5 vector (TcR, mob+), while the S. fredii SDRGA was cloned into this plasmid without FdGA. The pstb-LAFR5 vector contains a promoter sequence that has been shown to be highly and constitutively expressed in S. meliloti 102150, and was thus deemed appropriate for heterologous expression of the operon genes.
E. coli codon-optimized synthetic B. japonicum USDA110 gene constructs for CYP114 (blr2145), FdGA, SDRGA (blr2146), and five putative ferredoxin reductases (FdR1, mvrA/blr3831; FdR2, blr3195; FdR3. bll0100; FdR4, blr7998; and FdR5, paaE/blr2895) were ordered from Thermo Fisher scientific (via GeneArt™ gene synthesis). The synthetic CYP114 was cloned into pET101/D-TOPO (SpR) as per the manufacturer’s instructions, but with the stop codon included to prevent fusion with the C-terminal tag. The B. japonicum USDA110 SDRGA and FdGA were cloned separately into the pCDF-duet expression vector (SpR) with FdGA coexpressed with each of the putative FdR’s (Supplementary Table 7).
Triparental mating for movement of constructs into S. meliloti 1021
For expression in S. meliloti 1021, the pstb-LAFR5 constructs (Supplementary Table 7) were moved into the host cells through triparental mating. First, pstb-LAFR5 constructs were transformed into chemically competent E. coli MM294A cells51 using LB plates with Tc to select for positive colonies. Liquid cultures of the transformed MM294A cells along with the helper E. coli strain MT61652 (CmR) and the host, S. meliloti 1021 (SmR) were grown to mid-log phase, at which point 1 mL of each culture was centrifuged at 4000 x g to pellet the cells. Cell pellets were resuspended in 1 mL of sterile 0.85% NaCl to wash away media and antibiotics, after which the cells were again pelleted by centrifugation at 4000 x g. These cell pellets were then resuspended in 50 µL of sterile 0.85% NaCl and each culture was mixed in equal proportion. The mixtures were spotted in 10–30 µL aliquots onto non-selective LB/MC plates and allowed to incubate at 30° C for 1–2 days. The spots were then resuspended in 1 mL of sterile 0.85% NaCl, serially diluted, and plated on LB/MC plates containing Sm and Tc to select for S. meliloti 1021 (SmR) colonies that contain the pstb-LAFR5 construct (TcR). Positives were confirmed through colony PCR for the gene(s) of interest.
Substrates for heterologous expression
The ent-kaurene (1), ent-kaurenol (2), and ent-kaurenoic acid (4) substrates/standards were made through our metabolic engineering system in E. coli, as previously described53. Authentic ent-7α-hydroxykaurenoic acid (5), GA15 (8), and GA24 (9)-methyl ester were provided by Dr. Peter Hedden. GA12-aldehyde (6), GA12 (7), and GA9 (10) were purchased from OlChemIm Ltd.
Production of ent-kaurenal (3) was achieved through the oxidation of 2 with MnO2. Approximately 1 mg of 2 was dissolved in MeOH, and an excess (>1000-fold molar ratio) of activated MnO2 was added and stirred overnight with a magnetic stir bar and plate. The mixture of catalyst and compound(s) was filtered through Whatman filter paper (GE Healthcare Life Sciences) to remove MnO2, dried down under a gentle nitrogen stream, and resuspended in n-hexane for GC analysis. This process yielded nearly 100% conversion and recovery of 3 from 2.
GA12-aldehyde (6) was purchased from OlChemIm Ltd. for use as a substrate. However, this preparation contained a small amount of GA12 (7), which would potentially confound results, as 7 is the product expected from enzymatic activity with 619. Therefore, 2 mg of this substrate was further purified with an Agilent 1200 series HPLC equipped with a reverse-phase C-8 column (Kromasil® C8, 50×4.6 mm), autosampler, diode array UV detector, and fraction collector. The mobile phases utilized were acetonitrile and water, both with 0.1% formic acid to lower the pH to ~3, which leads to predominant protonation of carboxylic acids and better peak resolution. 6 was dissolved in MeOH, and this sample was injected in 50 µL aliquots at a flow rate of 0.5 mL/min (300 bar, 25°C) with an initial mobile phase of 50% acetonitrile in water which was held for 2 minutes. For the next 5 minutes, the mobile phase was increased to 100% acetonitrile. This was held for 16 minutes, at which point the mobile phase was switched back to 50% acetonitrile in water over a time of 1 minute, and this was held for 3 minutes. With this method, a single peak of 6 eluted at 14.3 minutes, and a fraction was collected from 14.0 to 14.5 minutes to recover purified product. With this purification, most of the contaminating 7 was removed, enabling use of this preparation of 6 as a substrate.
GA24 (9) was produced from 9-methyl ester for substrate feeding studies through base hydrolysis. Briefly, 9-methyl ester (~1 mg) was resuspended in 0.5 M KOH in methanol (3 mL) and incubated at 40° C for 16 hours. Water (10 mL) was used to quench the reaction, and this aqueous fraction was acidified to pH 3. The acidified aqueous fraction was extracted five times with ethyl acetate (10 mL), and this organic fraction was dried down under a gentle nitrogen stream and resuspended in MeOH. Hydrolysis was confirmed through GC-MS, both of the hydrolyzed product (i.e. disappearance of a peak on GC-MS due to loss of methyl ester derivatization) and re-methylated product to confirm the presence of 9.
Before incubation, GA15 (8) was hydrolyzed to preferentially form the free C-20 alcohol in contrast to the 19,20 δ-lactone, as the free alcohol form has been implicated as the major substrate of GA 20-oxidases in plant GA biosynthesis54. This was done by dissolving 8 in 0.5 M KOH with subsequent heating at 100°C for 2 hours, as previously described55. As with the plant system, we found more efficient turnover with hydrolyzed 8 in our cells expressing CYP112.
Substrates were dissolved in ethanol (1, 2, and 3) or methanol (4, 5, 6, 7, 8, 9, and 10) and stored at a concentration of 2 or 3 mg/mL.
Heterologous expression and substrate feeding studies
For S. meliloti 1021 heterologous expression and feeding with potential substrates, 5 mL culture tubes of liquid LB/MC with appropriate antibiotics (Sm for wild-type controls, Sm and Tc for expression constructs) were inoculated with individual colonies and allowed to grow to confluence, with shaking at 225 RPM for 1–2 days at 30 °C. The presence of the construct in the cells of the culture was confirmed with colony PCR using gene specific primers. 0.5 mL of the starter culture and 5 mL of phosphate buffer was then added to 50 mL LB/MC containing the appropriate antibiotics. These larger cultures were grown to log phase (OD600 0.5–09), and substrate (in EtOH or MeOH) mixed with DMSO in a 1:1 mixture was added into the culture to a final concentration of 10 µM. Additionally, riboflavin (1 mM) was added for enhanced metabolism, 5-aminolevulinic acid (1 mM) for enhanced heme synthesis, and FeCl3 (0.25 mM) for increased CYP and Fd functionality (i.e. supplying iron for the CYP heme and Fd)56. Substrates were incubated for three days in the cultures while shaken at 30 °C and 225 RPM. Once the GA operon enzymes were found to be functionally expressed, culture size was reduced to 10 mL and substrate concentration was reduced to 5 µM for subsequent experiments. Each substrate incubation which resulted in chemical transformation was repeated at least twice to ensure reproducibility. See Supplementary Table 9 for a list of all of the functionally characterized genes, along with their NCBI/GenBank and UniProtKB identifiers.
To determine if substrates were able to enter the CYP114-FdGA-expressing cells, cultures were incubated with substrate as described above, and following incubation, were centrifuged at 4000 x g to pellet cells away from aqueous media. The cell pellet was washed twice with an original culture volume of 0.85% NaCl, these aqueous washes were combined with the original media supernatant, and the cell pellet was resuspended in an original culture volume of 0.85% NaCl. Both the cell pellet fraction and the aqueous fraction were extracted and analyzed as described below (see Metabolite extraction, purification, and GC/MS analysis for heterologous expression studies).
In order to assess whether GA12-aldehyde (6) oxidation to GA12 (7) can be catalyzed by non-specific oxidoreductases in rhizobia or by CYP114 (+/−FdGA), 6 was incubated in non-inoculated media (supplied with antibiotics and expression supplements), wild-type/untransformed S. meliloti 1021, and S. meliloti cells expressing either CYP114 or CYP114-FdGA. Because trace amounts of 7 were still present in 6 substrate following HPLC purification, each incubation was run in triplicate, and potential increases in the amount of 7 as an enzymatic product were determined by dividing the ion extracted peak area (m/z = 300) of 7 by the ion extracted peak area (m/z = 300) of 6 in each sample and averaging this value between treatments (Supplementary Figure 7). Statistical analysis (Student’s t-test: Two-Sample Assuming Equal Variance) was performed in Microsoft Excel. The corresponding p-values are listed in Supplementary Figure 7, and the variances between the cultures incubations were reasonably similar (Sm1021=6.37×10−5; CYP114=3.09×10−4; CYP114-FdGA=1.11×10−4).
For E. coli expression and substrate feeding, 5 mL culture tubes of Terrific Broth (TB) media (12 g/L casein, 24 g/L extract, 0.4% glycerol) with Sp for selection were inoculated with individual colonies of E. coli C41 cells transformed with the construct of interest. These small cultures were allowed to grow to late log phase overnight at 37 °C and with 225 RPM shaking. 1 mL of this starter culture and 5 mL phosphate buffer was then added to 50 mL TB containing appropriate antibiotics. 50 mL cultures were grown to log phase at 37 °C with 225 RPM shaking, then moved to 16 °C with 225 shaking for one hour, after which expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and riboflavin (1 mM) and substrate (10 µM) were added. Cultures were incubated with substrate for 3 days at 16 °C and 225 RPM shaking.
Metabolite extraction, purification, and GC/MS analysis for heterologous expression studies
After three days of incubation with substrate, the cultures were acidified to pH 3 with 5.0 M HCl to neutralize the carboxylate groups on the targeted compounds, and these were then extracted three times with an equivalent volume of hexanes (for 1, 2, and 3 substrates) or ethyl acetate (for 4, 4-methyl ester, 5, 6, 7, 8, 9, and 10 substrates). The three extractions from each culture were combined in a round bottom flask and dried on a rotary evaporator. The residue was resuspended in 1 mL of ethyl acetate and loaded onto a ~1 mL silica gel column and purified with elution of hexane:ethyl acetate solutions. Briefly, this was done by adding 1 mL aliquots of elution solvent, starting with 100% hexane, and incrementally increasing the ethyl acetate concentration (by 5–10%) until reaching a final elution of 100% ethyl acetate. Distinct fractions were collected for each elution solvent, and these were dried down under a gentle nitrogen stream, dissolved in ethyl acetate, and derivatized with diazomethane in order to esterify any free acids. Derivatized samples were dried down under a stream of nitrogen, re-dissolved in n-hexane and analyzed with gas chromatography coupled to a mass spectrometer detector (GC-MS), using a 3900 Saturn GC with Saturn 2100T ion trap MS (Varian) equipped with an HP-5MS column (Agilent). For analysis of each sample, 1 µL was injected in splitless mode with a column flow of 1.2 mL/minute at an initial injector temperature of 250 °C and a column temperature of 50°C, which was held for 3 minutes. The column temperature was then increased to 300°C at a rate of 15°C/minute, and this temperature (300°C) was further held for 3 minutes. Mass spectra acquisition using electrospray ionization with a low mass ion limit of 90 m/z and a high mass ion limit of 650 m/z began at 13 minutes and continued until the end of the run. For optimal peak resolution, publication quality GC-MS runs were performed as described, but with a column temperature increase of 10°C/min and a mass range of 90–450 m/z. Potential products were compared to authentic standards and published chromatograms/mass spectra57 (www.planthormones.info/gibberellins.htm). Fractions from each individual sample that contained the GA substrate and products were combined for the final published chromatograms.
Development of B. japonicum and S. fredii knockout mutants
To delete the GA operon genes of interest in B. japonicum (cyp112/blr2144, cyp114/blr2145, fdGA, sdrGA/blr2146, and cyp117/blr2147) and S. fredii (cyp112, cyp114, fdGA, sdrGA, and cyp117) we utilized the well-established double-homologous recombination approach, which has been shown to be effective in a number of different bacterial species23,24. To begin, we cloned regions that were 500–1000 bp upstream and downstream of the gene of interest, including a 3’ linker on the upstream fragment that is complementary to the 5’ fragment, and a 5’ linker on the downstream fragment that is complementary to the 3’ fragment (Supplementary Table 8). These two fragments were fused using crossover-PCR, and appropriate restriction sites were added to the ends of this construct using unique primers. This construct was then ligated into the suicide vector pK19mobsacB (Supplementary Table 7), which is mobilizable and contains the Km resistance cassette for positive selection of colonies, along with the sacB gene, which results a restricted growth phenotype on sucrose for bacteria carrying the plasmid, and thus negative selection of colonies.
These plasmid constructs were transformed into chemically competent MM294A E. coli cells and moved into the rhizobia of interest through triparental mating (see Triparental mating for movement of constructs into S. meliloti 1021 for general outline of this procedure). Briefly, cultures of the “donor” MM294A cells, “helper” MT616 E. coli cells, and the “recipient” strain (either B. japonicum USDA110 or S. fredii NGR234) (Supplementary Table 7) were grown to late log phase, mixed in equal proportion, and spotted on non-selective agar media (AG for B. japonicum and TY for S. fredii) for 3–5 days (B. japonicum) or 1–2 days (S. fredii). These mixtures were suspended in sterile 0.85% NaCl, and serial dilutions were plated on the appropriate agar containing an antibiotic to select for the recipient (Tc at 10 ug/mL for B. japonicum and Rf for S. fredii) and Km to select for the first crossover integration of the suicide vector into the genome. Potential first crossover colonies were streaked out on Km-containing plates at least once in order to confirm their resistance, and these colonies were further confirmed with colony PCR using primers designed to amplify a region of the sacB gene. Positive strains were grown to late log phase in non-selective liquid media (thus allowing for the 2nd crossover event to occur), and serial dilutions were plated on the appropriate media containing 10% (w/v) sucrose. Possible double recombinants were streaked on Km along with on sucrose plates to ensure loss of the vector, resulting in restricted growth on Km and the ability to grow on sucrose. Colony PCR with primers designed upstream and downstream of the gene of interest was used to determine whether the 2nd recombination resulted in restoration to the wild-type or the deletion of the gene of interest (i.e. a reduction in amplicon size equal to the length of the gene indicated a successful deletion).
Isolation of B. japonicum or S. fredii knockout bacteroids
Soybean (Glycine max cv. Jack) plants inoculated with liquid cultures of B. japonicum knockout mutants, or alternatively cowpea (Vigna unguiculata cv. Blackeye) plants inoculated with liquid cultures of S. fredii knockout mutants, were grown in Leonard jars as described18. Symbiotic root nodules were obtained from plants around flowering and stored at −20 °C. Bacteroids were isolated from nodule homogenates by differential centrifugation as described19 and the bacteroid pellet, resuspended in 50 mM TES pH 7.5, 2.5 mM MgCl2, 1 mM KHPO4, was utilized for enzymatic assays.
Labeled substrates for bacteroid enzymatic assays
[14C4]GA12-aldehyde and ent-[14C4]7α-hydroxykaurenoic acid were synthesized from R-[2-14C]mevalonic acid (Amersham) by incubation with an endosperm preparation from Cucurbita maxima58,59. ent-[17-14C1]kaurenoic acid, [17-14C1]GA12, [17-14C1]GA15, [17-14C1]GA24, and [17-14C1]GA9 were obtained from Professor L. Mander (Australian National University, Canberra). ent-kaurenol (2) was prepared as described above.
Knockout bacteroid incubations with labeled GA precursors
14C-GA precursors (830–5000 Bq, 0.1–0.6 nmoles) were added as methanolic solutions to 2 mL of a bacteroid suspension prepared from 0.5 g of root nodules. After incubation for 3 days at 28 °C with shaking at 200 rpm, products were extracted and isolated from the reaction media. The suspension was centrifuged at 12,000 x g for 5 min and the supernatant extracted with ethyl acetate after acidification to pH 3.0. Products were further separated by HPLC on a C18 Symmetry column (5µm; 250×4 mm; Waters) in a Waters 600 HPLC instrument, after purification by solid phase extraction in C18 cartridges. A linear gradient of MeOH:H2O pH 3.0 (60:40→ 100:0; v/v) over 30 min was used for elution at a flow rate of 1 mL/min. Fractions (1 mL) were collected and the radioactivity measured by liquid scintillation counting.
Purified fractions containing 14C-labeled products were derivatized with ethereal diazomethane and N-methyl-N-trimethylsilyltrifluoroacetamide and identified by GC-MS as described60. Compounds were identified by comparison of their mass spectra with published spectra57. For GA9 17-nor-16-one and GA12 17-nor-16-one the spectra of the methyl esters of ketone and enol-TMS derivative were compared to those of authentic standards.
In these knockout-bacteroid feeding experiments, various oxidized kaurenoid or GA-derived compounds were identified that are unlikely to belong to the GA biosynthesis pathway. These would presumably correspond to reactions potentially catalyzed by non-pathway-related oxidoreductases due to blocked biosynthesis and/or intermediate accumulation. In correspondence with this, previous experimentation has found non-pathway related GAs such as GA9-norketone and 13-OH-kaurenoic acid, which do not seem to be relevant GA intermediates, in incubations with wild-type B. japonicum19. Also, as summarized in Supplementary Figure 3, greater substrate turnover was detected in B. japonicum bacteroid incubations than for those in S. fredii.
Molecular modeling
ent-kaurenoic acid (4) and GA12-aldehyde (6) were modeled using Jmol 13.0. Briefly, the simplified molecular-input line-entry system (SMILES) for each molecule were used to create a 3D structure in Jmol. Each molecule was computationally energy-optimized within Jmol and the distance from C-19 to C-20 and C-19 to C-6 was measured. The same measurements were made using 3D structures generated from the International Chemical Identifier (InChl) and Chemical Abstracts Service (CAS) number of each compound to confirm the measured distances.
CYP sequence analysis
CYPs involved in GA biosynthesis in plants (Arabidopsis thaliana KO/CYP701A3, KAO1/CYP88A3, and KAO2/CYP88A4), fungi (Gibberella fujikuroi KO/CYP503, KAO/CYP68A, and GA20ox/CYP68B) , and bacteria (Bradyrhizobium japonicum USDA110 CYP117, CYP114, and CYP112) were compared using the Needle EMBOSS (Needleman-Munsch algorithm) pairwise alignment tool available on the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) website (www.ebi.ac.uk/Tools/psa/emboss_needle) to determine the amino acid percent identity shared between each enzyme. The default parameters were used for this analysis, and are as follows: matrix=BLOSUM62, gap open=10, gap extend=0.5, end gap penalty=false, end gap open=10, end gap extend=0.5.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (GM109773 to R.J.P.) and the Iowa Soybean Association (grant to R.J.P.); a Discovery Grant and an Engage Grant, both from Natural Sciences and Research Engineering Council of Canada (NSERC; grants to T.C.C.), and an Ontario Graduate Scholarship (A.M.); the 20:20 Wheat© Integrated Strategic Programme at Rothamsted Research funded by the Biotechnology and Biological Sciences Research Council of the United Kingdom (support to P.H.); and Fondo Nacional de Desarrollo Cientifico y Tecnologico (grant 1150797 to M.C.R.).
Footnotes
Author Contribution
All authors provided significant contribution to this work.
R.S.N. performed all aspects of the heterologous expression assays, created many of the knockout bacterial strains, and wrote the manuscript and a majority of the supplemental information and prepared manuscript figures. X.L. aided in cloning related to the heterologous expression assays, and R.N. provided insightful data analysis. M.M. and M.C.R. performed the bacteroid incubation assays and analyzed the resulting extracts by HPLC, along with contributing to the manuscript and the supplemental information. A.M. and T.C.C. created several of the knockout bacterial strains and contributed to the supplemental information. P.H. analyzed extracts from the bacteroid incubation assays by GC-MS and contributed to the manuscript and supplemental information. R.J.P. conceived the project, aided in data analysis, and helped write the manuscript.
Competing Financial Interests Statement
The authors involved with this work declare no competing financial interests.
Supplementary Information
Supplementary Figures 1– 12 and Supplementary Tables 1– 9 are included in the Supplementary Information document.
REFERENCES
- 1.Hedden P, Sponsel V. A century of gibberellin research. J. Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 3.Bari R, Jones JDG. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi S, Gresshoff PM, Ferguson BJ. Mechanistic action of gibberellins in legume nodulation. J. Integr. Plant Biol. 2014;56:971–8. doi: 10.1111/jipb.12201. [DOI] [PubMed] [Google Scholar]
- 5.Peng J, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki A, et al. A mutant gibberellin-synthesis gene in rice. Nature. 2002;291:1–2. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 7.Bömke C, Tudzynski B. Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry. 2009;70:1876–1893. doi: 10.1016/j.phytochem.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 8.MacMillan J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 2002;20:387–442. doi: 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- 9.Tudzynski B. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 2005;66:597–611. doi: 10.1007/s00253-004-1805-1. [DOI] [PubMed] [Google Scholar]
- 10.Morrone D, et al. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum . FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 11.Magome H, et al. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1947–52. doi: 10.1073/pnas.1215788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya A, et al. Characterization of the fungal gibberellin desaturase as a 2-oxoglutarate-dependent dioxygenase and its utilization for enhancing plant growth. Plant Physiol. 2012;160:837–845. doi: 10.1104/pp.112.201756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters RJ. In: Isoprenoid Synth. Plants Microorg. New Concepts Exp. Approaches. Bach TJ, Rohmer M, editors. Springer; 2013. pp. 233–249. [Google Scholar]
- 14.Atzorn R, Crozier A, Wheeler CT, Sandberg G. Production of gibberellins and indole-3-acetic acid by Rhizobium phaseoli in relation to nodulation of Phaseolus vulgaris roots. Planta. 1988;175:532–538. doi: 10.1007/BF00393076. [DOI] [PubMed] [Google Scholar]
- 15.Delamuta JRM, et al. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 2013;63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- 16.Tully RE, Keister DL. Cloning and mutagenesis of a cytochrome P-450 locus from Bradyrhizobium japonicum that is expressed anaerobically and symbiotically. Appl. Environ. Microbiol. 1993;59:4136–4142. doi: 10.1128/aem.59.12.4136-4142.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tully RE, Van Berkum P, Lovins KW, Keister DL. Identification and sequencing of a cytochrome P450 gene cluster from Bradyrhizobium japonicum . Biochim. Biophys. Acta. 1998;1398:243–255. doi: 10.1016/s0167-4781(98)00069-4. [DOI] [PubMed] [Google Scholar]
- 18.Keister DL, Tully RE, Van Berkum P. A cytochrome P450 gene cluster in the Rhizobiaceae. J. Gen. Appl. Microbiol. 1999;45:301–303. doi: 10.2323/jgam.45.301. [DOI] [PubMed] [Google Scholar]
- 19.Méndez C, et al. Gibberellin oxidase activities in Bradyrhizobium japonicum bacteroids. Phytochemistry. 2014;98:101–109. doi: 10.1016/j.phytochem.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Hershey DM, Lu X, Zi J, Peters RJ. Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J. Bacteriol. 2014;196:100–106. doi: 10.1128/JB.01031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galibert F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti . Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 22.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems-biological variations of electron transport chains. Biochim. Biophys. Acta - Gen. Subj. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 23.Schäfer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 24.Sukdeo N, Charles TC. Application of crossover-PCR-mediated deletion-insertion mutagenesis to analysis of the bdhA-xdhA2-xdhB2 mixed-function operon of Sinorhizobium meliloti . Arch. Microbiol. 2003;179:301–304. doi: 10.1007/s00203-003-0532-9. [DOI] [PubMed] [Google Scholar]
- 25.Mander LN. The chemistry of gibberellins: an overview. Chem. Rev. 1992;92:573–612. [Google Scholar]
- 26.Silva LF. Construction of cyclopentyl units by ring contraction reactions. Tetrahedron. 2002;58:9137–9161. [Google Scholar]
- 27.Citron CA, Brock NL, Tudzynski B, Dickschat JS. Labelling studies on the biosynthesis of terpenes in Fusarium fujikuroi . Chem. Commun. (Camb) 2014;50:5224–6. doi: 10.1039/c3cc45982a. [DOI] [PubMed] [Google Scholar]
- 28.Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojas MC, Hedden P, Gaskin P, Tudzynski B. The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5838–5843. doi: 10.1073/pnas.091096298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertea CM, et al. Identification of intermediates and enzymes involved in the early steps of artemisin biosynthesis in Artemisia annua . Planta Med. 2005;71:40–47. doi: 10.1055/s-2005-837749. [DOI] [PubMed] [Google Scholar]
- 31.Teoh KH, Polichuk DR, Reed DW, Covello PS. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua . Botany. 2009;87:635–642. [Google Scholar]
- 32.Du L, et al. Characterization of a unique pathway for 4-cresol catabolism initiated by phosphorylation in Corynebacterium glutamicum . J. Biol. Chem. 2016;291:6583–6594. doi: 10.1074/jbc.M115.695320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao R, et al. Diterpene cyclases and the nature of the isoprene fold. Proteins Struct. Funct. Bioinforma. 2010;78:2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, et al. Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum . Sci. Rep. 2014;4:6214. doi: 10.1038/srep06214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson DR. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 36.Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli . Proc. Natl. Acad. Sci. U. S. A. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen NB, et al. Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nat. Commun. 2011;2:273. doi: 10.1038/ncomms1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortiz de Montellano PR, Nelson SD. Rearrangement reactions catalyzed by cytochrome P450s. Arch. Biochem. Biophys. 2011;507:95–110. doi: 10.1016/j.abb.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song ZL, Fan CA, Tu YQ. Semipinacol rearrangement in natural product synthesis. Chem. Rev. 2011;111:7523–7556. doi: 10.1021/cr200055g. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi K, et al. Endogenous diterpenes derived from ent-kaurene, a common gibberellin precursor, regulate protonema differentiation of the moss Physcomitrella patens . Plant Physiol. 2010;153:1085–1097. doi: 10.1104/pp.110.157909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navarro L, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 42.Yang DL, et al. Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant. 2008;1:528–537. doi: 10.1093/mp/ssn021. [DOI] [PubMed] [Google Scholar]
- 43.Wiemann P, et al. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu X, Hershey DM, Wang L, Bogdanove AJ, Peters RJ. An ent-kaurene-derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola. New Phytol. 2015;206:295–302. doi: 10.1111/nph.13187. [DOI] [PubMed] [Google Scholar]
- 45.Anderson JP, et al. Plants versus pathogens: An evolutionary arms race. Funct. Plant Biol. 2010;37:499–512. doi: 10.1071/FP09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 46.Buikema WJ, et al. Physical and genetic characterization of Rhizobium meliloti symbiotic mutants. J. Mol. Appl. Genet. 1983;2:249–260. [PubMed] [Google Scholar]
- 47.Trinick MJ. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J. Appl. Bacteriol. 1980;49:39–53. [Google Scholar]
- 48.Bellamine A, Mangla AT, Nes WD, Waterman MR. Characterization and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. U. S. A. 1999;96:8937–42. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You Z, Omura S, Ikeda H, Cane DE. Pentalenolactone biosynthesis: Molecular cloning and assignment of biochemical function to PtlF, a short-chain dehydrogenase from Streptomyces avermitilis, and identification of a new biosynthetic intermediate. Arch. Biochem. Biophys. 2007;459:233–240. doi: 10.1016/j.abb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones KM. Increased production of the exopolysaccharide succinoglycan enhances Sinorhizobium meliloti 1021 symbiosis with the host plant Medicago truncatula . J. Bacteriol. 2012;194:4322–4331. doi: 10.1128/JB.00751-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Backman K, Boyer HW. Tetracycline resistance determined by pBR322 is mediated by one polypeptide. Gene. 1983;26:197–203. doi: 10.1016/0378-1119(83)90190-7. [DOI] [PubMed] [Google Scholar]
- 52.Finan TM, Kunkel B, De Vos GF, Signer ER. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrone D, et al. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 2010;85:1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward JL, et al. Stereochemistry of the oxidation of gibberellin 20-alcohols, GA15 and GA44, to 20-aldehydes by gibberellin 20-oxidases. Chem. Commun. 1997:13–14. [Google Scholar]
- 55.Kamiya Y, Graebe JE. The biosynthesis of all major pea gibberellins in a cell-free system from Pisum sativum . Phytochemistry. 1983;22:681–689. [Google Scholar]
- 56.Furuya T, Arai Y, Kino K. Biotechnological production of caffeic acid by bacterial cytochrome P450 CYP199A2. Appl. Environ. Microbiol. 2012;78:6087–6094. doi: 10.1128/AEM.01103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Binks R, MacMillan J, Pryce RJ. Combined gas chromatography-mass spectrometry of the methyl esters of gibberellins A1 to A24 and their trimethylsilyl ethers. Phytochemistry. 1969;8:271–84. [Google Scholar]
- 58.Graebe JE, Hedden P, Gaskin P, MacMillan J. Biosynthesis of gibberellins A12, A15, A24, A36, and A37 by a cell-free system from Cucurbita maxima . Phytochemistry. 1974;13:1433–1440. [Google Scholar]
- 59.Urrutia O, Hedden P, Rojas MC. Monooxygenases involved in GA12 and GA14 synthesis in Gibberella fujikuroi . Phytochemistry. 2001;56:505–511. doi: 10.1016/s0031-9422(00)00381-2. [DOI] [PubMed] [Google Scholar]
- 60.Troncoso C, Cárcamo J, Hedden P, Tudzynski B, Rojas MC. Influence of electron transport proteins on the reactions catalyzed by Fusarium fujikuroi gibberellin monooxygenases. Phytochemistry. 2008;69:672–83. doi: 10.1016/j.phytochem.2007.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.