The idea that mismatched base pairs occur in cells and that such lesions trigger their own repair was suggested 50 years ago by Robin Holliday in the context of genetic recombination (1). Breakage and rejoining of DNA helices was known to occur during this process (2), with precision of rejoining attributed to formation of a heteroduplex joint, a region of helix where the two strands are derived from the different recombining partners. Holliday pointed out that if this heteroduplex region should span a genetic difference between the two DNAs, then it will contain one or more mismatched base pairs. He invoked processing of such mismatches to explain the recombination-associated phenomenon of gene conversion (1), noting that “If there are enzymes which can repair points of damage in DNA, it would seem possible that the same enzymes could recognize the abnormality of base pairing, and by exchange reactions rectify this.”
Direct evidence that mismatches provoke a repair reaction was provided by bacterial transformation experiments (3–5), and our interest in this effect was prompted by the Escherichia coli (E. coli) work done in Matt Meselson’s lab at Harvard. Using artificially constructed heteroduplex DNAs containing multiple mismatched base pairs, Wagner and Meselson (6) demonstrated that mismatches elicit a repair reaction upon introduction into the E. coli cell. They also showed that closely spaced mismatches, mismatches separated by a 1000 base pairs or so, are usually repaired on the same DNA strand. Based on this strand bias effect, Wagner and Meselson proposed that in addition to its role in genetic recombination, “…mismatch repair may act to correct mutations that arise as replication errors. If so, it may be that mismatch repair acts in a directed manner in conjunction with sister chromatid exchange or that it occurs with particularly high efficiency on newly synthesized DNA strands, possibly because of their undermethylation or because of a special relation to the replication complex.” This suggestion proved to be particularly insightful, and we now know from work in many labs that correction of DNA biosynthetic errors is a primary job of mismatch repair (7).
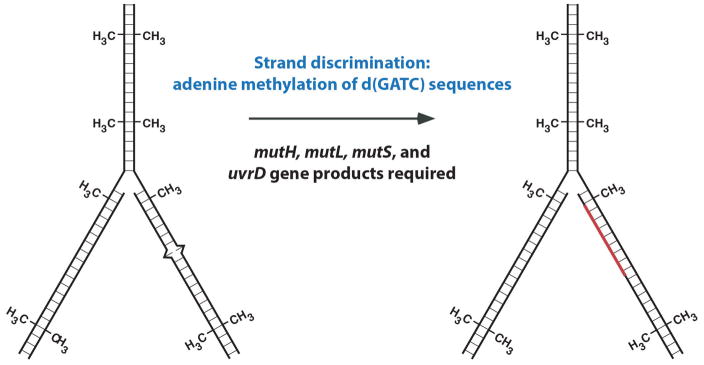
In order to function in this manner, the repair system must be able to do two things. It must recognize the mismatched base pair produced by the replication error, but it also has to identify the new DNA strand, which contains the mistake. Pat Pukkila in the Meselson lab showed that the strand direction of E. coli mismatch repair is dictated by the state of adenine methylation at d(GATC) sequences (8). Because this modification occurs after DNA synthesis, newly synthesized DNA exists transiently in an unmodified state, and it is this transient absence of methylation that directs repair to the new strand (Fig. 1). Consistent with the idea that mismatch repair contributes to replication fidelity, Miro Radman, Barry Glickman, and others (9–11) showed that the methyl-directed pathway depends on the products of four E. coli mutator genes: mutH, mutL, mutS, and uvrD. Inactivation of any of these genes increases mutation production in the E coli cell 50-to 100-fold, indicating the importance of this pathway in mutation avoidance and genetic stability.
FIGURE 1.
E. coli methyl-directed mismatch repair.
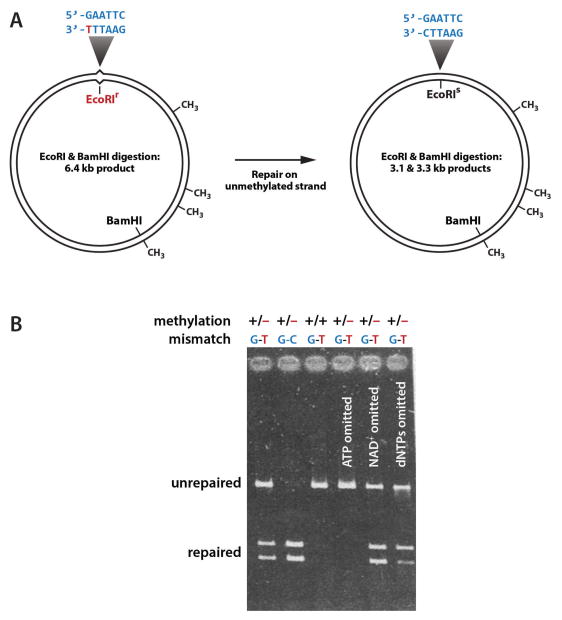
This is where we entered the picture. I was interested in how mismatches might be recognized and how the state of DNA methylation at one site on the helix directs mismatch repair elsewhere on the DNA. To address these questions, we needed a biochemical assay. Exploiting several tricks developed in Norton Zinder’s laboratory at Rockefeller University (12,13), A-Lien Lu built heteroduplex DNAs like that shown in Fig. 2A: circular molecules in which the strands are in defined states of d(GATC) methylation and which contain a G-T mismatch within the recognition site for EcoRI restriction endonuclease (14,15). Because the mismatch blocks DNA cleavage by EcoRI, digestion of this DNA with EcoRI and BamHI endonucleases yields a full-length linear product (Fig 2B, unrepaired). However, if repair occurs on the unmethylated strand, as predicted by the Meselson mechanism, the G-T mismatch will be corrected to a G-C base pair restoring EcoRI sensitivity, and digestion with EcoRI and BamHI will produce the two smaller DNA fragments (Fig 2B, repaired). In fact, incubation of this DNA with extracts prepared from broken E. coli cells converts it to an EcoRI sensitive form. As anticipated by Meselson and colleagues (8), this in vitro reaction is blocked when both DNA strands are methylated, and like in vivo methyl-directed repair (9–11), the in vitro reaction depends on functional products of the mutH, mutL, mutS and uvrD genes (14,15).
FIGURE 2.
Methyl-directed mismatch repair in E. coli cell extracts.
A. Assay for in vitro mismatch repair. Presence of the G-T mismatch within the EcoRI recognition sequence in unrepaired DNA blocks cleavage by this endonuclease. B. In vitro repair of heteroduplex shown in panel A. Residual heteroduplex repair occurring in the absence of exongenous dNTPs (right lane) is due to presence of the DNA biosynthetic precursors in the extract (18).
Panel A is adapted with permission from reference 15; panel B is adapted from reference 14.
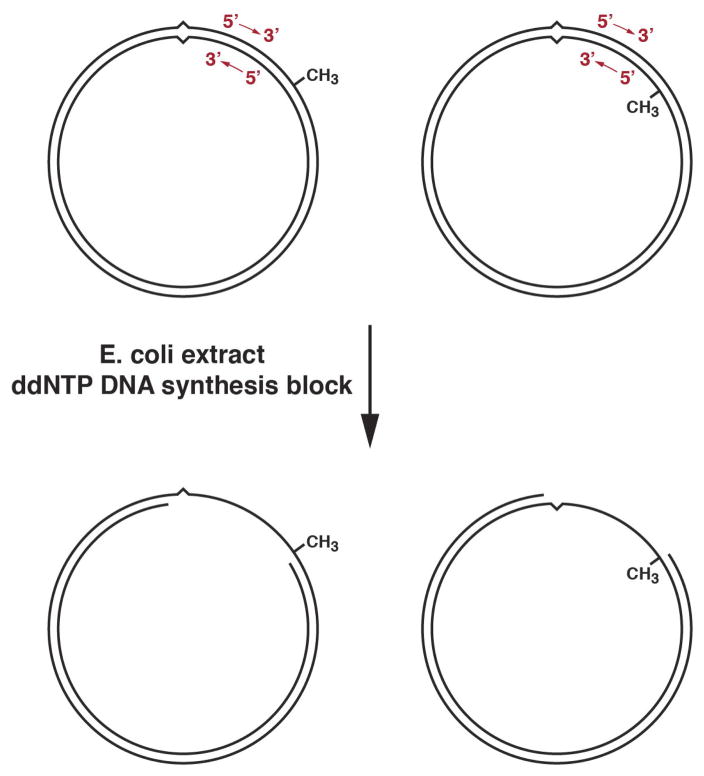
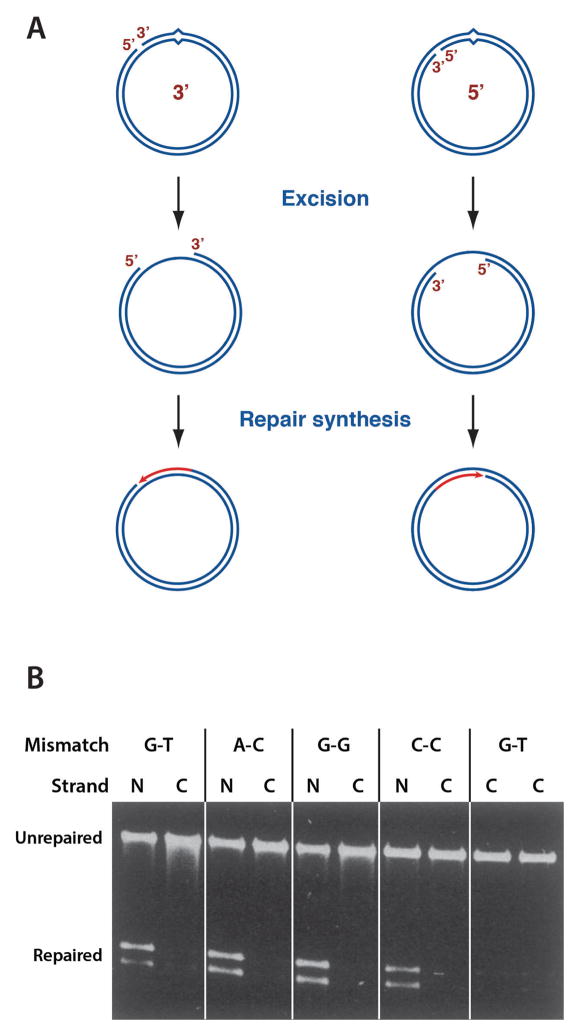
Michael Su, Bob Lahue, and Karin Au showed that this in vitro extract reaction supports repair of all of the base-base mismatches except C-C (16), and that at least one hemimethylated d(GATC) site is required for repair to occur (17). The latter finding prompted simplification of our substrates to molecules that contain a mismatch and a single d(GATC) site (Fig. 3, upper) separated by a thousand base pairs (shorter path). Essentially all of our subsequent work was done with molecules like these.
FIGURE 3.
Methyl-directed repair in E. coli extract supports bidirectional excision.
Incubation of 5′ (left) or 3′ (right) hemimethylated G-T heteroduplex DNA in E. coli extract in the presence if dideoxynucleoside-5′-triphosphates results in production of a single-strand gap that spans the shorter path between the two DNA sites (18,19).
A-Lien Lu demonstrated that the extract reaction is accompanied by DNA synthesis occurring on the unmethylated strand, suggesting an excision repair mechanism (14). Working in collaboration with Jack Griffith’s lab at the University of North Carolina, Michael Su and Michelle Grilley confirmed this to be the case. Incubation of hemimethylated heteroduplex DNA with E coli extract under conditions of DNA synthesis block resulted in the production of a single-strand gap spanning the shorter path between the two sites in the circular DNA (18,19), and this was true for both hemimethylated configurations (Fig. 3, bottom). Because the two strands of the helix are antiparallel, this indicated that there is no obligate polarity of the two DNA sites, and suggested that methyl-directed repair supports bidirectional excision.
To clarify how this pathway works we isolated the MutH, MutL, MutS, and UvrD proteins in pure form (20–23). We knew from the prior work of Peter Emmerson (24) that the uvrD gene product is DNA helicase II, an enzyme that unwinds the two strands of the helix in an ATP-dependent fashion. But the nature of the other three proteins was unknown.
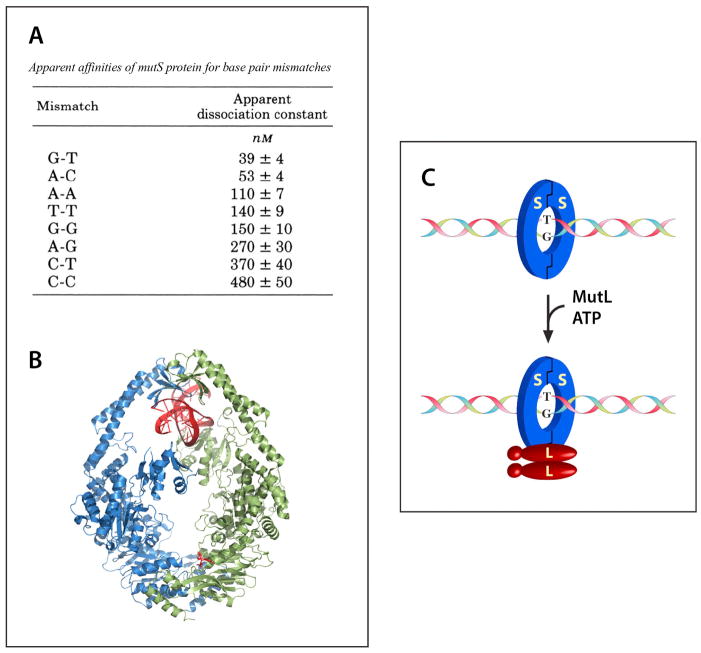
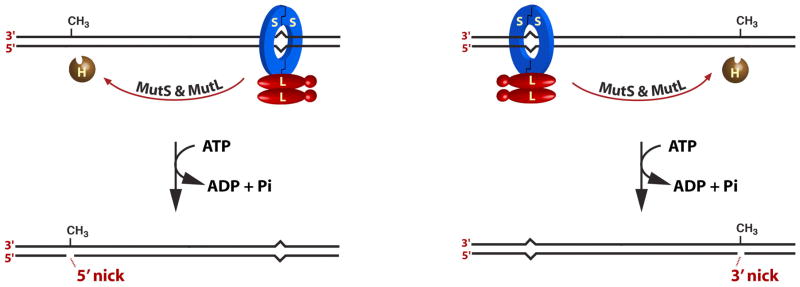
Michael Su showed that MutS is responsible for mismatch recognition (16,20; see Fig. 4A). The ultimate confirmation of this conclusion is the beautiful crystal structure of the E. coli MutS dimer bound to a G-T mismatch (Fig. 4B) that was solved in Titia Sixma’s laboratory at the Netherlands Cancer Institute (25). Michelle Grilley showed that MutL is recruited to the MutS-heteroduplex complex in an ATP-dependent reaction (Fig 4C), but does not otherwise alter the covalent nature of the helix (22). Kate Welsh and A-Lien Lu found that the MutH protein has a very tightly associated, but nearly dead (< 1 turnover/hour) endonuclease activity that incises DNA at an unmethylated d(GATC) sequence (21). As discussed below, assembly of the MutL-MutS-heteroduplex ternary complex leads to dramatic activation of this latent MutH d(GATC) endonuclease.
FIGURE 4.
Biological activities of MutS and MutL.
A. MutS binds mismatched base pairs. B. Crystal structure of the E. coli MutS dimer bound to a G-T mismatch was determined by Titia Sixma and colleagues (25). C. MutL is recruited to the MutS-mismatch complex in an ATP-dependent fashion.
Panel A is reproduced from reference 16; the image in panel B was provided by Titia Sixma with permission.
Because these four proteins are not sufficient to support mismatch repair, Bob Lahue and Karin Au used biochemical and genetic approaches to identify other required components, and they identified four: exonuclease I, which hydrolyzes single-stranded DNA with 3′ to 5′ polarity, the E. coli single strand DNA binding protein SSB, DNA polymerase III holenzyme, which functions in DNA replication, and DNA ligase (23). They showed that a system comprised of MutH, MutL, MutS, UvrD (DNA helicase II), exonuclease I (Exo I), SSB, DNA polymerase III holoenzyme, and DNA ligase was sufficient to reconstitute methyl-directed mismatch repair in vitro.
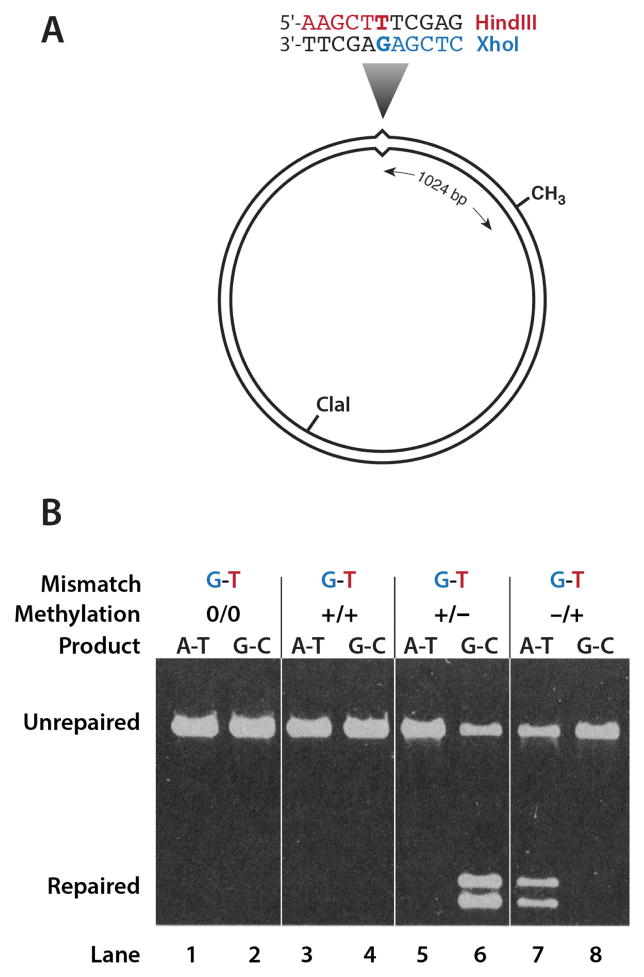
The assay used in these reconstitution experiments was a refined version of the one described in Fig. 1. In this example shown in Fig. 5A, a G-T mismatch is placed in overlapping recognition sites for two restriction enzymes, HindIII and XhoI. The mismatch blocks cleavage by both enzymes, but repair on the bottom strand generates an A-T base pair and a good HindIII site, while repair on the top strand produces a G-C base pair and a good XhoI site. This permits repair on either strand to be directly scored. A heteroduplex that lacks a d(GATC) site is not a substrate for the purified system (Fig. 5B, lanes 1 and 2), nor is a heteroduplex in which both strands are methylated (lanes 3 and 4). However, hemimethylated DNA is repaired. When the methyl group resides on the strand containing the mismatched G, repair is exclusively to G-C(lanes 5 and 6), and when the strand containing the mismatched T is methylated, repair is to A-T (lanes 7 and 8), as expected for methyl-directed correction. This reconstituted system supports repair of all base-base mismatches except C-C (23).
FIGURE 5.
Methyl-directed mismatch repair in a purified system.
A. The heteroduplex substrates used in these experiments contained a mismatched base pair within overlapping recognition sites for two restriction enzymes (16), which permits repair on either DNA strand to be monitored. B. Repair of a G-T heteroduplex is methyl-directed and requires presence of a hemimethylated d(GATC) site.
Panels A and B are reproduced from reference 23 with permission from AAAS.
As described above, analysis of the extract reaction suggested a bidirectional excision capability, which would presumably depend on both 3′ to 5′ and 5′ to 3′ exonucleases. This proved to be correct. Deani Cooper, Vickers Burdett and Celia Baitinger (26–28) showed that when the unmethylated d(GATC) sequence resides 5′ to the mismatch, in vitro repair absolutely depends on 5′ to 3′ excision by exonuclease VII (ExoVII) or RecJ exonuclease. Collaborative experiments with Susan Lovett′s lab showed that in vitro repair directed by a d(GATC) sequence located 3′ to the mismatch depends on 3′ to 5′ hydrolysis by Exo1 or ExoX (26,28), although ExoVII (which supports both 5′ to 3′ and 3′ to 5′ hydrolysis (29)) may also play a limited role in 3′-directed excision (28). Our original reconstitution experiments, which contained only ExoI (23), were successful because the DNA polymerase III holoenzyme preparations used in these early studies were contaminated with ExoVII (26).
Although extracts prepared from RecJ-ExoVII-ExoI-Exo X-E. coli cells are completely defective in both 3′-and 5′-directed mismatch repair, this strain displays only a 7-fold increase in mutation rate, substantially less than the 30-to 100-fold mutability increases observed with MutS-or UvrD-cells (28). This paradox was resolved with the demonstration that production of mismatched base pairs in RecJ-ExoVII-ExoI-Exo X-cells results in loss of viability in manner that depends on upstream action of MutH, MutL, MutS, and UvrD proteins (27). These findings confirm involvement of the four exonucleases in methyl-directed mismatch repair in the E. coli cell and suggest that reduced mutability of the exonuclease-deficient strain is due to under recovery of mutants as a consequence of chromosome loss.
Availability of the required set of purified E. coli proteins permitted us to address the mechanism of the methyl-directed reaction. Mismatch recognition by MutS leads to recruitment of MutL to the heteroduplex (Fig. 6). MutL serves to interface mismatch recognition by MutS to activation of downstream repair activities, one of which is MutH, the latent d(GATC) endonuclease mentioned above. Karin Au showed that methyl-directed repair initiates by activation of MutH endonuclease in a mismatch, MutS, and MutL dependent fashion (30). DNA incision by activated MutH is targeted to the unmethylated stand at a hemimethylated d(GATC) sequence and can occur either 5′ or 3′ to the mismatch, consistent with a bidirectional mechanism, and the resulting strand break that is the actual signal that directs mismatch repair to the unmethylated strand.
FIGURE 6.
MutH activation and initiation of methyl-directed mismatch repair.
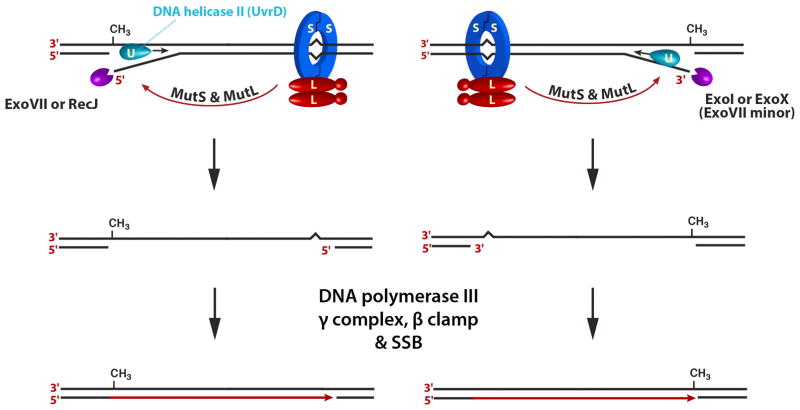
Assembly of the MutL-MutS-heteroduplex complex also activates the excision system, which is comprised of the uvrD gene product, DNA helicase II, and the four single-strand exonucleases mentioned above (Fig. 7). Vivian Dao and Miyuki Yamaguchi demonstrated that MutS and MutL load DNA helicase II at the MutH strand break with an orientation bias so that unwinding of the helix proceeds toward the mismatch (31,32). This loading bias is true for both heteroduplex orientations, without regard to location of the strand break 5′ or 3′ to the mismatch. The single-strand displaced by helicase unwinding is degraded by ExoVII or RecJ when the nick is located 5′ to the mismatch, and by ExoI or ExoX when the break is 3′ to the mispair. The single-strand gap produced in this manner is repaired by the components of DNA polymerase III holoenzyme, and although not shown in Fig. 7, DNA ligase restores covalent continuity to the product. Anna Pluciennik showed that function of the polymerase III holoenzyme components is restricted to the repair synthesis step of E. coli mismatch correction, and have no involvement in the MutH activation or excision steps of repair (33). This is noteworthy because the human pathway described below differs in this respect.
FIGURE 7.
Excision and repair synthesis steps of methyl-directed mismatch repair.
Methyl-directed repair clearly involves action at a distance, effective interaction of two DNA sites that in our substrates are separated by a thousand base pairs. But the mechanism by which this occurs has been a subject of debate for almost 20 years (34,35). The orientation-dependent loading of helicase II summarized in Fig. 7 indicates that the repair system can establish the relative orientation of the mismatch and d(GATC) site, which implies that signaling must occur along the helix contour between the two sites. There is good evidence from multiple labs that MutS and probably the MutL•MutS complex can move along the helix in an ATP-dependent manner (34–36), and we favor the idea that signaling between the two sites is mediated by this sort of movement. However, this point is not yet proven.
As our work on E. coli mismatch repair progressed, we were curious whether a similar stand-directed pathway might exist in higher cells. Work from multiple labs indicated that mismatches are rectified in eukaryotic cells (37–40), but there was no evidence for strand-directed repair. The problem we had in addressing this question was the lack of information concerning the nature of the biological strand signals. To circumvent this problem Jude Holmes, an MD/PhD student who began this work, exploited a finding we had made in the E. coli system described above, namely that the function of MutH and the hemimethylated d(GATC) strand signals in the bacterial reaction is provision of a strand break, which serves as the actual signal that directs methyl-directed mismatch repair. Jude constructed heteroduplex DNAs that contained a site-specific strand-specific nick (Fig. 8A) and found that these DNAs are subject to repair on the nicked DNA strand in extracts prepared from human (Fig. 8B) or Drosophila melanogaster cells (41). No significant repair occurs on the continuous strand, and when both strands are covalently continuous, repair is blocked. Furthermore, Woei-horng Fang demonstrated that DNA hydrolytic events occurring on these DNAs are largely restricted to the shorter path between the nick and the mismatch regardless of 5′ or 3′ orientation of the two DNA sites (Fig 8A). This is reminiscent of bidirectional methyl-directed repair, but as described below, the mechanism of the human reaction differs in fundamental ways from that of the E. coli pathway.
FIGURE 8.
Mismatch repair of nicked heteroduplexes in human cell extracts.
A. Schematic of substrate design and mechanism of repair deduced from extract experiments.
B. Mismatch repair in nuclear extracts of human cells is directed to the strand that contains a preexisting strand break (N). No significant repair occurs on the covalently continuous strand (C).
Panel B is reproduced from reference 41.
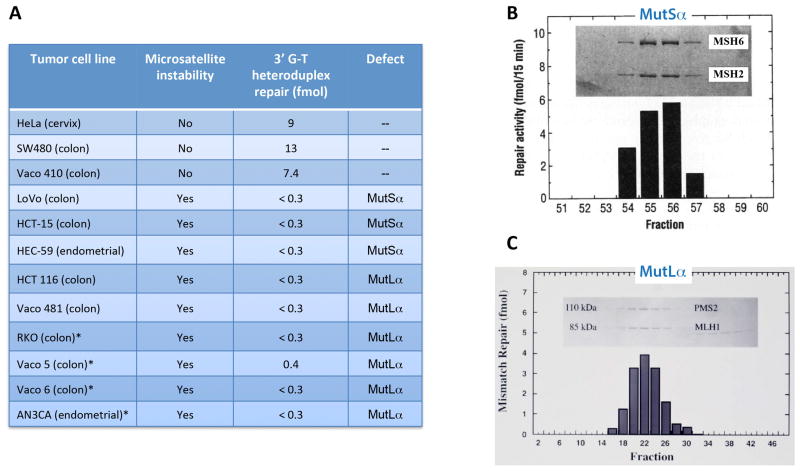
The development that permitted us to identify the key proteins involved in the initiation of human mismatch repair and to pursue the mechanism of the reaction was the publication in 1993 of two papers from de la Chapelle, Vogelstein, and Perucho laboratories (42,43). These papers showed that a high frequency of mutation within simple mononucleotide and dinucleotide repeat sequences, a phenotype called microsatellite instability, is characteristic of certain cancers, including tumors that occur in patients with Lynch syndrome (also called hereditary nonpolyposis colorectal cancer or HNPCC), a common hereditary cancer that accounts for about 5% of colon cancers, and about 15% of sporadic cancers with Lynch-like features (44,45).
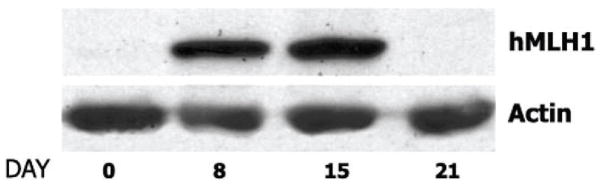
Because we knew that microsatellite mutations are common in E. coli mismatch repair mutants (46), it seemed plausible that these tumor cells might be defective in mismatch repair. We acquired a number of microsatellite unstable tumor cell lines, initially from Bert Vogelstein (Johns Hopkins University School of Medicine) and then from other sources (Fig. 9A). Guo-Min Li, Woei-horng Fang, Matt Longley, and Jim Drummond found each of these cell lines to be defective in mismatch repair (47–50). Availability of these repair-defective cells permitted us to identify and isolate the repair components lacking in these lines (Fig. 9B and C). Jim Drummond and Guo-Min Li showed several of these lines to be deficient in MutSα,a heterodimer of the two MutS homologs MSH2 and MSH6 (48,51), which proved to be the primary human mismatch recognition protein (52). Guo-Min Li identified the repair activity lacking in the other cell lines as MutLα, a heterodimer of the MutL homologs MLH1 and PMS2 (49). While we were pursuing these biochemical studies, several labs, primarily those of Richard Kolodner and Bert Vogelstein, were sequencing MutS and MutL homolog genes in Lynch families. They showed the mutations in these genes segregate with disease phenotype (53,54), and it is now quite clear that the majority of Lynch syndrome cancers are caused by defects in one of these two heterodimers (44,45). Inactivation of mismatch repair in these tumor cells increases the rate of mutation production 100 to 1000-fold (55–57), which is believed to play a direct role in the development of these cancers.
FIGURE 9.
Microsatellite unstable tumor cell lines are defective in mismatch repair and deficient in MutSα or MutLα.
A. Mismatch repair activity in microsatellite stable and unstable cell lines. B. Isolation of MutSα (MSH2-MSH6 heterodimer). C. Isolation of MutLα (MLH1-PMS2 heterodimer).
Panel B is reproduced from reference 48 with permission from AAAS. Panel C is reproduced from reference 49, Copyright 1995 National Academy of Sciences, U.S.A.
The last four tumor cell lines shown in Fig. 9A proved to be exceptional: each is derived from a sporadic cancer with microsatellite instability and each is defective in mismatch repair. Jim Drummond and Guo-Min Li showed that these cell lines fail to produce the MLH1 subunit of MutLα and that repair is restored to extracts in each case by addition of purified MutLα (50). However, we encountered skepticism when we shared these findings with some of our sequencing colleagues, who told us that they had sequenced the MLH1 gene in several of these lines, and it was normal. But Sandy Markowitz (Case Western Reserve University), with whom we were working on several of these cell lines, took us seriously, and collaborative experiments largely done in Sandy’s lab showed that the MLH1 gene in these cell lines is epigenetically silenced by CpG methylation within the promoter region (50). The gene is normal, but it is not transcribed and MLH1 polypeptide is not produced, resulting in a mismatch repair defect. The proof for this conclusion is shown in Fig. 10. Exposure of these microsatellite unstable, sporadic cancer cells to 5-azacytidine, which leads to transient cytosine demethylation, results in transient expression of MLH1. This effect is important in the clinics, where silencing of the MLH1 locus is believed to account for about 15% of colon cancers (58).
FIGURE 10.
5-azacytidine exposure results in transient MLH1 expression in AN3CA tumor cells. Cells were treated with 5-azacytidine for 24 hours on days 2 and 5. Levels of MLH1 and the actin loading control were determined by western blot.
The figure is reproduced with permission from reference 50, Copyright 1998 National Academy of Sciences, U.S.A.
In addition to MutSα (recognizes base-base and small insertion/deletion mismatches of 1 to about 3 extrahelical residues) and MutLα, genetic and biochemical studies have implicated six additional proteins in eukaryotic mismatch repair: MutSβ (MSH2-MSH3 heterodimer; prefers insertion/deletion mismatches of 2 to about 10 extrahelical residues (52,59–61)), exonuclease 1 (62–64), the single-strand DNA binding protein RPA (65,66), the PCNA sliding clamp (67), the clamp-loader RFC (68), and DNA polymerase δ (69). These 8 proteins are sufficient to reconstitute a minimal system that supports human bidirectional mismatch repair in vitro (68,70). We identified two strand-directed reactions, which are supported by subsets of these proteins, that provide insight into the mechanisms of human mismatch repair.
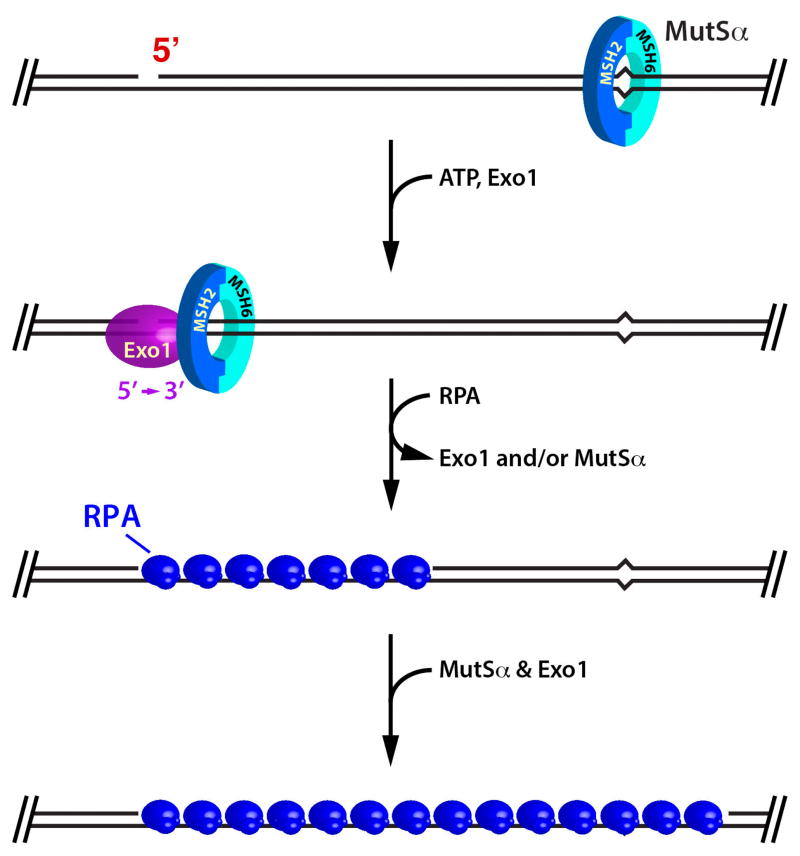
The simplest of these (Fig. 11) is in a sense a trivial example of strand direction. The only exonuclease definitively implicated in eukaryotic mismatch repair is exonuclease 1 (Exo1), which hydrolyzes duplex DNA with 5′ to 3′ polarity (62–64). Jochen Genschel demonstrated that Exo1 is activated by MutSα in a mismatch-dependent manner (64,71), and similar results have been obtained in Guo-Min Li’s laboratory (72). Like bacterial MutS, MutSα is capable of ATP-dependent movement along the helix (34–36), and we think it likely that such movement serves to couple action at the two DNA sites. Activation by MutSα renders Exo1 highly processive. Processive action of the MutSα-Exo1 complex is controlled by the RPA single-strand binding protein, which displaces the processive complex from the helix after removal of about 200 nucleotides (71,73). The resulting RPA-filled gap is a poor substrate for Exo1, and the enzyme cannot reload without the mismatch-dependent assistance of MutSα. This leads to an iterative cycle of removal of about 200 nucleotides per Exo1 reloading event that continues until the mismatch is removed, at which point excision is dramatically attenuated because MutSα can no longer promote Exo1 loading. MutLα is not required for this reaction, but it modestly enhances the mismatch dependence of excision by suppressing hydrolysis on mismatch-free homoduplex DNA, and poly[ADP-ribose] polymerase PARP-1 further potentiates mismatch dependence in this system (74).
FIGURE 11.
MutSα activation of Exo1 and control of processive action of the MutSα-Exo1 complex by RPA.
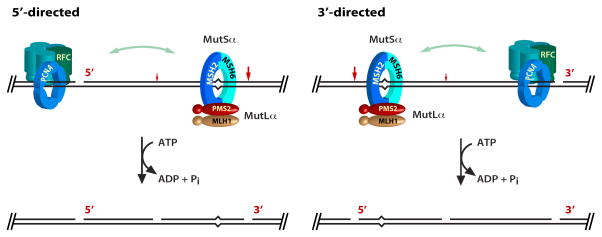
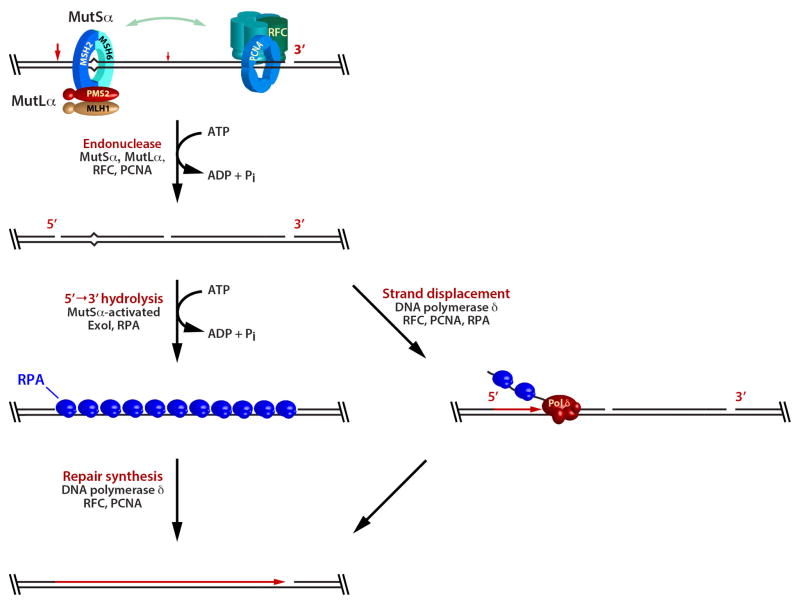
The second strand-directed reaction is more interesting (Fig. 12). Farid Kadyrov, Leo Dzantiev, and Nicoleta Constantin demonstrated that unlike E. coli MutL, human MutLα is a latent endonuclease that is activated in a manner that depends on a mismatch, a preexisting strand break, MutSα, the PCNA sliding clamp, and RFC, the clamp loader that places PCNA on the helix (75). Endonuclease action is strand-directed: it is targeted to the heteroduplex strand that contains the preexisting break. Incision by activated MutLα can occur at multiple sites on the pre-incised strand, but initial events appear to be biased to the distal side of the mismatch. This reaction occurs on both 5′ and 3′ heteroduplexes to yield molecules in which the mismatch is bracketed by 5′ and 3′ strand breaks. Farid Kadyrov showed that these 5′-termini serve as loading sites for two alternate modes of mismatch removal: hydrolytic excision by MutSα-activated Exo1 (75) or synthesis-driven strand displacement by DNA polymerase δ (Fig. 13). Other excision mechanisms may also be possible.
FIGURE 12.
MutLα is a strand-directed endonuclease that depends on a mismatch, a preexisting strand break, MutSα, PCNA, and RFC for activation.
FIGURE 13.
Mismatch removal from MutLα-incised heteroduplex DNA by MutSα-activated Exo1 (left) or synthesis-driven strand displacement by DNA polymerase δ (right).
We believe that endonuclease action is a primary function of MutLα in eukaryotic mismatch repair. The basis for this conclusion is shown in Fig 14. Farid identified a DQHA(X)2E(X)4E metal-binding, endonuclease active site motif within the C-terminal domain of the PMS2 subunit of MutLα (Fig 14, motif highlighted in red). Amino acid substitutions within this motif have no effect on stability of the MLH1-PMS2 heterodimer, MutLα ATPase activity, or assembly of the MutLα-MutSα-heteroduplex ternary complex, but they inactivate endonuclease function and abolish mismatch repair in human, yeast, and mouse cells (75–77). In mice, inactivation of MutLα endonuclease function is associated with strong cancer predisposition and a partial defect in immunoglobulin class switch recombination (77).
FIGURE 14.
MutLα endonuclease active site motif.
C-terminal PMS2 DQHA(X)2E(X)4E endonuclease active site motif is conserved in eukaryotic PMS2 homologs (S. cerevisiae PMS1 is a homolog of human PMS2) and in many bacterial MutL proteins, with the exception of MutL proteins from bacteria like E. coli that rely on d(GATC) methylation to direct mismatch repair. Amino acid residues shown in blue at the top of the figure correspond to substitution mutations used to assess involvement of the motif in MutLα function.
The figure is reproduced with from reference 75, Copyright 2006 with permission from Elsevier.
Although not present in members of the MLH1 family, the DQHA(X)2E(X)4E endonuclease motif is conserved in eukaryotic PMS2 homologs. It is also found in many bacterial MutL proteins (Fig. 14), and recent work has shown that like human and yeast MutLα, at least some of these bacterial MutL proteins function as endonucleases (78). The notable exceptions are MutL proteins from bacteria like E. coli and related organisms that rely on d(GATC) methylation to direct mismatch repair, where this motif is conspicuously absent. Like E. coli MutL (22,76), these latter proteins presumably lack endonuclease function. Thus two distinct mechanisms for strand-targeting of mismatch repair occur in nature.
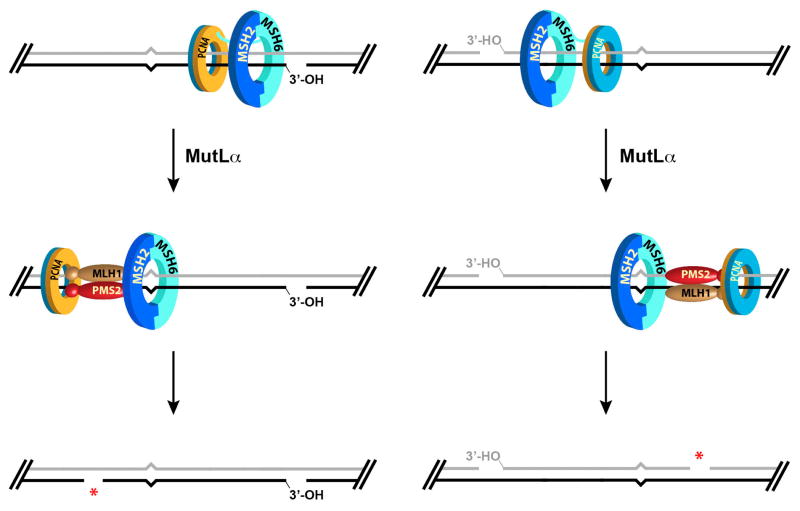
The mechanism of MutLα activation is complex, but Anna Pluciennik and Leo Dzantiev have clarified several of its features (79). They showed that the only function of the preexisting strand break within the heteroduplex (Fig. 12) is to provide a loading site for the PCNA sliding clamp, and that RFC involvement in MutLα activation is restricted to loading of the clamp onto the helix. Loaded PCNA plays two important roles in the reaction. Physical interaction of MutLα with the loaded clamp is required for endonuclease activation, and the orientation with which PCNA is loaded onto the helix determines strand direction of endonuclease action (79). These functions of the loaded clamp are illustrated in the model shown in Fig. 15. The two faces of the PCNA clamp are not equivalent, and RFC loads the clamp at a 3′-double strand-single strand junction with a unique orientation (depicted in Fig. 15 with gold face oriented toward the 3′ terminus). The Jiricny and Kolodner labs have shown that MutSα is tethered to the replication fork via physical interaction with PCNA (80,81), and both of these proteins are capable of movement along the helix (34–36,82). Mismatch recognition by MutSα triggers recruitment of MutLα, which is capable of interacting with both proteins. The idea is that the asymmetry of clamp loading is preserved in the MutLα-PCNA complex, and this serves to uniquely orient the MutLα endonuclease active site relative to two DNA strands of the helix.
FIGURE 15.
Strand direction of MutLα endonuclease action is determined by the orientation with which the PCNA sliding clamp is loaded onto the DNA helix.
When we began our work on human mismatch repair, we had no idea concerning the nature of the biological strand signals, although we and others had suggested that DNA termini that occur naturally during the course of DNA replication might suffice in this regard (41,67,83). The strand-directed reactions described above are consistent with this view. 5′ termini on the lagging strand at the replication fork would presumably support loading of MutSα-activated Exo1, and genetic studies from the Kolodner and Kunkel laboratories are compatible with this idea (81,84). Furthermore, strand direction of MutLα endonuclease action is controlled by the loading orientation of the PCNA clamp on the helix, which in turn is determined by 3′-termini on the leading and lagging strands at the fork. It therefore seems likely that 5′ and 3′ DNA termini that occur naturally during the course of DNA replication serve as default signals that direct mismatch repair in the eukaryotic cell.
Acknowledgments
I have been truly fortunate to share these mismatch repair studies with a group of outstanding graduate students and postdoctoral fellows, a number of whom have touched my life in ways other than science. I also thank the Howard Hughes Medical Institute and the National Institutes of Health for their generous support of this work over the years.
References
- 1.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 2.Meselson M, Weigle JJ. Chromosome brekage accompanying genetic recombination in bacteriophage. Proc Natl Acad Sci U S A. 1961;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ephrussi-Taylor H, Gray TC. Genetic studies of recombining DNA in pneumococcal transformation. J Gen Physiol. 1966;49(part 2):211–231. doi: 10.1085/jgp.49.6.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White RL, Fox MS. Genetic consequences of transfection with heteroduplex bacteriophage lambda DNA. Mol Gen Genet. 1975;141:163–171. doi: 10.1007/BF00267681. [DOI] [PubMed] [Google Scholar]
- 5.Wildenberg J, Meselson M. Mismatch repair in heteroduplex DNA. Proc Natl Acad Sci U S A. 1975;72:2202–2206. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner R, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976;73:4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiricny J. Postreplicative mismatch repair. Cold Spring Harbor perspectives in biology. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983;104:571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nevers P, Spatz H. Escherichia coli mutants uvrD uvrE deficient in gene conversion of lambda heteroduplexes. Mol Gen Genet. 1975;139:233–243. doi: 10.1007/BF00268974. [DOI] [PubMed] [Google Scholar]
- 10.Rydberg B. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 1978;52:11–24. doi: 10.1016/0027-5107(78)90091-x. [DOI] [PubMed] [Google Scholar]
- 11.Glickman BW, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vovis GF, Horiuchi K, Hartman N, Zinder ND. Restriction endonuclease B and f1 heteroduplex DNA. Nat New Biol. 1973;246:13–16. doi: 10.1038/newbio246013a0. [DOI] [PubMed] [Google Scholar]
- 13.Enea V, Vovis GF, Zinder ND. Genetic studies with heteroduplex DNA of bacteriophage f1. Asymmetric segregation, base correction, and implications for the mechanism of genetic recombination. J Mol Biol. 1975;96:495–509. doi: 10.1016/0022-2836(75)90175-8. [DOI] [PubMed] [Google Scholar]
- 14.Lu AL, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu AL, Welsh K, Clark S, Su SS, Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harbor symposia on quantitative biology. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- 16.Su SS, Lahue RS, Au KG, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 17.Lahue RS, Su SS, Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci U S A. 1987;84:1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su SS, Grilley M, Thresher R, Griffith J, Modrich P. Gap formation is associated with methyl-directed mismatch correction under conditions of restricted DNA synthesis. Genome. 1989;31:104–111. doi: 10.1139/g89-020. [DOI] [PubMed] [Google Scholar]
- 19.Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 20.Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 22.Grilley M, Welsh KM, Su SS, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 23.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 24.Hickson ID, Arthur HM, Bramhill D, Emmerson PT. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190:265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- 25.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G-T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 26.Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 27.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci U S A. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J Biol Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 29.Chase JW, Richardson CC. Exonuclease VII of Escherichia coli: mechanism of action. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- 30.Au KG, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 31.Dao V, Modrich P. Mismatch, MutS, MutL, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 33.Pluciennik A, Burdett V, Lukianova O, O’Donnell M, Modrich P. Involvement of the beta clamp in methyl-directed mismatch repair in vitro. J Biol Chem. 2009;284:32782–32791. doi: 10.1074/jbc.M109.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkel TA, Erie DA. DNA Mismatch Repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 35.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 36.Fishel R. Mismatch Repair. J Biol Chem. 2015;290:26395–26403. doi: 10.1074/jbc.R115.660142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muster-Nassal C, Kolodner R. Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986;83:7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glazer PM, Sarkar SN, Chisholm GE, Summers WC. DNA mismatch repair detected in human cell extracts. Mol Cell Biol. 1987;7:218–224. doi: 10.1128/mcb.7.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown TC, Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988;54:705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- 40.Brooks P, Dohet C, Almouzni G, Mechali M, Radman M. Mismatch repair involving localized DNA synthesis in extracts of Xenopus eggs. Proc Natl Acad Sci U S A. 1989;86:4425–4429. doi: 10.1073/pnas.86.12.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes J, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR, Petersen GM, Kinzler KW, Vogelstein B, de la Chapelle A. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 43.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 44.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 45.Rowley PT. Inherited susceptibility to colorectal cancer. Annu Rev Med. 2005;56:539–554. doi: 10.1146/annurev.med.56.061704.135235. [DOI] [PubMed] [Google Scholar]
- 46.Levinson G, Gutman GA. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987;15:5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 48.Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2•p160 heterodimer that restores mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 49.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci U S A. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iaccarino I, Palombo F, Drummond J, Totty NF, Hsuan JJ, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 52.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of hMutSbeta from human cells and comparison of the mismatch repair specificities of hMutSbeta and hMutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 53.Fishel R, Kolodner RD. Identification of mismatch repair genes and their role in the development of cancer. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 54.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 55.Eshleman JR, Lang EZ, Bowerfind GK, Parsons R, Vogelstein B, Willson JK, Veigl ML, Sedwick WD, Markowitz SD. Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 56.Glaab WE, Tindall KR. Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis. 1997;18:1–8. doi: 10.1093/carcin/18.1.1. [DOI] [PubMed] [Google Scholar]
- 57.Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacinto FV, Esteller M. Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis. 2007;22:247–253. doi: 10.1093/mutage/gem009. [DOI] [PubMed] [Google Scholar]
- 59.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 61.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSb, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 62.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 63.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 65.Lin YL, Shivji MK, Chen C, Kolodner R, Wood RD, Dutta A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem. 1998;273:1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 66.Ramilo C, Gu L, Guo S, Zhang X, Patrick SM, Turchi JJ, Li GM. Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Mol Cell Biol. 2002;22:2037–2046. doi: 10.1128/MCB.22.7.2037-2046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 68.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 69.Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 70.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 73.Genschel J, Modrich P. Functions of MutL{alpha}, RPA, and HMGB1 in 5′-directed mismatch repair. J Biol Chem. 2009;284:21536–21544. doi: 10.1074/jbc.M109.021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Kadyrov FA, Modrich P. PARP-1 enhances the mismatch-dependence of 5′-directed excision in human mismatch repair in vitro. DNA Repair (Amst) 2011;10:1145–1153. doi: 10.1016/j.dnarep.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 76.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Oers JM, Roa S, Werling U, Liu Y, Genschel J, Hou H, Jr, Sellers RS, Modrich P, Scharff MD, Edelmann W. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci U SA. 2010;107:13384–13389. doi: 10.1073/pnas.1008589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lenhart JS, Pillon MC, Guarne A, Biteen JS, Simmons LA. Mismatch repair in Gram-positive bacteria. Res Microbiol. 2016 doi: 10.1016/j.resmic.2015.08.006. in press. [DOI] [PubMed] [Google Scholar]
- 79.Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hedglin M, Kumar R, Benkovic SJ. Replication clamps and clamp loaders. Cold Spring Harbor perspectives in biology. 2013;5:a010165. doi: 10.1101/cshperspect.a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Claverys JP, Lacks SA. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986;50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liberti SE, Larrea AA, Kunkel TA. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair (Amst) 2013;12:92–96. doi: 10.1016/j.dnarep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]