Summary
The Echinacea genus is exemplary of over 30 plant families that produce a set of bioactive amides, called alkamides. The Echinacea alkamides may be assembled from two distinct moieties, a branched-chain amine that is acylated with a novel polyunsaturated fatty acid. In this study we identified the potential enzymological source of the amine moiety as a pyridoxal phosphate dependent decarboxylating enzyme that uses branched chain amino acids as substrate. This identification was based on a correlative analysis of the transcriptomes and metabolomes of 36 different E. purpurea tissues and organs, which expressed distinct alkamide profiles. Although no correlation was found between the accumulation patterns of the alkamides and their putative metabolic precursors (i.e., fatty acids and branched chain amino acids), isotope-labeling analyses supported the transformation of valine and isoleucine to isobutylamine and 2-methylbutylamine as reactions of alkamide biosynthesis. Sequence homology identified the pyridoxal phosphate dependent decarboxylase-like proteins in the translated proteome of E. purpurea. These sequences were prioritized for direct characterization by correlating their transcript levels with alkamide accumulation patterns in different organs and tissues, and this multi-pronged approach led to the identification and characterization of a branched-chain amino acid decarboxylase, which would appear to be responsible for generating the amine moieties of naturally occurring alkamides.
Keywords: Echinacea purpurea, fatty acids, metabolomics, alkamides, transcriptomics, specialized metabolism, amines
Introduction
Plant metabolism has been classically divided into primary and secondary (more recently called specialized) metabolism (Dudareva et al. 2010, Pichersky and Lewinsohn 2011, Schilmiller et al. 2012b). Metabolic processes that are common to all plants and are central to the bioenergetics of growth, development and the biosynthesis of essential structural components are classified as primary metabolism, whereas those that are distributed discretely among different phylogenetic clades of the plant kingdom that furnish the physiochemical phenotypic differences among the taxonomic groups are classified as specialized metabolism. The plant kingdom produces an overwhelming variety of specialized metabolites, many with highly interesting biological activities. Plant-derived, specialized metabolites are the basis for about 25% of today’s pharmaceuticals; commonly known examples include aspirin (acetylsalicylic acid, originally derived from salicin, the active ingredient in willow bark), morphine from poppy flowers, and digitalin (from purple foxglove, Digitalis purpurea). In this paper, we exploit current advances in plant functional genomics in combination with modern methods of metabolomics and metabolite labeling analysis to elucidate the biosynthesis of a class of bioactive secondary metabolites, the alkamides of the Echinacea genus, and discover key enzymes/genes required in this biosynthetic pathway.
Alkamides comprise about 200 chemically related compounds, and these occur in 33 plant families (Christensen and Lam 1991, Kashiwada et al. 1997, Parmar et al. 1997, Rios 2012). The alkamides of Echinacea have pharmacological activities that are consistent with the rich ethnobotanical history of Echinacea species (Wu et al. 2009); these plants were used by the Native Indian populations of North America as antidotes and remedies for many ailments (Gilmore 1977). The pharmacological activities of alkamides may result, in part, by mimicking the structurally related compound, anandamide, by targeting the cannabinoid receptors and acting as immunomodulators (Gertsch et al. 2004, Woelkart et al. 2005). Alkamides also display insecticidal activities (Jacobson 1971, Miyakado et al. 1989) and have been shown to affect plant growth (Campos-Cuevas et al. 2008, Ramirez-Chavez et al. 2004). In general, these compounds are alkyl or aryl amides, composed of an amine moiety acylated with a variety of different fatty acids (Fig. 1A).
Figure 1.
Alkamides of Echinacea. (A) Generalized chemical structure of Echinacea alkamides. An alkyl amine (blue chemical structure), with variable branching indicated by dashes, and a diversity of unsaturated fatty acids (red chemical structure) are connected via an amide bond (black). The common locations of carbon-carbon double and triple bonds are indicated by dashes. (B) Echinacea purpurea organs and tissues used for the transcriptomic and metabolomic analyses. Numbers identify each organ and tissue that was profiled and these are defined in Data S1. Sample #1–23, 34 and 35 were obtained from accessions PI 631313, and sample #24–33 and 36 were obtained from accession PI 633670. (C) Hierarchical clustering analysis of the distribution of alkamides among different E. purpurea organs and tissues. The dendrogram was generated with Java Treeview (Saldanha 2004), based upon the Pearson correlations of the relative accumulation patterns of 33 alkamide metabolites among 36 different E. purpurea organs and tissues.
In Echinacea, over 20 different alkamides have been characterized in which the amine moiety is either isobutylamine or 2-methylbutylamine, and the acyl moiety is between 11- and 16-carbon atoms in length with a trans-double bond situated at the 2-position (Bauer and Foster 1991, Bauer et al. 1989). Beyond the variation in chain length, the degree of unsaturation in the acyl moiety varies with additional double bonds, most frequently at carbons 4, 8 and/or 10, or occurrence of acetylenic bonds at the 8th and 10th positions. There is considerable natural variation in the types of alkamides that accumulate across different Echinacea species and accessions (Wu et al. 2004, Wu, et al. 2009) and the pattern of alkamide accumulation is dependent on plant growth and development (Bauer and Foster 1991, Bauer and Remiger 1989, Binns et al. 2002, Wu, et al. 2004, Wu, et al. 2009). In this study, we combined transcriptome and metabolite profiling, and isotopic labeling experiments to identify the key enzyme that probably catalyzes the initial reaction in the metabolic pathway required for the generation of the amine moiety of the Echinacea alkamides.
Results
Alkamide profiles of E. purpurea organs and tissues
The overarching strategy that was implemented to identify genetic elements that are required for the biosynthesis of Echinacea alkamides initially involved finding correlations among profiles of alkamides, potential metabolite precursors and the transcriptomes of selected organs that express different levels of alkamides. Thirty-six different E. purpurea organs and tissues that represent different stages of development were subjected to alkamide analysis (Figure 1B, Table 1). These analyses detected 371 analytes (Data S1), and MS analyses of these profiles enabled the accurate annotation of eight previously identified alkamides (Bauer et al. 1988b), which are listed in Table 2. In addition to these previously characterized alkamides, our analyses identified an additional 22 analytes as possible alkamides. This tentative identification was based on three mass-spectrometric attributes, which are common of amide-containing metabolites (Mudge et al. 2011). Representative mass-spectra are provide in Figure S4, and the attributes that lead to their identification as possible alkamides are: a) that the nominal m/z value of the parent ion is an odd number (i.e., (M-H)+ is even-numbered), indicative of a nitrogen-containing metabolite (McLafferty and Tureček 1993); b) the occurrence of a common signature base-ion due to the McLafferty-rearrangement (McLafferty 1959) of either an 2-methylbutylamine-containing amide (m/z 128; Figure S4A) or isobutylamine-containing amide (m/z 115; Figure S4B); and c) the occurrence of a mass-spectrometric fragment ion with an m/z value of 81, characteristic of diene structure at the omega-end of the acyl moiety, as occurs in many alkamides (e.g., Bauer Alkamide #8). Although these characteristics identified 22 analytes as possible alkamides, their exact chemical structures were not deduced.
Table 1.
E. purpurea tissues and organs used in this study, and accumulation of alkamide i4N-12:4Δ2E,4E,8Z,10E.
| Tissue and Organ name | Tissue and Organ Number# | i4N-12:4Δ2E,4E,8Z,10E (nmol/g) |
|---|---|---|
| Bracts of stage 1 flower | 1 | 27 ± 5 |
| Bracts of stage 2 flower | 2 | 5 ± 12 |
| Bracts of stage 3 flower | 3 | 4 ± 8 |
| Bracts of stage 4 flower | 4 | 14 ± 4 |
| Bracts of stage 5 flower | 5 | 48 ± 15 |
| Receptacle of stage 1 flowers | 6 | 54 ± 14 |
| Receptacle of stage 2 flowers* | 7 | 94 ± 13 |
| Receptacle of stage 3 flowers | 8 | 44 ± 33 |
| Receptacle of stage 4 flowers* | 9 | 25 ± 170 |
| Receptacle of stage 5 flowers | 10 | 121 ± 2 |
| Disc florets of stage 2 flower | 11 | 56 ± 15 |
| Disc florets of stage 3 flower* | 12 | 45 ± 6 |
| Disc florets of stage 4 flower* | 13 | 112 ± 133 |
| Disc florets of stage 5 flower* | 14 | 995 ± 2 |
| Petal of stage 2 flower | 15 | 48 ± 1 |
| Petal of stage 3 flower | 16 | 83 ± 52 |
| Petal of stage 4 flower*^ | 17 | 30 ± 21 |
| Petal of stage 5 flower*^ | 18 | 501 ± 38 |
| Flower Stage 1*^ | 19 | 19 ± 2 |
| Flower Stage2* | 20 | 13 ± 2 |
| Flower Stage3*^ | 21 | 119.1 ± 0.4 |
| Flower Stage4^ | 22 | 104 ± 3 |
| Flower Stage5*^ | 23 | 327 ± 9 |
| Inflorescence stem - apical section*^ | 24 | 21 ± 7 |
| Inflorescence stem - basal section*^ | 25 | 210 ± 80 |
| Immature cauline leaves | 26 | 24 ± 13 |
| Expanding cauline leaves* | 27 | 9 ± 4 |
| Fully expanded cauline leaves^ | 28 | 8 ± 4 |
| Expanding rosette leaves* | 29 | 16 ± 8 |
| Fully expanded rosette leaves | 30 | 7 ± 3 |
| Petiole of fully expanded cauline leaves*^ | 31 | 165 ± 159 |
| Petiole of expanding rosette leaves | 32 | 83 ± 32 |
| Petiole of fully expanded rosette leaves* | 33 | 202 ± 105 |
| Lateral Root*^ | 34 | 8 ± 1 |
| Basal Root*^ | 35 | 32 ± 13 |
| Entire root system *^ | 36 | 1200 ± 670 |
As defined in Figure 1b
samples subjected to transcriptomic analysis
samples analyzed for amino acid and fatty acid profiles
Table 2.
Short-hand nomenclature for Bauer alkamides a
| Common name | Scientific name | Structure | Abbreviated name |
|---|---|---|---|
| Bauer alkamide 2 | (2Z,4E)-N-isobutylundeca-2,4-diene-8,10-diynamide |
|
i4N-11:2Δ2Z,4E,8a,10a |
| Bauer alkamide 3 | (2E,4Z)-N-isobutyldodeca-2,4-diene-8,10-diynamide |
|
i4N-12:2Δ2E,4Z,8a,10a |
| Bauer alkamide 4 | (2E,4Z)-N-(2-methylbutyl)undeca-2,4-diene-8,10-diynamide |
|
ai5N-11:2Δ2E,4Z,8a,10a |
| Bauer alkamide 6 | (2E,7Z)-N-isobutyltrideca-2,7-diene-10,12-diynamide |
|
i4N-13:2Δ2E,7Z,10a,12a |
| Bauer alkamide 7 | (2E,4Z)-N-(2-methylbutyl)dodeca-2,4-diene-8,10-diynamide |
|
ai5N-12:2Δ2E,4Z,8a,10a |
| Bauer alkamide 8 | (2E,4Z,8Z,10E)-N-isobutyldodeca-2,4,8,10-tetraenamide |
|
i4N-12:4Δ2E,4E,8Z,10E |
| Bauer alkamide 9 | (2E,4E,8Z,10Z)-N-isobutyldodeca-2,4,8,10-tetraenamide |
|
i4N-12:4Δ2E,4E,8Z,10Z |
| Bauer alkamide 10 | (2E,4E,8Z)-N-isobutyldodeca-2,4,8-trienamide |
|
i4N-12:3Δ2E,4E,8Z |
| Bauer alkamide 11 | (2E,4E)-N-isobutyldodeca-2,4-dienamide |
|
i4N-12:2Δ2E,4E |
The Bauer alkamide numbering system is defined in Bauer et al., 1988a
Table 2 also introduces an improved shorthand nomenclature for naming the alkamides. The advantage of the suggested naming scheme is that it is simpler than the systematic IUPAC chemical naming system (also included in Table 2) yet it provides chemical information that is lost in the vernacular that has been used to name these molecules when they were first characterized by Bauer and colleagues (Bauer et al. 1988a). This abbreviated nomenclature is based on that widely used for fatty acids, and indicates the nature of the amine moiety of each alkamide followed by the nature of the fatty acid moiety. Thus, for example in the name “i4N-12:2Δ2E,4Z,8a,10a”, which is the suggested short-hand name for Bauer alkamide #3 (dodeca-2E,4Z-diene-8,10-diynoic acid isobutylamide), “i4N” indicates an isobutyl group is bonded to the amide nitrogen (indicated by the N) to a fatty acid of 12-carbon chain length that has two double bonds at positions 2 and 4, which are in the E and Z configuration, respectively, and the two acetylenic bonds indicated by the letter “a” in the suffix at positions 8 and 10 (i.e., “12:2Δ2E,4Z,8a,10a”). As with the short-hand nomenclature of fatty acids, only carbon-carbon double-bonds or triple-bonds are accounted for in the digits following the colon and the lack of a E/Z designation in a name would indicate that the configuration of the double bond is undetermined (for example, i4N-12:2Δ2,4,8a,10a designates any of four possible stereoisomers). The nitrogen designator in the name could be substituted in other classes of fatty acid derivatives (e.g., S for a thioester).
The most prevalent alkamide present among the surveyed E. purpurea tissues and organs is i4N-12:4Δ2E,4E,8Z,10E, which was previously annotated as Bauer Alkamide #8 (Bauer, et al. 1988b). In these extracts we also identified a number of different alkamides that eluted at different retention times, but their mass-spectra were consistent with i4N-12:4Δ2,4,8,10; these include for example Bauer Alkamide #9 (i4N-12:4Δ2E,4E,8Z,10Z). The abundance of alkamide i4N-12:4Δ2E,4E,8Z,10E (Bauer Alkamide #8) among the surveyed tissues and organs is presented in Table 1. This alkamide is most abundant in disc-florets (tissue sample #14; disc florets of a developmental stage 5 flower) and roots (sample #36; entire root system) and is at lowest abundance in the bracts (sample #2 and #3; bracts of a developmental stage 2 flower and bracts of a stage 3 flower, respectively). The difference in abundance of this alkamide among these organs is approximately 300-fold. The next most abundant alkamide is i4N-12:4Δ2E,4E,8Z,10Z, which accumulates at levels that are at most, 1/5th of that of i4N-12:4Δ2E,4E,8Z,10E. We used these abundance data as the “anchors” for correlating other genome expression data, including profiles of metabolites and transcripts.
Figures 1C and Figure S1 explore the correlations among the abundance of i4N-12:4Δ2,4,8,10 and the other alkamides in the 36 organs and tissues that were surveyed. The log-ratio plot shown in Figure S1 plots the relative abundance of the 33 detected alkamides and alkamide-like metabolites, normalized relative to each alkamide’s abundance in the lateral root sample (tissue sample #34; Fig. 1B). The numbered order on the y-axis is based on the organ and tissue samples that were subjected to analysis, and these are identified in Table 1 and Figure 1B. The lines that link the data-points in Figure S1, correspond to the color shading of the clades shown in Figure 1C. The fact that the lines joining the data-points on this graph (Fig. S1) are primarily parallel to each other indicate that most alkamides show a tissue/organ distribution that is highly correlated, as is confirmed by the Pearson’s correlation calculation. The Pearson’s correlation calculation, which is the basis for the hierarchical cluster tree shown in Figure 1C, demonstrate that the developmentally induced changes in the abundance of the majority of the alkamides and alkamide-like metabolites cluster within a single clade (shaded light-green), indicating that most alkamides show a tissue/organ distribution pattern that is highly correlated.
Correlations among alkamide profiles and potential precursors
Because alkamides are chemically composed of two moieties, an amine moiety acylated by a fatty acid-derived moiety, we hypothesized that the accumulation of potential precursor amino acids and precursor fatty acids may be correlated with the tissue distribution of the alkamides. We explored this potential by selecting a smaller set of tissue samples (13 samples) and analyzing their fatty acid and amino acid profiles. These 13 samples were selected to represent the widest range of alkamide content, from the lowest (leaf tissue, sample #28) to the highest levels (root tissue, sample # 36) of accumulation, representing a 300-fold range in abundance. These analyses revealed the accumulation patterns of 371 analytes, of which nearly 100 where chemically identified as amino acids (15), fatty acids (34), sterols (5), organic acids (1), alcohols (3), hydrocarbons (8), and alkamide and alkamide-like metabolites (33); but the chemical identities of the remaining 270 analytes were not determined (Data S1).
Pearson correlation coefficients were calculated from the metabolite abundance data of all the compounds detected among these 13 tissue samples analyzed. These coefficients were used to create the hierarchical clustering representation of the data (Figure S2). This dendrogram provides a visual representation of the abundance relationships among the 371 compounds as affected by the genetic developmental program that defined the 13-tissue samples. Those clades that are separated by a correlation coefficient value of greater than 0.7 are classified as belonging to the same accumulation cluster, and these calculations indicate that the accumulation patterns of the 371 compounds segregate into 21 separate clades, labeled as A-U.
The chemical nature of the components in each clade is identified in Data S1. All eight of the previously identified alkamides that occur in these samples clustered in clade U, which also contained the majority of the 13 alkamide-like metabolites that were detected in this study. The 18 amino acids that were detected clustered in five separate clades (C, F, I, N, and Q). Most significantly, the specific branched-chain amino acids (BCAAs) that are anticipated to be precursors of the amine moiety of the Echinacea alkamides, Val and Ile, clustered together in clade N, but this clade is statistically distinct from the alkamide-enriched clade U. The fatty acid profiling platform detected 169 analytes including 40 that were accurately annotated as fatty acids; other compounds detected by this platform included triterpenes and other carboxylic acids. Very few of the metabolites assessed by this analytical procedure clustered in the alkamide-containing clade, with the majority of the profiled fatty acids being distributed among eight different clades (C, F, H, I, J, N, O, and P). All these chemically identified fatty acids have chain lengths of 14-carbon atoms or longer; no fatty acid with a C12 acyl chain length is detectable in the alkamide-enriched clade U, C12 being the predominant acyl-chain length that is associated with the alkamides. Therefore, the results of these metabolite-profiling studies did not support our starting hypothesis that there is a metabolic correlation between the accumulation levels of the potential precursors (amino acids and fatty acids) and the accumulation of the final products of the alkamide biosynthesis pathway.
Isotope labeling experiments indicates the role of BCAAs in generating the amine moiety of alkamides
A series of isotopic tracer experiments were conducted to directly test whether BCAAs, specifically Val and Ile, are the precursors of the amine moiety of the alkamides and to identify the probable chemical transformations leading to that amine moiety. Initially, E. purpurea seedlings were cultured in media containing [U-13C6]glucose ([13C]Glc), and LC-MS and GC-MS analyses were used to identify the labeling patterns in Val and alkamides. These analyses revealed extensive penetration of the label into Val through the glycolytic metabolite pool, evidenced by the appearance of an M+5 signal for [13C5]valine, which was at an abundance level of 7.4% of the parent ion. GC/MS analysis revealed an M+10 signal for the m/z 167 ion of alkamide i4N-12:2Δ2E,4E,8Z,10E, which contains the isobutyl amine moiety plus the first six carbon atoms from the acyl moiety of the alkamide; this M+10 signal was detected due to the complete 13C labeling of the diagnostic electron-impact fragment.
The involvement of de novo BCAA biosynthesis was tested by conducting a parallel experiment in the presence of chlorsulfuron, an inhibitor of acetolactate synthase, which catalyzes the gateway reactions of this process. In this experiment, chlorsulfuron inhibited seedling growth by 50% over a 72 h period. Analyses of the Val from the plants that were fed [13C6]Glc showed only minor incorporation of label into the 13C3 and 13C2 fragments of Val (≤1% of the parent ion). Furthermore, alkamide production was reduced and signals above the M+6 ion were suppressed for the alkamide signature ion (the m/z 167 ion). Seedling growth and alkamide accumulation was restored to near wild-type levels by supplementing the culture medium with 15 mM Val and 15 mM Ile. Together, these experiments indicate that the amine moiety of this alkamide originates through Val and Ile biosynthesis. Furthermore, these experiments established an experimental system in which alkamide biosynthesis was dependent on externally provided BCAA precursors.
This BCAA-dependent in vivo alkamide biosynthesis system was further improved by the inclusion of methyl jasmonate (MeJA) in the media. Prior studies with E. pallida have shown that MeJA amplifies the in vivo production of alkamides (Binns et al. 2001). In our optimized conditions, MeJA treatment of E. purpurea seedlings increased the de novo accumulation of alkamides by 2-fold over a 72-h treatment period (Figure S3).
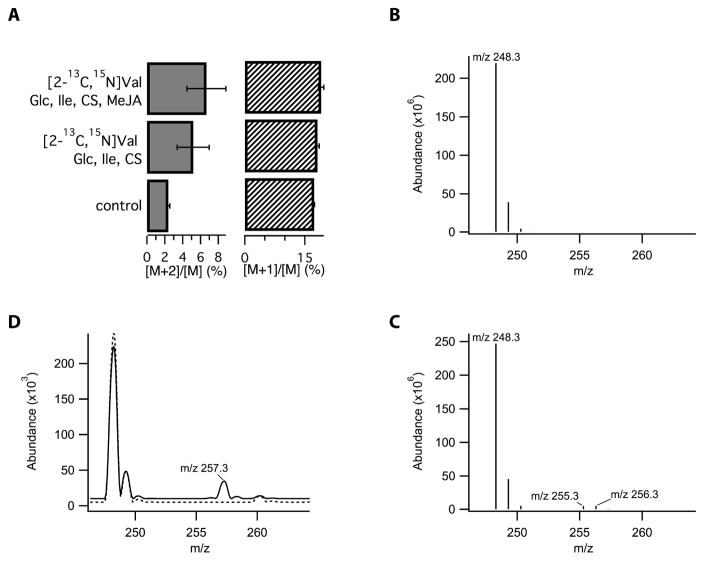
Using the MeJA-treated E. purpurea seedling system, several Val isotopomers were used to define the chemical constraints on the transformation of Val to the isobutyl amine moiety that is incorporated into the alkamides. The origin of the C1′-N bond of alkamides was established by feeding [2-13C,15N]Val to the seedlings. In these conditions, LC-MS analysis revealed that the newly formed M+2 isotopolog of i4N-12:2Δ2E,4E,8Z,10E accumulated to a level of 4.2% relative to the lower mass ions detected for seedlings provided only with the natural abundance Val precursor. This finding verified the integrity of the C-N bond that is incorporated into alkamides from the Val precursor (Fig. 2A).
Figure 2.
Enhanced isotopologue ratios in i4N-12:4Δ2E,4E,8Z,10E resulting from feeding of E. purpurea seedlings with isotopically labeled precursors. (A) Feeding of [2-13C/15N] valine significantly increased the abundance of M+2 versus M+1 species, indicating the retention of the C-N bond of the precursor, corrected for background Val transamination. MeJA and the presence of additional carbon sources, such as glucose (Glc), isoleucine (Ile), and chlorsulfuron (CS) had no significant effect on isotope incorporation. Isotopic envelope revealed by full-scan QqQ mass spectroscopy of i4N-12:4Δ2E,4E,8Z,10E isolated from seedlings incubated with (A) natural abundance valine and isoleucine and (B) media supplemented with [2H8]valine, unlabeled isoleucine, and in the presence of chlorsulfuron. Ions with m/z 256 and 255 correspond to the M+8 and M+7 ions resulting from incorporation of eight deuterium atoms from the decarboxylated form of [2H8]valine or the incorporation of seven deuterium atoms following the metabolic exchange of [2H8]valine with 2-ketoisopentanoate via transamination that results in the loss of one of the deuterium atoms. (C) Isotopic envelope from plants cultured in standard media (dashed line) or media supplemented with [2H9]isobutylamine hydrochloride (solid line); the M+9 ion at m/z 257 indicates that this precursor is incorporated into the alkamide without loss of any deuterium atoms.
The next labeling experiment probed the nature of the chemical transformation that generates the isobutylamine moiety from Val, by examining the fate of the hydrogen bonded to the C-2 of Val. Electrospray MS analysis of alkamide i4N-12:2Δ2E,4E,8Z,10E from seedlings fed fully deuterated-Val (i.e., [2H8]Val) exhibited two new M+7 and M+8 peaks at a ratio of 1:1.1 (Fig. 2B and C). The lower mass peak (i.e., M+7) can be ascribed to background transamination of valine, which is consistent with the [d7]Val that was observed in the mass spectra of free Val after 72 h incubation. This background transamination reaction is also consistent with the 0.8–1.7% increase in the M+1/M ion ratio that was observed in the alkamides of the earlier 13C-15N-Val labeling experiment (Fig. 2D). Incorporation of the entire complement of deuterium atoms from the precursor valine, particularly the deuteron at C-2, is consistent with a pyridoxal phosphate (PLP)-dependent decarboxylative mechanism that does not oxidize the C-2 carbon and thus does not break the C-2 to deuterium bond. This non-oxidative decarboxylation mechanisms generates the amine moiety of the alkamides and predicts that isobutylamine is an intermediate in this process. While isobutylamine itself was not observed to accumulate in Echinacea seedlings, the inclusion of 0.25 mM [2H8]isobutylammonium chloride in the growth medium resulted in the appearance of a peak in the LC-MS spectrum that was 9 atomic mass units larger than the parent ion labeled at 3.7% versus control seedlings (Fig. 2C). This is consistent with the incorporation of [2H8]isobutylamine into alkamides, and thus establishes the intermediacy of the amine in the alkamide biosynthetic pathway.
Finally, the use of isotopically labeled Ile provided analogous evidence for its involvement in the synthesis of 2-methylbutylamide alkamides. When seedlings exposed to chlorsufuron and MeJA were labeled with [15N]Ile, the [M+1] labeling increased from 18% for seedlings grown on identical medium with natural abundance Ile, to 35–90% for the 2-methylbutylamide alkamides, ai5N-12:2Δ2E,4E,8Z,10 and ai5N-13:2Δ2E,7Z,10a,12a. The enhanced labeling of these alkamides in the presence of MeJA is consistent with a short biosynthetic path from Ile through 2-methylbutylamine to the final alkamide products. This is a result similar to that obtained in the labeling of i4N-12:2Δ2E,4E,8Z,10E by [2H8]valine, which was activated by MeJA-treatment (increasing by five-fold, from 4.1% 20.1%), where significant labeling would be expected for if the Val-based pathway for the biosynthesis of isobutylamine alkamides has a small number of transformations from the amino acid precursor.
Transcriptomic analysis of Echinacea purpurea organs
Genes that may be involved in alkamide biosynthesis were identified in the Illumina mRNA-seq determined transcriptomes of different E. purpurea organs and tissues. These data were generated in collaboration with the Medicinal Plant Genomics Resource (http://medicinalplantgenomics.msu.edu). The collected datasets along with the metabolomics data were co-analyzed using the computational and statistical functionalities available at the Plant/Eukaryotic and Microbial Systems Resource (PMR) database (Wurtele et al. 2012).
Because there was no prior reference genome or transcriptome for Echinacea, the initial analyses were directed at assembling the short-read sequences into a reference transcriptome. An initial assembly of a reference transcriptome of E. purpurea was generated using RNA-sequencing from a cDNA library derived from root tissues of a single unique field-grown plant and a normalized cDNA library constructed from mature flower, primary stem (apical and intermediate), and leaves (immature and mature). To increase the representation of the reference transcriptome, unique reads from an array of 12 additional tissues and organs were used to supplement the initial assembly, and construction of a second and final transcriptome assembly (Table S1) (Góngora-Castillo et al 2012a; 2012b). In total, the final reference transcriptome assembly contained 44,422 loci (unigenes) representing 110,838 transcripts with an N50 contig size of 1,294 nucleotides (Table S2). As each assembled locus can contain multiple transcripts that may represent alternative splice forms, alleles, close paralogs, and/or homologs, downstream analyses utilized the “representative” transcript defined as the longest transcript for each locus. The 44,422 loci were functionally annotated using Pfam domain composition and alignments to UniRef sequences and the A. thaliana proteome, with 18,781 (42.2%) representative transcripts encoding a Pfam domain, 29,752 (66.9%) with a significant alignment to a UniRef entry, and 28,849 (64.1%) aligning to an A. thaliana protein.
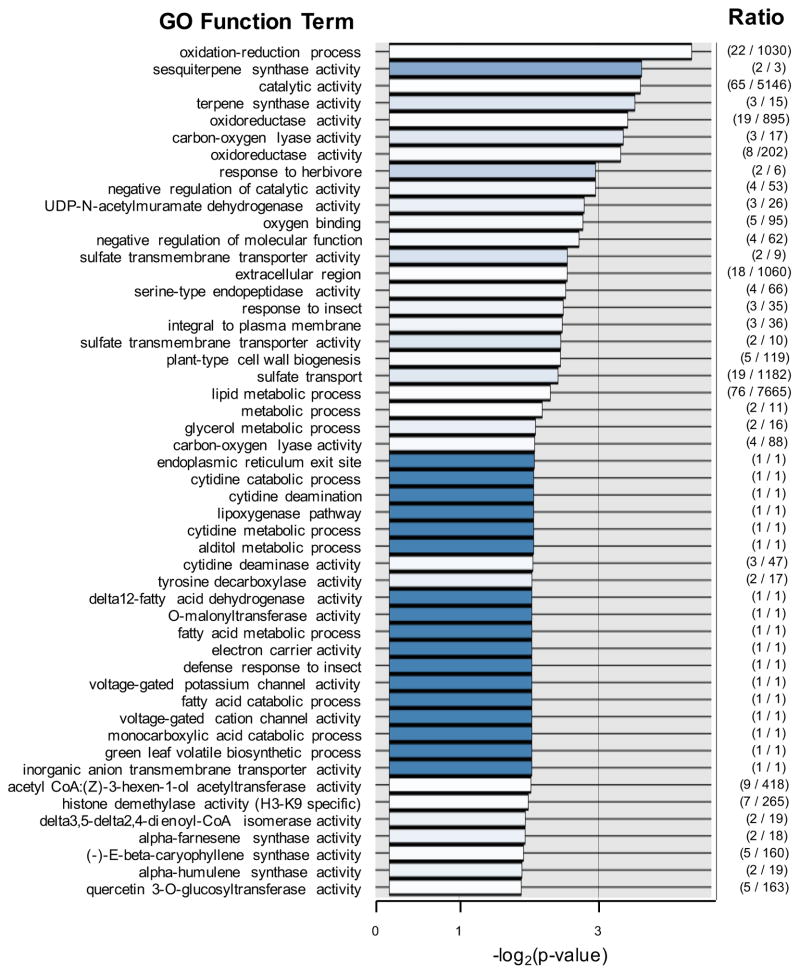
We further assessed the E. purpurea transcriptome of 20 different E. purpurea organs and tissues, and determined the expression profiles in these tissues/organs by aligning single end RNA-seq reads from each sample to the reference E. purpurea transcriptome (Data S2). The transcriptome profiles of the 20 metabolically diverse plant tissues were correlated with the distribution of the alkamide i4N-12:4Δ2E,4E,8Z,10E (the anchor alkamide) among these organ and tissue samples using the PMR database. These analyses identified 208 transcript contigs whose accumulation are highly correlated (correlation index above 0.7) to the anchor-alkamide distribution (Data S3). As with most genome annotations, BLAST-based homology analyses of these transcript contigs indicate that about 1/3 of them are related to genes functions are unknown. Among those that showed significant homology to Arabidopsis functional annotations, we utilized the GO Molecular Function annotation nomenclature to assign putative functions to the E. purpurea transcript contigs. These analyses indicate that the 208 transcripts that are co-expressed with alkamide accumulation are enriched in GO annotation functions associated with catalytic activity (42%), binding activity (31%), transcription (13%), and transporters (6%). Figure 3 presents a more detailed breakdown and distribution of GO Molecular Function terms that are associated with this group of transcripts whose expression is correlated with the distribution of the alkamide i4N-12:4Δ2E,4E,8Z,10E. Consistent with the potential role of fatty acid metabolism as the source of the acyl moiety of the alkamides, there are 9 GO Molecular Function terms that are significantly enriched in fatty acid and lipid metabolism functions in this group of transcripts.
Figure 3.
Functional annotation analysis of the transcriptome of E. purpurea in relation to the accumulation of alkamides. The transcriptomes of 20 different E. purpurea organs and tissues were sequenced in collaboration with the Medicinal Plant Genomics Resource (http://medicinalplantgenomics.msu.edu). Transcript sequences were annotated with GO functional annotations based on sequence homology with Arabidopsis genome annotations. The relative abundance of each, 13,431 unique E. purpurea reference transcripts was correlated with the abundance of the “anchor” alkamide, i4N-12:4Δ2E,4E,8Z,10E among the 20 different E. purpurea organs and tissues subjected to RNA-Seq analysis. The top 50 GO Molecular Function terms, and Biological Processes and Cellular Components terms were gathered from this correlation matrix, and the abundance of these terms was compared to their abundance in the entire 13,431-member reference transcriptome of E. purpurea. The enrichment of these GO functional terms in the alkamide-correlation matrix are sorted by increasing p-values (the order on the y-axis), and the x-axis plots the value of -log2(p-value). The blue-color gradient in each data-bar is proportional to the ratio of the genes annotation with the identified GO term in the alkamide-correlation matrix in relation to the number of genes with that GO term in the entire reference transcriptome (the absolute value of this ratio is shown in the last column).
Identification of E. purpurea BCAA decarboxylase
Amino acid decarboxylases catalyze PLP-dependent reactions (Hayashi 1995), and the Val and Ile-labeling experiments described above indicates a role for such an enzyme in the generation of the amine moiety of the alkamides. PLP-dependent enzymes have been structurally categorized into 5 families, the aspartate aminotransferase family (Class I), tryptophan synthase family (Class II), alanine racemase family (Class III), D-amino acid aminotransferase family (Class IV), and the glycogen phosphorylase family (Class V) (Eliot and Kirsch 2004). Only a single decarboxylase that can catalyze the decarboxylation of Val has been characterized, VlmD from Streptomyces viridifaciens (Garg et al. 2002), which is a Class II PLP-dependent enzyme. We used two correlative strategies to identify genetic elements within the sequenced transcriptome of E. purpurea that may encode for such an enzyme. One of these strategies is based on sequence homology shared among PLP-dependent amino acid decarboxylases, particularly the one biochemically confirmed Val decarboxylase (VlmD) (Garg, et al. 2002). The other strategy is based on the expectation that the expression of the genetic elements responsible for alkamide biosynthesis, including the expected decarboxylase, should correlate with the accumulation of the alkamide profiles.
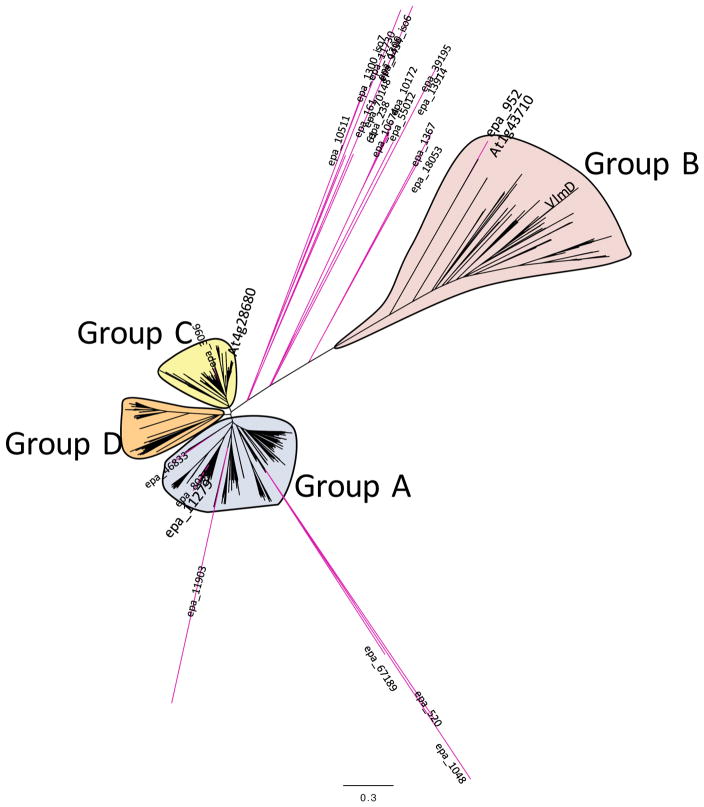
As a first step therefore, we searched the E. purpurea transcriptome data with NCBI Conserved Domain Database (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Marchler-Bauer et al. 2013) for genes homologous to VlmD, which identified 21 E. purpurea transcripts potentially encoding for such enzymes (Table 3 and Data S4). We subsequently searched GenBank for sequences that are homologous to VlmD and the ORFs encoded by the 21 E. purpurea VlmD-homologous transcripts. This resulted in the identification of 595 putative PLP-enzymes, which all belonged to the Class II tryptophan synthase family. The homology relationships among the putative E. purpurea PLP-dependent amino acid decarboxylases and Class II family, were explored via the sequence homology-based phylogenetic tree shown in Figure 4. These analyses categorized the 595 PLP-dependent enzymes into 4 distinct clades (Groups A-D) (Data S5). Group A consist of 247 sequences, which are primarily derived from photosynthetic organisms, including higher plants and algae, and they are annotated as tyrosine, DOPA, or L-aromatic amino acid decarboxylases. This clade contains 7 of the E. purpurea sequences putatively identified as PLP-dependent amino acid decarboxylases. Group B contains 101 sequences, which are sourced from Gram positive and Gram negative bacteria, a few lower plants and a biochemically characterized Arabidopsis serine decarboxylase (At1g43710) (Rontein et al. 2001). Most of these bacterial organisms appear to be pathogenic, and these sequences are richly annotated as histidine or glutamate decarboxylases. This clade also contains the biochemically characterized valine decarboxylase from S. viridifaciens (VlmD) and a single E. purpurea sequences (contig epa_952). The Group C clade contains 140 sequences, sourced from higher and lower plants, including one of the E. purpurea sequences (epa_3096) and an Arabidopsis tyrosine decarboxylase (At4g28680); the sequences in this clade are richly annotated as possible tyrosine, tryptophan or L-aromatic amino acid decarboxylases. The smallest clade (Group D) contains 92 sequences, most of which are putatively annotated as aromatic amino acid decarboxylases, and these are sourced from a wide variety of organisms, including bacteria, cyanobacteria, animals (e.g., mammals, primates, fish and birds), but not from plants. The majority of the E. purpurea sequences (12 sequences) are distinct from these four groups and map between the four clades.
Table 3.
Correlation of the expression of putative PLP-dependent decarboxylase mRNAs with the accumulation of iC4N-12:4Δ2E,4E,8Z,10E
| Sequence ID | Annotation | Pearson correlation coefficient |
|---|---|---|
| epa__11279 | Tryptophan decarboxylase | 0.80 |
| epa__952 | Serine decarboxylase | 0.28 |
| epa__70148 | Carboxy-lyase | 0.14 |
| epa__9434 | MYB transcription factor | 0.14 |
| epa__10511 | Glutamate decarboxylase | 0.01 |
| epa__11730 | Rab geranylgeranyl transferase type II beta subunit | −0.09 |
| epa__13914 | Diaminopimelate decarboxylase | −0.11 |
| epa__3096 | Tyrosine decarboxylase | −0.12 |
| epa__55012 | Cytokinin riboside 5′-monophosphate phosphoribohydrolase LOG3 | −0.13 |
| epa__8034 | Tryptophan decarboxylase | −0.16 |
| epa__520 | Arginine decarboxylase | −0.18 |
| epa__10674 | Carboxy-lyase | −0.20 |
| epa__1367 | Conserved gene of unknown function | −0.21 |
| epa__10172 | Carboxy-lyase | −0.22 |
| epa__1300 | Glutamate decarboxylase | −0.22 |
| epa__1048 | Arginine decarboxylase | −0.25 |
| epa__238 | Carboxy-lyase | −0.25 |
| epa__16161 | Decarboxylase | −0.26 |
| epa__11903 | Carboxy-lyase | −0.34 |
| epa__46833 | Tyrosine/dopa decarboxylase | NA |
| epa__18053 | Arginine decarboxylase | NA |
| epa__39195 | Diaminopimelate decarboxylase | NA |
| epa__67189 | Arginine decarboxylase | NA |
Figure 4.
Molecular phylogenetic analysis of putative PLP-dependent amino acid decarboxylases encoded by the E. purpurea transcriptome. The translated amino acid sequences of each E. purpurea contig are provided in Data S4. The biochemically authenticated bacterial valine decarboxylase (VlmD) and Arabidopsis serine decarboxylase encoded by locus At1g43710 is positioned within the Group B clade; this clade contains the protein encoded by E. purpurea contig epa_952 that was characterized as a serine decarboxylase. The E. purpurea contig epa_11279, whose transcript abundance positively correlates with the “anchor” alkamide, i4N-12:4Δ2E,4E,8Z,10E, is positioned in the Group A clade. The branch lengths of the tree represent the evolutionary distance among the sequences, calculated from 500 bootstrap trials, and the scale for this divergence distance unit is indicated.
The second correlative strategy used the PMR database to find correlations between the accumulation of alkamide i4N-12:4Δ2E,4E,8Z,10E among the 20 tissues and organs of E. purpurea with the expression patterns of the E. purpurea transcripts encoding the 21 putative PLP-dependent amino acid decarboxylases identified in Table 3. The expression pattern of each putative decarboxylase was extracted from the matrix of E. purpurea sequenced transcriptomes, which was determined from the identical samples used to obtain the metabolite abundance data. This calculation identified only a single putative PLP-dependent amino acid decarboxylase (contig Epa_11279) with a significant correlation to the accumulation pattern of alkamide i4N-12:4Δ2E,4E,8Z,10E (coefficient of 0.8), whereas all other potential PLP-enzyme candidates showed a correlation coefficient of below 0.3 (Table 3), including contig Epa_952, which is a candidate identified by homology to the biochemically confirmed Val decarboxylase from S. viridifaciens (VlmD) (Figure 4).
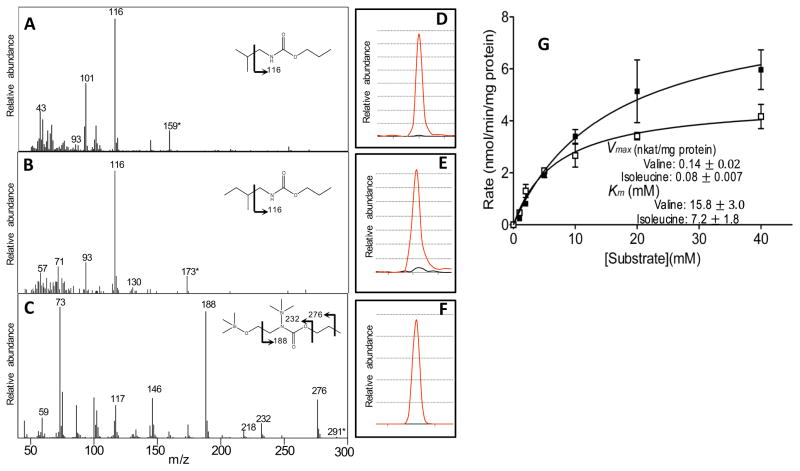
These correlation-based predictions of the identity of the Val decarboxylase responsible for alkamide biosynthesis were tested by heterologous expression in E. coli. Full-length Epa_952 (GenBank Accession #LT593931) and Epa_11279 (GenBank Accession #LT593930) ORFs were RT-PCR amplified from Echinacea RNA samples, and were cloned into an expression vector, and expressed in E. coli. The resulting purified recombinant proteins were used in in vitro assays using a variety of different amino acids as substrates, including valine and isoleucine. The products of these assays were evaluated for the accumulation of the appropriate primary amines, isobutylamine or 2-methylbutylamine, which would be the expected products from the decarboxylation of Val or Ile, respectively. These amine products were confirmed by comparing EI-MS fragmentation fingerprints and GC retention times to authentic standards. (Fig. 5A, B, D, E)
Figure 5.
Enzymatic characterization of recombinant PLP-dependent amino acid decarboxylases identified in the E. purpurea transcriptome (Epa_11279 and Epa_952). GC-MS identification of the enzymatic products of recombinant protein encoded by Epa_11279 incubated with valine or isoleucine, which led to the time-dependent appearance of isobutylamine (A) and 2-methybutylamine (B), respectively (all products were derivatized with propyl chloroformate prior to GC-MS analysis). The analogous incubation with serine of the recombinant protein encoded by Epa_952, led to the appearance of ethanolamine, which was derivatized with propyl chloroformate and silylated prior to GC-MS analysis (C). The abundance-trace of the molecular ion of each product (D–F), extracted from the corresponding GC-chromatogram (red-line), and the trace from the negative enzyme-control incubation (black-line). (G) Kinetic characterization of the recombinant E. purpurea BCAA decarboxylase (encoded by Epa_11279). Michaelis-Menten kinetic constants (Km and Vmax) for the BCAA decarboxylase were deduced using valine (■) or isoleucine (□) as the substrate. Data represents average of 3 determinations ± the standard error (see Methods for detail).
The recombinant protein encoded by Epa_952 did not support the enzymatic decarboxylation of either valine or isoleucine, but did catalyze the decarboxylation of serine to form ethanolamine (Fig. 5C and F). This finding is consistent with the high sequence homology between the Epa_952-coded protein and the serine decarboxylase of the Arabidopsis protein encoded by At1g43710 (Rontein, et al. 2001). We therefore conclude that Epa_952 encodes a E. purpurea serine decarboxylase. In contrast, Epa_11279 catalyzed the decarboxylation of both valine and isoleucine (Fig. 5A), resulting in the appearance of isobutylamine and 2-methybutylamine as reaction products in these incubations. These latter results therefore, establish that Epa_11279 encodes the BCAA decarboxylase that can generate the amine moieties of the alkamides.
Biochemical characterization of the Epa_11279-encoded amino acid decarboxylase
Computational translation of the Epa_11279 cDNA indicates that it encodes a protein of 503 amino acid residues. Alignment of this amino acid sequence with other biochemically characterized amino acid decarboxylases indicates that it belongs to the Group II amino acid decarboxylases (Sandmeier et al. 1994). Analysis of the sequence with the NCBI CDD program (Marchler-Bauer, et al. 2013) identified the eight conserved amino acid residues that bind the pyridoxal cofactor, which is a structural characteristic of Group II amino acid decarboxylases. This sequence alignment also identified the conserved catalytic lysine residue at position 320 (Figure 6), which forms an internal aldimine bond between the protein and the PLP cofactor (pyridoxal 5′-phosphate) (Hayashi 1995, Vaaler et al. 1986, Vaaler and Snell 1989). Additional computational analysis of the amino acid sequence with subcellular localization algorithm TargetP (Emanuelsson et al. 2000, Nakai and Kanehisa 1991), indicates that this decarboxylase is most likely a cytosolic enzyme.
Figure 6.
Sequence comparison of the biochemically characterized E. purpurea PLP-dependent enzymes with closely homologous decarboxylases. Identical residues are identified in red font and conservative substitutions are in blue font. The asterisk identifies the lysine residue, which forms an internal aldimine bond between the protein and the PLP cofactor, and this residue resides in the midst of an eight residue conserved motif that is a structural characteristic of Group II amino acid decarboxylases.
Experimental data concerning the substrate specificity and PLP-dependency of the isolated BCAA decarboxylase was explored by assaying the purified recombinant protein with different amino acid substrates (Table 4), and in the absence or presence of the cofactor (Table 5). The formation of isobutylamine and 2-methylbutylamine from valine and isoleucine respectively, was dependent on the presence of PLP; when PLP was omitted from the assay, the activity was 1.1% of that when it was included (Table 5). The enzyme was highly specific for valine and isoleucine, and there was very low activity with leucine; the turnover rate with leucine as a substrate is 4% of that for isoleucine and valine. The Km values for valine and isoleucine are 15.8 mM and 7.2 mM, with Vmax values of 0.14 nkat/mg protein and 0.08 nkat/mg protein, respectively (Figure 5G). These are values that are in a range similar to those of other group II amino acid decarboxylases (Garg, et al. 2002, Stevenson et al. 1990). There was no detectable decarboxylase activity with histidine, tyrosine or serine, which are substrates of the other known Group II amino acid decarboxylases (Eliot and Kirsch 2004, Sandmeier, et al. 1994).
Table 4.
Substrate specificity of recombinant E. purpurea BCAA decarboxylase (Epa_locus_11279)
| Substrate (5 mM) | Rate a (nkat/mg protein) |
|---|---|
| L-valine | 0.03 ± 0.003 |
| L-isoleucine | 0.035 ± 0.0016 |
| L-serine | <0.001 |
| L-leucine | <0.001 |
| L-histidine | <0.001 |
| L-tyrosine | <0.001 |
Data represents average of triplicate determinations ± standard error
These characterizations indicate that Epa_11279-encodes a type II BCAA decarboxylase with near equal activity with both valine and isoleucine. This enzyme appears to be unique in that it can decarboxylate all three BCAAs, with a strong preference for valine and isoleucine. The other reported decarboxylase that can act on BCAAs are from S. viridifaciens, which catalyze only the decarboxylation of valine (Garg, et al. 2002).
Spatial expression pattern of BCAA decarboxylase
The spatial expression pattern of the Echinacea BCAA decarboxylase was investigated at the mRNA and protein levels (Fig. 7). Protein and mRNA extracts were prepared from eight Echinacea organ samples, these being independent samplings of the eight organs that were used to determine the Illumina-based RNA-Seq transcriptome expression matrix and the alkamide metabolite profiles. The isolated mRNAs were used at templates to perform real-time PCR analysis, and the protein extracts were used to profile the accumulation of BCAA decarboxylase protein via immunological western-blot analysis.
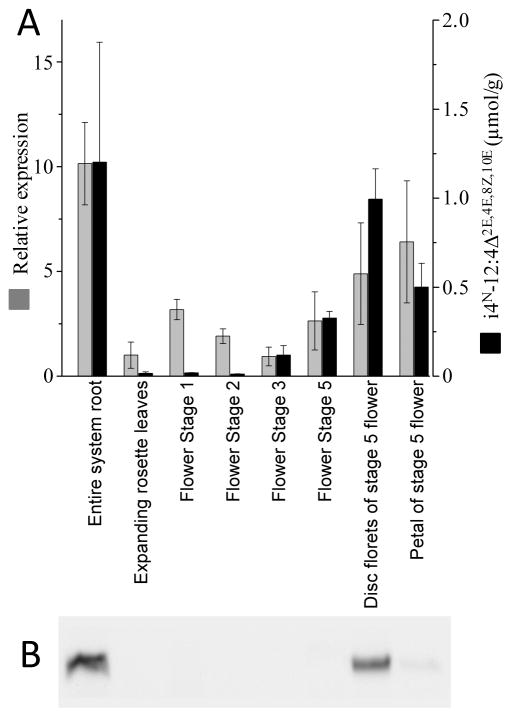
Figure 7.
Temporal and spatial expression of BCAA decarboxylase during E. purpurea growth and development. (A) The expression of the BCAA decarboxylase mRNA (Epa_11279) was measured by quantitative RT-PCR analysis, normalized relative to ubiquitin E2 mRNA (encoded by Epa_locus_12798). The accumulation of the anchor alkamide (i4N-12:4Δ2E,4E,8Z,10E) was determined GC-MS analysis. (B) Western blot analysis of the BCAA decarboxylase protein in extracts from the indicated organs and tissues.
Figure 7 compares these data, relative to the accumulation of the most abundant alkamide, i4N-12:4Δ2E,4E,8Z,10E. Roots are the organs that accumulate the largest concentration of the alkamide and they also show the highest expression of the BCAA decarboxylase at both the mRNA and protein levels. Although more detailed temporal studies would be required to elucidate the level at which BCAA decarboxylase expression is controlled, there appears to be a close correlation between the levels of the enzyme and the alkamide metabolic product, but such a correlation is less stringent when one compares the levels of the decarboxylase enzyme and mRNA. Namely, the disc florets and petals of stage 5 flowers show the second and third highest level of the alkamide and the decarboxylase enzyme, but during the development of the flowers (between stages 1 and 5), whereas the BCAA decarboxylase mRNA is quantifiable the level of the decarboxylase protein is below detection limit.
Discussion
Plants are renowned for their metabolic flexibility to generate many thousands of specialized metabolites that are non-uniformly distributed among discrete phylogenetic clades (Schilmiller et al. 2012a, Soltis and Kliebenstein 2015). Because of their sessile lifestyles, it has been suggested that plants have developed this metabolic diversity to enable a biochemical response to environmental stimuli, often exploiting the language of low molecular weight chemicals. Consistent with this hypothesis, plants possess a deep ancestral lineage providing access to a long evolutionary timeframe (500 MY for land plants, and longer if one considers photosynthetic eukaryotes) to exploit metabolic diversity in a myriad of ecological and geographical niches. In addition, plants provide a key interface between the abiotic and biotic worlds, and therefore drive the chemical diversity in the global ecosystem. Human civilizations have adapted and developed technologies to explore and manage this biochemical diversity (i.e., the basis for agronomic agriculture), which is exemplified by the selection and domestication plants for their unique adaptabilities, resilience and chemical constituents for such items as food, fiber, biofuel, medicinal and other products used to sustain and enhance human life (Fuller 2007, Heun et al. 2012, Olsen and Wendel 2013).
The traditional reductionist strategy for dissecting natural product based plant traits focuses on fractionation and analysis, with the aim of defining biochemical components that are responsible for the desirable trait (Cutler and Cutler 2000). In the modern era, advances in molecular-genetic and biochemical characterizations, particularly the technology associated with biochemical analysis of nucleic acids (exemplified by the emergence of genomics), and chromatography, mass spectrometry and NMR (exemplified by the emergence of proteomics and metabolomics) have provided new avenues for directly exploring such diverse biological traits. Initially, because of expense, these efforts focused on model genetic organisms (i.e., E. coli, Saccharomyces cerevisiae, Arabidopsis, etc.); however, as the cost of nucleic acid sequencing has plunged, genomics-based analytic technologies have been a major driver for deciphering biological diversity.
In this study we integrated these strategies and explored the biosynthesis of alkamides, which are a class of specialized metabolites that occur in at least 33 different plant families (Rios 2012). Alkamides are lipids made up of two moieties, an amine moiety that is acylated with a carboxylic acid moiety. A wide range of chemistries are associated with both moieties (e.g., aliphatic, cyclic or aromatic amines, and polyunsaturated and aromatic carboxylic acids) and these are integrated in the nearly 300 structurally diverse natural alkamides that have been characterized to date (Rios 2012). We selected to characterize the biosynthesis of the alkamides of Echinacea as a model. The Echinacea alkamides are relatively simple examples, being composed of either isobutylamine or 2-methylbutylamine moieties, which are acylated with variety of polyunsaturated or polyacetylenic fatty acids. In this study, we integrated genome-wide expression profiling with RNA-Seq technology, coupled with MS-based metabolite profiling and metabolic labeling experiments to discover and characterize how the amine moiety of these alkamides is generated. This specifically led to the isolation and characterization of a never before characterized plant BCAA decarboxylase. This decarboxylase is a pyridoxal-dependent enzyme that can utilize Val or Ile as substrates, and generate isobutylamine or 2-methylbutylamine, respectively. This characterization validated the in vivo labeling experiments, which established that Val and Ile are the direct precursors of isobutylamine and 2-methylbutylamine that constitute the amine moiety of the Echinacea alkamides. We propose that these two amines are subsequently acylated with fatty acid derivatives to generate the two types of alkamides that occur in the Echinacea genus.
This discovery highlights the need to integrate multiple datasets to narrow the genome search space for the correct identification of a target gene. We initially searched the translated Echinacea proteome (based upon the RNA-Seq data) for proteins that can catalyze the decarboxylation of amino acids. Using conserved motifs that are common to such PLP-dependent decarboxylases, and the sequences of the only known BCAA decarboxylases, initially led to the erroneous identification of a protein that ultimately proved to be a serine decarboxylase. It was only after we integrated alkamide-profiling data into the sequence homology datasets, and identified the decarboxylase sequence whose expression correlated with the accumulation of alkamides that we focused on the correct sequence that encodes the Echinacea BCAA decarboxylase. Ultimate biochemical proof of this enzymological identity was obtained by directly assaying the conversion of Val and Ile to the appropriate amine by the recombinantly produced pure Echinacea protein. The role of such a PLP-dependent BCAA decarboxylase in producing the amine moiety of the Echinacea alkamides is consistent with our isotopic labeling experiments, which established that BCAAs are converted to the amines, via a chemical mechanism that conserves the C-N bond of the amino acids, and the hydrogen positioned on the C-2 of the amino acids. Both of these constraints are satisfied by the PLP-dependent BCAA decarboxylase that we have isolated and characterized from E. purpurea.
The authentication of the role of a PLP-dependent decarboxylase that acts on amino acid substrates and generates the amine moiety of the Echinacea alkamides can be used as a basis to predict how the broader classes of alkamides are generated in other plant families. For example, in the broader Asteraceae family, alkamides that contain N-(3-methylbutyl), N-phenethyl and N-tyramido amine moieties have been characterized, and we would therefore predict that these are generated by the decarboxylation of leucine, phenylalanine and tyrosine, respectively. Evidence that supports this model for generating amides has also been gathered by studies of the alkamides of Acmella radicans, commonly called the “tooth herb” (Cortez-Espinosa et al. 2011). In the Solanaceae family, the capsaicinoids that occur in the Capsicum (peppers) genus are acylated derivatives of 4-hydroxy-3-methoxybenzylamine. A bioinformatics-based study has suggested that this amine may be generated from phenylalanine via a decarboxylase-independent process that couples the phenylpropanoid pathway with β-oxidation (Mazourek et al. 2009). This would indicate that two distinct evolutionary adaptations, one specific to the Asteraceae family and a second specific to the Solanaceae family, may have given rise to the alkamide biosynthesis trait.
Finally, this study is exemplary of the strength of integrating genomics datasets with metabolomics datasets to facilitate the accurate annotation of gene functions. Because of the relative simplicity by which genomics data can be generated, it is tempting to use only such datasets for the identification of functions by sequence homology. This study indicates however, the ease with which erroneous conclusions can be reached, which initially led to the identification of an Echinacea serine decarboxylase. Therefore by integrating genomics and metabolomics datasets using such computational resources as the Plant/Eukaryotic and Microbial Systems Resource (PMR) database (Wurtele, et al. 2012), enabled an accurate identification of the correct protein that catalyzes the specific BCAA decarboxylation reaction. PMR was initially established to integrate and allow the querying of combined genomics and metabolomics datasets of medicinally relevant plants (Hur et al. 2013); these are plants that express very diverse natural products and are usually poorly characterized at the molecular genetic level. In its current form, PMR not only integrates genomics and metabolomics data from medicinal plants, but also crop and model plant systems, microbes and animal sources. In addition to this study that identified an enzyme involved in the biosynthesis of Echinacea natural products, PMR has also been used to identify genetic elements in other areas of natural product biochemistry, including polyketides and alkaloids (Crispin et al. 2013, Fukushima et al. 2014, Giddings et al. 2011, Gongora-Castillo et al. 2012a, Yeo et al. 2013).
Methods
Plant material
Echinacea purpurea accessions PI 633670 and PI 631313 seeds (www.arsgrin.gov/npgs/acc/acc_queries.html) were obtain from the North Central Regional Plant Introduction Station (NCRPIS) (Ames, IA) of the US Department of Agriculture, Agricultural Research Service (USDA/ARS). Accession PI 633670 was grown in a growth chamber (Environmental Growth Chambers, Chagrin Falls, Ohio) in Sunshine soil mix SB 300 (Sungro, Belle Vue, WA) in 20-cm garden pots, at 25/19 °C, with 16 h of illumination at 110 μmol m−2 s−1. Accession PI 631313 was propagated in a greenhouse in RediEarth soil in 20-cm garden-pots, fertilized weekly with Miracle Grow. The greenhouse conditions were maintained at 25/12 °C with 13 h of illumination at 250 μmol m−2 s−1 of natural light supplemented with illumination by Na/Hg lamps.
The reference transcriptome sequence was determined from RNA isolated from a single field-grown plant (accession PI 633670) grown in silty clay loam soil at the NCRPIS (Ames, IA). Seeds for this plant were sown in April 2009, and aerial parts were harvested in August 2009, and the roots were harvested in May, 2010. Plant organs and tissues were dissected, quickly washed to remove soil contaminants, flash frozen in liquid N2 and stored at −80 °C.
Germination and growth of E. purpurea
E. purpurea seeds (Prairie Nursery, Westfield, WI) were cold stratified in a 10% aqueous solution of Plant Preservation Mixture (PPM; Plant Cell Technologies, Washington, DC) for 16 h at 4 °C. The solution was decanted and seeds were washed with 70% ethanol (aq.), 0.5% bleach solution containing 0.02% Tween-20, and autoclaved nanopure H2O until the odor of bleach was eliminated. The seeds were then placed on ½-strength MS agar plates containing sucrose (11 mM), MES (2.56 mM), pH 5.4, PPM (0.2%) and placed in an incubator at 22 °C with a 18 h light/6 h dark cycle. The light intensity was 100 μmol m−2 s−1 at the level of the plants. Germination was asynchronous and occurred between 10–21 days after imbibition. Seedlings for labeling experiments were used when the first leaf unfurled, which occurred over a 2–5 day-span (Figure S3).
Fatty acid analysis
Fatty acid analyses were conducted as previously described (Quanbeck et al. 2012). Frozen plant tissue (50–100 mg aliquot) was homogenized with 1 mL of 10% (w/v) barium hydroxide and 0.55 mL of 1,4-dioxane, containing 20 μg/mL nonadecanoic acid (Sigma Chemical Co., St. Louis, MO, USA) as an internal standard. The mixture was incubated at 110 °C for 24 h. After cooling the mixture was acidified with 6 M aqueous HCl, and fatty acids were recovered by extracting twice with hexane. The pooled hexane extracts were concentrated by evaporation under a stream of nitrogen gas. The recovered fatty acids were converted to methyl esters with HCl:methanol (1:5.25 v/v) at 80 °C for 60 min. The ester-containing extracts were concentrated with a stream of nitrogen gas, derivatized with BSTFA/TCMS (Sigma) (65 °C, 30 min), and subjected to GC-MS analysis.
Amino acid analysis
Amino acid analyses were conducted as previously described (Quanbeck, et al. 2012). Frozen plant tissue (50–100 mg aliquot) was homogenized with 10% TCA and following centrifugation at 10,000 ×g for 10 min, the supernatant was transferred to a glass vial for further purification using “EZ:faast” GC-FID kit for free amino acid analysis (Phenomenex, Torrance, CA). Norvaline (10 nmol) was used as the internal standard. For the isotopic labeling of amino acids, the derivatized amino acids were monitored with an Agilent 6410 triple-quadrupole mass spectrometer.
Alkamide analysis
Alkamides were extracted with a method developed by Hudaib et al. (2002). About 50–100 mg of frozen tissue (spiked with 34 nmol of the internal standard, (2Z,4E)-dodeca-2,4-dienoic acid isobutylamide) was homogenized and extracted using a mortar and pestle with two aliquots of 70% v/v methanol. After centrifugation at 3500 ×g for 5 min, the pooled supernatants were extracted with two aliquots of n-hexane-ethyl acetate (1:1 v/v). The organic phases were collected, pooled, and evaporated under a stream of nitrogen gas. The dried samples were dissolved in 1 ml of acetonitrile, and silylated by incubating at 70 °C for 20 min with 70 μl of N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% chlorotrimethylsilane (Sigma). After evaporating the excess reagents under a stream of N2 gas, the samples were dissolved in acetonitrile and subjected to GC-MS analysis.
Isotopic labeling experiments
Stable isotope incorporation experiments were performed by transferring seedlings that were unfurling their first leaf (Figure S3) from germination plates to sterile 90-mm Petri dishes each containing a 70-mm Whatman No. 1 filter disc and flooded with 6-mL of ½-strength MS containing MES (2.56 mM), pH 5.4, PPM (0.2% v/v). Chlorsulfuron (30 μM final, from a stock in 5 mM KPi buffer, pH 7.5), methyl jasmonate (10 μM from 0.5 M ethanol stock), amino acids (15 mM) and isobutylammonium chloride (0.25 mM) were added to the MS medium as required. Seedlings were incubated with the medium for 3 days, flash-frozen with liquid nitrogen and stored at −80 °C until analysis. Four seedlings were used in each of the isobutylammonium chloride feeding experiments, 5 seedlings were used for each of the [2-13C-15N]valine and [15N]isoleucine labelling experiments, and 10 seedlings were used for each the [d8]valine labelling experiments. All these experiments were repeated at least three times.
For LC-MS analysis, three frozen seedlings were homogenized and extracted using a mortar and pestle with two aliquots of 70% (v/v) aqueous methanol. After centrifugation at 2000 rpm for 5 min, the pooled supernatants were extracted with two 2-mL aliquots of hexane-EtOAc (1:1, v/v). The organic phases were collected and pooled for analysis. The MS parameters for triple quadrupole analysis (Agilent 6410) were as follows: MS2 full scan method, ESI positive mode, Fragmentor voltage: 135 V, Gas temperature: 300 °C, Gas flow: 10 L/min, Nebulizer pressure: 45 psi, Capillary: +/− 4000 V. The chromatographic parameters were: Waters XBridge-C18 column (4.6 × 150 mm, 5 μm particle size); Solvent A: H2O + 0.1% formic acid. Solvent B: CH3CN + 0.1% formic acid; Flow rate: 1 mL/min; 70 → 100% B over 10 min with a 1-min hold and then ramped back to 70% B over 3 min; post time: 2 min. Labeling was measured using the alkamide quasimolecular ion. Raw isotopic enhancement equals the sum of the intensities of isotopologue peaks in the envelope, less the intensity of M and a proportionate contribution due to M+1 and M+2, as determined from alkamides extracted from plants grown under natural abundance conditions with standard medium. Percentage isotopic enhancement is the (raw isotopic enhancement/total intensity for the envelope) x 100.
For GC/MS, the organic extracts were evaporated under a stream of N2 gas, dissolved in 1 mL of acetonitrile and silylated by incubating at 70 °C for 20 min with 70 μL of N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% chlorotrimethylsilane. After evaporating the excess reagents under a stream of N2 gas, the samples were dissolved in 200 μL of acetonitrile and subjected to GC/MS analysis.
Amino acid derivatization for isotopic analysis
Amino acid analysis was performed using the “EZ:faast” LC kit for free amino acid analysis, following the manufacturer’s protocol (Phenomenex; Torrance, CA). The derivatized amino acids were solubilized in 33 μL of 10 mM aqueous NH4HCO2 and 66 μL of 10 mM methanolic NH4HCO2 prior to LC/MS analysis.
Free amino acid analysis was performed with chromatography on an Agilent 6410 Triple Quadrupole MS. Samples were injected via the system autosampler and separated by an EZ:faast AAA-MS column (2.0 × 250 mm, 4 μm particle size). The chromatographic parameters were Solvent A: H2O + 10 mM NH4HCO2. Solvent B: CH3CN + 10 mM NH4HCO2, Flow rate: 0.25 mL/min; 68 → 83% B over 16 min; 83 → 68% B over 0.1 min; hold at 68% B for 2 min. The MS parameters were: MS2 full scan method, ESI positive mode, Fragmentor voltage: 110 V, Gas temperature: 350 °C, Gas flow: 7 L/min, Nebulizer pressure: 20 psi, Capillary: +/− 4000 V.
GC-MS analysis
GC-MS analyses were performed using either a 6890 Series gas chromatograph, equipped with a model 5973 Mass Detector or a 7890A GC equipped with a 5975C MS (Agilent Technologies, Palo Alto, CA) operating in the electron-impact ionization mode (70 eV). The analyses were carried out in splitless mode. Metabolites were separated on Agilent HP 5ms, 5% diphenyl/95% dimethyl polysiloxane capillary column (30 m × 0.25 mm, 0.25 μm film thickness). Helium was used as the carrier gas at a flow rate of 1.2 to 1.56 ml/min. Oven conditions: 80 °C, 2 min; 10 °C/min to 200 °C, hold 20 min; 3 °C/min to 250 °C, hold 5 min). The identification of compounds was facilitated by using Agilent Enhanced ChemStation version D.02.00.275 and the quantification was calculated by integrating the corresponding peak areas relative to the area of the internal standard.
RNA-sequencing and expression analyses
RNA was isolated from the E. purpurea tissues and organs as described previously (Gongora-Castillo et al. 2012b). An iterative assembly approach was used to generate the E. purpurea reference transcriptome as described previously (Gongora-Castillo, et al. 2012a). First, a normalized TruSeq RNA-seq library was constructed from equimolar amounts of pooled RNA isolated from mature flowers, primary stem (apical and intermediate) and leaves (immature and mature). The normalized library and a single TruSeq RNA-seq library made of root tissues were sequenced in the paired end mode to 120 nucleotides on the Illumina Genome Analyzer II platform. Reads were cleaned and assembled using Velvet/Oases using a kmer of 31 (Schulz et al. 2012). Second, to augment the initial transcriptome assembly, individual Illumina TruSeq libraries were constructed from 12 tissues and organs (Data S1) and 35 nucleotide single end reads were generated on an Illumina Genome Analyzer IIx. Single end reads were cleaned and aligned to the initial transcriptome assembly to identify reads not present in the assembled transcriptome. To generate the final assembly, paired end reads from the normalized cDNA library and root library were combined with all unique single end reads from 12 libraries (Data S1) and assembled using Velvet/Oases using a kmer of 27 (Schulz, et al. 2012). Low complexity sequences were filtered out and contaminants were identified based on BLASTX sequence similarity to bacterial, fungal, viral, viroid, arthropod, stramenopile or human sequences in UniRef. Peptides were predicted from the representative transcripts using ESTScan v 3.0.3 (Iseli et al. 1999), and Pfam domains were identified using HMMer (v 3.0) and Pfam 24.0 (Punta et al. 2012). Functional annotation was assigned using sequence similarity to UniRef entries, the predicted A. thaliana proteome, and Pfam domains as described previously.
To determine expression abundances in an atlas of E. purpurea tissues and organs, additional TruSeq RNA-seq libraries were constructed from eight additional tissues/organs (Table S1) and sequenced to 35 nucleotides in the single end mode as described above. In total, RNA-seq reads from 20 different tissues and organs (Table S1) were used to determine expression abundances (fragments per kilobase transcript per million mapped reads; FPKM) by aligning the single end reads to the final reference transcriptome using TopHat (Trapnell et al. 2009) and Cufflinks (Trapnell et al. 2010).
Transcriptomic sequences were also annotated with NCBI Conserved Domain Database (CDD.v2.26) (ftp.ncbi.nih.gov/pub/mmdb/cdd/(Marchler-Bauer, et al. 2013)) using ncbi-blast-2.2.24+-win64 package (ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.2.24/) with the following parameters: rpsblast -i querydatabase.fasta -d cdd -p F -e 1e-5 -o output.txt. The database of transcriptomic sequences for BLAST analysis was generated the following parameters: makeblastdb.exe -in -inputdatabase.fasta -parse_seqids -hash_index -dbtype nucl.
RNA-Seq Data Access
Raw RNAseq reads are available in the National Center for Biotechnology Information Sequence Read Archive under accession number SRP007464. The assembled E. purpurea transcriptome is available at ftp://ftp.plantbiology.msu.edu/pub/data/MPGR/Echinacea_purpurea/ and the Data Dryad Digital Repository under this doi (to be provided upon publication).
DNA Manipulation
DNA manipulation techniques, such as PCR amplification, plasmid preparation, DNA digestion and ligation, agarose gel electrophoresis, and genetic transformation were carried out by standard methods. The full-length Epa_11279 and Epa_952 cDNAs were PCR amplified (using primers P1, P2 and P3, P4 respectively, Table S3) and cloned into pENTR-D/TOPO (Invitrogen, Carlsbad, CA). The insert was recombined into the vector pET-54-DEST (EMD4Biosciences, Billerica, MA) using L/R-Clonase (Invitrogen).
RT-PCR Analysis
RNA was extracted from E. purpurea tissues using the RNease Plant Mini Kit (Qiagen Inc., Valencia, CA), then treated with RNase-free DNase (Invitrogen) to remove any remaining DNA. RNA was reverse-transcribed using a Reverse Transcription Kit according to the manufacturer’s instructions (Invitrogen). The relative abundance of the Epa_11279 cDNA was determined on a Bio-Rad iCycler iQ5 Real Time PCR System (Bio-Rad, Hercules, CA), using the primers listed in Table S3. Epa_12798, which encodes ubiquitin E2, was used as internal reference to normalize mRNA content.
Protein Methods
To over-express the putative BCAA decarboxylase in E. coli, an overnight culture of the strains harboring the appropriate expression vector in Arctic Express (Agilent Technologies) or BL21 (DE3) (Invitrogen) cells were inoculated into LB medium containing 100 μg/ml ampicillin and the cultures were grown at 37 °C to an A600 of 0.5–0.6. At this point, IPTG was added to a concentration of 0.1 or 0.4 mM, and growth was continued at 20 °C for further 8–10 h. The cells were harvested by centrifugation and cell pellets were suspended in 50 mM NaH2PO4, pH 8.0; 300 mM NaCl, and 5 mM imidazole and disrupted by sonication at 4 °C. Recombinant His-tagged protein was purified from the extracts using a nickel affinity-column chromatography (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions. Imidazole was removed from the purified protein solution by dialysis at 4 °C against a buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 5 mM dithiothreitol, 1 mM PLP, and 10% glycerol (v/v). The purified His-tagged recombinant protein (encoded by both Epa_11279 and Epa_952) was used to immunize mice to generate antisera. Purified protein preparations were routinely stored at −80 °C after freezing in liquid nitrogen to preserve enzymatic activity. Protein concentrations were determined by Bradford’s method (Bradford 1976) using bovine serum albumin as the standard. Protein extracts from plant tissues and immunoblot analyses were performed as described previously (Che et al. 2002, Li et al. 2011).
Enzyme Activity Assay
The amino acid decarboxylase assays were performed in 100 μl reaction buffer consisting of 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 5 mM dithiothreitol, 1 mM PLP, 10% glycerol (v/v), and variable concentrations of valine, isoleucine, serine, leucine, histidine, or tyrosine, at between 1 mM and 40 mM, and 400 μg/ml of purified recombinant decarboxylase protein. After incubation for 1 h at 37 °C, the reaction was stopped with equal volume of 20% TCA. The amine product was subsequently subjected to alkylation using the EZ:faast kit (Kugler et al. 2006) and the resultant amine derivatives were analyzed with an Agilent 7890A GC coupled to an Agilent 5975C MSD detector.
Statistical Analyses
Protein sequences of PLP-dependent amino acid decarboxylases were downloaded from the GenBank (www.ncbi.nlm.nih.gov/). The protein sequences were aligned using ClustalW (Thompson et al. 1994) using the default parameters. The rooted tree was constructed from aligned sequences using Neighbor-Joining distance method (Saitou and Nei 1987) with Jones-Taylor-Thornton (JTT) model (Jones et al. 1992) by Molecular Evolutionary Genetics Analysis 5.05 (MEGA5.05) (Tamura et al. 2011) software package. Alignment gaps were subjected to pairwise deletion. A bootstrap test with 1000 replicates was applied to further verify the phylogenetic tree.
Accumulation of metabolites detected in 13 different plant tissues and organs were subjected to hierarchical clustering analysis using CLUSTER (version 3.0) (de Hoon et al. 2004). The resulting dendogram was viewed with Java Treeview (Saldanha 2004). Statistical correlations between metabolite abundance and transcript abundance data were calculated using the functionalities of the Plant/Eukaryotic and Microbial Systems Resource (PMR) database (Wurtele, et al. 2012). This database (http://metnetdb.org/PMR/) and computational platform enables users to identify “non-obvious” genetic elements that regulate biological processes by statistical co-analysis of transcriptomics and metabolomics data. These analyses include real-time pairwise correlations of metabolites with transcripts from large datasets, which was used for this analysis. Specifically, PMR was used to calculate the pairwise Pearson correlation between alkamide metabolites with the entire RNA-seq data, using the datasets from the Echinacea tissues and organs whose transcriptomes were sequenced (Gongora-Castillo, et al. 2012b).
Supplementary Material
Metabolite levels in different organs and tissues of E. purpurea.
Expression abundances of the E. purpurea transcriptome in an atlas of 20 tissues and organs.
Correlations of E. purpurea transcripts with the accumulation of the alkamide, iC4N-12:4Δ2E,4E,8Z,10E.
Amino acid sequences of putative PLP-dependent enzymes encoded by Echinacea purpurea.
PLP-dependent enzyme sequences as phylogenetically classified in Figure 4.
Log ratio plot of the relative abundance of 33 different alkamides among 36 different organs and tissues
Hierarchical clustering analysis of the distribution of fatty acids, amino acids and alkamides among different E. purpurea organs and tissues.
Effect of incubating Echinacea seedlings with MeJA on the accumulation of alkamides.
Mass spectra of “possible new alkamides (PNA)”.
cDNA libraries and RNA-Seq data generated for E. purpurea.
Statistics for the E. purpurea reference transcriptome.
DNA-primers used in PCR reactions.
Acknowledgments
We thank members of the former Iowa Center for Research on Botanical Dietary Supplements, which was supported between 2002 and 2011 by the Office of Dietary Supplements and the National Institute for Environmental Health Sciences, National Institutes of Health (NIH) (grant number 5RC2GM092521). We acknowledge the W.M. Keck Metabolomics Research Laboratory (Iowa State University) for providing access to analytical instrumentation. This research was made possible by grant numbers 0919743 (BJN) and 0919938 (REM) from the National Science Foundation (NSF). Mass-spectrometry facilities at IUPUI were funded through NSF grant DBI-0821661 (REM). The authors declare no conflicts of interest.
References
- Bauer R, Foster S. Analysis of alkamides and caffeic acid derivatives from Echinacea simulata and E. paradoxa roots. Planta Med. 1991;57:447–449. doi: 10.1055/s-2006-960147. [DOI] [PubMed] [Google Scholar]
- Bauer R, Khan IA, Wagner H. TLC and HPLC Analysis of Echinacea pallida and E. angustifolia Roots1. Planta Med. 1988a;54:426–430. doi: 10.1055/s-2006-962489. [DOI] [PubMed] [Google Scholar]
- Bauer R, Remiger P. TLC and HPLC Analysis of Alkamides in Echinacea Drugs1,2. Planta Med. 1989;55:367–371. doi: 10.1055/s-2006-962030. [DOI] [PubMed] [Google Scholar]
- Bauer R, Remiger P, Wagner H. Alkamides from the roots of Echinacea purpurea. Phytochemistry. 1988b;27:2339–2342. [Google Scholar]
- Bauer R, Remiger P, Wagner H. Alkamides from the roots of Echinacea angustifolia. Phytochemistry. 1989;28:505–508. [Google Scholar]
- Binns SE, Inparajah I, Baum BR, Arnason JT. Methyl jasmonate increases reported alkamides and ketoalkene/ynes in Echinacea pallida (Asteraceae) Phytochemistry. 2001;57:417–420. doi: 10.1016/s0031-9422(00)00444-1. [DOI] [PubMed] [Google Scholar]
- Binns SE, Livesey JF, Arnason JT, Baum BR. Phytochemical variation in echinacea from roots and flowerheads of wild and cultivated populations. J Agr Food Chem. 2002;50:3673–3687. doi: 10.1021/jf011439t. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campos-Cuevas J, Pelagio-Flores R, Raya-Gonzalez J, Mendez-Bravo A, Ortiz-Castro R, Lopez-Bucio J. Tissue culture of Arabidopsis thaliana explants reveals a stimulatory effect of alkamides on adventitious root formation and nitric oxide accumulation. Plant Sci. 2008;174:165–173. [Google Scholar]
- Che P, Wurtele ES, Nikolau BJ. Metabolic and environmental regulation of 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis. Plant Physiol. 2002;129:625–637. doi: 10.1104/pp.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen LP, Lam J. Acetylenes and Related-Compounds in Heliantheae. Phytochemistry. 1991;30:11–49. [Google Scholar]
- Cortez-Espinosa N, Avina-Verduzco JA, Ramirez-Chavez E, Molina-Torres J, Rios-Chavez P. Valine and phenylalanine as precursors in the biosynthesis of alkamides in Acmella radicans. Nat Prod Commun. 2011;6:857–861. [PubMed] [Google Scholar]
- Crispin MC, Hur M, Park T, Kim YH, Wurtele ES. Identification and biosynthesis of acylphloroglucinols in Hypericum gentianoides. Physiol Plant. 2013;148:354–370. doi: 10.1111/ppl.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SJ, Cutler HG. Biologically active natural products: pharmaceuticals. Boca Raton, FL: CRC Press; 2000. [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E, Werck-Reichhart D, Lewinsohn E. Plant metabolism. Editorial. Mol Plant. 2010;3:1. doi: 10.1093/mp/ssp111. [DOI] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Mejia RF, Iwasa M, Kobayashi M, Hayashi N, Watanabe-Takahashi A, Narisawa T, Tohge T, Hur M, Wurtele ES, Nikolau BJ, Saito K. Metabolomic Characterization of Knockout Mutants in Arabidopsis: Development of a Metabolite Profiling Database for Knockout Mutants in Arabidopsis. Plant Physiol. 2014;165:948–961. doi: 10.1104/pp.114.240986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DQ. Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World. Ann Bot. 2007;100:903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RP, Ma Y, Hoyt JC, Parry RJ. Molecular characterization and analysis of the biosynthetic gene cluster for the azoxy antibiotic valanimycin. Mol Microbiol. 2002;46:505–517. doi: 10.1046/j.1365-2958.2002.03169.x. [DOI] [PubMed] [Google Scholar]
- Gertsch J, Schoop R, Kuenzle U, Suter A. Echinacea alkylamides modulate TNF-alpha gene expression via cannabinoid receptor CB2 and multiple signal transduction pathways. FEBS Lett. 2004;577:563–569. doi: 10.1016/j.febslet.2004.10.064. [DOI] [PubMed] [Google Scholar]
- Giddings LA, Liscombe DK, Hamilton JP, Childs KL, Dellapenna D, Buell CR, O’Connor SE. A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome p450 in Catharanthus Roseus. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.M111.225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore MR. Uses of Plants by the Indians of the Missouri River Region. Lincoln: University of Nebraska Press; 1977. [Google Scholar]
- Gongora-Castillo E, Childs KL, Fedewa G, Hamilton JP, Liscombe DK, Magallanes-Lundback M, Mandadi KK, Nims E, Runguphan W, Vaillancourt B, Varbanova-Herde M, Dellapenna D, McKnight TD, O’Connor S, Buell CR. Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS One. 2012a;7:e52506. doi: 10.1371/journal.pone.0052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongora-Castillo E, Fedewa G, Yeo Y, Chappell J, DellaPenna D, Buell CR. Genomic approaches for interrogating the biochemistry of medicinal plant species. Methods in enzymology. 2012b;517:139–159. doi: 10.1016/B978-0-12-404634-4.00007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H. Pyridoxal enzymes: mechanistic diversity and uniformity. J Biochem. 1995;118:463–473. doi: 10.1093/oxfordjournals.jbchem.a124931. [DOI] [PubMed] [Google Scholar]
- Heun M, Abbo S, Lev-Yadun S, Gopher A. A critical review of the protracted domestication model for Near-Eastern founder crops: linear regression, long-distance gene flow, archaeological, and archaeobotanical evidence. J Exp Bot. 2012;63:4333–4341. doi: 10.1093/jxb/ers162. [DOI] [PubMed] [Google Scholar]
- Hur M, Campbell AA, Almeida-de-Macedo M, Li L, Ransom N, Jose A, Crispin M, Nikolau BJ, Wurtele ES. A global approach to analysis and interpretation of metabolic data for plant natural product discovery. Nat Prod Rep. 2013;30:565–583. doi: 10.1039/c3np20111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings/… International Conference on Intelligent Systems for Molecular Biology; ISMB. International Conference on Intelligent Systems for Molecular Biology; 1999. pp. 138–148. [PubMed] [Google Scholar]
- Jacobson M. The unsaturated isobutilamides. In: Jacobson M, Crosby DG, editors. Naturally occurring insecticides. New York: Marcel Dekker; 1971. pp. 137–176. [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences: CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kashiwada Y, Ito C, Katagiri H, Mase I, Komatsu K, Namba T, Ikeshiro Y. Amides of the fruit of Zanthoxylum spp. Phytochemistry. 1997;44:1125–1127. [Google Scholar]
- Kugler F, Graneis S, Schreiter PP, Stintzing FC, Carle R. Determination of free amino compounds in betalainic fruits and vegetables by gas chromatography with flame ionization and mass spectrometric detection. J Agric Food Chem. 2006;54:4311–4318. doi: 10.1021/jf060245g. [DOI] [PubMed] [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ. Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol. 2011;155:293–314. doi: 10.1104/pp.110.165910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazourek M, Pujar A, Borovsky Y, Paran I, Mueller L, Jahn MM. A dynamic interface for capsaicinoid systems biology. Plant Physiol. 2009;150:1806–1821. doi: 10.1104/pp.109.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLafferty FW. Mass Spectrometric Analysis. Molecular Rearrangements. Analytical Chemistry. 1959;31:82–87. [Google Scholar]
- McLafferty FW, Tureček Fe. Interpretation of mass spectra. 4. Mill Valley, Calif: University Science Books; 1993. [Google Scholar]
- Miyakado M, Nakayama I, Ohno N. Insecticidal unsaturated isobutylamides from natural products of agrochemical leads. In: Arnason JT, Philogene BJR, Morand P, editors. Insecticides of plant origin. Washington: American Chemical Society; 1989. pp. 173–205. [Google Scholar]
- Mudge E, Lopes-Lutz D, Brown P, Schieber A. Analysis of alkylamides in Echinacea plant materials and dietary supplements by ultrafast liquid chromatography with diode array and mass spectrometric detection. J Agric Food Chem. 2011;59:8086–8094. doi: 10.1021/jf201158k. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Wendel JF. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol. 2013;64:47–70. doi: 10.1146/annurev-arplant-050312-120048. [DOI] [PubMed] [Google Scholar]
- Parmar VS, Jain SC, Bisht KS, Jain R, Taneja P, Jha A, Tyagi OD, Prasad AK, Wengel J, Olsen CE, Boll PM. Phytochemistry of the genus Piper. Phytochemistry. 1997;46:597–673. [Google Scholar]
- Pichersky E, Lewinsohn E. Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol. 2011;62:549–566. doi: 10.1146/annurev-arplant-042110-103814. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quanbeck SM, Brachova L, Campbell AA, Guan X, Perera A, He K, Rhee SY, Bais P, Dickerson JA, Dixon P, Wohlgemuth G, Fiehn O, Barkan L, Lange I, Lange BM, Lee I, Cortes D, Salazar C, Shuman J, Shulaev V, Huhman DV, Sumner LW, Roth MR, Welti R, Ilarslan H, Wurtele ES, Nikolau BJ. Metabolomics as a Hypothesis-Generating Functional Genomics Tool for the Annotation of Arabidopsis thaliana Genes of “Unknown Function”. Front Plant Sci. 2012;3:15. doi: 10.3389/fpls.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Chavez E, Lopez-Bucio J, Herrera-Estrella L, Molina-Torres J. Alkamides isolated from plants promote growth and alter root development in Arabidopsis. Plant Physiol. 2004;134:1058–1068. doi: 10.1104/pp.103.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios MY. Natural Alkamides: Pharmacology, Chemistry and Distribution. In: Vallisuta O, editor. Drug Discovery Research in Pharmacognosy. 2012. [Google Scholar]
- Rontein D, Nishida I, Tashiro G, Yoshioka K, Wu WI, Voelker DR, Basset G, Hanson AD. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem. 2001;276:35523–35529. doi: 10.1074/jbc.M106038200. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sandmeier E, Hale TI, Christen P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur J Biochem. 1994;221:997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Pichersky E, Last RL. Taming the hydra of specialized metabolism: how systems biology and comparative approaches are revolutionizing plant biochemistry. Current Opinion in Plant Biology. 2012a;15:338–344. doi: 10.1016/j.pbi.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Pichersky E, Last RL. Taming the hydra of specialized metabolism: how systems biology and comparative approaches are revolutionizing plant biochemistry. Curr Opin Plant Biol. 2012b;15:338–344. doi: 10.1016/j.pbi.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis NE, Kliebenstein DJ. Natural variation of plant metabolism: genetic mechanisms, interpretive caveats, evolutionary and mechanistic insights. Plant Physiol. 2015 doi: 10.1104/pp.15.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson DE, Akhtar M, Gani D. L-methionine decarboxylase from Dryopteris filix-mas: purification, characterization, substrate specificity, abortive transamination of the coenzyme, and stereochemical courses of substrate decarboxylation and coenzyme transamination. Biochemistry. 1990;29:7631–7647. doi: 10.1021/bi00485a013. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaaler GL, Brasch MA, Snell EE. Pyridoxal 5′-phosphate-dependent histidine decarboxylase. Nucleotide sequence of the hdc gene and the corresponding amino acid sequence. J Biol Chem. 1986;261:11010–11014. [PubMed] [Google Scholar]
- Vaaler GL, Snell EE. Pyridoxal 5′-phosphate dependent histidine decarboxylase: overproduction, purification, biosynthesis of soluble site-directed mutant proteins, and replacement of conserved residues. Biochemistry. 1989;28:7306–7313. doi: 10.1021/bi00444a024. [DOI] [PubMed] [Google Scholar]
- Woelkart K, Xu W, Pei Y, Makriyannis A, Picone RP, Bauer R. The endocannabinoid system as a target for alkamides from Echinacea angustifolia roots. Planta Medica. 2005;71:701–705. doi: 10.1055/s-2005-871290. [DOI] [PubMed] [Google Scholar]
- Wu L, Bae J, Kraus G, Wurtele ES. Diacetylenic isobutylamides of Echinacea: synthesis and natural distribution. Phytochemistry. 2004;65:2477–2484. doi: 10.1016/j.phytochem.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Wu L, Dixon PM, Nikolau BJ, Kraus GA, Widrlechner MP, Wurtele ES. Metabolic profiling of echinacea genotypes and a test of alternative taxonomic treatments. Planta Med. 2009;75:178–183. doi: 10.1055/s-0028-1112199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele E, Chappell J, Jones A, Celiz M, Ransom N, Hur M, Rizshsky L, Crispin M, Dixon P, Liu J, Widrlechner PM, Nikolau B. Medicinal Plants: A Public Resource for Metabolomics and Hypothesis Development. Metabolites. 2012;2:1031–1059. doi: 10.3390/metabo2041031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo YS, Nybo SE, Chittiboyina AG, Weerasooriya AD, Wang YH, Gongora-Castillo E, Vaillancourt B, Buell CR, Dellapenna D, Celiz MD, Jones AD, Wurtele ES, Ransom N, Dudareva N, Shaaban KA, Tibrewal N, Chandra S, Smillie T, Khan IA, Coates RM, Watt DS, Chappell J. Functional Identification of Valerena-1,10-diene Synthase, a Terpene Synthase Catalyzing a Unique Chemical Cascade in the Biosynthesis of Biologically Active Sesquiterpenes in Valeriana officinalis. The Journal of biological chemistry. 2013;288:3163–3173. doi: 10.1074/jbc.M112.415836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolite levels in different organs and tissues of E. purpurea.
Expression abundances of the E. purpurea transcriptome in an atlas of 20 tissues and organs.
Correlations of E. purpurea transcripts with the accumulation of the alkamide, iC4N-12:4Δ2E,4E,8Z,10E.
Amino acid sequences of putative PLP-dependent enzymes encoded by Echinacea purpurea.
PLP-dependent enzyme sequences as phylogenetically classified in Figure 4.
Log ratio plot of the relative abundance of 33 different alkamides among 36 different organs and tissues
Hierarchical clustering analysis of the distribution of fatty acids, amino acids and alkamides among different E. purpurea organs and tissues.
Effect of incubating Echinacea seedlings with MeJA on the accumulation of alkamides.
Mass spectra of “possible new alkamides (PNA)”.
cDNA libraries and RNA-Seq data generated for E. purpurea.
Statistics for the E. purpurea reference transcriptome.
DNA-primers used in PCR reactions.