Abstract
Engineering cardiac tissues and organ models remains a great challenge due to the hierarchical structure of the native myocardium. The need of integrating blood vessels brings additional complexity, limiting the available approaches that are suitable to produce integrated cardiovascular organoids. In this work we propose a novel hybrid strategy based on 3D bioprinting, to fabricate endothelialized myocardium. Enabled by the use of our composite bioink, endothelial cells directly bioprinted within microfibrous hydrogel scaffolds gradually migrated towards the peripheries of the microfibers to form a layer of confluent endothelium. Together with controlled anisotropy, this 3D endothelial bed was then seeded with cardiomyocytes to generate aligned myocardium capable of spontaneous and synchronous contraction. We further embedded the organoids into a specially designed microfluidic perfusion bioreactor to complete the endothelialized-myocardium-on-a-chip platform for cardiovascular toxicity evaluation. Finally, we demonstrated that such a technique could be translated to human cardiomyocytes derived from induced pluripotent stem cells to construct endothelialized human myocardium. We believe that our method for generation of endothelialized organoids fabricated through an innovative 3D bioprinting technology may find widespread applications in regenerative medicine, drug screening, and potentially disease modeling.
Keywords: Bioprinting, Cardiac tissue engineering, Vascularization, Heart-on-a-chip, Cardiovascular toxicity
1. Introduction
Engineering cardiac tissues poses a series of critical challenges that need to be addressed in order to translate basic research products from bench to clinical practice [1–3]. The engineered cardiac organoids coupled with microfluidic bioreactors (e.g. heart-on-chips) have also found increasing applications in functioning as enabling in vitro biomimetic models to study pathology, measure cardiotoxicity, and develop new therapeutics [3–13]. The first challenge in engineering cardiac organoids and their on-chip forms lies in the fact that mature cardiomyocytes exhibit limited self-renewing potential [14]. In this framework, induced pluripotent stem cells (iPSCs) hold great promise, due to their wide availability and the possibility to differentiate into multiple cell lineages including cardiomyocytes [3,11,15,16]. Second, the alignment of cardiomyocytes and their organization into bundles characterized by spontaneous and synchronous contraction further complicate the development of biologically relevant cardiac tissues [1–3,17,18]. Third, the generation of thick (cardiac) tissue constructs requires the introduction of microvascular networks in order to provide oxygen and nutrients, remove waste products, and eventually promote vessel anastomosis with the host vasculature [3,19,20].
Several approaches have so far been explored to generate functional tissue constructs including the myocardium [21–24]. For example, scaffold-free multicellular cardiac spheroids have been developed that could spontaneously and synchronously contract [21,22]. While the cardiac spheroids have served an important role in drug testing and have been widely used due to the ease of preparation, these constructs lack the directionality characteristic of the physiological myocardium, which is critical to maintain the long-term functionality of the engineered cardiac tissues. On the other side, scaffold-based techniques provide an ideal support for cell adhesion, distribution, and responses [12,18,25–27]. Importantly, the architecture of the scaffolds can be conveniently modulated in order to promote the biological relevance of the engineered tissues by tuning spatial organizations that mimic their in vivo counterparts [25]. In this context, Freed and co-workers demonstrated that anisotropic scaffolds bearing an accordion-like honeycomb structure could induce the generation of highly oriented cardiac fibers [26]. Radisic and colleagues developed a biowire approach to induce the differentiation and alignment of the cardiomyocytes from human pluripotent stem cells [27]. Healy and co-workers recently engineered aligned cardiac tissues by populating microfilament arrays with cardiomyocytes [12]. Our group has also recently developed hydrogel substrates with aligned ridges/grooves via photopatterning to improve the adhesion and alignment of cardiomyocytes [18]. Strategies have further been investigated to integrate blood vessels into engineered tissues including the myocardium [28–31]. For example, Leong and colleagues have provided a general and versatile strategy by using transwell-mediated layering of endothelial cells and tissue cells for drug testing [30,31]. However, generating volumetric cardiac tissues containing embedded endothelial networks remains challenging.
Bioprinting has recently emerged as a promising technology to produce geometrically defined structures in three dimensions (3D), significantly improving their physiological relevance through architectural mimicry of native tissues and organs [32,33]. Particularly, bioprinting overcomes major drawbacks of conventional scaffold-based approaches including limited control over the 3D structures of engineered tissues and thus reduced reproducibility. The bioprinting process is usually biocompatible, allowing for direct encapsulation of bioactive molecules and cells. Furthermore, bioprinting may enable vascularization of the engineered tissue constructs based on sacrificial methods [34–36] or direct deposition [37,38], providing additional versatility in producing vascularized cardiac organoids.
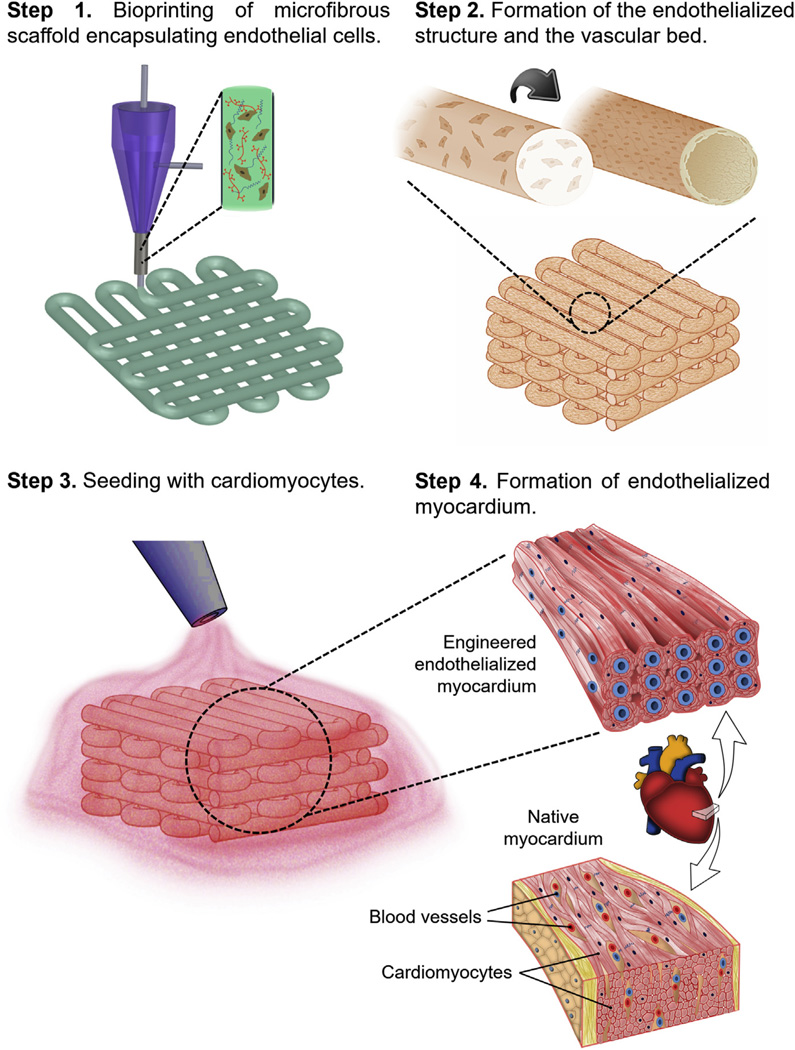
In this work we present a novel hybrid strategy based on 3D bioprinting, to engineer endothelialized myocardial tissues (Fig. 1). Based on the microfluidic technology that we developed in our previous work [37], we directly encapsulated endothelial cells within the bioprinted microfibrous lattices to inuce their migration towards the peripheries of the microfibers to form a layer of confluent endothelium. Different from our previous report, however, this 3D bioprinted endothelialized microfibrous scaffold, together with precisely controlled macroscale anisotropic architecture of the microfibers, was then seeded with cardiomyocytes to induce the formation of myocardium with improved alignment capable of spontaneous and synchronous contraction. When further combined with a specially designed microfluidic perfusion bioreactor, the resulting endothelialized-myocardium-on-a-chip platform was adopted to screen pharmaceutical compounds for their cardiovascular toxicity. Finally, we investigated the possibility to translate such a model to endothelialized human myocardium and their on-chip forms that were responsive to drugs using human iPSC (hiPSC)-derived cardiomyocytes.
Fig. 1.
Schematics showing the procedure of fabricating endothelialized myocardium using the 3D bioprinting strategy. Step 1: bioprinting of a microfibrous scaffold using a composite bioink encapsulating endothelial cells. Step 2: formation of the vascular bed through migration of HUVECs to the peripheries of the microfibers. Step 3: seeding of cardiomyocytes into the interstitial space of the endothelialized scaffold. Step 4: formation of engineered endothelialized myocardium structurally resembling the native myocardium.
2. Materials and methods
2.1. Cell culture
Human umbilical vein endothelial cells (HUVECs) and green fluorescent protein (GFP)-labeled HUVECs were obtained from Lonza and cultured in endothelial growth medium (EGM, Lonza). Neonatal rate cardiomyocytes were isolated from 2-day-old Sprague-Dawley rats following our established protocol approved by the Institutional Animal Care and Use Committee at the Brigham and Women's Hospital [39]. The cells were then maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10 vol% fetal bovine serum (FBS) and 1 vol% penicillin-streptomycin (P-S, all from ThermoFisher). The hiPSC-cardiomyocytes were purchased from Stem Cell Theranostics and maintained in RPMI-1640 medium containing 1× B27 supplement (ThermoFisher).
2.2. Bioprinting process
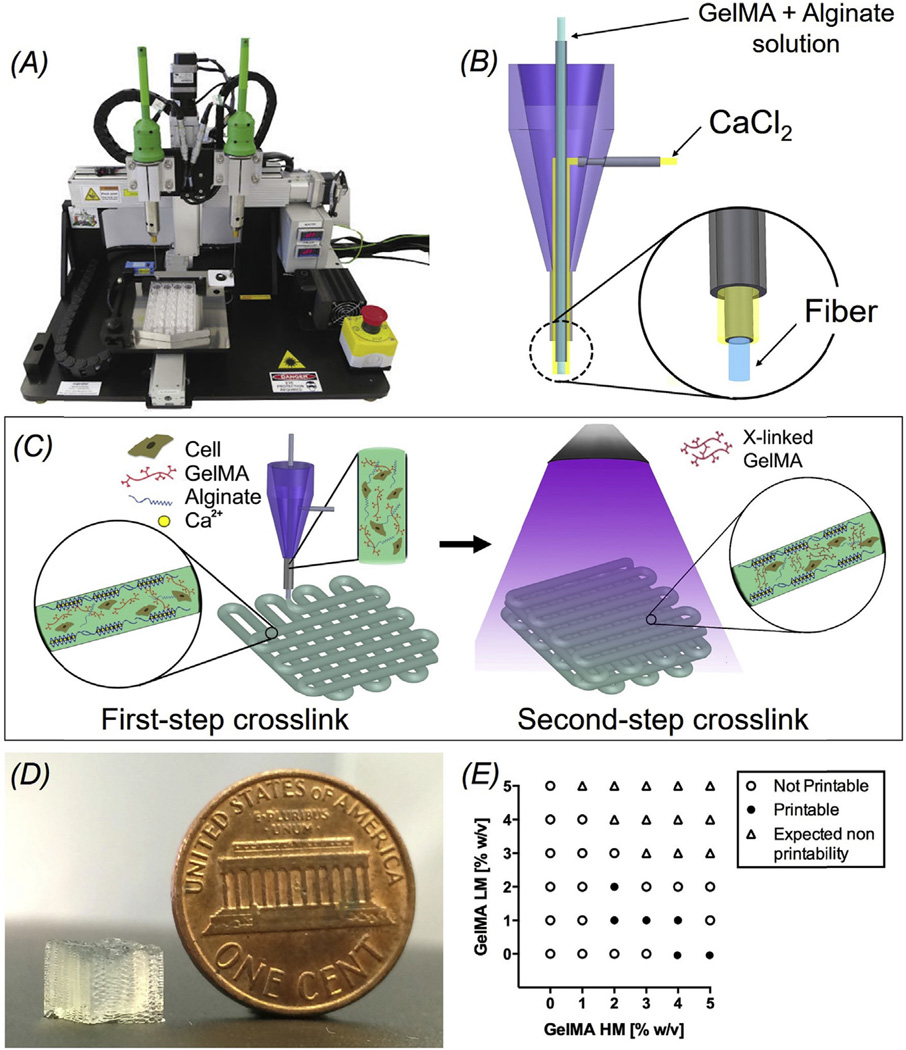
A commercial bioprinter (Organovo, NovoGen MMX) was used in combination with a custom-made coaxial nozzle assembled from syringe needles (Fig. 2B). An 18G needle (OD: 1270 µm; ID: 840 µm) was used as the sheath and a 27G needle (OD: 410 µm; ID: 210 µm) constituted the core. The needles were connected to a syringe pump for injection of the bioink and the CaCl2 solution through two poly(vinyl chloride) (PVC) tubes (Cole-Parmer). All the junctions were permanently fixed using epoxy glue. The extruder was mounted onto the pump head of the bioprinter by an in-house fabricated L-shaped plastic holder made of poly(methyl methacrylate) (PMMA).
Fig. 2.
(A) Photograph of an Organovo Novogen MMX bioprinter. (B) Schematic of the coaxial needle where the bioink is delivered from the core and the ionic crosslinking CaCl2 solution is sheathed on the side. (C) Schematic diagrams showing the two-step crosslinking process, where the alginate component is first physically crosslinked by the CaCl2 followed by chemical crosslinking of the GelMA component using UV illumination. (D) Photograph of a bioprinted cubic microfibrous scaffold (6-mm edge length). (E) Bioink optimization where conditions of printability and non-printability for different concentrations of GelMA-HM and GelMA-LM (with a constant alginate concentration of 4 w/v%) were analyzed.
The bioink used for the bioprinting consisted of a mixture of alginate (Sigma-Aldrich), gelatin methacryloyl (GelMA), and photoinitiator Irgacure 2959 (Ciba Specialty Chemicals) dissolved in 25 mM 2-[4–(2-hydroxyethyl) piperazin-1-yl] ethane sulfonic acid (HEPES buffer, pH 7.4, Sigma-Aldrich) containing 10 vol% FBS. The composition of the bioink enabled a dual-step crosslinking procedure. During the bioprinting process the ionic crosslinking of the alginate component of the bioink delivered through the core of the nozzle was first induced by exposing the extruded microfibers to a 0.3-M CaCl2 solution in HEPES buffer containing 10 vol% FBS, carried by the sheath. When the scaffold was printed, a stable gelation was then achieved by crosslinking GelMA via UV exposure. The sample was placed 7 cm away from the UV source (OmniCure; power: 800 mW) and crosslinked for 30 s [37]. In order to ensure continuous production of scaffolds avoiding clogging during the bioprinting, the composition of the bioink was optimized by maintaining the concentration of alginate constant at 4 w/v% while varying the relative concentrations of GelMA low degree of methacryloyl substitution (LM) and GelMA high degree of methacryloyl substitution (HM).
A MATLAB (Mathworks) code was written to automatically generate the G-code to control the bioprinter to deposit desired structures. Specifically, microfibrous scaffolds with a dimension of 5.5 × 3.5 × 0.75 mm3 were bioprinted through continuous deposition of one single continuous microfiber shaped in 3D for each scaffold. Printing was performed by using the same flow rate of 5 µL min−1 for both the bioink and the crosslinking solution and a deposition speed of 4 mm s−1.
2.3. Mechanical characterization of the bioprinted scaffolds
Compressive stress-strain measurements were performed to evaluate the elastic moduli of the scaffolds. The samples were bioprinted and allowed to swell for 6 h in phosphate buffered saline (PBS). The scaffolds were loaded onto an Instron 5943 equipped with a 10-N load cell. The compressions were carried out at a strain rate of 1 mm min−1 to 70% deformation, at room temperature. Elastic moduli of the scaffolds were derived from the regression of the first linear portion of the stress-strain curves (whereas the second linear portion relates to the property of the crosslinked bioink after compression of all the pores). Each measurement was performed in quintuplicate.
2.4. Bioprinting of HUVECs and seeding of cardiomyocytes
Both bioink and CaCl2 solutions were maintained at 37 °C before use. HUVECs were resuspended in the bioink at a concentration of 1 × 107 cells mL−1. This density of HUVECs has been previously optimized through our extensive preliminary experiments. Following the bioprinting and the subsequent crosslinking processes, the constructs was washed with PBS to remove the excess CaCl2. After washing, the constructs were cultured in EGM at 37 °C and 5 vol% CO2 throughout the entire period of up to 33 days. The medium was changed twice in the first day and then every 2 days afterwards.
Neonatal rat cardiomyocytes were seeded following the formation of a layer of confluent endothelium at the peripheries of the microfibers of the scaffolds, which generally occurred in 15 days. Freshly isolated cardiomyocytes were suspended in DMEM at a final concentration of 1 × 106 cells mL−1. The scaffolds were individually placed on top of a thin layer of hydrophobic PDMS. Approximately 40 µL of cell suspension was dropped to cover each scaffold, and incubated at 37 °C for 2 h to allow the cardiomyocytes to adhere onto the microfibers. The scaffolds were then gently washed and cultured in DMEM supplemented with 10 vol% FBS. The medium was changed every day in the first 2–3 days until the cardiomyocytes started beating, after which only ½ medium was exchanged every 2–3 days.
The seeding of hiPSC-cardiomyocytes followed the same procedure except for that a RPMI-1640 medium containing 1× B27 supplements was used for culturing.
2.5. Bioreactor design and fabrication
A microfluidic bioreactor was designed, optimized, and fabricated in order to construct the endothelialized-heart-on-a-chip device and study drug effects [40]. The designed bioreactor consisted of two hemi-chambers, composed by a pair of rigid supports made of PMMA and two complementary micro-featured gaskets made of PDMS. Both PMMA layers were rectangular (5 × 3.5 × 0.3 cm3), each one containing four clearance holes. These holes allowed for the mechanical compression of the PDMS layers (3 × 1.8 × 0.3 cm3) sandwiched in between through the use of four sets of screws and nuts to guarantee hydraulic tightness. The two micro-featured PDMS layers together formed the bioreactor chamber with a resulting thickness of 1 mm (which reduced to approximately 0.85 mm upon compression). The main chamber of the bioreactor was squared (7 × 7mm2), which featured four pillars that fixed the scaffold in place, avoiding its potential movement under flow. The width of the inlet channel of each bioreactor was slightly smaller than that of the outlet (0.65 mm and 1.3 mm, respectively), to reduce the chance of bubbles formation and retention during the perfusion. The inlet and outlet of each bioreactor were connected with Teflon microtubes (#30 AWG thin-wall tubing, Cole-Palmer), joined with a segment of silicone tubing to ensure sufficient oxygenation of the medium during culture. The silicone tubing was then fitted onto a peristaltic pump (MP2-4-PC Micro Peristaltic Pump, Elemental Scientific) to allow perfusion. A 5-mL reservoir was connected between the silicone tube and the inlet of the bioreactor to provide nutrients as well as to entrap bubbles. Epoxy was used to seal any possible source of leakage.
2.6. Computational simulations
Comsol Multiphysics (Version 4.3b) was used to simulate the flow rates and oxygen distribution within the chamber of the bioreactor. Since the bioreactor was sandwiched by two pieces of gas-impermeable PMMA frames, we considered the upper/lower boundaries as oxygen-impermeable (zero mass flux) while lateral boundaries permeable due to the exposure of PDMS [41]. The tissue constructs based on bioprinted fibrous scaffolds were modeled as a hydrogel with uniform volumetric oxygen consumption rate associated with the total number of cells. We assumed the oxygen consumption rates at around 1.18 × 10−4 mol s−1 cell−1 for HUVECs and 1.14 × 10−5 mol s−1 cell−1 for cardiomyocytes, respectively [42,43]. The numbers of the HUVECs and the cardiomyocytes were estimated to be 7.70 × 104 (considering the total volume of the microfibrous scaffold of approximately 7.7 µL at a cell density of 1 × 107 cells mL−1) and 4.00 × 105 per scaffold, respectively, according to the bioprinting/seeding conditions. The oxygen concentration at the inlet carried by the infusing medium was considered constant, uniform, and equal to the atmospheric concentration. The diffusion coefficient of oxygen in the culture medium and the hydrogel were approximated at 3.80 × 10−9 m2 s−1 and 2.30 × 10−9 m2 s−1, respectively [40,44]. A Michaelis-Menten kinetics was considered, assuming Michaelis constants of 0.55 × 10−3 mol m−3 and 6.88 × 10−3 mol m−3 for HUVECs and cardiomyocytes, respectively [42,43]. A flow rate of 50 µL min−1 was adopted after scaling down the flow rate of the blood in the heart according to the weights of the cardiac tissues [45]. A symmetry condition along the long axis of the bioreactor chamber was considered and a numerical grid consisting of about 3 × 107 tetrahedral elements was applied.
2.7. Immunofluorescence staining
Samples were fixed for 1 h at room temperature using 4 vol% paraformaldehyde (Sigma-Aldrich) in PBS. Cells were permeabilized by soaking the samples in 0.1 vol% Triton X-100 (Sigma-Aldrich) dissolved in PBS for 30 min while non-specific binding was inhibited using 10 vol% bovine serum albumin (BSA, Sigma-Aldrich) for 1 h at room temperature. Samples were then incubated for overnight at 4 °C in a solution containing primary antibodies at 1:200 dilution in 10 vol% BSA and 0.1 vol% Triton X-100 in PBS. In particular, rabbit polyclonal anti-CD31 (ab32457, Abcam), mouse monoclonal anti-sarcomeric α-actinin (ab9465, Abcam), and rabbit polyclonal anti-connecxin-43 (ab11370, Abcam) antibodies were used. Secondary antibodies were used at 1:200 dilution. For F-actin staining, samples were incubated for 30 min at room temperature in a solution of Alexa 488-phalloidin (A12379, ThermoFisher) at 1:40 dilution in 10 vol% BSA and 0.1 vol% Triton X-100 in PBS. Nuclei of the cells were stained by 4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher). Images were taken using a fluorescence optical microscope (Axio Observer D1, Zeiss) or a confocal fluorescence microscope (SP5 X MP, Leica).
2.8. Characterization of tissue constructs
Live/Dead staining was performed according to the manufacturer's instructions (ThermoFisher). Beating of the cardiomyocytes was observed using an optical microscope and analyzed using custom-coded MATLAB programs [46]. Monitoring of the beating behavior was performed every day until contractions were no longer observed. It should be noted that due to the 3D nature of the constructs and the vibration of the media during the video recording, the beating plots obtained are expected to only accurately reflect the frequencies of the beating but not other functions. The levels of secreted von Willebrand factor (vWF) were measured by an ELISA kit (ab189571, Abcam).
2.9. Statistics
When two groups were compared, statistical analyses were conducted using unpaired t-tests. When more than two groups were compared, ANOVA followed by a post-hoc test was performed. Statistical significance was determined at p < 0.05. A sample size of at least 3–5 scaffolds per group was used.
3. Results and discussions
3.1. Bioprinting 3D microfibrous scaffolds
Our novel 3D bioprinting approach allowed us to conveniently generate multilayer hydrogel microfibrous scaffolds using an Organovo Novogen MMX bioprinter (Fig. 2A), which was optimized through the implementation of a custom-designed coaxial nozzle for continuous extrusion of the bioink [37]. The internal needle, having a size of 210 µm (27G), was fed with the bioink composed of a mixture of hydrogel precursors, i.e. alginate, GelMA, and photoinitiator Irgacure 2959; the crosslinking solution, i.e. CaCl2, was simultaneously dispensed through the outer sheath flow using a needle with size of 840 µm (18G; Fig. 2B). This specially designed bioink was developed by modifying our recently developed protocol [37], featuring a sequential crosslinking mechanism that allows for stable production of 3D microfibrous scaffolds. As indicated in Fig. 2C, when the two fluids come into contact at the tip of the printhead, a temporary ionic gelation of the alginate component in the bioink occurs. This rapid gelation [47,48], leads to the formation of the microfibers and their deposition in the 3D space as programmed. More importantly, the constant wetting of the deposited microfibers by the co-extruded CaCl2 solution further induces physical crosslinking of the microfibers between adjacent layers to stabilize the structure. In addition, the CaCl2 solution continuously provided through the external sheath efficiently avoids scaffold dehydration during the bioprinting process. Subsequently, permanent chemical gelation of the microfibers is achieved by exposing the scaffold to light to photocrosslink the GelMA component of the bioink. It is noteworthy to highlight that the temporary ionic crosslinking of the alginate is critical in ensuring structural integrity of the bioprinted scaffolds, thus allowing for the generation of self-sustaining multilayered structures in a highly reproducible manner prior to chemical crosslinking (Fig. 2D). The alginate component of the bioink, may eventually be washed off from the printed scaffolds following GelMA crosslinking using a solution containing Ca2+-chelating agent such as ethylenediaminetetraacetic acid (EDTA), to promote cell adhesion and spreading [37].
The bioink composition was then optimized to obtain a viscosity level compatible with a constant extrusion flow, avoiding clogging of the bioink inside the nozzle while maintaining structural integrity of the bioprinted microfibrous tissue constructs. Multiple bioink compositions were assessed for their printability. In particular, the concentrations of the alginate and photoinitiator were maintained constant at 4 w/v% and 0.2 w/v%, respectively [37], while the relative concentrations of GelMA-HM and GelMA-LM were both varied in the range of 0–5 w/v% (Fig. 2E). The optimal composition of the bioink was found to include 3.5 w/v% GelMA-HM and 1 w/v% GelMA-LM, in addition to 4 w/v% alginate and 0.2 w/v% Irgacure 2959, and was used throughout the following experiments. A flow rate of 5 µL min−1 was adopted for both the bioink and the sheath crosslinking solution, while the extruder speed was set at 4 mm s−1. The diameter of the resulting microfibers after complete swelling reached approximately 150 µm, in comparison to a size of 120 µm immediately after bioprinting.
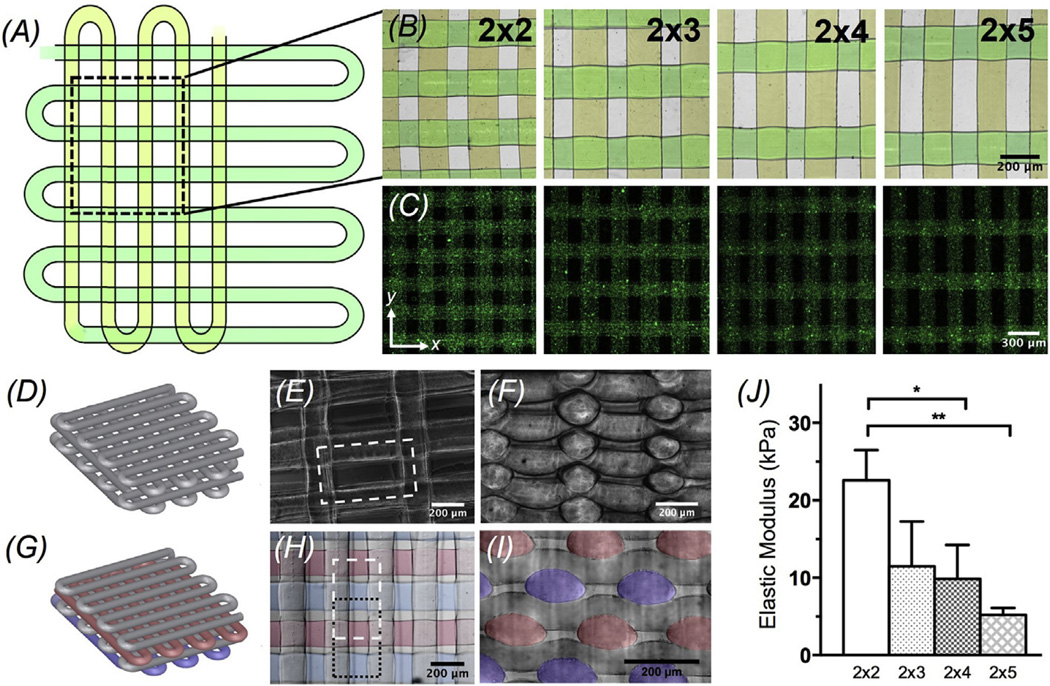
Most biological tissues in the body are anisotropic, particularly in the case of the myocardium where cardiomyocytes are uni-directionally aligned at cellular levels throughout the thickness of the tissue [1–3,49]. Therefore, we next demonstrated the capability of our technique to bioprint 3D microfibrous scaffolds with anisotropic arrangements. Rectangular scaffolds with an aspect ratio of the unit grid of 2 × 2 were used as the isotropic control, while such aspect ratio could be enlarged to 2 × 3, 2 × 4, and 2 × 5 to produce scaffolds with gradually increasing overall anisotropy at macroscale (Fig. 3A–C). Anisotropic scaffolds were fabricated by varying the distance between the fibers deposited in the y direction while keeping the distance constant in the x direction at 220 µm. The distance between the central axes of the adjacent microfibers deposited in the y direction was increased stepwise from 220 µm, i.e. the isotropic control with 2 × 2 aspect ratio of the unit grid, to 330 µm, 440 µm, and 550 µm, for anisotropic scaffolds with unit grids containing aspect ratios of 2 × 3, 2 × 4, and 2 × 5, respectively (Fig. 3A–B). Optical (Fig. 3B; the microfibers were pseudocolored in two different hues to mark the microfibers in the two perpendicular directions) and fluorescence (Fig. 3C; green fluorescent microbeads were added to the bioink prior to printing) micrographs of the bioprinted scaffolds demonstrated that the distance between the microfibers was in agreement with our predictions. The resulting spacing between the microfibers in the x direction was 226.3 ± 5.3 µm, whereas the spacing between those in the y direction became 229.3 ± 8.8 µm, 326.0 ± 7.7 µm, 451.0 ± 12.5 µm, and 563.0 ± 9.1 µm in the four types of scaffolds with increasing anisotropy, respectively.
Fig. 3.
(A) Top view single-layer schematic of the design of the bioprinted microfibrous scaffold and corresponding (B) brightfield (pseudocolored to match the schematic) and (C) fluorescence micrographs showing the bioprinted scaffolds with different aspect ratios of unit grids. (D) 3D schematic of a scaffold without offset and corresponding (E) top-view and (F) cross-sectional micrographs. (G) 3D schematic of a scaffold with offset and corresponding (H) top-view and (I) cross-sectional micrographs showing the offset between the layers. (J) Elastic moduli of the bioprinted scaffolds with different aspect ratios of unit grids.
It was found that however, in the bioprinted multi-layer scaffold the junction sites where the interlacing microfibers laid over each other slightly collapsed due to the compression incurred by the weight of the partially crosslinked bioink (Fig. 3D–F; 2 × 5). Such compression inevitably reduced the space between the microfibers in alternating layers (Fig. 3F), which might limit the seeding efficiency of the cardiomyocytes at a later stage. In order to maximize the surface area of the microfibrous scaffolds exposed to cardiomyocytes during the seeding process for their attachment, an offset between the alternating layers of the microfibers was further incorporated into the design of the bioprinting process. In this case, the overall layer of microfibers along the long axis of the unit grids was shifted by a distance of ½ unit grid, resulting in significantly increased surface-to-volume ratio of the microfibers without influencing the overall porosity of the scaffolds (Fig. 3G–I; 2 × 5). The microfibers in both the schematics and optical micrographs were labeled in red and blue in alternating layers to clearly demarcate the shift. The microfibers along the short axis of the unit grids were designed to remain in the same positions to support the three-dimensionality of the bioprinted scaffolds (Fig. 3G–I).
Mechanical properties of scaffolds with different aspect ratios of the unit grid were further characterized (Fig. 3J). Our results indicated that the elastic modulus of the bioprinted scaffolds was dependent on the distance between the adjacent microfibers caused by the difference in the aspect ratio of the unit grid, which is linked to the porosity of the scaffold. Theoretical porosities of the four types of bioprinted scaffolds were calculated to be 37.2%, 43.2%, 46.9%, and 49.4% for those with aspect ratios of unit grids of 2 × 2, 2 × 3, 2 × 4, and 2 × 5, respectively. The measured elastic modulus of the 2 × 2 scaffolds (22.6 ± 3.9 kPa) was higher than those of both scaffolds with aspect ratios of 2 × 4 (9.9 ± 4.4 kPa; p < 0.05) and 2 × 5 (5.2 ± 0.9 kPa; p < 0.01) for the unit grids, whereas the modulus of the 2 × 3 scaffolds was (11.5 ± 5.8 kPa). Our results are in good agreement with previously reported data, which showed compressive moduli on the order of 20–30 kPa for isotropic microfibrous scaffolds of similar structure and material [37].
3.2. Construction of endothelialized myocardium
Vascularization presents one of the most critical steps during the development of many functional tissue and organ systems since mature networks of blood vessels enable the transport of nutrient, oxygen, and wastes to/from the tissues [19,20,50,51]. This is particularly true for highly metabolically active organs including the heart [2,3]. A plethora of strategies have been recently developed to promote the vascularization of tissue constructs. Conventional approaches of vasculogenesis and angiogenesis relying on self-organization of endothelial cells into interconnected capillary structures are limited by their variability and efficiency [20,52]. Recently, bioprinting has emerged as a highly reproducible and versatile strategy to deposit sacrificial microfibers within hydrogel matrices; following template dissolution or removal, the hollow microchannel network could then be endothelialized to generate perfusable microvessels [34–36,53,54]. Here we further propose a hybrid technology combining guided self-assembly and 3D bioprinting to develop endothelialized tissue constructs by encapsulating endothelial cells within the GelMA-alginate bioink to fabricate scaffolds possessing a biomimetic anisotropic pattern [37]. These multilayer scaffolds could be subsequently cellularized by cardiomyocytes to generate the endothelialized myocardial constructs.
3.2.1. Endothelialization of the bioprinted microfibrous scaffolds
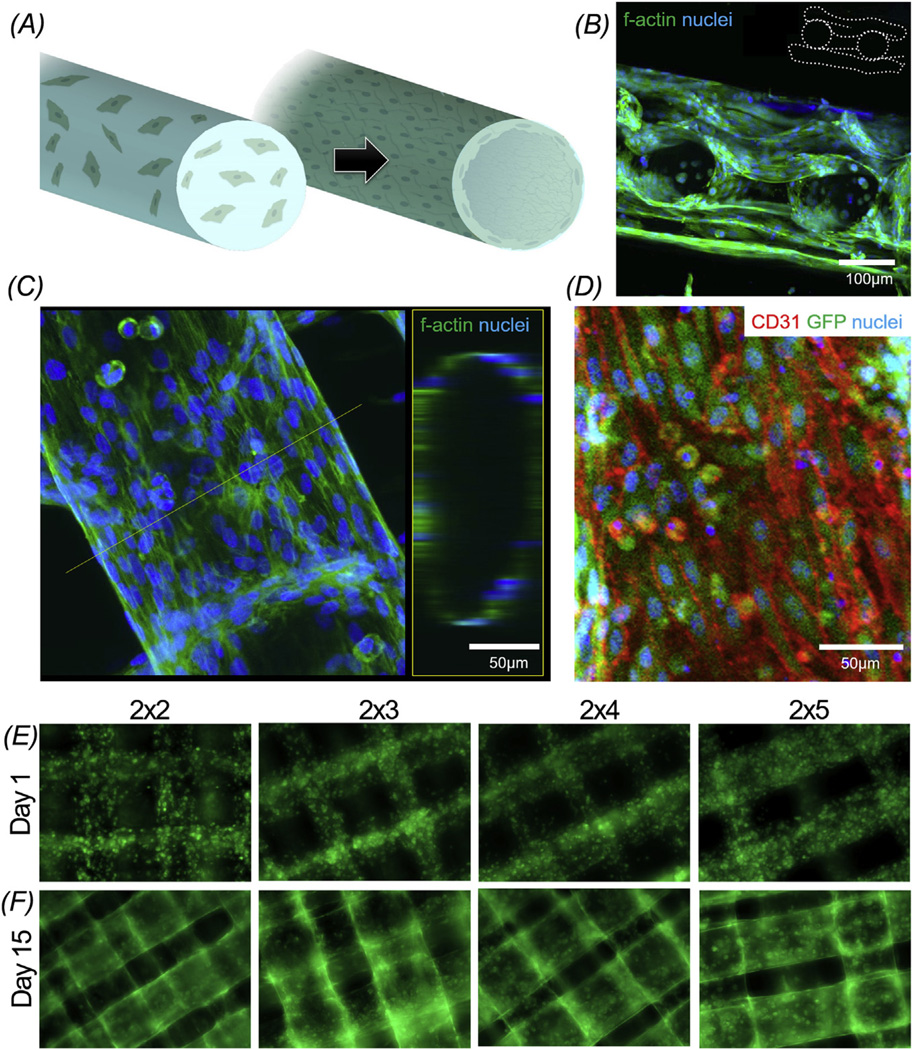
The bioink that we have designed possessed strong biocompatibility that readily allowed the embedment of cells during the bioprinting process. Interestingly, it was found that following the bioprinting of the scaffolds, the HUVECs, initially homogeneously dispersed within the microfibers, could gradually organize into a layer of confluent endothelium surrounding the microfibers in approximately 2 weeks of culture, potentially through migration towards the peripheries (Fig. 4A). Confocal image of the cross-section of a three-layer scaffold at Day 15 clearly revealed that the HUVECs concentrated at the borders of the microfibers, forming a pattern resembling the blood vessel walls (Fig. 4B). The magnified high-resolution confocal projection and reconstruction images of a single microfiber shown in Fig. 4C further confirmed the confluency of the HUVECs at the entire periphery of the microfiber at Day 15 of culture, resulting in 83.1± 5.4% hydrogel area occupation. In addition, the endothelial cells expressed surface marker CD31, forming tight junctions among adjacent cells in the confluent layer (Fig. 4D).
Fig. 4.
(A) Schematic representation showing the assembly of the encapsulated HUVECs inside the bioprinted microfibers into a confluent layer of endothelium. (B) Confocal fluorescence micrograph showing the cross-sectional view of a three-layer scaffold at Day 14, indicating the formation of the endothelium by the HUVECs. (C) High-resolution confocal fluorescence micrograph showing the distribution of the HUVECs in a single microfiber at Day 14. Left: projection view; Right: 3D rendering of the tubular structure at the position of the dotted line. (D) Confocal fluorescence micrograph showing the GFP-HUVECs in a single fiber for CD31, GFP, and nuclei. (E, F) Fluorescence micrographs showing the distribution and spreading of GFP-HUVECs in the bioprinted microfibrous scaffolds with different aspect ratios of unit grids at Day 1 and Day 15, respectively.
The chronological evolution of the formation of the endothelium by GFP-HUVECs inside the bioprinted microfibrous scaffolds is shown in Fig. 4E and F as well as Figure S1. The HUVECs were homogeneously distributed inside the microfibers at Day 1 post bioprinting (Fig. 4E); the HUVECs gradually migrated towards the peripheries of the microfibers at Day 9 (Fig. S1A), potentially driven by the intrinsic polarization tendency of these cells to stay at the liquid-matrix interfaces as well as the higher availability of nutrients and oxygen surrounding the microfibers [55]. We further discovered that the ionically crosslinked alginate component could dissolve in the culture medium and leach out from the bioprinted microfibers in approximately 5–10 days [37]. Since the microfibers were better ionically crosslinked for the alginate component at the peripheries during the bioprinting process, the release of alginate resulted in the formation of larger pores along the borders (Fig. S2; 7.33 ± 3.16 µm after alginate leach versus 2.44 ± 1.24 µm before alginate release), which further promoted the spreading and proliferation of the HUVECs, eventually forming well-patterned endothelium in approximately 15 days of culture (Fig. 4F). It was found that the density of the initially encapsulated HUVECs did not play a significant role in such a migration process, with migration observed across a wide density range of 1–15 × 106 cells mL−1. However, the density did impact the formation of the endothelialized structure as overly small cell densities could not result in the formation of intact endothelium while excessive cells led to aggregation within the microfibers. Therefore, a mid-range encapsulation density of the HUVECs of 10 × 106 cells mL−1 was used in this study.
At the shared borders between the overlapping microfibers, the HUVECs appeared to re-organize, defining an interconnected region between those of different layers (Fig. 4F and Fig. S1B). Importantly, the aspect ratios of the unit grid of the bioprinted microfibrous scaffolds did not remarkably alter their capability to endothelialize. However, it was found that the morphologies of the HUVECs did not significantly change during the rest of the culture for up to 33 days observed; in contrast, they gradually broke the boundaries of the microfibers and migrated out to the bottom of the microwells (Fig. S1B and C), likely due to the degradation of the GelMA and thus impaired integrity of the microfibrous structures [56,57]. As a consequence, the bioprinted microfibrous scaffolds embedded with HUVECs were maintained in culture for 15 days throughout the subsequent experiments. These findings suggested that our bioprinting technique using a bioink specially designed to possess a dual-step crosslinking mechanism allowed the formation of 3D endothelialized networks of any desired shape and architecture. The crossing microfibers in the bioprinted scaffolds possessing macroscale anisotropy should not affect final functionality of the endothelialized myocardium, since the microvascular network within the native contractile myocardium is not strictly aligned with the direction of the cardiomyocytes [58]. It should be further noted that, although the endothelialized microfibers were not hollow during the period analyzed, our bioprinted microfibrous network could provide excellent guidance for endothelialization in the entire volume. We expect that, when these endothelialized scaffolds are further embedded within tissue constructs, the degradation of the hydrogel in the interior may eventually open up the channels and form hollow lumens that will enhance the functionality of the spatially defined vascular network. However, the proof of such hypothesis needs further experimental validation and will be reported in our future work.
3.2.2. Construction of the myocardium
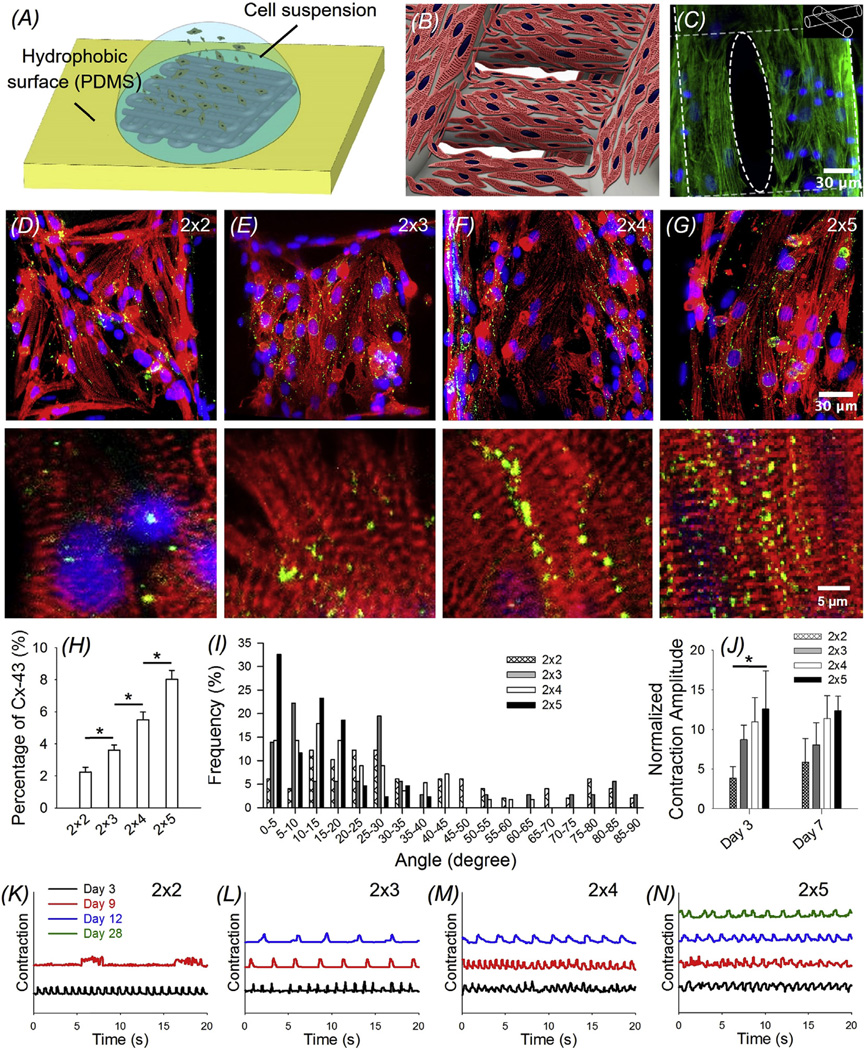
We next explored the possibility of employing the bioprinted microfibrous scaffolds as substrates for the construction of cardiac tissues. Neonatal rat cardiomyocytes were used as the model cells due to their abundant availability. The cells were seeded into bioprinted microfibrous scaffolds lodged on top of hydrophobic PDMS surfaces (Fig. 5A). Such a hydrophobic platform was adopted to ensure that cell suspensions for seeding could be directly laid over the scaffolds without spreading, thus leading to the attachment of a high density of the cardiomyocytes on the microfibers of the scaffolds (Fig. 5B). In particular, scaffolds with a dimension of 3.5 × 5.5 mm2 and 5 layers of interlacing microfibers were used to sufficiently recapitulate the three-dimensionality of the myocardium while minimizing the amount of cells required for the experiments. In order to simulate the conditions of endothelialized scaffolds, in these experiments where no HUVECs were encapsulated in the microfibers, the scaffolds were still incubated for 15 days in the medium prior to seeding for behavioral analysis of cardiomyocyte monoculture. Immediately post seeding, the density of the adhered cardiomyocytes was measured and no significant differences were observed among the scaffolds with different aspect ratios of the unit grid (2 × 2: 1883 ± 415 cells mm−2; 2 × 3: 1896 ± 651 cells mm−2; 2 × 4: 2020 ± 147 cells mm−2; 2 × 5: 1773 ± 335 cells mm−2). Myocardial constructs were then cultured for 3 days to allow for the maturation of the cardiomyocytes. The cardiomyocytes uniformly adhered onto the surface of the microfibers in the scaffolds, leaving space at the junction points formed between two crossing microfibers in adjacent layers (Fig. 5C). The cardiomyocytes adhered and spread on the surface of the microfibers across the entire thickness of the scaffolds due to a combinatory effect of gravity, diffusion, and capillary force during the seeding process.
Fig. 5.
(A) Schematic showing the seeding procedure of cardiomyocytes onto the bioprinted microfibrous scaffolds. (B) Schematic showing a scaffold seeded with neonatal rat cardiomyocytes. (C) F-actin (green) staining showing the distribution of the cardiomyocytes on the surface of the scaffold at the location where two microfibers of adjacent layers crossed. (D–G) Immunofluorescence staining of sarcomeric α-actinin (red) and connexin-43 (Cx-43, green) of cardiomyocytes seeded on bioprinted microfibrous scaffolds with different aspect ratios of unit grids, showing varying degrees of alignment of the cardiomyocytes. Lower panels: magnified images showing the sarcomeric banding. (H) Quantification of Cx-43 expression by the cardiomyocytes on the four types of scaffolds, plotted as percentages of area coverage calculated from the fluorescence images; *p < 0.005. (I) Quantification of the angle distribution of cardiomyocytes on the four types of bioprinted microfibrous scaffolds. (J) Quantification of the contraction amplitude of the four types of bioprinted myocardial constructs. *p < 0.05. (K–N) Beating analysis of the cardiac organoid on bioprinted scaffolds with different aspect ratios of unit grids. Note that the contraction amplitudes in (K–N) were all normalized to the same height for easy comparison across the samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Cardiomyocytes grown on the bioprinted microfibrous scaffolds strongly expressed proteins necessary for proper contractile function, i.e. sarcomeric α-actinin, and inter-cellular conductive function, i.e. connexin-43, as demonstrated by immunostaining (Fig. 5D–G). Indeed, the presence of organized sarcomeric banding and the formation of a large number of gap junctions provided evidence of the maturation of the cardiomyocytes and represented a critical basis for generating synchronous beating of the cardiac constructs [2,59–61]. In addition, connexin-43 expression was higher for cardiomyocytes on samples with macroscale anisotropy (2 × 3: 3.59± 0.32%; 2 × 4: 5.49± 0.48%; and 2 × 5: 8.02± 0.54%; area coverage) when compared to the isotropic controls (2 × 2: 2.23± 0.30%) (Fig. 5D–H). The alignment of the cardiomyocytes on bioprinted scaffolds with different aspect ratios of unit grids was further analyzed by quantifying the angles between individual cells and the long axis of the grids. An angle equal to 0° referred to cells that were perfectly aligned in the direction of long axis of the unit grids, while an angle of 90° indicated perpendicularly alignment of the cells, i.e. along the short axis of the unit grids. It was revealed that the cardiomyocytes aligned increasingly better even at the cellular level in the direction of the microfibers along the long axis as the macroscopic anisotropy of the bioprinted scaffolds was increased (Fig. 5D–G, I). As expected, the isotropic controls presented an almost uniform distribution of the orientation of the cardiomyocytes. On the contrary, scaffolds with the highest macroscale anisotropy (2 × 5) were characterized by cardiomyocytes with alignment angles all lower than 40°, and most angles fell within the range of 20°. This observation is in agreement with previous studies, where anisotropic scaffolds induced much higher alignment of cardiomyocytes than anisotropic controls [18,26].
The spontaneous beating of the cardiac tissue constructs started after 48 h of culture for scaffolds with aspect ratios of unit grids of 2 × 2 and 2 × 3, and after 72 h for 2 × 4 and 2 × 5 samples. The cardiomyocytes-populated scaffolds were beating synchronously and slight transient shrinkage in the length of the microfibers during the contractions could be observed (Mov. S1). We compared the contraction amplitudes of the constructs as a function of the aspect ratios of unit grids of the scaffolds. In particular, the distances between the nuclei of adjacent cells during contraction (D) and relaxation (D0) were measured. To compare the results across different samples, the difference between the two distances were further normalized against that during relaxation (i.e. (D0−D)/D0). The resulting value, denoted as the contraction amplitude, may partially reflect the contraction force generated by the cardiomyocytes [62], and is also associated with the intrinsic physical properties of the scaffolds. As shown in Fig. 5J, the contraction amplitude increased with the macroscale anisotropy of the scaffolds at both Day 3 (2 × 2: 3.82± 1.50%; 2 × 3: 8.70± 1.81%; 2 × 4: 10.98± 3.06%; and 2 × 5: 12.62± 4.79%) and Day 7 (2 × 2: 5.90± 3.00%; 2 × 3: 8.08± 2.77%; 2 × 4: 11.39± 2.89%; and 2 × 5: 12.39± 1.81%).
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.biomaterials.2016.09.003.
Beating frequencies overtime for scaffolds with different aspect ratios of unit grids (and thus the alignment and maturation of the cardiomyocytes) were subsequently quantified. Representative contraction plots of the cardiac tissue constructs at Days 3, 9, 12, and 28 are illustrated in Fig. 5K–N. In all four types of samples the beating of the cardiomyocytes was robust during the first few days, ranging from 55 to 75 beats per min (bpm) depending on the scaffolds, while the contraction of each individual construct was uniform. However, the beating of the cardiomyocytes on isotropic scaffolds (2 × 2) significantly slowed down with possible arrhythmia after 9 days (Fig. 5K). On the contrary, cardiomyocytes seeded on 2 × 3 and 2 × 4 scaffolds continued beating until Days 15–22, although the frequencies slightly decreased over the period (Fig. 5L–N). The longest contraction was observed for 2 × 5 scaffolds (3 out of 4 samples) for up to 28 days, while the beating frequency was still maintained at approximately 40 bpm. This result was in good agreement with cardiomyocyte alignment, connexin-43 expression, and contraction amplitude analyses. We hypothesize that the improvement in the beating of the cardiac constructs with the increase in the macroscale anisotropy of the scaffold might be related with the alignment of the cardiomyocytes at the cellular level. Indeed, in 2 × 2 and 2 × 3 scaffolds the cardiomyocytes showed insufficient alignment at cellular level, possibly leading to an early but immature phenotype with limited beating capacity. In contrast, for scaffolds with 2 × 4 and 2 × 5 aspect ratios of unit grids, the cardiomyocytes were able to fully align even at the cellular level in the direction along the long axis, so that delayed but more complete maturation and prolonged beating could be achieved [18]. In addition, the difference in the beating behaviors might be partially attributed to the slightly decreased elastic modulus of the scaffolds as the aspect ratio of the unit grids was increased (Fig. 3J). It is believed that, since the densities of the cardiomyocytes on each type of scaffolds did not differ significantly, the decreasing total amounts of cells on scaffolds with increased macroscale anisotropy should have not affected the beating behaviors of the scaffolds.
Overall, we demonstrated that by tuning the macroscale anisotropy of the bioprinted microfibrous scaffold we were able to obtain engineered cardiac organoids characterized by improved alignment at the cellular level, mimicking the bundled structure of the myocardium in vivo. It should be noted that, the presence of crossing microfibers partially cancelled the macroscopic anisotropy of the overall structure. This effect nevertheless, could be minimized in bioprinted scaffolds with higher macroscale anisotropy (e.g., for those with a 2 × 5 ratio of the unit grids the microfibers aligning in the direction of the long axis were 2.5 times more than the perpendicular ones, thus still maintaining a relatively high degree of anisotropy), which further enhanced the alignment of the cardiomyocytes at the cellular level. Since scaffolds with an aspect ratio of unit grids of 2 × 5 presented better results in terms of the maturation, alignment, and contraction of the cardiomyocytes, they were chosen for all subsequent experiments involving heart-on-a-chip and cardiotoxicity studies.
3.3. Microfluidic microbioreactor for on-chip integration of the bioprinted tissue constructs
3.3.1. Bioreactor design and assembly
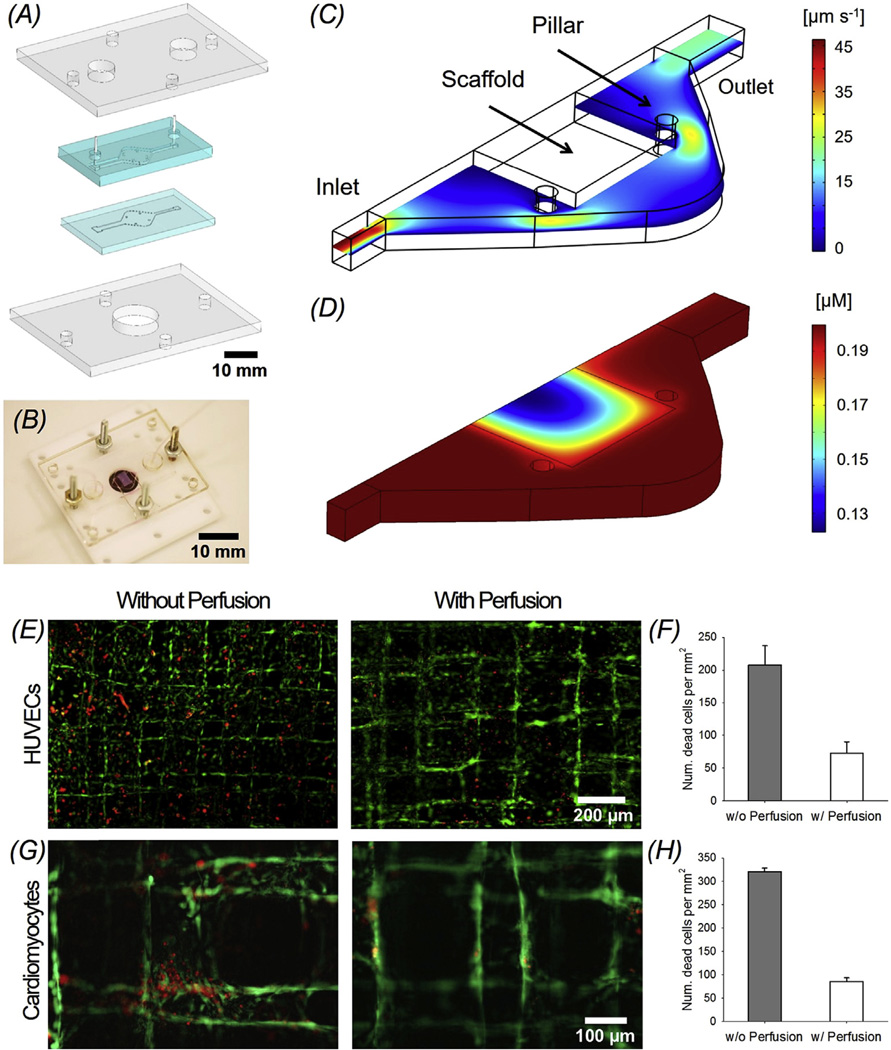
We developed an innovative resealable microbioreactor for perfusion culture to support the long-term viability of the endothelialized myocardium and in situ observation of the bioprinted endothelialized myocardial constructs, by modifying our recently published protocols [40]. As illustrated in Fig. 6A, the bioreactor was designed to possess two hemi-chambers embedded in a pair of PDMS gaskets, which were sandwiched between two rigid supports made of PMMA to ensure hydraulic tightness. The pair of micro-featured PDMS layers together formed the bioreactor chamber at a thickness of approximately 1 mm when closed together (which further reduced to approximately 0.85 mm upon compression), connected with the inlet and outlet channels on the two sides. The main chamber was squared (7 × 7 mm2) with tapered edges (Fig. 6A). Turbulent or stagnation zones and bubble formation could be avoided in such a design due to the progressive increase and decrease of the cross-sectional area of the chamber, as well as the presence of rounded corners. A set of four PDMS micropillars were also included into the design of the central chamber of each bioreactor to hold the scaffold in place, avoiding its potential displacement during the perfusion culture (Fig. 6A–B). Finally, the bioreactor featured a circular opening in the center of the PMMA support at the bottom to enable direct microscopic monitoring of the beating behavior of the cardiac tissue construct without the need of disassembling the bioreactor (Fig. 6A–B).
Fig. 6.
(A) Schematics showing exploded view of the design of the two-layer microfluidic bioreactor sandwiched by a pair of PMMA clamps. (B) Photograph of the bioreactor with an embedded bioprinted scaffold. (C, D) Simulation results of flow velocity and oxygen distribution, respectively, in the bioreactor chamber at a flow rate of 50 µL min−1 (E, F) Live/dead micrographs and quantified cell morbidity of bioprinted endothelialized scaffolds without and with perfusion in the bioreactors. (G, H) Live/dead micrographs and quantified cell morbidity of bioprinted cardiac organoids without and with perfusion in the bioreactors.
3.3.2. Computational simulations
Cardiomyocytes and endothelial cells are known to be sensitive to oxygen levels. A computational model was thus developed with the finite element method using COMSOL Multiphysics to simulate the flow velocity in the bioreactor as well as oxygen distribution within and at the vicinity of the tissue construct. A flow rate of 50 µL min−1 was adopted after scaling down the flow rate of the blood in the heart according to the weights of the heart and the engineered cardiac organoid [45]. As indicated in the simulation results in Fig. 6C, the flow rates were uniform inside the chamber of the bioreactor except for slightly increased speed at the inlet/outlet and the corners where the pillars were present. More importantly, at such a perfusion rate of 50 µL min−1, the oxygen concentrations across the volume of the scaffold were abundant. Indeed, even the center of the endothelialized myocardial construct could experience an oxygen level of approximately 0.12 mM, approximating the range required for optimal survival of cardiomyocytes and endothelial cells in engineered tissue constructs [62,63].
3.3.3. Effect of perfusion on cell viability
The effects of perfusion on individual cell types were investigated. The tissue constructs were first cultured under static conditions for 3 days prior to being transferred to the bioreactors. The cell viability was then assessed using live/dead staining. As shown in Fig. 6E, the HUVECs exhibited pronounced cellular mortality at Day 7 post culture in the bioreactor when no perfusion was applied with a large number of dead cells (red), whereas the viability of the cells was greatly improved when the scaffolds underwent perfusion culture at the flow rate of 50 µL min−1. The quantification result further revealed a significant difference between the two groups (Fig. 6F, p < 0.001). Similarly, the perfusion of the scaffolds during bioreactor culture also significantly reduced the total number of dead cardiomyocytes (Fig. 6G–H, p < 0.05). Consequently, we chose a perfusion rate of 50 µL min−1 for maintaining the bioprinted tissue constructs inside the bioreactor. It should be noted that, the use of the microbioreactors was not intended to simulate the flow patterns in the native myocardium; instead, the perfusion culture at the low flow rate could enhance the delivery of nutrients and oxygen to the embedded tissue constructs and support their long-term viability.
3.4. Endothelialized-myocardium-on-a-chip as an enabling cardiovascular drug screening platform
3.4.1. Generation of endothelialized-myocardium-on-a-chip model
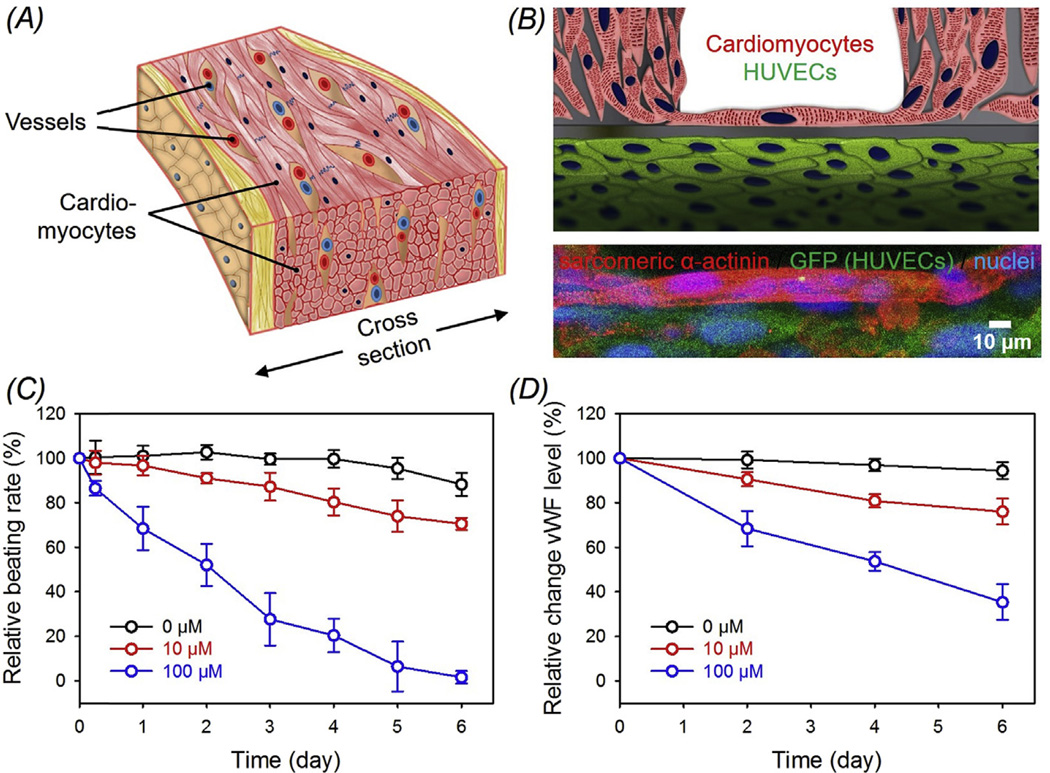
We have previously discussed the effects of bioink, scaffold architecture, and perfusion on the endothelialization and construction of cardiac tissues based on the bioprinted microfibrous scaffolds. A preliminary study on neonatal rat cardiomyocytes seeded onto HUVECs-laden bioprinted scaffolds was performed. Since both HUVECs and cardiomyocytes were present, we employed a 1:1 mixture of EGM and DMEM for the co-culture. This common medium did not seem to pose any adverse influence on the viability and functionality of both cell types. In fact, the presence of vascular endothelial growth factor (VEGF), which is a standard supplement of the endothelial growth medium and further continuously secreted by the HUVECs in the vascular network, could on the contrary, enhance the cardiac function in engineered cardiac organoids through upregulation of the connexin-43 expression as well as other contractile molecules [64]. A schematic of the native myocardium is shown in Fig. 7A, where blood vessels are embedded within a matrix of cardiomyocytes. From the schematic and magnified confocal fluorescence micrograph in Fig. 7B, it was clear that the endothelial cells aligned at the periphery of the bioprinted microfiber, whereas the aligned cardiomyocytes tightly attached on the outside, together assuming the configuration of an endothelialized myocardial tissue resembling the structure of its native counterpart. It should be noted that, although in the current work these bioprinted endothelialized microfibers were not hollow, perfusion may be achieved in future designs where sacrificial bioinks that can be later on removed are used during the bioprinting process. The endothelialized myocardial constructs showed uniform beating at a rate in the range of 50–70 bpm (Mov. S2) similar to monoculture, and the beating could extend to up to at least 2 weeks tested inside the bioreactor during perfusion culture.
Fig. 7.
(A) Schematic showing a native myocardium containing blood vessels embedded in a matrix of cardiomyocytes. (B) Schematic and high-resolution confocal fluorescence micrograph showing an endothelialized myocardial tissue formed by seeding neonatal rat cardiomyocytes onto the bioprinted endothelialized microfibrous scaffold after 15 days of pre-endothelialization. (C, D) Relative beating of the endothelialized myocardial tissues and the levels of vWF expression by the endothelial cells, respectively, upon treatment with different dosages of doxorubicin.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.biomaterials.2016.09.003.
Comparing to the existing engineered cardiac tissues [12,18,25–27], our bioprinting strategy possesses advantages. In our report, the size, shape, and architecture of the microfibrous scaffold can be conveniently controlled by programming the bioprinter. The ability to encapsulate endothelial cells that migrated and formed the confluent endothelium further provided the opportunity to engineer co-cultured models of myocardial tissues with an organized network of endothelial cells, closely mimicking the structural arrangement and to a certain extent recapitulating the functionality of their in vivo counterpart. To the best of our knowledge, prior literature on the construction of 3D endothelialized myocardium for both regeneration and in the on-chip formats has been rare, with most studies focusing on single cell type cultures [12,26,27] and/or simplified co-cultures with limited structural similarity [28,65], including those of our own works [18,52,66–71]. Most importantly, our methodology is a platform technology, where the bioprinted microfibrous network emulating the blood vessels can serve as a vascular bed for engineering any type of endothelialized tissue besides the myocardium demonstrated in our current work.
3.4.2. Endothelialized-myocardium-on-a-chip for cardiovascular drug testing
Drug discovery is a lengthy and expensive process [72]. Organ side-effects have posed a great challenge for drug development and have resulted in rapidly increasing drug attrition rate [3]. In particular, more than 15 drugs have been removed from the European and US markets over the past decade primarily due to toxicity concerns related with the cardiovascular system, contributing to half of the total drug retractions during this period of time [73,74]. We therefore expect that, our 3D endothelialized myocardium model fabricated using this innovative bioprinting technology, combined with physiological relevance through incorporation of the perfusable microfluidic bioreactor, will likely function to predict cardiovascular drug toxicity that could not be achieved using individual cell types alone.
We exposed our endothelialized-myocardium-on-a-chip model to treatment by a common anti-cancer drug, doxorubicin. Unlike other pre-existing models where only individual cell types were typically included [3,7,75–77], doxorubicin elicited dose-dependent responses towards both cardiomyocytes and endothelial cells in our model. The beating rate of the cardiomyocytes decreased to 70.5% and 1.62% (close to 0 bpm) at Day 6 post exposure to 10 µM and 100 µM drugs, respectively, while the control endothelialized myocardial organoids largely maintained a high relative beating rate at approximately 88.3% (Fig. 7C). This endurance towards doxorubicin of the cardiac tissues is comparable to our prior studies using two-dimensional (2D) monolayer cultures of cardiomyocytes [78], but with slight increment presumably due to the three-dimensionality of the model as well as the perfusion culture, both contributing to partially mitigated drug toxicity. Similarly, the levels of vWF secreted by the endothelial cells also reduced to 76.0% and 35.3% at Day 6 for constructs treated with doxorubicin at 10 µM and 100 µM, respectively, while the levels for the controls remained at >90% at all time points tested (Fig. 7D).
3.4.3. Endothelialized-human-myocardium-on-a-chip: a step forward towards personalized medicine
While prototype model has been optimized using neonatal rat cardiomyocytes due to their abundant availability, we finally demonstrated the proof-of-concept possibility to also include cardiomyocytes of human origin to construct an endothelialized-human-myocardium-on-a-chip model. In this case, hiPSC-cardiomyocytes were used as the source of cardiomyocytes. The hiPSCs represent a versatile cell source for obtaining a variety of mature cell types thanks to recent advancements on the stem cell technology [15,16,79,80]. These cells, obtainable from adult individuals, further opens up the door towards personalized medicine for the potential in constructing patient-derived organoids [15,16,79,80]. Prior studies have attempted to use hiPSC-cardiomyocytes and hiPSC-endothelial cells for investigating human cardiotoxicity caused by pharmaceutical compounds [13,81,82]. However, still only single cell types were analyzed individually in addition to the relatively simplified structures, comparing to our bioprinted endothelialized-human-myocardium-on-a-chip platform reported in this work to a better extent mimicking its in vivo counterpart.
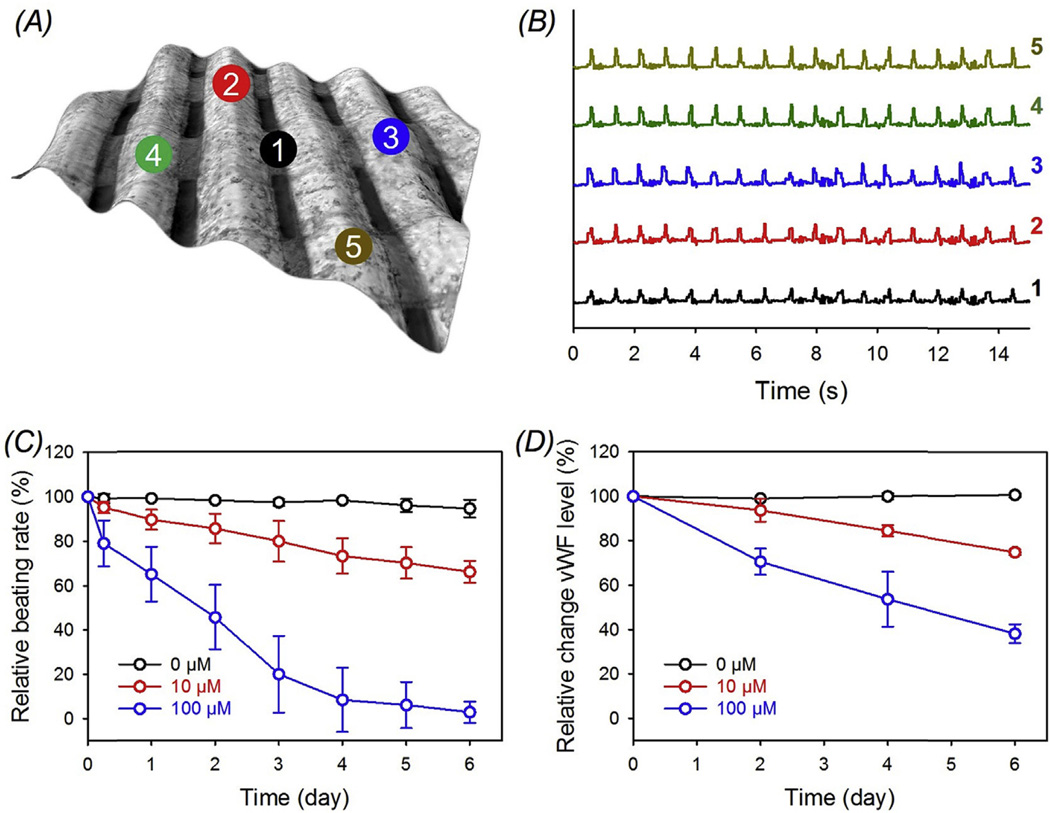
We maintained the same procedures for fabricating the vascular beds using bioprinted microfibrous scaffolds, but hiPSC-cardiomyocytes were subsequently seeded instead of the rat cells. Similarly, a common medium composed of 1:1 mixture of EGM and RPMI/B27 was adopted for the co-culture without imposing any adverse influence on both cell types. As expected, the resulting endothelialized human myocardial organoids presented uniform and highly synchronized beating across the entire scaffold (Fig. 8A and B). The beating rate lasted at approximately 60 bpm for up to 7–10 days when cultured in the perfusion bioreactor, followed by slight decrease over the remaining period. In addition, both hiPSC-cardiomyocytes and endothelial cells showed dose-dependent responses towards doxorubicin. The beating of the hiPSC-cardiomyocytes reduced from 94.5% for the control constructs to 66.0% and 2.78% (close to 0 bpm) for those treated with 10 µM and 100 µM doxorubicin, respectively (Fig. 8C). The human-derived myocardial organoids seemed to have slightly lower endurance at all time points analyzed in comparison to those of rat origin (Fig. 8C versus Fig. 7C). The toxicity of doxorubicin on the endothelial component of the constructs at different doses corresponded well with that observed for the rat myocardial organoids due to the same origin of the cell source (Fig. 8D versus Fig. 7D). Therefore, our endothelialized-myocardium-on-a-chip platform has provided a model for probing drug-induced cardiovascular toxicity with a translational potential for personalized drug screening in the future.
Fig. 8.
(A) Pseudo-3D brightfield micrograph showing an all-human endothelialized myocardial tissue formed by seeding hiPSC-cardiomyocytes onto the bioprinted endothelialized scaffold after 15 days of pre-endothelialization. (B) Beating plots of the different local regions indicated in (A). Note that the contraction amplitudes were all normalized to the same height for easy comparison across the samples. (C, D) Relative beating of the endothelialized human myocardial tissues and the levels of vWF expression by the endothelial cells, respectively, upon treatment with different dosages of doxorubicin.
4. Conclusions and perspectives
In summary, we have presented a novel strategy to construct endothelialized-myocardial-tissues by adopting an innovative bioprinting technology. The endothelial cells, encapsulated inside the microfibers composing the backbone of the scaffolds, gradually migrated towards the peripheries of the microfibers to form a layer of confluent endothelium. The assembly of the endothelial cells within the bioprinted microfibers resembling a blood vessel structure was enabled by the composite bioink featuring a dual-step crosslinking procedure and potentially facilitated by the intrinsic polarization tendency of these cells and presence of a nutrient gradient across the diameter of the microfibers. Importantly, when combined with a microfluidic perfusion bioreactor, the endothelialized-myocardium-on-a-chip model could be used as a platform for cardiovascular drug screening, where dose-dependent responses of both cardiomyocytes and endothelial cells were observed. While the proof-of-concept optimizations were conducted using neonatal rat cardiomyocytes as a model cell type, we also demonstrated that such system might be translated to human cardiac organoids through the application of hiPSC-cardiomyocytes, although more detailed mechanisms on drug-induced cardiovascular toxicity still remain to be examined.
We believe that the combination of bioprinting, microfluidics, and stem cells in our endothelialized-myocardium-on-a-chip platform would provide an enabling technology for the development of next-generation human organ models for not only engineering healthy and diseased myocardial surrogates, but more importantly for their use in personalized drug screening to mitigate drug-induced cardiovascular toxicity or improve treatment efficacy. This endothelialized-myocardium-on-a-chip platform would also enable testing of nanomedicine [4,5,83,84], such as the interactions between nanoparticles and the cardiac cells [85–87] as well as those between nanoparticles and the endothelium (e.g. nanoparticle-induced endothelial leakage, a non-toxic effect of nanoparticles on endothelial cells [31,88]). It should be noted that, although the bioprinted microfibrous structures in this work were not perfusable, we anticipate the perfusion of such an endothelialized network in the future upon usage of sacrificial bioinks that can be removed, to enhance the biomimetic properties of produced vascularized organoids, which is currently under investigation and will be reported in our future reports.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge funding by the Defense Threat Reduction Agency (DTRA) under Space and Naval Warfare Systems Center Pacific (SSC PACIFIC) Contract No. N66001-13-C-2027. The authors also acknowledge funding from the National Institutes of Health (AR057837, DE021468, D005865, AR068258, AR066193, EB022403, EB021148), the Air Force Office of Scientific Research Award #FA9550-15-1-0273, and the Presidential Early Career Award for Scientists and Engineers (PECASE). Y.S.Z. acknowledges the National Cancer Institute Pathway to Independence Award (1K99CA201603-01A1). The Italian Ministry of Health is greatly acknowledged for its support as well. The publication of this material does not constitute approval by the government of the findings or conclusions herein.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biomaterials.2016.09.003.
References
- 1.Chiu L, Iyer RK, Reis LA, Nunes SS, Radisic M. Cardiac tissue engineering: current state and perspectives. Front. Biosci. 2011;17:1533–1550. doi: 10.2741/4002. [DOI] [PubMed] [Google Scholar]
- 2.Chiu LL, Radisic M. Cardiac tissue engineering. Curr. Opin. Chem. Eng. 2013;2:41–52. [Google Scholar]
- 3.Zhang YS, Aleman J, Arneri A, Bersini S, Piraino F, Shin SR, et al. From cardiac tissue engineering to heart-on-a-chip: beating challenges. Biomed. Mater. 2015;10:034006. doi: 10.1088/1748-6041/10/3/034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, et al. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YS, Khademhosseini A. Seeking the Right context for evaluating nanomedicine: from tissue models in petri dishes to microfluidic organs-on-a-chip. Nanomedicine. 2015;10:685–688. doi: 10.2217/nnm.15.18. [DOI] [PubMed] [Google Scholar]
- 6.Huh D, Torisawa Y-s, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: organs-on-Chips. Lab. Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab. Chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selimović Š, Dokmeci MR, Khademhosseini A. Organs-on-a-chip for drug discovery. Curr. Opin. Pharmacol. 2013;13:829–833. doi: 10.1016/j.coph.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015;14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yum K, Hong SG, Healy KE, Lee LP. Physiologically relevant organs on chips. Biotechnol. J. 2014;9:16–27. doi: 10.1002/biot.201300187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Z, Koo S, Finnegan MA, Loskill P, Huebsch N, Marks NC, et al. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials. 2014;35:1367–1377. doi: 10.1016/j.biomaterials.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell stem cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Camci-Unal G, Annabi N, Dokmeci MR, Liao R, Khademhosseini A. Hydro-gels for cardiac tissue engineering. NPG Asia Mater. 2014;6:e99. [Google Scholar]
- 18.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, et al. Highly elastic micropatterned hydrogel for engineering functional cardiac tissue. Adv. Funct. Mater. 2013;23:4950–4959. doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouwkema J, Rivron NC, van Blitterswijk CA. Vascularization in tissue engineering. Trends Biotechnol. 2008;26:434–441. doi: 10.1016/j.tibtech.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Bae H, Puranik AS, Gauvin R, Edalat F, Carrillo-Conde B, Peppas NA, et al. Building vascular networks. Sci. Transl. Med. 2012;4:160ps23-ps23. doi: 10.1126/scitranslmed.3003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartholomä P, Gorjup E, Monz D, Reininger-Mack A, Thielecke H, Robitzki A. Three-dimensional in vitro reaggregates of embryonic cardiomyocytes: a potential model system for monitoring effects of bioactive agents. J. Biomol. Screen. 2005;10:814–822. doi: 10.1177/1087057105280070. [DOI] [PubMed] [Google Scholar]
- 22.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ. Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 23.Chia SL, Tay CY, Setyawati MI, Leong DT. Biomimicry 3D gastrointestinal spheroid platform for the assessment of toxicity and inflammatory effects of zinc oxide nanoparticles. Small. 2015;11:702–712. doi: 10.1002/smll.201401915. [DOI] [PubMed] [Google Scholar]
- 24.Kim B, Han G, Toley BJ, Kim C-k, Rotello VM, Forbes NS. Tuning payload delivery in tumour cylindroids using gold nanoparticles. Nat. Nanotechnol. 2010;5:465–472. doi: 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YS, Xia Y. Multiple facets for extracellular matrix mimicking in regenerative medicine. Nanomedicine. 2015;10:689–692. doi: 10.2217/nnm.15.10. [DOI] [PubMed] [Google Scholar]
- 26.Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat. Mater. 2008;7:1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Habib IHM, et al. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ. Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- 29.Sakaguchi K, Shimizu T, Okano T. Construction of three-dimensional vascularized cardiac tissue with cell sheet engineering. J. Control. Release. 2015;205:83–88. doi: 10.1016/j.jconrel.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Chia SL, Tay CY, Setyawati MI, Leong DT. Decoupling the direct and indirect biological effects of ZnO nanoparticles using a communicative dual cell-type tissue construct. Small. 2016;12:647–657. doi: 10.1002/smll.201502306. [DOI] [PubMed] [Google Scholar]
- 31.Setyawati MI, Mochalin VN, Leong DT. Tuning endothelial permeability with functionalized nanodiamonds. ACS Nano. 2016;10:1170–1181. doi: 10.1021/acsnano.5b06487. [DOI] [PubMed] [Google Scholar]
- 32.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 33.Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, et al. 25th anniversary article: engineering hydrogels for biofabrication. Adv. Mater. 2013;25:5011–5028. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- 34.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, et al. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab. Chip. 2014;14:2202–2211. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 36.Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo S-S, et al. Creating perfused functional vascular channels using 3D bio-printing technology. Biomaterials. 2014;35:8092–8102. doi: 10.1016/j.biomaterials.2014.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colosi C, Shin SR, Manoharan V, Massa S, Constantini M, Barbetta A, et al. Microfluidic bioprinting of heterogeneous 3D tissue constructs using low viscosity bioink. Adv. Mater. 2015;28:677–684. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Q, He Y, Fu J-z, Liu A, Ma L. Coaxial nozzle-assisted 3D bioprinting with built-in microchannels for nutrients delivery. Biomaterials. 2015;61:203–215. doi: 10.1016/j.biomaterials.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 39.Khademhosseini A, Eng G, Yeh J, Kucharczyk P, Langer R, Vunjak-Novakovic G, et al. Microfluidic patterning for fabrication of contractile cardiac organoids. Biomed. Microdevices. 2007;9:149–157. doi: 10.1007/s10544-006-9013-7. [DOI] [PubMed] [Google Scholar]
- 40.Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Bio-fabrication. 2016;8:014101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 41.Merkel TC, Bondar VI, Nagai K, Freeman BD, Pinnau I. Gas sorption, diffusion, and permeation in poly (dimethylsiloxane) J. Polym. Sci. Part B Polym. Phys. 2000;38:415–434. [Google Scholar]
- 42.Abaci HE, Truitt R, Tan S, Gerecht S. Unforeseen decreases in dissolved oxygen levels affect tube formation kinetics in collagen gels. Am. J. Physiol.-Cell Physiol. 2011;301:C431–C440. doi: 10.1152/ajpcell.00074.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radisic M, Deen W, Langer R, Vunjak-Novakovic G. Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers. Am. J. Physiol. - Heart Circ. Physiol. 2005;288:H1278–H1289. doi: 10.1152/ajpheart.00787.2004. [DOI] [PubMed] [Google Scholar]
- 44.Kaemmerer E, Melchels FPW, Holzapfel BM, Meckel T, Hutmacher DW, Loessner D. Gelatine methacrylamide-based hydrogels: an alternative three-dimensional cancer cell culture system. Acta Biomater. 2014;10:2551–2562. doi: 10.1016/j.actbio.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 45.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab. Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang YS, Ribas J, Nadhman A, Aleman J, Selimović Š, Lesher-Perez SC, et al. A cost-effective fluorescence mini-microscope for biomedical applications. Lab. Chip. 2015;15:3661–3669. doi: 10.1039/c5lc00666j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 48.Augst AD, Kong HJ, Mooney DJ. Alginate hydrogels as biomaterials. Macromol. Biosci. 2006;6:623–633. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 49.Tandon N, Cannizzaro C, Chao P-HG, Maidhof R, Marsano A, Au HTH, et al. Electrical stimulation systems for cardiac tissue engineering. Nat. Protoc. 2009;4:155–173. doi: 10.1038/nprot.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon JJ, West JL. Vascularization of engineered tissues: approaches to promote angiogenesis in biomaterials. Curr. Top. Med. Chem. 2008;8:300–310. doi: 10.2174/156802608783790983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi S-W, Zhang Y, MacEwan MR, Xia Y. Neovascularization in biodegradable inverse opal scaffolds with uniform and precisely controlled pore sizes. Adv. Healthc. Mater. 2012;2(1):145–154. doi: 10.1002/adhm.201200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin R-Z, Chen Y-C, Moreno-Luna R, Khademhosseini A, Melero-Martin JM. Transdermal regulation of vascular network bioengineering using a photo-polymerizable methacrylated gelatin hydrogel. Biomaterials. 2013;34:6785–6796. doi: 10.1016/j.biomaterials.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu W, DeConinck A, Lewis JA. Omnidirectional printing of 3D microvascular networks. Adv. Mater. 2011;23:H178–H183. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 54.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aung A, Theprungsirikul J, Lim HL, Varghese S. Chemotaxis-driven assembly of endothelial barrier in a tumor-on-a-chip platform. Lab. Chip. 2016;16(10):1886–1898. doi: 10.1039/c6lc00184j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutson CB, Nichol JW, Aubin H, Bae H, Yamanlar S, Al-Haque S, et al. Synthesis and characterization of tunable poly (ethylene glycol): gelatin methacrylate composite hydrogels. Tissue Eng. A. 2011;17:1713–1723. doi: 10.1089/ten.tea.2010.0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials. 2015;73:254–271. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carola R, Harley JP, Noback CR. Human Anatomy and Physiology. McGraw-Hill College. 1992 [Google Scholar]
- 59.Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, et al. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proct Natl. Acad. Sci. U. S. A. 2004;101:18129–18134. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim Sb, et al. Carbonnanotube-embedded hydrogel sheets for engineering cardiac constructs and bioactuators. ACS Nano. 2013;7:2369–2380. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kharaziha M, Shin SR, Nikkhah M, Topkaya SN, Masoumi N, Annabi N, et al. Tough and flexible CNT-polymeric hybrid scaffolds for engineering cardiac constructs. Biomaterials. 2014;35:7346–7354. doi: 10.1016/j.biomaterials.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Radisic M, Marsano A, Maidhof R, Wang Y, Vunjak-Novakovic G. Cardiac tissue engineering using perfusion bioreactor systems. Nat. Protoc. 2008;3:719–738. doi: 10.1038/nprot.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radisic M, Malda J, Epping E, Geng W, Langer R, Vunjak-Novakovic G. Oxygen gradients correlate with cell density and cell viability in engineered cardiac tissue. Biotechnol. Bioeng. 2006;93:332–343. doi: 10.1002/bit.20722. [DOI] [PubMed] [Google Scholar]
- 64.Iyer RK, Odedra D, Chiu LLY, Vunjak-Novakovic G, Radisic M. Vascular endothelial growth factor secretion by nonmyocytes modulates Connexin-43 levels in cardiac organoids. Tissue Eng. A. 2012;18:1771–1783. doi: 10.1089/ten.tea.2011.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madden LR, Mortisen DJ, Sussman EM, Dupras SK, Fugate JA, Cuy JL, et al. Proangiogenic scaffolds as functional templates for cardiac tissue engineering. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15211–15216. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin SR, Farzad R, Tamayol A, Manoharan V, Mostafalu P, Zhang YS, et al. A bioactive carbon nanotube-based ink for printing 2D and 3D flexible electronics. Adv. Mater. 2016;28:3280–3289. doi: 10.1002/adma.201506420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin SR, Zihlmann C, Akbari M, Assawes P, Cheung L, Zhang K, et al. Reduced graphene oxide-gelma hybrid hydrogels as scaffolds for cardiac tissue engineering. Small. 2016;12(27):3677–3689. doi: 10.1002/smll.201600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, et al. Functional human vascular network generated in photocrosslinkable gelatin methacrylate hydrogels. Adv. Funct. Mater. 2012;22:2027–2039. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, et al. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab. Chip. 2014;14:2202–2211. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, et al. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials. 2016;106:58–68. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W, Zhang YS, Bakht SM, Aleman J, Shin SR, Yue K, et al. Elastomeric free-form blood vessels for interconnecting organs on chip systems. Lab. Chip. 2016;16:1579–1586. doi: 10.1039/c6lc00001k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 73.Shah RR. Can pharmacogenetics help rescue drugs withdrawn from the market? 2006 doi: 10.2217/14622416.7.6.889. [DOI] [PubMed] [Google Scholar]
- 74.Schiller LR, Johnson DA. Balancing drug risk and benefit: toward refining the process of FDA decisions affecting patient care. Am. J. Gastroenterol. 2008;103:815–819. doi: 10.1111/j.1572-0241.2008.01818.x. [DOI] [PubMed] [Google Scholar]
- 75.Kaneko T, Nomura F, Hamada T, Abe Y, Takamori H, Sakakura T, et al. On-chip in vitro cell-network pre-clinical cardiac toxicity using spatiotemporal human cardiomyocyte measurement on a chip. Sci. Rep. 2014;4 doi: 10.1038/srep04670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marsano A, Conficconi C, Lemme M, Occhetta P, Gaudiello E, Votta E, et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab. Chip. 2016;16:599–610. doi: 10.1039/c5lc01356a. [DOI] [PubMed] [Google Scholar]
- 77.Ribas J, Sadeghi H, Manbachi A, Leijten J, Brinegar K, Zhang YS, et al. Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl. in vitro Toxicol. 2016;2(2):82–96. doi: 10.1089/aivt.2016.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropek DM, et al. A cell-based biosensor for real-time detection of cardiotoxicity using lensfree imaging. Lab. Chip. 2011;11:1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin. Pharmacol. Ther. 2011;89:655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- 80.Schenke-Layland K, Nerem RM. In vitro human tissue models – moving towards personalized regenerative medicine. Adv. Drug Del Rev. 2011;63:195–196. doi: 10.1016/j.addr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 81.Morimoto Y, Mori S, Takeuchi S. 3D human cardiac muscle on a chip: quantification of contractile force of human iPS-derived cardiomyocytes; 2015 28th IEEE International Conference on Micro Electro Mechanical Systems (MEMS), IEEE; 2015. pp. 566–568. [Google Scholar]
- 82.Sato H, Idiris A, Miwa T, Kumagai H. Microfabric vessels for embryoid body formation and rapid differentiation of pluripotent stem cells. Sci. Rep. 2016;6:31063. doi: 10.1038/srep31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tay CY, Muthu MS, Chia SL, Nguyen KT, Feng SS, Leong DT. Reality check for nanomaterial-mediated therapy with 3D biomimetic culture systems. Adv. Funct. Mater. 2016;26:4046–4065. [Google Scholar]
- 84.Jang HL, Zhang YS, Khademhosseini A. Boosting clinical translation of nanomedicine. Nanomedicine. 2016;11:1495–1497. doi: 10.2217/nnm-2016-0133. [DOI] [PubMed] [Google Scholar]
- 85.Vandergriff AC, Hensley TM, Henry ET, Shen D, Anthony S, Zhang J, et al. Magnetic targeting of cardiosphere-derived stem cells with ferumoxytol nanoparticles for treating rats with myocardial infarction. Biomaterials. 2014;35:8528–8539. doi: 10.1016/j.biomaterials.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 86.Han J, Kim B, Shin J-Y, Ryu S, Noh M, Woo J, et al. Iron oxide nanoparticle-mediated development of cellular gap junction crosstalk to improve mesenchymal stem cells' therapeutic efficacy for myocardial infarction. ACS Nano. 2015;9:2805–2819. doi: 10.1021/nn506732n. [DOI] [PubMed] [Google Scholar]
- 87.Ferreira MP, Ranjan S, Correia AM, Mäkilä EM, Kinnunen SM, Zhang H, et al. In vitro and in vivo assessment of heart-homing porous silicon nanoparticles. Biomaterials. 2016;94:93–104. doi: 10.1016/j.biomaterials.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 88.Setyawati MI, Tay CY, Chia SL, Goh SL, Fang W, Neo MJ, et al. Titanium dioxide nanomaterials cause endothelial cell leakiness by disrupting the homophilic interaction of VE-cadherin. Nat. Commun. 2013;4:1673. doi: 10.1038/ncomms2655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.