ABSTRACT
The salivary epithelium initiates as a solid mass of epithelial cells that are organized into a primary bud that undergoes morphogenesis and differentiation to yield bilayered acini consisting of interior secretory acinar cells that are surrounded by contractile myoepithelial cells in mature salivary glands. How the primary bud transitions into acini has not been previously documented. We document here that the outer epithelial cells subsequently undergo a vertical compression as they express smooth muscle α-actin and differentiate into myoepithelial cells. The outermost layer of polarized epithelial cells assemble and organize the basal deposition of basement membrane, which requires basal positioning of the polarity protein, Par-1b. Whether Par-1b is required for the vertical compression and differentiation of the myoepithelial cells is unknown. Following manipulation of Par-1b in salivary gland organ explants, Par-1b-inhibited explants showed both a reduced vertical compression of differentiating myoepithelial cells and reduced levels of smooth muscle α-actin. Rac1 knockdown and inhibition of Rac GTPase function also inhibited branching morphogenesis. Since Rac regulates cellular morphology, we investigated a contribution for Rac in myoepithelial cell differentiation. Inhibition of Rac GTPase activity showed a similar reduction in vertical compression and smooth muscle α-actin levels while decreasing the levels of Par-1b protein and altering its basal localization in the outer cells. Inhibition of ROCK, which is required for basal positioning of Par-1b, resulted in mislocalization of Par-1b and loss of vertical cellular compression, but did not significantly alter levels of smooth muscle α-actin in these cells. Overexpression of Par-1b in the presence of Rac inhibition restored basement membrane protein levels and localization. Our results indicate that the basal localization of Par-1b in the outer epithelial cells is required for myoepithelial cell compression, and Par-1b is required for myoepithelial differentiation, regardless of its localization.
KEYWORDS: apicobasal polarity, branching morphogenesis, differentiation, myoepithelial cells, Par-1b, Rac1, ROCK, submandibular gland
INTRODUCTION
Organogenesis of complex 3-dimensional organs during development requires coordination of morphogenesis and cellular differentiation. During embryonic development, the mouse submandibular salivary gland (SMG) undergoes a series of morphogenetic events that result in the spatial ordering of cells that are collectively referred to as the process of branching morphogenesis.1-3 Branching morphogenesis is a conserved developmental mechanism that is utilized by many other organs, including the lungs and mammary glands, to increase the epithelial surface area for either secretion or absorption.4,5 The embryonic mouse SMG serves as an ideal system for studying the molecular mechanisms governing branching morphogenesis and differentiation as glands can be grown ex vivo on a floating filter at the air/media interface and can be manipulated genetically or pharmacologically while recapitulating the normal developmental processes that occur in vivo.6 In mice, SMG development begins in utero on embryonic day 11 (E11), as a protrusion of a solid mass of epithelial cells from the oral epithelium into the surrounding mesenchyme to form a primary bud. Branching initiates at E12 as clefts, or invaginations, form in the primary bud.7,8 Rapid and iterative rounds of cleft formation and proliferation of newly formed epithelial buds characterize the early branching morphogenesis stages (E11-E14). During late development (E15-E18), secretory units begin to form that consist of differentiating secretory acinar cells surrounded by a layer of differentiating stellate, contractile myoepithelial cells, which are embedded in and participate in the assembly of the surrounding basement membrane, a specialized form of extracellular matrix that provides structural support as well as outside-in signals.9-12 While the myoepithelial cells are presumably critical for effective saliva secretion by the bilayered acinar units, little is understood regarding the molecular events required for myoepithelial cell differentiation.
Branching morphogenesis of the salivary gland is highly dependent upon the assembly of the epithelial basement membrane. The basal restriction of the serine-threonine kinase Par-1b (also known as MARK2 or EMK1) is required for the basal placement and assembly of the basement membrane by the outer cuboidal cells (OCCs), the first polarized cell layer to form in the epithelium.8 Which mature cell population the OCCs contribute to has not been clearly demonstrated. The PAR family of proteins were initially identified in C. elegans as partition defective mutants13-15 and are also critical players in establishment of apicobasal polarity in drosophila and mammalian epithelial cells.16,17 In previous work, we have shown that the basal localization of Par-1b is regulated by ROCK1 (Rho-associated coiled-coil containing kinase) to control the deposition and organization of the basement membrane surrounding the OCCs in a manner that is independent of its regulation of non-muscle myosin II.8,18 How Par-1b participates in the shape changes required for the development of bilayered acini has not been examined.
Actin dynamics are known to be critical for branching morphogenesis in the salivary gland19,20 and yet the control of actin dynamics is incompletely understood. One class of molecules that regulates many morphogenetic processes through regulation of the actin cytoskeleton are Rac proteins, which are a subfamily of the Rho family of small GTPases.21 Rac1 (Ras-related C3 botulinum substrate 1) is an ubiquitously expressed member of the Rho family of small GTPases21-23 and is the most abundantly expressed Rho GTPase in the mouse embryo. Rac2 and Rac3 are also expressed in the mouse SMG, but both are found at significantly lower levels than Rac1 based on microarray profiling (http://sgmap.nidcr.nih.gov/).24,25 Like other Rho GTPases, Rac1 acts as a molecular switch, cycling between a GTP-bound active signaling state and a GDP-bound inactive state. In Drosophila melanogaster, loss of Rac1 function disrupted salivary gland tube invagination,26 demonstrating Rac1's importance in salivary gland branching morphogenesis; however, this specific event does not occur in mammalian salivary glands. Since the Rac1 null mouse is embryonic lethal before the formation of the salivary gland,27,28 a function for Rac1 in mammalian salivary gland development has not previously been investigated.
In the current study, we investigated the roles of Rac, ROCK, and Par-1b in myoepithelial cell differentiation. SMG organ explants are a robust model system to study morphogenesis and cell differentiation, as we previously demonstrated that SMG organ explants undergo differentiation with smooth muscle α-actin being expressed during ex vivo culture.29 Here we describe and quantify the vertical compression of the OCCs and expression of SM α-actin during myoepithelial cell differentiation. We manipulated Par-1b in organ explants using targeted siRNA or Par-1b delivered with an adenoviral vector. siRNAs and pharmacological inhibitors were used to manipulate Rac levels and activity, and pharmacological inhibitors were used to inactivate ROCK. We examined changes in cellular morphology and quantified protein levels using immunocytochemistry (ICC) with confocal imaging and Western analysis, respectively. We report here that Par-1b is required for the morphological changes and differentiation of myoepithelial cells. The localization of Par-1b is controlled by both Rac and ROCK, and Par-1b levels are maintained by Rac in developing submandibular salivary glands during myoepithelial cell differentiation.
RESULTS
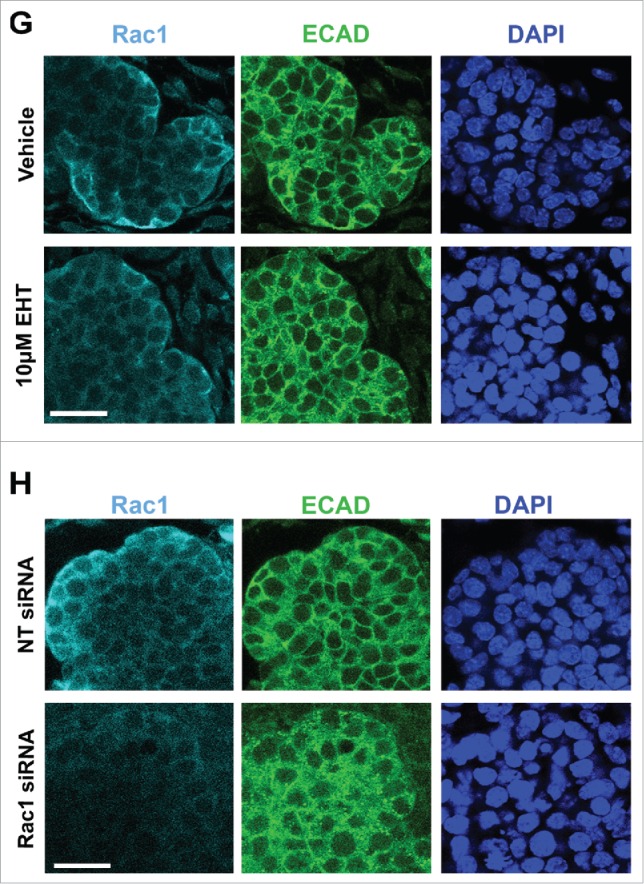
To investigate a role for Rac1 in the developing mouse submandibular salivary gland (SMG), we cultured embryonic day 13 (E13) SMG whole organ explants in which we manipulated Rac levels and activity. We used either a targeted siRNA construct to decrease Rac1 levels directly, or a pharmacological inhibitor that targets Rac family members, Rac1, Rac2, and Rac3 (EHT1864, or EHT), and inhibits their binding to GTP, rendering them inactive.30 We observed an inhibition of branching morphogenesis using brightfield imaging of live explants that were treated with either 10 µM EHT1864 (Fig. 1A) or 400 nM Rac1 siRNA (Fig. 1B) for 96 hours when compared to respective controls. Morphometric analysis through whole bud counting indicates that Rac inactivation using EHT resulted in a statistically significant reduction in branching morphogenesis when compared to controls at 72 and 96-hour time points (Fig. 1C). The reduction of branching morphogenesis using EHT is reversible, as branching morphogenesis was rescued following a washout of the inhibitor (data not shown). Rac1 siRNA treatment also resulted in decreased branching (Fig. 1D). Following treatment with Rac1 siRNA, western analysis shows a 46% reduction in Rac1 levels after 96 hours in culture, and no significant change in Rac1 levels was detected after 96 hours of EHT treatment when compared to vehicle control (Fig. 1E and F), as expected. Immunocytochemistry (ICC) of treated explants confirms that that the siRNA treatment significantly reduces the levels of Rac1 (Fig. 1H) and that there are no major changes to the levels of Rac1 after EHT treatment (Fig. 1G). Based on these results, in subsequent experiments we used EHT to inhibit Rac function. Together, these results indicate that Rac GTPase activity is required for branching morphogenesis of the SMG.
FIGURE 1.
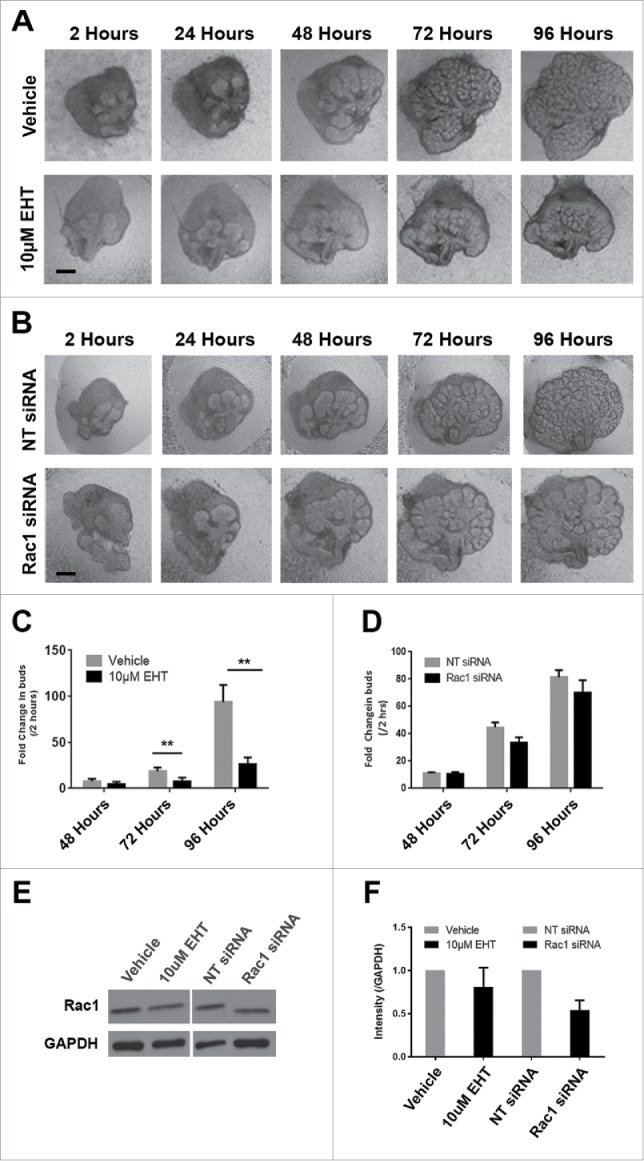
Decreased Branching Morphogenesis of the Mouse Submandibular Salivary Gland with Pharmacological Inactivation of Rac GTPase activity or siRNA Knockdown of Rac1. Representative brightfield images of E13 SMG organ explants cultured for 96 hours (A) with vehicle control or 10 µM EHT, (B) non-targeting (NT) siRNA or Rac-1 siRNA indicate Rac1 expression and activity are both required for proper branching morphogenesis of the developing SMG. Scale bars, 100 µm. (C) Morphometric analysis of control and 10 µM EHT-treated E13 SMGs show a statistically significant decrease in the number of buds in EHT-treated glands (**p ≤ 0 .01) and (D) morphometric analysis of Rac1 siRNA-treated glands show a decreasing trend in bud number relative to NT siRNA-treated glands. (E) Representative western blots and (F) quantification of Rac1 levels in glands treated with 10 µM EHT or Rac1 siRNA, normalized to GAPDH, and compared to controls shows a decrease after siRNA treatment (46% reduction) but not with EHT treatment, as expected (n ≥ 3 experiments) (n.s.). (G and H) Immunocytochemistry (ICC) for Rac1 (cyan) and ECAD (green), with DAPI (blue) staining for nuclei shows Rac1 localized primarily in the outer epithelial cells with no change in localization following 10 µM EHT treatment for 96 hours but a decrease in Rac1 levels within the ECAD+ epithelium after Rac1 siRNA treatment. Scale bars, 10 µm.
Since Rac inhibition leads to a significant decrease in branching morphogenesis, we examined the levels of apoptosis and cell proliferation within the glands following treatment with the Rac inhibitor. Following 48–72 hours of culture, glands were either fixed or lysed and both ICC and western analysis was performed to detect cleaved caspase 3 (CC3), an executioner caspase that is activated by cleavage by both extrinsic and intrinsic apoptosis pathways.31 Analysis of CC3 indicated that there was no difference in the levels of apoptosis in glands treated with the Rac inhibitor in comparison with vehicle controls, as measured with western analysis or quantification of ICC at 48–72 hours (data not shown). Since apoptosis appears not to be operative in Rac-mediated regulation of branching morphogenesis, we examined the levels of proliferation within the glandular epithelium by examining mitosis-specific phosphorylation on Serine 10 of histone H3 (pHH3) in glands treated with the Rac inhibitor and compared to vehicle control (Fig. 2A). Following treatment of glands with 10 µM EHT over 48 hours, quantification of levels of pHH3 normalized to total histone H3 and GAPDH by western analysis indicated that there was no change in EHT-treated glands relative to vehicle control (Fig. 2B and C). These data suggest that Rac does not globally regulate apoptosis or cell proliferation during early salivary gland branching morphogenesis.
FIGURE 2.
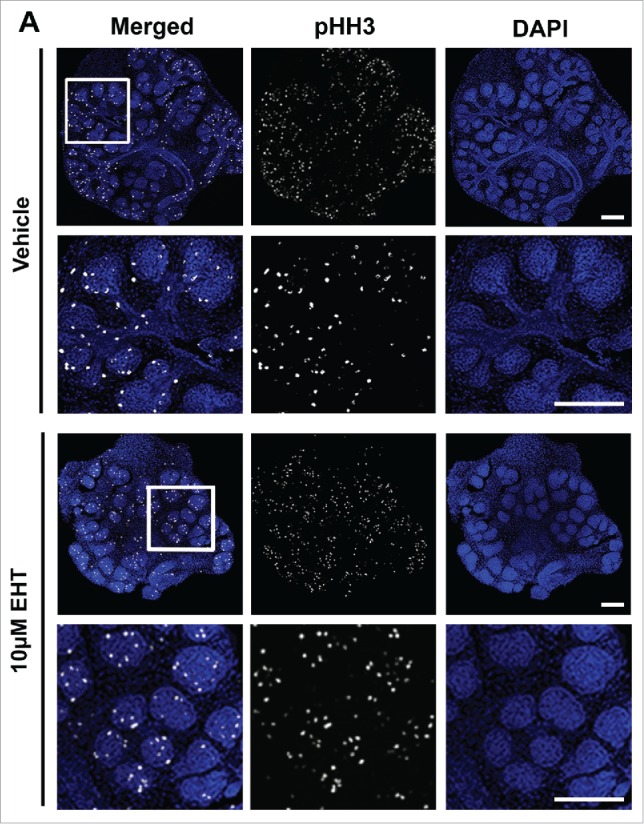
Epithelial Proliferation is Not Affected by the Inhibition of Rac GTPase. (A) ICC to detect histone H3 phosphorylated on Serine 10 (pHH3, white) and DAPI to stain nuclei (blue) following treatment with vehicle or 10 µM EHT for 48 hours shows no major change in pHH3 staining or localization of positive cells within the epithelium. Boxed area in the top panel shows the zoom in area for the bottom panel for each condition. All scale bars, 100 µm. (B and C) A representative protein gel blot and quantification (n ≥ 3) indicate no significant change in pHH3 with EHT-mediated inactivation of Rac1 when compared to total histone H3 levels and GAPDH. (D and E) Pixel intensity quantifications performed on 63X confocal images of E13 glands treated with vehicle or 10 µM EHT for 48 hours and subjected to ICC indicate no notable change in pHH3 levels in the epithelium but an increase in pHH3 in the mesenchymal compartment (p ≤0.05) when quantified separately (n ≥10 images per condition).
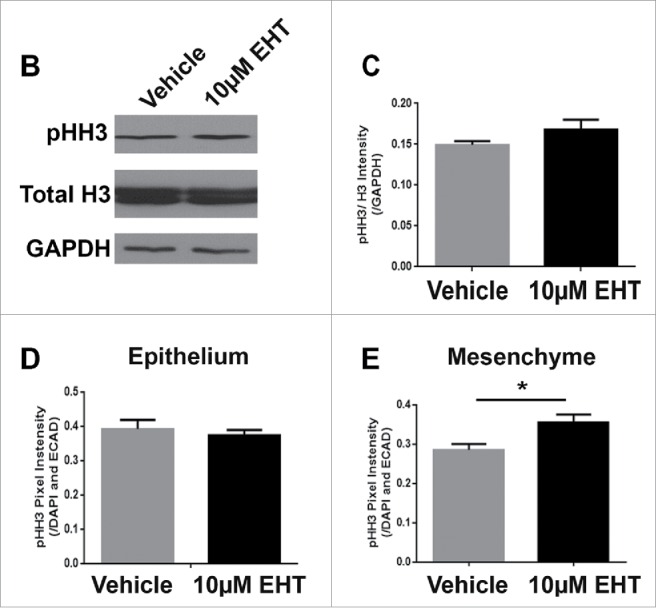
Since western analysis takes into account levels of pHH3 within both the mesenchyme and the epithelial compartments of the gland, we wanted to determine whether there was a change in the number of proliferative cells in either the epithelial or mesenchymal compartments in response to Rac inhibition. Following treatment of glands with 10 µM EHT over 48 hours and ICC to detect pHH3, no change in the pattern of pHH3 in the Rac inhibitor-treated glands vs vehicle control was observed. We used E-cadherin, an epithelial cell-cell adhesion protein to demark the epithelium. We quantified the pixel intensity of the pHH3 signal within the epithelial area independently of the mesenchymal area within RO1s and normalized to the DAPI pixel intensity. No significant difference in the pHH3 staining pattern or pixel quantifications was observed within the E-cadherin-positive epithelium (Fig. 2D), but there was an increase in the pHH3 signal within the mesenchyme after treatment with EHT for 48 hours when compared to the negative control (Fig. 2E). In addition, we quantified the average size of nuclei in both the epithelium and mesenchyme and used this number to estimate the number of nuclei within each sample from the total DAPI staining. While the nuclear size was larger in the EHT-treated glands than in controls, there was no significant change to the calculated number of cells in response to EHT treatment (data not shown). Together, these results indicate that the decreased size of the gland with Rac inhibition is not due to decreased proliferation in the epithelium or mesenchyme. The decrease in the number of buds and change in nuclear size in response to Rac1 inhibition is likely due to Rac's regulation of cellular morphogenesis, independent of proliferation and apoptosis.
Salivary gland branching morphogenesis requires basement membrane remodeling,8,32 leading us to question whether the basement membrane was disrupted by the inactivation of Rac. After inhibition of Rac with EHT for 96 hours, we performed an ICC to detect collagen IV, a major component of the basement membrane, and noted a significant loss in collagen IV (Fig. 3A) which was similar to the reduction in collagen IV following treatment with targeted Rac1siRNA for 96 hours (Supplementary Fig. 1). The loss of collagen IV was also detected via western analysis (Fig. 3B), which revealed a 48% reduction in collagen IV protein levels in glands treated with EHT for 48 hours (data not shown) or 96 hours (Fig. 3C). We also detected a marked decrease in laminin-111 and perlecan, two other major components of the basement membrane, after treatment with EHT for 96 hours when compared to controls (data not shown). These data taken together suggest that Rac1 directly or indirectly regulates the levels of and/or assembly of basement membrane proteins.
FIGURE 3.
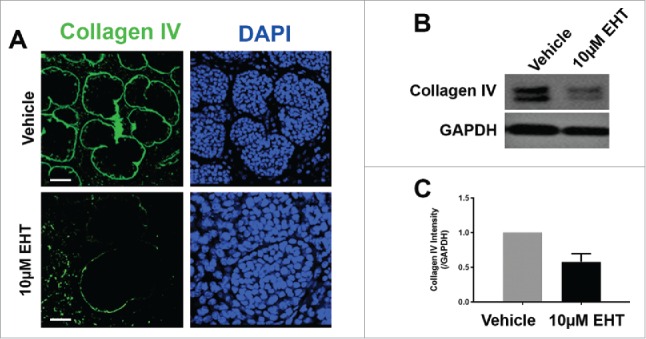
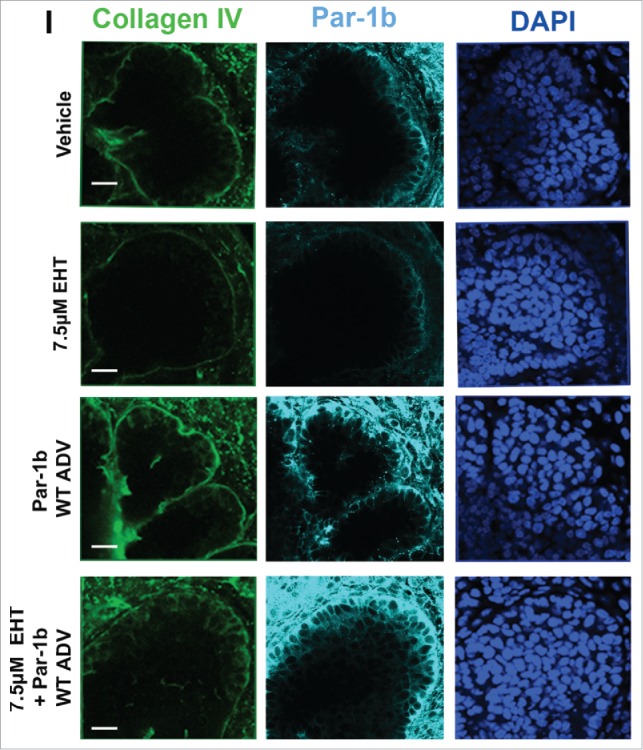
Rac GTPase Inactivation Results in Disruption of the Basement Membrane and Cellular Polarity. (A) ICC for collagen IV (green) and DAPI staining (blue) following 96-hour culture of E13 SMGs treated with vehicle or 10 µM EHT indicate Rac1 inactivation reduces collagen IV in the basement membrane surrounding epithelial buds. Scale bars, 10 µm. (B and C) Representative western analysis and quantification performed following 96-hour culture with or without 10 µM EHT demonstrates a slight loss in collagen IV levels (42% reduction) in E13 SMG with EHT treatment (p = 0.66, n ≥ 3 Experiments). (D) ICC following 96-hour culture with or without 10 µM EHT for Par-1b (cyan) and collagen IV (green) with DAPI staining (blue) demonstrates a loss in Par-1b levels in the outer epithelial cells with EHT treatment. Scale bars, 20 µm. (E and F). Western analysis for Par-1b following EHT treatment indicates a loss of total Par-1b within the gland (25% reduction), quantified relative to GAPDH (n ≥ 3). (G) SMGs were treated with either Par-1b or NT control siRNA, and Western analysis was performed to detect collagen IV and GAPDH. (H) Quantification of collagen IV levels demonstrated 60% reduction in Par-1b levels relative to GAPDH (n = 3). (I) E13 epithelial rudiments treated with vehicle or Par-1b WT adenovirus were recombined with untreated mesenchyme, and cultured for 96 hours either with or without 7.5 µM EHT. ICC was performed for collagen IV (green) and Par-1b (cyan), together with DAPI staining for nuclei (blue). Par-1b WT adenovirus treatment shows increased collagen IV levels relative to control, and glands treated with both WT Par-1b adenovirus and EHT show an intermediate phenotype indicating partial rescue of collagen IV with exogenous Par-1b in the presence of EHT. Scale bars, 10 µm.
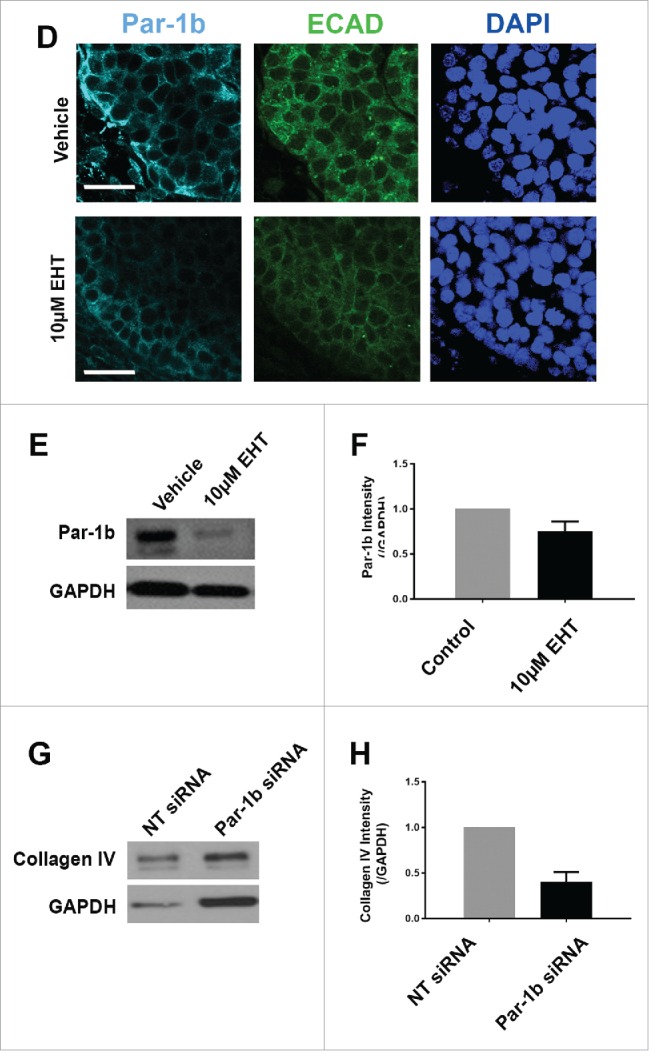
Since we previously reported that the basal localization of the polarity protein Par-1b is critical for the basal deposition of basement membrane by the OCC layer of epithelial cells in the developing SMG,8 we questioned whether Rac1 regulates Par-1b. In Rac-inhibited glands relative to controls, we identified a significant decrease in Par-1b protein levels via ICC as well as a mislocalization of the remaining Par-1b protein from its basal localization in the OCCs to a more lateral localization in these cells (Fig. 3D). To further confirm that Par-1b functions downstream of Rac, we performed western analysis following EHT treatment in E13 glands cultured for 96 hours and found a decrease of approximately 35% in Par-1b levels in the EHT-treated glands when compared to controls (Fig. 3E and F), similar to the Par-1b reductions detected at 48 hours (data not shown). These data suggest that Par-1b localization and protein levels are both regulated by Rac1 and that the disruption of basement membrane following Rac inhibition is likely to be Par-1b mediated. We previously demonstrated that Par-1b controls the localized deposition of multiple basement membrane proteins, including collagen IV and perlecan, after 24 hours of culture.8 To confirm that Par-1b controls basement membrane deposition after 96 hours in culture, we treated SMGs with either Par-1b or non-targeting (NT) control siRNA and examined collagen IV. Following culture, western analysis was performed and quantified, and we confirmed 60% loss of collagen IV levels following Par-1b siRNA treatment (Fig. 3G and H). The loss in collagen IV following Par-1b siRNA knockdown is greater than the loss following EHT treatment, likely due to the greater loss in Par-1b with siRNA knockdown. We then overexpressed Par-1b in EHT-treated SMG epithelial rudiments using an adenoviral construct and recombined the epithelium with mesenchyme (Fig. 3I). The loss of collagen IV in EHT-treated epithelial rudiments was partially rescued by Par-1b (Fig. 3I, bottom panel), further indicating that Rac1 functions upstream of Par-1b in basement membrane deposition.
Since loss of Rac1 and Par-1b affected basement membrane in the developing glandular epithelium, we questioned whether there was a defect in the maturation of the outer epithelial cells that are closely associated with the basement membrane. We harvested SMG tissue from embryos at 24 hour increments at E14, 15, 16, and 17, embedded the tissues in paraffin, and performed fluorescent immunohistochemistry (IHC) on the non-cultured glands to detect E-cadherin (ECAD) to demark epithelial cells and smooth muscle α-actin (SM α-actin), which has been used as a marker of the mature myoepithelial cell type in the mammary gland33,34 and of both immature and mature myoepithelial cells the salivary gland27-29 (Fig. 4). Consistent with our previous observations,35-37 from this time-course, we noted that the OCCs expressed ECAD but not SM α-actin at E14 and 15. By E16, many of the OCCs expressed SM α-actin, and a subset of cells were beginning to flatten. By E17, the SM α-actin-positive cells had assumed the flattened, stellate appearance typical of myoepithelial cells. This time-course data reveals that the OCCs undergo a morphological change and express SM-α-actin in vivo as they transition into myoepithelial cells.
FIGURE 4.
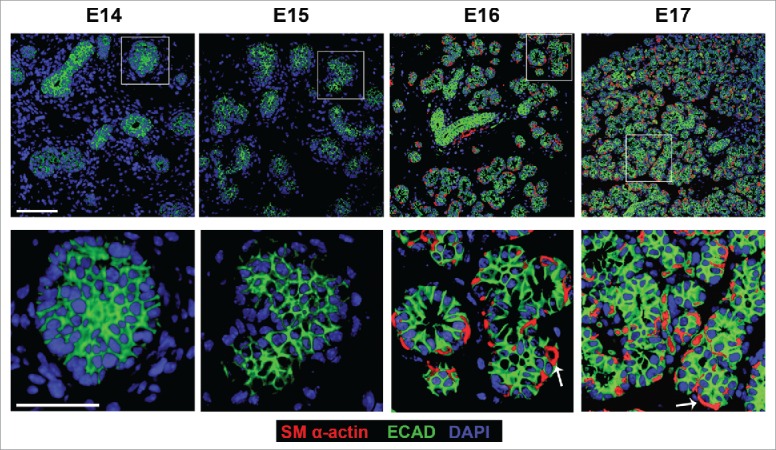
In Vivo Morphogenesis and Differentiation of the Myoepithelium in the Developing SMG. E14, E15, E16 and E17 SMGs were removed from embryos, immediately fixed, and subjected to ICC for SM α-actin (red), ECAD (green), and staining for DAPI (blue). The immature OCC epithelial cells do not express detectable SM α-actin until the E16 stage, coincident with the vertically compression that occurs as they differentiate into myoepithelial cells. Scale Bar, 100 µm top panel and 50 µm in bottom panel.
Since we previously noted that vertical compression occurs together with myoepithelial differentiation in organ explants after 96 hours of culture,35 we examined the effect of Rac inhibition on these cells. Interestingly, we noted that the inhibition of Rac signaling for 96 hours in organ explants appeared to prevent the compression of the OCC population that normally occurs in vivo and after 96 hours in culture (Fig. 5A, arrows); the cells in the EHT-treated glands remained cuboidal in shape (Fig. 5A). We saw a similar phenotype following 96 hours of treatment with a targeted Rac1 siRNA when compared to NT siRNA controls (Supplementary Fig. 1). We measured the height of SM α-actin-positive outer epithelial cells in control glands and in glands treated with EHT to confirm that the loss of Rac activation inhibited the morphological changes in the differentiating myoepithelial cells. Cells in the control glands were significantly more compressed than SM α-actin-positive cells in EHT-treated glands, indicating that loss of Rac activation inhibits the transition of SM α-actin-positive cells from the cuboidal shape of the immature cells toward the flattened shape of the differentiated myoepithelial cells (Fig. 5B). We therefore questioned whether there was a defect in the differentiation of the myoepithelium, as measured by levels of SM α-actin, with Rac inhibition using ICC and western analysis. We found that differentiation is markedly reduced with Rac inhibition as seen with the reduction in the levels of SM α-actin, detected by both ICC and western analysis (Fig. 5D-E), Since SM α-actin is not specific to the myoepithelial cells, we counted the number of SM α-actin+/ECAD+ cells +/− EHT and found a significant decrease in the number of these cells with Rac Inhibition (Fig. 5C). These data suggest that Rac GTPase activity is required for the morphological changes that occur during maturation of OCCs into myoepithelial cells.
FIGURE 5.
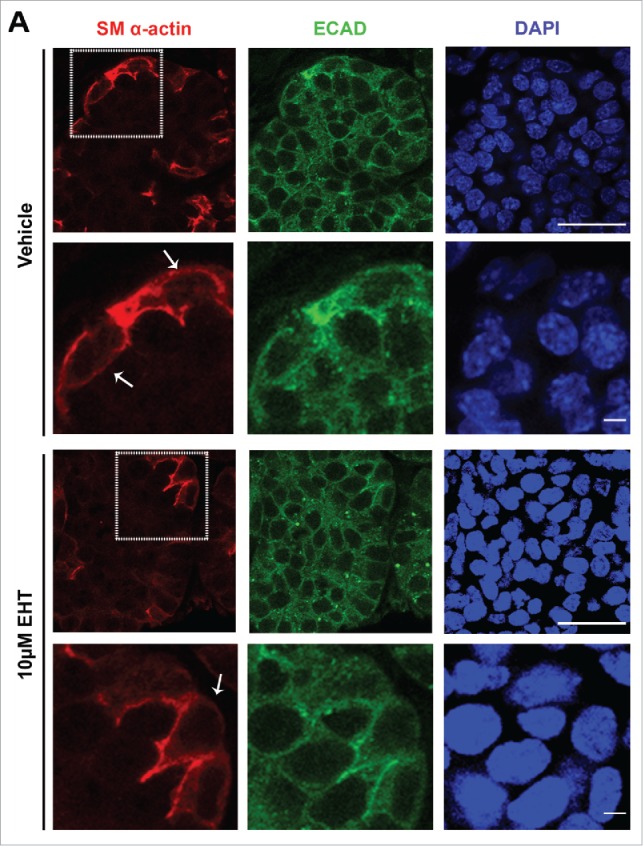
Rac GTPase Inhibition Disrupts the Morphogenesis and Differentiation of the Myoepithelium in the Developing SMG. (A) ICC for SM α-actin (red) and ECAD (green) with DAPI staining (blue) performed on E13 SMGs treated with 10µM EHT for 96 hours shows that the SM α-actin+ cells undergo a vertical compression after 96 hrs culture relative to the glands that were treated with EHT. Scale bar, 10µm (rows 1 and 3) and 2µm (rows 2 and 4). (B) The cell height (µm) of individual ECAD+/SM α-actin+ epithelial cells was quantified in glands treated with or without 10µM EHT for 96 hours. The height of these cells in the EHT-treated glands was significantly greater than in the control glands (***p ≤ 0.001, n = 45). (C) The number of SM α-actin+ cells within an epithelial ROI (n ≥ 35) was significantly decreased following EHT treatment when compared to controls (****p < 0.0001). (D-E) Representative western analysis and quantification (n ≤ 3) performed on whole E13 glands following 96-hour culture +/− EHT treatment indicated a decrease of approximately 40% in SM α-actin levels in EHT-treated relative to control glands.
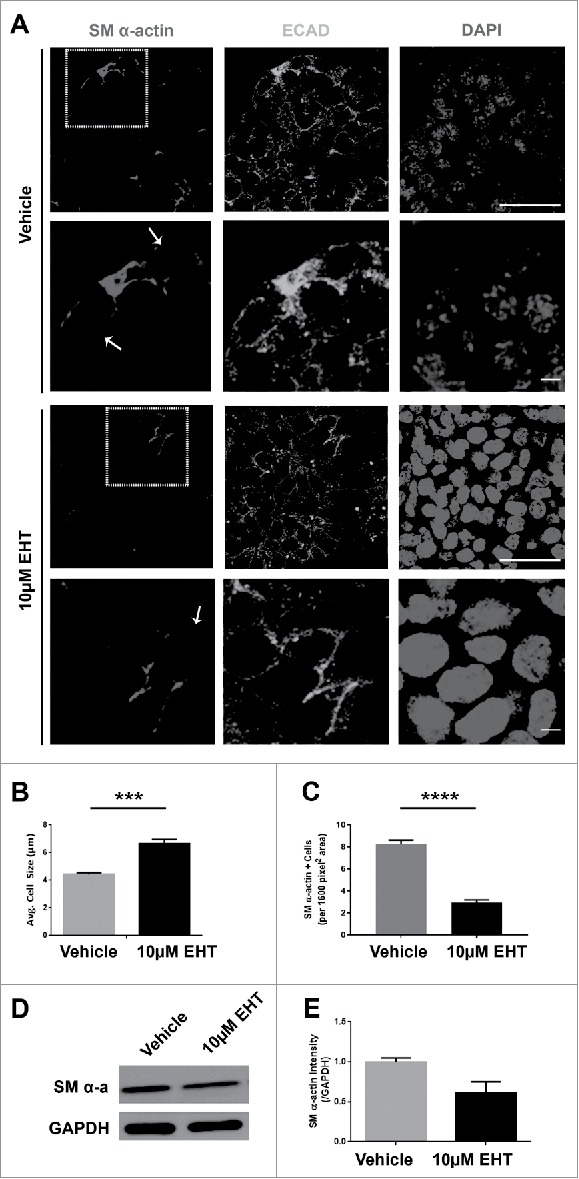
FIGURE 6.
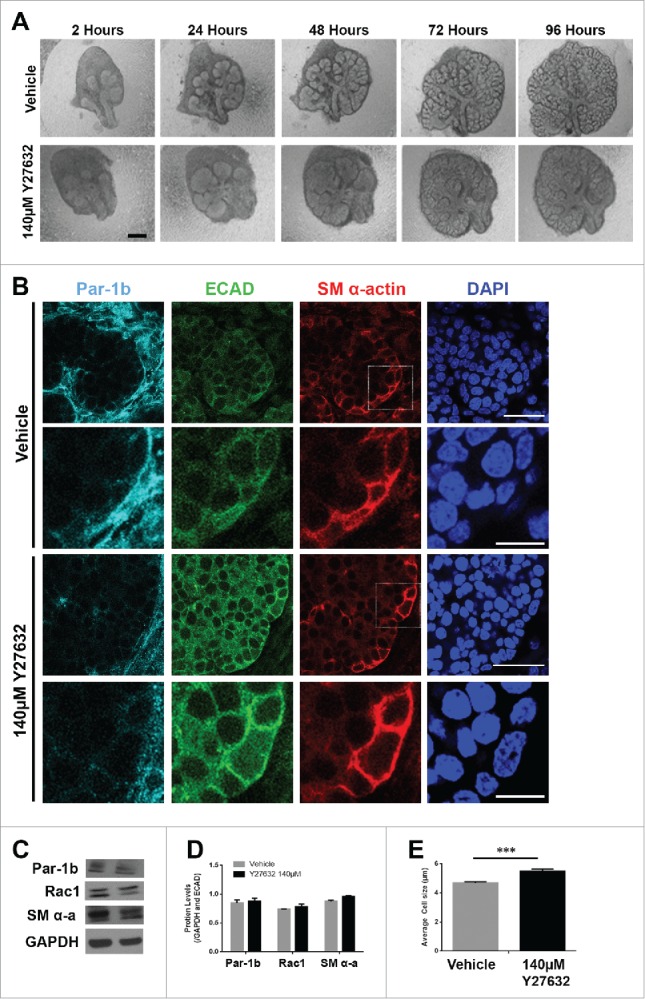
ROCK is Required for the Morphogenesis but Not the Differentiation of the Myoepithelial Cells in the Developing Mouse SMG (A) Representative brightfield images of E13 SMG explants cultured for 96 hours with vehicle control media or with 140µM Y27632 to inactivate ROCK. Scale bar, 100µm. (B) ICC was performed on E13 explants grown in culture for 96 hours and treated with vehicle or with 140µM Y27632 ECAD (green), Par-1b (cyan), and SM α-actin (red) with DAPI staining (blue). Y27632-treated glands show a mislocalization of Par-1b throughout the cytoplasm of ECAD+ cells rather than a basolateral restriction in these cells. Additionally, SM α-actin+ cells appeared to be less compressed with Y27632 treatment. Scale bars, 10µm top panels, 2µm bottom panels. Lower panels are zooms from boxed areas in top panels (C and D) Representative western analysis and quantification (n ≥ 3) following 96-hour culture +/− 140µM Y27632 treatment shows no significant change in the levels of Par-1b, Rac1, or SM α-actin. (E) The height (µm) of individual epithelial cells expressing SM α-actin (n ≥100 ) was measured in glands treated +/− 140µM Y27632 for 96 hours. Height of the ECAD+/SM α-actin+ cells in the glands treated with Y27632 was significantly larger than in the vehicle control-treated glands (*** p ≤ 0.001)
Given that Rac appears to function upstream of Par-1b and that the deposition of the basement membrane is reduced following knockdown of Par-1b after 96 hours in culture (Fig. 3G and H), we questioned whether ROCK1, also known to be an upstream regulator of Par-1b,8 regulates Par-1b at this time point and if it influences myoepithelial differentiation. We treated E13 SMGs with a pharmacological inhibitor for ROCK, Y27632,8 over a 96-hour culture period. Time course images show decreased branching morphogenesis, consistent with our previous report that ROCK inhibition inhibits branching at 24 hours (Fig. 6A).8 ICCs performed for Par-1b showed no major change to the levels of Par-1b in response to Y27632 treatment for 96 hours; however, the localization of Par-1b is markedly different in response to Y27632 at this time point, showing redistribution of Par-1b away from the basal side of the cell (Fig. 6B). To determine whether ROCK inhibition was also affecting the levels of Par-1b protein, we performed western analysis. Western analysis confirmed there was no significant change in the levels of Par-1b protein following 96 hours of Y27632 treatment in E13 SMG organ explants (Fig. 6C and D). Although ICC performed to detect SM α-actin as an indicator of myoepithelial cell differentiation showed a minor increase, there was no significant change to levels of SM α-actin (Fig. 6C and D) following Y27632 treatment. We also measured cell heights of SM α-actin-positive cells and confirmed that there was a statistically significant decrease in vertical compression of SM α-actin-positive cells in the Y27632-treated glands when compared to vehicle controls (Fig. 6E). Although Rho is known to regulate Rac, we found no evidence that ROCK regulates Rac1 levels in this context as Y27632 inhibition did not change levels of Rac1, as measured by western analysis (Fig. 6C and D), nor did EHT treatment affect ROCK1 levels (Supplementary Fig. 2). These results taken together indicate that ROCK regulates the localization of Par-1b, which may contribute to regulation of the vertical compression of the myoepithelial cells, but ROCK is not required for myoepithelial differentiation.
We next questioned whether Par-1b itself is required for the vertical compression of OCCs and/or myoepithelial differentiation. We knocked down Par-1b using a targeted siRNA (Fig. 7A) and confirmed the reduction in Par-1b levels by ICC and by quantification of western analysis (Fig. 7B and C). ICC to detect SM α-actin distribution in explants treated with Par-1b siRNA revealed a phenotype that very closely mimics that seen after inactivation of Rac (Fig. 5A). After treatment with targeted Par-1b siRNA, the height of the cells expressing SM α-actin was measured, and we found a significant reduction in vertical compression, consistent with results following inactivation of either Rac or ROCK (Fig. 5B and 6E). We also examined SM α-actin protein levels via western analysis, and results indicated a significant reduction in SM α-actin (Fig. 7B and D), indicating that Par-1b is also required for myoepithelial differentiation. These data indicate that Par-1b is required for both the vertical compression and differentiation of myoepithelial cells in the developing SMG.
FIGURE 7.
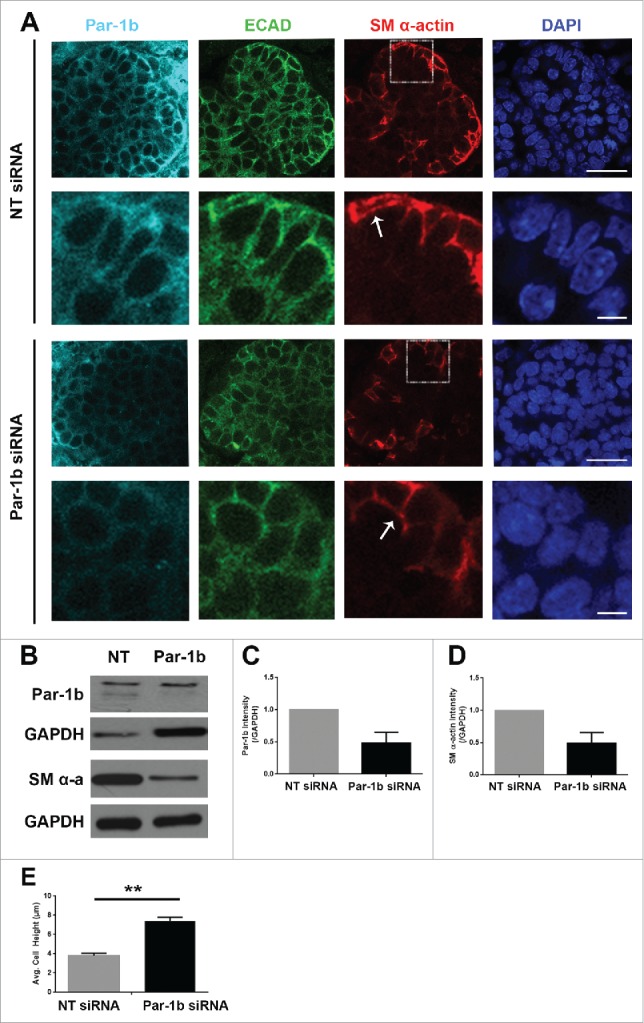
Par-1b is Required for the Morphogenesis and Differentiation of Myoepithelial Cells in the Developing Mouse SMG. (A) ICC was performed on E13 glands grown in culture for 96 hours that were treated with either NT siRNA or Par-1b siRNA (500nM) to detect Par-1b (cyan), ECAD (green), and SM α-actin (red), with DAPI staining (blue). The lower panel of images are zooms from boxed area in the top panel. A significant decrease in the number of SM α−actin-positive cells was observed following Par-1b siRNA treatment as well as an increase in ECAD+/SM α−actin+ cell height. Scale bars, 10µm top panels, 2µm bottom panels. (B) Western analysis was performed on whole glands following 96-hour culture with Par-1b or NT siRNA to detect the same proteins relative to GAPDH. Representative Westerns are shown. n ≥ 3 experiments for each blot. (C) Par-1b siRNA-induced knockdown was confirmed. (D) Par-1b siRNA treatment led to a reduction in the levels of SM α-actin relative to NT siRNA. (E) The height (µm) of individual epithelial cells expressing SM α-actin were measured in glands treated with NT siRNA or Par-1b siRNA for 96 hours. The height of the ECAD+/SM α-actin+ Par-1b siRNA-treated cells was significantly greater than that of ECAD+/SM α-actin+ NT siRNA-treated cells (n ≥ 25 cells/condition). (** p ≤ 0.01).
DISCUSSION
Myoepithelial differentiation has long been of interest in the cancer field due to the tumor-suppressive effects of myoepithelial cells in breast cancer38 and the prevalence of myoepithelial-derived tumors, but little is understood regarding the early development of this cell type in the salivary gland. Actin dynamics participate in myoepithelial differentiation,39 but the signaling pathways that control myoepithelial differentiation are not well characterized. In developing submandibular salivary glands in vivo and in cultured organ explants, the outer epithelial cell population undergoes a vertical compression, or flattening, as the cells begin to express SM α-actin, a marker of myoepithelial cell differentiation, and differentiate into myoepithelial cells. Here we report a function for Par-1b in the vertical compression and differentiation of the myoepithelial cell type in mammalian submandibular salivary glands. In organ explants with reduced levels of Par-1b, we noted a reduced level of SM α-actin and less vertical compression of the OCCs (Fig. 7A). We here identified Rac and ROCK as regulators of Par-1b localization to control myoepithelial vertical compression, and Rac regulation of Par-1b levels to control myoepithelial differentiation. Inhibition of Rac GTPase signaling showed a similar reduction in OCC vertical compression and SM α-actin levels. Since Rac inhibition decreased total levels of Par-1b protein and disrupted its basal membrane localization, we propose that Rac directly or indirectly regulates the localization and levels of Par-1b protein. Inhibition of ROCK, which we previously showed regulates Par-1b localization,8 showed only a mislocalization of Par-1b protein and lack of OCC compression but no change in SM α-actin. Overexpression of WT Par-1b in epithelial cells in the presence of Rac inhibitor restores basement membrane protein levels, indicating that Par-1b-mediated assembly of the basement membrane is likely functioning downstream of Rac and is required for myoepithelial vertical compression and/or differentiation (summarized in Fig. 8). Together, these data support a model that Par-1b localization to the basal membrane is a biological determinant required for the vertical compression of the differentiating myoepithelial cells and that Par-1b expression is also required for myoepithelial cell differentiation, regardless of its cellular localization.
FIGURE 8.
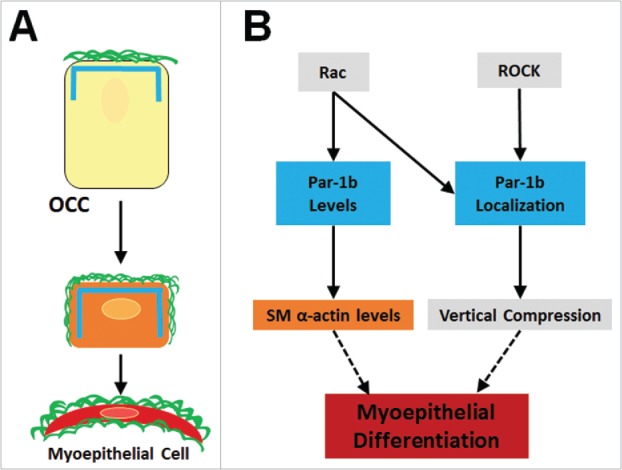
Summary of Myoepithelial Cell Differentiation and Morphogenesis. (A) Outer cuboidal cells (OCCS) in E13 SMGs localize Par-1b (blue lines) to the basal periphery, adjacent to the assembled basement membrane (green lines). Following 96-hour culture, untreated OCCs undergo vertical compression and begin to express the myoepithelial cell marker, SM α-actin (orange rectangle), and localization of Par-1b and basement membrane is extended laterally in as the cells undergo vertical compression and myoepithelial differentiation. (B) Working model for molecular control of myoepithelial cell differentiation. Par-1b localization is controlled directly or indirectly downstream of both Rac and ROCK. Localized Par-1b regulates vertical compression in differentiating myoepithelial cells. Par-1b protein levels are controlled directly or indirectly by Rac and maintain levels of SM α-actin, a myoepithelial marker.
The events leading to activation of Rac and ROCK signaling and subsequent activation of Par-1b during myoepithelial differentiation remain to be clarified. It is possible that inhibition of ROCK leads to inactivation of Rac1, although this is unlikely as we do not see all of the consequences of Rac1 inactivation following ROCK inhibition. We also cannot rule out that Rac acts on Par-1b through an intermediate signaling protein or mechanism. Previous work indicated that Rac1 may control Par-1b levels indirectly through its interactions with the apical PAR complex via its guanine exchange factor, T-lymphoma invasion and metastasis 1 (Tiam1), which can directly bind to Par3.40-42 Rac1 is recruited to Par-3 and can bind to Par-6, leading to activation of atypical protein kinase C (aPKC).43 Alternatively, Rho GTPase-mediated signaling pathways have previously been demonstrated to regulate morphogenetic behaviors in developing salivary glands. Rac but not ROCK also regulates the levels of Par-1b in the developing myoepithelial cells, and it is not known whether Rac increases levels of Par-1b through transcriptional mechanisms or by stabilizing Par-1b mRNA or protein. ROCK has been previously shown to be required for differentiation of myofibroblasts, including playing a role in ECM expression,44 and it is not clear how similar the signaling pathways are in myoepithelial cells. Since Rac and ROCK are both expressed in both the epithelium and mesenchyme, signaling within the mesenchymal cells may contribute to myoepithelial differentiation. Further investigation into the detailed mechanisms through which Rac and ROCK regulate Par-1b and contribute to myoepithelial differentiation is required.
Understanding how acinar structures are formed from progenitor cells is critical for the design of regenerative medicine strategies and for bioengineering of such structures. The mechanism through which myoepithelial cells are generated from a precursor cell having a cuboidal cell type is not fully understood. Although the outer epithelial cells are the first epithelial cell type to polarize during salivary gland development,8 we here report that as myoepithelial cells differentiate in vivo, these cells or a subpopulation of these cells assume a flattened morphology. Differentiation of the myoepithelial cells, although requiring Par-1b, does not appear to require basal positioning of Par-1b, but the basal localization of Par-1b appears to be required for myoepithelial cell vertical compression.
We hypothesize that the localization of Par-1b in the OCCs of the SMG is critical to the process of vertical compression. Par-1b is a kinase with many downstream targets. Recently, Par-1b was shown to phosphorylate RNF41, an E3 ubiquitin ligase for epithelial cells to basally localize laminin receptors and achieve apicobasal polarity.45 Par-1b localization is tightly restricted to the basal periphery of the OCCs in early glandular development, but as the gland undergoes many morphological changes, the tight basal restriction of Par-1b is lost. We hypothesize that this loss of basal restriction allows for the movements of dystroglycan and/or integrins around the lateral edges of the OCCs. This movement of basement membrane receptors would allow for the cell to make contact with the basement membrane over a broader surface area which may contribute to the vertical compression of the OCCs. Further investigation is required into the signaling mechanisms through which Par-1b regulates myoepithelial cell vertical compression and differentiation in the developing mouse submandibular salivary gland.
Experimental procedures
Ex vivo organ culture
Embryonic mouse submandibular salivary glands (SMGs) were dissected from timed-pregnant female mice (strain CD-1, Charles River Laboratories, Wilmington, MA) at embryonic day 12.5 or 13 (E12.5 or E13, with the day of plug discovery designated as E0), following protocols approved by the University at Albany IACUC committee. Embryonic tissues were microdissected as previously described46,47 and cultured at the air/media interface on Nuclepore Track-Etch polycarbonate membrane filters (0.2 μm pore size, Whatman) floating on 1:1 DMEM/Ham's F12 medium (F12) (Invitrogen) supplemented with 150 μg/ml Vitamin C, 50 μg/ml Transferrin and 1X pen/strep. Five or more intact SMGs or epithelial rudiments were tested in each condition with experiments repeated at least 3 times. For culture experiments lasting longer than 48 hours, the media was replaced at the 48 hour time point, including replacement of siRNA or inhibitors.
For recombination experiments, SMG epithelial rudiments removed from the surrounding mesenchyme were prepared and cultured after recombination with previously removed mesenchyme tissue pieces, as previously described.8,46 Briefly, dissected E13 epithelial rudiments and mesenchyme were separately incubated at room temperature in DMEM/F12 containing 0, or 2µl adenovirus construct in 100µl culture media for 2 hours. Epithelial rudiments were then washed in DMEM/F12 and recombined on top of Nuclepore filters with previously removed mesenchymes from > 3 glands. Recombination media consisted of 1:1 DMEM/Ham's F12 medium (F12) supplemented with 150 μg/ml Vitamin C, 50 μg/ml Transferrin, 1X pen/strep, 20ng/ml Epidermal Growth Factor (EGF), and 200ng/ml and Fibroblast Growth Factor 7 (FGF7).
Targeted siRNAs to Rac1 (sc-36352, Santa Cruz Biotechnology) or Par-1b (s65473, Abcam) were delivered to explants at 400nM or 500nM, respectively, using RNAiFect (Qiagen, 301605) according to the manufacturer's protocol. Target knockdown was confirmed by western analysis. Morphometric analyses are representative of experiments repeated at least 3 times, with at least 10 SMGs per condition in each experiment. The pharmacological Rac inhibitor, EHT1864 (Sigma)30,49 was dissolved in DMEM/F12 and used at either 7.5µM or 10µM for times indicated. The pharmacological ROCK inhibitor Y27632 (Calbiochem, 688000) was dissolved in DMEM/F12 and used at 140µM for times indicated. Par-1b wild type (Par-1b WT) adenovirus was a generous gift from Dr. Anne Meusch8 and was amplified in HEK293 cells and purified using cesium chloride density gradient centrifugation before infecting SMG rudiments in culture, as previously described.46
Brightfield images of glands for morphometric analysis were captured initially at 2 hours and every 24 hours thereafter using a Nikon Eclipse TS100 microscope equipped with a Canon EOS 450D digital camera using a 4X objective. Quantification of branching morphogenesis in whole glands is represented as the fold-change in number of buds at each time point divided by the number of buds at time zero (2 hours) (MetaMorph™ Version 61. MDS Analytical Technologies). Statistical analyses were completed using freeware (VassarStats) to perform a 2-tailed Student's t-test using the number of buds per time point.
Western analysis
Western analysis was performed as previously described.46 Briefly, cultured SMGs were lysed for total protein and protein concentrations of the resulting supernatants were determined using a Micro-BCA assay kit (Pierce, Rockford, IL). Western blots were developed on X-ray film in ECL or SuperSignal solution (Thermo Scientific), scanned (CanoScan, Canon, USA), and quantified using ImageJ software (Version 1.46r) or BioRad QuantityOne software (Version4.6.1). All western analyses were repeated with lysates from at least 3 independent experiments. A representative image from a single experiment and quantification averaged from at least 3 experiments are presented in each figure. Western analysis was performed for GAPDH (Fitzgerald Labs, 10R-G109A), Collagen IV (Millipore, AB769), Par-1b (Sigma, HPA038790), Rac1 (Millipore, 05–389), Smooth Muscle α-actin (Sigma, C6198), phosphorylated histone H3 (Ser 10) (Cell Signaling, 9701), and total histone H3 (Cell Signaling, 9715). Statistical analyses were completed using freeware (VassarStats) to perform 2-tailed Student's t-tests using the pixel intensities from digitized data.
Immunocytochemistry, immunohistochemistry, and fluorescent imaging
Whole-mount SMG immunocytochemistry (ICC) analysis was performed as previously described.18,46 Specimens were fixed in freshly prepared 2 or 4% paraformaldehyde containing 5% w/v sucrose, in 1x PBS for 20–30 minutes. Fixed samples were permeabilized, blocked, and exposed to primary antibodies and incubated overnight at 4°C. After washing, samples were incubated with donkey anti-species cyanine or Alexa Fluor dye-conjugated F(ab’)2 secondary antibodies (Jackson ImmunoResearch Laboratories) for 1 hour at room temperature, protected from light. Nuclei were stained with DAPI (Life Technologies, D1306) after secondary antibody treatment. The samples were mounted on slides with mounting media (Fluoro-Gel, Electron Microscopy Sciences) and glass coverslips before imaging. Laser scanning confocal fluorescent microscopy on immunocytochemistry (ICC) samples was performed using a Zeiss 710 or Zeiss 510 confocal microscope, and images were acquired at 20X or 63X magnification. Confocal images were processed in Zen (2012, Carl Zeiss Microscopy) and all confocal images within a given experiment were captured using the same laser intensity and gain settings so that intensities of each signal could be compared across samples. Antibodies used and their sources are as follows: Rac1 (Cytoskeleton, ARC03), Par-1b (Sigma, HPA038790), Smooth muscle α-actin (Sigma, CY3 Conjugated, A2574), ECAD (BD Biosciences, 610182 and C20824-1), Collagen IV (Millipore, AB769), phosphorylated histone H3 (Ser 10) (Cell Signaling, 9701).
Immunohistochemistry was performed as previously described.48 Briefly specimens were fixed in 10% neutral buffered saline and then moved to 70% ethanol until paraffin embedding. 0.5 μm sections were mounted on slides, deparaffinized, and immunostained with Smooth muscle α-actin, ECAD and DAPI. Images were captured using an Olympus IX-81 microscope with a 20X Plan Apo 0.75 NA objective and processed as previously described.48 Images were manually overlaid in Photoshop CS6 (13.0.1×64).
Quantification of immunocytochemistry
Quantification of fluorescent pixel intensity from a single equatorial projection of confocal images (n ≥ 3), encompassing either the entire 512×512 image or a region of interest (ROI) drawn around the epithelial bud within an image, was calculated using ImageJ software (FIJI version 1.49J10). The intensity of fluorescent staining was expressed as a ratio normalized to the intensity of DAPI and/or ECAD stain within the same image. Statistical analyses were completed using freeware (VassarStats) to perform a 2-tailed Student's t-tests using the pixel intensities from images (n ≥ 10).
Cell height measurements were performed on cells that were dually positive for E-cadherin and SM α-actin. Measurements of cell height were performed using Image J, and were made perpendicular to the cell edge in contact with the basement membrane (n ≥ 25). Measurements made in pixels were then converted to µm. Statistical analyses were completed using VassarStats to perform a 2-tailed Student's t-test using the height measurements in pixels from all measured cells (n ≥ 25).
The number of SM α-actin+ cells was counted in each of 2 ECAD+ 1600 square pixel regions of interest (ROI) per image for both EHT-treated and control tissues after 96 hours of culture. Averages of at least 35 ROIS per treatment were included in statistical analyses which were completed using VassarStats to perform a 2-tailed Student's t-test.
Supplementary Material
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Anne Meusch for the generous gift of the Par-1b adenovirus and Dr. Deirdre A. Nelson for helpful comments.
FUNDING
This work was supported by a NIH pre-doctoral Ruth L. Kirschstein National Research Service Award (F31DE023455) to E.M.G., a NIH post-doctoral Ruth L. Kirschstein National Research Service Award (F32DE020980) to S.S.; NIH grants R01 DE019244, RC1 DE020402, and R01DE022467 to M.L.; and NIH C06 RR015464 to University at Albany, SUNY.
ORCID
Melinda Larsen http://orcid.org/0000-0002-5026-2012
REFERNCES
- [1].Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation 2006; 74:349-64; PMID:16916374; http://dx.doi.org/ 10.1111/j.1432-0436.2006.00088.x [DOI] [PubMed] [Google Scholar]
- [2].Tucker AS. Salivary gland development. Semin Cell Dev Biol 2007; 18:237-44; PMID:17336109; http://dx.doi.org/ 10.1016/j.semcdb.2007.01.006 [DOI] [PubMed] [Google Scholar]
- [3].Grobstein C. Morphogenetic interaction between embryonic mouse tissues separated by a membrane filter. Nature 1953; 172(4384):869-70; PMID:1311121916613830 [DOI] [PubMed] [Google Scholar]
- [4].Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development 2006; 133:1611-24; PMID:16613830 [DOI] [PubMed] [Google Scholar]
- [5].Sakai T. Epithelial branching morphogenesis of salivary gland:exploration of new functional regulators. J Med Investig 2009; 56:234-8; http://dx.doi.org/ 10.2152/jmi.56.234 [DOI] [PubMed] [Google Scholar]
- [6].Sakai T, Onodera T. Embryonic Organ Culture. Curr Protoc Cell Biol 2008; 30:0-8 [DOI] [PubMed] [Google Scholar]
- [7].Brien LEO, Zegers MMP, Mostov KE. Building epithelial architecture: insights from three-dimensional culture models. Mol Cell Biol 2002; 3:1-7 [DOI] [PubMed] [Google Scholar]
- [8].Daley WP, Gervais EM, Centanni SW, Gulfo KM, Nelson DA, Larsen M. ROCK1-directed basement membrane positioning coordinates epithelial tissue polarity. Development 2012; 139:411-22; PMID:22186730; http://dx.doi.org/ 10.1242/dev.075366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Redman RS. Myoepithelium of salivary glands. Microsc Res Tech 1994; 27:25-45; PMID:8155903; http://dx.doi.org/ 10.1002/jemt.1070270103 [DOI] [PubMed] [Google Scholar]
- [10].Ianez RF, Buim ME, Coutinho-Camillo CM, Schultz R, Soares FA, Lourenço SV. Human salivary gland morphogenesis: Myoepithelial cell maturation assessed by immunohistochemical markers. Histopathology 2010; 57:410-7; PMID:20840670; http://dx.doi.org/ 10.1111/j.1365-2559.2010.03645.x [DOI] [PubMed] [Google Scholar]
- [11].Kandagal VS, Redder CP, Shetty S, Vibhute N, Ahamad S, Ingaleshwar P. Myoepithelial cells: Current perspectives in salivary gland tumors. Clin Cancer Investig J 2013; 2:101; http://dx.doi.org/ 10.4103/2278-0513.113624 [DOI] [Google Scholar]
- [12].Tamgadge S, Tamgadge A, Satheesan E, Modak N. Myoepithelial Cell – A Morphologic Diversity – A Review/n. Res Rev A J Dent 2013; 4:5-13 [Google Scholar]
- [13].Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 1995; 81:611-20; PMID:7758115; http://dx.doi.org/ 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- [14].Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 1995; 83:743-52; PMID:8521491; http://dx.doi.org/ 10.1016/0092-8674(95)90187-6 [DOI] [PubMed] [Google Scholar]
- [15].Hung TJ, Kemphues KJ. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 1999; 126:127-35; PMID:9834192 [DOI] [PubMed] [Google Scholar]
- [16].Hayashi K, Suzuki A, Ohno S, Mark MTAK. PAR-1 / MARK : a Kinase Essential for Maintaining the Dynamic State of Microtubules. Cell Struct Funct 2012; 25:21-5 [DOI] [PubMed] [Google Scholar]
- [17].Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, et al.. aPKC Acts Upstream of PAR-1b in Both the Establishment and Maintenance of Mammalian Epithelial Polarity. Current 2004; 14:1425-35; http://dx.doi.org/ 10.1016/j.cub.2004.08.021 [DOI] [PubMed] [Google Scholar]
- [18].Daley WP, Gulfo KM, Sequeira SJ, Larsen M. Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev Biol 2009; 336:169-82; PMID:19804774; http://dx.doi.org/ 10.1016/j.ydbio.2009.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spooner S, Faubion JONM. Collagen Involvement in Branching Morphogenesis and Salivary Gland of Embryonic. Synthesis (Stuttg) 1980; 102:84-102 [DOI] [PubMed] [Google Scholar]
- [20].Ray S, Fanti JA, Macedo DP, Larsen M. LIM Kinase regulation of cytoskeletal dynamics is required for salivary gland branching morphogenesis. Mol Biol Cell 2014; 25:2393-407; PMID:24966172; http://dx.doi.org/ 10.1091/mbc.E14-02-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hall A. Rho GTPases and the Actin Cytoskeleton. Science 1998; 279:509-14; PMID:9438836; http://dx.doi.org/ 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- [22].Corbetta S, Gualdoni S, Ciceri G, Monari M, Zuccaro E, Tybulewicz VLJ, de Curtis I. Essential role of Rac1 and Rac3 GTPases in neuronal development. FASEB J 2009; 23:1347-57; PMID:19126596; http://dx.doi.org/ 10.1096/fj.08-121574 [DOI] [PubMed] [Google Scholar]
- [23].Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. Rac, a Novel Ras-Related Family of Proteins That Are Botulinum Toxin Substrates. J Biol Chem 1989; 264:16378-82; PMID:2674130 [PubMed] [Google Scholar]
- [24].Wang L, Zheng Y. Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol 2007; 17:58-64; PMID:17161947; http://dx.doi.org/ 10.1016/j.tcb.2006.11.009 [DOI] [PubMed] [Google Scholar]
- [25].Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9:690-701; PMID:18719708; http://dx.doi.org/ 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- [26].Pirraglia C, Jattani R, Myat MM. Rac function in epithelial tube morphogenesis. Dev Biol 2006; 290:435-46; PMID:16412417; http://dx.doi.org/ 10.1016/j.ydbio.2005.12.005 [DOI] [PubMed] [Google Scholar]
- [27].Migeotte I, Grego-Bessa J, Anderson K V. Rac1 mediates morphogenetic responses to intercellular signals in the gastrulating mouse embryo. Development 2011; 138:3011-20; PMID:21693517; http://dx.doi.org/ 10.1242/dev.059766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Migeotte I, Omelchenko T, Hall A, Anderson K V. Rac1-dependent collective cell migration is required for specification of the anterior-posterior body axis of the mouse. PLoS Biol 2010; 8:e1000442; PMID:20689803; http://dx.doi.org/ 10.1371/journal.pbio.1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Larsen HS, Aure MH, Peters SB, Larsen M, Messelt EB, Galtung HK. Localization of AQP5 during development of the mouse submandibular salivary gland. J Mol Histol 2011; 42:71-81; PMID:21203896; http://dx.doi.org/ 10.1007/s10735-010-9308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem 2007; 282:35666-78; PMID:17932039; http://dx.doi.org/ 10.1074/jbc.M703571200 [DOI] [PubMed] [Google Scholar]
- [31].Salvesen GS, Dixit VM. Caspases: Intracellular Signaling by Proteolysis. Cell 1997; 91:443-6; PMID:9390553; http://dx.doi.org/ 10.1016/S0092-8674(00)80430-4 [DOI] [PubMed] [Google Scholar]
- [32].Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature 2003; 423:876-81; PMID:12815434 [DOI] [PubMed] [Google Scholar]
- [33].Deugnier M, Moiseyeva EP, Thiery JP, Glukhova M. Myoepithelial Cell Differentiation in the Developing Mammary Gland : Progressive Acquisition of Smooth Muscle Phenotype. Dev Dyn 1995; 117:107-17 [DOI] [PubMed] [Google Scholar]
- [34].Deugnier M, Teulière J, Faraldo MM, Thiery JP, Glukhova MA. The importance of being a myoepithelial cell. Breast Cancer Res 2002; 4:224-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Peters SB, Naim N, Nelson DA, Mosier AP, Cady NC, Larsen M. Biocompatible tissue scaffold compliance promotes salivary gland morphogenesis and differentiation. Tissue Eng Part A 2014; 20:1632-42; PMID:24410370; http://dx.doi.org/ 10.1089/ten.tea.2013.0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Peters SB, Nelson DA, Kwon HR, Koslow M, DeSantis KA, Larsen M. TGFβ signaling promotes matrix assembly during mechanosensitive embryonic salivary gland restoration. Matrix Biol 2015; 43:109-24; PMID:25652203; http://dx.doi.org/ 10.1016/j.matbio.2015.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nelson DA, Manhardt C, Kamath V, Sui Y, Santamaria-Pang A, Can A, Bello M, Corwin A, Dinn SR, Lazare M, et al.. Quantitative single cell analysis of cell population dynamics during submandibular salivary gland development and differentiation. Biol Open 2013; 2:439-47; PMID:23789091; http://dx.doi.org/ 10.1242/bio.20134309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, Richardson A, Violette S, Nikolskaya T, Nikolsky Y, et al.. Regulation of In Situ to Invasive Breast Carcinoma Transition. Cancer Cell 2008; 13:394-406; PMID:18455123; http://dx.doi.org/ 10.1016/j.ccr.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol 2006; 26:5797-808; PMID:16847332; http://dx.doi.org/ 10.1128/MCB.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nakayama M, Goto TM, Sugimoto M, Nishimura T, Shinagawa T, Ohno S, Amano M, Kaibuchi K. Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev Cell 2008; 14:205-15; PMID:18267089; http://dx.doi.org/ 10.1016/j.devcel.2007.11.021 [DOI] [PubMed] [Google Scholar]
- [41].Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol 2005; 7:262-9; PMID:15723052; http://dx.doi.org/ 10.1038/ncb1226 [DOI] [PubMed] [Google Scholar]
- [42].Mertens AEE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol 2006; 16:308-16; PMID:16650994; http://dx.doi.org/ 10.1016/j.tcb.2006.04.001 [DOI] [PubMed] [Google Scholar]
- [43].Munro EM. PAR proteins and the cytoskeleton: a marriage of equals. Curr Opin Cell Biol 2006; 18:86-94; PMID:16364625; http://dx.doi.org/ 10.1016/j.ceb.2005.12.007 [DOI] [PubMed] [Google Scholar]
- [44].Akhmetshina A, Dees C, Pileckyte M, Szucs G, Spriewald BM, Zwerina J, Distler O, Schett G, Distler JHW. Rho-associated kinases are crucial for myofibroblast differentiation and production of extracellular matrix in scleroderma fibroblasts. Arthritis Rheum 2008; 58:2553-64; PMID:18668558; http://dx.doi.org/ 10.1002/art.23677 [DOI] [PubMed] [Google Scholar]
- [45].Lewandowski KT, Piwnica-Worms H. Phosphorylation of the E3 ubiquitin ligase RNF41 by the kinase Par-1b is required for epithelial cell polarity. J Cell Sci 2014; 127:315-27; PMID:24259665; http://dx.doi.org/ 10.1242/jcs.129148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sequeira SJ, Gervais EM, Ray S, Larsen M. Genetic modification and recombination of salivary gland organ cultures. J Vis Exp 2013; 71:e50060; PMID: 23407326; http://dx.doi.org/22016182 10.3791/50060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Daley WP, Kohn JM, Larsen M. A focal adhesion protein-based mechanochemical checkpoint regulates cleft progression during branching morphogenesis. Dev Dyn 2011; 240:2069-83; PMID:22016182; http://dx.doi.org/ 10.1002/dvdy.22714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gervais EM, Desantis KA, Pagendarm N, Nelson DA, Enger T, Skarstein K, Liaaen Jensen J, Larsen M. Changes in the Submandibular Salivary Gland Epithelial Cell Subpopulations During Progression of Sjögren's Syndrome-Like Disease in the NOD/ShiLtJ Mouse Model. Anat Rec 2015; 298:1622-34; http://dx.doi.org/ 10.1002/ar.23190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Onesto C, Shutes A, Picard V, Schweighoffer F, Der CJ. Characterization of EHT186, a novel small molecule inhibitor of Rac family small GTPases. Methods Enzymol 2008; 439:111-29; PMID: 18374160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


